Abstract
Lysine-specific demethylase (LSD) 1 is an FAD-dependent demethylase that catalyzes the removal of methyl groups from lysine-4 in histone H3, thereby mediating gene repression. Here we tested the hypothesis that riboflavin deficiency causes a loss of LSD1 activity in HepG2 human hepatocarcinoma cells, leading to an accumulation of lysine-4-dimethylated histone H3 (H3K4me2) marks in the albumin promoter and aberrant upregulation of albumin expression. Cells were cultured in riboflavin-defined media providing riboflavin at concentrations representing moderately deficient (3.1 nmol/L), sufficient (12.6 nmol/L), and supplemented (301 nmol/L) cells in humans for 7 d. The efficacy of treatment was confirmed by assessing glutathione reductase activity and concentrations of reduced glutathione as markers of riboflavin status. LSD activity was 21% greater in riboflavin-supplemented cells compared with riboflavin-deficient and -sufficient cells. The loss of LSD activity was associated with a gain in the abundance of H3K4me2 marks in the albumin promoter; the abundance of H3K4me2 marks was ∼170% higher in riboflavin-deficient cells compared with sufficient and supplemented cells. The abundance of the repression mark, K9-trimethylated histone H3, was 38% lower in the albumin promoter of riboflavin-deficient cells compared with the other treatment groups. The expression of albumin mRNA was aberrantly increased by 200% in riboflavin-deficient cells compared with sufficient and supplemented cells. In conclusion, riboflavin deficiency impairs gene regulation by epigenetic mechanisms, mediated by a loss of LSD1 activity.
Introduction
Riboflavin is an essential precursor in the synthesis of the flavocoenzymes FMN and FAD (1). FMN and FAD, and their reduced forms FMNH2 and FADH2, serve as coenzymes in a large number of redox reactions and in a comparatively small number of reactions with no net redox change in mammalian intermediary metabolism (1–3). Representative examples of flavin-dependent pathways include reactions in electron transport chain, citric cycle, FA β-oxidation, glutathione homeostasis, and protein folding. In most cases, flavocoenzymes are not covalently attached to apoproteins but form noncovalent complexes (4). Flavin metabolites other than riboflavin, FMN, and FAD were identified in human plasma and urine by McCormick and coworkers (5–8), but these metabolites probably lack biologic activity.
Rather recently, a new FAD-dependent enzyme was identified: the nuclear amine oxidase homolog, lysine-specific demethylase (LSD)4 1 (9, 10). LSD1 demethylates lysine-4 in monomethylated (H3K4me1) and dimethylated (H3K4me2) histone H3, thereby removing gene activation marks and mediating gene repression (11, 12). H3K4me1 and H3K4me2 are enriched in chromatin adjacent to transcription start sites of active genes, whereas inactive gene promoters are characterized by low levels of H3K4 methylation marks (13). Consistent with its role as a transcriptional repressor, LSD1 forms a complex with the repressor element-1 silencing transcription factor (REST) corepressor and histone deacetylases 1 and 2, which synergize with LSD1 in gene repression by mediating the deacetylation of histone tails (14–16).
The loss of LSD1 precipitates strong phenotypes. For example, LSD1 knockout is lethal in embryonic mice (17). LSD1-deficient mouse embryonic stem cells are characterized by defects in differentiation, severe growth impairments due to increased cell death and impaired cell cycle progression, and global DNA hypomethylation (18).
To date, there is no published report regarding the impact of riboflavin nutrition on LSD1 activity. This knowledge gap has possible implications for disease prevention, because a variety of factors can cause riboflavin depletion, including alcohol consumption (19, 20), genetic defects of riboflavin transporter genes (19, 20), impaired conversion of riboflavin to its coenzyme forms in persons with thyroid hormone insufficiency (21), and abnormal riboflavin metabolism such as inhibition of the incorporation of riboflavin into FAD in response to treatment with tricyclic and tetracyclic compounds such as chlorpromazine, tetracycline, and adriamycin (22, 23). Note that current recommendations for riboflavin intake are largely based on studies using the FAD-dependent glutathione reductase as a marker of riboflavin nutrition, without considering the potentially more subtle FAD-dependent changes in the epigenome (24–26).
HepG2 human hepatocarcinoma cells were chosen as the model in this study because flavin-dependent pathways in these cells respond robustly to changes in riboflavin concentrations in culture media, when testing concentration that represent moderate deficiency, sufficiency, and supplementation in adults (27–29). The albumin gene (ALB) was chosen as a target locus because HepG2 cells produce albumin in large quantities and albumin plays crucial roles in serum osmolarity and transport of vitamins, FAs, and hormones (30). By using this model system, we tested the hypothesis that riboflavin deficiency causes a loss of LSD1 activity, leading to an enrichment of H3K4me2 marks in the ALB locus and an aberrantly high expression of ALB.
