Abstract
Pediatric high-grade glioma (HGG) is a devastating disease with a two-year survival of less than 20%1. We analyzed 127 pediatric HGGs, including diffuse intrinsic pontine gliomas (DIPGs) and non-brainstem HGGs (NBS-HGGs) by whole genome, whole exome, and/or transcriptome sequencing. We identified recurrent somatic mutations in ACVR1 exclusively in DIPG (32%), in addition to the previously reported frequent somatic mutations in histone H3, TP53 and ATRX in both DIPG and NBS-HGGs2-5. Structural variants generating fusion genes were found in 47% of DIPGs and NBS-HGGs, with recurrent fusions involving the neurotrophin receptor genes NTRK1, 2, or 3 in 40% of NBS-HGGs in infants. Mutations targeting receptor tyrosine kinase/RAS/PI3K signaling, histone modification or chromatin remodeling, and cell cycle regulation were found in 68%, 73% and 59%, respectively, of pediatric HGGs, including DIPGs and NBS-HGGs. This comprehensive analysis provides insights into the unique and shared pathways driving pediatric HGG within and outside the brainstem.
Although childhood and adult HGG share related histopathological characteristics, adult HGGs arise predominantly in the cerebral cortex, while childhood HGGs more frequently involve a broader spectrum of locations. There are also significant differences in molecular features between pediatric and adult HGG3,6-16. Histone H3 (H3F3A and HIST1H3B) pK27M mutations are frequent in DIPGs, which arise in the brainstem almost exclusively in children, and in pediatric HGGs in midline structures such as thalamus and cerebellum, while pG34R/V histone H3 mutations occur in pediatric HGGs of the cerebral cortex3-5,17. In contrast, histone H3 mutations are extremely rare in adult HGGs 3. HGGs arising in infants younger than 3 years of age have a better prognosis, and a lower frequency of TP53 mutations, suggesting that there may be age-dependent subgroups of HGG even within the pediatric population2. Thus, the selective pressures driving gliomagenesis in children vary with age and anatomical site.
To more comprehensively understand the pathways driving childhood glioma, we analyzed the genomic landscape of HGGs from 118 pediatric patients (127 tumors, 108 matched to germline DNA) consisting of 57 DIPGs and 70 non-brainstem HGGs (NBS-HGG) by whole genome (WGS) (n= 42), whole exome (n= 80) or transcriptome sequencing (n= 75) (Supplementary Tables 1-9). A total of 39,590 sequence mutations, including single nucleotide variations (SNVs) and small insertions or deletions, and 2,039 structural variations (SVs) were found by WGS while an additional 2,600 sequence mutations and 138 SVs were found by exome sequencing and transcriptome sequencing, respectively. Overall, the cohort showed a median background mutation rate of 9E-07 and a median of 22 SVs per genome (Supplementary Fig. 1). All SNVs and SVs found in WGS were verified experimentally by independent sequencing methods (Online Methods).
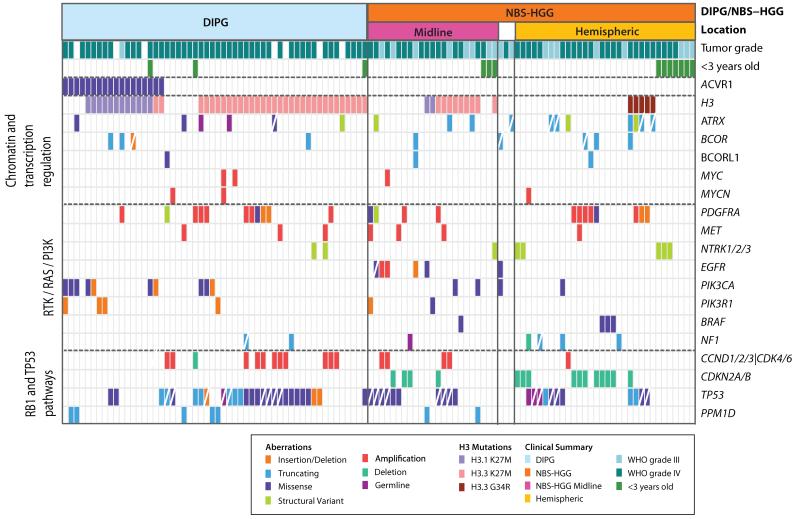
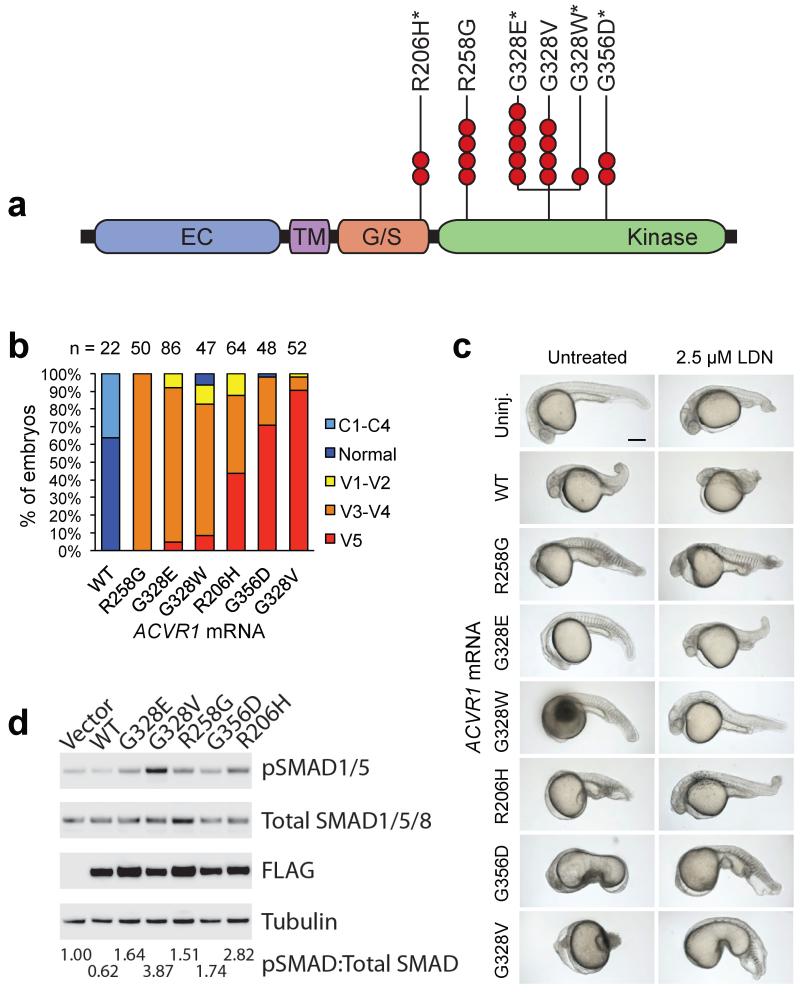
Among recurrent mutations in pediatric HGG, the most frequently mutated gene not previously identified in cancer was ACVR1 (also known as ALK2) encoding a BMP type I receptor (Fig. 1 and 2, Supplementary Fig. 2-3). Clonal missense ACVR1 mutations were found exclusively in DIPGs (32%), and were significantly associated with younger age, longer survival, and the presence of pK27M HIST1H3B (p<0.0000001), or PIK3CA or PIK3R1 mutations (p<0.005)(Fig. 1 and 2, Supplementary Fig. 3, Supplementary Tables 4 and 5). Four of these somatic ACVR1 mutations were the same as germline mutations previously identified in the autosomal dominant syndrome fibrodysplasia ossificans progressiva (FOP), in which aberrant cellular differentiation drives progressive heterotopic ossifications18,19. All residues impacted by mutation in DIPG cluster around either the inhibitory glycine/serine rich (G/S) domain or the ATP binding pocket of the kinase domain, and would be expected to shift the kinase to an active conformation (Figure 2 and Supplementary Fig. 3c)20. Indeed, mutations of these residues induced a weak gain of function20,21. A previous study showed that the R206H ACVR1 mutation caused a ventralized phenotype in zebrafish embryos, an indicator of BMP pathway activation22. We tested all of the ACVR1 mutations found in DIPG using this assay. Zebrafish embryos injected with ACVR1WT mRNA (WT) displayed a mild dorsalized phenotype consistent with BMP pathway inhibition, while all six ACVR1 mutants, shown in order of severity, exhibited varying degrees of ventralization with partial to complete loss of head and dorsal structures (Fig. 2b,c, Supplementary Fig. 3d,e). A moderate dose of LDN-193189 (LDN), a highly selective antagonist of the BMP pathway22,23, partially reversed the ventralization effects induced by ACVR1 mutants as can be seen by the rescue of dorsal head structures for R258G, G328E, G328W, R206H and the reduced severity of ventralization for G356D and G328V (Fig. 2c). Expression of ACVR1 mutants in mouse primary astrocyte cultures caused increased levels of phospho-SMAD1/5, a downstream indication of active BMP signaling, with varying magnitude (Fig. 2d). LDN also effectively blocked signaling to phospho-SMAD1/5 downstream of the mutant ACVR1 in primary astrocytes (Supplementary Fig. 3f).
