Abstract
Background
The increased reports of ESBL dissemination from various centres in south western, Nigeria and the recent emergence of carbapenem resistant bacteria prompted the conception of this study.
Objectives
To demonstrate the relationship between high molecular weight plasmids and the expression of antibiotic multi-resistance including ESBL and carbapenemase.
Methods
We investigated 97 isolates of selected organisms consisting of 67 E. coli and 30 Klebseilla spp for the presence of plasmids expressing ESBL including carbapenem-hydrolysing enzymes. Beta-lactamase was determined using acidometric method, while ESBL and carbapenemase activity was determined using the double-disk diffusion test as well as the Modified Hodge test (MHT). Plasmid profiles of ESBL and carbapenemase positive isolates were determined according to standard protocols.
Results
An ESBL prevalence rate of 21.6% and carbapenem- resistance rate of 9.3% was recorded. Antibiotic susceptibility profile of ESBL isolates showed 100.0% resistance against Amoxicillin, Cotrimoxazole and Erythromycin. Moderate susceptibility was recorded against the Quinolone class of antibiotics; Meropenem remained the most active antibiotic against ESBL isolates with 62.5% against E. coli and 60% against K. pneumoniae. The plasmid profiles of our study isolates ranged from 11.8kbp to 35.5kbp.
Conclusion
Due to the relationship between high molecular weight plasmids and multi-drug resistance, we hereby recommend regular molecular surveillance of this form in our study setting.
Keywords: Carbapenem-resistance, ESBL isolates, Plasmid profile, Abeokuta
Introduction
There has been a global increase in the occurrence of carbapenem, hydrolysing â-lactamases among bacterial isolates from different clinical sites1. Majority of the bacteria isolates that harbour this enzyme are of the class enterobacteriaceae, which are able to inhabit the human intestinal tract as commensals2, these group of bacteria are able to spread easily between humans through hand carriage and contaminated food and water causing large outbreaks2, and are also capable of acquiring genetic material through horizontal gene transfer3. The first carbapenemase producer in Enteriobacteriaceae (NmcA) was identified in 1993 from an isolate of Enterobacter clocae1. Cabapenemases are mainly of the Ambler class A penicillinase or Ambler class B metallo-enzyme groups, or class D enzymes4, there is also the rare chromosome encoded class C enzymes. All these groups of enzymes are capable of hydrolyzing virtually all the â-lactam antibiotics including the 3rd generation cephalosporins and carbapenems.
In Nigeria there have been reports of carbapenemase producing clinical isolates of enteric bacteria particularly among E. coli and Klebsiella spp5. In addition regular reports of various infections with ESBL organisms have been recorded in our environment; there has also been laboratory evidence of co-acquisition of muti-resistant plasmids in clinical isolates of E. coli and other enteric organisms from Abeokuta2–3. This has most undoubtedly put Abeokuta on the global antibiotic multi-drug resistant map. Previous reports have indicated the usefulness of plasmid profiling in categorising and determining the extent of antibiotic multi-drug resistance and risk of epidemic spread3,6–7. These reports have shown that community colonisation and other reservoir sources such as food animals are capable of conjugative spread of these R-plasmids with ESBL and carbapenemase activity within the hospital community to cause nosocomial outbreaks2, the implication of this on the community and National public health system can be enormous because of the cost of treating affected patients and managing outbreaks. Therefore the essence of maintaining a log of Epidemiological record of multi-resistant isolates to include their possible plasmid profiles, conjugative activity and molecular epidemiology is essential in our study setting.
It was this need of basic molecular evidence of transmissible genetic material capable of conferring carbapenemase and or ESBL activity on their carrier bacteria isolates that prompted the inception of this study. Our objective was therefore to demonstrate the relationship between high molecular weight plasmids and the expression of antibiotic multi-resistance including ESBL and carbapenemase.
Methods
Isolates
A total of 97 isolates were recovered from various clinical samples consisting of 67 E. coli and 30 Klebsiella spp. Isolates were recovered using standard Microbiological techniques and identified by gram reaction and biochemical tests8–9.
