Abstract
Pharmacogenetics is frequently cited as an area for initial focus of the clinical implementation of genomics. Through the PG4KDS protocol, St. Jude Children’s Research Hospital pre-emptively genotypes patients for 230 genes using the Affymetrix Drug Metabolizing Enzymes and Transporters (DMET) Plus array supplemented with a CYP2D6 copy number assay. The PG4KDS protocol provides a rational, stepwise process for implementing gene/drug pairs, organizing data, and obtaining consent from patients and families. Through August 2013, 1559 patients have been enrolled, and 4 gene tests have been released into the electronic health record (EHR) for clinical implementation: TPMT, CYP2D6, SLCO1B1, and CYP2C19. These genes are coupled to 12 high-risk drugs. Of the 1016 patients with genotype test results available, 78% of them had at least one high-risk (i.e., actionable) genotype result placed in their EHR. Each diplotype result released to the EHR is coupled with an interpretive consult that is created in a concise, standardized format. To support-gene based prescribing at the point of care, 55 interruptive clinical decision support (CDS) alerts were developed. Patients are informed of their genotyping result and its relevance to their medication use through a letter. Key elements necessary for our successful implementation have included strong institutional support, a knowledgeable clinical laboratory, a process to manage any incidental findings, a strategy to educate clinicians and patients, a process to return results, and extensive use of informatics, especially CDS. Our approach to pre-emptive clinical pharmacogenetics has proven feasible, clinically useful, and scalable.
Keywords: Pharmacogenetics, pharmacogenomics, electronic health record, clinical decision support, personalized medicine
INTRODUCTION
Pharmacogenetics is frequently cited as an area for initial focus of the clinical implementation of genomics [Meyer, 2004; Veenstra et al., 2010; Green and Guyer 2011; Manolio and Green 2011; Voora and Ginsburg 2011; Wang et al., 2011]. A growing number of clinically actionable pharmacogenetic variants exists, and for at least 10 gene/drug pairs, the Clinical Pharmacogenetics Implementation Consortium (CPIC), a group established by the National Institutes of Health's Pharmacogenomics Research Network (http://www.pgrn.org) and the Pharmacogenomics Knowledge Base (PharmGKB, http://www.pharmgkb.org), has combined genomic and clinical information to establish clinical practice guidelines for gene-based prescribing [Relling and Klein 2011; Swen et al., 2011]. To be immediately available to guide prescribing decisions at any time, pharmacogenetic test results should be available pre-emptively [Altman 2013, Altman et al., 2013; Shuldiner et al., 2013]. The availability of high-quality genotyping arrays focused on pharmacogenes at a reasonable cost facilitates this pre-emptive approach [Altman et al., 2013].
St. Jude Children’s Research Hospital (St. Jude) provides comprehensive inpatient and outpatient care for children with catastrophic diseases, focusing on childhood cancer, infectious diseases, and sickle cell disease. St. Jude provides all home prescriptions, inpatient, and outpatient medications to approximately 4,200 patients each year. Since the early 1980s, the hospital’s pharmaceutical department has operated both the clinical and laboratory aspects of the clinical pharmacokinetics service, which provides therapeutic drug monitoring for a variety of drugs administered to patients with catastrophic illnesses in a clinical research setting. St. Jude has a history of research and discovery in pharmacogenetics and genomics [Evans and Relling, 1999; Evans and McLeod, 2003; Cheok and Evans, 2006; Evans et al., 2013]. Our translational research experience combined with clinical and laboratory expertise in therapeutic drug monitoring have formed the foundation of our efforts to implement pharmacogenetics as standard of care for our patients. Unique aspects of St. Jude have allowed us to overcome many barriers to the implementation of pharmacogenetics and serve as a model for others [Shuldiner et al., 2013].
Clinical pharmacogenetics services at St. Jude initially focused on single gene tests through the clinical pharmacokinetics laboratory. We initially began by providing two pharmacogenetic tests (TPMT and CYP2D6) directed to patients who were highly likely to receive drugs affected by these genes [Relling et al., 2006; Crews, Cross et al., 2011]. In 2011, our efforts expanded substantially through the introduction of the PG4KDS protocol (http://www.stjude.org/pg4kds) to pre-emptively genotype patients for multiple genes. This paper describes the successful pre-emptive implementation of array-based pharmacogenetic testing. Essential elements for success that can be applied to other settings are described below.
DEVELOPMENT OF PG4KDS
The goal of the PG4KDS protocol is to establish processes for using pharmacogenetic tests in the electronic health record (EHR) to pre-emptively guide prescribing. We elected to implement array-based clinical pharmacogenetics in the context of a clinical trial for several reasons. The PG4KDS protocol allows for a rational, stepwise process for implementing each gene, organizing and reporting data, and obtaining consent from patients and families. Because results are selectively entered into the EHR and communicated to patients (or their parents or guardians) using systematic procedures, the consent process focuses on procedures to withhold and share results, including managing incidental findings [Clayton, 2008; Wolf et al., 2008]. During the informed consent process, patients and families must select whether they would like to be contacted when a gene test result is added to their medical record, and patients over the age of 18 choose whether they wish to be informed of incidental findings relating to disease risk. It is important to highlight that a clinical research approach is not essential for implementation of a hospital-wide pre-emptive pharmacogenetics program. Other large scale implementations of pre-emptive pharmacogenetics have been successful in the context of routine clinical care [Pulley et al., 2012].
Institutional support and coordination
St. Jude has a longstanding commitment to rapidly translating new laboratory discoveries into routine patient care across many areas. Genomics, including pharmacogenetics, has been a particular focus for many years. For example, St. Jude’s leadership consistently implemented laboratory discoveries to advance the treatment for children with acute lymphoblastic leukemia (ALL) providing a paradigm that illustrates the stepwise implementation of genomic medicine over many years [Evans et al., 2013]. Further, the implementation of genomics and individualized medicine is specifically noted in the 2010–2015 St. Jude strategic plan, with the objective to be a model center for translating biomedical discoveries into innovative treatment strategies for childhood cancer, sickle cell disease and other catastrophic diseases in children.
Because the institutional environment exists where clinicians embrace the implementation of innovative treatment strategies, we were able to readily obtain collaboration and support from across the organization, including physicians of all primary clinical services (Oncology, Hematology, Infectious Diseases) and our Information Sciences department to develop EHR and clinical decision support (CDS) functions. Several other aspects of the organization supported the development of PG4KDS, including the Family Advisory Council, which provided crucial input on patient and family perceptions of pharmacogenetics and the protocol, and the St. Jude Ethics Committee, which provided feedback and guidance on managing and reporting incidental findings.