Materials and Methods
Cell cultures.
HepG2 human hepatocarcinoma cells (passage number 5; American Type Culture Collection) were cultured in riboflavin-defined Roswell Park Memorial Institute–1640 medium (HyClone), as described previously (29, 31). Riboflavin concentrations in the culture medium were adjusted to 3.1 nmol/L (denoted “deficient”), 12.6 nmol/L (denoted “sufficient”), and 301 nmol/L (denoted “supplemented”), taking into account the residual concentrations of riboflavin, FMN, and FAD in dialyzed FBS. Cells were cultured in riboflavin-sufficient medium for 7 d before transfer into the riboflavin-defined medium and continued to culture for 7 d before analysis. This protocol was chosen on the basis of previous studies suggesting that HepG2 cells achieve new steady state concentrations of flavins within 4 d of culture in riboflavin-defined media and that cell proliferation was adversely affected after 10 d of culture in riboflavin-deficient media (27, 28). Cells cultured in riboflavin-sufficient medium were designated as the control group.
Glutathione metabolism.
Glutathione reductase is an FAD-dependent enzyme, and both the activity of glutathione reductase and the concentration of reduced glutathione are robust markers of riboflavin status (29, 31). After 7 d in riboflavin-defined media, HepG2 cells were harvested and lysed for assessment of glutathione metabolism. Glutathione reductase activity was quantified in cell lysates containing 0.5 mg protein, as described previously (32). One unit of glutathione reductase activity is defined as the change of absorbance at 340 nm/0.5 mg protein in 10 min of incubation. The concentration of reduced glutathione in lysates was determined colorimetrically by using the 5,5-dithiobis(2-nitrobenzoic acid) reduction assay at 412 nm, as described previously (33).
LSD activity.
LSD activity was measured by using the demethylase (LSD-type) activity assay kit (Cayman Chemical) according to the manufacturer’s instructions. One unit of LSD activity is defined as the ratio of sample fluorescence to background fluorescence. This assay does not distinguish between the 2 LSDs in the human proteome, LSD1 and LSD2 (34).
Western blot analysis.
LSD1 expression was quantified by Western blot, as described previously (29), by using rabbit polyclonal anti-human LSD1 (Abcam). GAPDH was probed by using anti-GAPDH (Santa Cruz Biotechnology) and used as a loading control. Data were quantified by gel densitometry analysis.
Chromatin immunoprecipitation assay.
The enrichment of LSD1, H3K4me2, and lysine-9-trimethylated histone H3 (H3K9me3) in the promoter region of the human ALB gene (−166 to +58) was assessed by chromatin immunoprecipitation (ChIP) assay as described previously (35). Immunoprecipitations were performed with specific antibodies to LSD1, H3K4me2 (Abcam), H3K9me3 (Abcam), H3 (Abcam), and rabbit IgG (Santa Cruz Biotechnology). Nuclear chromatin extracts without immunoprecipitation were used as an input control. Precipitation of chromatin with nonspecific rabbit IgG was used as a negative control. Histone H3 occupancy was used to normalize H3K4me2 and H3K9me3 occupancy in the ALB promoter.
qRT-PCR.
The abundance of mRNA coding for LSD1 and albumin was quantified by qRT-PCR using Power SYBR Green PCR Master Mix (Applied Biosystems), as described previously (36) (Supplemental Table 1). The relative amount of each gene was normalized by using the housekeeping gene GAPDH.
Amplicons from ChIP assays were analyzed by using the PerfeCTa SYBR Green FastMix (Quanta Biosciences), as described previously (37). The relative occupancy of histones in the ALB promoter was calculated as described (38), and values are reported as the percentage of input DNA.
Statistical analysis.
Data were distributed normally and variances were homogenous, as assessed by Kolmogorov-Smirnov normality test and Bartlett test, respectively. Significance was assessed by 1-factor ANOVA and Fisher’s protected least significant difference post hoc test (39). All analyses and data points are based on 3 biologically independent repeats. StatView 5.0.1 (SAS Institute) was used to perform all calculations. Differences were considered significant if P < 0.05. Data are expressed as means ± SDs.
Results
Treatment efficacy.
