ABSTRACT
The fungal pathogen Cryptococcus neoformans has several virulence factors, among which the most important is a polysaccharide capsule. The size of the capsule is variable and can increase significantly during infection. In this work, we investigated the relationship between capsular enlargement and the cell cycle. Capsule growth occurred primarily during the G1 phase. Real-time visualization of capsule growth demonstrated that this process occurred before the appearance of the bud and that capsule growth arrested during budding. Benomyl, which arrests the cells in G2/M, inhibited capsule growth, while sirolimus (rapamycin) addition, which induces G1 arrest, resulted in cells with larger capsule. Furthermore, we have characterized a mutant strain that lacks a putative G1/S cyclin. This mutant showed an increased capacity to enlarge the capsule, both in vivo (using Galleria mellonella as the host model) and in vitro. In the absence of Cln1, there was a significant increase in the production of extracellular vesicles. Proteomic assays suggest that in the cln1 mutant strain, there is an upregulation of the glyoxylate acid cycle. Besides, this cyclin mutant is avirulent at 37°C, which correlates with growth defects at this temperature in rich medium. In addition, the cln1 mutant showed lower intracellular replication rates in murine macrophages. We conclude that cell cycle regulatory elements are involved in the modulation of the expression of the main virulence factor in C. neoformans.
IMPORTANCE
Cryptococcus neoformans is a pathogenic fungus that has significant incidence worldwide. Its main virulence factor is a polysaccharide capsule that can increase in size during infection. In this work, we demonstrate that this process occurs in a specific phase of the cell cycle, in particular, in G1. In agreement, mutants that have an abnormal longer G1 phase show larger capsule sizes. We believe that our findings are relevant because they provide a link between capsule growth, cell cycle progression, and virulence in C. neoformans that reveals new aspects about the pathogenicity of this fungus. Moreover, our findings indicate that cell cycle elements could be used as antifungal targets in C. neoformans by affecting both the growth of the cells and the expression of the main virulence factor of this pathogenic yeast.
INTRODUCTION
Cryptococcus neoformans is a facultative intracellular fungal pathogen that causes meningitis, primarily in immunocompromised patients (1, 2). Its incidence increased significantly in association with the HIV pandemic, and it is estimated to cause more than 650,000 deaths per year, mainly in developing areas (3).
The best-characterized virulence factors of C. neoformans are a polysaccharide capsule (reviewed in reference 4), melanin production (5, 6), and the ability to grow at body temperature (7). During infection, C. neoformans undergoes changes that contribute to its persistence. These changes include capsule enlargement and the appearance of titan/giant cells (8–10).
A characteristic feature of the capsule is its ability to enlarge in response to certain environmental conditions (11–16). This process is believed to be essential for yeast survival in the host, because capsule growth confers protection against host defense mechanisms, including oxidative stress (17). Moreover, capsule enlargement has been correlated with increased intracranial pressure during human cryptococcal meningitis (18). Although capsule enlargement is an early response of the pathogen during infection, little is known about its regulation and the underlying molecular mechanisms that regulate this process. Recent articles have highlighted the importance of some transcription factors (such as Ada2, Rim101, and Gat201) in this process (19–21), indicating that capsule growth occurs through the induction of a gene expression rearrangement. In addition, the structurally conserved Pbs2-Hog1 mitogen-activated protein (MAP) kinase cascade controls morphological differentiation and virulence factors in serotype A C. neoformans (22).
In this study, we investigated the hypothesis that capsule enlargement in C. neoformans is coordinated with the cell cycle. This idea was hypothesized from different pieces of evidence. Under conditions of capsule growth, there is normally a decrease in the growth rate of the yeast, which suggests that factors that elongate some phases of the cell cycle result in capsule enlargement. In addition, we hypothesize that capsule enlargement occurs mainly in G1, prior to the emergence of the bud, because the addition of more polysaccharide during the S/G2/M phases might interfere with budding and the subsequent separation of the bud from the mother cell. Furthermore, several findings from the literature indicate that there is a correlation between capsule size and cell body size (11, 23), and the fact that cell body growth occurs mainly in G1 suggests capsule growth would occur in the same cell cycle phase.
Cell cycle control has been extensively studied in Saccharomyces cerevisiae (24, 25), Schizosaccharomyces pombe (26), and Ustilago maydis (27). Cell cycle progression depends on the activity of a cyclin-dependent kinase (Cdk1/Cdc28/Cdc2), which remains low during G1 and increases its kinase activity during the rest of the phases (28). The transition between G1 and S is regulated by specific G1/S cyclins (29, 30). However, little is known about the cell cycle regulation in C. neoformans. This yeast replicates by budding, and during the exponential phase, budding and DNA synthesis occur simultaneously (31). However, at the end of the exponential phase, budding is delayed and cells are arrested at G1 or G2 phases (32, 33).
To test our hypothesis, we have investigated and modulated cell cycle progression during capsule growth. In addition, we have investigated capsular phenotypes in a C. neoformans mutant lacking a putative cyclin. Our results indicate that capsule growth is linked to cell cycle progression and, in particular, to the G1 phase. Therefore, we conclude that the link between cell cycle elements and capsule growth reveals new aspects about the biology of this virulence factor.
RESULTS
Analysis of DNA content during capsule growth.
We first investigated whether capsule growth changed during the cell cycle progression of C. neoformans. Consistent with our initial hypothesis, capsule enlargement occurred mainly during the G1 phase, so we expected that the cells would arrest in this phase after transfer to capsule-inducing medium. As shown in Fig. 1A, cells grown overnight at 30°C in Sabouraud medium were mainly in the G2 phase. However, when cells were transferred to capsule-inducing medium, we observed an arrest in G1 (shown as “n” in Fig. 1A).
FIG 1 .
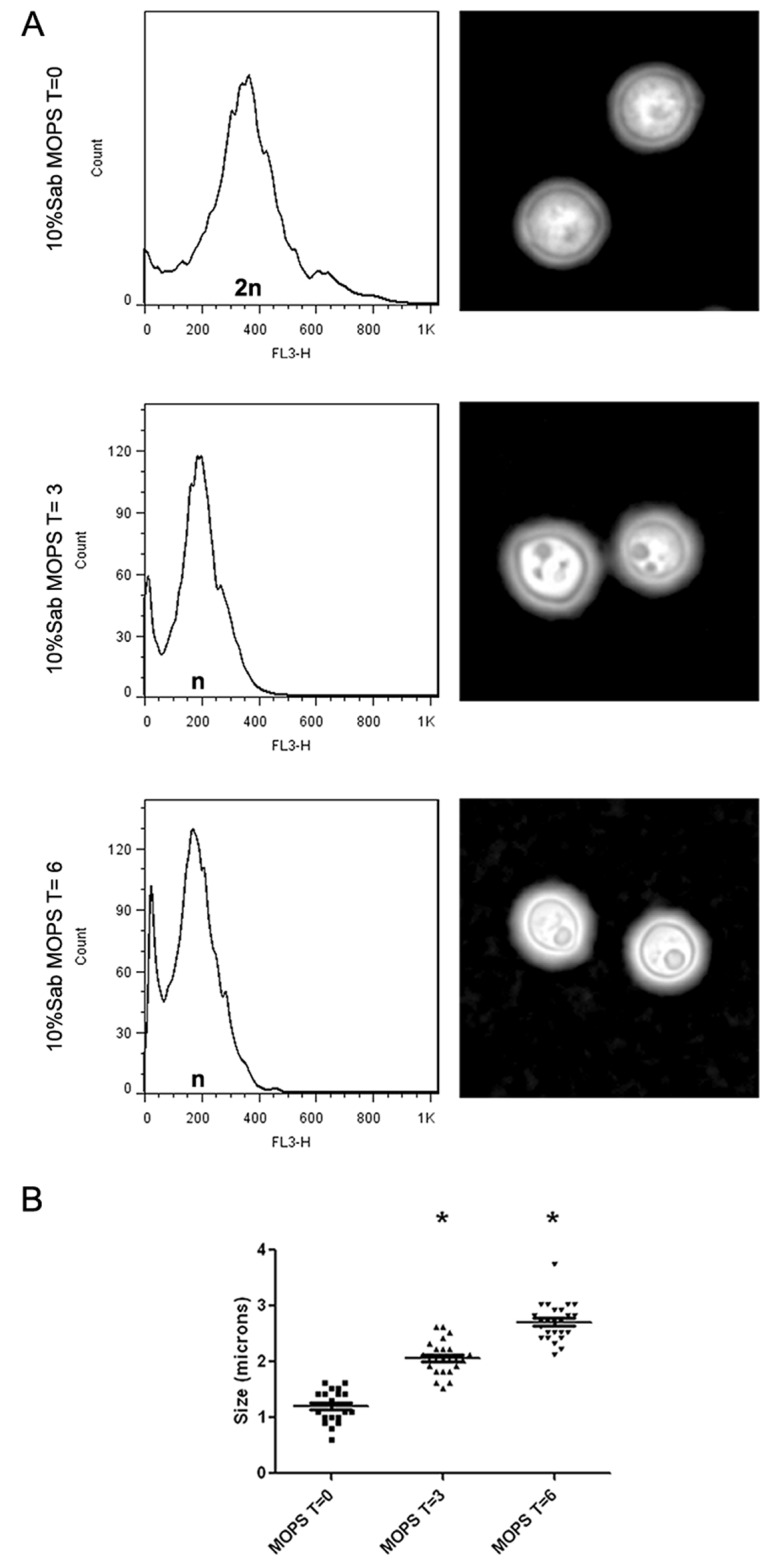
Flow cytometry analysis of cell cycle progression during capsule enlargement. (A) Samples were taken for flow cytometry analysis at the time indicated after PI staining (see Materials and Methods). Micrographs of cells suspended in India ink were taken in parallel. Sab, Sabouraud medium. (B) Capsule sizes at the different time points.
Capsule sizes were measured in parallel at the different time points to confirm capsule enlargement in the capsule-inducing medium. Capsule growth was already noticeable after 3 h of incubation under these conditions, and the process continued after 6 h (P < 0.05) (Fig. 1B).
Capsule enlargement process is altered in the presence of cell cycle inhibitors.