In addition to the supportive environment that already existed, some new governance and infrastructure were developed to support PG4KDS. First, a multidisciplinary Pharmacogenetic Oversight Committee (POC) was established in 2011. The committee is chaired by the principal investigator of the PG4KDS study, and it includes the clinical co-investigators of the PG4KDS protocol, physician representatives from all the clinical departments of the institution, a pathologist external to the study, the clinical decision support officer, and an external advisor. The POC provides oversight for the PG4KDS study and determines which gene test results should be placed in the EHR, what constitutes priority (high-risk) diplotypes, which drugs should be linked to genetic test results, and preferred methods of notification. The committee meets at least quarterly, and it is established as a subcommittee of the hospital’s Pharmacy and Therapeutics Committee, which reports to the Medical Executive Committee. Disagreements have not occurred to date, but should they occur in the future the oversight committee is positioned to gain consensus first through the Pharmacy and Therapeutics Committee, and if necessary through the Medical Executive Committee. Second, a clinical pharmacogenetics coordinator role was established, which is filled by a pharmacist with clinical expertise in our patient population and in pharmacogenetics. Finally, we established a clinical pharmacogenetic residency training program accredited by the American Society of Health-System Pharmacists, and the resident is engaged in all facets of PG4KDS.
Genotyping
A crucial first step was to select the right testing platform conducted by a laboratory with the expertise to perform pharmacogenetic testing. Our approach is to genotype DNA using a high-throughput, clinically-oriented genotyping platform. Genotyping for 230 genes that includes 1936 loci relevant to pharmacogenomics is performed by the Medical College of Wisconsin (MCW) in a Clinical Laboratory Improvement Amendments (CLIA)-certified laboratory using the Affymetrix Drug Metabolizing Enzymes and Transporters (DMET) Plus array (Affymetrix, Santa Clara, CA) [Sissung et al., 2010], supplemented with a CYP2D6 copy number determination using a quantitative PCR test. The DMET array itself detects a deletion of CYP2D6 (*5/*5). In order to further characterize the complex structure of this gene, we perform additional qPCR for copy number analysis using the following commercially available probes (Life Technologies Inc): Hs00010001_cn, Hs04502391_cn, and Hs04083572_cn. These probes allow for a more detailed dissection. In particular, we describe heterozygous deletions, duplications as well as the potential presence of a hybrid allele. We demonstrated outstanding concordance between the DMET platform and orthogonal genotyping methods [Fernandez et al., 2012]. Because genotyping technology continues to evolve, PG4KDS allows flexibility to change to other genotyping methodologies over the course of the protocol.
Process to manage and report incidental findings
The management and communication of incidental or secondary findings identified through genetic testing is a challenge for the clinical implementation of genomics [Wolf et al., 2008]. In consultation with the institutional Ethics Committee, we defined an incidental finding in PG4KDS as one where: (a) the finding should carry a substantially higher risk than baseline in order to be considered significant; (b) the knowledge of the incidental finding would in some way be actionable (e.g., more frequent mammography, follow-up blood tests, prophylactic antibiotics, or genetic counseling would be possibly indicated if the finding were known); and (c) the incidental finding contributes information that is above and beyond what would be expected to be found in the course of normal health care (e.g., if an incidental finding related to risk of type II diabetes is of modest odds ratio compared to a well-recognized and easily measured risk factor, such as being overweight, it may not be worthy of bringing to the attention of the patient).
Because our criteria for an incidental finding are well defined and relatively stringent, we do not allow parents of minor children enrolled on PG4KDS the option of refusing this information on their child’s behalf. If a finding is identified relating to the risk of disease, but the intervention to prevent, detect earlier, or treat that disease does not likely need to be taken until the patient reaches 18 years of age, the results are not reported until the patient reaches the age of majority and can give informed consent regarding incidental findings. All patients on the study who reach 18 years of age are contacted to give informed consent as an adult, and at that time, patients can choose whether they want to continue to participate and to have pharmacogenetic results placed in their EHR, and whether they want to be informed of incidental findings related to disease risk. If a patient elects to be informed of incidental findings relating to disease risk, the patient’s attending physician (assisted by a genetic counselor, as appropriate) will discuss the meaning of any incidental result with the patient and review recommended actions. Overall, our approach to incidental findings recognizes that the return of selected incidental findings in coordination with the patient’s primary physician is consistent with the recent guidance for reporting incidental findings in clinical exome and genome sequencing from the American College of Medical Genetics and Genomics (ACMG) [Green, Berg, et al., 2012]. However, the ACMG list of genes to be returned is not relevant to PG4KDS because the DMET array does not test for these genes.
THE PG4KDS PROCESS
Gene/drug pairs are prioritized for migration to the EHR based on a variety of criteria: inclusion in guidelines by CPIC or other professional organizations, FDA labeling recommendations, evidence of reimbursement for genetic testing for that drug’s use, the availability of a stand-alone CLIA-approved test for the individual gene, and the publication of clinical trials linking drug effects to functional pharmacogenetic loci. The St. Jude POC makes final recommendations on the clinical implementation of gene/drug pairs. To systematically introduce each new gene/drug pair, we have created standard operating procedures and use a checklist that tracks the major steps to complete before implementing each gene/drug pair.
Patient Consent and Education
Patient consent for PG4KDS is obtained by a team of research nurses with extensive experience in enrolling patients on pharmacogenetic research studies. To avoid additional venipuncture whenever possible, a blood sample is collected at a convenient time when the patient is having venipuncture performed for another purpose. The patient education process and decisions regarding return of results for each patient is initiated during the patient consent process. In addition to providing consent materials in St. Jude’s standard format and adhering to the organization’s established consent procedures, we collaborated with St. Jude’s Family Advisory Council to produce a video (http://www.stjude.org/pg4kds) that provides further detail about the PG4KDS protocol and pharmacogenetics, including differences between obtaining genetic information to guide safe medication use as compared to information about the risk of developing certain diseases, placing test results in the medical record, the advantages and disadvantages of participation, and a description of the consent process. This video is not a mandatory component of the consent process, but it is a supplement to the consent document and the individual face to face enrollment conversation. The video can be viewed before or after the consent process, and it is also available online for general education purposes to participants and others interested in more information about pharmacogenetic testing.
