SUMMARY
Fetal oocyte attrition (FOA) is a conserved but poorly understood process of elimination of over two-thirds of meiotic prophase I (MPI) oocytes before birth. We now implicate retrotransposons LINE-1 (L1), activated during epigenetic reprogramming of the embryonic germline, in FOA in mice. We show that wild-type fetal oocytes possess differential nuclear levels of L1ORF1p, a L1-encoded protein essential for L1 ribonucleoprotein particle (L1RNP) formation and L1 retrotransposition. We demonstrate that experimental elevation of L1 expression correlates with increased MPI defects, FOA, oocyte aneuploidy and embryonic lethality. Conversely, reverse transcriptase (RT) inhibitor AZT has a profound effect on the FOA dynamics and meiotic recombination, and implicates an RT-dependent trigger in oocyte elimination in early MPI. We propose that FOA serves to select oocytes with limited L1 activity and therefore best suited for the next generation.
INTRODUCTION
Fetal oocyte attrition (FOA) is the process of elimination of ~80% of the initial pool of human oocytes by the time of birth (Baker, 1963; Kurilo, 1981). This process is not unique to humans and has been observed in primates and extensively documented in several rodent species (Baker, 1966; Beaumont and Mandl, 1962; Burgoyne and Baker, 1985; Ioannou, 1964; McClellan et al., 2003). In addition, oocyte loss is observed in invertebrates suggesting a possibility of ancient evolutional origin of FOA (Matova and Cooley, 2001). In mice, fetal oocyte loss occurs continuously throughout the meiotic prophase I (MPI) and appears to require, at least in part, apoptotic mechanisms (Bergeron et al., 1998; Ene et al., 2013; Ghafari et al., 2007; McClellan et al., 2003; Morita et al., 2000). However, despite the apparent evolutional conservation of FOA, questions of the molecular basis and rationale (if any) for oocyte purging remain open (Hartshorne et al., 2009). Over the years, a few scenarios have been considered but none have been firmly ruled out or confirmed experimentally to date (Tilly, 2001). These include “death by neglect”, “death by defect” and “death by self-sacrifice” that correspond to proposed roles of growth factors, meiotic checkpoints and cyst organization of the embryonic oogenesis (Barlow et al., 1998; Lei and Spradling, 2013; Morita et al., 1999; Morita et al., 2001; Pepling and Spradling, 2001).
Over the past decade, DNA methylation remodeling of the embryonic germline has become recognized as an important aspect of germ cell development and differentiation (Lees-Murdock and Walsh, 2008; Popp et al., 2010; Seisenberger et al., 2012). The erasure of repressive DNA methylation creates a window of opportunity for expression of transposable elements (TEs) whose intact and mutated copies constitute ~40% of the mouse genome (Bourc'his and Bestor, 2004; Hajkova et al., 2002; Walsh et al., 1998; Waterston et al., 2002). At least two mechanisms, DNA methylation and PIWI-interacting RNAs (piRNAs), are required to efficiently silence TEs de novo (Aravin and Bourc'his, 2008; Bourc'his and Bestor, 2004). Studies of mouse mutants lacking piRNAs demonstrated the essential role of these small RNAs in transcriptional and post-transcriptional transposon control (Aravin et al., 2008; Kuramochi-Miyagawa et al., 2008). Interestingly, upregulation of transposons is particularly detrimental to MPI male germ cells (Aravin et al., 2009; Carmell et al., 2007; Ollinger et al., 2010; Shoji et al., 2009; Soper et al., 2008). This observation is important since the onset of DNA methylation reprogramming and transposon derepression just precede sex determination of primordial germ cells, which is manifested as the cell-cycle arrest of prospermatogonia and the meiotic entry of oocytes (Seisenberger et al., 2012; Western, 2009). Therefore, by analogy with lethality of piRNA- or DNA methylation-deficient spermatocytes, massive elimination of fetal oocytes could be a product of the concurrency of transposon derepression and meiotic initiation (Figure 1A). While none of the reported mouse mutants lacking piRNA machinery have been described to exhibit female infertility, a prior study linked extensive global DNA demethylation in the Hells/LSH mutant with MPI defects and derepression of IAP elements, which otherwise elude extensive DNA methylation reprogramming (De La Fuente et al., 2006; Lane et al., 2003). In this work we set out to examine in details the impact of retrotransposons on viability and quality of fetal oocytes in mice.
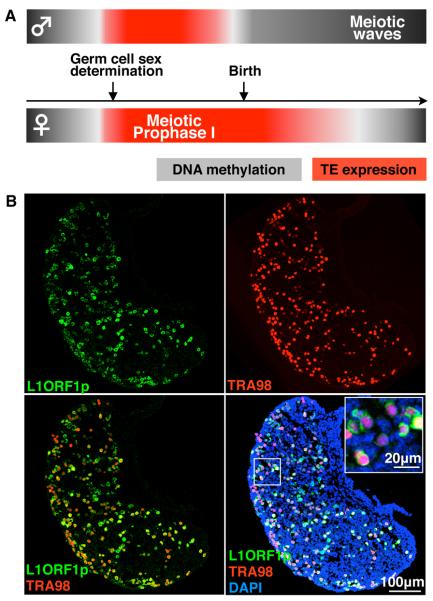
Figure 1. L1 Expression in Meiotic Prophase I Fetal Oocytes.
(A) Meiotic prophase I in oocytes coincides with epigenetic reprogramming of the germ cell genome and activation of retrotransposon expression.
(B) Single and merged channels of single confocal sections of E18.5 WT ovary showing L1ORF1p (green) expression in TRA98-positive (red) fetal oocytes counterstained with DAPI (blue).
See also Figure S1.
RESULTS
Mutation of Mael Increases L1 Expression and Enhances Fetal Oocyte Attrition
We reasoned that expression of transposable elements throughout MPI could contribute to FOA (Figure 1A). To begin to test this hypothesis, we first used immunofluorescence to assess fetal oocyte expression of two classes of retrotransposons active in the mouse genome - non-LTR retrotransposons L1 and endogenous retroviruses IAP (Goodier and Kazazian, 2008). Based on immunostaining for L1ORF1p, a L1-encoded protein that is a component of L1 ribonucleoprotein particles (L1RNPs) with an essential role in L1 retrotransposition (Doucet et al., 2010; Martin, 2006; Martin et al., 2008), L1 elements were found to be expressed in all MPI oocytes of the fetal ovary (Figure 1B). In contrast, we did not detect IAP GAG protein expression until later in oogenesis (Figure S1). This is consistent with a prior report of IAPs being resistant to epigenetic reprogramming (Lane et al., 2003). Given these observations and persistence of L1 activity in the human genome (Beck et al., 2010; Brouha et al., 2003; Kazazian, 2004), we focused our subsequent studies on L1 elements.
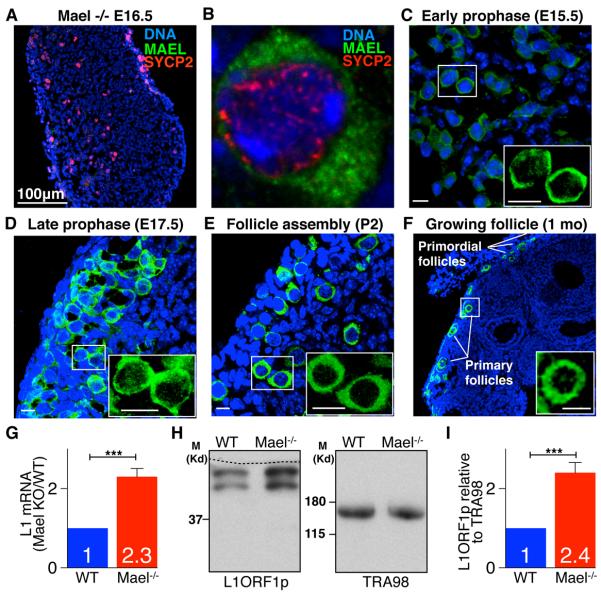
To assess the effect of increased L1 expression on viability of fetal oocytes, we first studied mice lacking maelstrom homolog (Drosophila) (Mael) gene that encodes a protein with an evolutionarily conserved role in transposon silencing (Aravin et al., 2009; Lim and Kai, 2007; O'Donnell et al., 2008; Sienski et al., 2012; Soper et al., 2008). Using anti-MAEL antibodies (Soper et al., 2008), we detected MAEL expression in fetal oocytes, primordial and primary follicles (Figures 2A–2F). Mael-null embryonic day 15.5 (E15.5) ovaries contained a 2.3- and 2.4-fold excess of L1 mRNA and L1ORF1p, respectively, above levels already present in the wild type (WT) (Figures 2G–2I). RNA-Seq analysis of WT and Mael-null E15.5 ovaries revealed abundant expression of L1 elements of active families and their 2-fold increase in the Mael mutant (Figure S2A). We also observed increased expression of genes associated with cell death and transcripts from contaminating hematopoietic cells but did not see altered expression of genes with established roles in germ cells or meiosis (Figure S2B, Table S1 and Experimental Procedures).
Figure 2. MAEL is Required for L1 Regulation in Fetal Ovaries.
(A) Section of E16.5 Mael-mutant ovary section probed with MAEL antibody.