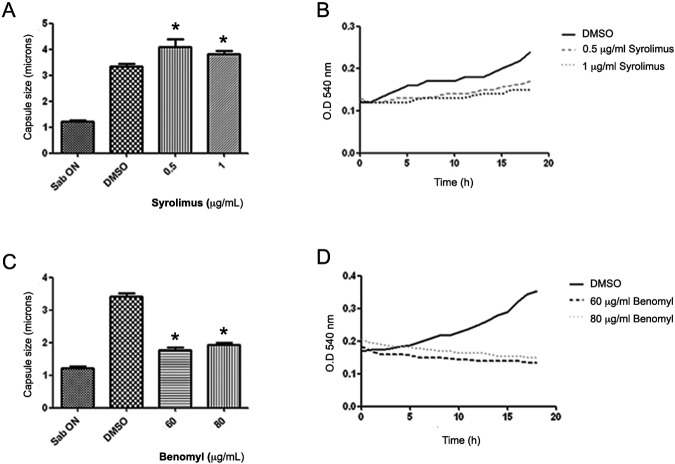
We studied the effect of cell cycle inhibitors on capsule growth. We first used sirolimus (rapamycin), which is an inhibitor of the TOR pathway and produces G1 arrest. Sirolimus promoted capsule enlargement of C. neoformans in capsule-inducing medium (Fig. 2A). For both concentrations tested (0.5 and 1 µg/ml), the capsule sizes were significantly larger in cells exposed to sirolimus relative to the control condition (capsule-inducing medium with dimethyl sulfoxide [DMSO]) (P < 0.05). To ensure that sirolimus was inhibiting cell cycle progression, we performed growth curve experiments under the same conditions and observed a significant defect in the growth rate at the concentrations used (Fig. 2B).
FIG 2 .
Capsule enlargement in the presence of cell cycle inhibitors. Bars show the means ± standard deviations (SD) of the capsule sizes measured from cultures incubated with different concentrations of sirolimus (rapamycin) (A) or benomyl (C). *, P < 0.05. (B and D) Growth curves of C. neoformans in capsule-inducing medium with the different concentrations of sirolimus (B) and benomyl (D). Cultures containing DMSO were used as controls.
We also used benomyl, an inhibitor of microtubule polymerization that induces M-phase arrest. Benomyl inhibited capsule enlargement of C. neoformans under capsule-inducing conditions (Fig. 2C) (P < 0.05). In addition, benomyl inhibited yeast growth at both concentrations tested (Fig. 2D). To discard the possibility that inhibition of capsule growth was not a consequence of cell death induced by the drug, we measured the viability of the cells after benomyl treatment by propidium iodide (PI) staining. We found that approximately 80% of the cells were alive after incubation with benomyl at the highest concentration tested (data not shown), indicating that lack of capsule growth was not a consequence of cell death.
Capsule growth visualization by time-lapse microscopy.
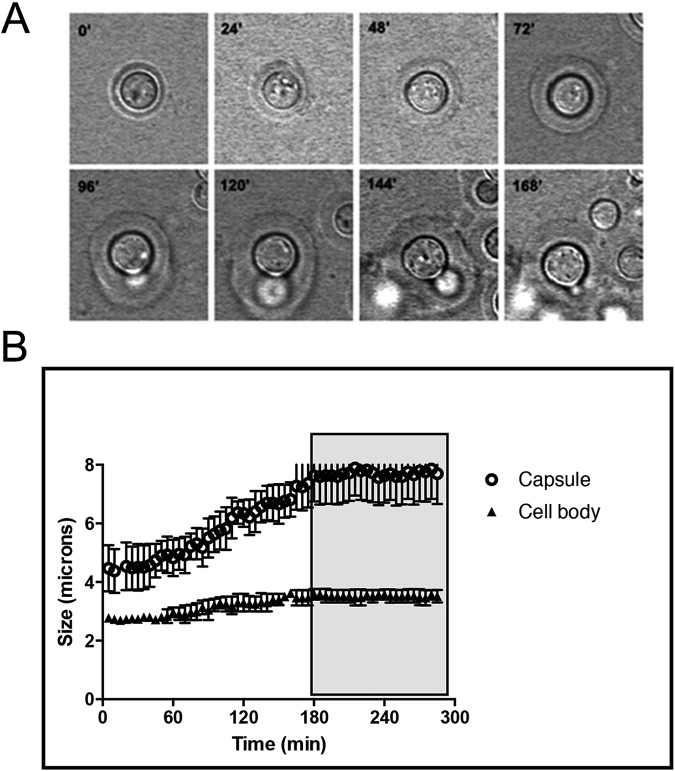
According to our hypothesis, capsule growth should stop or diminish during budding, so we used time-lapse microscopy to visualize capsule growth. The capsule, however, is not visible under regular conditions given its high water content and similar refractive properties to that of the medium. Different methods have been described to visualize the capsule by light microscopy, with India ink negative staining being the easiest and most widely used in the literature. We tried to induce capsule growth in medium containing India ink, but we observed that under these conditions, capsular enlargement was impaired (result not shown), suggesting that the India ink interferes with the process. Another way to visualize the capsule is based on a phenomenon known as the capsular “quellung” reaction (34–37), which involves a change in the capsule’s optical properties (i.e., refraction index) following binding of specific monoclonal antibodies (MAbs) to the capsular polysaccharide. The resulting Ab-coated capsule can be readily visualized using Nomarski microscopy (also known as differential interference contrast [DIC]). Given that the IgM MAb 13F1 induced a capsular reaction without significantly affecting the mechanical properties of the capsule (38), this antibody was used to visualize capsule enlargement as a function of time (Fig. 3A). When cells were transferred to capsule-inducing medium, enlargements of the cell and capsule were observed at 3.6 and 8.8 nm/min, respectively (Fig. 3B; see Movie S1 in the supplemental material). When the capsule reached a certain size (approximately 6 µm), the cell cycle progressed with the subsequent appearance of the bud and a decrease in capsule growth (Fig. 3B). Interestingly, after budding, the mother cells with enlarged capsule underwent a new round of cell cycle with no further cell or capsule growth (see Movie S1).
FIG 3 .
Capsule growth visualization by time-lapse microscopy. Capsule enlargement was observed based on the capsular “quellung” reaction produced by the 13F1 MAb. (A) Sequence of frames showing capsule and cell body enlargement. (B) The rate of capsule and cell body growth was determined by linear regression analysis of lengths as a function of time. The capsule enlargement speed was 8.8 nm/min, and the cell body enlargement speed was 3.6 nm/min. A plateau in capsule and cell body size is observed following yeast replication (boxed in gray).
Identification of G1/S cyclins in C. neoformans.
Next, we investigated capsular phenotypes in mutants affected in cell cycle regulation. For this purpose, we identified G1/S cyclins in C. neoformans by looking for homologues of the corresponding CLN1 gene from Ustilago maydis (39). Since this organism is also a basidiomycete, we reasoned that the C. neoformans cell cycle proteins were more homologous to this organism than to those from members of the ascomycetes, such as Saccharomyces cerevisiae, Schizosaccharomyces pombe, or Candida albicans. After performing a BLAST comparison using the Cln1p from U. maydis against the C. neoformans genome database available at the Broad Institute, we identified the open reading frame (ORF) CNAG_06092, which was already annotated as a putative cyclin gene. We obtained the corresponding mutant from a library of targeted disruptants available at the ATCC (20).
We reconstituted the wild-type (wt) gene in this mutant. For this purpose, the wild-type gene was fused to the Neo marker, which confers resistance to Geneticin, using fusion PCR (see Materials and Methods). The DNA construct was integrated into the genome by biolistic transformation. In this way, we continued this study with the wild-type strain (H99), the cyclin mutant, and the corresponding reconstituted strain.
Capsule enlargement in vitro.
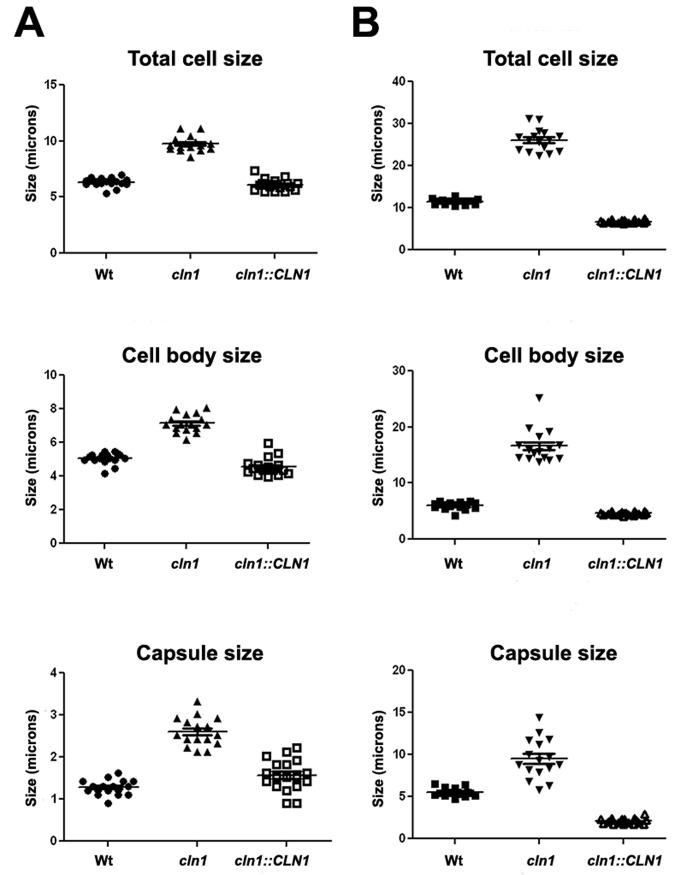
We first investigated whether capsule size was affected in the cyclin mutant. For this purpose, we measured the total cell size, the cell body size (delimited by the cell wall), and the capsule size after growth under regular conditions (Sabouraud liquid medium) or in capsule-inducing medium (10% Sabouraud medium [pH 7.3]). As shown in Fig. 4, the cln1 mutant had larger cell body size than the wild-type and reconstituted strains. The mutant had also significantly larger capsule size than the other two strains, suggesting that, in fact, capsule size was coordinated with cell cycle progression.
FIG 4 .
Morphogenesis of wt, cln1, and cln1::CLN1 strains from C. neoformans. (A and B) Distribution of total cell size, cell body size, and capsule diameter of C. neoformans cells grown in Sabouraud medium (A) and in 10% Sabouraud medium buffered at pH 7.3 with 50 mM MOPS (B).
