Abstract
Trovafloxacin (TVX) is a drug that has caused idiosyncratic, drug-induced liver injury (IDILI) in humans. In a murine model of IDILI, otherwise nontoxic doses of TVX and the inflammagen lipopolysaccharide (LPS) interacted to produce pronounced hepatocellular injury. The liver injury depended on a TVX-induced, small but significant prolongation of tumor necrosis factor-α (TNF) appearance in the plasma. The enhancement of TNF expression by TVX was reproduced in vitro in RAW 264.7 murine macrophages (RAW cells) stimulated with LPS. The current study was designed to identify the molecular target of TVX responsible for this response in RAW cells. An in silico analysis suggested a favorable binding profile of TVX to eukaryotic topoisomerase II-α (TopIIα), and a cell-free assay revealed that TVX inhibited eukaryotic TopIIα activity. Topoisomerase inhibition is known to lead to DNA damage, and TVX increased the DNA damage marker phosphorylated histone 2A.X in RAW cells. Moreover, TVX induced activation of the DNA damage sensor kinases, ataxia telangiectasia mutated (ATM) and Rad3-related (ATR). The ATR inhibitor NU6027 [6-(cyclohexylmethoxy)-5-nitrosopyrimidine-2,4-diamine] prevented the TVX-mediated increases in LPS-induced TNF mRNA and protein release, whereas a selective ATM inhibitor [2-(4-morpholinyl)-6-(1-thianthrenyl)-4H-pyran-4-one (KU55933)] was without effect. TVX prolonged TNF mRNA stability, and this effect was largely attenuated by NU6027. These results suggest that TVX can inhibit eukaryotic topoisomerase, leading to activation of ATR and potentiation of TNF release by macrophages, at least in part through increased mRNA stability. This off-target effect might contribute to the ability of TVX to precipitate IDILI in humans.
Introduction
Idiosyncratic, drug-induced liver injury (IDILI) is an adverse response to numerous pharmaceuticals. IDILI is responsible for approximately 13% of all cases of acute liver failure (Ostapowicz et al., 2002) and for many of the Food and Drug Administration-imposed restrictions on drug use (Lasser et al., 2002). Although typically rare, these reactions cause significant morbidity and mortality and pose a financial burden to pharmaceutical companies when offending drugs must be withdrawn from the market (Shaw et al., 2007). Although drugs from several pharmaceutical classes have been associated with human IDILI, many are antibiotics. For example, in the class of broad-spectrum, fluoroquinolone antibiotics, trovafloxacin (TVX), ciprofloxacin (CPX) and moxifloxacin (MOX) have caused IDILI in human patients, whereas levofloxacin (LVX) has shown little to no such liability (Leitner et al., 2010).
Several hypotheses have emerged to explain IDILI, yet none has been proven conclusively (Shaw et al., 2010). One hypothesis states that a transient inflammatory episode can interact with a normally nontoxic dose of a drug to bring about liver injury. Rodent models of IDILI based on this inflammatory stress hypothesis have been developed for several drugs, including TVX, sulindac, amiodarone, and others (Roth and Ganey, 2011). In these models, drugs associated with IDILI in humans synergize with an inflammagen such as lipopolysaccharide (LPS) to precipitate hepatotoxicity. At the doses used in these models, LPS exposure prompts an early increase in tumor necrosis factor-α (TNF) in the plasma but no liver necrosis. IDILI-associated drugs do not by themselves cause liver injury or TNF expression, but coadministration of drug with LPS causes a small prolongation of the LPS-stimulated TNF appearance that is critical to the pathogenesis of liver injury in cotreated animals (Shaw et al., 2007, 2009a; Zou et al., 2009; Lu et al., 2012).
An example is a murine model involving TVX/LPS coexposure. TVX is not hepatotoxic in mice even when given at large doses. However, when mice were cotreated with TVX and an otherwise nontoxic dose of LPS, pronounced hepatocellular necrosis occurred. Interestingly, this hepatotoxic interaction with LPS did not occur upon cotreatment with LVX. The liver injury from LPS/TVX cotreatment was absent in TNF receptor knockout mice or when TNF was neutralized by etanercept treatment (Shaw et al., 2007, 2009b). Importantly, when etanercept was administered at the peak of LPS-stimulated TNF appearance to prevent the prolongation of TNF appearance in TNF/LPS-cotreated mice, liver injury was prevented. Thus, although the prolongation was relatively brief and the increase was minor in magnitude compared with that which occurred from LPS alone, it was required for hepatotoxicity (Shaw et al., 2007, 2009b).
Examination of the TVX-LPS interaction in the murine model in vivo did not reveal a specific molecular target of TVX. The enhancement of LPS-stimulated TNF release by TVX could arise from a direct effect of the drug on TNF-producing cells in the liver. Indeed, pretreatment of murine RAW 264.7 cells (RAW cells) with TVX potentiated LPS-induced TNF release (Poulsen et al., 2014). Thus, the influence of TVX on LPS-stimulated TNF appearance that occurs in vivo was recapitulated in a macrophage cell line, thereby providing an in vitro system that can be employed to evaluate mechanisms of the LPS-drug interaction.
The antibiotic activity of the fluoroquinolones derives from their ability to inhibit bacterial topoisomerases and gyrases (Brighty and Gootz, 1997). Interestingly, in addition to their ability to inhibit prokaryotic topoisomerases, the fluoroquinolones TVX, CPX, and MOX have weak inhibitory activity against eukaryotic topoisomerase II-α (TopIIα) (Barrett et al., 1989; Albertini et al., 1995; Herbold et al., 2001; Reuveni et al., 2008). It is well recognized that inhibiting (“poisoning”) topoisomerases can lead to DNA damage (Ryan et al., 1991; Drummond et al., 2011). DNA damage prompts intracellular signaling involving activation of kinases that might enhance TNF expression. Accordingly, we tested the hypothesis that potentiation of LPS-induced TNF production in RAW cells by TVX results from topoisomerase inhibition and the consequent DNA damage response.
Materials and Methods
Chemicals and Inhibitors.
All chemicals and reagents were purchased from Sigma-Aldrich (St. Louis, MO) unless stated otherwise. Antibiotic/antimycotic and Dulbecco’s modified Eagle’s medium were purchased from Life Technologies (Grand Island, NY). KU55933 [2-(4-morpholinyl)-6-(1-thianthrenyl)-4H-pyran-4-one] was purchased from Tocris Bioscience (Bristol, UK).
In Silico Docking of TVX and LVX to Topoisomerase II-α.
