Abstract
Although CDKN2A is the most frequent high-risk melanoma susceptibility gene, the underlying genetic factors for most melanoma-prone families remain unknown. Using whole exome sequencing, we identified a rare variant that arose as a founder mutation in the telomere shelterin POT1 gene (g.7:124493086 C>T, Ser270Asn) in five unrelated melanoma-prone families from Romagna, Italy. Carriers of this variant had increased telomere length and elevated fragile telomeres suggesting that this variant perturbs telomere maintenance. Two additional rare POT1 variants were identified in all cases sequenced in two other Italian families, yielding a frequency of POT1 variants comparable to that of CDKN2A mutations in this population. These variants were not found in public databases or in 2,038 genotyped Italian controls. We also identified two rare recurrent POT1 variants in American and French familial melanoma cases. Our findings suggest that POT1 is a major susceptibility gene for familial melanoma in several populations.
Approximately 10% of cutaneous malignant melanoma (CMM) cases occur in a familial setting1. Established high-penetrance melanoma susceptibility genes are CDKN2A on chromosome 9p21 and CDK4 on 12q14. Germline mutations of the CDKN2A gene have been described in approximately 20-40% of familial melanoma kindreds2-6 whereas mutations of CDK4 are quite rare with only seventeen published families worldwide7,8. Recently, germline mutations in BAP1 (BRCA1-associated protein 1, located on 3p21) were reported to predispose to melanocytic tumors including uveal and cutaneous melanoma9. In addition, a germline mutation in the promoter of the telomerase reverse transcriptase (TERT) gene, which encodes the catalytic subunit of telomerase, demonstrated disease co-segregation in an informative melanoma-prone family10, suggesting that this gene may also be a rare high-penetrance CMM susceptibility gene. Together, these genes account for melanoma susceptibility in a small proportion of melanoma-prone families.
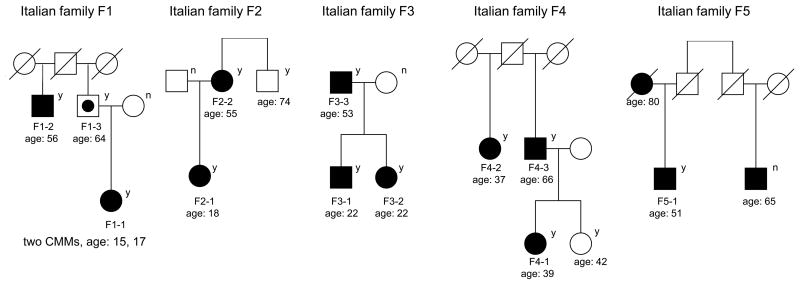
To identify additional high-penetrance susceptibility genes for familial CMM, we performed whole exome sequencing in 101 CMM cases or obligate carriers (assuming dominant inheritance) in 56 unrelated melanoma-prone families without CDKN2A/CDK4 mutations recruited from the Romagna area in Italy. After filtering out common and non-segregating variants (Online Methods), the co-segregating rare variant that was seen at highest frequency in multiple families was a missense rare variant in Protection Of Telomeres 1 (POT1, g.7:124493086 C>T; Ser270Asn, NP_056265); this variant was observed in all CMM cases/obligate carriers (N=11) from four Italian families (F1-4, Fig. 1, Supplementary Table 1). This variant was also seen in one of two cases examined in a bilineal Italian family (F5, Fig. 1). The affected individual without the POT1 variant may have inherited the melanoma risk allele from the other side of the family, but the parental origin of the variant could not be determined. Targeted sequencing performed on all available subjects in the five families with Ser270Asn confirmed the variant in CMM cases. The mode of transmission is consistent with an autosomal dominant inheritance with incomplete penetrance. We genotyped the variant in 2,038 Italian controls and none carried the variant, resulting in a significantly different frequency of this variant in familial CMM cases compared to controls (P=1.2×10-11 after accounting for the relatedness among familial cases using a chromosome-based exact test, Online Methods). We further genotyped this variant in 1,824 Italian CMM cases (Online Methods), and this variant was only found in a single sporadic melanoma case from Romagna, Italy (Supplementary Table 1). This variant was not seen in dbSNP, The 1000 Genomes Project (1,092 subjects), NHLBI GO Exome Sequencing Project (ESP, including exomes from up to 4,300 subjects of European ancestry and 2,200 subjects of African American ancestry), NCI DCEG Cancer Sequencing Working Group database (290 and 200 individuals of European and Asian ancestry, respectively from high-risk families of non-melanoma malignancies), an in-house McGill exome database (1,307 individuals of European ancestry with no cancers, unpublished data), and 878 CMM cases and 1,451 controls from Spain (Online Methods).
Figure 1. Pedigrees of cutaneous malignant melanoma (CMM)-prone families with the Ser270Asn variant (g.7:124493086 C>T) in POT1.
Solid squares and circles: CMM cases; Circle with a dot in the center: obligate gene carrier; Circles: females; Squares: males. “y” indicates a variant carrier and “n” indicates a non-carrier. “age” is the age at diagnosis for CMM cases and age at exam for unaffected family members.
Since all subjects carrying the Ser270Asn variant were from Romagna, Italy, we verified the relatedness among Ser270Asn carriers using genome-wide SNP genotyping data (Online Methods). The kinship analysis did not detect cryptic relatedness across families at a threshold of 0.05 (Supplementary Fig. 1). Interestingly, the analysis revealed a haplotype encompassing the POT1 region encoding Ser270Asn shared by all carriers, suggesting that the Ser270Asn variant was inherited from the same common ancestor. By long range phasing and modeling the distribution of the length of shared haplotypes encoding Ser270Asn, we estimated the age of the most recent common ancestor carrying the variant to be 9.5 (standard deviation=3.9) generations (Online Methods, Supplementary Fig. 2).
POT1 is a component of the telomeric shelterin complex that directly binds with high specificity to single-stranded (ss) telomeric repeats11. POT1 prevents inappropriate processing of exposed chromosome ends by DNA damage response pathways and regulates telomerase function, thereby playing a critical role in maintaining telomere integrity and regulating telomere length12. POT1 contains two oligonucleotide/oligosaccharide-binding (OB) folds in the N terminus that bind to the telomeric overhang. The first OB fold interacts with the first six nucleotides of the telomeric repeat, whereas the second binds and protects the 3′ ssDNA13,14. Most reported POT1 somatic mutations15, including those found in chronic lymphocytic leukemia (CLL)16, affect the OB folds of POT1, indicating the importance of this region in human cancers. The novel POT1 variant we identified, Ser270Asn, is located in the OB2 domain and appears to be in very close proximity with DNA binding sites based on the crystal structure (Fig. 2, Supplementary Fig. 3)13,14. The variant is highly conserved among vertebrates and predicted to be deleterious by most computational programs we evaluated (Fig. 2, Supplementary Table 2).