(B) Cytoplasmic MAEL (green) localization in WT E15.5 oocytes. SYCP2 (red) marks chromosome axes.
(C–F) WT ovaries of indicated developmental stages and ages probed with MAEL antibody. MAEL expression is observed in fetal oocytes throughout meiotic prophase I and in primordial, and primary follicles. Insets - MAEL single channel (green) close-up views. Bars represent 10 μm.
(G) qRT-PCR analysis of L1 mRNA levels relative to expression of germ cell-specific Mvh. (Mean ± St.dev. from three independent measurements are shown, t=10.82, p=0.0004). (H) Western blot detection of L1ORF1p relative to TRA98 expression in E15.5 ovaries based on results of analysis of three biological replicates. Dashed line marks the cut line of the membrane. (I) Quantification of Western blot data shown in (H). (Mean ± St.dev. from three independent measurements are shown, t =9.31, p=0.0007). Two-tailed unpaired Student's t-test, ***-P<0.001 (G, I).
To evaluate the effect of the Mael mutation on FOA, we quantified oocytes in E15.5, E18.5 and postnatal day 2 (P2) WT and Mael-mutant ovaries. At E15.5, when all oocytes have entered MPI, ovaries of both genotypes had comparable oocyte numbers (Figure 3A, Tables S2A and S2B). However, of the initial WT and Mael-null E15.5 populations, only 55% and 18% survived to E18.5, and 40% and 14% to P2, respectively (Figure 3A, Tables S2A and S2B). Therefore, upregulation of L1 expression in Mael-null fetal oocytes correlated with their reduced viability.
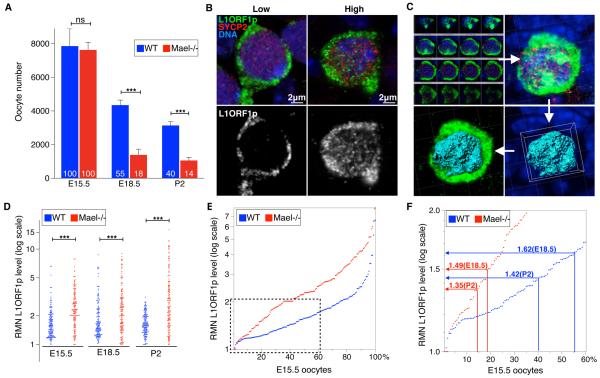
Figure 3. Viability of Wild-type and Mael-null Oocytes Correlates with Nuclear L1ORF1p Levels.
(A) Quantification of WT and Mael-null oocytes. Mean ± St.dev. from at least three independent measurements are shown (two-tailed unpaired Student's t-test, ns-P>0.05; ***-P<0.001).
(B) Variable (low and high) nuclear L1ORF1p levels (green) in E15.5 WT oocytes. SYCP2 (red) marks the synaptonemal complex.
(C) Representation of 3D reconstruction from single confocal sections, and generation of the nuclear surface (cyan) within which green signal intensity is quantified to determine L1ORF1p RMN levels in oocyte nuclei using Imaris Software.
(D) Dot-plot of RMN L1ORF1p levels in WT and Mael-null oocytes (two-tailed Kolmogorov-Smirnov test, ***-P<0.001).
(E) E15.5 WT and Mael-null oocytes ranked according to their RMN L1ORF1p levels.
(F) Chart relating RMN L1ORF1p levels in WT and Mael-null E15.5 oocytes to percentage of their survival to E18.5 and P2. Boxed area in (E) is shown in detail in (F).
See also Table S2.
Differential Accumulation of L1ORF1p in Fetal Oocyte Nuclei
We next wanted to understand how widespread derepression of L1 expression in the female germline could lead to elimination of not all but only a subpopulation of fetal oocytes. Since L1 must reach the oocyte genome to retrotranspose and cause DNA damage during this process (Belgnaoui et al., 2006; Gasior et al., 2006; Soper et al., 2008), we examined nuclear levels of L1ORF1p, an RNA-binding chaperone protein and essential component of L1RNPs (Martin, 2006; Martin et al., 2008; Martin et al., 2005), in fetal oocytes. Unexpectedly, we observed that oocytes of the same age and genotype exhibited significant variation of nuclear levels of L1ORF1p immunofluorescence (Figure 3B). To further characterize this variation, we developed an approach to determine relative mean nuclear (RMN) L1ORF1p levels in individual fetal oocytes (Figure 3C and Experimental Procedures). Using this approach, we quantified nuclear L1ORF1p levels in WT and Mael-null E15.5, E18.5 and P2 oocytes (Figure 3D, Tables S2C and S2D). This analysis revealed broad ranges of nuclear L1ORF1p accumulation even in the starting population of WT and Mael-null E15.5 oocytes with maximum values of 6.62 and 7.92 RMN units, respectively. In surviving WT E18.5 oocytes (55% of the starting E15.5 population), L1ORF1p levels peaked 8.7 RMN units compared to the maximum of 9.0 RMN units in Mael-null oocytes that survived to E18.5 (18% of the starting E15.5 population). Despite comparable ranges of L1ORF1p nuclear accumulation between the two genotypes, statistical analysis established that Mael-null E15.5 and E18.5 oocyte populations were distinct from their respective WT counterparts and contained more oocytes with higher nuclear L1ORF1p levels (Tables S2C and S2D). By P2, however, the maximum nuclear L1ORF1p level in WT oocytes (40% of the starting E15.5 population) decreased to 3.13 RMN units. In contrast, L1ORF1p levels remained high in viable Mael-null P2 oocytes (14% of the starting E15.5 population). These results established that WT and Mael-mutant fetal and early postnatal ovaries contained highly heterogeneous populations of MPI oocytes characterized by differential and dynamic accumulation of L1ORF1p in their nuclei.
The unexpected variation of L1ORF1p levels as early as E15.5 could arise from any combination of differential DNA demethylation and derepression of 11,000 full-length L1 elements in the mouse genome (Goodier et al., 2001) or asynchronous onset of epigenetic reprogramming. To reconcile oocyte viability data with differential L1ORF1p nuclear accumulation in both genotypes, we hypothesized that the developmental fate of a fetal oocyte (survival versus elimination) could be determined by its nuclear L1 content. This idea is based on prior evidence of increased DNA damage and meiotic defects following L1 activation (Belgnaoui et al., 2006; Gasior et al., 2006; Soper et al., 2008) and increased L1 expression and FOA in the absence of Mael (Figures 2G–I and 3A). A simplest scenario was that L1ORF1p nuclear levels (hence L1 activity) in an E15.5 oocyte determine the probability of its survival to E18.5 or P2. To test this model, we ranked E15.5 oocytes of both genotypes (representing starting oocyte populations) according to their RMN L1ORF1p values and intersected percent survival of E15.5 oocytes to E18.5 and P2 with corresponding RMN L1ORF1p curves (Figure 3E and 3F). This operation identified close maximum RMN values for WT and Mael-null oocytes corresponding to their survival to E18.5 (1.62 and 1.49 RMN units, respectively) and P2 (1.42 and 1.35 RMN units, respectively). This analysis suggested that WT and Mael-null E15.5 oocytes with L1ORF1p levels above the threshold of 1.6 RMN units were first to accumulate lethal levels of L1 activity before E18.5 while E15.5 oocytes with RMN L1ORF1p values from 1.35 to 1.6 units survived past E18.5 but were killed by P2. This analysis strongly supported our working hypothesis that L1 could play a role in FOA by causing elimination of oocytes with excessive L1 activity. Furthermore, in agreement with prior studies of L1 derepression on meiotic cells, this analysis predicted the appearance of non-meiotic DNA double-stranded breaks (DSBs) in WT fetal oocytes and increased incidence of meiotic defects in Mael-null fetal oocytes overexpressing L1.
Non-meiotic DNA Double-stranded Breaks in WT Fetal Oocytes
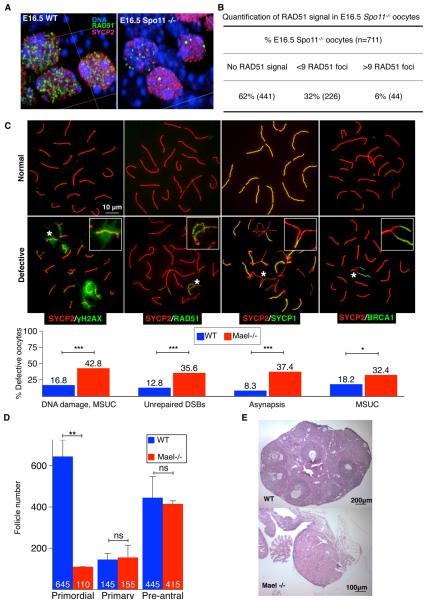
We have previously shown that derepression of L1 elements is accompanied by the appearance of SPO11-independent, non-meiotic DSBs (Romanienko and Camerini-Otero, 2000; Soper et al., 2008). By analogy with spermatocytes lacking L1 silencing, our observations made in fetal oocytes led us to test the prediction that Spo11-null but otherwise WT fetal oocytes should possess non-meiotic DSBs. Indeed, antibodies to RAD51 recombinase, a RecA homolog, revealed that 38% of E16.5 Spo11-null oocytes contained RAD51 foci indicative of non-meiotic DSBs (Figure 4A and 4B) (Moens et al., 1997). A majority of RAD51-positive oocytes (32% of all oocytes scored) possessed fewer than 9 RAD51 foci while 6% of oocytes contained numerous DSBs (Figure 4B). The presence of Spo11-independent DNA breaks in fetal oocytes, independently observed recently by others (Carofiglio et al., 2013), strongly supports our working hypothesis of a role for L1 elements in FOA.