Genotyping, Result Management, and Incorporation into the EHR
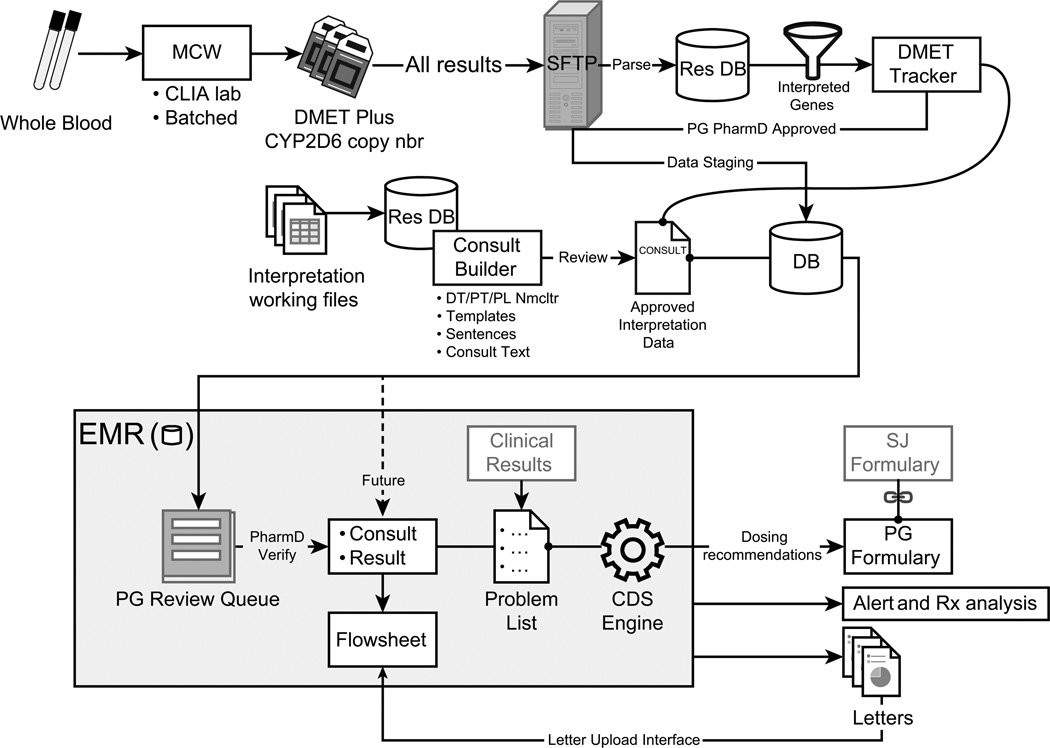
After a patient agrees to enroll in the study, the PG4KDS protocol standardizes the process for genotyping, managing and interpreting results, and incorporating the result in the EHR, including CDS (Fig. 1). After collection, the sample is processed and shipped to the laboratory at MCW. Once a patient’s test results are available, a gene-specific diplotype translation report is sent to St. Jude via Secure File Transfer Protocol. The results are managed and posted to the EHR through the use of two in-house custom web-based applications (DMET Tracker and Consult Builder). Standard functions of St. Jude’s EHR (Millennium system, Cerner Corporation, Kansas City, MO) are used to post results and deploy CDS.
Figure 1. The PG4KDS Data Flow and Result Process.
CDS = clinical decision support; CLIA = Clinical Laboratory Improvement Amendments; DB = database; DMET = Drug Metabolizing Enzymes and Transporters; DT = diplotype; EMR= Electronic Medical Record; MCW = Medical College of Wisconsin; PG = pharmacogenetics; PL Nmcltr = problem list nomenclature; PT = phenotype; Res DB = research database; Rx = prescription; SJ = St. Jude; SFTP = Secure File Transfer Protocol.
Diplotype results are parsed from the translation report files and stored in a research database. Results for selected genes are made available for review through a web-based curation application called DMET Tracker. This application was created to assess the quality of pharmacogenetic test results and control the transfer of results from the research database to the patient’s EHR. Informaticists check incoming genetic test results for consistency with existing results, possible sample handling errors, and result duplication. Pharmacists from the pharmacogenetics service manually review each result and approve or reject them for transfer into a pharmacogenetic review queue in the EHR. As each diplotype result is approved, it is coupled with an interpretive consult that explains the result and clinical significance of the diplotype in a concise and standardized format. From the review queue, a second independent clinical pharmacist inspects the genotype and its interpretive consult before verifying them and making them viewable in the EHR.
Predefined interpretive consults for each result are created using the Consult Builder application. This custom built web-based application provides templates, reusable text and versioning to produce and maintain consults in a consistent way across genes and diplotypes. The text of each consult includes five standard sections with a placeholder for patient-specific comments to be manually added by the verifying pharmacist as needed. These five standard sections include a phenotype assignment section, an interpretation of the diplotype, a dosing recommendation section, an activity score section when appropriate, and a link to the PG4KDS webpage for more information [Hicks et al., 2012]. For example, for CYP2D6 and TPMT, Consult Builder contains a library of over 200 predefined clinical pharmacogenetic consults, which we have further translated into 7 possible CYP2D6 and 4 possible TPMT phenotypes; that are then categorized as “routine” or “priority”.
In order to facilitate finding pharmacogenetic information in the patient’s medical record, a pharmacogenetics tab was created in the EHR. This tab displays all the pharmacogenetic relevant information that has been posted to a patient’s medical record. Up to 999 items can be viewed in this tab irrespective of when the test was performed, which is different from most EHR laboratory flow sheets that are organized by test date. The “lifetime” nature of genomic tests dictates that they be displayed for the lifetime of the patient.
Priority Phenotypes Prompt Additional Action
Additional actions are taken for priority (high-risk) phenotypes which are defined as results that will require either a dosage modification, the use of an alternative agent, or additional patient monitoring. A gene-specific problem list entry is automatically populated for these phenotypes, and serves as the unambiguous trigger that drives point of care alerts at the time of prescribing and dispensing. When a high-risk medication is ordered for a patient with a priority phenotype problem list entry, an alert is presented to the clinician with specific recommendations to guide prescribing such as dose reduction or advice to select an alternative medication. Because current vocabularies widely used in EHRs (e.g., SNOMED) do not adequately differentiate various phenotypes for priority (high-risk) results, we created our own problem list nomenclature (Table I). We have previously described the development and use of active CDS for pharmacogenetics in further detail [Bell et al., 2013]. When a problem list entry is added to the EHR, an automated email containing the patient’s name, medical record number, gene name and phenotype is sent to the patient’s primary attending physician, nurse practitioner, and fellow. Finally, during the result verification process, the pharmacist also performs a thorough medication reconciliation to determine if the patient is currently receiving a medication affected by the priority (high-risk) genotype. If the patient is receiving such a medication, action is taken to modify drug therapy based on the pharmacogenetic information (Table II).
Table I.