The docking routine of TVX and LVX onto TopIIα consisted of 1) ligand optimization, 2) protein preparation, and 3) protein-ligand docking. A brief description of each procedure follows. 1) The three-dimensional geometries of TVX and LVX were optimized using density functional theory, employing the B3LYP/6-31G basis set, and calculations were carried out with the Gaussian 03 software package (Vreven et al., 2003). Open Babel was used to transform optimized geometries to Mol2 format for subsequent processing (Guha et al., 2006). 2) Experimental coordinates of the X-ray crystallographic structure of TopIIα (PDB ID 1ZXN, chains A and B) were downloaded from Protein Data Bank. Sybyl-X 2.0 Suite (SYBYL-X 2.0, Molecular modeling software 2012; Tripos, St. Louis, MO) was employed to prepare protein structures for molecular docking. During this process, ligands and water molecules were removed, side chains were repaired, and hydrogen atoms were added to the protein. The binding sites for the ligands on TopIIα were defined utilizing MGL Tools 1.5.0 (Sanner et al., 1999) by forming a box with the dimensions 86 × 70 × 90 Å, engulfing the whole protein structure, using a grid point spacing of 1.0 Å and center grid boxes of 63.249, 3.440, and 58.618, in X, Y, and Z coordinates, respectively. 3) Molecular docking methods were employed to model the ability of TVX and LVX structures to form complexes with TopIIα. Protein-ligand docking calculations were performed with AutoDock Vina 1.0 program (Trott and Olson, 2010). All calculations with AutoDock Vina included 20 number modes, an energy range of 1.5, and exhaustiveness equal to 25. Five hundred docking runs were executed for each ligand, saving the best-obtained pose for each one. The average binding affinity for these poses was computed as the affinity value for a given predicted complex. As the docking procedure allowed the identification of several binding sites within the same protein, in silico affinities, measured as kilocalories per mole, were presented for each theoretical binding site suggested by AutoDock Vina.
Topoisomerase Decatenation Assay.
TopIIα isoform activity was analyzed in the presence of vehicle (VEH) or TVX at various concentrations using etoposide as a positive control with the Human Toposiomerase II Assay Kit (TopoGEN Inc, Port Orange, FL). Briefly, 1 unit of human TopIIα was incubated with 200 ng kinetoplastid DNA (kDNA) in the presence of VEH or TVX in complete assay buffer at 37°C for 30 minutes. One unit of topoisomerase is defined as the amount of enzyme required to separate the highly catenated kDNA substrate at 37°C for 30 minutes. The reaction was stopped using the stop buffer provided, and the reaction products were loaded onto a 1% agarose gel for analysis of topoisomerase activity.
Cell Culture.
RAW 264.7 macrophage-like cells (American Type Culture Collection, Manassas, VA) were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% heat-inactivated fetal bovine serum (FBS) and 1% antibiotic/antimycotic (Life Technologies) at 37°C in 5% CO2. Cells were harvested by detachment with a sterile spatula and plated at a density of 4 × 104 cells per well in 24-well plates (Costar, Lowell, MA) for cytokine release and RNA isolation or 1.5 × 105 cells per well in 6-well plates (Costar). Twenty-four hours after plating, cells were synchronized by replacing medium with 0.5% FBS-containing medium. After overnight incubation, cells were exposed to drug. Exposure to TVX (100 μM) was not cytotoxic to RAW cells (3.6 ± 0.6% release of lactate dehydrogenase over 6 hours) in the absence or presence of LPS.
Quantitative Polymerase Chain Reaction.
Total RNA was isolated from RAW cells using TRIzol reagent (Life Technologies). cDNA was prepared with the iScript cDNA synthesis kit using 1 μg of isolated RNA (Bio-Rad Laboratories, Hercules, CA). The expression level of TNF was analyzed using the StepOne Real-Time PCR machine and SYBR Green reagents for amplicon detection (Applied Biosystems, Foster City, CA). Expression level was normalized to β-actin. TNF mRNA stability was assessed by treating cells with TVX or an equal volume of 0.1N KOH VEH for 1 hour before adding 5 μg/ml actinomycin D (ActD) to stop transcription. RNA was isolated at 15-minute intervals after the addition of ActD and converted to cDNA, and TNF mRNA was measured and normalized to β-actin mRNA.
PCR primers used were mouse TNF [5′-TCTCATGCACCACCATCAAGGACT-3′ (forward) and 5′-ACCACTCTCCCTTTGCAGAACTCA-3′ (reverse)] and mouse β-actin [5′–TGTGATGGTGGGAATGGGTCAGAA-3′ (forward) and 5′–TGTGGTGCCAGATCTTCTCCATGT-3′ (reverse)].
Western Analysis.
Cells were lysed in radioimmunoprecipitation assay buffer supplemented with HALT protease and phosphatase inhibitor cocktail (Thermo Scientific, Waltham, MA). Protein concentration in cell isolates was determined by the bicinchonic assay. Western analyses were performed by loading 20 μg of protein on precast NuPAGE SDS-PAGE gels (Life Technologies) using all NuPAGE reagents. Samples were separated on precast 12% gels. Separated proteins were transferred to polyvinylidene difluoride membranes (Millipore, Billerica, MA) for 1 hour at 4°C. Membranes were blocked in 5% BSA dissolved in Tris-buffered saline plus 0.1% Tween 20 and then probed for phospho-(Ser/Thr) ATM/ATM and Rad3-related (ATR) substrate (pATM/ATR substrate) or phospho-histone 2A.X (Ser139) (Cell Signaling Technology, Boston, MA). Membranes were then stripped with Restore Western blot stripping agent (Thermo Scientific) and reprobed for Lamin B1 (Abcam, Cambridge, MA).
Measurement of TNF Concentration.
For determination of TNF protein in culture medium, an enzyme-linked immunosorbent assay was performed (BD Biosciences, San Jose, CA). Cell culture medium was withdrawn at various times and stored at −20°C until the time of analysis. Ninety-six-well plates were coated with an anti-TNF capture antibody in a coating buffer overnight at 4°C. Medium was diluted to remain within standard curve concentrations.
Studies with Inhibitors.
Inhibitors KU55933 (Hickson et al., 2004), NU6027 [6-(cyclohexylmethoxy)-5-nitrosopyrimidine-2,4-diamine] (Peasland et al., 2011), and wortmannin (Powis et al., 1994) were dissolved in dimethylsulfoxide at a stock concentration of 10 mM and diluted to final concentrations in 0.5% FBS-containing medium. Inhibitors or an equivalent volume of dimethylsulfoxide vehicle were added at the moment RAW cells were exposed to VEH or TVX, unless noted otherwise.
Statistical Analysis.
A one- or two-way analysis of variance was performed on data sets with Tukey’s post hoc test applied for comparisons among groups. The criterion for significance was P < 0.05.
Results
TVX Interaction with TopIIα: In Silico Analysis.
