ABSTRACT
Growing evidence shows that gut microbes are key factors involved in the regulation of energy homeostasis, metabolic inflammation, lipid metabolism, and glucose metabolism. Therefore, gut microbiota modulations caused by selectively fermented oligosaccharides or probiotic bacteria constitute an interesting target in the physiopathology of obesity. However, to date, no probiotic yeast has been investigated in this context. Therefore, our study aimed to evaluate the impact of the most-studied probiotic yeast (i.e., Saccharomyces boulardii Biocodex) on obesity and associated metabolic features, such as fat mass development, hepatic steatosis, and low-grade inflammation, in obese mice. S. boulardii was administered daily by oral gavage to leptin-resistant obese and type 2 diabetic mice (db/db) for 4 weeks. We found that S. boulardii-treated mice exhibited reduced body weight, fat mass, hepatic steatosis, and inflammatory tone. Interestingly, these effects of S. boulardii on host metabolism were associated with local effects in the intestine. S. boulardii increased cecum weight and cecum tissue weight but also induced dramatic changes in the gut microbial composition at the phylum, family, and genus levels. These gut microbiota changes in response to S. boulardii may also be correlated with the host metabolism response. In conclusion, this study demonstrates for the first time that S. boulardii may act as a beneficial probiotic treatment in the context of obesity and type 2 diabetes.
IMPORTANCE
To date, no probiotic yeast have been investigated in the context of obesity and type 2 diabetes. Here we found that type 2 diabetic and obese mice (db/db) treated with Saccharomyces boulardii exhibited reduced body weight, fat mass, hepatic steatosis, and inflammatory tone. These effects on host metabolism were associated with local effects in the intestine. Importantly, by using pyrosequencing, we found that S. boulardii treatment induces changes of the gut microbiota composition at the phylum, family, and genus levels. Moreover, we found that gut microbiota changes in response to S. boulardii were correlated with several host metabolism responses.
INTRODUCTION
Growing evidence supports that gut microbiota-host interactions control energy homeostasis, glucose metabolism, and lipid metabolism (1–4). We and others have shown that the gut microbiota influences whole-body metabolism by affecting energy balance (1–3) and metabolic inflammation associated with obesity and related disorders (5, 6). However, the exact roles of specific microorganisms present in the gut remain poorly defined. Among the different strategies available to modify the gut microbiota in the context of obesity and type 2 diabetes, compelling evidence suggests that oral supplementation with selectively fermented oligosaccharides (i.e., prebiotics, arabinoxylans, and resistant starches) improves these metabolic disorders via several mechanisms (7–12). Moreover, the use of probiotic bacteria has also been suggested (3, 13–19). Strikingly, to our knowledge, the role of probiotic yeast in the modulation of obesity and associated related disorders has never been investigated. The most-studied probiotic yeast is Saccharomyces cerevisiae var. boulardii Biocodex (S. boulardii), and this yeast has been widely investigated and used for the prevention of antibiotic-associated diarrhea (20). S. boulardii differs from other strains by several physiological and metabolic characteristics. For instance, the optimum growth temperature of S. boulardii is approximately 37°C, and other strains of S. cerevisiae prefer lower temperatures (between 30 and 33°C) for growth (21). In addition, S. boulardii is resistant to low pH and is highly tolerant to bile acids (22). S. boulardii has been widely characterized, and its beneficial roles have been associated with specific mechanisms, such as specific antitoxin effects, antimicrobial activities, a trophic effect on the gut mucosa, an improved immune response (20), and increased production of butyrate (23), which is a short-chain fatty acid (SCFA) known for its impact on intestinal functions (24).
Although it is well established that S. boulardii improves gut health, the potential roles of S. boulardii in obesity, associated hepatic disorders, and metabolic inflammation are unknown.
Thus, this study had the following aims: (i) to elucidate the impact of S. boulardii on obesity, fat mass development, hepatic steatosis, and low-grade inflammation in leptin-resistant obese and type 2 diabetic mice (db/db) and (ii) to investigate the influence of S. boulardii treatment on the taxonomic profile of the mouse gut microbiota by using high-throughput sequencing analysis.
RESULTS
S. boulardii administration reduces body weight gain and fat mass in obese and type 2 diabetic mice.
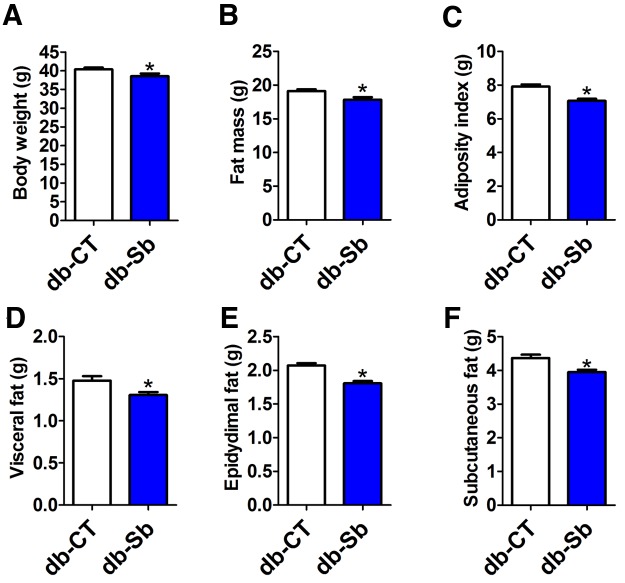
Leptin-resistant obese and type 2 (ic>db/db) diabetic mice develop fatty livers associated with severe obesity and type 2 diabetes (25). Here, we found that after 4 weeks of daily oral gavage with S. boulardii, treated (db-Sb) mice exhibited a modest but significant decrease in body weight (Fig. 1a) compared to vehicle-treated mice (db-CT). The body weight gain was about 15% lower in S. boulardii-treated animals (10.13 ± 0.56 g in db-CT mice versus 8.71 ± 0.98 g in db-Sb mice [means ± standard errors of the means, or SEM]; P > 0.05). This effect was accompanied by a significantly reduced whole-body fat mass (Fig. 1b) and adiposity index (Fig. 1c), which was assessed by weighing the main fat depots (visceral, epididymal, and subcutaneous) (Fig. 1d to f). This effect was not associated with any changes in food intake (cumulative food intake per mouse. 140.97 ± 6.01 g in db-CT mice versus 149.02 ± 3.81 g in db-Sb mice; P > 0.05).
FIG 1 .
S. boulardii administration reduces body weight gain and fat mass in obese and type 2 diabetic mice. Body weight (a), fat mass measured by nuclear magnetic resonance (b), the adiposity index (c), visceral adipose tissue weight (d), epididymal adipose tissue weight (e), and subcutaneous adipose tissue (f) were measured in db/db mice treated with the vehicle (saline; db-CT; n = 15) or S. boulardii (db-Sb; n = 15). Data are means ± SEM. *, P < 0.05 according to Student’s t test.
S. boulardii administration reduces hepatic steatosis in obese and type 2 diabetic mice.
We found that S. boulardii significantly reduced liver weight (Fig. 2a). To identify if this decrease might be attributed to the fat content, total lipids were extracted from the liver. We found that S. boulardii significantly decreased total hepatic lipid content in db-Sb mice compared to db-CT mice (Fig. 2b). These effects were not associated with changes in fasted glycemia (487 ± 22 mg/dl in db-CT versus 489 ± 18 mg/dl in db-Sb; P > 0.05) and fasted insulinemia (8.7 ± 0.9 µg/liter in db-CT versus 7.5 ± 0.8 µg/liter in db-Sb; P > 0.05).
FIG 2 .