Problem List Entries Created to Date.
| Gene | Problem List Entry |
|---|---|
| TPMT | TPMT - Intermediate Activity |
| TPMT - Low or Absent Activity | |
| TPMT - Possible Intermediate Activity | |
| CYP2D6 | CYP2D6 - Possible Ultra-rapid Metabolizer |
| CYP2D6 - Ultra-rapid Metabolizer | |
| CYP2D6 - Intermediate Metabolizer | |
| CYP2D6 - Possible Intermediate Metabolizer | |
| CYP2D6 - Poor Metabolizer | |
| CYP2D6 - Possible Poor Metabolizer | |
| SLCO1B1 | SLCO1B1 - Possible Intermediate Function |
| SLCO1B1 - Intermediate Function | |
| SLCO1B1 - Possible Low Function | |
| SLCO1B1 - Low Function | |
| CYP2C19 | CYP2C19 - Ultra-rapid Metabolizer |
| CYP2C19 - Intermediate Metabolizer | |
| CYP2C19 - Poor Metabolizer | |
| CYP2C19 - Possible Poor Metabolizer | |
Table II.
Summary of Drug Dosing Recommendations Based on the Phenotype for 4 Genes and 12 Drugs Implemented as of August, 2013.
| Drug | Gene | Phenotype | Dosing Recommendation |
|---|---|---|---|
| Codeine | CYP2D6 | Ultra-rapid metabolizer | Change drug |
| Extensive metabolizer | No change | ||
| Intermediate metabolizer | No change | ||
| Poor metabolizer | Change drug | ||
| Tramadol | CYP2D6 | Ultra-rapid metabolizer | Change drug |
| Extensive metabolizer | No change | ||
| Intermediate metabolizer | No change | ||
| Poor metabolizer | Change drug | ||
| Oxycodone | CYP2D6 | Ultra-rapid metabolizer | Change drug |
| Extensive metabolizer | No change | ||
| Intermediate metabolizer | No change | ||
| Poor metabolizer | Change drug | ||
| Amitriptyline | CYP2D6 | Ultra-rapid metabolizer | Change drug |
| Extensive metabolizer | No change | ||
| Intermediate metabolizer | Reduce dose | ||
| Poor metabolizer | Reduce dose/Change drug | ||
| Ondansetron | CYP2D6 | Ultra-rapid metabolizer | Change drug |
| Extensive metabolizer | No change | ||
| Intermediate metabolizer | No change | ||
| Poor metabolizer | No change | ||
| Fluoxetine | CYP2D6 | Ultra-rapid metabolizer | Change drug |
| Extensive metabolizer | No change | ||
| Intermediate metabolizer | No change | ||
| Poor metabolizer | Reduce dose | ||
| Paroxetine | CYP2D6 | Ultra-rapid metabolizer | Change drug |
| Extensive metabolizer | No change | ||
| Intermediate metabolizer | No change | ||
| Poor metabolizer | Reduce dose | ||
| Simvastatin | SLCO1B1 | High function | No change |
| Intermediate function | Reduce dose/change drug | ||
| Low function | Change drug | ||
| Clopidogrel | CYP2C19 | Ultra-rapid metabolizer | No change |
| Extensive metabolizer | No change | ||
| Intermediate metabolizer | Change drug | ||
| Poor metabolizer | Change drug | ||
| Mercaptopurine Thioguanine Azathioprine | TPMT | High (normal) function | No change |
| Intermediate function | Reduce dose | ||
| Low or absent function | Reduce dose/change drug |
For more detailed recommendations consult the corresponding CPIC guideline (http://www.pharmgkb.org/page/cpic) or other information on PharmGKB (http://www.pharmgkb.org).
Patient Specific Communication
Finally, in order to inform patients of their genotype test result and to describe how it pertains to their medication use, letters are sent to the patients who have elected to receive them. The patient letter templates were shared with the Family Advisory Council, and their feedback was incorporated into the final templates. The information and format of each letter is reviewed by the hospital’s patient education committee, and the final format of the letters contains the edits suggested by this committee. The letters are mailed to their home address and include gene-specific information sheets called “Do You Know…” (DYK) sheets. The electronic copies of the letters are posted to the patient’s medical record and the DYK sheets are also available on the St. Jude website under the patient resources section for easy patient access (http://www.stjude.org/dyk-medications). We are also currently developing a pharmacogenetic result section within the patient web portal of the St. Jude EHR.
PATIENT AND CLINICIAN EDUCATION
PG4KDS has prompted us to expand education on pharmacogenetics across St. Jude for patients and clinicians. Pharmacogenetic information now extends throughout all of our patient education materials. For example, in order to standardize patient education for medications, St. Jude’s Patient Education Committee develops drug specific patient medication sheets, which are provided to patients when they pick up their prescriptions from the pharmacy. For the 12 medications that have been implemented as part of PG4KDS, the medication sheets used across the hospital for all patients now include a section explaining that the use of the medication is influenced by genetics (www.stjude.org/med-info).
For clinicians, our education efforts have focused on general education across the clinical staff as well as specific advanced education for pharmacists. With every new gene/drug pair implementation, clinicians are provided educational material through various methods to allow them to learn how to use genetic information when prescribing. When a new gene/drug pair is released to the EHR for the first time, an email is sent to all St. Jude clinicians to inform them of the availability of the results and corresponding CDS. An overview of new gene/drug pairs is published in each issue of the hospital’s quarterly newsletter on drug therapy. The webpage for the PG4KDS protocol is also updated to include information about the new pair. Information on dosing recommendations according to pharmacogenetic status has been added to the St. Jude formulary, with specific dosing recommendations for prescribing medications according to the patient’s gene-specific phenotype. Presentations on pharmacogenetics are routinely scheduled at various conferences within the organization.
Because pharmacists at St. Jude lead the interpretation and use of pharmacogenetic information, we have created a structured education and competency process for all pharmacists tailored to the individual’s job functions. All pharmacists are provided with education material and tested on their ability to perform basic interpretation of a phenotype with corresponding drug-specific dosing recommendations. Clinical pharmacy specialists are also expected to be competent to determine a patient’s expected phenotype when provided with a diplotype result. Pharmacist education on new gene/drug pairs is provided at routine department meetings before formal competencies are assigned. Pharmacogenetic competencies are now part of the initial competencies that pharmacists must complete when they are hired at St. Jude. Retraining to retain competency is performed every three years. The research nurses who enroll patients on the protocol also undergo an extensive education process and pass a competency related to pharmacogenetics, including how to discuss genomic tests with patients and families.
PG4KDS PROGRESS TO DATE
When the protocol opened in May 2011, our efforts focused on two genes: TPMT and CYP2D6. These two well-established pharmacogenes where already implemented to some extent to individualize drug therapy in our patients with ALL [Crews, Cross et al., 2011]. Our prioritization of new gene/drug pair implementation has relied heavily on the availability of CPIC guidelines, whose priorities closely matched those of the POC. We first prioritized implementation of agents that are routinely prescribed at St. Jude, but we have quickly expanded to gene/drug pairs less frequently used in our patient population, including SLCO1B1/simvastatin and CYP2C19/clopidogrel.