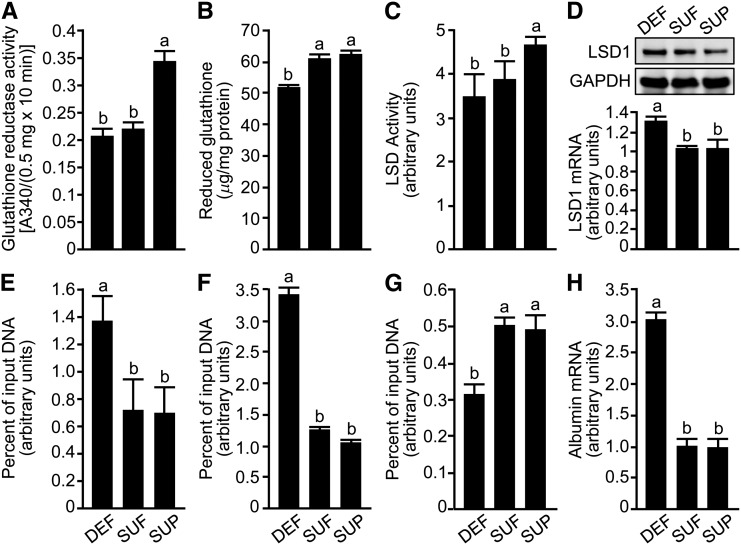
HepG2 cells showed the expected response to alterations in riboflavin concentrations in culture media, i.e., glutathione reductase activity was higher in riboflavin-supplemented cells compared with deficient and sufficient cells, and the concentrations of reduced glutathione were lower in riboflavin-deficient cells compared with sufficient and supplemented cells (Fig. 1A, B). These findings are consistent with efficacy of riboflavin treatment in HepG2 cell cultures in previous studies (27, 29).
FIGURE 1.
Effects of riboflavin concentrations in culture media on LSD1-dependent gene regulation in HepG2 cells. Glutathione reductase activity (A); reduced glutathione (B); LSD activity (C); expression of LSD1 mRNA (D) and protein (insert in panel D); enrichment of LSD1 (E), H3K4me2 (F), and H3K9me3 (G) in the albumin promoter; and albumin mRNA abundance (H). Values are means ± SDs, n = 3. Means not sharing a common letter are significantly different for the same variable, P < 0.05. DEF, deficient; H3K4me2, lysine-4-dimethylated histone H3; H3K9me3, lysine-9-trimethylated histone H3; LSD, lysine-specific demethylase; SUF, sufficient; SUP, supplemented.
LSD activity and expression.
LSD activity depended on the concentration of riboflavin in cell culture media. When HepG2 cells were cultured in riboflavin-defined medium for 7 d, LSD activity was 21% greater in riboflavin-supplemented cells compared with deficient and sufficient cells (Fig. 1C).
The expression of LSD1 mRNA was ∼30% higher in riboflavin-deficient cells than in sufficient and supplemented cells (Fig. 1D). LSD1 protein abundance followed a similar pattern (in arbitrary units of gel densitometry): 9.44 ± 0.87 for deficient cells vs. 6.81 ± 0.87 for sufficient cells vs. 3.93 ± 0.94 for supplemented cells (P < 0.05 for all possible comparisons).
Transcriptional regulation of ALB.
Riboflavin deficiency caused an aberrant expression of albumin in HepG2 cells. The binding of LSD1 to the ALB promoter was ∼90% greater in riboflavin-deficient cells than in sufficient and supplemented cells after 7 d of culture (Fig. 1E). Despite the increase in LSD1 binding, the abundance of H3K4me2 and H3K9me3 marks was 170% higher and 38% lower, respectively, in the albumin promoter in riboflavin-deficient cells compared with sufficient and supplemented cells (Fig. 1F, G). (See the Discussion for a role of LSD1 in the regulation of H3K9me3 repression marks.) These changes in epigenetic activation and repression marks were associated with albumin mRNA levels that were 200% higher in riboflavin-deficient cells compared with sufficient and supplemented cells (Fig. 1H).
Discussion
Although it was recognized 9 y ago that LSD1 is an FAD-dependent enzyme (9), this is, to the best of our knowledge, the first study to demonstrate the importance of riboflavin nutrition in the regulation of the H3K4me-dependent gene expression. Our studies provide unambiguous evidence that LSD1 expression and chromatin binding is higher in riboflavin-deficient liver cells compared with sufficient and supplemented cells. This upregulation was not sufficient to compensate for the depletion of the flavocoenzyme FAD and holo-LSD1 in riboflavin-deficient cells. Our studies suggest that the loss of LSD1 activity causes aberrant gene regulation in riboflavin-deficient cells, as assessed by the accumulation of H3K4me2 gene activation marks in the ALB promoter and increased ALB expression in riboflavin-deficient cells.