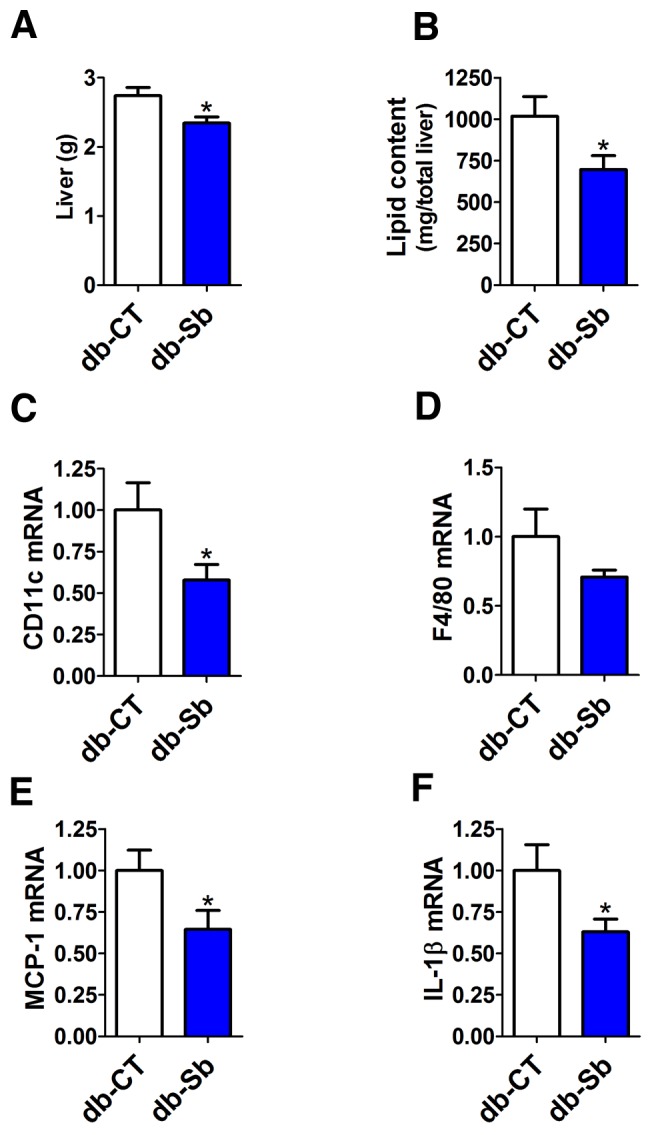
S. boulardii administration reduces liver weight, hepatic steatosis, and hepatic inflammatory markers in obese and type 2 diabetic mice. Liver weight (a), total lipid liver content (b), and inflammatory marker mRNA expression of CD11c (c), F4/80 (d), MCP-1 (e), and IL-1β (f) measured in db/db mice treated with the vehicle (saline; db-CT; n = 15) or S. boulardii (db-Sb; n = 15). mRNA data are expressed relative to results in db-CT mice. Data are means ± SEM. *, P < 0.05 according to Student’s t test.
S. boulardii administration decreases hepatic and systemic inflammation.
Evidence suggests that obesity is associated with the development of inflammatory liver diseases, such as nonalcoholic fatty liver disease and nonalcoholic steatohepatitis (26, 27). We have also previously demonstrated that the gut microbiota contributes to the development of hepatic steatosis and inflammation (7, 28, 29). In the present study, we found that the decreased hepatic steatosis observed in db-Sb mice was associated with a marked decrease in liver macrophage infiltration markers, as shown by the 50% decrease in cluster of differentiation 11c (CD11c) and F4/80 mRNA levels as well as the reduced expression (to approximately 40%) of monocyte chemoattractant protein 1 (MCP-1) mRNA (Fig. 2c to e). In accordance with the lower expression of macrophage infiltration markers, we found that S. boulardii treatment reduced liver interleukin-1β (IL-1β) mRNA levels by approximately 37% compared to vehicle-treated mice (Fig. 2f). In addition to the reduced hepatic inflammation, we found that systemic markers of inflammation were reduced following S. boulardii treatment. Plasma cytokine concentrations of IL-6 and IL-4 were significantly reduced by approximately 2-fold (Fig. 3a and c) in db-Sb mice compared to db-CT mice. IL-1β was reduced by approximately 40% (P = 0.06), and tumor necrosis factor alpha (TNF-α) was reduced by approximately 20% (P = 0.12) (Fig. 3b and d).
FIG 3 .
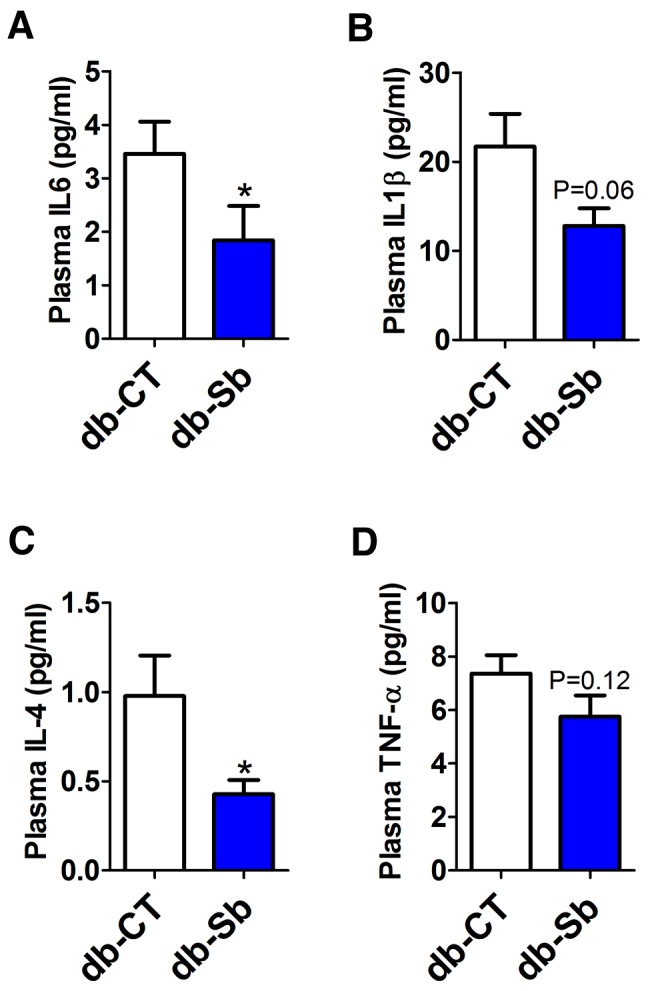
S. boulardii administration reduces plasma cytokines in obese and type 2 diabetic mice. Plasma cytokine concentrations of IL-6 (a), IL-1β (b), IL-4 (c), and TNF-α (d) measured in the plasma of db/db mice treated with the vehicle (saline; db-CT; n = 15) or S. boulardii (db-Sb; n = 15). Data are means ± SEM. *, P < 0.05 according to Student’s t test.
S. boulardii significantly increases cecum weight.
The gut mucosa is subjected to a constant and rapid cellular turnover (30, 31), and S. boulardii has been shown to exert a trophic effect on gut mucosa (32, 33). Here we found that S. boulardii significantly increased cecum weight and cecal tissue weight, thereby suggesting a trophic effect on this tissue (Fig. 4a and b).
FIG 4 .