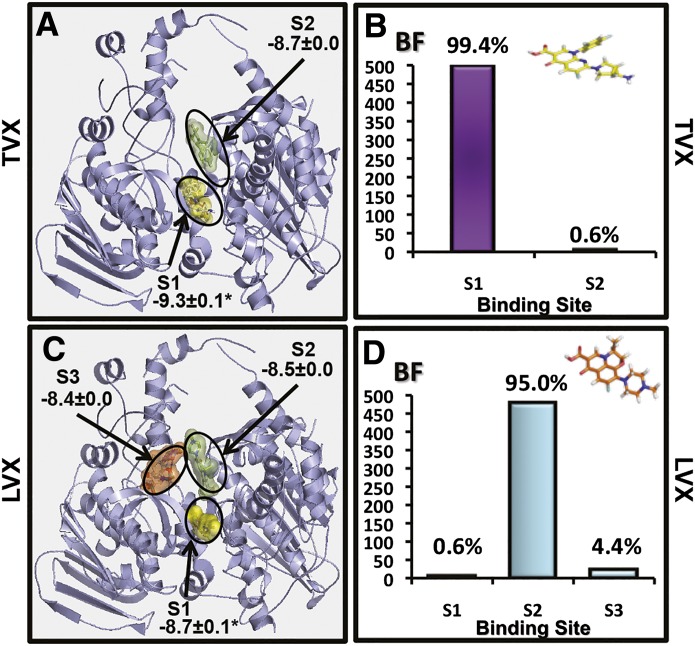
Human TopIIα was selected for analysis because it is the eukaryotic homolog to prokaryotic DNA gyrase and topoisomerase IV (Bates et al., 2011; Drummond et al., 2011). TVX binding to eukaryotic TopIIα occurred at two binding sites (Fig. 1A), the most frequently occupied of which (99.4%) was the one with the greatest predicted affinity (−9.3±0.0 kcal/mol) (Fig. 1B). In contrast, LVX was predicted to bind to TopIIα at three sites (Fig. 1C). The site most frequently occupied by LVX (95%) (Fig. 1D) differed from that to which TVX was most commonly bound. In addition, the predicted affinity for LVX binding to TopIIα (−8.5±0.0 kcal/mol) was smaller than that observed for TVX. These results indicate that TVX is predicted to bind to eukaryotic TopIIα and does so at a different site and with greater affinity than LVX.
Fig. 1.
In silico analysis of TVX binding to TopIIα. (A) Theoretical binding sites for TVX on TopIIα molecule are shown with binding affinities displayed in kcal/mol. (B) Theoretical frequency of occupation (BF; the number of docking runs in which the drug bound to a site) for TVX in sites 1 and 2. (C) Theoretical binding sites for LVX on TopIIα molecule are shown with binding affinities displayed in kcal/mol. (D) Theoretical frequency of occupation for LVX in sites 1–3. For explanation of analysis and calculations, see Materials and Methods.
TVX Inhibits TopIIα-Dependent Decatenation of kDNA.
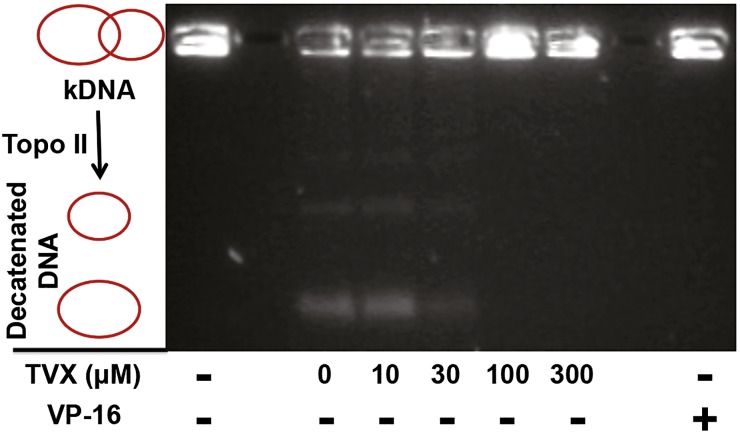
The ability of TVX to inhibit TopIIα-dependent decatenation of kDNA was evaluated in a cell-free assay (Fig. 2). In this assay, decatenation of kDNA by TopIIα results in two distinct DNA catenates of different molecular weights that migrate through the agarose gel, whereas kDNA remains in the loading wells. In the absence of TopIIα (lane labeled kDNA), the kDNA does not migrate. As a positive control, VP-16, a potent inhibitor of human TopIIα, completely prevented kDNA decatenation. Although 10 μM TVX did not affect TopIIα, the presence of TVX at concentrations of 30–300 μM decreased decatenated DNA products and increased kDNA retention compared with the control lane (0 μM TVX) containing only TopIIα. This indicated that TVX could inhibit eukaryotic topoisomerase at concentrations near those attained in the plasma during TVX therapy (Teng et al., 1996).
Fig. 2.
Effect of TVX on TopIIα activity. A reaction mixture containing kDNA in the absence (1st lane on the left) or presence (all other lanes) of TopIIα was incubated with 0 (VEH), 10, 30, 100, or 300 μM TVX or 10 μM VP-16. After 30 minutes, the reaction was quenched, and samples were separated on a 1% agarose gel and stained with ethidium bromide to visualize DNA decatenate migration. Image is representative from an n = 3.
TVX Increases DNA Lesions in RAW 264.7 Cells.
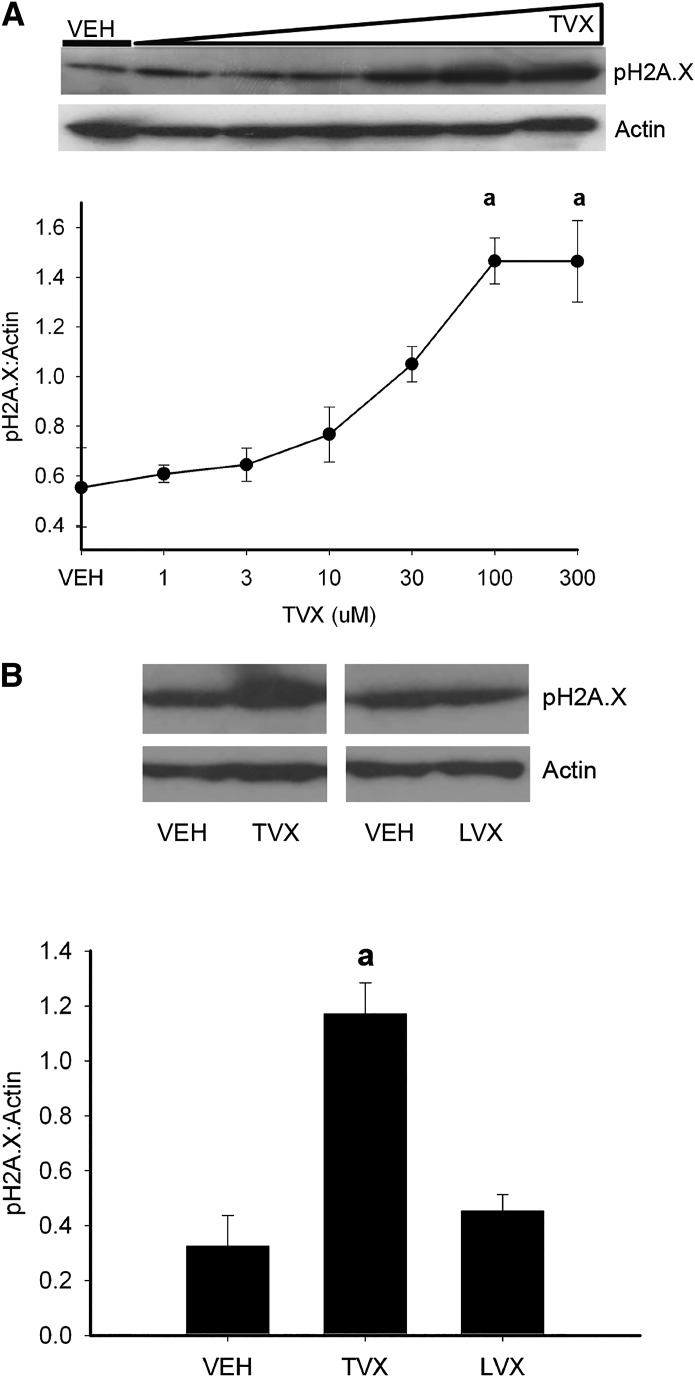
Poisoning of topoisomerase activity in cells leads to several outcomes, one of which is the formation of double-stranded lesions in DNA (Ryan et al., 1991; Drummond et al., 2011). Phosphorylated histone 2A.X (pH2A.X) is a sensitive marker of DNA lesions. When lesions occur, H2A.X is phosphorylated rapidly by the damage-sensing kinases ATM and ATR (Kastan et al., 2000). After a 2-hour incubation of RAW cells with TVX, pH2A.X increased in a concentration-dependent manner (Fig. 3A). LVX, however, did not increase pH2A.X in RAW cells over the same duration of exposure (Fig. 3B).