Fig. 1. Recurrent genetic alterations in pediatric high-grade glioma.
Genetic alterations detected in 19 genes, including ACVR1, and genes most recurrently mutated in the pathways indicated on the left, are displayed according to the color key shown below. Diagonal white line indicates loss of the wild-type allele, or male patient for ATRX, which is X-linked. H3F3A (H3.3) and HIST1H3B (H3.1) mutations are grouped together into the category H3. Structural variants involving NTRK1, NTRK2, or NTRK3, and copy number variants of components of the CyclinD1, D2, D3, or CDK4, CDK6 G1 checkpoint complex are grouped together as NTRK1/2/3 or CCND1/2/3/CDK4/6, respectively. Tumor subgroup (DIPG or NBS-HGG), location of NBS-HGGs (midline versus tumors in cerebral hemispheres), and tumor grade are indicated. White boxes for location or tumor grade indicates information not available. < 3 y.o denotes less than 3 years of age. Mutations for 112 HGGs are shown. Four hypermutator samples and 11 samples for which only RNA-seq data was available, were excluded from this summary. Data is shown in tabular form in Supplementary Table 9.
Fig. 2. ACVR1 mutations in DIPG activate BMP signaling.
a. Missense ACVR1 substitutions in DIPG were clustered in the glycine/serine rich domain (G/S) or kinase domain. Each red circle indicates a DIPG carrying the specified mutation, and an * indicates mutations previously found as germline mutations in individuals with FOP. The extracellular domain (EC) and transmembrane domain (TM) did not contain mutations.
b. ACVR1 mutations ventralize zebrafish embryos. Graph shows the percentage of embryos exhibiting a dorsalized or ventralized phenotype. Embryos injected with wild-type ACVR1 mRNA (WT) showed a dorsalized phenotype, while embryos injected with mutant ACVR1 mRNA showed a ventralized phenotype (increasing severity from left to right). R258G had the least severe effect, resulting only in the V3-V4 ventralized phenotype, whereas G328V had the most severe effect with 90% of embryos showing the V5 ventralized phenotype. The number of embryos examined is shown on top.
c. Representative phenotype images of zebrafish embryos injected with the indicated ACVR1 mRNA. Untreated mutants R258G, G328E, G328W, and R206H have little to no dorsal structures, and G356D and G328V are more severely affected. Treatment with LDN-193189 (LDN) reversed the ventralization effects in the ACVR1 mutants, as can be seen by the partial rescue of dorsal structures (i.e. head) for R258G (100%, n=20), G328E (83%, n=23), G328W (100%, n=20), R206H (100%, n=27), and the reduced severity of ventralization without the formation of dorsal structures for G356D and G328V. Scale bar is 200 μm.
d. ACVR1 mutations drive increased levels of phospho-SMAD1/5 in primary astrocyte cultures. Western blots from lysates of primary astrocytes isolated from brainstem of neonatal Tp53 conditional knockout mice, transduced with retroviruses expressing FLAG-tagged ACVR1 wild-type, or indicated mutants, and serum starved for 2 hours. Quantitation of the ratio of phospho-SMAD/Total SMAD normalized to the empty vector control is shown below.
The recurrent and clonal activating mutation of ACVR1 in 32% of DIPGs provides strong evidence that it is an oncogenic driver in this disease. However, germline ACVR1 mutation in the genetic syndrome FOP is not associated with cancer predisposition, indicating that ACVR1 mutation likely provides a selective advantage in the presence of other critical mutations, rather than driving tumor initiation. Consistent with this hypothesis, all 6 of the DIPG-associated ACVR1 mutants failed to render Tp53-null astrocytes tumorigenic when implanted into brain (not shown). BMP signaling is associated with contrasting effects dependent on context, driving astrocytic differentiation, or proliferation of early hindbrain progenitors24, and inducing differentiation of medulloblastoma25, but driving either a differentiation or proliferative response in glioblastoma stem cells, related in part to the epigenetic state of the cell26,27. These context-dependent consequences likely drive the exclusive association between ACVR1 mutation and DIPG.
Principal Component Analysis using the 1000 most variable probesets shows that HGG samples clustered by tumor location with no segregation of DIPGs by their ACVR1 mutation status (Supplementary Fig. 3g). Genes involved in regulation of immune system processes were significantly different between DIPGs with and without ACVR1 mutation (Fisher’s exact test, p=0.0002, FDR=0.26%) (Supplementary Table 10).
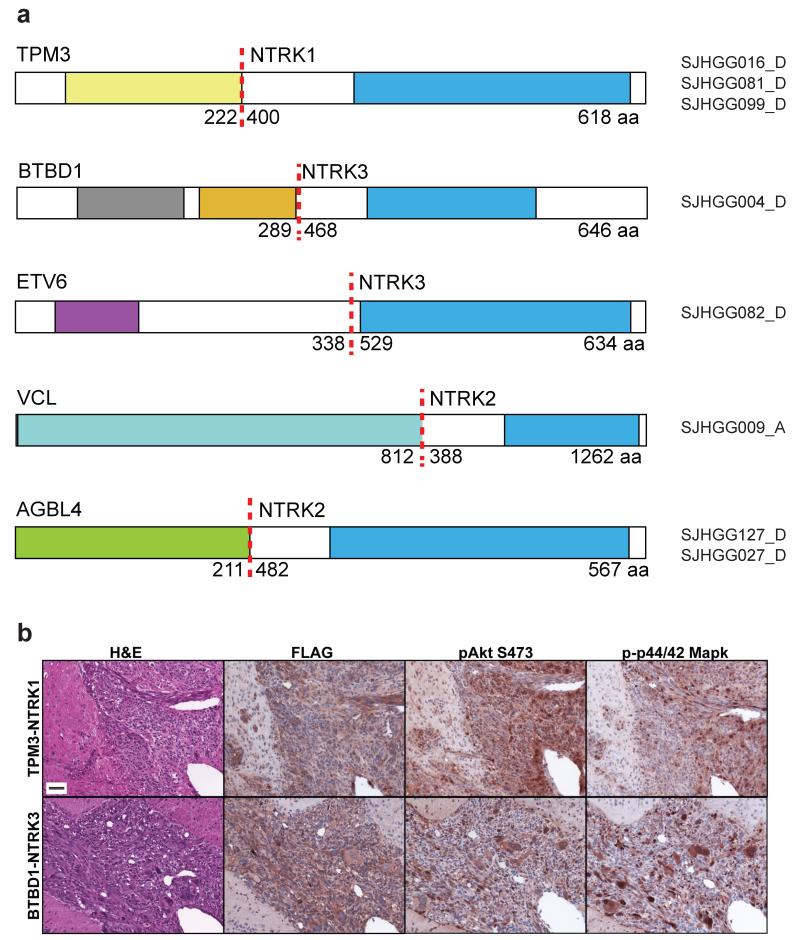
Structural variants generating fusion genes are common, and were identified in 47% of pediatric HGG, in equal proportions of DIPGs and NBS-HGGs, from transcriptome and WGS data. Gene fusions involving the kinase domain of each of the three neurotrophin receptor (NTRK) genes and five different N-terminal fusion partners, were identified in 4% of DIPGs and 10% of NBS-HGGs. Notably, 40% (4/10) of NBS-HGGs in children younger than 3 years old contained an NTRK fusion gene (Fig. 1 and 3, Supplementary Tables 7 and 8). The NTRK receptors transduce a wide range of developmental signals in the nervous system ranging from induction of neurite outgrowth, differentiation, neuronal survival or death28,29. NTRK fusion genes have recently been identified at low frequency in low-grade pediatric astrocytomas as well as adult glioblastomas30-32. Two of the five NTRK fusions found in our cohort, TPM3-NTRK1 and ETV6-NTRK3, were previously identified in other cancer types and shown to be oncogenic33-37.