Figure 4. Non-meiotic DSBs and Meiotic Prophase I Defects in Fetal Oocytes.
(A) A 3D-rendered image of a section of a WT and Spo11-null E16.5 ovary immunostained for RAD51 (green) and SYCP2 (red).
(B) Quantification of RAD51 foci in Spo11-null E16.5 oocytes.
(C) Representative images of pachytene meiotic spreads and quantification of defective patterns of DNA damage, DNA repair, synapsis and MSUC, assessed respectively with γH2AX, RAD51, SYCP1, BRCA1 antibodies (green). SYCP2 (red) marks synaptonemal complex lateral elements. Insets – close-ups of abnormal patterns indicated with asterisks. (Two-tailed Fisher's exact test, *-P<0.05; ***-P<0.001).
(D) Follicle counts in P19 WT and Mael−/− ovaries. Lack of primordial follicles is apparent in P19 Mael-mutant ovaries. No effect of Mael deletion is observed on the number of primary and pre-antral follicles. Mean ± St.dev. from at least three independent biological measurements are shown (two-tailed unpaired Student's t-test, ns-P>0.05; **-P<0.01).
(E) Hematoxylin/Eosin-stained sections of ovaries of a 1 year-old WT and Mael−/− females.
DNA Damage and Asynapsis in Mael-null Oocytes
We next assessed the impact of the Mael deletion (hence associated L1 overexpression) on MPI. We first compared the distribution of MPI substages in WT and Mael-null E18.5 ovaries and concluded that the Mael mutation did not alter the progression of MPI (Figure S3A and Table S3). Labeling of nuclear spreads of E18.5 pachytene oocytes for γH2AX, a marker of double-stranded breaks (DSBs) and unpaired meiotic chromatin, (Turner et al., 2005) identified that 42.8% of Mael-null oocytes harbored such defects compared to 16.8% in the WT (Figure 4C and Table S4A). Antibodies to RAD51 revealed unrepaired DSBs in 35.6% of Mael-null compared to 12.8% of WT oocytes (Figure 4C and Table S4A). Co-staining for synaptonemal complex markers SYCP1 and SYCP2 revealed incomplete homologous chromosome synapsis in 37.4% of Mael-null E18.5 oocytes compared to 8.3% in the wild-type (Yang and Wang, 2009) (Figure 4C and Table S4A). We also labeled fetal oocytes for BRCA1, a component of the synapsis quality control mechanism known as meiotic silencing of unpaired chromatin (MSUC) (Ichijima et al., 2011; Mahadevaiah et al., 2008; Schimenti, 2005; Turner, 2007; Turner et al., 2004; Turner et al., 2005). This analysis allows positive identification of even small areas of chromosome asynapsis but is limited to oocytes with asynapsis of fewer than two or three homologous chromosomes (Kouznetsova et al., 2009). This analysis revealed MSUC in 18.2% of WT E18.5 oocytes suggesting the presence of small areas of asynapsis that could not be reliably identified by SYCP1/SYCP2 co-staining (Figure 4C, S3C–S3F and Table S4A). In the Mael mutant, BRCA1 antibody labeled 32.4% of E18.5 oocytes which when compared to SYCP1/SYCP2 staining suggested a higher extent of asynapsis in the presence of elevated L1 expression (Figures 4C and S3C–S3F, and Table S4A). Overall, this analysis demonstrated that up to 18% of WT and 40% of Mael-null E18.5 oocytes possessed clear evidence of meiotic defects including unrepaired DSBs and asynapsis, known as primary triggers for oocyte elimination during the perinatal and early postnatal period (Di Giacomo et al., 2005).
Consistent with DNA damage- and asynapsis-activated oocyte loss during the perinatal period, the number of primordial follicles was severely reduced in P19 Mael-null ovaries (Figure 4D and Table S4B) (Di Giacomo et al., 2005). Furthermore, we did not observe signs of folliculogenesis in ovaries of 1 year-old mice (Figure 4E). Thus, increased L1 expression in Mael-mutant fetal oocytes is associated with extensive DNA damage, MPI defects, a severely reduced ovarian follicle reserve and shortened female reproductive lifespan.
Crossover and Meiotic Chromosome Segregation Defects in Mael-null Oocytes
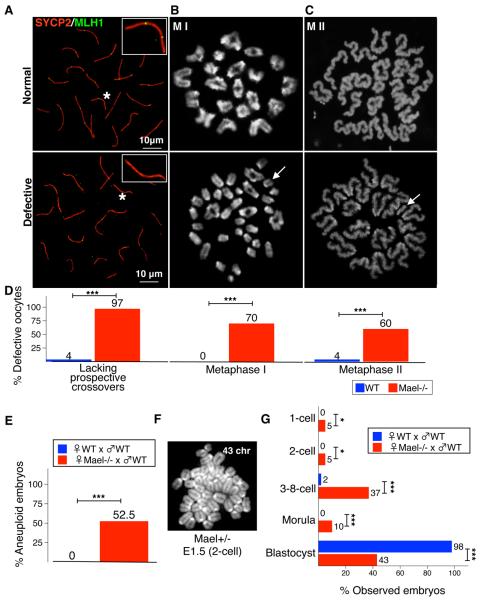
A crucial outcome of MPI is the formation of crossovers that are essential for the precise meiotic chromosome segregation. To assess the effect of the Mael mutation and associated increased L1 expression on crossover formation, we examined the localization of mismatch repair protein MLH1 normally recruited to 1–3 foci on each synaptonemal complex (Baker et al., 1996). Unexpectedly, we found that 97% of all Mael-null pachytene E18.5 oocytes with fully synapsed homologous chromosomes were lacking MLH1 foci on at least one chromosome bivalent (Figures 5A, 5D, S4A–S4C, Tables S4A, S4C and S4D). This defect could not be attributed to reduced Mlh1 mRNA and protein levels (Figure S4E–S4G). Consistent with this finding, 70% of metaphase I Mael-null oocytes possessed univalent chromosomes (Figure 5B and 5D, and Table S4A) while the remaining 30% mutant oocytes exhibited a significant reduction of the average number of chiasmata per oocyte (20.9±1.7 compared to 23.4±1.5 in the WT) (Table S4C). Among ovulated metaphase II Mael-null oocytes, 60% had abnormal karyotypes (Figure 5C and 5D, and Table S4A) and their fertilization with WT sperm resulted in 52.5% embryonic aneuploidy at the 2-cell stage (Figure 5E and 5F, and Table S5A, and 57% lethality before the blastocyst stage (Figure 5G and Table S5B). These observations suggest that elevated L1 expression might interfere with crossover formation leading to meiotic chromosome segregation errors in the adult ovary.
Figure 5. Crossover Defects, Aneuploidy and Premature Ovarian Oocyte Depletion in the Mael-null Mutant.
(A) Representative images and quantification of prospective crossovers identified using MLH1 antibody (green) in WT and Mael-null E18.5 pachytene nuclei. Insets - close-up views of synaptonemal complexes (identified with asterisks) with two and none MLH1 foci.
(B, C) Representative images and quantification of karyotypes in metaphase I and metaphase II WT and Mael-null oocytes. Arrow in defective MI oocyte (B) identifies one of 30 chromosomal univalents present in this sample. Arrow in defective MII oocyte (C) identifies a single extra chromosome.
(D) Quantification of defective oocytes in (A–C).
(E) Quantification of aneuploid karyotypes of 2-cell embryos obtained from Mael-null and control WT oocytes.
(F) Representative image of an aneuploid karyotype (43 chromosomes) of an E1.5 Mael+/− embryo.
(G) Staging of Mael+/− embryos developed from Mael-null oocytes and control WT embryos collected at E3.5 (1C - 1-cell, 2C - 2-cell, 3-8C - 3- to 8-cell arrested embryos, M - Morula, and B - Blastocyst). Two-tailed Fisher's exact test, ns-P>0.05; **-P<0.01, ***-P<0.001 (D, E and I).