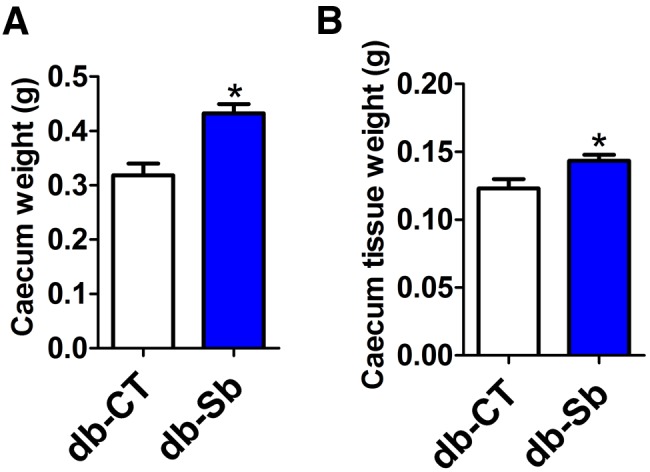
S. boulardii administration increases cecum weight and cecum tissue weight in obese and type 2 diabetic mice. Total cecum weight (a) and cecum tissue weight (b) were measured in db/db mice treated with the vehicle (salinel db-CT; n = 15) or S. boulardii (db-Sb; n = 15). Data are means ± SEM. *, P < 0.05 according to Student’s t test.
S. boulardii profoundly affects the gut microbial community at different taxonomic levels.
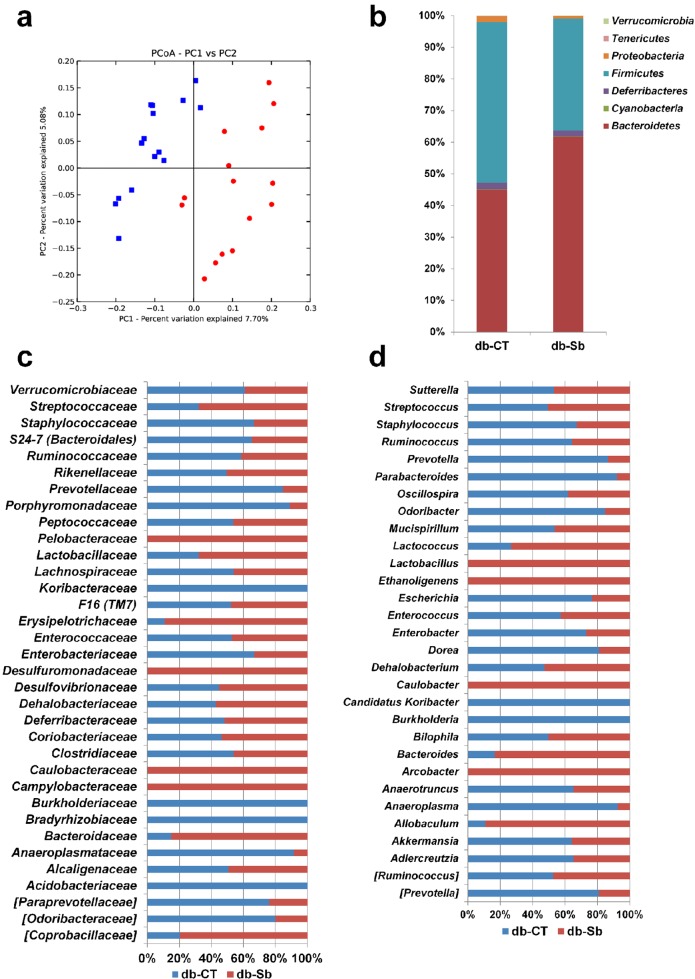
We first quantified the abundance of total yeast cells as well as total Saccharomyces cells. The number of yeast cells reached 6.57 ± 0.09 log10 cells/g of cecal content in db-CT mice and 8.21 ± 0.17 log10 cells/g of cecal in db-Sb mice; P = 1 × 10−15). The abundance of total Saccharomyces was 5.85 ± 0.09 log10 cells/g of cecal content in db-CT mice and 8.08 ± 0.22 log10 cells/g of cecal content in db-Sb mice (P = 5.9 × 10−17), thereby showing that S. boulardii administration increased by about 2 logs the abundance of Saccharomyces yeast cells in the cecal content of mice. Principal coordinates analysis (PCoA) showed that the overall gut microbial community was significantly modified by S. boulardii treatment (Fig. 5a). We previously demonstrated that db/db mice present an altered gut microbiota composition that is characterized by a decrease in the abundance of the phylum Bacteroidetes, an increase of Firmicutes, and a dramatic increase in Proteobacteria compared to lean mice (25). In the present study, we found that S. boulardii treatment profoundly affected the abundance of different phyla. For instance, we found that S. boulardii was associated with a significant increase in Bacteroidetes (by approximately 37%) and a significant decrease in the abundance of Firmicutes (by 30%) compared to db-CT mice (Fig. 5b; see also Table S2 in the supplemental material). Both Proteobacteria and Tenericutes were profoundly affected by the treatment, as we found decreases of 55 and 57%, respectively. These results suggested that S. boulardii changes the gut microbial community by affecting the relative fractional abundance of the four main phyla (Fig. 5b; see also Table S2). At the family level, we found several important modifications of the gut microbiota composition. Among the 34 families identified, 5 of them were significantly changed following S. boulardii treatment, after correction via a false-discovery rate (FDR) test according to the Benjamini-Hochberg procedure (Fig. 5c; see also Table S3 in the supplemental material). The major differences were observed at the level of the dominant families, as follows: the Bacteroidaceae family was increased by 6-fold in db-Sb mice, and Porphyromonadaceae was decreased by 8-fold in db-Sb mice (Fig. 5c; see also Table S3).
FIG 5 .
S. boulardii administration changes the gut microbiota composition at different taxonomic levels. (a) PCoA results for the gut bacterial community, based on the weighted Unifrac analysis of the different OTUs in db-CT (red dots) and db-Sb (blue squares) mice. Phyla (b), families (c), and genera (d) were detected in the cecal contents of db/db mice treated with the vehicle (saline; db-CT; n = 15) or S. boulardii (db-Sb; n = 15). Undetected taxa are not represented in the graphic. In panels c and d, each column is set at 100% to illustrate the proportion of each taxa among the two groups; the presence of only one color indicates that the taxa was present only in this group of mice. The significant changes and raw values of each taxa are shown in the supplemental material.
Among the 30 genera detected, Bacteroides was the most abundant, with a mean abundance of 8.3% across all samples. In the S. boulardii treatment group, this genus was increased by 400%. Conversely, the following genera were decreased in S. boulardii-treated mice: Anaeroplasma (−92%), Anaerotruncus (−47%) Dorea (−77%), Odoribacter (−82%), Oscillospira (−38%), Parabacteroides (−91%), Prevotella (−76%), and Ruminococcus (−44%) (Fig. 5d; see also Table S4 in the supplemental material).
S. boulardii-induced modifications of the gut microbiota correlates with metabolic parameters.
We performed a Spearman correlation analysis corrected by a false-discovery rate test according to the Benjamini-Hochberg procedure in order to evaluate the potential link between significant changes in gut microbiota composition induced by S. boulardii and host metabolism (Fig. 6).
FIG 6 .
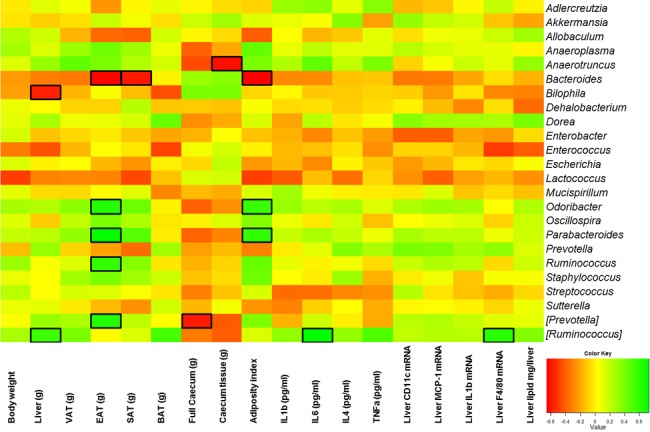
Specific genera are correlated with several host markers. The heat map shows the Spearman r correlations between the bacterial genera detected in the cecal contents of db/db mice treated with the vehicle (saline; db-CT; n = 15) or S. boulardii (db-Sb; n = 15). Square cells depict significant differences in treated and control animals following a Spearman correlation analysis. *, P < 0.05. BAT, brown adipose tissue; EAT, epididymal adipose tissue; SAT, subcutaneous adipose tissue; VAT, visceral adipose tissue weight.