Fig. 3. Structural variants generate oncogenic chimeric NTRK fusion proteins.
a. All fusions included the C-terminal kinase domain from NTRK1, NTRK2 or NTRK3 (blue). N-terminal fusion partners include the tropomyosin domain (yellow) of TPM3, an actin-binding protein fused to NTRK1, the BTB/POZ dimerization domain (gray), and Kelch domain (orange) from the topoisomerase I-interacting protein BTBD1, or the pointed protein-protein interaction domain (purple), of the ETS transcription factor ETV6, fused to NTRK3. The N-terminus of the actin-binding protein Vinculin (light blue, VCL) was fused to NTRK2, and the N-terminus (green) of the ATP/GTP binding protein AGBL4 was fused to NTRK2. The functional carboxypeptidase domain of AGBL4 is not present in the fusion protein. For each fusion protein, the dotted red line shows the fusion point, with the amino acid of the N-terminal and C-terminal fusion partners breakpoint indicated. The full-length of the fusion protein is shown on the right end.
b. NTRK fusion proteins induce high-grade astrocytomas. Tp53-null mouse primary astrocytes isolated from neonatal cortex or brainstem were transduced with FLAG-tagged TPM3-NTRK1 (top row) or BTBD1-NTRK3 (lower row) respectively, and implanted into mouse brain. Representative (of 7 independent mice per construct) H&E stains show pleomorphic tumor cells, many with features of astrocytic differentiation and high mitotic activity. Tumors induced by BTBD1-NTRK3 showed the frequent presence of giant cells reminiscent of giant cell glioblastoma. Immunohistochemical analysis showed expression of FLAG-tagged NTRK fusion proteins, and elevated phospho-Akt, and phospho-p42/44 Mapk in tumor relative to surrounding normal tissue. Scale bar=50μm.
To test the ability of NTRK fusion genes to drive glioma formation, we implanted Tp53-null primary mouse astrocytes transduced with retrovirus expressing FLAG-tagged TPM3-NTRK1 or BTBD1-NTRK3 into mouse brain. Both NTRK fusions induced high-grade astrocytomas with very short latency and complete penetrance (Fig. 3b and Supplementary Fig.4). The resulting tumors showed elevated levels of phospho-Akt and phospho-p42/44 Mapk, downstream indicators of PI3K and MAPK pathway activation.
Although NTRK activating fusions were specifically found at high frequency in infant tumors, activation of RTK/RAS/PI3K signaling through other mutations was frequent across the entire cohort, occurring in 69% of DIPGs and 67% of NBS-HGGs (Fig. 1, Supplementary Fig. 5 and Online Methods). In contrast to previous reports detecting EGFRvIII expression in pediatric HGG38,39, we only detected EGFRvIII in one out of the 85 tumors analyzed by WGS or RNA seq.
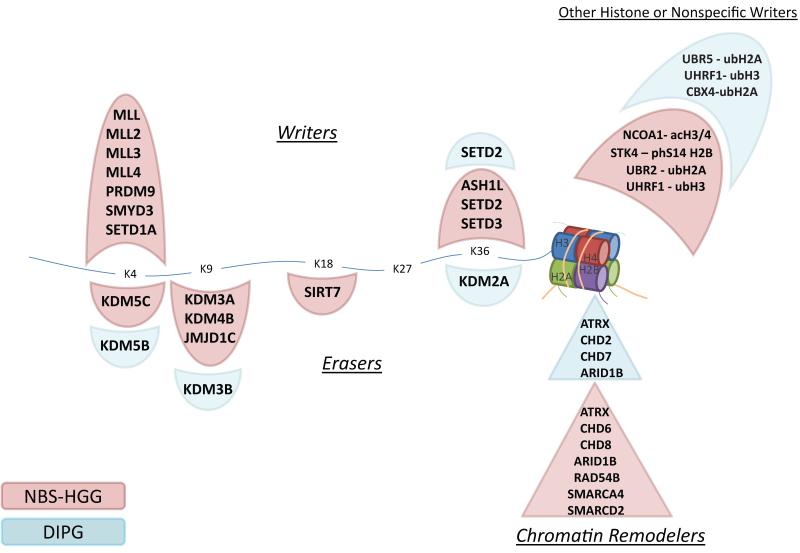
In addition to recurrent histone H3, ATRX and SETD2 mutations reported previously,3-5,17,40 frequent mutations of other histone writers and erasers, and chromatin remodeling genes, were also detected (Fig. 4 and Supplementary Fig. 5). Interestingly, mutations in histone H3 writers were significantly more frequent in NBS glioblastomas (p=0.007), while mutations in histone erasers were not significantly different between DIPG and NBS-HGG (p=0.3). Although only ATRX mutations were highly recurrent, collectively this group of genes involved in epigenetic regulation was targeted by mutation in 22% of DIPGs and 48% of NBS-HGG, excluding histone H3 mutations (Fig. 4 and Supplementary Fig. 5). These mutations were often concurrent with missense mutations in histone H3. Indeed, including histone H3 mutations, 91% of DIPGs, 70% of midline NBS-HGGs, and 48% of hemispheric NBS-HGGs contain mutations in histone and/or this subgroup of epigenetic regulators. pK27M H3.3 or H3.1 mutation results in a dominant loss of H3K27me3 in the entire cellular pool of histone H341-44. Mutations that modulate H3K27me3 by targeting components of the PRC2 complex that methylates H3K27, or the H3K27 demethylases KDM6A and KDM6B, are found in other tumor types45-50. However, there were no such mutations across the entire HGG cohort, including DIPGs with wild-type histone H3, supporting the unique selective advantage of pK27M mutation in pediatric HGG (Fig. 4). Mutations in transcriptional regulators that impact the epigenetic landscape were also found, including focal amplifications of MYC and MYCN, transcription factors that act to amplify levels of expressed genes across the genome51,52, and truncating mutations in the transcriptional co-repressors BCOR and BCORL1 (Fig. 1).
Fig. 4. Pediatric HGG mutations in histone modifiers or chromatin regulators.
Mutations identified in NBS-HGG (pink) or DIPG (blue) are shown. Genetic alterations were identified in proteins that attach (writers, shown above) or remove (erasers, shown below) post-translational modifications of lysines (K4, K9, K18, K27, K36) in the tail of histone H3, as well as proteins involved in modifications of other histones, or chromatin remodelers. Notably, there were no mutations in writers or erasers of K27, which is directly mutated at high frequency in DIPG, and to a lesser degree in NBS-HGG.
There was an enormous range in the complexity of somatic mutations driving pediatric HGG. The 10 NBS-HGGs in children under three years old showed significantly lower mutation rates than the rest of the cohort (p<0.0001), suggesting that only a small number of driver mutations is required in tumors from this age group (Supplementary Fig. 7 and Fig. 5). ETV6-NTRK3 was identified as one of only two non-silent alterations in SJHGG082, a glioblastoma from a one month-old patient. Notably, NTRK fusion genes, including two of the fusions found here, were identified in multiple tumor types, including those from very young children such as congenital fibrosarcoma, as well as in papillary thyroid cancer, an adult disease30,33-36. The high frequency of NTRK fusion genes in NBS-HGGs from children younger than three, the paucity of additional mutations in these tumors, and the rapid tumor onset in our experimental glioma model, strongly suggest that these fusion genes are potent oncogenic drivers in early postnatal brain tumor development.