Time-lapse microscopy revealed differences in G1 length and budding time in the cln1 mutant.
G1/S cyclin mutants showed a delay in the initiation of DNA replication (40), so we investigated if the mutant also had a delay in the appearance of the bud. For this purpose, we performed live imaging and measured the time of cell growth and budding. We focused on nascent daughter cells, not on mother cells, since the latter cells have already reached the size that induces cell cycle progression and the G1 phase is almost absent after the separation of the bud (41, 42). The analysis of these videos showed that more than 50% of the cells in the mutant strain population had defects in cell cycle progression. Daughter cells showed aberrant shapes and remained attached to the mother cell, and thus, it was difficult to quantify the time required for these daughter cells to bud again. However, the other 50% of the daughter cells managed to completely separate from the mother cells and showed a significant delay in the appearance of the new bud compared to the wild-type strain (61 ± 3 min versus 98 ± 12 min; P < 0.05) (see Movies S2 and S3 in the supplemental material).
The cln1 mutant showed increased vesicle production.
Capsule synthesis has been related to the secretion of vesicles with capsular polysaccharide and other components inside (43, 44). Therefore, we investigated whether cln1 had an enhanced ability to secrete vesicles during capsule growth. We isolated and estimated the amount of vesicles from the supernatant of the cultures by using a fluorometric assay (43, 44). Using this approach, we observed that cln1 produced more extracellular vesicles than the wt and reconstituted strains (Fig. 5A). In parallel, we quantified the amount of sterol present in the vesicle preparation using high-performance thin-layer chromatography (HPTLC) as an independent parameter that reflects vesicle secretion. In agreement with the fluorometric assay, we found that cln1 had higher sterol content than the wild-type and reconstituted strains (Fig. 5B and C).
FIG 5 .
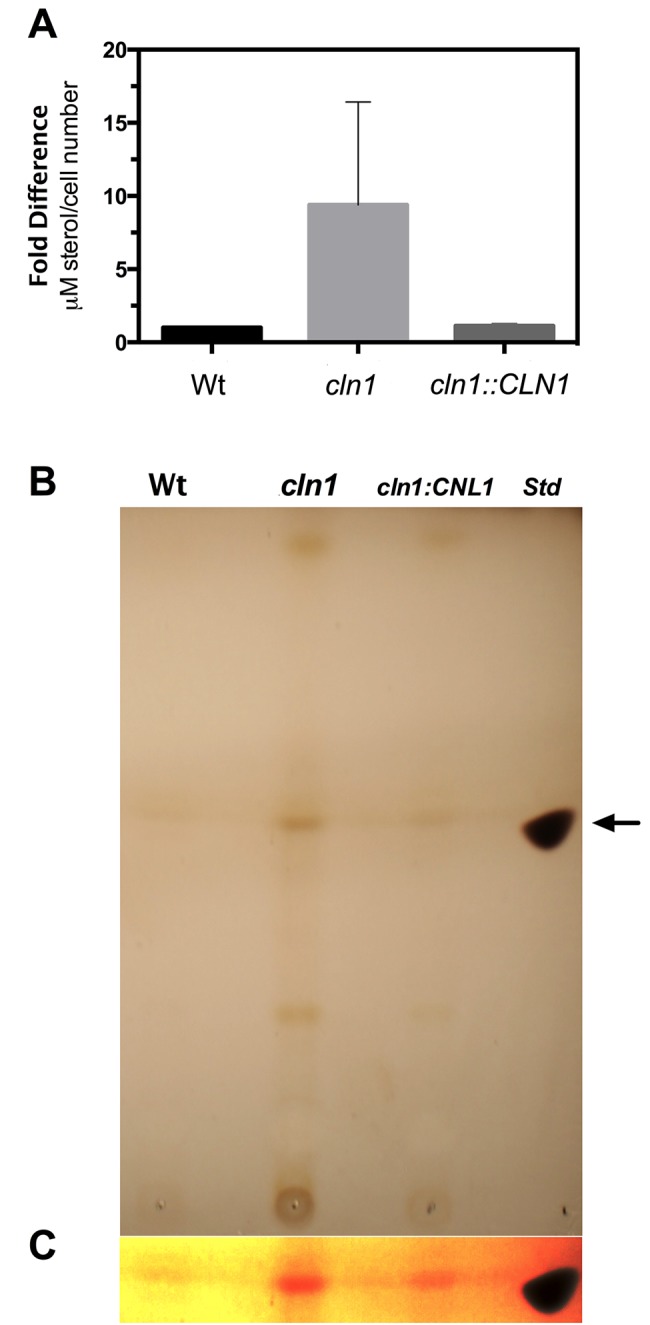
The cln1 mutant shows increased production of extracellular vesicles. (A) Comparative analysis of vesicle production based on an indirect sterol-based vesicle quantification of fractions obtained from the wt, mutant, and reconstituted C. neoformans strains, using a fluorometric assay kit. Error bars represent standard deviations of 3 independent experiments. (B) Comparative analysis of the sterol content in vesicle preparations using HPTLC. An arrow indicates the migration of an ergosterol standard (Std). The result is representative of 2 independent analyses showing similar results. (C) Contrast image of the area highlighted with the arrow in panel B to emphasize the differences in band intensity among strains.
Analysis of differentially accumulated proteins in cln1 during capsule enlargement.
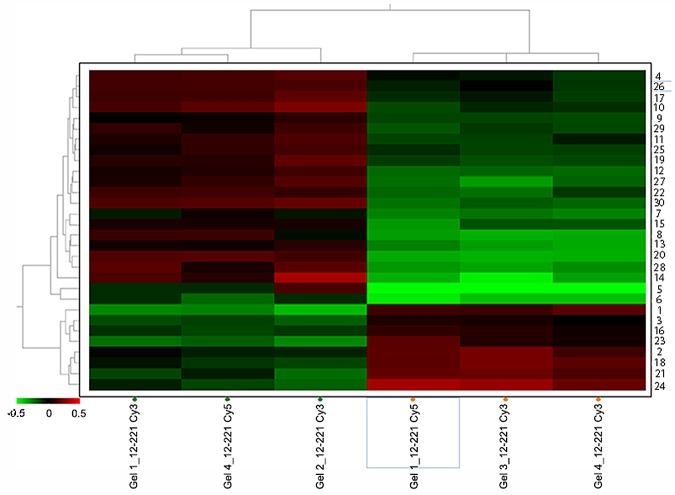
To investigate the mechanisms by which the cln1 mutant produces larger capsules relative to the wild-type strain, we compared the accumulation of proteins in these two strains during the capsule enlargement process. We performed protein extraction from wt and cln1 cells incubated under capsule-inducing conditions for 6 h (see Materials and Methods). The analysis was performed with 3 independent replicates. Protein extracts were processed by difference gel electrophoresis (DIGE). A difference of ±1.5-fold change in protein accumulation was considered statistically significant (P < 0.05). All possible proteins and their relative abundance were represented in a heat map (Fig. 6). Thirty possible proteins were automatically matched. Twenty-six proteins out of 30 could be cut using ImageQuant v 5.1 software (GE Healthcare) after the gel had been stained with colloidal Coomassie blue (CCB). These 26 proteins were imported to the Oracle database, and 19 of them were identified by mass spectrophotometry with a 4800 matrix-assisted laser desorption ionization–tandem time of flight (MALDI-TOF/TOF) mass spectrometer. Finally, sequences were identified by comparison to Cryptococcus neoformans database available at the Broad Institute.
FIG 6 .
Relative abundance of proteins accumulated in the cln1 mutant under capsule-inducing conditions. The upper dendrogram shows the relationship among individual gels, while the left dendrogram shows the relationship among proteins. Hierarchical clustering analysis (HCA) was performed to obtain the values of abundance of every spot, which were then represented in a heat map. Different red spots show a positive relative abundance (cln1 mutant/H99 ratio), while different green spots show a negative relative abundance.
As shown in Table 1, some proteins involved in meiosis and budding were less abundant in the cln1 mutant than in the wt strain (H99). However, proteins such as malate synthase (which is involved in the glyoxylate cycle and allows the yeast to get energy from fatty acids) and other proteins related to glucose and xylose metabolism (i.e., transketolase and aldoketolase) were more abundant in the cln1 mutant than in the wt strain.
TABLE 1 .
List of proteins identified by mass spectrophotometrya
| Spot | Protein identification | cln1 mutant/H99 ratio (fold change) | ORF |
|---|---|---|---|
| 1040 | Transketolase | 3.74 | CNAG_07445 |
| 617 | Heat shock proteín 90 | 3.59 | CNAG_06150 |
| 2002 | Wos2 (p21) | 2.76 | CNAG_07558 |
| 861 | Malate synthase | 2.54 | |
| 2228 | Phosphopyruvate hydrogenase | 2.37 | CNAG_03072 |
| 1427 | Aldoketoreductase | 1.94 | CNAG_01257 |
| 974 | Chaperone | 1.92 | CNAG_01750 |
| 1222 | Ornithine carbamyltransferase | 1.8 | CNAG_02813 |
| 1062 | Aminotransferase | 1.79 | CNAG_02853 |
| 860 | Malate synthase | 1.7 | CNAG_05653 |
| 1149 | Adenylsuccinate synthase | 1.68 | CNAG_02858 |
| 2227 | Phosphogluconate dehydrogenase | 1.67 | CNAG_07561 |
| 598 | Heat shock protein | 1.59 | CNAG_05199 |
| 915 | UTP-glucose-1-phosphate uridyltransferase | 1.51 | CNAG_02748 |
| 1133 | Methione adenosyltransferase | −1.52 | CNAG_00418 |
| 584 | Heat shock protein 70 | −1.55 | CNAG_01727 |
| 532 | Heat shock protein | −1.69 | CNAG_06443 |
| 1473 | Budding-related protein | −2.08 | CNAG_04194 |
| 531 | Meiosis-related protein | −2.71 | CNAG_00995 |
The first and second columns indicate the proteins. The third column shows the fold change in accumulation of the protein between wt and cln1 strains. The last column is the corresponding ORF for each protein when a BLAST protein-protein comparison was performed in the C. neoformans database of the Broad Institute
Virulence and capsule variations of the cln1 strain during G. mellonella infection.