We found that several markers were positively or negatively associated with body weight, fat mass, cecum weight, hepatic steatosis, or inflammatory markers. For instance, we found that the adiposity index and specific adipose tissue weights were significantly associated with several genera. Odoribacter, Parabacteroides, Prevotella, and Ruminococcus were all positively associated with the adiposity index or epididymal adipose tissue (EAT) weight, whereas Bacteroides was inversely associated with fat mass (Fig. 6). We found that cecum weight was negatively correlated with Prevotella, and Anaerotruncus. Among the different genera affected by S. boulardii, we found that Bilophila was the only genus that was negatively correlated with liver tissue weight.
In contrast, Ruminococcus was positively associated with these parameters and with F4/80 mRNA expression (Fig. 6).
DISCUSSION
This study demonstrated that S. boulardii administration in obese and type 2 diabetic mice profoundly modifies host metabolism and is associated with changes in the gut microbial composition. S. boulardii-treated mice exhibited reduced fat mass, hepatic steatosis, and inflammatory tone, thereby suggesting that S. boulardii may also act as a beneficial probiotic treatment in the context of obesity and type 2 diabetes. To our knowledge, this study is the first high-throughput study that has analyzed the effects of this yeast on the gut microbiota as well as the first study that has shown an impact of S. boulardii on metabolic disorders associated with cardiometabolic risk factors, such as fat mass development, steatosis, and inflammation. Nevertheless, numerous studies have already shown a protective effect of S. boulardii in different models associated with inflammation (e.g., inflammatory bowel diseases, colitis, intestinal infections, and hepatic injury) (20, 34–36). We did not find any changes in food intake between groups. This observation suggested that S. boulardii modulates energy homeostasis via a mechanism other than energy intake. Importantly, we found that S. boulardii treatment reduced hepatic and systemic inflammation. Because liver lipid accumulation is associated with liver and systemic inflammation, one may postulate that the decreased inflammatory tone may be related to the lower liver and whole-body fat accumulation. However, the impacts of S. boulardii on both fat mass and body weight were approximately 10%, and the inflammatory markers were reduced by 40 to 50%. Therefore, we suggest that S. boulardii contributed to the reduced inflammation by a putative gut-to-liver axis. Given that this treatment has been previously associated with an improved gut barrier function (20, 32, 34–38), we may not exclude that S. boulardii improved the gut barrier function in this model. Regarding intestinal integrity, we found that S. boulardii treatment increased both cecum and cecal tissue weights, thereby suggesting a trophic effect of the yeast on the intestinal epithelial cells. These results were also consistent with previous studies that showed that S. boulardii exerts a trophic effect on the intestinal epithelium via several molecular mechanisms (33, 37),
We and others previously demonstrated that the gut microbiota contributes to the development of hepatic steatosis, hepatic inflammation, and systemic inflammation, but the impact of S. boulardii on the gut microbiota is poorly defined (7, 28, 39–43). Thus, we decided to determine the impact of S. boulardii on the abundance of total yeast and the genus Saccharomyces but also on the gut microbiota composition by using a high-throughput sequencing method. We found that S. boulardii administration increased by about 160-fold the abundance of total Saccharomyces in the cecal content, whereas the total number of yeast cells was increased by about 40-fold, thereby increasing the relative proportion of Saccharomyces cells per total yeast cells from 18.9% to 73.9%. Thus, this result suggests that the relatively lower increase in total yeast cells observed upon S. boulardii treatment might be explained by a modification of the abundance of other yeasts. However, this hypothesis merits further investigation.
Here, we found that S. boulardii significantly changed the gut microbiota composition with an increased proportion of Bacteroidetes and a decreased amount of the phyla Firmicutes, Proteobacteria, and Tenericutes. These phyla have been previously associated with obesity and type 2 diabetes in mice, with a higher abundance of Firmicutes, Proteobacteria, and Tenericutes as well as a lower abundance of Bacteroidetes (8, 25, 44–46). Moreover, we found that S. boulardii treatment affected several genera that have been previously associated with diabetes and inflammation in db/db mice (i.e., Odoribacter, Ruminococcus, and Prevotella) (25). Thus, we speculate that, in response to S. boulardii, the gut microbiota may contribute to the host metabolism response. However, the relationships that exist between S. boulardii and specific microbes remain unknown. S. boulardii has been shown to modify the production of SCFAs, such as butyrate (23). Evidence suggests that butyrate may contribute to the regulation of several functions at the level of the gut barrier but also to energy homeostasis (47–49) and hepatic steatosis (50). Further investigation is required to understand whether the positive effects observed upon S. boulardii treatment are mediated through butyrate- or SCFA-dependent mechanisms. In the present study, we found that 7 families among the 34 identified were significantly affected by the treatment. At the genera level, 9 of the 30 genera identified were affected by S. boulardii treatment. Importantly, most of them are poorly characterized and could be novel bacteria to study in the future in the context of obesity, because we cannot rule out that these specific changes in genera are involved in the beneficial effects of S. boulardii on host metabolism. Because some of the genera affected by S. boulardii are correlated with metabolic parameters, we postulate that these specific changes in the gut microbiota may contribute to the beneficial effects of S. boulardii on host metabolism. However, whether these genera directly contribute to the phenotype warrants further investigation. For instance, we found a decrease in Prevotella in S. boulardii-treated mice and a positive correlation between Prevotella and adipose tissue weight (EAT). These data were in accordance with data reported elsewhere in the literature, because Zhu et al. found an increase in these bacteria in human obese and nonalcoholic steatohepatitis patients, thereby suggesting a link between the presence of these bacteria and fat mass (51).
Interestingly, our results showed that Bacteroides was dramatically increased by S. boulardii treatment. Moreover, we found an inverse correlation between Bacteroides and the fat mass, suggesting a potential beneficial effect of this bacterium in host physiology. This result was in accordance with our recent study that showed that a prebiotic-enriched diet is associated with an increase in Bacteroides compared to a high-fat diet (52), and this result was also in accordance with a second study that revealed that treatment with alkaloid berberine, a plant that prevents obesity, is associated with an increase in this genus (53). In addition to its probiotic effect and immunomodulatory properties, S. boulardii could act as a prebiotic, which would explain the impressive increase of the Bacteroides genus in db-Sb mice. The cell walls of yeasts are made up of various proportions of β-glucans (54). These polysaccharides are poorly digested by the host due to a lack of specific enzymatic tools necessary for their digestion, but they can be fermented by intestinal bacteria (55). The genus Bacteroides has been recognized for a long time for its ability to metabolize this particular class of polysaccharides and could benefit greatly from an additional presence of this compound in the intestine for growth (56–58). Whether this specific effect on Bacteroides is restricted to the strain S. boulardii requires further investigation.