Conditional Expression of a L1 Element ORFeus Enhances Fetal Oocyte Attrition and Meiotic Defects
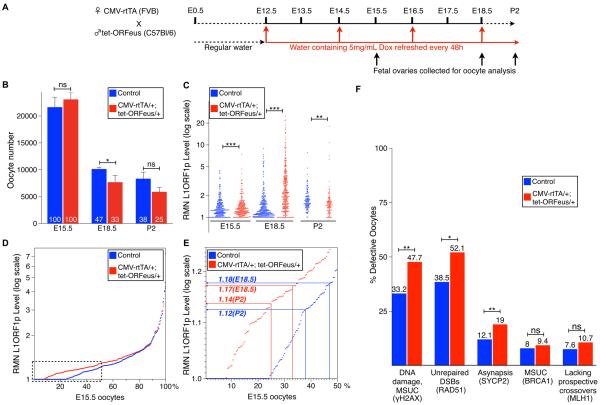
We next asked if Mael-mutant phenotypes could be reproduced by directly increasing L1 expression in fetal oocytes. To substantially increase L1 expression over an already high background level of expression of an as many as 11,000 full-length endogenous L1 elements in the mouse genome (Goodier et al., 2001) (Figure 1B), we used a recently described mouse transgene carrying a tetracycline-controlled L1 element (ORFeus) (Figure 6A) (O'Donnell et al., 2013). Consistent with our earlier results, doxycycline-induced CMV-rtTA/+; tet-ORFeus/+ (FVBB6F1) fetal ovaries contained 14 and 13 percentage points fewer oocytes at E18.5 and P2, respectively, compared to the control group comprising non-double transgenic littermates (Figure 6B, Tables S6A–S6C and Experimental Procedures). Quantification of L1ORF1p revealed the appearance of E15.5 CMV-rtTA/+; tet-ORFeus/+ oocytes with increased nuclear levels of the protein that peaked at E18.5 but become depleted by P2 (Figure 6C, Tables S6D and S6E). Despite a potentially slight delay in the onset of ORFeus expression, RMN L1ORF1p values in the E15.5 control and double transgenic populations were nonetheless significantly distinct to reveal a correlation between the survival of E15.5 oocytes to E18.5 and P2 with their nuclear L1ORF1p levels (Figure 6D and 6E).
Figure 6. Effects of Doxycycline-inducible tet-ORFeus Expression on Fetal Oocytes.
(A) Diagram of experimental induction of tet-ORFeus expression.
(B) Quantification of oocytes in doxycycline-induced non-double transgenic control and CMV-rtTA/+; tet-ORFeus/+ embryos. Mean ± St.dev. from at least three independent biological measurements are shown (two-tailed unpaired Student's t-test, ns-P>0.05; *-P<0.05; ***-P<0.001).
(C) Dot-plot of RMN L1ORF1p levels in non-double transgenic control and CMV-rtTA/+; tet-ORFeus/+ doxycycline-induced oocytes. Each dot represents the RMN level in an individual oocyte nucleus. (two-tailed Kolmogorov-Smirnov test, *-P<0.05; **-P<0.01; ***-P<0.001).
(D) E15.5 non-double transgenic control and CMV-rtTA/+; tet-ORFeus/+ oocytes ranked according to their RMN L1ORF1p levels. Boxed area is shown in detail in (E).
(E) Chart relating RMN L1ORF1p levels in non-double transgenic control and CMV-rtTA/+; tet-ORFeus/+ E15.5 oocytes to percentage of their survival to E18.5 and P2.
(F) Quantification of E18.5 non-double transgenic control and CMV-rtTA/+; tet-ORFeus/+ doxycycline-induced oocytes exhibiting evidence of DNA damage and MSUC (by γH2AX staining), persistent DSBs (by RAD51 foci), asynapsis (by abnormal SYCP2 staining), MSUC (by BRCA1 staining) and lacking MLH1 foci. (Two-tailed Fisher's exact test, ns-P>0.05; *-P<0.05; **-P<0.01).
To assess the effect of ORFeus overexpression on MPI, we first compared the distribution of MPI substages in doxycycline-induced control and CMV-rtTA/+; tet-ORFeus/+ E18.5 ovaries and determined that ORFeus overexpression did not alter the MPI progression (Figure S3B). We next examined both groups of E18.5 oocytes for the presence of MPI defects. Consistent with prior results of others (Koehler et al., 2006), we observed an increased baseline level of meiotic defects in oocytes of mixed genetic background. Nevertheless, doxycycline-induced E18.5 CMV-rtTA/+; tet-ORFeus/+ oocytes still accumulated RMN L1ORF1p levels sufficient to further increase DNA damage and chromosome asynapsis in oocytes surviving to E18.5 (Figure 6F and Table S6F). Subsequent labeling of E18.5 CMV-rtTA/+; tet-ORFeus/+ oocytes for BRCA1 suggested that ORFeus overexpression caused extensive chromosome asynapsis as indicated by a lower percent of MSUC-positive oocytes (9.4%) compared to 19% of asynapsed oocytes as determined by SYCP2 staining (Figure 6F and Table S6F). At the same time, ORFeus overexpression did not cause a statistically significant increase of oocytes lacking MLH1 foci on at least one chromosome bivalent (Figure 6F, Tables S6F and S6G). Perhaps the mechanism underlying the inhibitory effect of L1 on MLH1 localization to prospective crossover sites requires stronger L1 expression than offered by a single ORFeus transgene. Nonetheless, considered as a whole, the analysis showed that direct overexpression of a single L1 element in the fetal ovary had substantially enhanced oocyte elimination and reinforced the idea that L1 activity contributes to FOA.
Reverse Transcriptase Inhibitor AZT Alters Dynamics of Fetal Oocyte Attrition
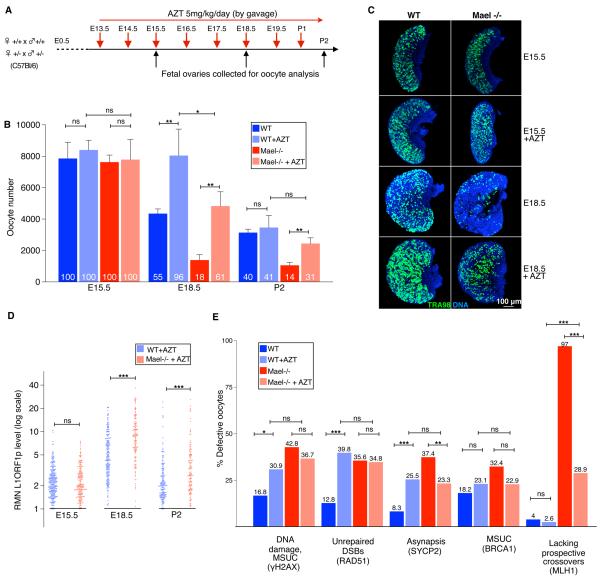
To test the key prediction of our working hypothesis that inhibition of L1 activity should reduce FOA, we administered pregnant female mice with azidothymidine (AZT), a nucleoside analog inhibitor of reverse transcriptases (RTs) including L1ORF2p, which has independent RT and endonuclease activities (Dai et al., 2011; Feng et al., 1996; Jones et al., 2008) (Figure 7A). We first determined that AZT treatment did not perturb MPI progression or induce oocyte proliferation (Figures S3A and S5). Quantification of the oocyte content of AZT-treated WT and Mael-null ovaries revealed a profound yet complex effect of the drug on oocyte viability (Figure 7B, Tables S7A–S7C). In contrast to untreated controls, AZT-treated WT E18.5 ovaries retained practically an entire starting oocyte population (96%). Likewise, 61% of the starting oocyte population survived in the AZT-treated E18.5 Mael-mutant ovaries. The effect of AZT on E18.5 oocyte survival was sufficiently robust to be apparent in whole-mount oocyte staining of WT and Mael-null E18.5 ovaries (Figure 7C). This result strongly supported the idea of a role of L1 in FOA and suggested that an RT-dependent intermediate or step during L1 retrotransposition triggers oocyte elimination in the early MPI (E15.5 – E18.5), before the engagement of checkpoints monitoring the completion of meiotic DNA recombination and synapsis starting at E18.5.
Figure 7. Fetal Oocyte Attrition and Meiotic Prophase I Progression in AZT-treated Mice.
(A) Diagram of AZT treatment of pregnant WT and Mael+/− mutant females.
(B) Quantification of untreated and AZT-treated WT and Mael-null oocytes. Mean ± St.dev. from at least three independent measurements are shown (two-tailed unpaired Student's t-test, ns-P>0.05; ***-P<0.001; ** - P<0.01, *-P<0.05).
(C) Whole mount TRA98 (green) labeling of fetal oocytes in untreated and AZT-treated WT and Mael-null ovaries.
(D) Dot-plot of RMN L1ORF1p levels in AZT-treated WT and Mael-null oocytes (two-tailed Kolmogorov-Smirnov test, ns-P>0.05, ***-P<0.001).
(E) Quantification of untreated and AZT-treated WT and Mael-null E18.5 oocytes exhibiting evidence of DNA damage and MSUC (by γH2AX staining), persistent DSBs (by RAD51 foci), asynapsis (by abnormal SYCP2 staining), MSUC (by BRCA1 staining) and lacking MLH1 foci. (Two-tailed Fisher's exact test, ns-P>0.05; ***-P<0.001; ** - P<0.01, *-P<0.05).
See also Figures S3 and S5, and Tables S3 and S7.
Quantification of L1ORF1p immunofluorescence in AZT-treated WT and Mael-null E18.5 oocytes revealed the presence of predicted but previously undetected oocytes with L1ORF1p levels greatly exceeding those observed in untreated ovaries (Figure 7D, Tables S7D–S7F). L1ORF1p nuclear levels peaked at 20.7 and 36.8 RMN units in AZT-treated WT and Mael-null E18.5 oocytes, respectively. Since practically all AZT-treated WT oocytes survived to E18.5, L1ORF1p nuclear accumulation in WT fetal oocytes has a maximum of about 20 RMN units. In light of this data and an up to 2.4-fold increase in L1 expression levels in the Mael-null ovaries (Figure 2D–2I), AZT-treated Mael-null oocytes should have peaked at 46 RMN units but in reality did not exceed the maximum of 36.8 RMN units. Most likely, the concentration of AZT used in our experiments (5mg/kg/day) was insufficient to block RT in Mael-null oocytes with most L1 expression resulting in elimination of 39% of the starting oocyte population. Taken together, these observations support the idea of preferential elimination of fetal oocytes with excessive levels of nuclear L1ORF1p during FOA.