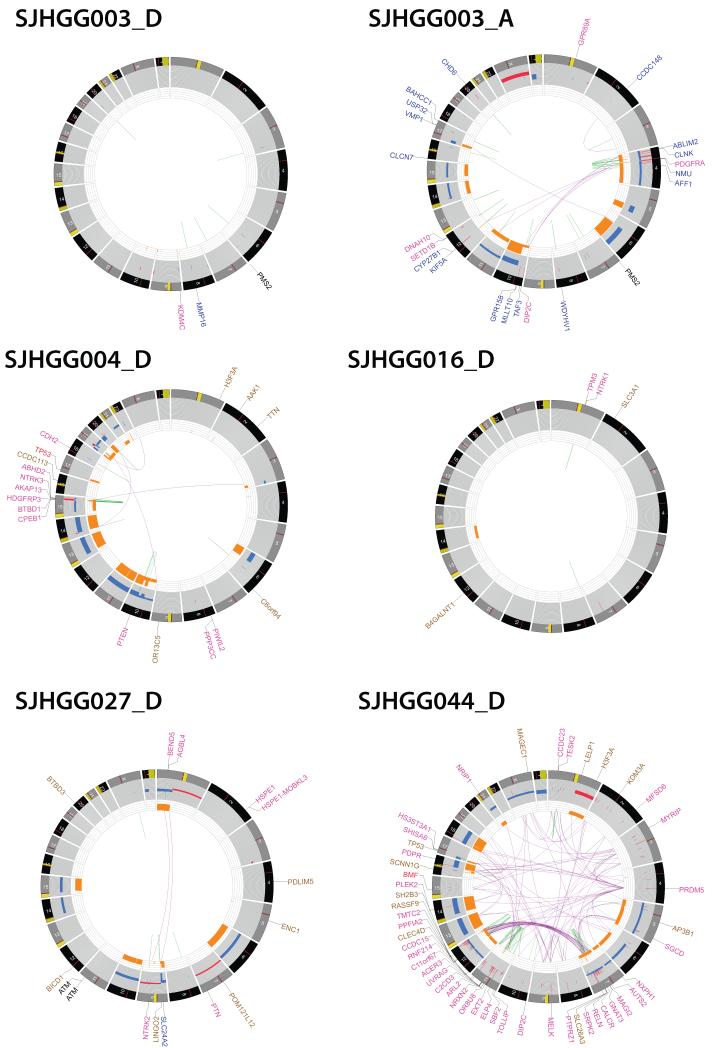
Fig. 5. CIRCOS plots showing the range of structural alterations in pediatric HGG.
CIRCOS plots display the genome by chromosome in a circular plot, and depict structural genetic variants, including DNA copy number alterations, intra- and inter-chromosomal translocations, and non-sequence mutations. Loss of heterozygosity, orange; amplification, red; deletion, blue Sequence mutations in RefSeq genes: missense SNVs, brown; indels, red; splice site SNVs, blue, Genes at SV breakpoints: genes involved in in-frame fusions, pink; others, blue.
Patient SJHGG003 carried a germline PMS2 mutation, and developed two independent tumors, first a hemispheric malignant glioneuronal tumor (SJHGG003_D), and 2 years later a DIPG (SJHGG003_A). The SNVs and indels for these two cases were too numerous to include on the plot for these two cases. The hypermutator tumor with more than 800,000 somatic mutations had an extremely stable genome (SJHGG003_D), while the second tumor with approximately 100-fold fewer SNVs, carried typical genomic copy number and structural abnormalities as seen in other HGGs (SJHGG003_A), thus demonstrating the broad range and complexity of mutations associated with germline PMS2 mutation. SJHGG016_D is an infant NBS-HGG with a TPM3-NTRK1 fusion and very stable genome. SJHGG027_D is an NBS-HGG from a patient with A-T, showing a relatively stable genome. This tumor sample was collected prior to radiotherapy. SJHGG004_D, is a DIPG with chromothripsis driving BTBD1-NTRK3 fusion, shown in more detail in Supplementary Fig. 8a. SJHGG044_D is an NBS-HGG showing dramatic chromothripsis. For the examples of chromothripsis, the names of genes disrupted by structural variants were too numerous to display.
Four tumors, three with matched normal DNA, were scored as hypermutators, with an extremely high background mutation rate, including more than 800,000 somatic mutations in SJHGG111, in which biallelic germline PMS2 mutations were identified (Supplementary Fig. 7). Patient SJHGG003 carried a heterozygous germline PMS2 mutation, and developed a grade IV hemispheric malignant glioneuronal tumor (MGNT) and a separate DIPG. Although both tumors independently acquired different somatic inactivating mutations in the remaining PMS2 allele, the basal mutation rate in the first tumor arising in this patient, the MGNT (SJHGG003_D) was nearly 100-fold higher than in the DIPG that arose 2 years later (SJHGG003_A), demonstrating the potential range in tumor mutation burden associated with inherited mismatch repair mutations (Supplementary Fig. 7, Fig. 5). Hypermutator tumors were excluded from the evaluation of mutation frequency. Seven patients carried germline mutations in known cancer predisposition genes, including TP53, PMS2, MSH6 and NF1 (Supplementary Table 11).
Thirteen tumors (31%) analyzed by WGS had chromothripsis53, shown by complex re-arrangements with multiple inter-connecting breakpoints corresponding to genomic segments of oscillating copy number states (Supplementary Results, Supplementary Table 12, Fig. 5 and Supplementary Fig. 8-9). Among our cohort, chromothripsis resulted in oncogenic re-arrangement including BTBD1-NTRK3 fusion, re-arrangement/amplification of PDGFRA and EGFR (Supplementary Fig. 8). Nearly half of all samples showing chromothripsis were collected prior to adjuvant therapy, indicating that the mechanism was, at least in some cases, independent of DNA-damaging therapeutics. SJHGG027_D, an NBS-HGG arising in a child with ataxia telangiectasia (A-T), had a relatively stable genome despite a compromised DNA damage checkpoint due to the absence of functional ATM (Fig. 5, and Supplementary Fig. 7). Multiple subclones were identified in almost all HGG tumors. A founder clone, or a descendant of a founder clone in the diagnosis tumor could seed the development of relapsed or autopsy tumor (Supplementary Results and Supplementary Fig. 10).
We identified TERT promoter mutations in only 2% of DIPGs and 3% of NBS-HGGs, in strong contrast to the frequency in 86% of adult primary glioblastomas54.
The genomic landscape of pediatric HGGs also includes frequent mutations in common cancer pathways, consistent with previous reports3,7-9,11,12,15,16. TP53 mutations occurred in 42% of pediatric HGG and were mutually exclusive with truncating mutations in the TP53-induced phosphatase PPM1D, previously shown to impair the TP53-dependent G1 checkpoint (Fig. 1)55. The G1 checkpoint regulators CCND1, 2 and 3, CDK4 and CDK6, were predominantly amplified in DIPG, while CDKN2A homozygous deletion was restricted to NBS-HGGs (Fig. 1). Taken together, mutations impacting cell cycle regulation, including the TP53 and RB pathways, were found in 59% of pediatric HGG (Fig. 1, Supplementary Fig. 5).
This global view of the genetic landscape of pediatric HGG defines critical pathways driving a devastating spectrum of childhood brain tumors, and identifies high frequency mutations in potential therapeutic targets; ACVR1 in DIPGs, and NTRK fusions in infant NBS-HGGs.
Online Methods
Patient cohorts and sample details
High grade gliomas (HGGs) (WHO Grade III and IV) were requested from the St. Jude Children’s Research Hospital tissue resource core facility, and from the Institute of Cancer Research/Royal Marsden Hospital with approval for genome sequence analysis in accordance with St Jude Institutional Review Board (IRB) approval for the Pediatric Cancer Genome Project (PCGP), and the Clinical Research and Development Board of the Royal Marsden Hospital and the United Kingdom Children’s Cancer and Leukemia Group research ethics approval. Detailed clinicopathologic and sequencing information is in Supplementary Table 1. There was a significant association between gender and tumor subtype, with 63% female DIPG patients and 63% male NBS-HGG patients (p=0.004).
The study cohort comprised 127 (57 DIPGs and 70 NBS-HGGs) tumors (54 DIPGs and 54 NBS-HGG with matching germline samples) from 118 patients in two cohorts: a cohort for whole genome sequencing (WGS, n=42, 20 DIPGs and 22 NBS-HGGs), a cohort for evaluating the frequency of abnormalities using whole exome sequencing (WES, n=80, 36 DIPGs and 44 NBS-HGGs). Six tumors including their matched germline samples, including two hypermutator tumors (SJHGG003_D and SJHGG111_D) and four non-hypermutator tumors (SJHGG003_A, SJHGG008_A, SJHGG019_E and SJHGG022_D) were sequenced by both whole genome and whole exome sequencing. Among these tumors, 75 (31 DIPGs and 44 NBS-HGGs) were characterized by RNA-seq. In addition, 12 tumors (3 DIPGs and 9 NBS-HGGs) were characterized by RNA-seq only for structural variant discovery.