In conclusion, our results demonstrated that S. boulardii intervention in mice may change the gut microbiota and reduce fat mass, hepatic steatosis, systemic inflammation, and hepatic inflammation in obese and type 2 diabetic mice. We identified a novel potential therapeutic role of S. boulardii treatment that profoundly affects numerous host metabolic parameters. Moreover, this is the first study that has provided a deep analysis of the gut microbiota modulations that occur after S. boulardii supplementation. In addition, we observed putative correlations between genera and several metabolic markers. Thus, our results provide new insights into the complex relationships that exist between S. boulardii yeast and several taxa on metabolism in the context of metabolic inflammation and obesity.
MATERIALS AND METHODS
Mice and treatment.
A set of 6-week-old db/db mice (n = 15/group) (BKS.Cg-Dock7m +/+Lepdb/J; Jackson Laboratory, Bar Harbor, ME) were housed in a controlled environment (12-h daylight cycle; lights off at 6 p.m.) in groups of two or three mice/cage. The mice were fed a control diet (CT; AIN93M; Research Diet, New Brunswick, NJ). Saccharomyces boulardii was provided by Biocodex (France). S. boulardii was suspended in sterile saline and immediately administered by oral gavage (120 mg; db-Sb), and the control group (db-CT) received the same volume of sterile saline solution. The treatment was continued for 4 weeks. Body composition was assessed by using a 7.5-MHz time domain nuclear magnetic resonance (TD-NMR) apparatus (LF50 Minispec; Bruker, Rheinstetten, Germany). The experiment was approved by and performed in accordance with the guidelines of the local ethics committee. Housing conditions were specified by the Belgian Law of 29 May 2013 regarding the protection of laboratory animals (agreement number LA1230314).
Tissue sampling.
The animals were anesthetized with isoflurane (Isoba; Schering-Plough Animal Health, Uxbridge, Middlesex, United Kingdom) before exsanguination and tissue sampling, and the mice were then killed by cervical dislocation. Visceral (corresponding to the mesenteric fat), brown, epididymal, and subcutaneous (corresponding to the inguinal and fat pads located on the lower back) adipose tissues were precisely dissected and weighed. The adiposity index corresponds to the sum of the different adipose tissue weights (visceral, epididymal, subcutaneous, and brown). Liver tissue was weighed, snap-frozen in liquid nitrogen, and stored at −80°C until further analysis. The cecum was weighed, and the cecal content was collected for microbiota analyses, immersed in liquid nitrogen, and stored at −80°C until further analysis. Cecal tissue was washed in cold saline, dried, and weighed.
Plasma cytokine measurement.
Plasma contents of IL-1, IL-6, IL-4, and TNF-α were determined in duplicate by using Bio-Plex Pro cytokine assays kit (Bio-Rad, Nazareth, Belgium) and measured using a Luminex instrument (Bio-Plex; Bio-Rad) following the manufacturer’s instructions.
RNA preparation and real-time qPCR analysis.
Total RNA was prepared from tissues by using TriPure reagent (Roche). Quantification and integrity analysis of total RNA were performed by analyzing 1 µl of each sample in an Agilent 2100 bioanalyzer (RNA 6000 Nano kit). cDNA was prepared by reverse transcription of 1 µg of total RNA by using a reverse transcription system kit (Promega, Leiden, The Netherlands). Real-time PCR was performed with the StepOnePlus real-time PCR system and software (Applied Biosystems, Den Ijssel, The Netherlands) and Mesa Fast quantitative PCR (qPCR; Eurogentec, Seraing, Belgium) for detection according to the manufacturers’ instructions. RPL19 RNA was chosen as the housekeeping gene. All samples were performed in duplicate in a single 96-well reaction plate, and data were analyzed according to the 2−ΔCT method. The identity and purity of the amplified product were assessed by melting curve analysis at the end of amplification. The primer sequences for the targeted mouse genes are presented in Table S1 in the supplemental material.
Liver lipid content.
Total lipids were measured in the liver tissue after an extraction in CHCl3-methanol (MeOH) according to the method of Folch et al. (59), adapted as previously described (60). Briefly, 100 mg of liver tissue was homogenized in 1 ml of phosphate buffer (pH 7.4) by using an Ultra-Turrax instrument (IKA, T10 basic; IMLAB, Boutersem, Belgium) until complete tissue lysis. Lipids were extracted by mixing 125 µl of lysate sample with 1 ml of CHCl3-MeOH (2:1). The chloroform phase was evaporated under nitrogen flux, and the dried residue was weighed to determine the total lipid content.
DNA isolation from mouse cecal samples.
Metagenomic DNA was extracted from the cecal contents by using a QIAamp-DNA stool minikit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions and the adapted procedure previously described (61). The quantity and the quality of the DNA extracted from the samples were checked before sending the samples for sequencing.
Sequencing.
The 16S rRNA gene from the cecal microbiota of the mice was amplified using the universal Eubacterial primers as follows: 27Fmod (5′ AGRGTTTGATCMTGGCTCAG 3′) and 519Rmodbio (5′-GTNTTACNGCGGCKGCTG-3’). Purified amplicons were sequenced utilizing Roche 454 FLX titanium instruments and reagents following the manufacturer’s guidelines.
Sequencing was performed at MR DNA (Shallowater, TX).
The Q25 sequence data derived from the sequencing process were analyzed with the QIIME 1.7 pipeline. In summary, sequences were depleted of bar codes and primers. Sequences of <200 bp or >1,000 bp were then removed, and sequences with ambiguous base calls and with homopolymer runs exceeding 6 bp were also removed. Sequences were denoised, and operational taxonomic units (OTUs) were generated. Moreover, chimeras were removed. OTUs were defined by clustering at 3% divergence (97% similarity). Final OTUs were taxonomically classified using BLASTn against a curated GreenGenes database. PCoA was generated with QIIME using the unweighted UniFrac distance matrix between the samples (62, 63).
Yeast cell quantification.
Yeast cells were quantified using the primers YEASTF (5′ GAGTCGAGTTGTTTGGGAATGC 3′) and YEASTR (5′ TCTCTTTCCAAAGTTCTTTTCATCTTT 3′) following the method described by Hierro et al. (64). Saccharomyces was quantified using primers SC1 (5′ GAAAACTCCACAGTGTGTTG 3′) and SC2 (5′ GCTTAAGTGCGCGGTCTTG 3′) according to the method described by Zott et al. (65). Detection was achieved with the StepOnePlus real-time PCR system and software (Applied Biosystems) and Mesa Fast qPCR (Eurogentec) according to the manufacturer’s instructions. Each assay was performed in duplicate in the same run. The cycle threshold (CT) of each sample was then compared with a standard curve (performed in triplicate) made by diluting genomic DNA (5-fold serial dilution) extracted from a pure culture of S. boulardii (Biocodex). The data are expressed as the log10 of bacteria per g of cecal content.
Statistical analysis.
Data are expressed as means ± SEM unless otherwise indicated. Differences between two groups were assessed using the unpaired two-tailed Student’s t test. Data were analyzed using GraphPad Prism version 5.00 for Windows (GraphPad Software, San Diego, CA). Data related to the gut microbiota were analyzed using JMP 8.0.1 (SAS Institute, Inc., Cary, NC) and R 3.0.2 (The R Foundation) with the RStudio 0.97.310 package and gplots for the heat map. The results were considered statistically significant at P level of <0.05. Correlation results were corrected by an FDR test according to the Benjamini-Hochberg procedure, with an α of <0.05.
SUPPLEMENTAL MATERIAL
Primer sequences
Phyla enriched or depleted following S. boulardii treatment. Data are means ± SEM. Only significant (P < 0.05) differences between the two groups are indicated. P values are based on a 2-sample t test assuming equal variances. *, P values considered nonsignificant when adjusted by an FDR-controlling method.
Families enriched or depleted following S. boulardii treatment. Data are means ± SEM. Only significant (P < 0.05) differences between the two groups are indicated. P values are based on a 2-sample t test assuming equal variances. *, P values considered nonsignificant when adjusted by an FDR-controlling method.