Despite the strong protective effect of AZT on early (E15.5–E18.5) MPI oocytes, we found that AZT-treated WT and Mael-mutant P2 ovaries contained 41% and 31% of the starting oocyte populations, respectively. A sharp drop in oocyte viability was not unexpected as AZT does inhibit the endonuclease activity of L1ORF2p. Up to 40% of AZT-treated WT E18.5 oocytes exhibited MPI defects, which is consistent with the percent of eliminated E18.5 oocytes in untreated WT ovaries (Figure 7E, Tables S7G and S7H). Likewise, in addition to 39% of Mael-null oocytes eliminated by E18.5, up to 37% of AZT-treated E18.5 oocytes possessed elevated DNA damage and asynapsis. These defects were the likely triggers of checkpoint activation and a sharp decline in oocyte numbers in the early postnatal period (Figure 7B, Tables S7A and S7B) (Di Giacomo et al., 2005).
Finally, we examined the effect of AZT treatment on the number and distribution of MLH1 foci. In a stark contrast to untreated Mael-null E18.5 oocytes 97% of which lacked MLH1 foci on at least one chromosome bivalent, we found that only 28.9% of AZT treated Mael-null E18.5 oocytes exhibited such a defect (Figure 7E, Tables S7G–S7J). The mean number of MLH1 foci in Mael-null E18.5 oocytes increased from 9.97±5.32 without treatment to 22.76±6.24 in the presence of AZT (Figure S5 and Table S7I). Furthermore, quantification of MLH1 foci in AZT-treated E18.5 WT oocytes revealed a statistically significant increase in the mean number of MLH1 foci from 26.55±5.94 to 28.83±4.9 (Figure S5 and Tables S7I, S7J). These findings suggest that an RT-dependent intermediate of L1 retrotransposition interferes with crossover formation not only in the absence of Mael but also under basal conditions of L1 expression in WT MPI oocytes.
Considered as a whole, the results of analysis of WT and Mael-mutant ovaries treated with AZT are highly consistent with the hypothesis that L1 activity plays a role in FOA and suggest that intermediates of L1 retrotransposition such as L1 RNA-DNA hybrids might trigger elimination of early MPI oocytes.
DISCUSSION
In this work, we examined a role of retrotransposon L1 in FOA in mice. We reasoned that expression of L1 elements following their activation in the course of epigenetic reprogramming could be detrimental to viability of MPI fetal oocytes. We have uncovered differential accumulation of nuclear L1ORF1p, hence L1RNPs, among WT fetal oocytes. By studying fetal oocytes of WT and L1-overexpressing mice we were able to establish a correlation between fetal oocyte nuclear L1ORF1p levels and oocyte viability. We used two different strategies to increase L1 expression in fetal oocytes to assess if high levels of L1 associate with enhanced MPI defects and oocyte elimination. A null mutation in the Mael gene allowed an up to 2.4-fold increase in global L1 expression which resulted in a 3-fold reduction in the number of fetal oocytes at birth. Induction of expression of a single L1 element ORFeus at the onset of MPI has also resulted in reduced oocyte survival which can be correlated with nuclear L1ORF1p levels. While not as strong as the Mael-mutant phenotype, the effect of ORFeus overexpression is quite significant especially in light of the fact that the mouse genome is estimated to encode 11,000 full-length L1 elements (Goodier et al., 2001). A central role of L1 in FOA is also strongly supported by results of experiments using RT inhibitor AZT whose administration had an immediate positive effect on the dynamics of FOA in WT and Mael-mutant ovaries. Furthermore, AZT treatment caused a 3-fold increase in the number of Mael-null oocytes with normal localization of MLH1 protein to prospective crossover sites. Overall, our data support the “death by defect” model of FOA (Tilly, 2001) with both reverse transcriptase and endonuclease activities of L1ORF2p contributing to oocyte elimination. AZT-dependent protection of oocytes during early MPI (E15.5–E18.5) suggests that L1 RNA-DNA hybrids, formed in the process of target-primed retrotransposition of L1 elements, could trigger FOA. This unexpected finding and the presence of non-meiotic DSBs agree with an earlier observation that both domains of L1ORF2p contributed individually to cytotoxicity of expression of human L1 element in HeLa cells (Wallace et al., 2008).
Our work has yielded two additional significant findings. First, we observed differential nuclear accumulation of L1ORF1p among otherwise genetically identical oocytes including in the wild type. The mechanism behind this observation remains to be elucidated. Perhaps the onset or extent of DNA demethylation of the mouse genome varies between individual primordial germ cells and MPI oocytes leading to differential expression of L1 elements (Kagiwada et al., 2013; Ohno et al., 2013). L1 mobilization during early embryogenesis might also lead to a greater diversity of genotypes than had been anticipated previously (Kano et al., 2009). Second, our data suggest that L1 activity in fetal oocytes might contribute to meiotic chromosome segregation errors in the adult ovary. The mechanism of this phenomenon is not yet clear but the rescue of the MLH1 localization defect by AZT implicates newly synthesized L1 cDNA or RNA-DNA intermediates in this process, perhaps by diverting mismatch repair machinery from sites of prospective crossovers to numerous sites of L1 insertions spread throughout the genome. This observation suggests that the maternal bias of meiotic chromosome segregation errors might arise from the concurrency of developmental timing of MPI and epigenetic reprogramming in fetal oocytes. Considered together, our results lead us to conclude that L1 activity could be a potent determinant of both the size and quality of a mammalian ovarian oocyte reserve.
Finally, with respect to the rationale behind the massive loss of oocytes, we propose that FOA selects oocytes with low L1 activity and, therefore, best suited for the next generation. In the long run, limited exposure to L1 might result in novel L1 insertions and, therefore, negative effects of L1-driven oocyte demise might be balanced by positive effects of increased genetic diversity.
EXPERIMENTAL PROCEDURES
Animals
ES cell targeting, Mael-mutant mice derivation, backcross to C57BL/6J genetic background for >10 generations and genotyping were described previously (Soper et al., 2008). Spo11 mutant mice were the gift of Dr. Camerini-Otero (NIDDK) (Romanienko and Camerini-Otero, 2000). Transgenic CMVrtTA/+ females in FVB genetic background (kindly provided by H.Varmus) (Sotillo et al., 2007) were mated with transgenic tet-ORFeus/+ males in C57BL/6J genetic background (O'Donnell et al., 2013). All experimental procedures were performed in compliance with ethical regulations and approved by the IACUC of Carnegie Institution for Science.
Oocyte and follicle counting
We scored all oocytes present in every fifth cryosection throughout the entire ovary. To obtain an estimate of the total number of oocytes per ovary, the numbers scored per sections was multiplied by 5 (Tilly, 2003). For statistical analysis we compared average number of oocytes per ovary counted in pairs of ovaries coming from embryos from different litters as indicated in supplemental tables (Tables S2A and S2B, S6A–S6C, S7A–S7C). To quantify follicles in postnatal ovaries, we scored follicles as “primordial” if they contained an intact oocyte with a visible nucleolus surrounded by a single layer of fusiform-shaped granulosa cells. We scored follicles as “primary” if they consisted of an intact, enlarged oocyte with a visible nucleolus and a single layer of cuboidal granulosa cells. We scored follicles as “preantral/antral” if they contained an oocyte with a visible nucleolus and more than one layer of granulosa or thecal cells. For statistical analysis we compared average number of follicles per ovary counted in pairs of ovaries coming from females from different litters as indicated in supplemental tables (Table S4B).
Induced ORFeus expression
To induce tet-ORFeus expression, doxycycline hyclate (Sigma-Aldrich, Cat#:D9891) (5mg/mL in drinking water supplemented with 1% sucrose) was provided ad libitum in opaque bottles to 2–6 months old transgenic CMV-rtTA/+ females mated with transgenic male tet-ORFeus/+ starting at 12.5 days post coitum (dpc) and refreshed every 48 hours. Treated females were dissected at 15.5 or 18.5 dpc, or P2 for subsequent analysis of ovaries. The effect of ORFeus activation on oocyte fate were assessed by comparing embryonic ovaries from female double transgenic (CMV-rtTA/+; tet-ORFeus/+) with their female littermates non DOX-inducible non-double transgenics (+/+; tet-ORFeus/+), (CMV-rtTA/+; +/+), (+/+; +/+) (O'Donnell et al., 2013). An independent one-sample t-test (Table S6C) demonstrates that the number of oocytes did not differ between different non-double transgenic littermates.
AZT administration
To inhibit L1 reverse transcriptase in embryonic oocytes, pregnant 2–6 months old Mael+/− females, mated with male Mael+/− males, received 5mg/kg/day AZT (Sigma Aldrich, Cat#. A2169) by gavage starting at 12.5 days post coitum (dpc). Treated females were dissected at 15.5 or 18.5 dpc, or P2 for subsequent analysis of oocytes. The route and doses of AZT used in this study were based on previous reports (Dai et al., 2011; Sharpe et al., 1988; Sharpe et al., 1987; Toltzis et al., 1991).