Figure 2. Structural illustrations of rare variants in POT1.
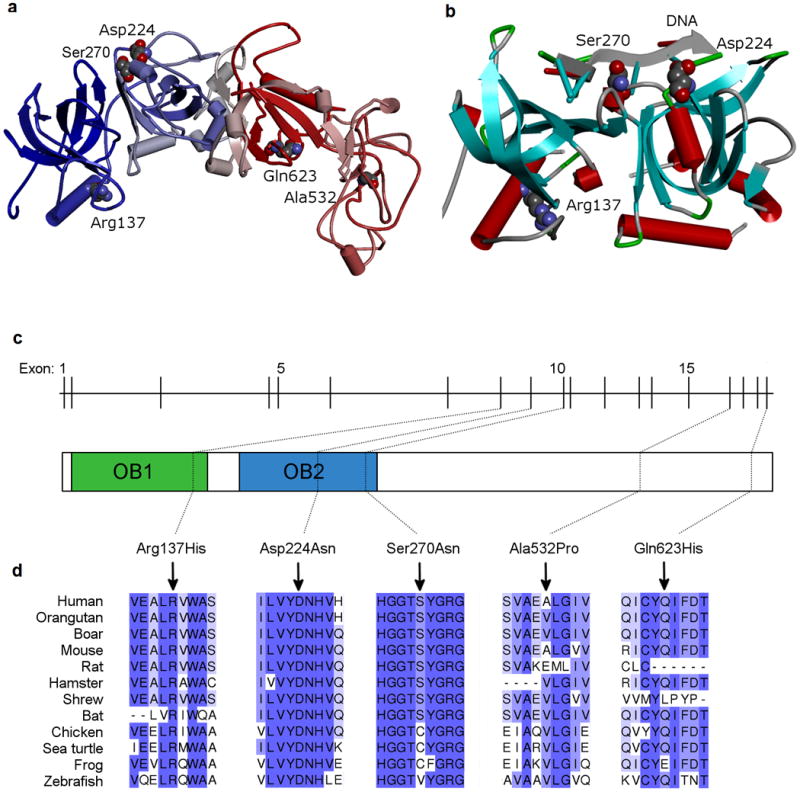
(a) Homology-based 3D model of hPOT1 (UniProt; Q9NUX5, POTE1_HUMAN) constructed using Phyre228,29. Secondary structural domains are schematically shown as cylinders (alpha helix), arrows (beta-sheet) and tubes (loop regions). The top templates used for modeling are 1XJV_A, 1ph4_A, 1jb7_A, and 1k8g_C13,28-30. The N-terminal sequence end (blue) is the same as the experimentally derived structure (1XJV_A) and the C-terminal end regions (red) are highly similar to the telomere binding proteins 1ph4 and 1jb7. (b) Schematic representation of the N-terminal region (OB1 and OB2 domains) of hPOT1 protein (Research Collaboratory for Structural Bioinformatics (RCSB) Protein Data Bank (PDB) ID, 1XJV). Helical motifs are shown as cylinders (red) and beta-sheets as arrows pointing in the direction of C-terminus. The coil and the turn region segments are displayed as tubes. The three residues in the OB1/OB2 domains containing missense variants are shown in Corey-Pauling-Koltun (CPK) mode. The two affected amino acids (Ser270Asn and Asp224Asn) are in close proximity to the bound telomeric single-stranded DNA (ssDNA) decamer. Discovery Studio (v. 3.5, Accelrys Inc.) was used for visualization and display. (c) Schematic of POT1 genomic structure and conserved OB domains. POT1 is composed of 19 exons spanning approximately 108,000 bases of genomic sequence on chromosome 7q31.33. (d) Amino acid conservation among POT1 homologs. Higher percent identity at a given amino acid position is indicated by a deeper purple color. The positions of the variants identified in this study are indicated relative to NP_056265.
Telomere length influences the requirement for telomere maintenance in human tumorigenesis17. To determine the impact of Ser270Asn on telomere length and integrity, we compared telomere length in peripheral blood mononuclear cell (PBMC) DNA from CMM cases carrying the variant to age-matched CMM cases without the variant (“controls”) by telomere restriction fragment analysis (Fig. 3a). In agreement with previous reports on POT1 mutations11,16,18, we found that in four unrelated melanoma carriers of the Ser270Asn variant from whom we had PBMCs, telomeres appeared to have increased length and heterogeneity (evident by telomere signals at higher and lower molecular weight), indicating that this variant perturbs telomere length maintenance. Quantitative telomere-fluorescence in situ hybridization (Q-FISH) analysis of metaphase spreads from ex vivo stimulated PBMCs further confirmed this observation (Fig 3b). Compared to the age-matched control, the Ser270Asn variant carrier also had a significant increase in the average number of fragile telomeres (P<0.0001, Fig 3b), the aberrant structures that are found in cells deficient in shelterin proteins and/or other proteins required for telomere replication19-22. The Ser270Asn variant did not lead to chromosome breaks, fusions, and chromosomes/chromatids without detectable telomere signals. Moreover, it did not alter the length of 3′-overhang or telomere sister chromatid exchanges, affect telomerase activity, or induce a DNA damage response at telomeres indicated by the presence of telomere dysfunction-induced foci23 (data not shown). These findings indicate that this rare variant perturbs the telomere maintenance rather than causes direct DNA damage.
Figure 3. Telomere length in PBMC from individuals with POT1-Ser270Asn variant.