We also discovered that the loss of catalytically active LSD1 was associated with a 38% decrease in H3K9me3 repression marks in the ALB promoter. This observation can be ascribed to the observed upregulation of LSD1 expression and promoter binding in riboflavin-deficient cells, along the following lines of reasoning. Unambiguous evidence suggests that LSD1 recruits Jumonji C domain–containing protein 2 (JMJD2C) to gene promoters; JMJD2C has trimethyl lysine demethylase activity, does not depend on FAD as coenzyme, and may catalyze the demethylation of H3K9me3 (40). We propose that high concentrations of apo-LSD1 to the ALB promoter cause local accumulation of JMJD2C, which then leads to the erasure of H3K9me3 marks. Collectively, the gain of the gene activation mark H3K4me2 and the loss of the gene repression mark H3K9me3 synergize in the transcriptional activation of ALB in riboflavin-deficient cells.
Note that ALB was chosen as a model, because it is expressed in large quantities by liver cells, is responsible for ∼75% of plasma osmolarity, and hyperalbuminemia may lead to congestive heart failure and hypertension (41). Moreover, previous DNA microarray studies are consistent with aberrant gene regulation in riboflavin-deficient HepG2 cells (29).
The effects of riboflavin described in this study represent concentrations of riboflavin in the plasma of normal adults, spanning the range of moderately deficient pregnant women to users of over-the-counter riboflavin supplements (42, 43). It was not necessary to use physiologically irrelevant treatments such as zero riboflavin to elicit meaningful effects on LSD activity and gene regulation. We acknowledge the possibility that cells other than liver cells might be less susceptible to loss of flavin-dependent enzyme activities than liver cells. For example, previous studies suggest that HepG2 cells are more susceptible than human lymphoid Jurkat cells, when using FAD-dependent protein folding as marker (27, 32).
We propose that this study depicts the effects of riboflavin depletion on gene regulation by LSD1, rather than LSD2, based on the following rationale. Although both LSD1 and LSD2 may catalyze demethylation of H3K4me1/2 through an FAD-dependent reaction (44), previous studies suggest that LSD2 may repress genes through a mechanism independent of its demethylase activity (45), suggesting partial functionality of LSD2 in states of riboflavin deficiency. LSD2 binds to coding regions, as opposed to promoters, where it synergizes with euchromatic histone methyltransferases 1 and 2 (EHMT1/2) and the histone-lysine N-methyltransferase, nuclear SET domain-containing protein 3 (NSD3) in the regulation of transcriptional elongation (34).
A few uncertainties remain and warrant further investigation. First, we did not formally assess whether apo- and holo-LSD1 bind to the ALB promoter with similar affinity. Such studies will be feasible only when antibodies become available that distinguish between apo- and holo-LSD1. Second, histone demethylases other than LSD1 do not depend on FAD as a coenzyme and might rescue FAD-deficient cells regarding H3K4me2 demethylation events. Examples include the H3K4me2 demethylases JARID1A, JARID1B, JARID1C, and JARID1D, which belong to the Jumonji AT-rich interactive domain subfamily of Jumonji C domain–containing proteins (46–51).
Even though uncertainties remain, this study provides unambiguous evidence for a role of riboflavin nutrition in gene regulation by epigenetic mechanisms. Future studies will need to assess the extent by which moderate riboflavin deficiency causes aberrant gene regulation and disease in humans.
Supplementary Material
Acknowledgments
D.L. conducted the experiments, analyzed the data, and cowrote the manuscript; J.Z. designed the research, cowrote the manuscript, analyzed the data, and had primary responsibility for the final content. Both authors read and approved the final manuscript.
Footnotes
Abbreviations used: ALB, albumin gene; EHMT, euchromatic histone methyltransferase; H3K4me1, lysine-4-monomethylated histone H3; H3K4me2, lysine-4-dimethylated histone H3; H3K9me3, lysine-9-trimethylated histone H3; JARID, Jumonji AT-rich interactive domain; JMJD2C, Jumonji C domain-containing protein 2; LSD, lysine-specific demethylase; NSD3, nuclear SET domain-containing protein 3; REST, repressor element-1 silencing transcription factor.