Through August 2013, 1559 out of 1605 patients (97%) approached have consented to enroll on the PG4KDS protocol. Most of the patients who declined participation did not provide a reason, but seven patients noted privacy or general concerns about gene-based research. None of the patients who declined participation noted concerns about participation due to incidental findings, including the protocol requirement that any incidental findings will be provided to parents of minor children. The mean age of the enrolled patients was 9.8 years (range 53 days to 52 years); 51% were white (including Hispanics and non-Hispanics), 42% were black, 4% were of mixed race and 3% of unknown race. We have found that 1506 of 1559 patients (97%) have requested an individualized letter be sent with their genotype test result when each gene result is moved into the EHR.
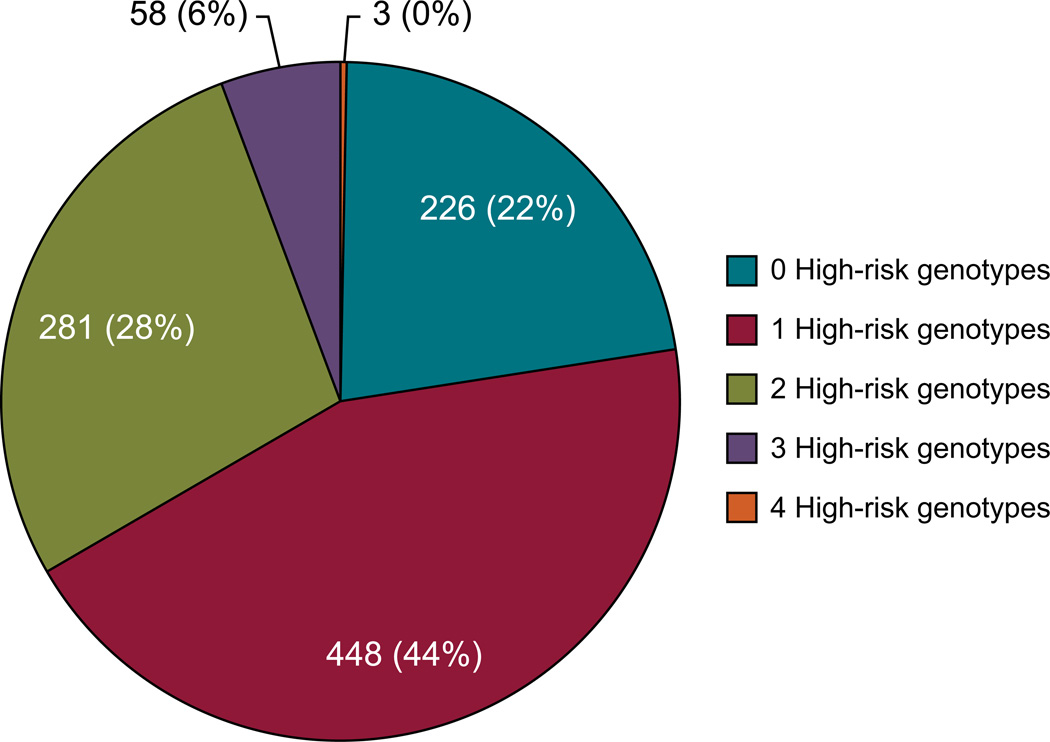
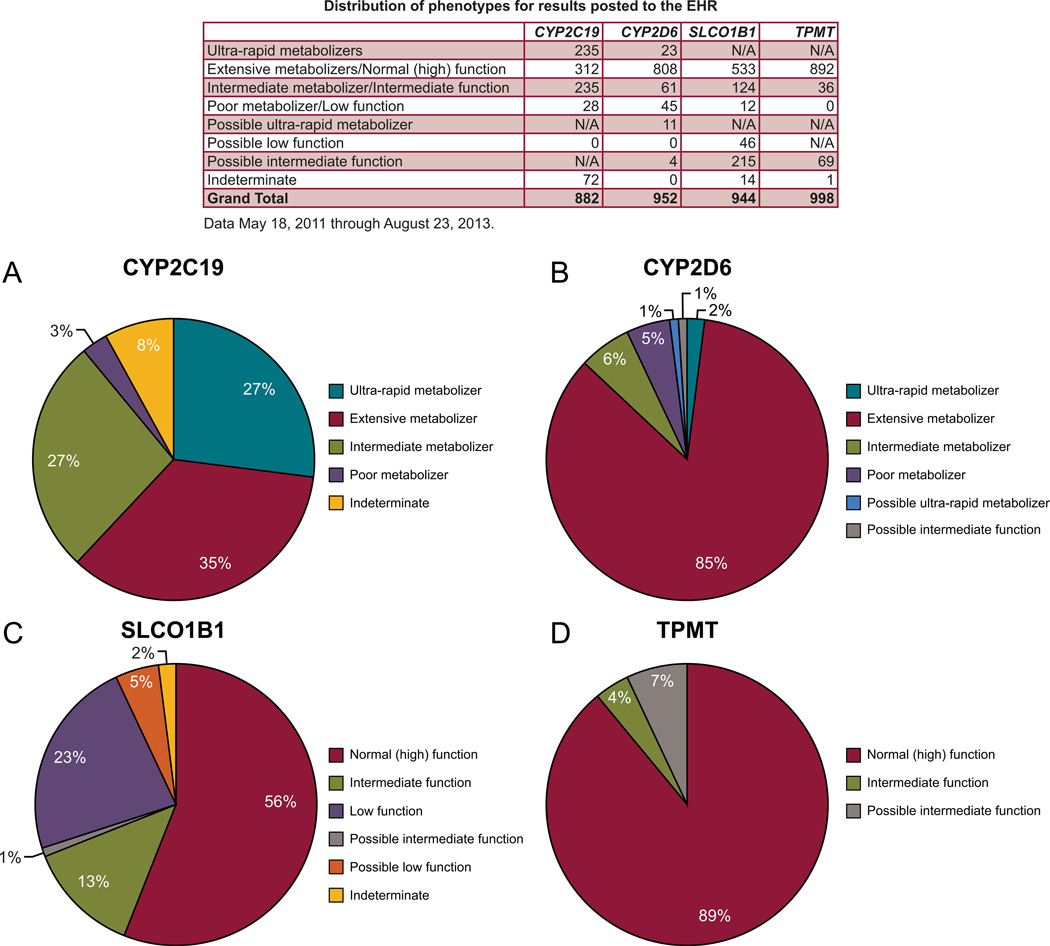
The results of 4 genes have been moved into the medical record: TPMT, CYP2D6, SLCO1B1, and CYP2C19. They are coupled to 12 high-risk drugs (Table II) and 55 clinical decision support rules. The distribution of high-risk phenotypes observed on our protocol, which matches the phenotypic distribution available in the medical literature [Crews, Gaedigk, et al., 2011; Wilke et al., 2012; Relling et al., 2013; Scott et al., 2013]; is outlined in figures 2 and 3. Of the 1016 patients with genotype results available, 78% of patients had at least one high risk (i.e., actionable) genotype result in their EHR. In the past 12 months, the turnaround time (defined by the time the sample is sent to the laboratory to time that results are sent back to St. Jude through the FTP server) ranged from 20 to 137 days. Because this testing is part of a research protocol and done for pre-emptive purposes (i.e. in most cases, there is no need for a faster turnaround time because high-risk drug prescribing is usually not pending the result of a genotype), no attempt has been made to optimize the turnaround time (TAT). We estimate that a practical TAT could be optimized to approximately 14–21 days should a faster TAT need to be achieved.