Fig. 3.
TVX-induced DNA damage in RAW cells. (A) RAW cells were exposed to VEH or TVX (1–300 μM) for 2 hours. pH2A.X was assessed in protein extracts by Western blot. Signals for pH2A.X were densitized and normalized to actin. (B) RAW cells were exposed to TVX (100 μM) or LVX (300 μM) for 2 hours. pH2A.X induction was assessed in protein extracts. Signals for pH2A.X were densitized and normalized to actin. Blots are representative from a minimum of n = 3. aSignificantly different from VEH, P < 0.05.
TVX Activates ATM/ATR-Dependent Signaling.
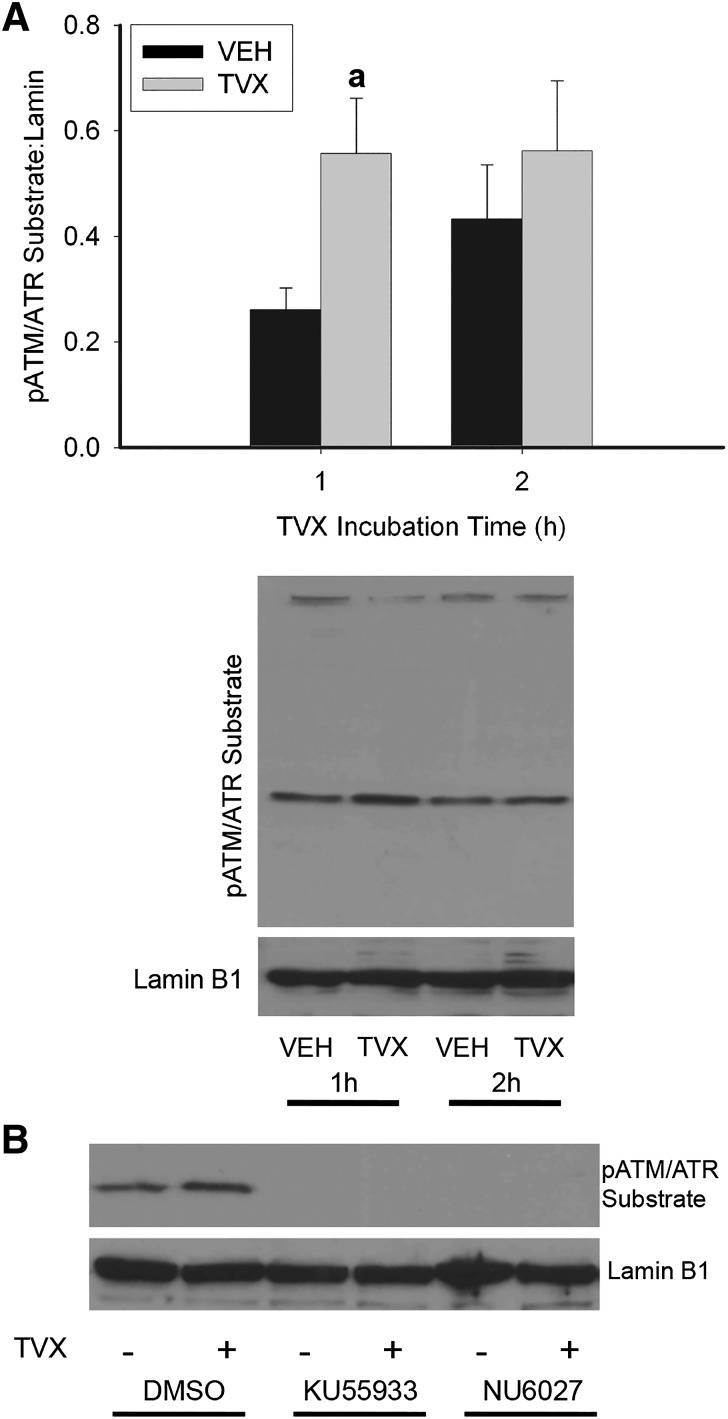
ATM and ATR are phosphoinositide 3-kinases (PI3Ks) that share a common minimal phosphorylation motif on protein substrates; i.e., a serine or threonine residue is phosphorylated if the amino acid occurs between leucine and glutamine (Kim et al., 1999). Incubation of RAW cells with TVX for 1 hour increased phosphorylation of a substrate containing the minimal ATM/ATR phosphorylation motif (Fig. 4A). This increase was absent after a 2-hour exposure to TVX. KU55933, a selective ATM inhibitor, and NU6027, an ATR-signaling inhibitor (Peasland et al., 2011), each prevented generation of the pATM/ATR substrate in VEH- or TVX-exposed RAW cells (Fig. 4B), indicating that ATM- and ATR-dependent signaling was activated by TVX.
Fig. 4.
ATM and ATR activation by TVX in RAW cells. (A) RAW cells were exposed to VEH or TVX (100 μM) for 1 or 2 hours. pATM/ATR substrate motif was assessed in isolated protein extracts by Western analysis, and signal was densitized and normalized to lamin B1. (B) RAW cells were exposed to VEH or TVX (100 μM) and to ATM inhibitor KU55933 (1 μM), ATR inhibitor NU6027 (10 μM), or their dimethylsulfoxide (DMSO; 0.05%) vehicle for 1 hour. pATM/ATR substrate motif was assessed in isolated protein extracts by Western analysis and normalized to lamin B1. Blots are representative from a minimum of n = 3. aSignificantly different from VEH, P < 0.05.