Tumor was available from diagnosis and relapse in 5 cases (SJHGG019_E/S, SJHGG024_D/R, SJHGG033_D/R, SJHGG112_D/R, SJHGG115_D/R), or diagnosis and autopsy in 3 cases (SJHGG001_D/A, SJHGG002_D/A, SJHGG093_D/A). One patient developed two independent tumors, a hemispheric malignant glioneuronal tumor (SJHGG003_D), and then a subsequent independent DIPG (SJHGG003_A).
Histopathology was centrally reviewed by DWE, an experienced neuropathologist, and MRI images of DIPG cases were reviewed by a pediatric neuro-oncologist (AB). DNA and RNA was extracted as previously described 56.
Whole-genome, whole exome and transcriptome sequencing and analysis
WGS, WES, and RNA-seq were performed as previously described 46,57. For WGS, WES and RNA-Seq, paired-end sequencing was performed using the Illumina GAIIx or HighSeq platform with 100bp read length. The WGS data are deposited at the European Bioinformatics Institute (EBI) with accession number: WGS mapping, coverage and quality assessment, Single nucleotide variations (SNVs)/indel detection, tier annotation for sequence mutations, prediction of deleterious effects of missense mutations, and identification of loss of heterozygosity (LOH) were described previously 46,57. The reference human genome assembly GRCh37-lite was used for mapping all samples. The mapping statistics and coverage of each tumor on different sequencing platforms were summarized in Supplementary Table 2 and Supplementary Figure 1.
SNVs were classified into the following four tiers, as previously described 46,57: a) Tier 1: coding synonymous, nonsynonymous, splice-site, and non-coding RNA variants; b) Tier 2: conserved variants (cutoff: conservation score ≥ 500, based on either the phastConsElements28way table or the phastConsElements17way table from the UCSC genome browser, and variants in regulatory regions annotated by UCSC annotation (Regulatory annotations included are targetScanS, ORegAnno, tfbsConsSites, vistaEnhancers,eponine, firstEF, L1 TAF1 Valid, Poly(A), switchDbTss, encodeUViennaRnaz, laminB1, cpgIslandExt); c) Tier 3: variants in non-repeat masked regions; d) Tier 4: the remaining SNVs.
All tier 1, tier2 and tier3 sequence mutations (including SNVs, indels and SVs) discovered in non-hypermutator WGS samples were validated by custom capture platform. The overall validation rate is 93%, with a median validation rate 95% per sample. All tier1 SNVs in WES cohort were also validated by custom capture. For all gene coding indels found in WES samples, we performed the validation with MiSeq platform and the validation rate was 92% (167/182). Four non-hypermutator WGS samples (SJHGG003_A, SJHGG008_A, SJHGG019_E and SJHGG022_D) were subjected to exome sequencing so the overlapping SNVs/indels were regarded as validated. The validated and high-quality variations for tiers 1-3 mutations in non-hypermutator tumors are summarized in Supplementary Table 3, 4, 5.
For two hypermutator WGS samples (SJHGG003_D and SJHGG111_D), the exome sequencing served as validation. SNVs found in both WGS and WES were regarded as validated. The validated and high-quality variations for tier 1 mutations in hypermutator tumors have been deposited at the PCGP Explorer website (http://pcgpexplore.org/).
CNVs were identified by evaluating the difference in read depth between each tumor and its matching normal using a novel algorithm, CONSERTING (COpy Number SEgmentation by Regression Tree In Next-Gen sequencing; Chen et al, manuscript in preparation). The results are reported in Supplementary Table 6.
Structural variations in WGS were analyzed using CREST and annotated as previously described 58,46. Custom capture was used to validate somatic SVs found in WGS samples. The results are reported in Supplementary Table 7. Paired-end reads from RNA-seq were aligned to the following 4 database files using BWA (0.5.5) aligner59: (1) the human GRCh37-lite reference sequence, (2) RefSeq, (3) a sequence file representing all possible combinations of non-sequential pairs in RefSeq exons, (4) AceView database flat file downloaded from UCSC representing transcripts constructed from human ESTs. The mapping results from (2) to (4) were aligned to human reference genome coordinates. The final BAM file was constructed by selecting the best alignment in the four databases. SV detection was carried out using CICERO, a novel algorithm that uses de novo assembly to identify structural variation in RNASeq (Li et al, manuscript in preparation). For the structural variants detected in RNA-seq, we validated with MiSeq sequencing. Primer pairs were designed (with Primer3) to bracket the genomic regions containing putative SVs. The SVs found by RNA-seq are reported in Supplementary Table 8.
Microarray copy number and expression analysis
Affymetrix SNP6.0 arrays were analyzed as described 8. Candidate targets of focal amplification or deletion were identified from minimum common regions with copy number greater than 5 or less than 0.8. For DIPG samples, we also identified candidate targets of focal gain or loss as described 8. Briefly, we derived minimum common regions for recurrent focal gains (copy number > 2.3) or recurrent focal deletions (copy number < 1.7) found in at least two tumors or were classified as a single focal gain or deletion. Regions associated with known CNVs were removed. All remaining regions with less than 60 genes were manually inspected for cancer/glioma-related genes, and candidate targets of focal gain or deletion were selected. Focal amplifications of MYC, MYCN,PDGFRA, MET, CDK4, CDK6, CCND1, CCND2, and CCND3, as well as focal deletions of CDKN2A and CDKN2B from SNP data were included in Fig. 1 because many samples sequenced only by exome lacked this copy number data.
Affymetrix U133v2 expression array data was available for 71 HGG, including 32 DIPG samples, nine with ACVR1 mutation 8. Principal Component Analysis was done using GeneMaths with top 1000 most variable probes selected based on median absolute deviation (MAD) score. The differentially expressed genes between DIPGs with and without ACVR1 mutation were selected with linear models (limma/R, p-value < 0.001), and enrichment of GO terms among the selected genes was evaluated using DAVID Bioinformatics Resource (http://david.abcc.ncifcrf.gov/).
Frequency of mutations in pathways
To summarize samples altered in RTK/RAS/PI3K pathways, we collected the genes from KEGG, Ingenuity, and NCI-Nature Protein Interaction Database, and limited to the following genes that have at least one somatic mutation in our study cohort: 1) RTK: PDGFRA, KIT, EGFR, MET, CSF1R, FGFR1, FGFR3, FLT1, IGF1R, INSR, NRTK1, NTRK2, NTRK3; 2) PIP3 regulation: PTEN, PIK3CA, PIK3R1; 3) Ligand: FGF3, FGF5, FIGF, PDGFA, VEGFC; and 4) downstream effector: PLCG2, PLEKHA2, YES1, SGK1, G6PC, GNB1, GNGT1, MLST8, PHLPP1, PKN2, PPP2R2D, PRKAA2, PRKCA, PRKCZ, RAC1, RPS6, RPTOR, SGK3, STK11, INPP5D, SOS1, AKT3, JAK1, KRAS, MAP2K1, TSC2, GAB2, PPM1L, NF1, BRAF, RASGRF2, RASSFS. To summarize the samples altered in cell cycle regulation, we included the following genes: TP53, TP73, CCND1, CCND2, CDK1, CDK6, CDKN1B, CDKN2A, CDKN2B, CDKN2C, RB1, CDK4, CCND3, RBL1, CDC27. For DNA repair genes, we included the following: ATR, ATM, BRCA1, RBBP8, RAD51, BRCA2, ERCC2, ERCC3, ERCC8, LIG4, RAD23A, XAB2, MSH6, LIG1, LIG3, LIG4, POLD1, POLE, RAD23A, RAD50, RAD54B, RUVBL2, SETX, PMS2, UVRAG. To summarize samples altered in histone modifications and chromatin regulators, we limited our analysis to the following genes: 1) Histone writer: MLL, MLL2, MLL3, MLL4, PRDM9, SMYD3, SETD1A, SETD2, SETD3, ASH1L; 2) Histone eraser: JMJD1C, KDM2A, KDM3A, KDM3B, KDM4B, KDM5B, KDM5C, SIRT7; 3) Chromatin remodeler: ATRX, SMARCA4, BRWD1, CBX4, CHD2, CHD4, CHD6, CHD7, CHD8, RAD54B; 4) Other writers: UHRF1, NCOA1, STK4, UBR2, UBR5. Therefore our estimates of the numbers of mutations impacting these pathways are conservative.
TERT promoter mutation analysis
Due to the high GC content at the TERT promoter regions, there was poor coverage (×3x on average) in WGS at the two recurrently mutated sites (chr5.1295228 and chr5.1295250). Therefore, a portion of the TERT promoter (HG19 coordinates, chr5: 1295151-1295347) was amplified by PCR from tumors and matched normal and sequenced by Sanger Sequencing using primers in Supplementary Table 13 to check for two previously described TERT promoter SNVs. Sequence was analyzed using SNPDetector 60 and manual review was performed using Consed61.