Beta lactamase testing
Beta-lactamase production was determined by the starch iodide acidometric method as described by Odugbemi et al.10 and further identified by the modified method of Stokes and Ridgway11.
ESBL detection
Beta lactamase positive isolates were further tested for the production of ESBL enzymes by the disc diffusion method using the NCCLS recommendations for non-fastidious bacteria12–13, by placing ceftazidime (30µg), cefotaxime (30µg) disc alone and placing the same discs in combination with Clavunalic acid (10µg) (Oxiod UK) placed 20mm apart. A differed of >5mm between the zones of inhibition of the discs in combination with clavunalic acid and those alone were considered positive for ESBL production. Control isolates used were E. coli ATTC 25922 and Klebseilla pneumonia ATCC 70603.
Carbapenemase detection
Detection of carbapenemase enzyme activity was done following the modified hodge test (MHT), as described by Landman et al.14. Briefly 0.5 McFarland suspension of each tested isolate was inoculated on fresh Muller-hinton agar and a 10µg Imipenem disk (Oxoid UK) was placed at the center of the plate touching the edge of each streaked isolate and incubated for 18–24hrs at 37°c. Interpretation was done with accordance with CLSI recommendations15.
Antibiotic susceptibility testing
Antibiotic susceptibility testing was done following the Kirby-bauer technique for disk diffusion9. Antibiotics tested include Amoxicillin (30µg), Cotrimoxazole (10µg), Erythromycin (10µg), Tetracyclin (30µg),Gentamycin (10µg), Amoxy-clav (30µg), Ofloxacin (10µg), Nitrofurantion(30µg), Cefuroxime (30µg), Ceftazidime (30µg) and Meropenem(10µg). The results from the zone of inhibition were interpreted according to CLSI guideline15.
Plasmid profiling
Plasmid DNA from each tested ESBL and carbapenemase producing isolate was extracted using the alkaline lysis method previously described by Akinduti et al.3 and Bradford et al.16. Extracted DNA plasmids were electropherosed on 0.8% Agarose gel, and stained with 14µl/g Ethidium bromide, gel pictures were photograghed with a Polariod camera under the view of a UV transilluminator. Molecular weights and distances were determined according to Kim et al.17.
Results
About 97 isolates were recovered from various clinical sites consisting 67 isolates of Escherichia coli and 30 Klebseilla spp. Table 1 shows the age and gender distribution of tested isolates in relation to carbapenemase and ESBL production.
Table 1.
Age and Gender distribution of ESBL and Carbapenemase producing strains of Escherichia coli and Klebsiella spp in Abeokuta, Nigeria
| Characteristic | No. tested (%) | Â-lac pos (%) | ESBL pos (%) | Carb pos (%) |
| Age (years) | ||||
| >1–14 | 27(27.8) | 12(44.4) | 9(33.3) | 3(11.1) |
| 15 – 44 | 52(53.6) | 18(34.6) | 9(17.3) | 4(7.7) |
| 45 and above | 18(18.6) | 9(50.0) | 3(16.7) | 2(11.1) |
| Gender | ||||
| Male | 44(45.4) | 18(40.9) | 9(20.5) | 4(9.1) |
| Female | 53(54.6) | 21(39.6) | 12(22.6) | 5(9.4) |
| Total | 97(100.0) | 39(40.2) | 21(21.6) | 9(9.3) |
Table 2 shows the distribution of ESBL and carbapenemase producing strains of Escherichia coli and Klebisella spp in relation to sample type/anatomical site. From the table, urine was the most frequently isolated sample site with 43 (44.3%), followed by Blood 13 (13.3%), and lastly CSF 3 (3%).
Table 2.
Distribution of ESBL and Carbapenemase producers in relation to Sample type/Anatomical site
| Sample type | No. tested (%) | Â-lact producer (%) | ESBL producer (%) | Carb producer (%) |
| Blood | 13(13.4) | 3(23.0) | 2(15.4) | 1(7.7) |
| Urine | 43(44.3) | 20(46.5) | 7(16.3) | 3(7.0) |
| Csf | 3(3.0) | 1(3.3) | 0(0.0) | 0(0.0) |
| Genitals | 4(4.1) | 2(50.0) | 2(50.0) | 0(0.0) |
| Others | 34(35.2) | 13(38.2) | 10(29.4) | 5(14.7) |
| Total | 97(100.0) | 39(40.2) | 29(29.9) | 9(9.3) |
Table 3 shows the Antibiotic susceptibility profile of ESBL producers to the most commonly used antibiotics including carbapenem.