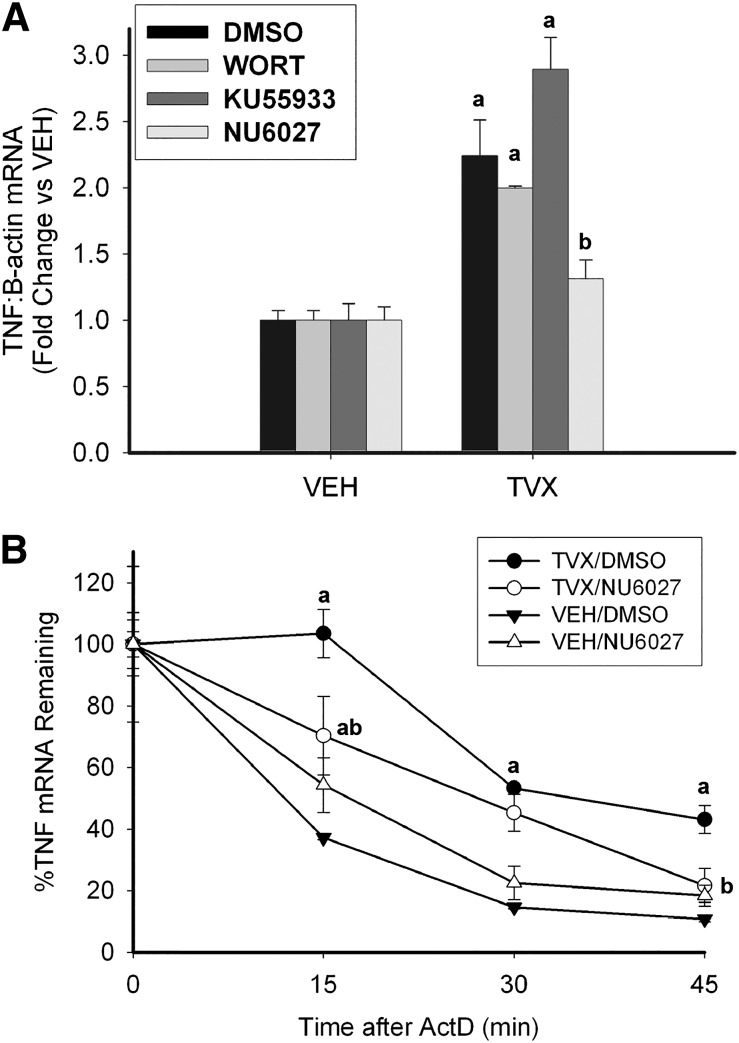
TVX Increases TNF mRNA in an ATR-Selective Manner.
As noted above, TNF is a critical factor in the pathogenesis of liver injury in TVX/LPS cotreated mice, and TVX increases TNF expression in LPS-stimulated RAW cells in vitro (Shaw et al., 2007; Poulsen et al., 2014). The influence of ATM and ATR activation on TVX-dependent TNF expression in RAW cells was assessed. TVX increased TNF mRNA after a 2-hour exposure to the drug (Fig. 5A). The increase in TNF mRNA was reduced by NU6027 but not by KU55933 or by the nonselective PI3K inhibitor wortmannin (WORT).
Fig. 5.
ATR-dependent expression of TNF mRNA in response to TVX. (A) RAW cells were coexposed to VEH or TVX (100 μM) and to WORT (1 μM), KU55933 (1 μM), NU6027 (10 μM), or their dimethylsulfoxide (DMSO; 0.05%) vehicle for 2 hours. TNF mRNA was assessed by reverse-transcription polymerase chain reaction. Values are expressed as fold of VEH/DMSO ± S.E.M. or VEH/Inhibitor ± S.E.M., n = 3–6. aP < 0.05 versus VEH group with same inhibitor, bP < 0.05 versus TVX/DMSO. (B) RAW cells were exposed to TVX (100 μM) or its VEH and to NU6027 (10 μM) or its DMSO (0.05%) vehicle for 1 hour before addition of ActD (5 μg/ml), and RNA was isolated at indicated times after ActD. TNF mRNA was normalized to the t = 0 value for each group. Values are expressed as %TNF remaining ± S.E.M., n = 6. aP < 0.05 versus VEH/DMSO at the same time; bP < 0.05 versus TVX/DMSO at the same time.
One way that increases in mRNA can occur is by stabilization of the transcript. To address this possibility, RAW cells were exposed to TVX for 1 hour before adding ActD to prevent RNA synthesis. This time of ActD addition was chosen because it coincides with the TVX-mediated increase in ATR signaling (Fig. 4A) but precedes the increase in TNF mRNA (seen at 2 hours; Fig. 5A). TVX markedly increased the stability of TNF mRNA (Fig. 5B), and NU6027 prevented this increased stability. This result suggested that the TVX-mediated increase in TNF mRNA depicted in Fig. 5A was likely due to an ATR-dependent stabilization of TNF transcripts.
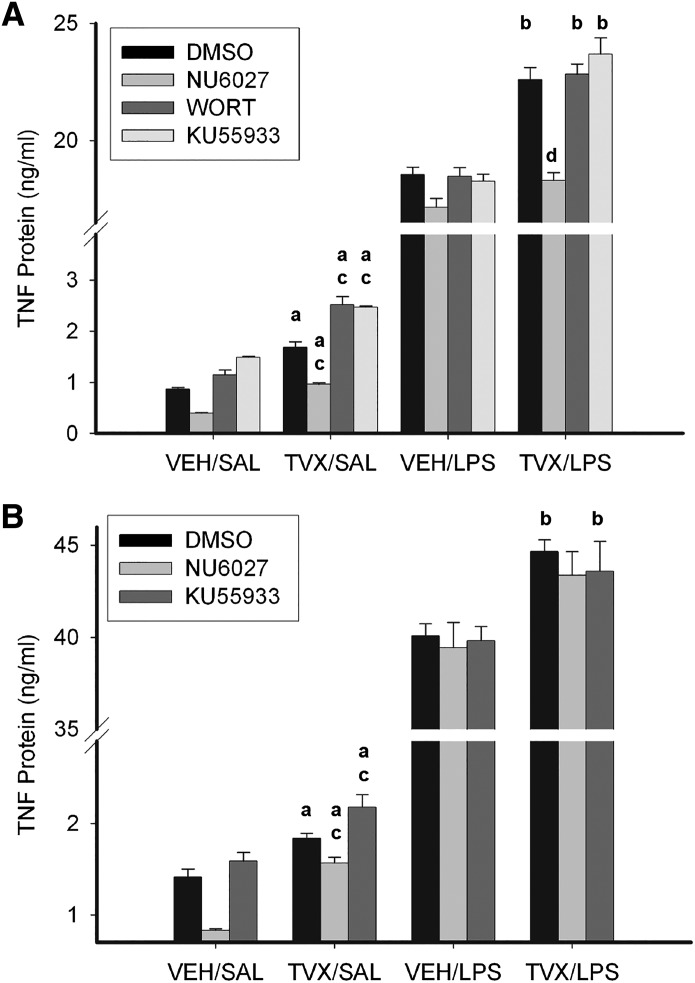
TVX Increases LPS-Induced TNF Protein Release in an ATR-Dependent Manner.
The role of ATR in the TVX-mediated increase in TNF release from RAW cells was assessed next. As expected, LPS stimulated the release of TNF from RAW cells (Fig. 6). TVX-pretreatment increased TNF release 3 hours after saline or LPS exposure (Fig. 6A, black bars). As found with TNF mRNA (Fig. 5), the increase in TNF protein release was insensitive to KU55933 or WORT but was reduced by NU6027. In cells cotreated with TVX and LPS, NU6027 reduced TNF release to the level stimulated by LPS alone. Unlike the results after 3 hours, TNF release 6 hours after LPS exposure was largely unaffected by NU6027 (Fig. 6B). At this time, NU6027 reduced the increase in TNF due to exposure to TVX alone but did not prevent the increase in LPS-induced TNF release caused by TVX pretreatment. Taken together, the results suggested that the TVX-mediated increase in LPS-induced TNF release depended on ATR at 3 but not 6 hours after LPS exposure.
Fig. 6.