Genera enriched or depleted following S. boulardii treatment. Data are means ± SEM. Only significant (P < 0.05) differences between the two groups are indicated. P values are based on the 2-sample t test assuming equal variances. *, P values considered nonsignificant when adjusted by an FDR-controlling method.
ACKNOWLEDGMENTS
We thank M.-E. Le Guern, E. Fargier, P. Agricole, T. Duparc, N. Salazar, and H. Plovier for helpful discussions and criticisms. We thank B. Es Saadi and R. Selleslagh for excellent technical assistance.
P.D.C. is a research associate at FRS-FNRS (Fonds de la Recherche Scientifique), Belgium. A.E. is a doctoral fellow at FRS-FNRS, Belgium. P.D.C. is the recipient of support from FNRS and FRSM (Fonds de la Recherche Scientifique Médicale, Belgium) and ARC (Action de Recherche Concertée). P.D.C. is a recipient of an ERC Starting Grant 2013 (European Research Council, starting grant 336452-ENIGMO).
This study was in part financially supported by Biocodex (France).
Footnotes
Citation Everard A, Matamoros S, Geurts L, Delzenne NM, Cani PD. 2014. Saccharomyces boulardii administration changes gut microbiota and reduces hepatic steatosis, low-grade inflammation, and fat mass in obese and type 2 diabetic db/db mice. mBio 5(3):e01011-14. doi:10.1128/mBio.01011-14.
REFERENCES
- 1. Claus SP, Ellero SL, Berger B, Krause L, Bruttin A, Molina J, Paris A, Want EJ, de Waziers I, Cloarec O, Richards SE, Wang Y, Dumas ME, Ross A, Rezzi S, Kochhar S, van BP, Lindon JC, Holmes E, Nicholson JK. 2011. Colonization-induced host-gut microbial metabolic interaction. mBio 2:e00271-10 . 10.1128/mBio.00271-10 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cani PD. 2014. Metabolism in 2013: the gut microbiota manages host metabolism. Nat. Rev. Endocrinol. 10:74–76. 10.1038/nrendo.2013.240 [DOI] [PubMed] [Google Scholar]
- 3. Petschow B, Doré J, Hibberd P, Dinan T, Reid G, Blaser M, Cani PD, Degnan FH, Foster J, Gibson G, Hutton J, Klaenhammer TR, Ley R, Nieuwdorp M, Pot B, Relman D, Serazin A, Sanders ME. 2013. Probiotics, prebiotics, and the host microbiome: the science of translation. Ann. N. Y. Acad. Sci. 1306:1–17. 10.1111/nyas.12303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cox LM, Blaser MJ. 2013. Pathways in microbe-induced obesity. Cell. Metab. 17:883–894. 10.1016/j.cmet.2013.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Everard A, Cani PD. 2013. Diabetes, obesity and gut microbiota. Best Pract. Res. Clin. Gastroenterol. 27:73–83. 10.1016/j.bpg.2013.03.007 [DOI] [PubMed] [Google Scholar]
- 6. Tremaroli V, Bäckhed F. 2012. Functional interactions between the gut microbiota and host metabolism. Nature 489:242–249. 10.1038/nature11552 [DOI] [PubMed] [Google Scholar]
- 7. Cani PD, Possemiers S, Van de Wiele T, Guiot Y, Everard A, Rottier O, Geurts L, Naslain D, Neyrinck AM, Lambert DM, Muccioli GG, Delzenne NM. 2009. Changes in gut microbiota control inflammation in obese mice through a mechanism involving GLP-2-driven improvement of gut permeability. Gut 58:1091–1103. 10.1136/gut.2008.165886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Everard A, Lazarevic V, Derrien M, Girard M, Muccioli GM, Neyrinck AM, Possemiers S, Van HA, Francois P, de Vos WM, Delzenne NM, Schrenzel J, Cani PD. 2011. Responses of gut microbiota and glucose and lipid metabolism to prebiotics in genetic obese and diet-induced leptin-resistant mice. Diabetes 60:2775–2786. 10.2337/db11-0227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, Guiot Y, Derrien M, Muccioli GG, Delzenne NM, de Vos WM, Cani PD. 2013. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc. Natl. Acad. Sci. U. S. A. 110:9066–9071. 10.1073/pnas.1219451110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Neyrinck AM, Van Hée VF, Piront N, De Backer F, Toussaint O, Cani PD, Delzenne NM. 2012. Wheat-derived arabinoxylan oligosaccharides with prebiotic effect increase satietogenic gut peptides and reduce metabolic endotoxemia in diet-induced obese mice. Nutr. Diabetes 2:e28. 10.1038/nutd.2011.24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Aziz AA, Kenney LS, Goulet B, Abdel-Aal E-S. 2009. Dietary starch type affects body weight and glycemic control in freely fed but not energy-restricted obese rats. J. Nutr. 139:1881–1889. 10.3945/jn.109.110650 [DOI] [PubMed] [Google Scholar]
- 12. Zhou J, Martin RJ, Tulley RT, Raggio AM, McCutcheon KL, Shen L, Danna SC, Tripathy S, Hegsted M, Keenan MJ. 2008. Dietary resistant starch upregulates total GLP-1 and PYY in a sustained day-long manner through fermentation in rodents. Am. J. Physiol. Endocrinol. Metab. 295:E1160–E1166. 10.1152/ajpendo.90637.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Delzenne NM, Neyrinck AM, Bäckhed F, Cani PD. 2011. Targeting gut microbiota in obesity: effects of prebiotics and probiotics. Nat. Rev. Endocrinol. 7:639–646. 10.1038/nrendo.2011.126 [DOI] [PubMed] [Google Scholar]
- 14. Molinaro F, Paschetta E, Cassader M, Gambino R, Musso G. 2012. Probiotics, prebiotics, energy balance, and obesity: mechanistic insights and therapeutic implications. Gastroenterol. Clin. North Am. 41:843–854. 10.1016/j.gtc.2012.08.009 [DOI] [PubMed] [Google Scholar]
- 15. Aronsson L, Huang Y, Parini P, Korach-André M, Håkansson J, Gustafsson JÅ, Pettersson S, Arulampalam V, Rafter J. 2010. Decreased fat storage by Lactobacillus paracasei is associated with increased levels of angiopoietin-like 4 protein (ANGPTL4). PLoS One 5:e13087. 10.1371/journal.pone.0013087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Arora T, Anastasovska J, Gibson G, Tuohy K, Sharma RK, Bell J, Frost G. 2012. Effect of lactobacillus acidophilus NCDC 13 supplementation on the progression of obesity in diet-induced obese mice. Br. J. Nutr. 108:1–8. 10.1017/S0007114511006957 [DOI] [PubMed] [Google Scholar]
- 17. Fåk F, Bäckhed F. 2012. Lactobacillus reuteri prevents diet-induced obesity, but not atherosclerosis, in a strain dependent fashion in Apoe−/− mice. PLoS One 7:e46837. 10.1371/journal.pone.0046837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chen JJ, Wang R, Li XF, Wang RL. 2011. Bifidobacterium longum supplementation improved high-fat-fed-induced metabolic syndrome and promoted intestinal Reg I gene expression. Exp. Biol. (Maywood) 236:823–831. 10.1258/ebm.2011.0.010339 [DOI] [PubMed] [Google Scholar]
- 19. Gauffin Cano P, Santacruz A, Moya Á, Sanz Y. 2012. Bacteroides uniformis CECT 7771 ameliorates metabolic and immunological dysfunction in mice with high-fat-diet induced obesity. PLoS One 7:e41079. 10.1371/journal.pone.0041079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. McFarland LV. 2010. Systematic review and meta-analysis of Saccharomyces boulardii in adult patients. World J. Gastroenterol. 16:2202–2222. 10.3748/wjg.v16.i18.2202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Graff S, Chaumeil JC, Boy P, Lai-Kuen R, Charrueau C. 2008. Influence of pH conditions on the viability of Saccharomyces boulardii yeast. J. Gen. Appl. Microbiol. 54:221–227. 10.2323/jgam.54.221 [DOI] [PubMed] [Google Scholar]