Figure 2. Patients Grouped by Number of High-risk Genotypes (N=1016).
Of the total number of patients genotyped, 78% have at least one high-risk genotype that warrants making medication therapy adjustments. “Possible” phenotypes are defined as genetic test results that cannot distinguish between two statuses, but are partially informative. In these cases, either the most actionable phenotype or the most likely phenotype is labeled as possible. [Color figure can be viewed in the online issue, which is available at http://onlinelibrary.wiley.com/journal/xxx].
Figure 3. Distribution of Phenotypes on the PG4KDS Protocol.
Percentages for CYP2C19 (A), CYP2D6 (B), SLCO1B1 (C), and TPMT (D) are displayed. The table lists the number of patients with each observed phenotype. “Possible” phenotypes are defined as genetic test results that cannot distinguish between two statuses, but are partially informative. In these cases, either the most actionable phenotype or the most likely phenotype is labeled as possible. For example a patient with a TPMT result of “*1/*3A,*3B/*3C” is labeled as having a possible intermediate TPMT phenotype because it is > 100,000 times more likely than a low or absent function phenotype, and the test cannot distinguish whether the variants identified are on the same allele (*1/*3A, intermediate function phenotype) or on two separate alleles (3B/*3C, low or absent function phenotype). [Color figure can be viewed in the online issue, which is available at http://onlinelibrary.wiley.com/journal/xxx].
FUTURE DIRECTIONS AND CONCLUSION
While each new gene/drug pair added to the EHR comes with its unique challenges, in the near term, we plan to implement gene-based prescribing involving multiple genes (e.g., warfarin with VKORC1 and CYP2C9, amitriptyline with CYP2D6 and CYP2C19), which adds some complexity to our established processes [Johnson et al., 2013]. We plan to implement at least 8 new gene test results into the EHR over the next three years (e.g., DPYD, UGT1A1, G6PD), along with additional drugs, based partly on the output from CPIC over the next several years. In addition, we will continue to update how many interventions are made (e.g. how many prescribing decisions are altered) based on the pre-emptive genotyping approach. A greater challenge may occur if new discoveries prompt a patient’s assigned phenotype to change after we have documented the result in the EHR and provided them with their phenotypic information. By only providing drug dose recommendations in the interruptive CDS alerts, we have established some flexibility to be responsive to changing information through new CDS alerts. Although we expect to manage such updates at our current scale, eventually a single well curated knowledge base for genomic medicine that can be linked to EHRs is needed that will help manage the evolving knowledge.
Through PG4KDS, St. Jude successfully implemented pre-emptive pharmacogenetics in over 1000 patients. Key elements of success include strong and broad-based institutional support, a knowledgeable clinical laboratory, a process to manage return of results and incidental findings, extensive use of informatics, the EHR and CDS, and broad clinician education efforts. All of these functions are needed for the successful clinical implementation of genomics at any institution. Our approach to pre-emptive clinical pharmacogenetics has proven feasible, clinically useful, and scalable.
ACKNOWLEDGEMENTS
The authors wish to thank the participating clinical and laboratory staff, patients, and their parents and especially our Pharmaceutical Sciences and Infectious Disease Department research nurses: Sheri Ring, Lisa Walters, Terri Kuehner, Paula Condy, Margaret Edwards, Melinda Wood, Shannon Gibbs, Pamela Finnie and Lennie Lott.
Funding
Supported by NCI grants CA 36401, CA 21765, NIH/NIGMS Pharmacogenomics Research Network (U01 GM92666, UO1 HL105918), and by the American Lebanese Syrian Associated Charities (ALSAC).
Biographies
James M. Hoffman, Pharm.D., MS, BCPS is an Associate Member in Pharmaceutical Sciences and the Medication Outcomes & Safety Officer at St. Jude Children’s Research Hospital. He is also an associate professor at the University Of Tennessee College Of Pharmacy. He earned both his undergraduate and doctoral degree from the Philadelphia College of Pharmacy at the University of the Sciences in Philadelphia. He completed a MS degree at the University of Wisconsin-Madison, and training at the University of Wisconsin Hospital and Clinics. His primary interests include medication safety, clinical decision support, and the clinical implementation of pharmacogenetics.
Cyrine E. Haidar, Pharm.D., BCPS, BCOP is the Clinical Pharmacogenetics Coordinator at St. Jude Children’s Research Hospital. She is also an assistant professor at the University Of Tennessee College Of Pharmacy. Dr. Haidar earned both her B.S. in pharmacy and her Pharm.D. degrees from the Lebanese American University. She completed a pharmacy practice residency at Hackensack University Medical Center and a pediatric oncology specialty residency at St. Jude Children’s Research Hospital. Her research interests include the clinical implementation of pharmacogenetics.
Mark R. Wilkinson, BS, is a senior research database analyst in the Department of Pharmaceutical Sciences at St. Jude Children’s Research Hospital. He serves as the lead analyst for software application and database development, data analysis, and data quality control for departmental initiatives.
Kristine R. Crews, Pharm.D., BCPS is Translational Research Laboratory Director in the Pharmaceutical Sciences department at St. Jude Children’s Research Hospital and is Director of the first ASHP-Accredited PGY2 Residency in Clinical Pharmacogenetics. Dr. Crews earned both her B.S. in pharmacy and her Pharm.D. degrees from Rutgers University. She completed a pharmacy practice residency and a clinical pharmacokinetics specialty residency at the University Of Kentucky Chandler Medical Center and completed a fellowship in clinical pharmacokinetics and pharmacodynamics at the University of North Carolina and Glaxo Wellcome, Inc. Her research interests include the clinical implementation of pharmacogenetics to individualize treatment regimens for children with cancer.