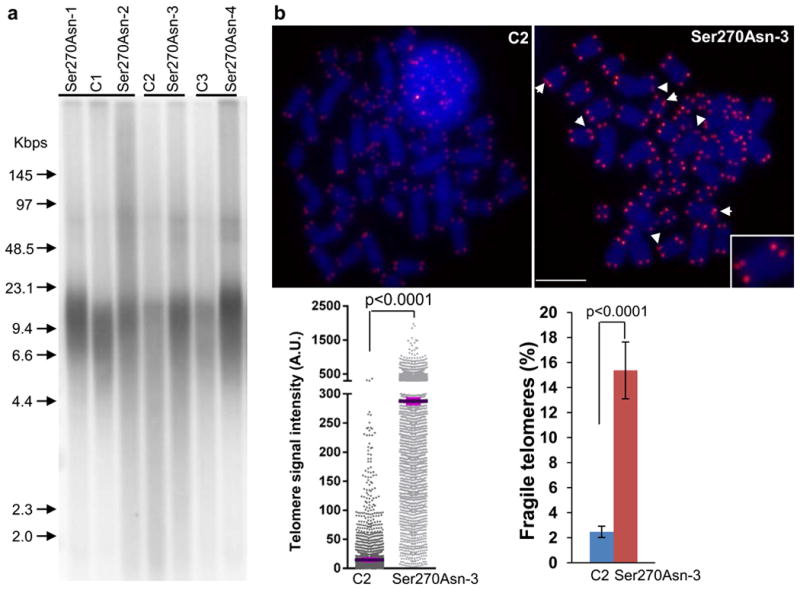
(a) telomere restriction fragment (TRF) analysis of PBMC. A representative blot shows telomere restriction fragments in each POT1 variant carrier (n=4) and the age-matched control (C, melanoma cases without POT1 variants, n=3). A labeled molecular weight marker (shown in kilobases) is used to determine telomere length. DNA was separated in a CHEF DR-II pulsed-field apparatus at 5 V/cm at an angle of 120° with switching times ramped from 1 to 15 s for 15 hours. Telomere length and heterogeneity are demonstrated by telomere signal intensities at higher and lower molecular weight. (b) Q-FISH analysis of ex vivo stimulated PBMC. Representative metaphase spreads of age-matched control and POT1 variant carrier showing DAPI staining (blue) and telomere fluorescence signals (red). Arrows: fragile telomeres (enlarged view demonstrated in the box). Left panel: quantitative measurement of telomere signal intensity in a jitter plot displaying complete distribution of telomeres with diverse signal intensity. Purple bars denote mean telomere signal intensity in a single control (C2) and a single Ser270Asn carrier (Ser270Asn-3). Additional variant carriers are described in Supplementary Fig. 6. Telomere signal intensity is depicted in arbitrary units (A.U). Right panel: Percentage of fragile telomeres in C2 and Ser270Asn-3. At least 30 metaphases / sample were counted. Error bars indicate standard deviation. P values were obtained from the Wilcoxon rank-sum test. The experiments were repeated three times on each sample.
Exome sequencing analysis of Italian families also identified two other rare missense substitutions in POT1 (g.7:124464052 C>G [Gln623His] and g.7:124503540 C>T [Arg137His]) that were found in all cases sequenced in two families (F6 and F7, Supplementary Fig. 4). Both missense variants are absent from public databases and 3,489 genotyped controls (2,038 Italian and 1,451 Spanish), and are predicted to be deleterious by most algorithms (Supplementary Table 2). Arg137His affects the alpha-helix in the OB1 domain and could play a key role in the structural integrity of the protein fold and indirectly influence DNA binding when mutated (Fig. 2, Supplementary Fig. 3). Gln623His affects the C terminus of POT1, which contains the TPP1 binding region. TPP1 (encoded by ACD) is another shelterin component and forms heterodimer with POT1. POT1-TPP1 binding increases the affinity of POT1 for ssDNA by 10-fold and enables the recruitment of telomerase to the overhang24,25. The carriers of Arg137His and Gln623His showed slightly but significantly increased telomere intensity signals (Supplementary Fig. 5) and telomere fragility (Supplementary Fig. 6) in PBMCs compared to age-matched controls.
We also evaluated copy number variations (CNVs) using exome data in these families (Online Methods) and observed no CNV disrupting POT1.
To assess the overall genetic burden due to rare variants in the POT1 gene, we sequenced POT1 exons in 768 CMM cases and 768 controls collected from a multicentric melanoma case-control study from Italy (Online Methods), and we found that CMM cases showed a significant increase in the burden of all rare variants (<1% in ESP and 1000 Genome) compared to controls (31 carriers among cases and 15 carriers among controls, OR=2.1, 95% CI=1.1-4.2, P=0.024, Fisher's exact test; Supplementary Table 3). The difference was more significant when we restricted the analysis to exonic variants (16 carriers among cases and 3 carriers among controls, OR=5.4, 95% CI=1.5-29.2, P=0.0021).
Subsequently, we used exome and targeted sequencing data obtained from familial CMM cases or sporadic cases with MPMs in three other populations to further evaluate POT1 variants: 68 families (139 cases) from the United States, 234 families (267 cases) and 157 MPMs from France, and three families (10 cases) from Spain. We identified two novel recurrent germline missense variants (g.7:124499043 C>T, Asp224Asn and g.7:124469308 C>G, Ala532Pro). The Asp224Asn variant was observed in four of five CMM cases from an American family (Supplementary Fig. 4, Supplementary Table 1) and was found in only one out of 6,500 subjects in ESP. This variant was also seen in a sporadic MPM case from Genoa, Italy. Like Ser270Asn, Asp224Asn is located in the OB2 domain near DNA binding sites (Fig. 2). It is predicted to be deleterious by all evaluated variant prediction programs (Supplementary Table 2). The Ala532Pro variant was identified in a familial case (the only case with DNA in the family) from a three-case family from France and one French case with five melanomas and one basal cell carcinoma (Supplementary Fig. 4, Supplementary Table 1), and was not reported in ESP. Ala532Pro is predicted to be damaging only by SIFT and PolyPhen-2, but is located near a splice junction and is predicted to disturb normal splicing. Both Asp224Asn and Ala532Pro were technically validated and neither was observed in the NCI DCEG or McGill in-house sequencing databases. Other rare POT1 variants identified in a single French familial or MPM case, all of which were confirmed by Sanger sequencing, are shown in Supplementary Table 4. No rare disease co-segregating POT1 variant was found in the Spanish CMM families.
Since POT1 was reported to frequently undergo somatic mutations in chronic lymphocytic leukemia (CLL)16, we also examined POT1 exome sequencing data obtained from 51 lymphoproliferative disorder (LPD) families from the US including 171 LPD cases affected with CLL (N=70), Non-Hodgkin's lymphoma (N=16), Hodgkin's lymphoma (N=46), or Waldenström macroglobulinemia (N=39) (Online Methods). No rare variants in POT1 were identified in the LPD subjects.