Table 3.
Antibiotic susceptibility profile of ESBL producers to various antibiotics
| Antibiotic n (%) | |||||||||
| Isolate | Amox | Cot | Gen | Ery | Nit | Oflox | Am/Clv | Caz | Mero |
| E. coli (n=16) | 0 | 0 | 6(37.5) | 0 | 9(47.4) | 9(47.4) | 3(18.8) | 7(43.8) | 10(62.5) |
| Kleb spp (n=5) | 0 | 0 | 1(20) | 0 | 3(60) | 1(20) | 1(20) | 2(40) | 3(60) |
Amox-Amoxicillin, Cot-Cotrimoxazole, Gen-Gentamycin, Ery-Erythromycin, Nit-Nitrofurantoin, Oflox-Ofloxacin, Am/Clv-Amoxicillin/Clavunalate, Caz-Ceftraxone, Mero-Meropenem
Table 4 shows the average plasmid sizes of R-plasmids of ESBL and carbapenemase isolates of Escherichia coli and Klebseilla spp, in relation to anatomical site. From the table below Klebseilla spp recorded a higher average plasmid weight of 22.1±1.8 as compared with E. coli 20.71±1.8. With catheter isolates recording the highest plasmid weight 24.3kb
Table 4.
Average plasmid sizes of R-plasmids of ESBL and Carbapenemase producing isolates of Escherichia coli and Klebsiella spp
| Average plasmid Size (Kb) | ||
| Samples | E. coli | Kleb. Spp |
| Blood(n=3) | 14.6 | 19.9 |
| Urine(n=10) | 15.8 | 21.7 |
| C.S.F(n=0) | 0.0 | 0.0 |
| E.C.S(n=2) | 13.8 | 0.0 |
| Catheter(n=3) | 24.3 | 0.0 |
| Average wt | 20.7±2.3 | 22.1±1.8 |
Discussion
The presence and dissemination of ESBL isolates of E. coli and Klebseilla spp has been established in Abeokuta, Southwest Nigeria3,18. Recent reports indicate an increasing upsurge of this pathogen in Abeokuta; with data bordering on epidemic values particularly in institutionalised settings (Motayo unpublished data). Our current study was targeted at determining molecular evidence of transmissible plasmids responsible for ESBL and carbapenemase resistance by plasmid profiling of isolates. In our current study, 97 isolates comprising 67 E. coli and 30 Klebsiella spp. were tested with an ESBL prevalence rate of 21.6% and carbapenemase resistance rate of 9.3%. This is higher than a previous report by Olowe and Aboderin18 which reported a prevalence rate of 9.0% for ESBL producers at FMC, Abeokuta. It is lower than a similar report from the same study environment which reported a rate of 27.3% ESBL prevalence among enteric isolates3. Our results are also far lower than the report of Iroha et al.19 which reported a prevalence of 56.6% ESBL production in Klebsiella spp. isolates from blood and urine in ESUTH Enugu Southeastern Nigeria. Iroha et al.19 also reported a 59.4% ESBL production in E. coli isolates from blood and urine in ESUTH Enugu Southeastern Nigeria. This shows the extent of epidemic spread the ESBL resistance phenotype is taking particularly in Southern Nigerian Hospital settings.
From our generated data, there was no statistically significant difference of â-lactamase producers in relation to gender (p>0.05). There was significantly higher prevalence of both ESBL and carbapenemase phenotype in the age extremes. The distribution of ESBL and carbapenemase resistant isolates in relation to anatomical site revealed that the highest prevalence of ESBL resistance was from genital samples (50.0%). This was followed by urine (16.3%) and lastly blood with 15.4%. Our data reveals a high degree of ESBL resistance among isolates from blood. This has serious clinical consequences, as these resistance conferred on this isolates in blood will increase risk of medical complication such as sepsis, renal failure and death if appropriate antibiotic is not administered quickly20. Also ESBL resistance often leads to increased hospital length of stay and financial burden in terms of cost of treatment.