Analysis of L1ORF1p nuclear signal
We co-immunolabelled cryosections of embryonic ovaries for L1ORF1p and germ cell marker (SYCP2 or TRA98), and counterstained with DAPI. We performed confocal imaging using SP5 confocal microscope (Leica, Exton, PA). The 3-D reconstruction of generated Z-stacked images was performed using Imaris Software (Bitplane). For each 3-D reconstructed image we determined the 3-D DAPI-rendered surfaces in individual oocyte nuclei and collected mean intensity of green signals inside surfaces, corresponding to the mean intensity of nuclear oocyte L1ORF1p signal. To eliminate the background signals, in parallel to oocyte nuclei analysis, on each image we determine the 3D DAPI-rendered surfaces of 3 ovarian somatic cell nuclei (with different location on the image) and collected mean intensity of green signals inside somatic cell nuclear surfaces. For further analysis, we determined the relative mean nuclear (RMN) L1ORF1p signals of oocytes for each image by normalizing nuclear mean intensity in each examined oocyte to the average of nuclear mean intensity of L1ORF1p signal of 3 ovarian somatic nuclei from the same image. We collected RMN from at least 99 oocytes in different images of ovaries from at least 3 different embryos for each embryonic stage and genotype.
RNA-Seq analysis
We purified total RNA from E15.5 WT and Mael−/− embryonic ovaries using TRIzol (Invitrogen). We depleted total RNA samples from ribosomal RNA using the Ribo-Zero™ rRNA Removal Kit (Epicentre) accordingly to the manufacturer's instructions. rRNA removal was visually assessed by Agilent 2100 Bioanalyzer (Agilent) RNA electropherograms. Library construction, cluster generation and sequencing on HiSeq 2000 (Illumina) were performed according to manufacturer's protocols. We obtained a total of 8.2 – 8.9 x107 filtered 100-bp reads for each ovarian sample. We used TopHat and Cufflinks to align reads to mouse reference genome NCBI37/mm9 and to identify differentially expressed genes (Trapnell et al., 2009; Trapnell et al., 2010). To account for the relatively complex cellular composition of the fetal ovary and anticipated variation in the number of fetal oocytes between samples, we focused on two WT and two mutant samples with best Pearson's r correlation coefficients (Figure S2B). Transcripts with “significant” Cuffdiff scores, FPKM count ≥ 5 and a ≥ 2-fold increase in expression in all four Mael mutant - WT pairwise comparisons were considered reliably altered. By these criteria, only one gene (besides Mael) was identified as downregulated (Table S1A) and 120 genes as upregulated in Mael-mutant ovaries compared to WT (Table S1B). By the same criteria, no known germ-cell or meiotic genes were found to be expressed differentially.
For analysis of expression of L1 elements belonging to young L1 families, we aligned the reads to 2383 intact, full-length L1 insertions in the mouse genome from the L1Base (http://l1base.molgen.mpg.de) (Penzkofer et al., 2005; Zemojtel et al., 2007). We used Bowtie software (Langmead et al., 2009) and allowed for 2 mismatches. To account for the cellular complexity of WT and Mael-mutant ovaries analyzed and compared we normalized count of mapped reads to the level of expression of a germ cell marker, Mouse Vasa Homolog (MVH) and a housekeeping gene Actin beta.
Data Deposition
High throughput sequencing data have been submitted to NCBI Short Read Archive under accession number SRP040979.
Gene Ontology Analysis
Gene Ontology analysis of genes upregulated in Mael-null ovaries was done using GOEAST software (Tables S1C and S1D) (Zheng and Wang, 2008). GO analysis is consistent with upregulation of apoptotic genes and contamination with cells of the hematopoietic lineage.
Statistical Analysis
We performed all measurements independently at least in triplicate. We analyzed random sample populations from at least 3 different samples from 3 random animals. Sample size populations are indicated in Supplemental Tables. The genotyping to allocate animals to different experimental groups were performed after sample processing. The random sample size populations used in our analyses are representative of the entire population with a 90 to 95% confidence interval and 95% confidence level (Creative Research Systems survey software http://www.surveysystem.com/sscalc.htm). Scoring of samples for analyses of meiotic prophase I progression were performed with blinding genotypes. We assessed differences between quantitative variables for normal populations of small and large sizes with the two-tailed unpaired Student's t-test (Figure 2G and 2I, Tables S2B, S4B, S4C, S4D, S5A, S5B, S6B, S6C, S6G, S7B, S7C, S7I, S7J) and with the one-sample t-test (Table S6C) (QuickCalcs http://www.graphpad.com/quickcalcs/index.cfm); two-tailed Kolmogorov-Smirnov test for non-normal populations of large size (Tables S2D, S6E, S7E, S7F) (http://www.physics.csbsju.edu). We assessed qualitative variables with two-tailed Fisher's exact test of populations of small and large sizes (Tables S4A, S5B, S6F, S7G, S7H) and chi-square test (Table S3B) (QuickCalcs http://www.graphpad.com/quickcalcs/index.cfm). Throughout the report statistical significance is indicated as (*) for p<0.05, (**) for p<0.01, (***) for p<0.001 and (ns) for p>0.05.
Details of the other standard methods and reagents are described in the Supplemental Experimental Procedures.
Supplementary Material
Highlights
Differential nuclear retrotransposon L1 levels in meiotic prophase I fetal oocytes
Increased L1 expression precipitates oocyte genome damage and meiotic defects
AZT implicates RNA/DNA hybrids in triggering oocyte death and chiasma failure
L1 activity underlies fetal oocyte attrition, reduced ovarian reserves and aneuploidy
ACKNOWLEDGEMENTS
We thank M. Halpern for manuscript critique, N. Ingolia, A. Pinder and F. Tan for RNA-Seq assistance; D. Camerini-Otero for Spo11 mice, P.J. Wang, B.R. Cullen, and R. Kanaar for antibodies. The work was supported by the endowment of Carnegie Institution for Science, by a CPRIT R1101 Award (K.A.O'D) and by the NIH grant GM40367 (S.L.M.). S.M. was a recipient of the EMBO long-term (ALTF 543-2006) and McClintock fellowships. G.W.H. was a recipient of Hollaender fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION Supplemental Information includes Supplemental Experimental Procedures, 5 Supplemental figures and 7 Supplemental tables and can be found with this article online at http://
AUTHOR CONTRIBUTIONS S.M. and A.B. conceived and designed the experiments. S.M. performed all experiments. S.M. and A.B. analyzed the data and wrote the paper. G.W.H., S.L.M. and K.A.O'D contributed expertise and reagents.
The authors declare no conflict of interests.