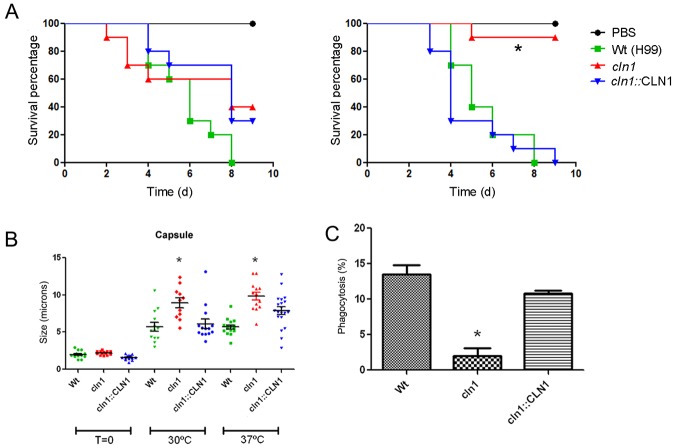
Since the cln1 strain showed a temperature-sensitive growth defect, we tested its potential virulence in Galleria mellonella (Fig. 7A). We chose this model because virulence can be assessed at different temperatures (45). The cyclin mutant strain was completely avirulent at 37°C (P < 0.05). In contrast, at lower temperature, the mutant was as virulent as the wild-type strain (P > 0.05). Interestingly, cln1 yeast cells recovered after 3 days of infection from larvae incubated at 30 and 37°C had a larger cell and capsular size than the wt strain (Fig. 7B). In all cases, the cln1::CLN1 strain restored the virulence phenotype.
FIG 7 .
In vivo model of the effect of cln1. (A) Survival curves of G. mellonella infected with 106 cells of C. neoformans strains at 30°C (left) and 37° C (right) (B) Capsule sizes of yeast cells recovered from G. mellonella after 3 days of infection at 30 and 37°C. (C) Phagocytosis was performed at a 2:1 ratio of yeast cells to macrophages (see Materials and Methods). Bars show the mean ± SD phagocytosis of C. neoformans cells after 2 h at 37°C. *, P <0.05.
We also tested the degree of phagocytosis using the G. mellonella model. After 2 h of the administration of the yeasts to the larvae, hemocytes showed reduced phagocytosis of cln1 strain compared to the wt and cln1::CLN1 strains (1% ± 0.7% versus 19% ± 3% and 13% ± 1%; P < 0.05) (Fig. 7C).
Macrophage and C. neoformans interaction.
Phagocytosis assays were also performed with the macrophage-like cell line RAW 264.7. Cells from the cln1 mutant were less efficiently phagocytosed by murine macrophages than the wt strain (41% ± 6% versus 88% ± 2%, respectively). Furthermore, intracellular replication was determined using time-lapse microscopy. The Cryptococcus neoformans wt strain was able to replicate within 30 to 40% of infected macrophages. In contrast, in macrophages infected with the cln1 mutant, we only observed fungal replication in <10% of the cases.
DISCUSSION
Our study provides evidence that capsule enlargement in C. neoformans is coordinated with the cell cycle. We hypothesized that C. neoformans enlarges its capsule during G1 phase for the following reasons. Several studies have already shown that capsule enlargement is induced in C. neoformans in low-iron medium (13), in the presence of CO2 (15), in serum (11), in diluted Sabouraud medium (12), or in the presence of mannitol (14). Under each of these conditions, yeast cells grow more slowly—probably because they spend more time in G1 phase. Furthermore, it is possible that the addition of capsule during S/G2/M phases could interfere with the process of budding and daughter cell separation, thus providing another reason for linking capsule growth to G1 phase of the cell cycle. Moreover, it has been observed that during capsule growth, there is a correlation between the cell body size and capsule size (11, 23), suggesting that these two processes are in fact coordinated.
Using different techniques (flow cytometry and live imaging), we confirmed the relationship between capsule growth and G1 arrest. Interestingly, we observed that after capsule enlargement and bud separation, mother cells progressed normally through the cell cycle (fast appearance of a new bud with a very short G1 phase), with the absence of further capsule growth. This result suggests that the G1/S checkpoint is controlled, not only by cell body size but also by capsule size after capsule enlargement stimulation. This regulation might be dependent on the TOR pathway, since sirolimus, which also causes G1 arrest (46), enhanced capsule growth. In budding yeast, the TOR pathway detects nutrient conditions and promotes cell growth and proliferation, which is also connected to cell size (47). Inhibition of the TOR pathway by sirolimus leads to arrest cryptococcal cells in G1 phase, and thus cryptococcal cells have more time to enlarge their capsules. These results were confirmed by using another cell cycle inhibitor, such as benomyl. This drug has been described to arrest cells in M phase due to the loss of microtubule function (48). Our results demonstrate a link between cell cycle and capsule growth, and open a new perspective to understand the regulation of this process.
We also investigated capsular phenotypes in a mutant that had a G1/S cyclin disrupted (CLN1) and which was available at the ATCC. Parallel studies have phenotypically confirmed the role of this gene as coding for a G1/S cyclin (31). In preliminary experiments, this mutant manifests an elongated G1 phase and exhibited a larger capsule relative to the wt strain under capsule-inducing conditions, confirming that capsule growth was coordinated with the cell cycle. The cln1 mutant produces more extracellular vesicles than wt and reconstituted strains, which confirmed the hypercapsular phenotype of cln1, since it is known that transport of vesicles in C. neoformans is linked to capsule production (44).
Besides, the cln1 mutant also manifested a defect in the width of the neck between the mother and the bud, which was significantly narrower in the WT strain than in the cyclin mutant (31). This finding had been already reported and could also explain the delay in budding since it restricts the movement of plasma membrane proteins and cell wall material (31). However, our results should not be interpreted as if any condition that enlarged G1 phase results in capsule growth, since an inducing signal is required (i.e., mannitol, serum, CO2, or nutrient deprivation). In this regard, little is known about the stimuli that trigger capsule induction, and further studies are required to understand the changes that occur in the cell during capsule growth.
Our initial characterization of the mutant using time-lapse microscopy demonstrated that the mutant had an elongated G1 phase, supporting its role in the regulation of the transition from G1 to S phase. In budding yeasts, the G1 phase in the mother cell is very short because it has already reached the proper size for cell division. However, the daughter cell of the cln1 mutant exhibits a longer G1 phase, until it reaches a cell size that triggers the initiation of the cell cycle (Start) (41, 42, 49). Furthermore, we observed a significant delay in budding in the cln1 mutant strain compared to wt or reconstituted strains, which is in agreement with previous findings (50, 51). This phenomenon could imply that cln1 mutation may cause a delay in a later part of the cell cycle, so that buds grew large before they separate (52, 53). Alternatively, it could mean that enlarged mother cells require more time to duplicate all of their cytoplasmic content during budding because of the larger size of the cells.
Cell cycle and G1 cyclins are also involved in the morphogenesis and differentiation of different yeast species, such as Candida albicans, Saccharomyces cerevisiae, and Ustilago maydis (39, 54, 55). Our results are in agreement with these findings, since capsule enlargement can be considered in C. neoformans as a morphological transition that involves multiple cellular changes, in particular, induction of changes in gene expression that result in a dramatic accumulation of new polysaccharide in the capsule (20, 21, 56–58). Cyclins are a highly conserved family of proteins that are involved in multiple processes in the cell through regulation of specific protein kinases. During cell cycle progression, cyclins bind and regulate the activity of the cyclin-dependent protein kinases (CDKs). There are several cyclins that control different parts of the cell cycle through activation of appropriate CDK partners (59). However, it has been reported that these cyclins could be involved in different signaling pathways, including fungal development, toxin metabolism, and pathogenicity (39, 60). Absence of Cln1 results in a longer G1 phase, suggesting that this protein is involved in the regulation of this transition. However, full identification of the biochemical function of this protein will require further studies to elucidate whether this protein binds to Cdk1 or to another protein kinase.
Proteomic analysis showed that some proteins related to meiosis and budding were in smaller amounts in the cln1 mutant than in the wt strain. In the cln1 mutant, proteins related to the glyoxylate cycle and glucose metabolism were more abundant than in the wt strain, which suggests that cln1 cells are more metabolically active. This result is in agreement with the idea that G1 is the phase the most metabolically active of the cell cycle. Therefore, our results suggest a model in which factors that elongate G1 allow cells to stay in G1 for a longer time and thus increase their capsules.
The protein Wos2 was also more abundant in the cln1 mutant than in the wt strain. This protein was described as a homolog of P23 in Schizosaccharomyces pombe that is involved in cell cycle progression, so its expression decreases when cells enter stationary phase or are grown under nutrient limitation conditions (61). Besides, different heat shock proteins appeared differently accumulated. Some of them were more abundant in the cln1 mutant than in the wt strain, such as the heat shock protein Hsp90, which is involved in the response to heat shock and is required for the viability of the cells (62). In contrast, other heat shock proteins appeared less abundant in the cln1 mutant than in the wt strain, such as Hsp70, which has been linked to the expression of laccase in C. neoformans (63).
The cyclin mutant exhibited virulence defects, especially at 37°C, which are related to its reduced growth at this temperature. Interestingly, yeast cells recovered from infected larvae showed an enlarged capsule compared to those observed in vitro (45, 64). This result correlates with the fact that cryptococcal cells can remain in the host in a latent state without being eliminated (reviewed in reference 65). Cryptococcus neoformans is an intracellular fungal pathogen, which means it can persist in hosts inside macrophages (66–70). However, we have observed that the cln1 mutant is poorly phagocytosed by Galleria hemocytes, possibly because its larger size impairs its internalization by phagocytic cells, which correlates with a defect in virulence. Furthermore, this hypothesis was confirmed by the results observed from the in vitro interaction between murine macrophages and wt and cln1 mutant strains of C. neoformans. Besides, the reduced capability of the cln1 mutant to replicate within macrophages could explain its decrease in virulence. During the interaction between the cln1 mutant and murine macrophages, extrusion was only observed in one case, which leads us to think that, finally, macrophages are able to control the infection and eliminate the phagocytosed yeast cells.
In summary, our results demonstrate a link between cell cycle and the regulation of the size of the main virulence factor of C. neoformans. We propose a model for capsule growth in which capsule induction stimulus is followed by a transient arrest in G1 that induces capsular enlargement until a certain size is reached, which then triggers the progression of the cell cycle. We have also demonstrated that the absence of Cln1 results in reduced virulence, so cryptococcal factors that regulate the cell cycle could offer a good target to develop new antifungal compounds.