The current study highlights an emerging trend of carbapenemase resistance in our study setting with 7.7% prevalence in urine isolates and 7.0% in isolates from blood cultures. These posses a potential public health threat, as only few antibiotics has been shown to be effective against carbapenem-resistant bacteria. And these are not available in our environment. The value reported in our study is higher than that of a recent study done at Abeokuta, which reported a carbapenemase-resistance rate of 2.3%3.
Antibiotic susceptibility profile of the ESBL positive isolates revealed a 100.0% resistance against amoxicillin, cotrimoxazole and erythromycin. Our result is in agreement with several previous reports on high level of â-lactam and macrolide resistance in enteriobacteriaceae from our region2,5,18. Moderate susceptibility was recorded against the quinolone class of antibiotics (ofloxacin) with 47.4% in E. coli and 20.0% for Klebsiella spp. This shows that there is still some degree of efficacy displayed by this brand of antibiotics despite its wide misuse21. Nitrofurantoin, a bacteriostatic agent displayed a fairly good susceptibility profile against our ESBL isolates. The only limitation to this is that, it is only prescribed mainly against urinary tract infections and under specific conditions because of its toxic effects22.
Poor susceptibility was recorded by amoxicillin/clavunalate, with E. coli having only 18.8%. Average susceptibility was recorded for ceftrazone with E. coli having 48.8% susceptibility. This is in agreement with previous reports which highlighted the various degrees of intermediate susceptibility displayed by some classes of ESBL producing bacteria such as the AmpC and OXA types7,23. Naturally, the carbapenem drug meropenem recorded the highest susceptibility level (62.5%). This shows that this class of drug still remains the drug of choice in treating severe infections with ESBL agents. The only challenge remains lack of availability and high cost of procuring these drugs. Although our study reveals an emerging danger with carbapenem-resistance of up to 40.0%, greater attention has to be paid to regular surveillance of antibiotic drug resistance.
From the plasmid profiling of isolates, the study revealed a very high plasmid profile ranging from 11.8kbp to 35.5kbp. This is higher than previous reports on R-plasmids in south-western Nigeria. For instance, in a study done at Lagos University Teaching Hospital by Adenipekun et al.7, plasmid sizes ranged from 3.0kbp to 4.9kbp. The relatively large plasmid sizes observed in this study can be directly attributable to multiple genes coding for various resistant phenotypes. This results in resistance to 2 or more classes of antibiotics as observed in this current study. Previous reports such that of Kim et al.17 have supported this finding, and molecular evidence have shown this to be common in most R-plasmids showing resistance to 2 or more classes of antibiotics24.
Conclusion
This study has revealed the presence of isolates with very high molecular weight plasmids harbouring ESBL and carbapenemase-resistant genes. It also showed the presence of possibly co-acquired macrolide and other unidentified resistant genes. The public health implication of continuous dissemination of such multi-resistant bacteria with high frequency transmissible DNA cannot be overlooked. With the increased attention being paid to stricter infection control practices across our country2,25, it is pertinent to note that health authorities as a matter of urgency should include molecular epidemiology in its infection control policy. This is in order to detect changing patterns and emergence of new resistance genes before wide spreads dissemination occurs.
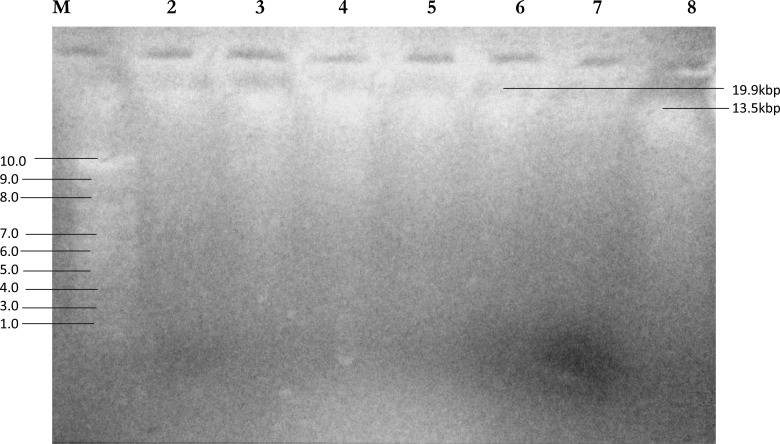
Figure 1.