In summary, we have identified POT1 as a susceptibility gene for familial melanoma in several populations. This was further supported by POT1 findings in melanoma-prone families from the UK and Australia reported in the same issue of this Journal26. In particular, we have identified germline rare variants of POT1 in Romagna, Italy, which showed a frequency comparable to that of CDKN2A mutations among melanoma-prone families in this population27. Together with the finding of TERT as a potential susceptibility gene in melanoma, our findings suggest that genes involved in telomere maintenance may play important roles in melanoma development.
URLs
PLINK, http://pngu.mgh.harvard.edu/∼purcell/plink;
UCSC genome browser, http://genome.ucsc.edu;
KAVIAR, http://db.systemsbiology.net/kaviar;
dbSNP, http://www.ncbi.nlm.nih.gov/projects/SNP;
The 1000 Genomes Project, http://www.1000genomes.org;
CoNIFER , http://conifer.sourceforge.net;
Regulome DB, http://regulome.stanford.edu;
PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/;
SIFT, http://sift.jcvi.org;
PROVEAN, http://provean.jcvi.org/index.php;
MutationAssessor, http://mutationassessor.org/;
MutationTaster, http://www.mutationtaster.org/;
GERP, http://mendel.stanford.edu/SidowLab/downloads/gerp/;
PhastCons , http://compgen.bscb.cornell.edu/phast/phastCons-HOWTO.html;
PHAST, http://compgen.bscb.cornell.edu/phast;
ProPhylER, http://www.prophyler.org/;
BEAGLE, http://faculty.washington.edu/browning/beagle/beagle.html;
GATK, http://www.broadinstitute.org/gatk/;
ESP, http://evs.gs.washington.edu/EVS/;
VariantGPS, http://variantgps.nci.nih.gov;
Jalview http://www.jalview.org;
Cancer Genomics Research, http://cgf.nci.nih.gov/
Online Methods
Subjects and families
Italian cutaneous malignant melanoma (CMM), American CMM, Spanish CMM, and lymphoproliferative disorder (LPD) families are participants in Institutional Review Board (IRB)-approved studies at the National Cancer Institute (NCI). Detailed descriptions of these studies have been reported previously27,31,32. In brief, the Italian CMM family study evaluated in the exome sequencing and POT1 sequencing/genotyping analyses included 101 CMM cases/obligate carriers and 198 unaffected individuals from 56 CDKN2A/CDK4-negative families with 2-5 affected relatives who were recruited at the Dermatology Unit of Maurizio Bufalini Hospital in Cesena, North-Eastern Italy. The American CMM family study was comprised of 68 CDKN2A/CDK4-negative families with 2-13 affected individuals who were ascertained through health care professionals or self-referral. All study participants were of European ancestry and exome sequencing data was based on 139 CMM cases and six unaffected individuals. The LPD families had two or more patients affected with the same lymphoproliferative disorders, including chronic lymphocytic leukemia (CLL, N=70), Waldenström macroglobulinemia (WM, N=39), Hodgkin lymphoma (HL, N=46), and non-Hodgkin lymphoma (NHL, N=16) and were accrued through health care professionals or self-referral. Exome sequencing was based on 171 patients or obligate carriers and five unaffected individuals from 51 families. The French melanoma families and sporadic cases with multiple primary melanomas (MPMs) were recruited through a national network of French Dermatology and Oncogenetic clinics that constitute the French Familial Melanoma Study Group and the MELARISK collection. This study was approved by Institutional Review Boards and has been described in detail previously33. The French data investigated in the present study included two sets of CDKN2A/CDK4-negative patients that were sequenced independently: the first set contained 22 melanoma-prone families (45 cases) and four sporadic melanoma cases with MPMs with whole exome sequencing conducted at McGill University, Canada; the second set included 212 melanoma families (222 cases) and 153 MPMs, for whom POT1 was sequenced at the Cancer Genomics Research (CGR) Laboratory, NCI. The Spanish family study included three families with seven affected and ten unaffected individuals, with whole exome sequencing conducted at CGR.
The additional 2,702 CMM cases included in the POT1 genotyping were derived from a multicentric case-control study from Italy (N=1,824)34 and a case-control study from Valencia, Spain (N=878)35. Unaffected individuals were controls from the two melanoma case-control studies (511 Italian, 1,451 Spanish) and 1,527 individuals without a personal history of cancer who were recruited as controls for the Environment And Genetics in Lung cancer Etiology (EAGLE), a population-based case-control study of lung cancer conducted in Italy 36,37. A subset of these subjects, including 768 cases and 768 controls from Italy, were further sequenced for the coding region of POT1. The subjects were selected based on their residence within or in the proximity of the Romagna area where Ser270Asn, the rare variant that arose as a founder mutation, was found.
Of note, for all studies, familial CMM cases included individuals with single or multiple primary melanomas. Sporadic CMM cases with multiple primary melanomas were counted separately - and defined as “MPMs” - to emphasize their potential genetic enrichment.
All studies were approved by IRBs at NCI and/or local institutions. All subjects provided written informed consent.
Exome sequencing
Whole exome sequencing for the Italian CMM, American CMM, Spanish CMM, and LPD families was performed at CGR, DCEG, NCI. The detailed procedures are presented in Supplementary Note and have been described previously38. Briefly, for each sample, 1.1 μg genomic DNA extracted from blood was used for exome sequence capture, which was performed with NimbleGen's SeqCap EZ Human Exome Library v2.0 or v3.0 (Roche NimbleGen, Inc., Madison, WI, USA). Pools of captured DNA were then paired-end sequenced using an Illumina HiSeq following Illumina-provided protocols for 2x100 paired-end sequencing. Each exome was sequenced to high-depth in order to achieve a minimum threshold of 80% of coding sequence (CDS) covered with at least 15 reads, based on the UCSC hg19 “known gene” transcripts. According to the NIH policy, the exome sequencing data used for discovery will be released to the database of Genotype and Phenotype (dbGaP).
Whole exome sequencing of the French subjects was performed at McGill University and Genome Quebec Innovation Centre. Briefly, 3 μg of genomic DNA was used to perform exome capture with Agilent SureSelect kit following the manufacturer's instructions. 100bp paired-end sequencing was performed on Illumina HiSeq2000 platform. We generated approximately 10Gb of sequence for each sample such that at least 90% of the coding sequence were covered by at least 10 reads.