Statistical analysis
Association between ACVR1 mutation and age at diagnosis was analyzed by the Kruskal-Wallis test: H = 6.62 DF = 1 P = 0.010 (adjusted for ties). Association of ACVR1 mutation with survival of DIPG patients was quantified by the log rank test: Chisq= 10.1 on 1 degrees of freedom, p= 0.00149. Based on a two-variable COX model for DIPG patients where age at diagnosis (as a continuous variable) and ACVR1 mutation status were included simultaneously, it appears that age at diagnosis was not significantly associated with survival within this cohort (p-val =0.55), but ACVR1 status is (p-val=0.0046). The co-occurrence of ACVR1 mutations with other mutations was quantified by Fisher’s exact test: ACVR1 co-occurrence with pK27M HIST1HB: p-Value = 0.0000001, and ACVR1 co-occurrence with PIK3CA and PIK3R1 mutations: p-Value = 0.0049702. The association between gender and tumor subtype (DIPG vs NBS-HGG) was quantified by Fisher’s exact test. The association between tier 1 mutation rate and tumors in children less than 3 years of age was quantified by Kruskal-Wallis Test (H=13.42, DF=1, p<0.0001). The association between mutations in H3 writers and NBS-HGGs was quantified by Pearson Chi-square (by monte-carlo), p=0.0007.
To identify samples with extremely high and low mutation rates, we used least median squares (LMS) as a method for robust estimation of the center of the data and outlier detection 62. Basically, LMS identifies the shortest interval that covers at least 50% of the data. This interval represents the densest “bulk” of the data and outliers are detected by comparing the data values to a normal distribution with the same IQR. Thus, the LMS interval will not be influenced by outliers and it can be used to detect outliers.
Germline mutation analysis
We identified germline variants as previously described 63. In this study, we implemented additional filters: 1) we only kept genes listed in Cancer Gene Consensus or genes involved in DNA damage/repair; 2) Only nonsense or splice SNVs and indels were kept, except a few missense mutations for TP53.
Zebrafish embryo injections
ACVR1 wild-type or mutant cDNA was cloned into pCS2 and mRNA was generated using the mMESSAGE mMACHINE Kit (Life Technologies). Zebrafish embryos of the AB strain were microinjected with approximately 20 pg of ACVR1 wild-type or mutant mRNA at the one-cell stage. Embryos were inspected at 24 hours post fertilization (hpf) and scored as ventralized (Classes V1-V5) or dorsalized (Classes C1-C4) based upon published criteria 23, 22. For chemical rescue, 2.5 μM LDN-193189 (Stemgent) was added to embryos at about 3 hpf.
Primary astrocyte transductions and in vivo tumorigenesis assays
A FLAG tag was incorporated by PCR immediately before the termination codon in cDNA encoding the full open reading frame of wild-type or mutant ACVR1, TPM3-NTRK1 and BTBD1-NTRK2 and cloned into the retroviral vectors MSCV-IRES-mCherry (MIC) 64, which was modified by inserting an adapter into a blunted EcoRI site to generate a Gateway cloning site, or MSCV-IRES-Puro (MIP) 65. Sequence of all constructs was verified. Retrovirus was produced by co-transfecting 293T cells along with helper plasmids as previously described 66.
Mouse experiments were approved by the Institutional Animal Care and Use Committee and are in compliance with national and institutional guidelines. Tp53-null primary mouse astrocytes (PMAs) were isolated from cortex or brainstem of postnatal day 2 GFAP-cre;Trp53loxP/loxP (Tp53 conditional knockout mice)67,68 and transduced with retroviruses as previously described 69. PMA cultures isolated from multiple mice were pooled for retroviral transductions to control for potential variation among primary cultures. For tumorigenesis studies, at passage one, Tp53-null PMAs were transduced with retroviruses generated from MIC vectors expressing wild-type or mutant ACVR1 or TPM3-NTRK1 or BTBD1-NTRK2 and implanted into immunodeficient mice as described 70. When mice displayed brain tumor symptoms, tumors were dissected, fixed in 4% paraformaldehyde in PBS at 4°C overnight, then processed, embedded in paraffin, and cut into 5 μm sections. Hematoxylin and eosin stained sections from all collected tumors were evaluated by a clinical neuropathologist (DWE). Immunohistochemistry was performed with microwave antigen retrieval, primary antibodies against p-Akt Ser 473(Cell Signaling #9271, 1:50), p-p42/44 Mapk Thr202/Tyr204 (Cell Signaling #4270, 1:400), or FLAG (Sigma Aldrich, F1804, 1:100), biotinylated secondary antibodies and horseradish peroxidase-conjugated streptavidin (Elite ABC, Vector Labs), detected with NovaRED substrate (Vector Labs) and counterstained with hematoxylin (Vector Labs). For TPM3-NTRK1, BTBD1-NTRK2 and empty vector controls, 7 mice were implanted for each construct. For ACVR1, empty vector control, wild-type, G328E, R258G or R206H ACVR1 (7 mice each construct), or G356D (6 mice) were implanted. A few of the immunodeficient mice were euthanized when they became ill without showing brain tumor symptoms (One G328E at 97 days post-implantation, one R258G at 217 days, 2 G356D at 139 and 190 days, and one WT at 212 days). The remaining mice were euthanized between 219 to 222 days postimplantation. Brains were formalin fixed and paraffin embedded, sectioned, stained with H&E and evaluated for ACVR1-driven brain tumor growth both by analysis of H&E sections in the area surrounding the implantation site, as well as by immunohistochemistry for the mCherry marker expressed as part of the bicistronic message with ACVR1. None of the mice showed brain tumor symptoms, and no histological evidence of ACRV1-driven tumor formation was detected in the entire cohort.
Western Blotting
For Western blotting, Tp53-null PMAs were isolated from neonatal brainstem and transduced at passage one with MIP retroviruses expressing wild-type or mutant ACVR1. 48 hours later, cells were selected with 2.5 μg/ml puromycin for 48 hours. For serum starvation, cells were washed twice with PBS, then incubated for 2 hours in media without serum or growth factors. For treatment with LDN-193189, cells were incubated in standard astrocyte growth media 69 with vehicle (DMSO) or LDN-193189 (1μM). Cells were lysed in RIPA buffer with protease and phosphatase inhibitors (Roche). 20μg of protein was resolved on 4-12% NuPAGE Bis-Tris gels, transferred to nitrocellulose membranes, and detected with antibodies targeting p-SMAD1/5 (Cell Signaling, 9516, 1:1000), total SMAD1/5/8 (Santa Cruz, sc-6031, 1:500), p-p38MAPK (Invitrogen, 44-684G, 1:1000), total p38MAPK (Cell Signaling, 9212, 1:1000), FLAG (Sigma-Aldrich, F1804, 1:1000) and tubulin (Santa Cruz, sc-23948, 1:1000). Following incubation with the appropriate HRP-conjugated secondary antibody, membranes were exposed to chemiluminescent substrate (SuperSignal West Dura, 34076, Thermo Scientific). Images were obtained with the Odyssey Imaging System (LI-COR Biosciences).
RT-PCR Validation of TRK Fusions
Random-primed cDNA was generated by reverse transcriptase from tumor RNA and used for PCR to identify the fusion gene of interest. PCR products were analyzed by Sanger sequencing. Primers are listed in Supplementary Table 13.
Statistical evaluation of chromothripsis in HGG tumors analyzed by WGS
Chromothripsis was described as localized chromosome shattering and repair occuring in a single event. The initial criterion is oscillation between 2 main CNV states 53, which were found in 15 HGG tumors in this study. Most recently, Korbel and Campbell 71 proposed four potential criteria for assessing chromothripsis: 1) clustering of breakpoints; 2) randomness of DNA fragment joins; 3) randomness of DNA fragment order; and 4) ability to walk the derivative chromosome. Since randomness of DNA fragment order (Criterion 3) was not entirely valid, even in Korbel and Campbell’s own analysis, we decided not to evaluate this feature. For the 13 tumors in Supplementary Table 12, we performed Bartlett’s goodness-of-fit test for exponential distribution to assess whether the distribution of SV breakpoints in each tumor departs from the null hypothesis of random distribution. A significant departure from random distribution supports clustering of SV breakpoints. To evaluate whether there is any bias in the DNA fragment joints categorized by the SV types (i.e. deletion, tandem duplication, head-to-head re-arrangements and tail-to-tail re-arrangements), we applied goodness-of-fit test separately for inter- and intra-chromosomal events with a minimum of 5 SVs. A significant p value suggests biased fragment joins, which would not support chromothripsis. When both inter- and intra-chromosomal data are available, we reported the lower p value to represent a more conservative assessment of the random distribution for DNA fragment joints.