Agarose gel electrophoresis of plasmid DNA recovered from the hospital isolates pathogens. Lane 1; DNA marker (M): 10.0, 9.0, 8.0, 7.0, 6.0, 5.0, 4.0, 3.0 kb, lane 2, 3, 4, 5, 6, 7 and 8 are the plasmid DNA bands of the isolates.
Figure 2.

Aculture plate showing positive samples using the Modified Hodge test for Carbapenemase resistance testing on our our isolates (At the center is Imipenem antibiotic disk 10ìg).
References
- 1.Nordman P, Naas TP, Poriel L. Global spread of carbapenemase-producing enteriobacteriacea. Emerging Infectious Diseases. 2011;17(10) doi: 10.32101/eid1710.110655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Motayo BO, Ogiogwa IJ, Okerentugba PO, Innocent-Adiele HC, Nwanze JC, Onoh CC, Okonko IO. Antimicrobial resistance profile of Extra-intestinal E. coli infections in a Southwestern Nigerian City. Journal of Microbiology Research. 2012;2(5):141–144. [Google Scholar]
- 3.Akinduti PA, Oluwadun BA, Iwalokun BA, Oluwaseun E, Onagbesan KO. Clonal dissemination of blaTEM â-lactamase strains among Enteric isolates in Abeokuta, Nigeria. Research Journal of Microbiology. 2011;6(12):919–925. [Google Scholar]
- 4.Mulvey MR, Grant JM, Plewes K, Roscoe D, Boyd DA. New Delhi Matallo-â-Lactamase in Klebsiella pnuemoniae and Escherichia coli, Canada. Emerging Infectious Diseases. 2011;17(1):103–106. doi: 10.3201/eid1701.101358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Akinduti PA, Oluwaseun E, Motayo BO, Adeyakinu AF. Emerging Multidrug resistant Ampc Beta-Lactamase and Carbapenemase enteric Isolates in Abeokuta. Nature and Science. 2012;10(7):70–74. [Google Scholar]
- 6.Schawaber MJ, Navon-Venezia S, Scwartz D, Carmeli Y. High levels of Antimicrobial coresistance among Extended spectrum beta-lactamase producing Enteriobacteriaceae Antimocrob. Agents. Chemotherapy. 2005;49:2137–2139. doi: 10.1128/AAC.49.5.2137-2139.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adenipekun EO, Aibinu IE, Daini OA, Ogunledun A, Ajekigbe AT, Adelowotan OA, et al. Occurrence of Beta-Lactamase resistance among Isolates from Cancer patients in Lagos, Nigeria. Researcher. 2009;1(6):1–6. [Google Scholar]
- 8.Cheesbrough M. Tropical Health Technology. Low priced Edition. Doddington, Cambridgeshire, England: 2003. Medical Laboratory Manual; pp. 20–35. [Google Scholar]
- 9.Cheesbrough M. Medical Laboratory manual for Tropical Countries. ELBS edition. Vol. 32. Cambridge: University Press; 1996. Microbiology; pp. 26–58. [Google Scholar]
- 10.Odugbemi TO, Hafiz S, McEntergart MG. Penicillinase-producing Neisseria gonorrhoeae: Detection by starch iodide paper techniques. British Medical Journal. 1977;2:500. doi: 10.1136/bmj.2.6085.500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stokes EJ, Ridgeway GL. Clinical Bacteriology. 5th Edn. London: Edward Arnold Ltd; 1990. p. 391. ISBN: 9780815182641. [Google Scholar]
- 12.National Committee for Clinical Laboratory Standards (NCCLS), author Performance standards for Antimicrobial susceptibility Testing; Twelfth International Supplement. Wayne, PA: National Committee for Clinical Laboratory Standards; 2002. M100-S12 (M2) [Google Scholar]
- 13.Livermore DM, Brown DFJ. Detection of â-lactamase-mediated resistance. Journal of Antimicrob Chemotherapy. 2001;48:59–64. doi: 10.1093/jac/48.suppl_1.59. [DOI] [PubMed] [Google Scholar]