Donald K. Baker, Pharm.D., MBA is the Clinical Decision Support Officer in the Information Sciences Department at St. Jude Children’s Research Hospital. He is responsible for the development and optimization of all the custom and vendor supplied clinical decision support in the St. Jude electronic health record. He earned his PharmD at the University of Tennessee and an MBA at the University of Memphis.
Nancy Kornegay, MBA, is a senior database administrator at St. Jude Children’s Research Hospital. She is responsible for the archival storage and management of all sampling, drug dosing, clinical and research pharmacokinetic and laboratory data for the Pharmaceutical Sciences Department.
Wenjian Yang, Ph.D., is a bioinformatics analyst and developer at St. Jude Children’s Research Hospital. He obtained PhD in statistics at University of Memphis. His research interests include discovering genetic predictors in treatment and biology of leukemia.
Ching-Hon Pui, M.D., is the Chair of the Department of Oncology at St. Jude Children’s Research Hospital, the co-leader of the Hematological Malignancies Program at St. Jude Cancer Center, and an American Cancer Society Professor. His primary research interest is in the biologic study and treatment of childhood leukemias. He translates laboratory discoveries to the development of "Total Therapies" for children with leukemia.
Ulrike M. Reiss, M.D., is the Director of the division of Clinical Hematology and Director of the Hemophilia Treatment Center at St. Jude Children’s Research Hospital. She is an associate faculty member at St. Jude. She trained at the University of Heidelberg and the Children’s Hospital in Oakland, California. Her clinical research focuses on bleeding disorders, thrombosis and bone marrow failure syndromes.
Aditya H. Gaur, MD., is an associate faculty member in the Department of Infectious Diseases at St. Jude Children’s Research Hospital in Memphis, TN. He obtained his pediatric training at the South Gujarat University in Surat, Gujarat, India and pediatric infectious disease training at St. Jude Children’s Research Hospital. His primary clinical interests are in Pediatric HIV infection, diagnosis and treatment of infections in immunocompromised children as well as the epidemiology of infectious diseases.
Scott C. Howard, MD, MS, a full member in the Leukemia/Lymphoma division at St. Jude Children’s Research Hospital. He is the director of Outcome Evaluation and the Morocco Programs for St. Jude’s International Outreach Program, and a professor at the University of Tennessee College of Medicine. His primary interests are in translating state-of-the-art pediatric cancer treatments to countries with limited resources, and treatment and research for children with Hodgkin lymphoma and acute lymphoblastic leukemia.
William E. Evans, Pharm.D., is the director and chief executive officer at St. Jude Children’s Research Hospital, he holds the Donald Pinkel Chair of Childhood Cancer Treatment at St. Jude and is a Professor of Pediatrics and Pharmacy at the University of Tennessee Colleges of Medicine and Pharmacy. He trained at the University of Tennessee. His primary interests are focused on the pharmacogenomics of anticancer agents in children with an emphasis on childhood acute lymphoblastic leukemia.
Ulrich Broeckel, MD, is Professor of Pediatrics, Medicine and Physiology and the Section Chief of Genomic Pediatrics at the Medical College of Wisconsin and the Children’s Research Institute. He graduated from the University of Heidelberg, and trained at the University of Regensburg Germany and the Medical College of Wisconsin where he completed his post-doctoral fellowship in physiology and genetics. His primary research interests are genetics of complex diseases and the application of genetics and genomics in clinical care.
Mary V. Relling, Pharm.D., is the chair of the Department of Pharmaceutical Sciences at St. Jude Children’s Research Hospital. She is one of the principal investigators within NIH’s Pharmacogenomics Research Network and co-founder of CPIC, the Clinical Pharmacogenetics Implementation Consortium. She is also a professor at the University of Tennessee in the Colleges of Medicine and Pharmacy. She earned her undergraduate B.S. degree from the University Of Arizona College Of Pharmacy and her doctoral degree from the University Of Utah College Of Pharmacy. Her primary interests are in treatment of childhood leukemia and clinical implementation of pharmacogenetics.
Footnotes
Conflict of Interest: Dr. Mary V. Relling and Dr. William E. Evans receive royalties from licensing TPMT genotyping, Prometheus Labs.
REFERENCES
- 1.Altman R. Personal genomic measurements: the opportunity for information integration. Clin Pharmacol Ther. 2013;93:21–23. doi: 10.1038/clpt.2012.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altman RB, Whirl-Carrillo M, Klein TE. Challenges in the pharmacogenomic annotation of whole genomes. Clin Pharmacol Ther. 2013;94:211–213. doi: 10.1038/clpt.2013.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bell GC, Crews KR, Wilkinson MR, Haidar CE, Hicks JK, Baker DK, Kornegay NM, Yang W, Cross SJ, Howard SC, Freimuth RR, Evans WE, Broeckel U, Relling MV, Hoffman JM. Development and use of active clinical decision support for preemptive pharmacogenomics. J Am Med Inform Assoc. 2013 doi: 10.1136/amiajnl-2013-001993. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheok MH, Evans WE. Acute lymphoblastic leukaemia: a model for the pharmacogenomics of cancer therapy. Nat Rev Cancer. 2006;6:117–129. doi: 10.1038/nrc1800. [DOI] [PubMed] [Google Scholar]
- 5.Clayton EW. Incidental findings in genetics research using archived DNA. J Law Med Ethics. 2008;36:286–291. doi: 10.1111/j.1748-720X.2008.00271.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crews KR, Gaedigk A, Dunnenberger HM, Klein TE, Shen DD, Callaghan JT, Kharasch ED, Skaar TC. Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines for codeine therapy in the context of cytochrome P450 2D6 (CYP2D6) genotype. Clin Pharmacol Ther. 2011;91:321–326. doi: 10.1038/clpt.2011.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crews KR, Cross SC, McCormick JN, Baker DB, Molinelli AR, Mullins R, Relling MV, Hoffman JM. Development and implementation of a pharmacist-managed clinical pharmacogenetics service. Am J Health Syst Pharm. 2011;68:143–150. doi: 10.2146/ajhp100113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Evans WE, Relling MV. Pharmacogenomics: translating functional genomics into rational therapeutics. Science. 1999;286:487–491. doi: 10.1126/science.286.5439.487. [DOI] [PubMed] [Google Scholar]