MATERIALS AND METHODS
Strains and culture conditions.
Cryptococcus neoformans var. grubii strain H99 (71) and the cln1 mutant strain CNAG_06092, obtained from a collection of mutants deposited at the ATCC by H. D. Madhani (20), were used in this study. The strains were routinely grown in liquid Sabouraud medium (Oxoid, Ltd., Basingstoke, Hampshire, England) at 30°C or 37°C with moderate shaking (150 rpm). To induce capsule growth, the cells were transferred to 10% Sabouraud medium buffered at pH 7.3 with 50 mM MOPS (morpholinepropanesulfonic acid) buffer (Sigma-Aldrich, St. Louis, MO) at 30°C or 37°C with shaking (12). Murine macrophage-like cells (72) were grown in feeding medium, which contains Dulbecco’s modified Eagle’s medium (Lonza, Verbiers, Belgium) supplemented with 10% heat-inactivated fetal bovine serum (FBS) (HyClone-Perbio), 10% NCTC medium (Sigma-Aldrich, Steinheim, Germany) and 1% nonessential amino acids (Sigma-Aldrich, Steinheim, Germany). Macrophages were regularly maintained at 37°C in a 5% CO2-enriched atmosphere.
Nuclear staining and FACS analysis.
Measurement of DNA content by flow cytometry was performed as described in reference 52 with minor modifications. Aliquots were taken at different time intervals from 10% Sabouraud medium in MOPS buffer and pelleted by centrifugation. Cells were suspended in 70% ethanol and kept overnight at 4°C for fixation. The cells were washed twice with distilled water and finally suspended in 2 ml of a mixture of 20 mM sodium citrate, 50 mM EDTA, and 0.45 mM sorbitol (pH 5.5). After 1 h of incubation at 30°C, RNase (Sigma-Aldrich, St. Louis, MO) was added at a final concentration of 10 µg/ml, and the tubes were kept at 30°C for 2 h.
Propidium iodide was added to a final concentration of 5 µg/ml, and samples were taken to the cytometer. Samples without propidium iodide were used in parallel as negative controls. Stained cells were observed under the fluorescence microscope, and DNA content was measured using a FacsCalibur flow cytometer (BD Biosciences, Woburn, MA). Flow cytometry data were processed using CellQuest (BD Biosciences) and FlowJo (Tree Star, Inc., Ashland, OR) software.
Capsule growth in the presence of cell cycle inhibitors.
Cryptococcus neoformans strain H99 cells were grown in 5 ml of Sabouraud medium at 30°C with moderate agitation. Cells were washed and transferred to 10% Sabouraud medium in MOPS buffered at pH 7.3 containing sirolimus (Sigma-Aldrich, St. Louis, MO) at different concentrations (0, 0.5, and 1 µg/ml), which induces G1 arrest. In addition, benomyl (Sigma-Aldrich, St. Louis, MO), which causes M arrest, was also tested for its effect on capsule growth. Cells were grown in Sabouraud liquid medium at 30°C overnight and then washed and incubated in 10% Sabouraud medium in MOPS (pH 7.3) containing different benomyl concentrations (0, 60, and 80 µg/ml). Cultures were incubated at 30°C overnight, and suspensions of India ink were photographed and measured as explained above. DMSO was added to the control cultures (without drugs). In addition, propidium iodide (5 µg/ml) was added to the cells after incubation with the drugs to verify that any possible effect on capsule growth was not due to lack of viability. Heat-killed cells (65°C, 1 h) were used as a positive control.
To confirm that the drugs had an effect on the cells, we performed growth curves in 10% Sabouraud liquid medium buffered in MOPS at pH 7.3 containing the different concentrations of the drugs mentioned above in a 96-well plate (Costar, NY) during 18 h at 30°C using an iEMS spectrophotometer (Thermofisher). The optical density at 540 nm (OD540) was measured every hour, and graphs were plotted using Graph Pad Prism 5.
In vivo capsular growth visualization using time-lapse microscopy.
Cells from the H99 strain were grown overnight in 10 ml of Sabouraud medium at 30°C under constant agitation. The cells were then washed 3 times with phosphate-buffered saline (PBS), and the density was enumerated using a hemocytometer. Approximately 104 cells were placed in a Lab-Tek chambered coverglass (Thermo, Fisher Scientific, Rochester, NY) containing 100 µl of capsule-inducing medium (10% Sabouraud medium buffered in 50 mM MOPS) and supplemented with 20 µg/ml of IgM MAb 13F1 (73). The chamber slide was placed in a temperature-controlled microscope chamber adjusted to 37°C. Image acquisition was done at 5-min intervals with a 40× differential inference contrast (DIC) objective in an SP5 confocal inverted microscope equipped with a camera. Images were processed using Leica Microsystems and ImageJ software. Capsule and cell body dimensions were determined from time-lapse microscopy images using Adobe Photoshop. Capsule radial length was calculated by subtracting the length of the cell body from the diameter of the whole cell, capsule included. Determination of the capsule and cell body growth rate was done by linear regression of a length versus time plot.
Identification of G1/S cyclins by homology with the corresponding cyclin from Ustilago maydis.
The Cln1 protein sequence from Ustilago maydis (39) was used to perform a BLAST comparison against the C. neoformans H99 strain genome deposited at the Broad Institute (http://www.broadinstitute.org/annotation/genome/cryptococcus_neoformans/MultiHome.html). In this way, we identified ORF CNAG_06092, which was already annotated as coding for a putative cyclin. Moreover, this gene has been characterized as encoding a G1/S cyclin protein (31) and consequently was denominated CLN1.
Reconstitution of the CnCLN1 gene.
The reconstituted strain (cln1::CLN1) was created by biolistic DNA delivery. Genomic DNA from wild-type C. neoformans var. grubii containing the full-length CnCLN1 gene was amplified by PCR using Phusion high-fidelity DNA polymerase (Finnzymes, Espoo, Finland) using the primers NEOCLN1 (5′ GTCATAGCTGTTTCCTGGAGCAGGTCTCCTCAACGTCTT 3′) and CLN13′3 (5′ AAGTATCACCGTCCAGTTCGTG 3′). The neomycin resistance marker (74) was amplified from the pPzp plasmid (75) using the primers CLN1MKRf (5′ CTTAGCCGTCTCATAACGCGACCCAGTCACGACGTTGTA 3′) and NEOCLN1r (5′ AAGACGTTGAGGAGACCTGCTCCAGGAAACAGCTATGAC 3′). The C. neoformans gene cassette was created by fusion PCR (76, 77) using the CLN1MKRf and CLN13′3 primers. Biolistic transformation was performed as in reference 76 using a Bio-Rad PDS 1000/He Biolistic PDS machine. Colonies were selected on YPD plates containing 100 µg/ml of Geneticin, and integration of the gene was confirmed by PCR.
Estimation of G1-phase length by real-time microscopy.
Suspensions of C. neoformans strains were prepared in Sabouraud liquid medium at 104/ml from overnight cultures. Wells from a 96-well plate were coated with 50 µl of a stock solution of MAb 18B7 at a final concentration of 0.2 µg/ml and incubated for 1 h at room temperature. Then mAb18B7 was removed, 100 µl from the yeast suspension was added, and the mixture was incubated at 30°C under a Leica DMI 4000B microscope. Photographs were taken every 2 or 3 min using the 20× objective. The videos generated by the Leica software were exported as AVI documents and processed with ImageJ software (NIH; http://rsb.info.nih.gov/ij). The G1 phase was calculated by counting the frames from the appearance of the first daughter cell until the same daughter cell began to bud again.
Analysis of extracellular vesicle production.
The wild type, cln1 mutant, and reconstituted strains were grown in 1-liter cultures of capsule-inducing medium under constant agitation for 2 days at 30°C. Culture supernatants, containing extracellular vesicles, were cleared by centrifugation at 5,000 × g (25 min, 4°C). Supernatants were then collected and passed through a 0.8-µm-pore filter membrane. The filtered volume was centrifuged at 100,000 × g for 1 h at 4°C using a 45 Ti rotor. Vesicle pellets were washed 3 times with sterile-filtered cold PBS, with the sample centrifuged each time at 100,000 × g for 1 h at 4°C.
Vesicle production was assayed by measuring the concentration of sterols (structural components and markers of fungal extracellular vesicles and vesicle membranes [44]) by a quantitative fluorometric kit (Amplex red sterol assay kit; Molecular Probes) and by high-performance thin-layer chromatography (HPTLC).
For sterol quantification using the Amplex kit, the resulting pellets were suspended in 500 µl of PBS and processed according to the manufacturer’s instructions. The sterol concentration was normalized by culture’s cell density.
For HPTLC, the vesicle pellets were extracted with 3 sample volumes of methanol-chloroform (1:1). The mixtures were homogenized by sonication. The lower phase (chloroform) was recovered, dried, and suspended in methanol for analysis by HPTLC. The volume of methanol used to resuspend the lipid material was proportional to the cell density of the cultures. Thirty microliters of the samples was loaded into HPTLC silica plates (Si 60F254s; LiChrospher, Merck, Germany) and separated using the mobile-phase hexane-ether-acetic acid (80:40:2 [vol/vol/vol]). Sterol spots were identified by spraying the plate with a ferric chloride solution and heating at 100°C for 3 to 5 min.
Proteomics.
To investigate the different patterns of protein accumulation between the cln1 mutant and wt strain, we performed proteomics. Yeast cells were grown in 10 ml of Sabouraud liquid medium overnight at 30°C. Then the cells were transferred to capsule-inducing medium (10% Sabouraud in 50 mM MOPS [pH 7.3]) at a final cell density of 107 cells/ml. After 6 h of incubation at 30°C, cultures were centrifuged at 3,500 × g for 10 min, and the pelleted cells were suspended in protein lysis buffer (5 mM EDTA, 1× Complete protease inhibitor cocktail [Roche, Indianapolis, IN] in TE buffer [10 mM Tris-HCl at pH 8.1, mM EDTA]). Next, two aliquots of 1 ml were separated in 2-ml tubes, and 425- to 600-µm Ø glass beads (Sigma-Aldrich, St. Louis, MO) were added. Cell rupture was carried in a Fast-Prep for 6 cycles of 20 s with 4-min intervals in ice. Finally, tubes were centrifuged for 10 min at 4°C, and supernatants were collected and kept at 4°C until protein determination using the Bradford protein assay (Bio-Rad, Munich, Germany) was performed.