- 14.Landman D, Salvani S, Bratu S, Quale J. Evaluation of techniques for detection of Carbapenem-resistant Klebseilla pnuemoniae in stool surveillance cultures. Journal of Clinical Microbiology. 2005;43:5639–5641. doi: 10.1128/JCM.43.11.5639-5641.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clinical Laboratory Standard Institute, author. Performance standards for Antimicrobial disk Susceptibility test. 1. Vol. 26. Wayne, P.A: 2006. pp. 11–23. [Google Scholar]
- 16.Bradford PA, Urban CE, Idemyor V, Rasmussen BA, Bush K. Multiple resistant K. Pnuemoniae strains from 2 Chicago Hospitals: Identification of the extended spectrum TEM-12 and TEM-10 Ceftaxidime -hydrolysing beta-lactamases in a single isolate. Jour nal of Antimicro Agents Chemotherapy. 1994;35:7–22. doi: 10.1128/aac.38.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim YK, Pai H, Lee HJ, Park SE, Choi EH. Bloodstream infections by ESBL producing Escherichia coli and Klebsiella pneumoniae in Children: Epidemiology and clinical outcome. Antimicrob Agents Chemotherapy. 2002;46:1481–1491. doi: 10.1128/AAC.46.5.1481-1491.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Olowe AO, Aboderin BW. Detection of Extended Spectrum â-Lactamase producing strains of Escherichia coli and Klebsiella sp in a Tertiary Health Centre in Ogun State. International Journal of Tropical Medicine. 2010;5(3):62–64. [Google Scholar]
- 19.Iroha IR, Amadi ES, Orji AE, Nwuzo AC, Ejike-Ugwu PC. Detection of Plasmid Borne Extended spectrum beta lactamase Enzymes from Blood and Urine of Gram-Negative Bacteria from a University teaching Hospital in Nigeria. Current Research in Bacteriology. 2010;3:77–83. [Google Scholar]
- 20.Boaz Adegboro. Microbiology. Ibadan University press; 2010. Enteriobacteriacea. [Google Scholar]
- 21.Nkang AO, Okonko IO, Mejeha OK, Babalola ET, Adewale OG, Motayo BO, et al. Survey of the efficacy and quality of some brands of Antibiotics sold in Calabar Metropolis, South-south region of Nigeria. Scientific Research and Essay. 2010;5(4):395–406. [Google Scholar]
- 22.Okonko IO, Donbraye-Emmanuel OB, Ijandipe LA, Ogun AA, Adedeji AO, Udeze AO. Antibiotics Sensitivity and Resistance patterns of Uropathogens to Nitrofurantoin and Nalidixic Acid in Pregnant Women with Urinary Tract Infections in Ibadan, Nigeria. Middle-East Journal of Science Research. 2009;4(2):105–109. [Google Scholar]
- 23.Bradford PA. Extended spectrum beta-lactamases in the 21st century: Characterisation, epidemiology and detection of this important resistant threat. Clinical Microbiology Reviews. 2001;14:933–951. doi: 10.1128/CMR.14.4.933-951.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grover SS, Sharma S, Chattopadhya D, Kapoor H, Pasha ST, Singh S. Phenotypic and Genotypic detection of ESBL mediated Cephalosporin resistance in Klebsiella pnuemoniae: Emergence of high resistance against cefepime, the 4th generation cephalosporin. Journal of Infection. 2006;54:279. doi: 10.1016/j.jinf.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 25.Okeshola AO, Makanjuola O. Resistance to 3rd generation cephalosporins and other antibiotics by enteriobacteriaceae in Western Nigeria. American Journal of Infectious Diseases. 2009;5:17–20. [Google Scholar]