REFERENCES
- Aravin AA, Bourc'his D. Small RNA guides for de novo DNA methylation in mammalian germ cells. Genes Dev. 2008;22:970–975. doi: 10.1101/gad.1669408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Bourc'his D, Schaefer C, Pezic D, Toth KF, Bestor T, Hannon GJ. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Molecular cell. 2008;31:785–799. doi: 10.1016/j.molcel.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin AA, van der Heijden GW, Castaneda J, Vagin VV, Hannon GJ, Bortvin A. Cytoplasmic compartmentalization of the fetal piRNA pathway in mice. PLoS Genet. 2009;5:e1000764. doi: 10.1371/journal.pgen.1000764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker SM, Plug AW, Prolla TA, Bronner CE, Harris AC, Yao X, Christie DM, Monell C, Arnheim N, Bradley A, et al. Involvement of mouse Mlh1 in DNA mismatch repair and meiotic crossing over. Nat Genet. 1996;13:336–342. doi: 10.1038/ng0796-336. [DOI] [PubMed] [Google Scholar]
- Baker TG. A Quantitative and Cytological Study of Germ Cells in Human Ovaries. Proc R Soc Lond B Biol Sci. 1963;158:417–433. doi: 10.1098/rspb.1963.0055. [DOI] [PubMed] [Google Scholar]
- Baker TG. A quantitative and cytological study of oogenesis in the rhesus monkey. Journal of anatomy. 1966;100:761–776. [PMC free article] [PubMed] [Google Scholar]
- Barlow C, Liyanage M, Moens PB, Tarsounas M, Nagashima K, Brown K, Rottinghaus S, Jackson SP, Tagle D, Ried T, et al. Atm deficiency results in severe meiotic disruption as early as leptonema of prophase I. Development. 1998;125:4007–4017. doi: 10.1242/dev.125.20.4007. [DOI] [PubMed] [Google Scholar]
- Beaumont HM, Mandl AM. A quantitative and cytological study of oognonia and oocytes in the foetal and nonatal rat. Proc R Soc Lond B Biol Sci. 1962;155:557–579. [Google Scholar]
- Beck CR, Collier P, Macfarlane C, Malig M, Kidd JM, Eichler EE, Badge RM, Moran JV. LINE-1 retrotransposition activity in human genomes. Cell. 2010;141:1159–1170. doi: 10.1016/j.cell.2010.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgnaoui SM, Gosden RG, Semmes OJ, Haoudi A. Human LINE-1 retrotransposon induces DNA damage and apoptosis in cancer cells. Cancer Cell Int. 2006;6:13. doi: 10.1186/1475-2867-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergeron L, Perez GI, Macdonald G, Shi L, Sun Y, Jurisicova A, Varmuza S, Latham KE, Flaws JA, Salter JC, et al. Defects in regulation of apoptosis in caspase-2-deficient mice. Genes & development. 1998;12:1304–1314. doi: 10.1101/gad.12.9.1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourc'his D, Bestor TH. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature. 2004;431:96–99. doi: 10.1038/nature02886. [DOI] [PubMed] [Google Scholar]
- Brouha B, Schustak J, Badge RM, Lutz-Prigge S, Farley AH, Moran JV, Kazazian HH., Jr. Hot L1s account for the bulk of retrotransposition in the human population. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:5280–5285. doi: 10.1073/pnas.0831042100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgoyne PS, Baker TG. Perinatal oocyte loss in XO mice and its implications for the aetiology of gonadal dysgenesis in XO women. J Reprod Fertil. 1985;75:633–645. doi: 10.1530/jrf.0.0750633. [DOI] [PubMed] [Google Scholar]
- Carmell MA, Girard A, van de Kant HJ, Bourc'his D, Bestor TH, de Rooij DG, Hannon GJ. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell. 2007;12:503–514. doi: 10.1016/j.devcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Carofiglio F, Inagaki A, de Vries S, Wassenaar E, Schoenmakers S, Vermeulen C, van Cappellen WA, Sleddens-Linkels E, Grootegoed JA, Te Riele HP, et al. SPO11-independent DNA repair foci and their role in meiotic silencing. PLoS genetics. 2013;9:e1003538. doi: 10.1371/journal.pgen.1003538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai L, Huang Q, Boeke JD. Effect of reverse transcriptase inhibitors on LINE-1 and Ty1 reverse transcriptase activities and on LINE-1 retrotransposition. BMC Biochem. 2011;12:18. doi: 10.1186/1471-2091-12-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La Fuente R, Baumann C, Fan T, Schmidtmann A, Dobrinski I, Muegge K. Lsh is required for meiotic chromosome synapsis and retrotransposon silencing in female germ cells. Nat Cell Biol. 2006;8:1448–1454. doi: 10.1038/ncb1513. [DOI] [PubMed] [Google Scholar]
- Di Giacomo M, Barchi M, Baudat F, Edelmann W, Keeney S, Jasin M. Distinct DNA-damage-dependent and -independent responses drive the loss of oocytes in recombination-defective mouse mutants. Proc Natl Acad Sci U S A. 2005;102:737–742. doi: 10.1073/pnas.0406212102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doucet AJ, Hulme AE, Sahinovic E, Kulpa DA, Moldovan JB, Kopera HC, Athanikar JN, Hasnaoui M, Bucheton A, Moran JV, et al. Characterization of LINE-1 ribonucleoprotein particles. PLoS genetics. 2010;6 doi: 10.1371/journal.pgen.1001150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ene AC, Park S, Edelmann W, Taketo T. Caspase 9 is constitutively activated in mouse oocytes and plays a key role in oocyte elimination during meiotic prophase progression. Developmental biology. 2013;377:213–223. doi: 10.1016/j.ydbio.2013.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q, Moran JV, Kazazian HH, Jr., Boeke JD. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. 1996;87:905–916. doi: 10.1016/s0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- Gasior SL, Wakeman TP, Xu B, Deininger PL. The human LINE-1 retrotransposon creates DNA double-strand breaks. J Mol Biol. 2006;357:1383–1393. doi: 10.1016/j.jmb.2006.01.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghafari F, Gutierrez CG, Hartshorne GM. Apoptosis in mouse fetal and neonatal oocytes during meiotic prophase one. BMC Dev Biol. 2007;7:87. doi: 10.1186/1471-213X-7-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodier JL, Kazazian HH., Jr. Retrotransposons revisited: the restraint and rehabilitation of parasites. Cell. 2008;135:23–35. doi: 10.1016/j.cell.2008.09.022. [DOI] [PubMed] [Google Scholar]
- Goodier JL, Ostertag EM, Du K, Kazazian HH., Jr. A novel active L1 retrotransposon subfamily in the mouse. Genome research. 2001;1:1677–1685. doi: 10.1101/gr.198301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajkova P, Erhardt S, Lane N, Haaf T, El-Maarri O, Reik W, Walter J, Surani MA. Epigenetic reprogramming in mouse primordial germ cells. Mech Dev. 2002;117:15–23. doi: 10.1016/s0925-4773(02)00181-8. [DOI] [PubMed] [Google Scholar]
- Hartshorne GM, Lyrakou S, Hamoda H, Oloto E, Ghafari F. Oogenesis and cell death in human prenatal ovaries: what are the criteria for oocyte selection? Mol Hum Reprod. 2009;15:805–819. doi: 10.1093/molehr/gap055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichijima Y, Ichijima M, Lou Z, Nussenzweig A, Camerini-Otero RD, Chen J, Andreassen PR, Namekawa SH. MDC1 directs chromosome-wide silencing of the sex chromosomes in male germ cells. Genes & development. 2011;25:959–971. doi: 10.1101/gad.2030811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannou JM. Ooegenesis in the Guinea-Pig. Journal of embryology and experimental morphology. 1964;12:673–691. [PubMed] [Google Scholar]
- Jones RB, Garrison KE, Wong JC, Duan EH, Nixon DF, Ostrowski MA. Nucleoside analogue reverse transcriptase inhibitors differentially inhibit human LINE-1 retrotransposition. PloS one. 2008;3:e1547. doi: 10.1371/journal.pone.0001547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagiwada S, Kurimoto K, Hirota T, Yamaji M, Saitou M. Replication-coupled passive DNA demethylation for the erasure of genome imprints in mice. The EMBO journal. 2013;32:340–353. doi: 10.1038/emboj.2012.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kano H, Godoy I, Courtney C, Vetter MR, Gerton GL, Ostertag EM, Kazazian HH., Jr. L1 retrotransposition occurs mainly in embryogenesis and creates somatic mosaicism. Genes Dev. 2009;23:1303–1312. doi: 10.1101/gad.1803909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazazian HH., Jr. Mobile elements: drivers of genome evolution. Science. 2004;303:1626–1632. doi: 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- Koehler KE, Schrump SE, Cherry JP, Hassold TJ, Hunt PA. Near-human aneuploidy levels in female mice with homeologous chromosomes. Curr Biol. 2006;16:R579–580. doi: 10.1016/j.cub.2006.07.018. [DOI] [PubMed] [Google Scholar]
- Kouznetsova A, Wang H, Bellani M, Camerini-Otero RD, Jessberger R, Hoog C. BRCA1-mediated chromatin silencing is limited to oocytes with a small number of asynapsed chromosomes. Journal of cell science. 2009;122:2446–2452. doi: 10.1242/jcs.049353. [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Totoki Y, Toyoda A, Ikawa M, Asada N, Kojima K, Yamaguchi Y, Ijiri TW, et al. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev. 2008;22:908–917. doi: 10.1101/gad.1640708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurilo LF. Oogenesis in antenatal development in man. Human genetics. 1981;57:86–92. doi: 10.1007/BF00271175. [DOI] [PubMed] [Google Scholar]
- Lane N, Dean W, Erhardt S, Hajkova P, Surani A, Walter J, Reik W. Resistance of IAPs to methylation reprogramming may provide a mechanism for epigenetic inheritance in the mouse. Genesis. 2003;35:88–93. doi: 10.1002/gene.10168. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome biology. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lees-Murdock DJ, Walsh CP. DNA methylation reprogramming in the germ line. Epigenetics. 2008;3:5–13. doi: 10.4161/epi.3.1.5553. [DOI] [PubMed] [Google Scholar]