Bioinformatic analysis
Details of the bioinformatic analysis pipeline are presented in Supplementary Note and were previously described38. Briefly, sequencing reads were aligned to hg19 reference genome using the Novoalign software version 2.07.14 (http://www.novocraft.com). Variant discovery and genotype calling of multi-allelic substitutions, insertions and deletions was performed on all individuals globally using the UnifiedGenotyper module from Genome Analysis Toolkit (GATK)39 with the minimum call quality parameter set to 30. An in-house NCI DCEG database containing variants presented in 490 exomes (200 Asians and 290 of European ancestry) from subjects in melanoma-unrelated cancer families sequenced in parallel with our CMM and LPD families was used as an additional control dataset. We excluded variants that were observed more than once in The 1000 Genomes Project or NHLBI Exome Sequencing Project (ESP), or more than once in our in-house exome database. We applied a dominant genetic model which required the variant to be present in all affected people within a family (including the single case sequenced in the family), or all but one affected subjects in families with >3 cases sequenced to reduce the impact of phenocopies or sequencing errors.
Variant annotation
We used computational tools including PolyPhen-2, SIFT, Provean, MutationAssessor, and MutationTaster to predict the potential impact of sequence variants on protein function. We also obtained conservation scores using GERP, PhastCons, and PhyloP, as well as ProPhylER to predict mutation impact based on evolutionary constraint analyses.
Chromosome-based exact test
We developed a chromosome-based exact test to test the association between CMM and the Ser270Asn variant by appropriately accounting for the relatedness for familial CMM cases within the same families. The details of this approach are presented in Supplementary Note. Briefly, we first conducted a kinship analysis, which did not reveal cryptic relatedness across families using a kinship coefficient cutoff of 0.05. We then inferred IBD at the Ser270Asn locus using SNP genotypes based on exome sequencing data to identify independent chromosomes and the number of patients carrying each chromosome in each family at the locus. Under H0 of no association, we randomly chose 5 chromosomes (from N=4,229 total independent chromosomes), assigned them as Ser270Asn variant allele, and counted the number of patients carrying the variant allele (denoted as M). The p-value of the chromosome-based exact test is defined as the probability P(M≥ M0|H0), where M0 =11, which is the observed number of familial CMM cases carrying the Ser270Asn variant allele (the obligate gene carrier was not included in this analysis).
Estimation of kinship coefficients across Italian CMM families
We estimated kinship coefficients to confirm the relationship for family members and to detect cryptic relatedness. In a first analysis, we merged the genotype of Italian CMM families and the EAGLE controls for 27,674 common autosomal SNPs overlapping between exomic sequencing (CMM families) and Illumina HumanHap550 BeadChip (EAGLE controls), and estimated the kinship coefficient using PLINK (v.1.07)40. In a second analysis, we genotyped the subjects carrying POT1 rare variants using Illumina OmniExpress Arrays with higher density to refine the estimates of kinship coefficients for these POT1 variant carriers using 306,684 common SNPs overlapping between the two Illumina genotyping platforms.
Estimation of the age of the most recent common ancestor carrying the Ser270Asn variant
To investigate whether the variants were independent or inherited from a common ancestor, we performed haplotype analysis using BEAGLE41, combining genotype data of all Ser270Asn carriers and EAGLE controls to improve the haplotyping accuracy. We next estimated the age of the most recent common ancestor (MRCA) of the Ser270Asn variant based on the length of the haplotypes shared by the carriers. Because close relatives are from the same lineage and do not provide additional information, we included only the proband from each of the five families with the Ser270Asn variant as well as the single sporadic case who also carried Ser270Asn. For a subject with one parent genotyped, we resolved the phase for SNPs homozygous in the subject and heterozygous in the parent or vice versa. For subjects with no parent genotyped, we followed the concept of “surrogate parent” 42 to identify subjects who shared a long stretch of identity by descent (IBD) and resolved the phase for the stretch. For a given pair of subjects, the length of shared IBD was determined using the SNPs with resolved phase. This approach provided an accurate estimate for the length of shared haplotypes with error typically less than 50kb in the boundary, which has a minimal effect on the inference of the age of the MRCA. Given the haplotype sharing structure, we modified the likelihood framework for sparse microsatellite markers43 to estimate the age of the MRCA and the standard deviation.
Copy number variation detection from exomic sequencing data
CoNIFER (v0.2.2)44 was used to analyze the sequence data on chromosome 7 for subjects in the Italian families with exome sequencing data. Briefly, reads per kilobase per million (RPKM) for each exon was calculated for each subject, which was normalized across subjects to derive ZRPKM (a z-score of PRKM with mean of zero and variance of one). Singular value decomposition (SVD) was then performed on ZRPKM values and the top components (six in our analysis), which typically represent sequencing artifacts, were removed. The resulting SVD-ZRPKM values were used for detecting copy number variations (CNVs). CNVs were called if average SVD-ZRPKM was greater than 1.5 (duplications) or less than -1.5 (deletions) for any interval. We detected 35 CNVs on chromosome 7, including 26 duplications and nine deletions in 29 subjects, none of which disrupted POT1.
POT1 crystal structure
Homology-based 3D model of hPOT1 (UniProt; Q9NUX5, POTE1_HUMAN) was constructed using Phyre228,29. The POT crystal structure of the N-terminal region (OB1 and OB2 domains) of hPOT1 protein (Research Collaboratory for Structural Bioinformatics (RCSB) Protein Data Bank (PDB) ID, 1XJV) was visualized using Discovery Studio (v. 3.5, Accelrys Inc.). Multiple sequence alignments were generated for homologous POT1 protein sequences using T-Coffee45 to evaluate conservation. Alignments were generated with the following sequences: NP_056265.2 (Homo sapiens), NP_001127526.1 (Pongo abelii), NP_001131107.1 (Sus scrofa), NP_598692.1 (Mus musculus), NP_001019493.1 (Rattus norvegicus), EGW09411.1 (Cricetulus griseus), ELW65533.1 (Tupaia chinensis), ELK29766.1 (Myotis davidii), NP_996875.1 (Gallus gallus), EMP32613.1 (Chelonia mydas), NP_998876.1 (Xenopus tropicalis), NP_001232889.1 (Danio rerio). Jalview46 was used to visualize and format the alignment.
POT1 gene sequencing
A custom set of Ampliseq™ primers from Life Technologies was used to amplify the coding region of the POT1 gene at CGR, DCEG, NCI. The primers were created using the primer design engine at www.ampliseq.com. 40ng of genomic DNA was used for each sample. The Ampliseq™ process was performed according to the official process documentation found on the Ion Community (www.ioncommunity.lifetechnologies.com). The samples were quality checked for proper amplicon length and quantity on the Agilent Bioanalyzer and were then sequenced according to the current Ion Torrent template and sequencing preparation documentation. Each PGM Ion 316 sequencing run consisted of 96 barcoded samples. The resulting sequencing data yielded coverage for each amplicon >300X in depth.