Effect of ATM and ATR inhibition on TVX-mediated increases in LPS-induced TNF release from RAW cells. RAW cells were pretreated with VEH or TVX (100 μM) and with NU6027 (10 μM), WORT (1 μM), KU55933 (1 μM), or their dimethylsulfoxide (DMSO; 0.05%) vehicle for 2 hours, after which time medium was replaced with one containing saline (SAL) or LPS (10 ng/ml) without inhibitors. TNF protein release was assessed at 3 (A) and 6 (B) hours after LPS exposure. Values are means ± S.E.M. from 3 to 6 separate experiments, each performed in triplicate. aSignificantly different from VEH/SAL with same inhibitor treatment, P < 0.05; bsignificantly different from VEH/LPS with same inhibitor treatment, P < 0.05; csignificantly different from TVX/SAL/DMSO group, P < 0.05; dsignificantly different from TVX/LPS/DMSO group, P < 0.05.
Discussion
Global gene expression analysis of livers from rats or primary hepatocytes treated with TVX suggested that TVX selectively targets eukaryotic topoisomerases, an off-target effect for a prokaryotic topoisomerase poison (Waring et al., 2006; Liguori et al., 2008). Additionally, TVX affected chromosomal expression patterns in a manner similar to the known eukaryotic topoisomerase poisons etoposide and doxorubicin, suggesting further that TVX might act as a topoisomerase poison in mammalian cells (Reymann and Borlak, 2008). In silico binding analysis (Fig. 1) suggested that TVX binds favorably to eukaryotic TopIIα. Moreover, TVX prevented human TopIIα-dependent decatenation of kinetoplastid DNA in a concentration-dependent manner (Fig. 2). Taken together, these results raised the possibility that the IDILI liability associated with TVX might be attributed to off-target poisoning of eukaryotic topoisomerase. Topoisomerase inhibition in cells can lead to double-strand DNA breaks generated from topoisomerase-DNA covalent complexes (Liu et al., 1983; Ryan et al., 1991; Li et al., 2010; Drummond et al., 2011). pH2A.X was chosen as a sensitive marker of double-strand DNA breaks. TVX increased pH2A.X in a concentration-dependent manner in RAW cells after a 2-hour incubation (Fig. 3A).
TVX is associated with IDILI in humans, whereas LVX does not share this liability (Leitner et al., 2010). In mice, TVX/LPS coexposure precipitated hepatotoxicity, but LVX/LPS did not (Shaw et al., 2007). The results in silico (Fig. 1) suggested that TVX has the capacity to bind to TopIIα in a distinct manner compared with LVX. Unlike TVX, LVX did not increase pH2A.X (Fig. 3B) or increase TNF expression (Poulsen et al., 2014). Thus, the difference in the association with IDILI for these two drugs was reflected in their abilities to cause modest DNA damage and augment TNF expression as well as to interact with LPS in mice to precipitate liver injury.
A recent screen of novel bacterial type II topoisomerase inhibitors in murine L5178Y lymphoma cells used pH2A.X as an indicator of topoisomerase inhibition (Smart and Lynch, 2012). A substantial proportion (22/63) of the novel inhibitors, as well as CPX and MOX, modestly increased pH2A.X in mammalian cells, and this increase coincided with a greater than sixfold increase in mutation frequency, suggesting that many bacterial topoisomerase inhibitors, including fluoroquinolones like CPX and MOX, can induce weak genotoxic effects in eukaryotic cells (Smart and Lynch, 2012). The phosphorylation of H2A.X that we observed with TVX was quite modest compared with the effect of potent eukaryotic DNA damaging agents (Anderson and Osheroff, 2001; Smart and Lynch, 2012). This modest, otherwise nontoxic damage is consistent with in vivo studies with TVX in the sense that TVX was nontoxic even at large doses in mice in the absence of a concurrent inflammatory stress (Shaw et al., 2007).
Taken together, the results in Figs. 1–3 suggest that TVX poisons topoisomerase in RAW cells and that this leads to DNA damage that does not result in cell death. Accordingly, it is likely that the TVX-induced DNA damage is within the capacity of the cells to repair, but the signaling activated in response to DNA lesions predisposes cells to enhance synthesis of TNF in response to LPS. DNA lesions activate several mediators and intracellular signaling pathways in a coordinated and dynamic manner that is referred to as the DNA damage response (DDR) (Ciccia and Elledge, 2010). ATM and ATR are rapidly activated kinases that are critical to the DDR (Kastan et al., 2000). Treatment with TVX caused an increase in phosphorylation of an epitope in proteins (Fig. 4A) that is a target for both ATM and ATR (Kim et al., 1999). This occurred prior to pH2A.X generation (Fig. 3), and ATM and ATR inhibitors prevented TVX-induced phosphorylation of this epitope (Fig. 4B), suggesting that TVX exposure activated ATM and ATR. The results in Figs. 1–4 support a scenario in which TVX poisons eukaryotic topoisomerase, damages DNA, and activates DDR kinases.
A large amount of evidence supports a link between induction of DNA damage and upregulation of cytokine expression. For example, potent eukaryotic topoisomerase poisons doxorubicin and etoposide, as well as the antimetabolite 5-fluorouracil, can increase cytokine expression in murine macrophages in vitro and in mice in vivo (Wood et al., 2006; Elsea et al., 2008). The increase in TNF mRNA caused by TVX in RAW cells was sensitive to inhibition of ATR but unaffected by inhibition of ATM and to nonselective PI3K inhibition by WORT (Fig. 5A). ATR is far less sensitive to inhibition by WORT than ATM or DNA-PK (Sarkaria et al., 1998), so it was likely that the enhanced TNF expression was downstream of ATR-mediated signaling. Thus, although several PI3Ks, including ATM, ATR, and DNA-PK, are activated in response to DNA lesions (Ciccia and Elledge, 2010), only ATR was implicated in the enhancement of TNF expression by TVX. TNF mRNA rapidly degrades in the absence of an inflammatory stimulus (Deleault et al., 2008). Interestingly, treatment with TVX stabilized TNF mRNA prior to LPS exposure (Fig. 5B), and NU6027 markedly reduced this effect, suggesting that TVX-dependent ATR activation stabilizes TNF mRNA.
Because ATR was implicated in the TVX-induced increase in TNF mRNA (Fig. 5), the involvement of ATR in LPS-induced TNF protein release was examined. KU55933 and NU6027 were included only during the period of exposure to TVX and were removed before addition of LPS or saline. This was done to determine if the critical TVX-induced signaling changes occurred prior to LPS exposure. TVX pretreatment enhanced LPS-induced TNF protein release within 3 hours after LPS addition, and this increase was prevented by NU6027 (Fig. 6A). When NU6027 was added after LPS addition, the TVX-mediated potentiation of TNF release was not prevented (Supplemental Fig. 1). Accordingly, the critical ATR activation must have occurred during the TVX pretreatment period, not after LPS addition. Elimination of the LPS-TVX interaction by NU6027 was evident 3 hours after LPS addition but not at 6 hours (Fig. 6B). One potential explanation for the lack of effect at 6 hour is that ATR could be activated after withdrawal of NU6027-containing medium if the DNA damage persists during the time of LPS exposure.