- Lei L, Spradling AC. Mouse primordial germ cells produce cysts that partially fragment prior to meiosis. Development. 2013;140:2075–2081. doi: 10.1242/dev.093864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim AK, Kai T. Unique germ-line organelle, nuage, functions to repress selfish genetic elements in Drosophila melanogaster. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:6714–6719. doi: 10.1073/pnas.0701920104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevaiah SK, Bourc'his D, de Rooij DG, Bestor TH, Turner JM, Burgoyne PS. Extensive meiotic asynapsis in mice antagonises meiotic silencing of unsynapsed chromatin and consequently disrupts meiotic sex chromosome inactivation. J Cell Biol. 2008;182:263–276. doi: 10.1083/jcb.200710195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SL. The ORF1 protein encoded by LINE-1: structure and function during L1 retrotransposition. J Biomed Biotechnol. 2006;2006:45621. doi: 10.1155/JBB/2006/45621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SL, Bushman D, Wang F, Li PW, Walker A, Cummiskey J, Branciforte D, Williams MC. A single amino acid substitution in ORF1 dramatically decreases L1 retrotransposition and provides insight into nucleic acid chaperone activity. Nucleic acids research. 2008;36:5845–5854. doi: 10.1093/nar/gkn554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SL, Cruceanu M, Branciforte D, Wai-Lun Li P, Kwok SC, Hodges RS, Williams MC. LINE-1 retrotransposition requires the nucleic acid chaperone activity of the ORF1 protein. Journal of molecular biology. 2005;348:549–561. doi: 10.1016/j.jmb.2005.03.003. [DOI] [PubMed] [Google Scholar]
- Matova N, Cooley L. Comparative aspects of animal oogenesis. Developmental biology. 2001;231:291–320. doi: 10.1006/dbio.2000.0120. [DOI] [PubMed] [Google Scholar]
- McClellan KA, Gosden R, Taketo T. Continuous loss of oocytes throughout meiotic prophase in the normal mouse ovary. Dev Biol. 2003;258:334–348. doi: 10.1016/s0012-1606(03)00132-5. [DOI] [PubMed] [Google Scholar]
- Moens PB, Chen DJ, Shen Z, Kolas N, Tarsounas M, Heng HH, Spyropoulos B. Rad51 immunocytology in rat and mouse spermatocytes and oocytes. Chromosoma. 1997;106:207–215. doi: 10.1007/s004120050241. [DOI] [PubMed] [Google Scholar]
- Morita Y, Manganaro TF, Tao XJ, Martimbeau S, Donahoe PK, Tilly JL. Requirement for phosphatidylinositol-3'-kinase in cytokine-mediated germ cell survival during fetal oogenesis in the mouse. Endocrinology. 1999;140:941–949. doi: 10.1210/endo.140.2.6539. [DOI] [PubMed] [Google Scholar]
- Morita Y, Maravei DV, Bergeron L, Wang S, Perez GI, Tsutsumi O, Taketani Y, Asano M, Horai R, Korsmeyer SJ, et al. Caspase-2 deficiency prevents programmed germ cell death resulting from cytokine insufficiency but not meiotic defects caused by loss of ataxia telangiectasia-mutated (Atm) gene function. Cell death and differentiation. 2001;8:614–620. doi: 10.1038/sj.cdd.4400845. [DOI] [PubMed] [Google Scholar]
- Morita Y, Perez GI, Paris F, Miranda SR, Ehleiter D, Haimovitz-Friedman A, Fuks Z, Xie Z, Reed JC, Schuchman EH, et al. Oocyte apoptosis is suppressed by disruption of the acid sphingomyelinase gene or by sphingosine-1-phosphate therapy. Nat Med. 2000;6:1109–1114. doi: 10.1038/80442. [DOI] [PubMed] [Google Scholar]
- O'Donnell KA, An W, Schrum CT, Wheelan SJ, Boeke JD. Controlled insertional mutagenesis using a LINE-1 (ORFeus) gene-trap mouse model. Proceedings of the National Academy of Sciences of the United States of America. 2013 doi: 10.1073/pnas.1302504110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell KA, Burns KH, Boeke JD. A descent into the nuage: the maelstrom of transposon control. Developmental cell. 2008;15:179–181. doi: 10.1016/j.devcel.2008.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno R, Nakayama M, Naruse C, Okashita N, Takano O, Tachibana M, Asano M, Saitou M, Seki Y. A replication-dependent passive mechanism modulates DNA demethylation in mouse primordial germ cells. Development. 2013;140:2892–2903. doi: 10.1242/dev.093229. [DOI] [PubMed] [Google Scholar]
- Ollinger R, Reichmann J, Adams IR. Meiosis and retrotransposon silencing during germ cell development in mice. Differentiation. 2010;79:147–158. doi: 10.1016/j.diff.2009.10.004. [DOI] [PubMed] [Google Scholar]
- Penzkofer T, Dandekar T, Zemojtel T. L1Base: from functional annotation to prediction of active LINE-1 elements. Nucleic acids research. 2005;33:D498–500. doi: 10.1093/nar/gki044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepling ME, Spradling AC. Mouse ovarian germ cell cysts undergo programmed breakdown to form primordial follicles. Developmental biology. 2001;234:339–351. doi: 10.1006/dbio.2001.0269. [DOI] [PubMed] [Google Scholar]
- Popp C, Dean W, Feng S, Cokus SJ, Andrews S, Pellegrini M, Jacobsen SE, Reik W. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature. 2010;463:1101–1105. doi: 10.1038/nature08829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanienko PJ, Camerini-Otero RD. The mouse Spo11 gene is required for meiotic chromosome synapsis. Mol Cell. 2000;6:975–987. doi: 10.1016/s1097-2765(00)00097-6. [DOI] [PubMed] [Google Scholar]
- Schimenti J. Synapsis or silence. Nature genetics. 2005;37:11–13. doi: 10.1038/ng0105-11. [DOI] [PubMed] [Google Scholar]
- Seisenberger S, Andrews S, Krueger F, Arand J, Walter J, Santos F, Popp C, Thienpont B, Dean W, Reik W. The dynamics of genome-wide DNA methylation reprogramming in mouse primordial germ cells. Molecular cell. 2012;48:849–862. doi: 10.1016/j.molcel.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpe AH, Hunter JJ, Ruprecht RM, Jaenisch R. Maternal transmission of retroviral disease: transgenic mice as a rapid test system for evaluating perinatal and transplacental antiretroviral therapy. Proceedings of the National Academy of Sciences of the United States of America. 1988;85:9792–9796. doi: 10.1073/pnas.85.24.9792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpe AH, Jaenisch R, Ruprecht RM. Retroviruses and mouse embryos: a rapid model for neurovirulence and transplacental antiviral therapy. Science. 1987;236:1671–1674. doi: 10.1126/science.3037694. [DOI] [PubMed] [Google Scholar]
- Shoji M, Tanaka T, Hosokawa M, Reuter M, Stark A, Kato Y, Kondoh G, Okawa K, Chujo T, Suzuki T, et al. The TDRD9-MIWI2 complex is essential for piRNA-mediated retrotransposon silencing in the mouse male germline. Dev Cell. 2009;17:775–787. doi: 10.1016/j.devcel.2009.10.012. [DOI] [PubMed] [Google Scholar]
- Sienski G, Donertas D, Brennecke J. Transcriptional silencing of transposons by piwi and maelstrom and its impact on chromatin state and gene expression. Cell. 2012;151:964–980. doi: 10.1016/j.cell.2012.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soper SF, van der Heijden GW, Hardiman TC, Goodheart M, Martin SL, de Boer P, Bortvin A. Mouse maelstrom, a component of nuage, is essential for spermatogenesis and transposon repression in meiosis. Dev Cell. 2008;15:285–297. doi: 10.1016/j.devcel.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotillo R, Hernando E, Diaz-Rodriguez E, Teruya-Feldstein J, Cordon-Cardo C, Lowe SW, Benezra R. Mad2 overexpression promotes aneuploidy and tumorigenesis in mice. Cancer cell. 2007;11:9–23. doi: 10.1016/j.ccr.2006.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilly JL. Commuting the death sentence: how oocytes strive to survive. Nat Rev Mol Cell Biol. 2001;2:838–848. doi: 10.1038/35099086. [DOI] [PubMed] [Google Scholar]
- Tilly JL. Ovarian follicle counts--not as simple as 1, 2, 3. Reprod Biol Endocrinol. 2003;1:11. doi: 10.1186/1477-7827-1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toltzis P, Marx CM, Kleinman N, Levine EM, Schmidt EV. Zidovudine-associated embryonic toxicity in mice. The Journal of infectious diseases. 1991;163:1212–1218. doi: 10.1093/infdis/163.6.1212. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature biotechnology. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner JM. Meiotic sex chromosome inactivation. Development. 2007;134:1823–1831. doi: 10.1242/dev.000018. [DOI] [PubMed] [Google Scholar]
- Turner JM, Aprelikova O, Xu X, Wang R, Kim S, Chandramouli GV, Barrett JC, Burgoyne PS, Deng CX. BRCA1, histone H2AX phosphorylation, and male meiotic sex chromosome inactivation. Curr Biol. 2004;14:2135–2142. doi: 10.1016/j.cub.2004.11.032. [DOI] [PubMed] [Google Scholar]
- Turner JM, Mahadevaiah SK, Fernandez-Capetillo O, Nussenzweig A, Xu X, Deng CX, Burgoyne PS. Silencing of unsynapsed meiotic chromosomes in the mouse. Nature genetics. 2005;37:41–47. doi: 10.1038/ng1484. [DOI] [PubMed] [Google Scholar]
- Wallace NA, Belancio VP, Deininger PL. L1 mobile element expression causes multiple types of toxicity. Gene. 2008;419:75–81. doi: 10.1016/j.gene.2008.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh CP, Chaillet JR, Bestor TH. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nature genetics. 1998;20:116–117. doi: 10.1038/2413. [DOI] [PubMed] [Google Scholar]
- Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Western P. Foetal germ cells: striking the balance between pluripotency and differentiation. Int J Dev Biol. 2009;53:393–409. doi: 10.1387/ijdb.082671pw. [DOI] [PubMed] [Google Scholar]
- Yang F, Wang PJ. The mammalian synaptonemal complex: a scaffold and beyond. Genome dynamics. 2009;5:69–80. doi: 10.1159/000166620. [DOI] [PubMed] [Google Scholar]
- Zemojtel T, Penzkofer T, Schultz J, Dandekar T, Badge R, Vingron M. Exonization of active mouse L1s: a driver of transcriptome evolution? BMC Genomics. 2007;8:392. doi: 10.1186/1471-2164-8-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q, Wang XJ. GOEAST: a web-based software toolkit for Gene Ontology enrichment analysis. Nucleic acids research. 2008;36:W358–363. doi: 10.1093/nar/gkn276. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.