Rare variant burden test
To assess the overall genetic burden due to rare variants in POT1, we counted subjects carrying any rare variant in POT1 and tested the difference between CMM cases and controls using Fisher's exact test. This test was based on POT1 gene sequencing using Ion Torrent performed on 768 cases and 768 controls from Italy. We excluded variants and genotypes with low quality scores (< 30) and we only included variants that were called by both the Torrent Variant Caller and GATK to ensure the accuracy of the burden test. This QC step was blind to case/control status. We first included all variants with minor allele frequency less than 1% in ESP and 1000 Genome, regardless of their predicted functions. We then repeated the analysis restricted to exonic variants. P values were two-sided. Based on simulation studies, we estimated that 768 cases and 768 controls had statistical power=0.8 to identify an OR=2.3 for differences in the frequency of rare variant carriers between cases and controls, for an overall rare variant allele frequency =0.01, and an OR=5 for rare variant allele frequency=0.002.
Genotyping of POT1 variants
TaqMan® assay (Life Technologies, Grand Island, NY) validation and SNP genotyping was performed at CGR, DCEG, NCI. The detailed procedures are described in the Supplementary Note. Briefly, loci that produced successful TaqMan® assay designs were validated using the 102 Variant GPS population panel and the 262 HapMap control panel (Coriell Cell Repositories, Camden, NJ). Assay validation was based on concordance (>99%) with reported HapMap genotypes (when available). SNP Genotyping was performed using 9700 Thermal Cycler (Life Technologies) and endpoint reads were evaluated using the 7900HT Sequence Detection System (Life Technologies). Cluster analysis was performed using SDS v2.2.2 software (Life Technologies).
Telomere Restriction Fragment (TRF) Analysis
Genomic DNA from peripheral blood mononuclear cell (PBMC) was digested with HinfI and RsaI and analyzed by standard Southern blotting using a 32P-α-dCTP labeled telomere probe under highly stringent conditions as previously described47. The experiments were repeated three times on each sample.
Quantitative telomere-fluorescence in situ hybridization (Q-FISH or telomere-FISH) and chromosome orientation FISH (CO-FISH)
PBMCs were briefly cultured with RPMI 1640 medium (Gibco BRL Life Technologies) supplemented with 2mM L-glutamine, 25mM HEPES, and 20% fetal calf serum in the presence of 1 to 10% interleukin 2 (Advanced Biotechnologies) and phytohemagglutinin P (5 μg/mL Sigma). For detecting Telomere Sister Chromatid Exchange (T-SCE), PBMCs were sub-cultured in medium containing a 3:1 ratio of BrdU/BrdC (Sigma) at a final concentration of 1 × 10-5 M and collected around 24 hours. Colcemid (0.1 μg/ml) was added 4 hours before harvest. Cells were then used for Q-FISH and CO-FISH analyses, as described previously48-50. Briefly, for Q-FISH, metaphase spreads were hybridized with Cy3-labeled PNA (CCCTAA)3 (Panagene). Images were captured using Cytovision software (Applied Imaging Corp.) on a fluorescence microscope (Axio2; Carl Zeiss, Germany), followed by quantification of individual telomere fluorescence signals using the TFL-Telo software (a kind gift from P. Lansdorp, Vancouver, BC). Metaphases were scored for chromosome breaks, fusions, fragile telomeres (a chromatid with ≥2 telomere signals), and SFEs or STL (chromosomes or chromatids without detectable telomere signals). For CO-FISH, cells were hybridized with Alexa Fluor 488-labeled (TTAGGG)3 and Cy3-labeled PNA (CCCTAA)3 probes. A chromosome with more than two telomeric DNA signals by both probes was scored as T-SCE positive. For each individual, the data were collected from at least 30 cells.
Telomere Dysfunction-Induced Focus (TIF) assay
EBV immortalized cells derived from cryopreserved lymphocytes were spun onto Cytospin microscopic slides at 200 rpm for 3 minutes. Cells were fixed in 2% paraformaldehyde, permeabilized with 0.5% triton X-100, and blocked with 10% FBS for 1-2 hours. Cells were stained with a rabbit anti-γH2AX antibody (1:200, Santa Cruz) 51, followed by Alexa 488-labeled secondary antibody (1:500; Molecular Probes). For combined telomere FISH, slides were fixed in 2% paraformaldehyde and then used to perform telomere-FISH. Z-stack images were captured on a fluorescence microscope (Axiovert 200M; Carl Zeiss). The experiments were repeated three times on each sample.
Telomeric G-overhang measurement
Two methods were used to measure the length and the abundance of G-overhangs. The length of the telomeric G-overhang was measured by telomere overhang protection assay as described 52. The relative abundance of G-overhangs was measured by non-denaturing in-gel hybridization as described 52 with minor modifications. Briefly, one microgram genomic DNA was resuspended in 15 μl of gp32 protection buffer, and treated with 1 μg of RNase A and with or without Exo1 at 37°C for 1 hr. Samples were size fractionated on a 0.8% agarose gel in 0.5× TBE at 8 V/cm. The gel was then dried at 50°C for 1 hr. The dried gel was rinsed with 2XSSC, and then hybridized with the high specificity C-rich probe at 42°C overnight. After two washes with 0.1% SDS-0.1XSSC for 15min each, the gel was then exposed to a phosphorimager screen, and signals were acquired by Storm860 PhosphorImager and quantified by ImageQuant software. The gel was then denatured in 0.5 M NaOH-1.5 M NaCl for 1hr at room temperature, neutralized in 1.5 M NaCl-0.5 M Tris-HCl (pH 8.0) for 15 min and then hybridized to C-rich probe again at 42°C overnight. After two washes with 0.1× SSC-0.1% SDS for 15 min each, the gel was exposed to a PhosphorImager screen. Relative amounts of overhangs were calculated by normalizing signals from the native gel with signals from denatured gel. The experiments were repeated three times on each sample.
Cell lysate preparation and telomerase assays
S-100 extracts from PBMCs were prepared and 5 and 1 μg cell extracts were assayed for the presence of telomerase activity using the telomere repeat amplification protocol (TRAP) as previously described 53. RNase A treatment was used as negative controls.