Protein samples were analyzed at the Proteomics Facility at the UCM-UPM (a member of the ProteoRed-ISCIII network). Samples were purified with the 2D-Clean Up cleaning kit (GE Healthcare). Bradford quantification was confirmed by 10% SDS-PAGE, and samples were stained with colloidal Coomassie blue (CCB).
Next, the 4 replicates of 100 µg from each sample (cln1 mutant and H99) were mixed and loaded in a two-dimensional (2D)-PAGE gel. To visualize the proteins, gels were stained with CCB and scanned. Half of the biological replicates were stained with Cy3 and the other half with Cy5. Then the gels were scanned in a Typhoon Trio fluorescent scanner (GE Healthcare) using filters corresponding to each fluorochrome (excitation/emission: Cy3, 532/580; Cy5, 633/670; Cy2, 488/520 nm). Once scanned, gels were stained with CCB.
Images were cut with ImageQuant v 5.1 software (GE Healthcare) and imported to the Oracle database. After the automatic matching of the putative proteins, manual corrections and matching were carried out. Different statistical analyses were performed, such as principal-component analysis and cluster analysis, to obtain the final number of proteins to be identified by mass spectrophotometry in a 4800 MALDI-TOF/TOF mass spectrometer. The identification of picks was performed using the NCBI database with taxonomic restriction in yeast.
Galleria mellonella survival experiments.
Galleria mellonella larvae (Alcotan, Valencia, Spain) were infected as described in references 45 and 64. Briefly, larvae were selected to weigh between 0.3 and 0.5 g and to be free of any dark marks. Then the pro-leg area was cleaned with 70% ethanol using a swab. The larvae were inoculated with 10 µl of a yeast suspension prepared at 108/ml in PBS containing 50 µg/ml of ampicillin by an injection using a 26-gauge needle with Hamilton syringes. The syringes were prepared by cleaning them with diluted bleach and ethanol. After injection, caterpillars were incubated in 90-mm plastic petri dishes (Soria Genlab, SA, Madrid, Spain) at 37 or 30°C, and the number of dead caterpillars was scored every day. A group of 20 larvae were inoculated with PBS with 50 µg/ml of ampicillin in each experiment to monitor killing due to physical injury, and another group of 20 caterpillars without any manipulation were used in parallel as untreated controls.
Isolation of C. neoformans cells from G. mellonella.
To isolate the yeasts from G. mellonella, larvae were smashed using cell strainers with a 100-µm pore size (BD Falcon, Erembodegem, Belgium) and 5-ml syringe plungers (BD Plastipak, Madrid, Spain) (64). Homogenates were collected in 1 ml of PBS, and samples were washed twice and suspended in 150 µl of PBS.
Fungal cells were suspended in India ink (Remel Bactidrop, Lenexa, KS), observed by microscopy, and photographed using a Leica DMI 3000B microscope. In parallel, pictures of the cryptococcal cells grown overnight in Sabouraud liquid medium were taken to control for cell size prior to infection (T = 0). Cell and capsule sizes were measured using Adobe Photoshop 7.0 (San Jose, CA). Total cell size was defined as the diameter of the complete cell, including the capsule. Capsule size was calculated as the difference between the diameter of the total cell and the cell body diameter, defined by the cell wall.
In vivo phagocytosis assays.
Yeast cells were grown in liquid Sabouraud medium as described above and stained with Calcofluor white (Sigma-Aldrich, St. Louis, MO) at 10 µg/ml for 30 min at 37°C. After incubation, cells were washed twice with PBS and suspended at 108 cells/ml. Larvae were infected with 10 µl of the inoculum (106 cells) and incubated at 37°C. After 2 h, hemolymph was collected in microcentrifuge tubes containing 100 µl of PBS and centrifuged at 1,500 × g for 3 min. Pelleted hemocytes were suspended in 200 µl of PBS and placed on coverslips for 20 min to allow the cells to adhere. Coverslips were placed on the slides with Fluoromount G (Southern Biotech), and the number of hemocytes with internalized C. neoformans cells was enumerated using a Leica DMI 3000B fluorescence microscope. Phagocytosis was expressed as the percentage of hemocytes that contained yeast cells. Experiments were performed on different days in triplicates.
Phagocytosis with murine macrophages and Giemsa staining.
Phagocytosis using the murine macrophage cell line RAW 264.7 and Giemsa staining was done as described in reference 78. Using a Leica DMI 3000B microscope, 5 pictures per well were taken to count the total number of macrophages and the number of macrophages with intracellular yeasts. The phagocytosis percentage was calculated as the number of infected macrophages divided by the number of total macrophages multiplied per 100.
Live imaging of the interaction between murine macrophages and C. neoformans.
After phagocytosis experiments using RAW 264.7 macrophages, performed as described above, the nonphagocytosed yeasts were removed by extensive washing with macrophage feeding medium. The 96-well plate (MatTek, Ashland) was placed under a Leica SP5 confocal microscope, and pictures were taken using a 20× objective in a 5% CO2 environment at 37°C. This motorized microscope allows the taking pictures of different wells in the same experiment. In that sense, two videos for the wt strain (H99) and 2 videos for the cln1 mutant were obtained in parallel. Pictures were taken every 3 min for around 12 h. The videos generated by the Leica software were exported as AVI documents and processed with ImageJ software (NIH) (http://rsb.info.nih.gov/ij). The final videos were generated by merging 8 frames per s, with each frame taken every 3 min, which means that 1 s from the video is equivalent to 24 min of the experiment.
Statistical analysis.
Survival data from G. mellonella experiments were analyzed by the Kaplan-Meier method using Graph Pad Prism 5 (La Jolla, CA). Every experiment was repeated three times, and the results were similar among all experiments. Scatter plot analysis of cell sizes was done with Graph Pad Prism 5 (La Jolla, CA), and statistical differences were assessed with a t test. A P value of <0.05 was considered significant. The t test was also performed to evaluate mean differences among the wt, cln1, and cln1::CLN1 strains regarding the duration of the G1 phase and the percentage of phagocytosis.
SUPPLEMENTAL MATERIAL
Real-time imaging of capsule growth. The movie shows time-lapse microscopy images at 5 frames per s of C. neoformans undergoing capsule enlargement followed by yeast budding. Image acquisition was performed at 5-min intervals over a total period of 395 min using a 63× DIC objective. The scale bar represents 5 µm. Download
The movie shows a representative cell of the H99 strain in Sabouraud liquid medium at 30°C in which the replication rate and G1 lapse were calculated. In this case, pictures were taken every 2 min. The interval between when the first bud emerges until that “daughter cell” buds again corresponds to the G1 lapse. In this case, the duration of the G1 phase is 61 min (28 pictures). Download
The movie shows a representative cell of the cln1 mutant strain in Sabouraud liquid medium at 30°C in which the replication rate and G1 lapse were calculated. In this case, the pictures were taken every 3 min. The interval between when the first bud emerges until that “daughter cell” buds again corresponds to the G1 lapse. In this case, the duration of the G1 phase is 98 min (31 pictures). Download
ACKNOWLEDGMENTS
O.Z. is funded by grants SAF2008-03761 and SAF2011-25140 from the Spanish Ministry for Economics and Competitivity. R.G.-R. is supported by an FPI fellowship (reference BES-2009-015913) from the Spanish Ministry of Science and Innovation. N.T.-C. is supported by an FPI fellowship (reference BES-2012-051837) from the Spanish Ministry for Economics and Competitivity. A.C. is supported by NIH grants HL059842-3, A1033774, A1052733, and AI033142. R.J.B.C. is supported by T32 AI07506 (NIH/NIAID).
Footnotes
Citation García-Rodas R, Cordero RJB, Trevijano-Contador N, Janbon G, Moyrand F, Casadevall A, Zaragoza O. 2014. Capsule growth in Cryptococcus neoformans is coordinated with cell cycle progression. mBio 5(3):e00945-14. doi:10.1128/mBio.00945-14.