The effect of TVX on cytokine expression has been addressed in two other studies (Khan et al., 1998; Purswani et al., 2000). In both of these studies, TVX decreased TNF expression in LPS-pretreated cells (Khan et al., 1998; Purswani et al., 2000), contrasting with the increase identified in this study. A key difference in those studies is that TVX was added to monocytes or peripheral mononuclear cells after stimulation with LPS, whereas in our study TVX was present only before LPS addition. In both of the studies in which TVX decreased TNF mRNA and protein release, the results were attributed to TVX acting as a topoisomerase inhibitor in eukaryotic cells (Khan et al., 1998; Purswani et al., 2000). Accordingly, the difference between the results could be due to temporal differences in TVX exposure relative to LPS. As the results from the current study indicate, the TVX-mediated DDR and consequent activation of ATR before LPS exposure appears to be critical for the TVX-mediated increase in LPS-induced TNF release.
In summary, the results of in silico, cell-free, and cultured cell approaches indicate that TVX, but not LVX, can decrease topoisomerase activity and induce DNA damage at concentrations that approach those occurring in patients treated with TVX (Teng et al., 1996). TVX activated ATM/ATR-dependent signaling, and ATR played a critical role in mediating increased TNF mRNA stability and LPS-induced TNF protein release from macrophages. The results from this study uncovered a previously unknown role for the DDR and specifically ATR in increasing TNF expression by macrophages exposed to a modest genotoxic stimulus. The results suggest that topoisomerase inhibition might contribute to IDILI caused by TVX and perhaps other fluoroquinolone antibiotics.
Supplementary Material
Abbreviations
- ActD
actinomycin D
- ATM
ataxia telangiectasia mutated
- ATR
ATM and Rad3-related
- CPX
ciprofloxacin
- DDR
DNA damage response
- FBS
fetal bovine serum
- IDILI
idiosyncratic, drug-induced liver injury
- kDNA
kinetoplastid DNA
- KU55933
2-(4-morpholinyl)-6-(1-thianthrenyl)-4H-pyran-4-one
- LPS
lipopolysaccharide
- LVX
levofloxacin
- MOX
moxifloxacin
- NU6072
6-(cyclohexylmethoxy)-5-nitrosopyrimidine-2,4-diamine
- pATM/ATR substrate
phosphorylated-(Ser/Thr) ATM/ATR substrate
- pH2A.X
phosphorylated histone 2A.X
- PI3K
phosphoinositide 3-kinase
- RAW cells
RAW 264.7 murine macrophages
- TNF
tumor necrosis factor-α
- TopIIα
eukaryotic topoisomerase II-α
- TVX
trovafloxacin
- VEH
vehicle
- WORT
wortmannin
Authorship Contributions
Participated in research design: Poulsen, Olivero-Verbel, Beggs, Ganey, Roth.
Conducted experiments: Poulsen, Olivero-Verbel, Beggs.
Performed data analysis: Poulsen, Olivero-Verbel.
Wrote or contributed to the writing of the manuscript: Poulsen, Olivero-Verbel, Ganey, Roth.
Footnotes
This study was supported by the National Institutes of Health National Institute of Diabetes and Digestive and Kidney Diseases [Grant R01-DK061315]. K.L.P. was supported by the National Institutes of Health National Institute of Environmental Health Sciences Training Grant [Grant T32-ES007255].
 This article has supplemental material available at jpet.aspetjournals.org.
This article has supplemental material available at jpet.aspetjournals.org.
References
- Albertini S, Chételat AA, Miller B, Muster W, Pujadas E, Strobel R, Gocke E. (1995) Genotoxicity of 17 gyrase- and four mammalian topoisomerase II-poisons in prokaryotic and eukaryotic test systems. Mutagenesis 10:343–351 [DOI] [PubMed] [Google Scholar]
- Anderson VE, Osheroff N. (2001) Type II topoisomerases as targets for quinolone antibacterials: turning Dr. Jekyll into Mr. Hyde. Curr Pharm Des 7:337–353 [DOI] [PubMed] [Google Scholar]
- Barrett JF, Gootz TD, McGuirk PR, Farrell CA, Sokolowski SA. (1989) Use of in vitro topoisomerase II assays for studying quinolone antibacterial agents. Antimicrob Agents Chemother 33:1697–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates AD, Berger JM, Maxwell A. (2011) The ancestral role of ATP hydrolysis in type II topoisomerases: prevention of DNA double-strand breaks. Nucleic Acids Res 39:6327–6339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brighty KE, Gootz TD. (1997) The chemistry and biological profile of trovafloxacin. J Antimicrob Chemother 39 (Suppl B): 1–14 [DOI] [PubMed] [Google Scholar]
- Ciccia A, Elledge SJ. (2010) The DNA damage response: making it safe to play with knives. Mol Cell 40:179–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deleault KM, Skinner SJ, Brooks SA. (2008) Tristetraprolin regulates TNF TNF-alpha mRNA stability via a proteasome dependent mechanism involving the combined action of the ERK and p38 pathways. Mol Immunol 45:13–24 [DOI] [PubMed] [Google Scholar]
- Drummond CJ, Finlay GJ, Broome L, Marshall ES, Richardson E, Baguley BC. (2011) Action of SN 28049, a new DNA binding topoisomerase II-directed antitumour drug: comparison with doxorubicin and etoposide. Invest New Drugs 29:1102–1110 [DOI] [PubMed] [Google Scholar]
- Elsea CR, Roberts DA, Druker BJ, Wood LJ. (2008) Inhibition of p38 MAPK suppresses inflammatory cytokine induction by etoposide, 5-fluorouracil, and doxorubicin without affecting tumoricidal activity. PLoS ONE 3:e2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guha R, Dutta D, Jurs PC, Chen T. (2006) Local lazy regression: making use of the neighborhood to improve QSAR predictions. J Chem Inf Model 46:1836–1847 [DOI] [PubMed] [Google Scholar]
- Herbold BA, Brendler-Schwaab SY, Ahr HJ. (2001) Ciprofloxacin: in vivo genotoxicity studies. Mutat Res 498:193–205 [DOI] [PubMed] [Google Scholar]
- Hickson I, Zhao Y, Richardson CJ, Green SJ, Martin NM, Orr AI, Reaper PM, Jackson SP, Curtin NJ, Smith GC. (2004) Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res 64:9152–9159 [DOI] [PubMed] [Google Scholar]
- Kastan MB, Lim DS, Kim ST, Xu B, Canman C. (2000) Multiple signaling pathways involving ATM. Cold Spring Harb Symp Quant Biol 65:521–526 [DOI] [PubMed] [Google Scholar]
- Khan AA, Slifer TR, Remington JS. (1998) Effect of trovafloxacin on production of cytokines by human monocytes. Antimicrob Agents Chemother 42:1713–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim ST, Lim DS, Canman CE, Kastan MB. (1999) Substrate specificities and identification of putative substrates of ATM kinase family members. J Biol Chem 274:37538–37543 [DOI] [PubMed] [Google Scholar]
- Lasser KE, Allen PD, Woolhandler SJ, Himmelstein DU, Wolfe SM, Bor DH. (2002) Timing of new black box warnings and withdrawals for prescription medications. JAMA 287:2215–2220 [DOI] [PubMed] [Google Scholar]