Supplementary Material
Acknowledgments
This work was supported by the Intramural Research Program of NIH, National Cancer Institute (NCI), Division of Cancer Epidemiology and Genetics. The samples from the Instituto Valenciano de Oncologia were retrieved from the Biobanco del Instituto Valenciano de Oncologia. The Genoa collection was partly supported by Università degli Studi di Genova Progetti di Ricerca di Ateneo PRA 2012-2013 and IRCCS Azienda Ospedaliera Universitaria San Martino – IST Istituto Nazionale per la Ricerca sul Cancro, 5 per 1000 per la Ricerca Corrente. The French MELARISK collection was partly supported by a PHRC (Programme Hospitalier de Recherche Clinique) grant [AOM-07-195]. This project has also been funded in part with federal funds from NIH, NCI, under Contract No. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
Author Contributions: JS, XRY, MR, XH, AMG, LRG, JNS, FD, AV, HM, MTL performed and interpreted genetics analyses; BB, SAS, SR performed functional prediction analyses; DC, MCF, PG, BBdP, EN, MFA, WB, LP, PQ, JBR, ZGC, FJ, KP, GBS, GL, MAT, MTL directed the clinical work for the melanoma cases from Italy, Spain, US, and France; NEC, MLM directed the clinical work for the LPD cases; MY, MAT, SJC directed all sequencing analyses at NCI DCEG CGR; JH, MC, ZW, XZ and The NCI DCEG Cancer Genomics Research Laboratory conducted the whole exome/target sequencing and genotyping analyses for the Italian, American, Spanish and part of the French melanoma cases and controls; The NCI Cancer Sequencing Working Group examined and enrolled non-melanoma and non-LPD families for whole exome sequencing analyses; The French Familial Melanoma Study Group examined and enrolled melanoma cases and controls from France; ML, YR, MF conducted whole exome sequencing and genotyping for the French melanoma cases and other non-cancer diseases; PLH, PM, CP, AE oversaw sample preparation for the laboratory studies; JY, HV, WC, YL performed functional analyses of POT1; MTL designed the overall study; XRY, JS, BB, AMG, MTL drafted the manuscript; all authors contributed to the final manuscript.
Competing Financial Interests: The authors declare no competing financial interests.
References
- 1.Goldstein AM, Tucker MA. Genetic epidemiology of cutaneous melanoma: a global perspective. Arch Dermatol. 2001;137:1493–6. doi: 10.1001/archderm.137.11.1493. [DOI] [PubMed] [Google Scholar]
- 2.Cannon-Albright LA, et al. Assignment of a locus for familial melanoma, MLM, to chromosome 9p13-p22. Science. 1992;258:1148–52. doi: 10.1126/science.1439824. [DOI] [PubMed] [Google Scholar]
- 3.Hussussian CJ, et al. Germline p16 mutations in familial melanoma. Nat Genet. 1994;8:15–21. doi: 10.1038/ng0994-15. [DOI] [PubMed] [Google Scholar]
- 4.Gruis NA, et al. Homozygotes for CDKN2 (p16) germline mutation in Dutch familial melanoma kindreds. Nat Genet. 1995;10:351–3. doi: 10.1038/ng0795-351. [DOI] [PubMed] [Google Scholar]
- 5.Goldstein AM. Familial melanoma, pancreatic cancer and germline CDKN2A mutations. Hum Mutat. 2004;23:630. doi: 10.1002/humu.9247. [DOI] [PubMed] [Google Scholar]
- 6.Eliason MJ, et al. Population-based prevalence of CDKN2A mutations in Utah melanoma families. J Invest Dermatol. 2006;126:660–6. doi: 10.1038/sj.jid.5700094. [DOI] [PubMed] [Google Scholar]
- 7.Zuo L, et al. Germline mutations in the p16INK4a binding domain of CDK4 in familial melanoma. Nat Genet. 1996;12:97–9. doi: 10.1038/ng0196-97. [DOI] [PubMed] [Google Scholar]
- 8.Puntervoll HE, et al. Melanoma prone families with CDK4 germline mutation: phenotypic profile and associations with MC1R variants. J Med Genet. 2013;50:264–70. doi: 10.1136/jmedgenet-2012-101455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wiesner T, et al. Germline mutations in BAP1 predispose to melanocytic tumors. Nat Genet. 2011;43:1018–21. doi: 10.1038/ng.910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horn S, et al. TERT promoter mutations in familial and sporadic melanoma. Science. 2013;339:959–61. doi: 10.1126/science.1230062. [DOI] [PubMed] [Google Scholar]
- 11.Loayza D, De Lange T. POT1 as a terminal transducer of TRF1 telomere length control. Nature. 2003;423:1013–8. doi: 10.1038/nature01688. [DOI] [PubMed] [Google Scholar]
- 12.Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu Rev Genet. 2008;42:301–34. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- 13.Lei M, Podell ER, Baumann P, Cech TR. DNA self-recognition in the structure of Pot1 bound to telomeric single-stranded DNA. Nature. 2003;426:198–203. doi: 10.1038/nature02092. [DOI] [PubMed] [Google Scholar]
- 14.Lei M, Podell ER, Cech TR. Structure of human POT1 bound to telomeric single-stranded DNA provides a model for chromosome end-protection. Nat Struct Mol Biol. 2004;11:1223–9. doi: 10.1038/nsmb867. [DOI] [PubMed] [Google Scholar]
- 15.Forbes SA, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39:D945–50. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramsay AJ, et al. POT1 mutations cause telomere dysfunction in chronic lymphocytic leukemia. Nat Genet. 2013;45:526–30. doi: 10.1038/ng.2584. [DOI] [PubMed] [Google Scholar]
- 17.Taboski MA, et al. Long telomeres bypass the requirement for telomere maintenance in human tumorigenesis. Cell Rep. 2012;1:91–8. doi: 10.1016/j.celrep.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kendellen MF, Barrientos KS, Counter CM. POT1 association with TRF2 regulates telomere length. Mol Cell Biol. 2009;29:5611–9. doi: 10.1128/MCB.00286-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martinez P, et al. Increased telomere fragility and fusions resulting from TRF1 deficiency lead to degenerative pathologies and increased cancer in mice. Genes Dev. 2009;23:2060–75. doi: 10.1101/gad.543509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sfeir A, et al. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell. 2009;138:90–103. doi: 10.1016/j.cell.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Badie S, et al. BRCA2 acts as a RAD51 loader to facilitate telomere replication and capping. Nat Struct Mol Biol. 2010;17:1461–9. doi: 10.1038/nsmb.1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gu P, et al. CTC1 deletion results in defective telomere replication, leading to catastrophic telomere loss and stem cell exhaustion. EMBO J. 2012;31:2309–21. doi: 10.1038/emboj.2012.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takai H, Smogorzewska A, de Lange T. DNA damage foci at dysfunctional telomeres. Curr Biol. 2003;13:1549–56. doi: 10.1016/s0960-9822(03)00542-6. [DOI] [PubMed] [Google Scholar]