- 9.Evans WE, McLeod HL. Pharmacogenomics-drug disposition, drug targets, and side effects. N Engl J Med. 2003;348:538–549. doi: 10.1056/NEJMra020526. [DOI] [PubMed] [Google Scholar]
- 10.Evans WE, Crews KR, Pui CH. A health-care system perspective on implementing genomic medicine: pediatric acute lymphoblastic leukemia as a paradigm. Clin Pharmacol Ther. 2013;94:224–229. doi: 10.1038/clpt.2013.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fernandez CA, Smith C, Yang W, Lorier R, Crews KR, Kornegay N, Hicks JK, Stewart CF, Kawedia DJ, Ramsey LB, Liu C, Evans WE, Relling MV, Broeckel U. Concordance of DMET Plus genotyping results with those of orthogonal genotyping methods. Clin Pharmacol Ther. 2012;92:360–365. doi: 10.1038/clpt.2012.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Green ED, Guyer MS. Charting a course for genomic medicine from base pairs to bedside. Nature. 2011;470:204–213. doi: 10.1038/nature09764. [DOI] [PubMed] [Google Scholar]
- 13.Green RC, Berg JS, Grody WW, Kalia SS, Korf BR, Martin CL, McGuire A, Nussbaum RL, O’Daniel JM, Ormond KE, Rehm HL, Watson MS, Williams MS, Biesecker LG. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med. 2013;15:565–574. doi: 10.1038/gim.2013.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hicks JK, Crews KR, Hoffman JM, Kornegay NM, Wilkinson MR, Lorier R, Stoddard A, Yang W, Smith C, Fernandez CA, Cross SJ, Haidar C, Baker DK, Howard SC, Evans WE, Broeckel U, Relling MV. A clinician-driven automated system for integration of pharmacogenetic interpretations into an electronic medical record. Clin Pharmacol Ther. 2012;92:563–566. doi: 10.1038/clpt.2012.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson JA, Klein TE, Relling MV. Clinical implementation of pharmacogenetics: more than one gene at a time. Clin Pharmacol Ther. 2013;93:384–385. doi: 10.1038/clpt.2013.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Manolio TA, Green ED. Genomics reaches the clinic: from basic discoveries to clinical impact. Cell. 2011;147:14–16. doi: 10.1016/j.cell.2011.09.012. [DOI] [PubMed] [Google Scholar]
- 17.Meyer UA. Pharmacogenetics-decades of therapeutic lessons from genetic diversity. Nat Rev Genet. 2004;5:669–676. doi: 10.1038/nrg1428. [DOI] [PubMed] [Google Scholar]
- 18.Pulley JM, Denny JC, Peterson JF, Bernard GR, Vnencak-Jones CL, Ramirez AH, Delaney JT, Bowton E, Brothers K, Johnson K, Crawford DC, Schildcrout J, Masys DR, Dilks HH, Wilke RA, Clayton EW, Shultz E, Laposata M, McPherson J, Hirjis JN, Roden DM. Operational implementation of prospective genotyping for personalized medicine: the design of the Vanderbilt PREDICT project. Clin Pharmacol Ther. 2012;92:97–95. doi: 10.1038/clpt.2011.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Relling MV, Pui CH, Cheng C, Evans WE. Thiopurine methyltransferase in acute lymphoblastic leukemia. Blood. 2006;107:843–844. doi: 10.1182/blood-2005-08-3379. [DOI] [PubMed] [Google Scholar]
- 20.Relling MV, Klein TE. CPIC: Clinical Pharmacogenetics Implementation Consortium of the pharmacogenomics research network. Clin Pharmacol Ther. 2011;89:464–467. doi: 10.1038/clpt.2010.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Relling MV, Gardner EE, Sandborn WJ, Schmiegelow K, Pui CH, Yee SW, Stein CM, Carrillo M, Evans WE, Hicks JK, Schwab M, Klein TE. Clinical pharmacogenetics implementation consortium guidelines for thiopurine methyltransferase genotype and thiopurine dosing: 2013 update. Clin Pharmacol Ther. 2013;93:324–325. doi: 10.1038/clpt.2013.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scott SA, Sangkuhl K, Stein CM, Hulot JS, Mega JL, Roden DM, Klein TE, Sabatine MS, Johnson JA, Shuldiner AR. Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines for cytochrome P450-2C19 (CYP2C19) genotype and clopidogrel therapy: 2013 Update. Clin Pharmacol Ther. 2013;94:317–323. doi: 10.1038/clpt.2013.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shuldiner AR, Relling MV, Peterson JF, Hicks K, Freimuth RR, Sadee, Pereira NL, Roden DM, Johnson JA, Klein TE. The Pharmacogenomics Research Network translational pharmacogenetics program: Overcoming challenges of real-world implementation. Clin Pharmacol Ther. 2013;94:207–210. doi: 10.1038/clpt.2013.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sissung TM, English BC, Venzon D, Figg WD, Deeken JF. Clinical pharmacology and pharmacogenetics in a genomics era: the DMET platform. Pharmacogenomics. 2010;11:89–103. doi: 10.2217/pgs.09.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Swen JM, Nijenhuis M, de Boer A, Grandia L, Maitland-van der Zee AH, Mulder H, Rongen GA, van Schaik RH, Schalekamp T, Touw DJ, van der Weide, Wilffert B, Deneer VH, Guchelaar HJ. Pharmacogenetics: from bench to byte-an update of guidelines. Clin Pharmacol Ther. 2011;89:662–673. doi: 10.1038/clpt.2011.34. [DOI] [PubMed] [Google Scholar]
- 26.Veenstra DL, Roth JA, Garrison LP, Ramsey SD, Burke W. A formal risk-benefit framework for genomic tests: facilitating the appropriate translation of genomics into clinical practice. Genet Med. 2010;12:686–693. doi: 10.1097/GIM.0b013e3181eff533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Voora D, Ginsburg GS. Institutional Profile: A hub for bench-to-bedside pharmacogenomic-based research. Pharmacogenomics. 2011;12:1095–1098. doi: 10.2217/pgs.11.62. [DOI] [PubMed] [Google Scholar]
- 28.Wang L, McLeod HL, Weinshilboum RM. Genomics and drug response. N Engl J Med. 2011;364:1144–1153. doi: 10.1056/NEJMra1010600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilke RA, Ramsey LB, Johnson SG, Maxwell WD, McLeod HL, Voora D, Krauss RM, Roden DM, Feng Q, Cooper-Dehoff RM, Gong L, Klein TE, Wadelius M, Niemi M. The clinical pharmacogenomics implementation consortium: CPIC guideline for SLCO1B1 and simvastatin-induced myopathy. Clin Pharmacol Ther. 2012;92:112–117. doi: 10.1038/clpt.2012.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wolf SM, Lawrenz FP, Nelson CA, Kahn JP, Cho MK, Clayton EW, Fletcher JG, Georgieff MK, Hammerschmidt D, Hudson K, Illes J, Kapur V, Keane MA, Koenig BA, Leroy BS, McFarland EG, Paradise J, Parker LS, Terry SF, Van Ness B, Wilfond BS. Managing incidental findings in human subjects research: analysis and recommendations. J Law Med Ethics. 2008;36:219–248. doi: 10.1111/j.1748-720X.2008.00266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]