- Leitner JM, Graninger W, Thalhammer F. (2010) Hepatotoxicity of antibacterials: Pathomechanisms and clinical. Infection 38:3–11 [DOI] [PubMed] [Google Scholar]
- Li Y, Luan Y, Qi X, Li M, Gong L, Xue X, Wu X, Wu Y, Chen M, Xing G, et al. (2010) Emodin triggers DNA double-strand breaks by stabilizing topoisomerase II-DNA cleavage complexes and by inhibiting ATP hydrolysis of topoisomerase II. Toxicol Sci 118:435–443 [DOI] [PubMed] [Google Scholar]
- Liguori MJ, Blomme EA, Waring JF. (2008) Trovafloxacin-induced gene expression changes in liver-derived in vitro systems: comparison of primary human hepatocytes to HepG2 cells. Drug Metab Dispos 36:223–233 [DOI] [PubMed] [Google Scholar]
- Liu LF, Rowe TC, Yang L, Tewey KM, Chen GL. (1983) Cleavage of DNA by mammalian DNA topoisomerase II. J Biol Chem 258:15365–15370 [PubMed] [Google Scholar]
- Lu J, Jones AD, Harkema JR, Roth RA, Ganey PE. (2012) Amiodarone exposure during modest inflammation induces idiosyncrasy-like liver injury in rats: role of tumor necrosis factor-alpha. Toxicol Sci 125:126–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostapowicz G, Fontana RJ, Schiødt FV, Larson A, Davern TJ, Han SH, McCashland TM, Shakil AO, Hay JE, Hynan L, et al. U.S. Acute Liver Failure Study Group (2002) Results of a prospective study of acute liver failure at 17 tertiary care centers in the United States. Ann Intern Med 137:947–954 [DOI] [PubMed] [Google Scholar]
- Peasland A, Wang LZ, Rowling E, Kyle S, Chen T, Hopkins A, Cliby WA, Sarkaria J, Beale G, Edmondson RJ, et al. (2011) Identification and evaluation of a potent novel ATR inhibitor, NU6027, in breast and ovarian cancer cell lines. Br J Cancer 105:372–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulsen KL, Albee RP, Ganey PE, Roth RA. (2014) Trovafloxacin Potentiation of Lipopolysaccharide-Induced Tumor Necrosis Factor Release from RAW 264.7 Cells Requires Extracellular Signal-Regulated Kinase and c-Jun N-Terminal Kinase. J Pharmacol Exp Ther 349:185–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powis G, Bonjouklian R, Berggren MM, Gallegos A, Abraham R, Ashendel C, Zalkow L, Matter WF, Dodge J, Grindey G, et al. (1994) Wortmannin, a potent and selective inhibitor of phosphatidylinositol-3-kinase. Cancer Res 54:2419–2423 [PubMed] [Google Scholar]
- Purswani M, Eckert S, Arora H, Johann-Liang R, Noel GJ. (2000) The effect of three broad-spectrum antimicrobials on mononuclear cell responses to encapsulated bacteria: evidence for down-regulation of cytokine mRNA transcription by trovafloxacin. J Antimicrob Chemother 46:921–929 [DOI] [PubMed] [Google Scholar]
- Reuveni D, Halperin D, Shalit I, Priel E, Fabian I. (2008) Quinolones as enhancers of camptothecin-induced cytotoxic and anti-topoisomerase I effects. Biochem Pharmacol 75:1272–1281 [DOI] [PubMed] [Google Scholar]
- Reymann S, Borlak J. (2008) Topoisomerase II inhibition involves characteristic chromosomal expression patterns. BMC Genomics 9:324–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth RA, Ganey PE. (2011) Animal models of idiosyncratic drug-induced liver injury—current status. Crit Rev Toxicol 41:723–739 [DOI] [PubMed] [Google Scholar]
- Ryan AJ, Squires S, Strutt HL, Johnson RT. (1991) Camptothecin cytotoxicity in mammalian cells is associated with the induction of persistent double strand breaks in replicating DNA. Nucleic Acids Res 19:3295–3300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanner MF, Duncan BS, Carrillo CJ, Olson AJ. (1999) Integrating computation and visualization for biomolecular analysis: an example using python and AVS. Pac Symp Biocomput 1999:401–412 [DOI] [PubMed] [Google Scholar]
- Sarkaria JN, Tibbetts RS, Busby EC, Kennedy AP, Hill DE, Abraham RT. (1998) Inhibition of phosphoinositide 3-kinase related kinases by the radiosensitizing agent wortmannin. Cancer Res 58:4375–4382 [PubMed] [Google Scholar]
- Shaw PJ, Beggs KM, Sparkenbaugh EM, Dugan CM, Ganey PE, Roth RA. (2009a) Trovafloxacin enhances TNF-induced inflammatory stress and cell death signaling and reduces TNF clearance in a murine model of idiosyncratic hepatotoxicity. Toxicol Sci 111:288–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw PJ, Ganey PE, Roth RA. (2009b) Tumor necrosis factor alpha is a proximal mediator of synergistic hepatotoxicity from trovafloxacin/lipopolysaccharide coexposure. J Pharmacol Exp Ther 328:62–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw PJ, Ganey PE, Roth RA. (2010) Idiosyncratic drug-induced liver injury and the role of inflammatory stress with an emphasis on an animal model of trovafloxacin hepatotoxicity. Toxicol Sci 118:7–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw PJ, Hopfensperger MJ, Ganey PE, Roth RA. (2007) Lipopolysaccharide and trovafloxacin coexposure in mice causes idiosyncrasy-like liver injury dependent on tumor necrosis factor-alpha. Toxicol Sci 100:259–266 [DOI] [PubMed] [Google Scholar]
- Smart DJ, Lynch AM. (2012) Evaluating the genotoxicity of topoisomerase-targeted antibiotics. Mutagenesis 27:359–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng R, Liston TE, Harris SC. (1996) Multiple-dose pharmacokinetics and safety of trovafloxacin in healthy volunteers. J Antimicrob Chemother 37:955–963 [DOI] [PubMed] [Google Scholar]
- Trott O, Olson AJ. (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31:455–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vreven T, Morokuma K, Farkas O, Schlegel HB, Frisch MJ. (2003) Geometry optimization with QM/MM, ONIOM, and other combined methods. I. Microiterations and constraints. J Comput Chem 24:760–769 [DOI] [PubMed] [Google Scholar]
- Waring JF, Liguori MJ, Luyendyk JP, Maddox JF, Ganey PE, Stachlewitz RF, North C, Blomme EA, Roth RA. (2006) Microarray analysis of lipopolysaccharide potentiation of trovafloxacin-induced liver injury in rats suggests a role for proinflammatory chemokines and neutrophils. J Pharmacol Exp Ther 316:1080–1087 [DOI] [PubMed] [Google Scholar]
- Wood LJ, Nail LM, Perrin NA, Elsea CR, Fischer A, Druker BJ. (2006) The cancer chemotherapy drug etoposide (VP-16) induces proinflammatory cytokine production and sickness behavior-like symptoms in a mouse model of cancer chemotherapy-related symptoms. Biol Res Nurs 8:157–169 [DOI] [PubMed] [Google Scholar]
- Zou W, Beggs KM, Sparkenbaugh EM, Jones AD, Younis HS, Roth RA, Ganey PE. (2009) Sulindac metabolism and synergy with tumor necrosis factor-alpha in a drug-inflammation interaction model of idiosyncratic liver injury. J Pharmacol Exp Ther 331:114–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.