Details of Tumor Purity and Tumor Heterogeneity Estimations, and Tumor Evolution Analysis are included in the Supplementary Note.
Supplementary Material
Acknowledgments
This work was supported by the St. Jude Children’s Research Hospital – Washington University Pediatric Cancer Genome Project, ALSAC of St Jude Children’s Research Hospital, and by grants from the NIH (P01 CA096832 to SJB, JZ, and DWE, and R01 CA135554 to SJB), The Cure Starts Now Foundation and Smile for Sophie Forever Foundation, and by Tyler’s Treehouse and Musicians Against Childhood Cancer. We thank the St Jude Children’s Research Hospital Tissue Resource Facility, and Brittney Gordon, Melissa Johnson, Shantel Brown and Christopher Calabrese in the St Jude Small Animal Imaging Core for expert assistance with intracranial implantations. We thank Dr. Eileen Shore and Andria Culbert (University of Pennsylvania) for helpful advice with antibody selection.
Footnotes
Accession number: All WGS, exome and transcriptome data have been deposited at the European Genome-phenome Archive (EGA), which is hosted by the European Bioinformatics Institute (EBI) under EGAS0000100192.
Author contributions
SJB, Jinghui Zhang, AKD, BSP, JE, LD, ERM, RKW, MRT, JRD, and DWE designed experiments or supervised research, AG, AB, CW, FAB, and CJ provided samples or clinical data, GW, AKD, BSP, SLR, BJ, YL, XZ, CQ, XC, Junyuan Zhang, JE, ME, XM, CL, PN, EH, MR, SP, TL, AO-T, RH, RK, MP, PG, JB, LW, HLM, KB, BV, DY, JCR, KO, RSF, LLF, LD, ERM, RKW, MRT, JRD, SWE, Jinghui Zhang, and SJB performed experiments, analyzed data or prepared tables and figures, DWE completed all pathological evaluations, SJB, Jinghui Zhang, GW and AKD wrote the manuscript with contributions from DWE, JRD and MRT.
The authors have no competing financial interests.
References
- 1.Gottardo NG, Gajjar A. Chemotherapy for malignant brain tumors of childhood. J Child Neurol. 2008;23:1149–59. doi: 10.1177/0883073808321765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pollack IF, et al. Age and TP53 mutation frequency in childhood malignant gliomas: results in a multi-institutional cohort. Cancer Res. 2001;61:7404–7. [PubMed] [Google Scholar]
- 3.Schwartzentruber J, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482:226–31. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- 4.Wu G, et al. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat Genet. 2012;44:251, 3. doi: 10.1038/ng.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khuong-Quang DA, et al. K27M mutation in histone H3.3 defines clinically and biologically distinct subgroups of pediatric diffuse intrinsic pontine gliomas. Acta Neuropathol. 2012;124:439–47. doi: 10.1007/s00401-012-0998-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Faury D, et al. Molecular profiling identifies prognostic subgroups of pediatric glioblastoma and shows increased YB-1 expression in tumors. J Clin Oncol. 2007;25:1196–208. doi: 10.1200/JCO.2006.07.8626. [DOI] [PubMed] [Google Scholar]
- 7.Qu HQ, et al. Genome-wide profiling using single-nucleotide polymorphism arrays identifies novel chromosomal imbalances in pediatric glioblastomas. Neuro Oncol. 2010;12:153–63. doi: 10.1093/neuonc/nop001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paugh BS, et al. Genome-wide analyses identify recurrent amplifications of receptor tyrosine kinases and cell-cycle regulatory genes in diffuse intrinsic pontine glioma. J Clin Oncol. 2011;29:3999–4006. doi: 10.1200/JCO.2011.35.5677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paugh BS, et al. Integrated molecular genetic profiling of pediatric highgrade gliomas reveals key differences with the adult disease. J Clin Oncol. 2010;28:3061–8. doi: 10.1200/JCO.2009.26.7252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schiffman JD, et al. Oncogenic BRAF mutation with CDKN2A inactivation is characteristic of a subset of pediatric malignant astrocytomas. Cancer Res. 2010;70:512–9. doi: 10.1158/0008-5472.CAN-09-1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Puget S, et al. Mesenchymal transition and PDGFRA amplification/mutation are key distinct oncogenic events in pediatric diffuse intrinsic pontine gliomas. PLoS One. 2012;7:e30313. doi: 10.1371/journal.pone.0030313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zarghooni M, et al. Whole-genome profiling of pediatric diffuse intrinsic pontine gliomas highlights platelet-derived growth factor receptor alpha and poly (ADP-ribose) polymerase as potential therapeutic targets. J Clin Oncol. 2010;28:1337–44. doi: 10.1200/JCO.2009.25.5463. [DOI] [PubMed] [Google Scholar]
- 13.Parsons DW, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–12. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–8. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bax DA, et al. A distinct spectrum of copy number aberrations in pediatric high-grade gliomas. Clin Cancer Res. 2010;16:3368–77. doi: 10.1158/1078-0432.CCR-10-0438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barrow J, et al. Homozygous loss of ADAM3A revealed by genome-wide analysis of pediatric high-grade glioma and diffuse intrinsic pontine gliomas. Neuro Oncol. 2011;13:212–22. doi: 10.1093/neuonc/noq158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sturm D, et al. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell. 2012;22:425–37. doi: 10.1016/j.ccr.2012.08.024. [DOI] [PubMed] [Google Scholar]
- 18.Shore EM, Kaplan FS. Role of altered signal transduction in heterotopic ossification and fibrodysplasia ossificans progressiva. Curr Osteoporos Rep. 2011;9:83–8. doi: 10.1007/s11914-011-0046-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shore EM, et al. A recurrent mutation in the BMP type I receptor ACVR1 causes inherited and sporadic fibrodysplasia ossificans progressiva. Nat Genet. 2006;38:525–7. doi: 10.1038/ng1783. [DOI] [PubMed] [Google Scholar]
- 20.Chaikuad A, et al. Structure of the bone morphogenetic protein receptor ALK2 and implications for fibrodysplasia ossificans progressiva. J Biol Chem. 2012;287:36990–8. doi: 10.1074/jbc.M112.365932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bagarova J, et al. Constitutively Active ALK2 Receptor Mutants Require Type II Receptor Cooperation. Mol Cell Biol. 2013;33:2413–24. doi: 10.1128/MCB.01595-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen Q, et al. The fibrodysplasia ossificans progressiva R206H ACVR1 mutation activates BMP-independent chondrogenesis and zebrafish embryo ventralization. J Clin Invest. 2009;119:3462–72. doi: 10.1172/JCI37412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cannon JE, Upton PD, Smith JC, Morrell NW. Intersegmental vessel formation in zebrafish: requirement for VEGF but not BMP signalling revealed by selective and non-selective BMP antagonists. Br J Pharmacol. 2010;161:140–9. doi: 10.1111/j.1476-5381.2010.00871.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bond AM, Bhalala OG, Kessler JA. The dynamic role of bone morphogenetic proteins in neural stem cell fate and maturation. Dev Neurobiol. 2012;72:1068–84. doi: 10.1002/dneu.22022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao H, Ayrault O, Zindy F, Kim JH, Roussel MF. Post-transcriptional down-regulation of Atoh1/Math1 by bone morphogenic proteins suppresses medulloblastoma development. Genes Dev. 2008;22:722–7. doi: 10.1101/gad.1636408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Piccirillo SG, et al. Bone morphogenetic proteins inhibit the tumorigenic potential of human brain tumour-initiating cells. Nature. 2006;444:761–5. doi: 10.1038/nature05349. [DOI] [PubMed] [Google Scholar]
- 27.Lee J, et al. Epigenetic-mediated dysfunction of the bone morphogenetic protein pathway inhibits differentiation of glioblastoma-initiating cells. Cancer Cell. 2008;13:69–80. doi: 10.1016/j.ccr.2007.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harel L, Costa B, Fainzilber M. On the death Trk. Dev Neurobiol. 2010;70:298–303. doi: 10.1002/dneu.20769. [DOI] [PubMed] [Google Scholar]