- 22. Edwards-Ingram L, Gitsham P, Burton N, Warhurst G, Clarke I, Hoyle D, Oliver SG, Stateva L. 2007. Genotypic and physiological characterization of Saccharomyces boulardii, the probiotic strain of Saccharomyces cerevisiae. Appl. Environ. Microbiol. 73:2458–2467. 10.1128/AEM.02201-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schneider SM, Girard-Pipau F, Filippi J, Hebuterne X, Moyse D, Hinojosa GC, Pompei A, Rampal P. 2005. Effects of Saccharomyces boulardii on fecal short-chain fatty acids and microflora in patients on long-term total enteral nutrition. World J. Gastroenterol. 11:6165–6169 http://www.wjgnet.com/1007-9327/11/6165.asp [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Leonel AJ, Alvarez-Leite JI. 2012. Butyrate: implications for intestinal function. Curr. Opin. Clin. Nutr. Metab. Care 15:474–479. 10.1097/MCO.0b013e32835665fa [DOI] [PubMed] [Google Scholar]
- 25. Geurts L, Lazarevic V, Derrien M, Everard A, Van Roye M, Knauf C, Valet P, Girard M, Muccioli GG, Francois P, de Vos WM, Schrenzel J, Delzenne NM, Cani PD. 2011. Altered gut microbiota and endocannabinoid system tone in obese and diabetic leptin-resistant mice: impact on apelin regulation in adipose tissue. Front. Microbiol. 2:149. 10.3389/fmicb.2011.00149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lanthier N, Molendi-Coste O, Cani PD, van Rooijen N, Horsmans Y, Leclercq IA. 2011. Kupffer cell depletion prevents but has no therapeutic effect on metabolic and inflammatory changes induced by a high-fat diet. FASEB J. 25:4301–4311. 10.1096/fj.11-189472 [DOI] [PubMed] [Google Scholar]
- 27. Lanthier N, Molendi-Coste O, Horsmans Y, van Rooijen N, Cani PD, Leclercq IA. 2010. Kupffer cell activation is a causal factor for hepatic insulin resistance. Am. J. Physiol. Gastrointest. Liver Physiol. 298:G107–G116. 10.1152/ajpgi.00391.2009 [DOI] [PubMed] [Google Scholar]
- 28. Cani PD, Amar J, Iglesias MA, Poggi M, Knauf C, Bastelica D, Neyrinck AM, Fava F, Tuohy KM, Chabo C, Waget A, Delmée E, Cousin B, Sulpice T, Chamontin B, Ferrières J, Tanti JF, Gibson GR, Casteilla L, Delzenne NM, Alessi MC, Burcelin R. 2007. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes 56:1761–1772. 10.2337/db06-1491 [DOI] [PubMed] [Google Scholar]
- 29. Cani PD, Bibiloni R, Knauf C, Waget A, Neyrinck AM, Delzenne NM, Burcelin R. 2008. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes 57:1470–1481. 10.2337/db07-1403 [DOI] [PubMed] [Google Scholar]
- 30. Vereecke L, Beyaert R, van Loo G. 2011. Enterocyte death and intestinal barrier maintenance in homeostasis and disease. Trends Mol. Med. 17:584–593. 10.1016/j.molmed.2011.05.011 [DOI] [PubMed] [Google Scholar]
- 31. Cliffe LJ, Humphreys NE, Lane TE, Potten CS, Booth C, Grencis RK. 2005. Accelerated intestinal epithelial cell turnover: a new mechanism of parasite expulsion. Science 308:1463–1465. 10.1126/science.1108661 [DOI] [PubMed] [Google Scholar]
- 32. Jahn HU, Ullrich R, Schneider T, Liehr RM, Schieferdecker HL, Holst H, Zeitz M. 1996. Immunological and trophical effects of Saccharomyces boulardii on the small intestine in healthy human volunteers. Digestion 57:95–104. 10.1159/000201320 [DOI] [PubMed] [Google Scholar]
- 33. Buts JP, De Keyser N, Marandi S, Hermans D, Sokal EM, Chae YH, Lambotte L, Chanteux H, Tulkens PM. 1999. Saccharomyces boulardii upgrades cellular adaptation after proximal enterectomy in rats. Gut 45:89–96. 10.1136/gut.45.1.89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Duman DG, Kumral ZN, Ercan F, Deniz M, Can G, Cağlayan Yeğen B. 2013. Saccharomyces boulardii ameliorates clarithromycin- and methotrexate-induced intestinal and hepatic injury in rats. Br. J. Nutr. 110:493–499. 10.1017/S000711451200517X [DOI] [PubMed] [Google Scholar]
- 35. Martins FS, Vieira AT, Elian SD, Arantes RM, Tiago FC, Sousa LP, Araújo HR, Pimenta PF, Bonjardim CA, Nicoli JR, Teixeira MM. 2013. Inhibition of tissue inflammation and bacterial translocation as one of the protective mechanisms of Saccharomyces boulardii against Salmonella infection in mice. Microbes Infect. 15:270–279. 10.1016/j.micinf.2012.12.007 [DOI] [PubMed] [Google Scholar]
- 36. Justino PF, Melo LF, Nogueira AF, Costa JV, Silva LM, Santos CM, Mendes WO, Costa MR, Franco AX, Lima AA, Ribeiro RA, Souza MH, Soares PM. 2014. Treatment with Saccharomyces boulardii reduces the inflammation and dysfunction of the gastrointestinal tract in 5-fluorouracil-induced intestinal mucositis in mice. Br. J. Nutr. 111:1–11. 10.1017/S0007114513004248 [DOI] [PubMed] [Google Scholar]
- 37. Buts JP, De Keyser N. 2010. Transduction pathways regulating the trophic effects of Saccharomyces boulardii in rat intestinal mucosa. Scand. J. Gastroenterol. 45:175–185. 10.3109/00365520903453141 [DOI] [PubMed] [Google Scholar]
- 38. Fooks LJ, Gibson GR. 2003. Mixed culture fermentation studies on the effects of Synbiotics on the human intestinal pathogens Campylobacter jejuni and Escherichia coli. Anaerobe 9:231–242. 10.1016/S1075-9964(03)00043-X [DOI] [PubMed] [Google Scholar]
- 39. Cani PD, Neyrinck AM, Fava F, Knauf C, Burcelin RG, Tuohy KM, Gibson GR, Delzenne NM. 2007. Selective increases of bifidobacteria in gut microflora improve high-fat-diet-induced diabetes in mice through a mechanism associated with endotoxaemia. Diabetologia 50:2374–2383. 10.1007/s00125-007-0791-0 [DOI] [PubMed] [Google Scholar]
- 40. Bäckhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, Semenkovich CF, Gordon JI. 2004. The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl. Acad. Sci. U. S. A. 101:15718–15723. 10.1073/pnas.0407076101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Axling U, Olsson C, Xu J, Fernandez C, Larsson S, Ström K, Ahrné S, Holm C, Molin G, Berger K. 2012. Green tea powder and Lactobacillus plantarum affect gut microbiota, lipid metabolism and inflammation in high-fat fed C57BL/6J mice. Nutr. Metab. (Lond) 9:105. 10.1186/1743-7075-9-105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Le Roy T, Llopis M, Lepage P, Bruneau A, Rabot S, Bevilacqua C, Martin P, Philippe C, Walker F, Bado A, Perlemuter G, Cassard-Doulcier AM, Gérard P. 2013. Intestinal microbiota determines development of non-alcoholic fatty liver disease in mice. Gut 62:1787–1794. 10.1136/gutjnl-2012-303816 [DOI] [PubMed] [Google Scholar]