REFERENCES
- 1. Heitman J, Kozel T, Kwon-Chung K, Perferct J, Casadevall A. 2011. Cryptococcus. From human pathogen to model yeast. ASM Press, Washington, DC [Google Scholar]
- 2. Mitchell TG, Perfect JR. 1995. Cryptococcosis in the era of AIDS—100 years after the discovery of Cryptococcus neoformans. Clin. Microbiol. Rev. 8:515–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Park BJ, Wannemuehler KA, Marston BJ, Govender N, Pappas PG, Chiller TM. 2009. Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. AIDS 23:525–530. 10.1097/QAD.0b013e328322ffac [DOI] [PubMed] [Google Scholar]
- 4. Zaragoza O, Rodrigues ML, De Jesus M, Frases S, Dadachova E, Casadevall A. 2009. The capsule of the fungal pathogen Cryptococcus neoformans. Adv. Appl. Microbiol. 68:133–216. 10.1016/S0065-2164(09)01204-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Williamson PR. 1997. Laccase and melanin in the pathogenesis of Cryptococcus neoformans. Front. Biosci. 2:e99–e107 [DOI] [PubMed] [Google Scholar]
- 6. Casadevall A, Rosas AL, Nosanchuk JD. 2000. Melanin and virulence in Cryptococcus neoformans. Curr. Opin. Microbiol. 3:354–358. 10.1016/S1369-5274(00)00103-X [DOI] [PubMed] [Google Scholar]
- 7. Casadevall A, Perfect J. 1998. Cryptococcus neoformans. ASM Press, Washington, DC [Google Scholar]
- 8. Zaragoza O, García-Rodas R, Nosanchuk JD, Cuenca-Estrella M, Rodríguez-Tudela JL, Casadevall A. 2010. Fungal cell gigantism during mammalian infection. PLoS Pathog. 6:e1000945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Okagaki LH, Strain AK, Nielsen JN, Charlier C, Baltes NJ, Chrétien F, Heitman J, Dromer F, Nielsen K. 2010. Cryptococcal cell morphology affects host cell interactions and pathogenicity. PLoS Pathog. 6:e1000953. 10.1371/journal.ppat.1000953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Feldmesser M, Kress Y, Casadevall A. 2001. Dynamic changes in the morphology of Cryptococcus neoformans during murine pulmonary infection. Microbiology 147:2355–2365 http://mic.sgmjournals.org/content/147/8/2355.abstract [DOI] [PubMed] [Google Scholar]
- 11. Zaragoza O, Fries BC, Casadevall A. 2003. Induction of capsule growth in Cryptococcus neoformans by mammalian serum and CO2. Infect. Immun. 71:6155–6164. 10.1128/IAI.71.11.6155-6164.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zaragoza O, Casadevall A. 2004. Experimental modulation of capsule size in Cryptococcus neoformans. Biol. Proced. Online 6:10–15. 10.1251/bpo68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Vartivarian SE, Anaissie EJ, Cowart RE, Sprigg HA, Tingler MJ, Jacobson ES. 1993. Regulation of cryptococcal capsular polysaccharide by iron. J. Infect. Dis. 167:186–190. 10.1093/infdis/167.1.186 [DOI] [PubMed] [Google Scholar]
- 14. Guimarães AJ, Frases S, Cordero RJ, Nimrichter L, Casadevall A, Nosanchuk JD. 2010. Cryptococcus neoformans responds to mannitol by increasing capsule size in vitro and in vivo. Cell. Microbiol. 12:740–753. 10.1111/j.1462-5822.2010.01430.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Granger DL, Perfect JR, Durack DT. 1985. Virulence of Cryptococcus neoformans. Regulation of capsule synthesis by carbon dioxide. J. Clin. Invest. 76:508–516. 10.1172/JCI112000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Anna EJ. 1979. Rapid in vitro capsule production by cryptococci. Am. J. Med. Technol. 45:585–588 [PubMed] [Google Scholar]
- 17. Zaragoza O, Chrisman CJ, Castelli MV, Frases S, Cuenca-Estrella M, Rodríguez-Tudela JL, Casadevall A. 2008. Capsule enlargement in Cryptococcus neoformans confers resistance to oxidative stress suggesting a mechanism for intracellular survival. Cell. Microbiol. 10:2043–2057. 10.1111/j.1462-5822.2008.01186.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Robertson EJ, Najjuka G, Rolfes MA, Akampurira A, Jain N, Anantharanjit J, von Hohenberg M, Tassieri M, Carlsson A, Meya DB, Harrison TS, Fries BC, Boulware DR, Bicanic T. 2014. Cryptococcus neoformans ex vivo capsule size is associated with intracranial pressure and host immune response in HIV-associated cryptococcal meningitis. J. Infect. Dis. 209:74–82. 10.1093/infdis/jit435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. O’Meara TR, Norton D, Price MS, Hay C, Clements MF, Nichols CB, Alspaugh JA. 2010. Interaction of Cryptococcus neoformans Rim101 and protein kinase A regulates capsule. PLoS Pathog. 6:e1000776. 10.1371/journal.ppat.1000776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu OW, Chun CD, Chow ED, Chen C, Madhani HD, Noble SM. 2008. Systematic genetic analysis of virulence in the human fungal pathogen Cryptococcus neoformans. Cell 135:174–188. 10.1016/j.cell.2008.07.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Haynes BC, Skowyra ML, Spencer SJ, Gish SR, Williams M, Held EP, Brent MR, Doering TL. 2011. Toward an integrated model of capsule regulation in Cryptococcus neoformans. PLoS Pathog. 7:e1002411. 10.1371/journal.ppat.1002411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bahn YS, Kojima K, Cox GM, Heitman J. 2005. Specialization of the HOG pathway and its impact on differentiation and virulence of Cryptococcus neoformans. Mol. Biol. Cell 16:2285–2300. 10.1091/mbc.E04-11-0987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zaragoza O, Telzak A, Bryan RA, Dadachova E, Casadevall A. 2006. The polysaccharide capsule of the pathogenic fungus Cryptococcus neoformans enlarges by distal growth and is rearranged during budding. Mol. Microbiol. 59:67–83. 10.1111/j.1365-2958.2005.04928.x [DOI] [PubMed] [Google Scholar]
- 24. Nandakumar H, Shankaramba KB. 1990. Massive sequestration of the upper jaw: a case report. Br. J. Oral Maxillofac. Surg. 28:55–56. 10.1016/0266-4356(90)90014-C [DOI] [PubMed] [Google Scholar]
- 25. Dirick L, Böhm T, Nasmyth K. 1995. Roles and regulation of Cln-Cdc28 kinases at the start of the cell cycle of Saccharomyces cerevisiae. EMBO J. 14:4803–4813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Forsburg SL, Nurse P. 1991. Identification of a G1-type cyclin puc1+ in the fission yeast Schizosaccharomyces pombe. Nature 351:245–248. 10.1038/351245a0 [DOI] [PubMed] [Google Scholar]
- 27. García-Muse T, Steinberg G, Perez-Martín J. 2004. Characterization of B-type cyclins in the smut fungus Ustilago maydis: roles in morphogenesis and pathogenicity. J. Cell Sci. 117:487–506. 10.1242/jcs.00877 [DOI] [PubMed] [Google Scholar]
- 28. Stern B, Nurse P. 1996. A quantitative model for the cdc2 control of S phase and mitosis in fission yeast. Trends Genet. 12:345–350. 10.1016/S0168-9525(96)80016-3 [DOI] [PubMed] [Google Scholar]
- 29. Wittenberg C, Reed SI. 2005. Cell cycle-dependent transcription in yeast: promoters, transcription factors, and transcriptomes. Oncogene 24:2746–2755. 10.1038/sj.onc.1208606 [DOI] [PubMed] [Google Scholar]
- 30. Berman J. 2006. Morphogenesis and cell cycle progression in Candida albicans. Curr. Opin. Microbiol. 9:595–601. 10.1016/j.mib.2006.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Virtudazo EV, Kawamoto S, Ohkusu M, Aoki S, Sipiczki M, Takeo K. 2010. The single Cdk1-G1 cyclin of Cryptococcus neoformans is not essential for cell cycle progression, but plays important roles in the proper commitment to DNA synthesis and bud emergence in this yeast. FEMS Yeast Res. 10:605–618 [DOI] [PubMed] [Google Scholar]
- 32. Takeo K, Tanaka R, Miyaji M, Nishimura K. 1995. Unbudded G2 as well as G1 arrest in the stationary phase of the basidiomycetous yeast Cryptococcus neoformans. FEMS Microbiol. Lett. 129:231–235. 10.1111/j.1574-6968.1995.tb07585.x [DOI] [PubMed] [Google Scholar]
- 33. Ohkusu M, Hata K, Takeo K. 2001. Bud emergence is gradually delayed from S to G2 with progression of growth phase in Cryptococcus neoformans. FEMS Microbiol. Lett. 194:251–255. 10.1111/j.1574-6968.2001.tb09478.x [DOI] [PubMed] [Google Scholar]
- 34. Neill JM, Castillo CG, Smith RH, Kapros CE. 1949. Capsular reactions and soluble antigens of Torula histolytica and of Sporotrichum schenckii. J. Exp. Med. 89:93–106. 10.1084/jem.89.1.93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mukherjee J, Cleare W, Casadevall A. 1995. Monoclonal antibody mediated capsular reactions (Quellung) in Cryptococcus neoformans. J. Immunol. Methods 184:139–143. 10.1016/0022-1759(95)00097-T [DOI] [PubMed] [Google Scholar]
- 36. MacGill TC, MacGill RS, Casadevall A, Kozel TR. 2000. Biological correlates of capsular (quellung) reactions of Cryptococcus neoformans. J. Immunol. 164:4835–4842. 10.4049/jimmunol.164.9.4835 [DOI] [PubMed] [Google Scholar]
- 37. Evans EE, Garcia C, Kornfeld L, Seeliger HP. 1956. Failure to demonstrate capsular swelling in Cryptococcus neoformans. Proc. Soc. Exp. Biol. Med. 93:257–260. 10.3181/00379727-93-22725 [DOI] [PubMed] [Google Scholar]
- 38. Cordero RJ, Bergman A, Casadevall A. 2013. Temporal behavior of capsule enlargement by Cryptococcus neoformans. Eukaryot. Cell 12:1383–1388. 10.1128/EC.00163-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Castillo-Lluva S, Pérez-Martín J. 2005. The induction of the mating program in the phytopathogen Ustilago maydis is controlled by a G1 cyclin. Plant Cell 17:3544–3560. 10.1105/tpc.105.036319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Irniger S, Nasmyth K. 1997. The anaphase-promoting complex is required in G1 arrested yeast cells to inhibit B-type cyclin accumulation and to prevent uncontrolled entry into S-phase. J. Cell Sci. 110:1523–1531 [DOI] [PubMed] [Google Scholar]
- 41. Johnston GC, Pringle JR, Hartwell LH. 1977. Coordination of growth with cell division in the yeast Saccharomyces cerevisiae. Exp. Cell Res. 105:79–98. 10.1016/0014-4827(77)90154-9 [DOI] [PubMed] [Google Scholar]