- 29.Thiele CJ, Li Z, McKee AE. On Trk--the TrkB signal transduction pathway is an increasingly important target in cancer biology. Clin Cancer Res. 2009;15:5962–7. doi: 10.1158/1078-0432.CCR-08-0651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang J, et al. Whole-genome sequencing identifies genetic alterations in pediatric low-grade gliomas. Nat Genet. 2013;45:602–12. doi: 10.1038/ng.2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones DT, et al. Recurrent somatic alterations of FGFR1 and NTRK2 in pilocytic astrocytoma. Nat Genet. 2013;45:927–32. doi: 10.1038/ng.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Frattini V, et al. The integrated landscape of driver genomic alterations in glioblastoma. Nat Genet. 2013;45:1141–9. doi: 10.1038/ng.2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lannon CL, Sorensen PH. ETV6-NTRK3: a chimeric protein tyrosine kinase with transformation activity in multiple cell lineages. Semin Cancer Biol. 2005;15:215–23. doi: 10.1016/j.semcancer.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 34.Li Z, et al. ETV6-NTRK3 fusion oncogene initiates breast cancer from committed mammary progenitors via activation of AP1 complex. Cancer Cell. 2007;12:542–58. doi: 10.1016/j.ccr.2007.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eguchi M, et al. Fusion of ETV6 to neurotrophin-3 receptor TRKC in acute myeloid leukemia with t(12;15)(p13;q25) Blood. 1999;93:1355–63. [PubMed] [Google Scholar]
- 36.Butti MG, et al. A sequence analysis of the genomic regions involved in the rearrangements between TPM3 and NTRK1 genes producing TRK oncogenes in papillary thyroid carcinomas. Genomics. 1995;28:15–24. doi: 10.1006/geno.1995.1100. [DOI] [PubMed] [Google Scholar]
- 37.Cetinbas N, et al. Mutation of the salt bridge-forming residues in the ETV6 SAM domain interface blocks ETV6-NTRK3 induced cellular transformation. J Biol Chem. 2013 doi: 10.1074/jbc.M113.475301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bax DA, et al. EGFRvIII deletion mutations in pediatric high-grade glioma and response to targeted therapy in pediatric glioma cell lines. Clin Cancer Res. 2009;15:5753–61. doi: 10.1158/1078-0432.CCR-08-3210. [DOI] [PubMed] [Google Scholar]
- 39.Li G, et al. Expression of epidermal growth factor variant III (EGFRvIII) in pediatric diffuse intrinsic pontine gliomas. J Neurooncol. 2012;108:395–402. doi: 10.1007/s11060-012-0842-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fontebasso AM, et al. Mutations in SETD2 and genes affecting histone H3K36 methylation target hemispheric high-grade gliomas. Acta Neuropathol. 2013;125:659–69. doi: 10.1007/s00401-013-1095-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chan KM, et al. The histone H3.3K27M mutation in pediatric glioma reprograms H3K27 methylation and gene expression. Genes Dev. 2013;27:985–90. doi: 10.1101/gad.217778.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lewis PW, et al. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science. 2013;340:857–61. doi: 10.1126/science.1232245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Venneti S, et al. Evaluation of Histone 3 Lysine 27 Trimethylation (H3K27me3) and Enhancer of Zest 2 (EZH2) in Pediatric Glial and Glioneuronal Tumors Shows Decreased H3K27me3 in H3F3A K27M Mutant Glioblastomas. Brain Pathol. 2013 doi: 10.1111/bpa.12042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bender S, et al. Reduced H3K27me3 and DNA hypomethylation are major drivers of gene expression in K27M mutant pediatric high-grade gliomas. Cancer Cell. 2013;24:660–72. doi: 10.1016/j.ccr.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 45.Fullgrabe J, Kavanagh E, Joseph B. Histone onco-modifications. Oncogene. 2011;30:3391–403. doi: 10.1038/onc.2011.121. [DOI] [PubMed] [Google Scholar]
- 46.Zhang J, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012;481:157–63. doi: 10.1038/nature10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Robinson G, et al. Novel mutations target distinct subgroups of medulloblastoma. Nature. 2012;488:43–8. doi: 10.1038/nature11213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dubuc AM, et al. Aberrant patterns of H3K4 and H3K27 histone lysine methylation occur across subgroups in medulloblastoma. Acta Neuropathol. 2013;125:373–84. doi: 10.1007/s00401-012-1070-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jones DT, et al. Dissecting the genomic complexity underlying medulloblastoma. Nature. 2012;488:100–5. doi: 10.1038/nature11284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pugh TJ, et al. Medulloblastoma exome sequencing uncovers subtypespecific somatic mutations. Nature. 2012;488:106–10. doi: 10.1038/nature11329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nie Z, et al. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell. 2012;151:68–79. doi: 10.1016/j.cell.2012.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lin CY, et al. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 2012;151:56–67. doi: 10.1016/j.cell.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stephens PJ, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Killela PJ, et al. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc Natl Acad Sci U S A. 2013;110:6021–6. doi: 10.1073/pnas.1303607110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kleiblova P, et al. Gain-of-function mutations of PPM1D/Wip1 impair the p53-dependent G1 checkpoint. J Cell Biol. 2013;201:511–21. doi: 10.1083/jcb.201210031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Torchia EC, Boyd K, Rehg JE, Qu C, Baker SJ. EWS/FLI-1 induces rapid onset of myeloid/erythroid leukemia in mice. Mol Cell Biol. 2007;27:7918–34. doi: 10.1128/MCB.00099-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang J, et al. A novel retinoblastoma therapy from genomic and epigenetic analyses. Nature. 2012;481:329–34. doi: 10.1038/nature10733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang J, et al. CREST maps somatic structural variation in cancer genomes with base-pair resolution. Nat Methods. 2012;8:652–4. doi: 10.1038/nmeth.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang J, et al. SNPdetector: a software tool for sensitive and accurate SNP detection. PLoS Comput Biol. 2005;1:e53. doi: 10.1371/journal.pcbi.0010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gordon D, Abajian C, Green P. Consed: a graphical tool for sequence finishing. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- 62.Rousseeuw P. Least Median Squares Regression. Journal of the American Statistical Association. 1984;79:871–880. [Google Scholar]
- 63.Holmfeldt L, et al. The genomic landscape of hypodiploid acute lymphoblastic leukemia. Nat Genet. 2013;45:242–52. doi: 10.1038/ng.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Szymczak AL, et al. Correction of multi-gene deficiency in vivo using a single ‘self-cleaving’ 2A peptide-based retroviral vector. Nat Biotechnol. 2004;22:589–94. doi: 10.1038/nbt957. [DOI] [PubMed] [Google Scholar]
- 65.Kawauchi D, et al. A mouse model of the most aggressive subgroup of human medulloblastoma. Cancer Cell. 2012;21:168–80. doi: 10.1016/j.ccr.2011.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Persons DA, et al. Enforced expression of the GATA-2 transcription factor blocks normal hematopoiesis. Blood. 1999;93:488–99. [PubMed] [Google Scholar]
- 67.Bajenaru ML, et al. Astrocyte-specific inactivation of the neurofibromatosis 1 gene (NF1) is insufficient for astrocytoma formation. Mol Cell Biol. 2002;22:5100–13. doi: 10.1128/MCB.22.14.5100-5113.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jonkers J, et al. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat Genet. 2001;29:418–25. doi: 10.1038/ng747. [DOI] [PubMed] [Google Scholar]
- 69.Fraser MM, et al. Pten loss causes hypertrophy and increased proliferation of astrocytes in vivo. Cancer Res. 2004;64:7773–9. doi: 10.1158/0008-5472.CAN-04-2487. [DOI] [PubMed] [Google Scholar]
- 70.Endersby R, Zhu X, Hay N, Ellison DW, Baker SJ. Nonredundant functions for Akt isoforms in astrocyte growth and gliomagenesis in an orthotopic transplantation model. Cancer Res. 2011;71:4106–16. doi: 10.1158/0008-5472.CAN-10-3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Korbel JO, Campbell PJ. Criteria for inference of chromothripsis in cancer genomes. Cell. 2013;152:1226–36. doi: 10.1016/j.cell.2013.02.023. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.