- 43. Pachikian BD, Essaghir A, Demoulin JB, Catry E, Neyrinck AM, Dewulf EM, Sohet FM, Portois L, Clerbaux LA, Carpentier YA, Possemiers S, Bommer GT, Cani PD, Delzenne NM. 2013. Prebiotic approach alleviates hepatic steatosis: implication of fatty acid oxidative and cholesterol synthesis pathways. Mol. Nutr. Food Res. 57:347–359. 10.1002/mnfr.201200364 [DOI] [PubMed] [Google Scholar]
- 44. Ley RE, Bäckhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. 2005. Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. U. S. A. 102:11070–11075. 10.1073/pnas.0504978102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. 2006. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444:1027–1031. 10.1038/nature05414 [DOI] [PubMed] [Google Scholar]
- 46. Turnbaugh PJ, Bäckhed F, Fulton L, Gordon JI. 2008. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe 3:213–223. 10.1016/j.chom.2008.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Brahe LK, Astrup A, Larsen LH. 2013. Is butyrate the link between diet, intestinal microbiota and obesity-related metabolic diseases? Obes. Rev. 14:950-959 [DOI] [PubMed] [Google Scholar]
- 48. Puertollano E, Kolida S, Yaqoob P. 2014. Biological significance of short-chain fatty acid metabolism by the intestinal microbiome. Curr. Opin. Clin. Nutr. Metab. Care 17:139–144. 10.1097/MCO.0000000000000025 [DOI] [PubMed] [Google Scholar]
- 49. Vidrine K, Ye J, Martin RJ, McCutcheon KL, Raggio AM, Pelkman C, Durham HA, Zhou J, Senevirathne RN, Williams C, Greenway F, Finley J, Gao Z, Goldsmith F, Keenan MJ. 2014. Resistant starch from high amylose maize (HAM-RS2) and Dietary butyrate reduce abdominal fat by a different apparent mechanism. Obesity (Silver Spring) 22:344–348. 10.1002/oby.20501 [DOI] [PubMed] [Google Scholar]
- 50. Mattace Raso G, Simeoli R, Russo R, Iacono A, Santoro A, Paciello O, Ferrante MC, Canani RB, Calignano A, Meli R. 2013. Effects of sodium butyrate and its synthetic amide derivative on liver inflammation and glucose tolerance in an animal model of steatosis induced by high fat diet. PLoS One 8:e68626. 10.1371/journal.pone.0068626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhu L, Baker SS, Gill C, Liu W, Alkhouri R, Baker RD, Gill SR. 2013. Characterization of gut microbiomes in nonalcoholic steatohepatitis (NASH) patients: a connection between endogenous alcohol and NASH. Hepatology 57:601–609. 10.1002/hep.26093 [DOI] [PubMed] [Google Scholar]
- 52. Everard A, Lazarevic V, Gaia N, Johansson M, Stahlman M, Backhed F, Delzenne NM, Schrenzel J, Francois P, Cani PD. 2014. Microbiome of prebiotic-treated mice reveals novel targets involved in host response during obesity. ISME J., in press. 10.1038/ismej.2014.45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhang X, Zhao Y, Zhang M, Pang X, Xu J, Kang C, Li M, Zhang C, Zhang Z, Zhang Y, Li X, Ning G, Zhao L. 2012. Structural changes of gut microbiota during berberine-mediated prevention of obesity and insulin resistance in high-fat diet-fed rats. PLoS One 7:e42529. 10.1371/journal.pone.0042529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Manners DJ, Masson AJ, Patterson JC. 1973. The structure of a beta-(1 leads to 3)-D-glucan from yeast cell walls. Biochem. J. 135:19-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hughes SA, Shewry PR, Gibson GR, McCleary BV, Rastall RA. 2008. In vitro fermentation of oat and barley derived beta-glucans by human faecal microbiota. FEMS Microbiol. Ecol. 64:482–493. 10.1111/j.1574-6941.2008.00478.x [DOI] [PubMed] [Google Scholar]
- 56. Salyers AA, Palmer JK, Wilkins TD. 1977. Laminarinase (beta-glucanase) activity in Bacteroides from the human colon. Appl. Environ. Microbiol. 33:1118–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Matsushita O, Russell JB, Wilson DB. 1990. Cloning and sequencing of a Bacteroides ruminicola B(1)4 endoglucanase gene. J. Bacteriol. 172:3620–3630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Larsbrink J, Rogers TE, Hemsworth GR, McKee LS, Tauzin AS, Spadiut O, Klinter S, Pudlo NA, Urs K, Koropatkin NM, Creagh AL, Haynes CA, Kelly AG, Cederholm SN, Davies GJ, Martens EC, Brumer H. 2014. A discrete genetic locus confers xyloglucan metabolism in select human gut Bacteroidetes. Nature 506:498–502. 10.1038/nature12907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Folch J, Lees M, Sloane Stanley GH. 1957. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 226:497–509 [PubMed] [Google Scholar]
- 60. Geurts L, Everard A, le Ruyet P, Delzenne NM, Cani PD. 2012. Ripened dairy products differentially affect hepatic lipid content and adipose tissue oxidative stress markers in obese and type 2 diabetic mice. J. Agric. Food Chem. 60:2063–2068. 10.1021/jf204916x [DOI] [PubMed] [Google Scholar]
- 61. Dewulf EM, Cani PD, Claus SP, Fuentes S, Puylaert PG, Neyrinck AM, Bindels LB, de Vos WM, Gibson GR, Thissen JP, Delzenne NM. 2013. Insight into the prebiotic concept: lessons from an exploratory, double blind intervention study with inulin-type fructans in obese women. Gut 62:1112–1121. 10.1136/gutjnl-2012-303304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72:5069–5072. 10.1128/AEM.03006-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, Andersen GL, Knight R, Hugenholtz P. 2012. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 6:610–618. 10.1038/ismej.2011.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hierro N, Esteve-Zarzoso B, Gonzalez A, Mas A, Guillamon JM. 2006. Real-time quantitative PCR (QPCR) and reverse transcription-QPCR for detection and enumeration of total yeasts in wine. Appl. Environ. Microbiol. 72:7148–7155. 10.1128/AEM.00388-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zott K, Claisse O, Lucas P, Coulon J, Lonvaud-Funel A, Masneuf-Pomarede I. 2010. Characterization of the yeast ecosystem in grape must and wine using real-time PCR. Food Microbiol. 27:559–567. 10.1016/j.fm.2010.01.006 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primer sequences
Phyla enriched or depleted following S. boulardii treatment. Data are means ± SEM. Only significant (P < 0.05) differences between the two groups are indicated. P values are based on a 2-sample t test assuming equal variances. *, P values considered nonsignificant when adjusted by an FDR-controlling method.
Families enriched or depleted following S. boulardii treatment. Data are means ± SEM. Only significant (P < 0.05) differences between the two groups are indicated. P values are based on a 2-sample t test assuming equal variances. *, P values considered nonsignificant when adjusted by an FDR-controlling method.
Genera enriched or depleted following S. boulardii treatment. Data are means ± SEM. Only significant (P < 0.05) differences between the two groups are indicated. P values are based on the 2-sample t test assuming equal variances. *, P values considered nonsignificant when adjusted by an FDR-controlling method.