- 24.Wang F, et al. The POT1-TPP1 telomere complex is a telomerase processivity factor. Nature. 2007;445:506–10. doi: 10.1038/nature05454. [DOI] [PubMed] [Google Scholar]
- 25.Xin H, et al. TPP1 is a homologue of ciliate TEBP-beta and interacts with POT1 to recruit telomerase. Nature. 2007;445:559–62. doi: 10.1038/nature05469. [DOI] [PubMed] [Google Scholar]
- 26.Robles-Espinoza CD, et al. POT1 loss-of-function variants predispose to familial melanoma. Nat Genet. 2014 doi: 10.1038/ng.2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Landi MT, et al. Genetic susceptibility in familial melanoma from northeastern Italy. J Med Genet. 2004;41:557–66. doi: 10.1136/jmg.2003.016907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Horvath MP, Schultz SC. DNA G-quartets in a 1.86 A resolution structure of an Oxytricha nova telomeric protein-DNA complex. J Mol Biol. 2001;310:367–77. doi: 10.1006/jmbi.2001.4766. [DOI] [PubMed] [Google Scholar]
- 29.Theobald DL, Schultz SC. Nucleotide shuffling and ssDNA recognition in Oxytricha nova telomere end-binding protein complexes. EMBO J. 2003;22:4314–24. doi: 10.1093/emboj/cdg415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Classen S, Ruggles JA, Schultz SC. Crystal structure of the N-terminal domain of Oxytricha nova telomere end-binding protein alpha subunit both uncomplexed and complexed with telomeric ssDNA. J Mol Biol. 2001;314:1113–25. doi: 10.1006/jmbi.2000.5191. [DOI] [PubMed] [Google Scholar]
- 31.Tucker MA, et al. A natural history of melanomas and dysplastic nevi: an atlas of lesions in melanoma-prone families. Cancer. 2002;94:3192–209. doi: 10.1002/cncr.10605. [DOI] [PubMed] [Google Scholar]
- 32.Liang XS, et al. Common genetic variants in candidate genes and risk of familial lymphoid malignancies. Br J Haematol. 2009;146:418–23. doi: 10.1111/j.1365-2141.2009.07790.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chaudru V, et al. Influence of genes, nevi, and sun sensitivity on melanoma risk in a family sample unselected by family history and in melanoma-prone families. J Natl Cancer Inst. 2004;96:785–95. doi: 10.1093/jnci/djh136. [DOI] [PubMed] [Google Scholar]
- 34.Bodelon C, et al. On the interplay of telomeres, nevi and the risk of melanoma. PLoS One. 2012;7:e52466. doi: 10.1371/journal.pone.0052466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maccioni L, et al. Variants at chromosome 20 (ASIP locus) and melanoma risk. Int J Cancer. 2013;132:42–54. doi: 10.1002/ijc.27648. [DOI] [PubMed] [Google Scholar]
- 36.Landi MT, et al. A genome-wide association study of lung cancer identifies a region of chromosome 5p15 associated with risk for adenocarcinoma. Am J Hum Genet. 2009;85:679–91. doi: 10.1016/j.ajhg.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Landi MT, et al. Environment And Genetics in Lung cancer Etiology (EAGLE) study: an integrative population-based case-control study of lung cancer. BMC Public Health. 2008;8:203. doi: 10.1186/1471-2458-8-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ballew BJ, et al. Germline mutations of regulator of telomere elongation helicase 1, RTEL1, in Dyskeratosis congenita. Hum Genet. 2013;132:473–80. doi: 10.1007/s00439-013-1265-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–8. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Browning BL, Browning SR. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am J Hum Genet. 2009;84:210–23. doi: 10.1016/j.ajhg.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kong A, et al. Detection of sharing by descent, long-range phasing and haplotype imputation. Nat Genet. 2008;40:1068–75. doi: 10.1038/ng.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Genin E, Tullio-Pelet A, Begeot F, Lyonnet S, Abel L. Estimating the age of rare disease mutations: the example of Triple-A syndrome. J Med Genet. 2004;41:445–9. doi: 10.1136/jmg.2003.017962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krumm N, et al. Copy number variation detection and genotyping from exome sequence data. Genome Res. 2012;22:1525–32. doi: 10.1101/gr.138115.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Notredame C, Higgins DG, Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302:205–17. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 46.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–91. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hathcock KS, Hodes RJ, Weng NP. Analysis of telomere length and telomerase activity. Curr Protoc Immunol. 2004;Chapter 10:30. doi: 10.1002/0471142735.im1030s62. Unit 10. [DOI] [PubMed] [Google Scholar]
- 48.Zijlmans JM, et al. Telomeres in the mouse have large inter-chromosomal variations in the number of T2AG3 repeats. Proc Natl Acad Sci U S A. 1997;94:7423–8. doi: 10.1073/pnas.94.14.7423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Y, et al. An increase in telomere sister chromatid exchange in murine embryonic stem cells possessing critically shortened telomeres. Proc Natl Acad Sci U S A. 2005;102:10256–60. doi: 10.1073/pnas.0504635102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bailey SM, Goodwin EH, Cornforth MN. Strand-specific fluorescence in situ hybridization: the CO-FISH family. Cytogenet Genome Res. 2004;107:14–7. doi: 10.1159/000079565. [DOI] [PubMed] [Google Scholar]
- 51.Vallabhaneni H, O'Callaghan N, Sidorova J, Liu Y. Defective repair of oxidative base lesions by the DNA glycosylase Nth1 associates with multiple telomere defects. PLoS Genet. 2013;9:e1003639. doi: 10.1371/journal.pgen.1003639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chai W, Shay JW, Wright WE. Human telomeres maintain their overhang length at senescence. Mol Cell Biol. 2005;25:2158–68. doi: 10.1128/MCB.25.6.2158-2168.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu Y, et al. The telomerase reverse transcriptase is limiting and necessary for telomerase function in vivo. Curr Biol. 2000;10:1459–62. doi: 10.1016/s0960-9822(00)00805-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.