- 42. Hartwell LH, Unger MW. 1977. Unequal division in Saccharomyces cerevisiae and its implications for the control of cell division. J. Cell Biol. 75:422–435. 10.1083/jcb.75.2.422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rodrigues ML, Nakayasu ES, Oliveira DL, Nimrichter L, Nosanchuk JD, Almeida IC, Casadevall A. 2008. Extracellular vesicles produced by Cryptococcus neoformans contain protein components associated with virulence. Eukaryot. Cell 7:58–67. 10.1128/EC.00370-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rodrigues ML, Nimrichter L, Oliveira DL, Frases S, Miranda K, Zaragoza O, Alvarez M, Nakouzi A, Feldmesser M, Casadevall A. 2007. Vesicular polysaccharide export in Cryptococcus neoformans is a eukaryotic solution to the problem of fungal trans-cell wall transport. Eukaryot. Cell 6:48–59. 10.1128/EC.00318-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mylonakis E, Moreno R, El Khoury JB, Idnurm A, Heitman J, Calderwood SB, Ausubel FM, Diener A. 2005. Galleria mellonella as a model system to study Cryptococcus neoformans pathogenesis. Infect. Immun. 73:3842–3850. 10.1128/IAI.73.7.3842-3850.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zinzalla V, Graziola M, Mastriani A, Vanoni M, Alberghina L. 2007. Rapamycin-mediated G1 arrest involves regulation of the Cdk inhibitor Sic1 in Saccharomyces cerevisiae. Mol. Microbiol. 63:1482–1494. 10.1111/j.1365-2958.2007.05599.x [DOI] [PubMed] [Google Scholar]
- 47. Jorgensen P, Nishikawa JL, Breitkreutz BJ, Tyers M. 2002. Systematic identification of pathways that couple cell growth and division in yeast. Science 297:395–400. 10.1126/science.1070850 [DOI] [PubMed] [Google Scholar]
- 48. Hoyt MA, Totis L, Roberts BT. 1991. S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell 66:507–517. 10.1016/0092-8674(81)90014-3 [DOI] [PubMed] [Google Scholar]
- 49. Moore SA. 1984. Synchronous cell growth occurs upon synchronizing the two regulatory steps of the Saccharomyces cerevisiae cell cycle. Exp. Cell Res. 151:542–556. 10.1016/0014-4827(84)90402-6 [DOI] [PubMed] [Google Scholar]
- 50. Moffat J, Andrews B. 2004. Late-G1 cyclin-CDK activity is essential for control of cell morphogenesis in budding yeast. Nat. Cell Biol. 6:59–66. 10.1038/ncb1078 [DOI] [PubMed] [Google Scholar]
- 51. Lew DJ, Reed SI. 1995. Cell cycle control of morphogenesis in budding yeast. Curr. Opin. Genet. Dev. 5:17–23. 10.1016/S0959-437X(95)90048-9 [DOI] [PubMed] [Google Scholar]
- 52. Lew DJ, Marini NJ, Reed SI. 1992. Different G1 cyclins control the timing of cell cycle commitment in mother and daughter cells of the budding yeast S. cerevisiae. Cell 69:317–327. 10.1016/0092-8674(92)90412-6 [DOI] [PubMed] [Google Scholar]
- 53. Benton BK, Tinkelenberg AH, Jean D, Plump SD, Cross FR. 1993. Genetic analysis of Cln/Cdc28 regulation of cell morphogenesis in budding yeast. EMBO J. 12:5267–5275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Loeb JD, Kerentseva TA, Pan T, Sepulveda-Becerra M, Liu H. 1999. Saccharomyces cerevisiae G1 cyclins are differentially involved in invasive and pseudohyphal growth independent of the filamentation mitogen-activated protein kinase pathway. Genetics 153:1535–1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Chapa y Lazo B, Bates S, Sudbery P. 2005. The G1 cyclin Cln3 regulates morphogenesis in Candida albicans. Eukaryot. Cell 4:90–94. 10.1128/EC.4.1.90-94.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zaragoza O. 2011. Multiple disguises for the same party: the concepts of morphogenesis and phenotypic variations in Cryptococcus neoformans. Front. Microbiol. 2:181. 10.3389/fmicb.2011.00181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Maxson ME, Dadachova E, Casadevall A, Zaragoza O. 2007. Radial mass density, charge, and epitope distribution in the Cryptococcus neoformans capsule. Eukaryot. Cell 6:95–109. 10.1128/EC.00306-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Maxson ME, Cook E, Casadevall A, Zaragoza O. 2007. The volume and hydration of the Cryptococcus neoformans polysaccharide capsule. Fungal Genet. Biol. 44:180–186. 10.1016/j.fgb.2006.07.010 [DOI] [PubMed] [Google Scholar]
- 59. Choi YE, Goodwin SB. 2011. Gene encoding a c-type cyclin in Mycosphaerella graminicola is involved in aerial mycelium formation, filamentous growth, hyphal swelling, melanin biosynthesis, stress response, and pathogenicity. Mol. Plant Microbe Interact. 24:469–477. 10.1094/MPMI-04-10-0090 [DOI] [PubMed] [Google Scholar]
- 60. Shim WB, Woloshuk CP. 2001. Regulation of fumonisin B(1) biosynthesis and conidiation in Fusarium verticillioides by a cyclin-like (C-type) gene, FCC1. Appl. Environ. Microbiol. 67:1607–1612. 10.1128/AEM.67.4.1607-1612.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Muñoz MJ, Bejarano ER, Daga RR, Jimenez J. 1999. The identification of Wos2, a p23 homologue that interacts with Wee1 and Cdc2 in the mitotic control of fission yeasts. Genetics 153:1561–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Burnie JP, Carter TL, Hodgetts SJ, Matthews RC. 2006. Fungal heat-shock proteins in human disease. FEMS Microbiol. Rev. 30:53–88. 10.1111/j.1574-6976.2005.00001.x [DOI] [PubMed] [Google Scholar]
- 63. Zhang S, Hacham M, Panepinto J, Hu G, Shin S, Zhu X, Williamson PR. 2006. The Hsp70 member, Ssa1, acts as a DNA-binding transcriptional co-activator of laccase in Cryptococcus neoformans. Mol. Microbiol. 62:1090–1101. 10.1111/j.1365-2958.2006.05422.x [DOI] [PubMed] [Google Scholar]
- 64. García-Rodas R, Casadevall A, Rodríguez-Tudela JL, Cuenca-Estrella M, Zaragoza O. 2011. Cryptococcus neoformans capsular enlargement and cellular gigantism during Galleria mellonella infection. PLoS One 6:e24485. 10.1371/journal.pone.0024485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Dromer F, Casadevall A, Perfect J, Sorrell T. 2011. Cryptococcus neoformans: latency and disease, p 431–441 In Joseph Heitman TRK, Kwon-Chung KJ, Perfect JR, Casadevall A. (ed), Cryptococcus. From human pathogen to model yeast. ASM Press, Washington, DC [Google Scholar]
- 66. Voelz K, Johnston SA, May RC. 2011. Intracellular replication and exit strategies, p 441–451 In Joseph Heitman TRK, Kwon-Chung KJ, Perferct JR, Casadevall A. (ed), Cryptococcus. From human pathogen to model yeast. ASM, Washington [Google Scholar]
- 67. Mcquiston T, Del Poeta M. 2011. The interaction of Cryptococcus neoformans with host macrophages and neutrofils, p 373–387 In Joseph Heitman TRK, Kwon-Chung KJ, Perfect JR, Casadevall A. (ed), Cryptococcus. From human pathogen to model host. ASM Press, Washington, DC [Google Scholar]
- 68. García-Rodas R, Zaragoza O. 2012. Catch me if you can: phagocytosis and killing avoidance by Cryptococcus neoformans. FEMS Immunol. Med. Microbiol. 64:147–161. 10.1111/j.1574-695X.2011.00871.x [DOI] [PubMed] [Google Scholar]
- 69. Feldmesser M, Tucker S, Casadevall A. 2001. Intracellular parasitism of macrophages by Cryptococcus neoformans. Trends Microbiol. 9:273–278. 10.1016/S0966-842X(01)02035-2 [DOI] [PubMed] [Google Scholar]
- 70. Feldmesser M, Kress Y, Novikoff P, Casadevall A. 2000. Cryptococcus neoformans is a facultative intracellular pathogen in murine pulmonary infection. Infect. Immun. 68:4225–4237. 10.1128/IAI.68.7.4225-4237.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Perfect JR, Lang SD, Durack DT. 1980. Chronic cryptococcal meningitis: a new experimental model in rabbits. Am. J. Pathol. 101:177–194 [PMC free article] [PubMed] [Google Scholar]
- 72. Raschke WC, Baird S, Ralph P, Nakoinz I. 1978. Functional macrophage cell lines transformed by Abelson leukemia virus. Cell 15:261–267. 10.1016/0092-8674(78)90101-0 [DOI] [PubMed] [Google Scholar]
- 73. Mukherjee J, Casadevall A, Scharff MD. 1993. Molecular characterization of the humoral responses to Cryptococcus neoformans infection and glucuronoxylomannan-tetanus toxoid conjugate immunization. J. Exp. Med. 177:1105–1116. 10.1084/jem.177.4.1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Azie N, Neofytos D, Pfaller M, Meier-Kriesche HU, Quan SP, Horn D. 2012. The PATH (Prospective Antifungal Therapy) Alliance registry and invasive fungal infections: update 2012. Diagn. Microbiol. Infect. Dis. 73:293–300. 10.1016/j.diagmicrobio.2012.06.012 [DOI] [PubMed] [Google Scholar]
- 75. Covert SF, Kapoor P, Lee M-H, Briley A, Nairn CJ. 2001. Agrobacterium tumefaciens-mediated transformation of Fusarium circinatum. Mycol. Res. 105:259–264. 10.1017/S0953756201003872 [DOI] [Google Scholar]
- 76. Moyrand F, Fontaine T, Janbon G. 2007. Systematic capsule gene disruption reveals the central role of galactose metabolism on Cryptococcus neoformans virulence. Mol. Microbiol. 64:771–781. 10.1111/j.1365-2958.2007.05695.x [DOI] [PubMed] [Google Scholar]
- 77. Kavanagh K. 2007. Medical mycology. Cellular and molecular techniques. Wiley, West; Sussex, United Kingdom [Google Scholar]
- 78. Zaragoza O, Taborda CP, Casadevall A. 2003. The efficacy of complement-mediated phagocytosis of Cryptococcus neoformans is dependent on the location of C3 in the polysaccharide capsule and involves both direct and indirect C3-mediated interactions. Eur. J. Immunol. 33:1957–1967. 10.1002/eji.200323848 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Real-time imaging of capsule growth. The movie shows time-lapse microscopy images at 5 frames per s of C. neoformans undergoing capsule enlargement followed by yeast budding. Image acquisition was performed at 5-min intervals over a total period of 395 min using a 63× DIC objective. The scale bar represents 5 µm. Download
The movie shows a representative cell of the H99 strain in Sabouraud liquid medium at 30°C in which the replication rate and G1 lapse were calculated. In this case, pictures were taken every 2 min. The interval between when the first bud emerges until that “daughter cell” buds again corresponds to the G1 lapse. In this case, the duration of the G1 phase is 61 min (28 pictures). Download
The movie shows a representative cell of the cln1 mutant strain in Sabouraud liquid medium at 30°C in which the replication rate and G1 lapse were calculated. In this case, the pictures were taken every 3 min. The interval between when the first bud emerges until that “daughter cell” buds again corresponds to the G1 lapse. In this case, the duration of the G1 phase is 98 min (31 pictures). Download