References
- 1.Pinto JT, Rivlin RS. Riboflavin (vitamin B2). In: Zempleni J, Suttie JW, Gregory III JF, Stover PJ, editors. Handbook of vitamins. 5th ed. Boca Raton (FL): Taylor and Francis; 2013. p. 191–265. [Google Scholar]
- 2.Bornemann S. Flavoenzymes that catalyse reactions with no net redox change. Nat Prod Rep. 2002;19:761–72. [DOI] [PubMed] [Google Scholar]
- 3.Henriques BJ, Olsen RK, Bross P, Gomes CM. Emerging roles for riboflavin in functional rescue of mitochondrial beta-oxidation flavoenzymes. Curr Med Chem. 2010;17:3842–54. [DOI] [PubMed] [Google Scholar]
- 4.Janin J, Chothia C. Role of hydrophobicity in the binding of coenzymes. Appendix. Translational and rotational contribution to the free energy of dissociation. Biochemistry. 1978;17:2943–8. [DOI] [PubMed] [Google Scholar]
- 5.Chastain JL, McCormick DB. Clarification and quantitation of primary (tissue) and secondary (microbial) catabolites of riboflavin that are excreted in mammalian (rat) urine. J Nutr. 1987;117:468–75. [DOI] [PubMed] [Google Scholar]
- 6.Chastain JL, McCormick DB. Flavin catabolites: identification and quantitation in human urine. Am J Clin Nutr. 1987;46:830–4. [DOI] [PubMed] [Google Scholar]
- 7.Chastain JL, McCormick DB. Characterization of a new flavin metabolite from human urine. Biochim Biophys Acta. 1988;967:131–4. [DOI] [PubMed] [Google Scholar]
- 8.Zempleni J, Galloway JR, McCormick DB. The identification and kinetics of 7a-hydroxyriboflavin (7-hydroxymethylriboflavin) in blood plasma from humans following oral administration of riboflavin supplements. Int J Vitam Nutr Res. 1996;66:151–7. [PubMed] [Google Scholar]
- 9.Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–53. [DOI] [PubMed] [Google Scholar]
- 10.Culhane JC, Cole PA. LSD1 and the chemistry of histone demethylation. Curr Opin Chem Biol. 2007;11:561–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kouzarides T, Berger SL. Chromatin modifications and their mechanism of action. In: Allis CD, Jenuwein T, Reinberg D, editors. Epigenetics. Cold Spring Harbor (NY): Cold Spring Harbor Press; 2007. p. 191–209. [Google Scholar]
- 12.Forneris F, Binda C, Vanoni MA, Mattevi A, Battaglioli E. Histone demethylation catalysed by LSD1 is a flavin-dependent oxidative process. FEBS Lett. 2005;579:2203–7. [DOI] [PubMed] [Google Scholar]
- 13.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–37. [DOI] [PubMed] [Google Scholar]
- 14.Hakimi MA, Bochar DA, Chenoweth J, Lane WS, Mandel G, Shiekhattar R. A core-BRAF35 complex containing histone deacetylase mediates repression of neuronal-specific genes. Proc Natl Acad Sci USA. 2002;99:7420–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.You A, Tong JK, Grozinger CM, Schreiber SL. CoREST is an integral component of the CoREST—human histone deacetylase complex. Proc Natl Acad Sci USA. 2001;98:1454–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Humphrey GW, Wang Y, Russanova VR, Hirai T, Qin J, Nakatani Y, Howard BH. Stable histone deacetylase complexes distinguished by the presence of SANT domain proteins CoREST/kiaa0071 and Mta-L1. J Biol Chem. 2001;276:6817–24. [DOI] [PubMed] [Google Scholar]
- 17.Wang J, Scully K, Zhu X, Cai L, Zhang J, Prefontaine GG, Krones A, Ohgi KA, Zhu P, Garcia-Bassets I, et al. Opposing LSD1 complexes function in developmental gene activation and repression programmes. Nature. 2007;446:882–7. [DOI] [PubMed] [Google Scholar]
- 18.Wang J, Hevi S, Kurash JK, Lei H, Gay F, Bajko J, Su H, Sun W, Chang H, Xu G, et al. The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nat Genet. 2009;41:125–9. [DOI] [PubMed] [Google Scholar]
- 19.Pinto J, Huang YP, Rivlin RS. Mechanisms underlying the differential effects of ethanol on the bioavailability of riboflavin and flavin adenine dinucleotide. J Clin Invest. 1987;79:1343–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horvath R. Update on clinical aspects and treatment of selected vitamin-responsive disorders II (riboflavin and CoQ 10). J Inherit Metab Dis. 2012;35:679–87. [DOI] [PubMed] [Google Scholar]
- 21.Cimino JA, Jhangiani S, Schwartz E, Cooperman JM. Riboflavin metabolism in the hypothyroid human adult. Proc Soc Exp Biol Med. 1987;184:151–3. [DOI] [PubMed] [Google Scholar]
- 22.Pinto J, Huang YP, Rivlin RS. Inhibition of riboflavin metabolism in rat tissues by chlorpromazine, imipramine, and amitriptyline. J Clin Invest. 1981;67:1500–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dutta P, Raiczyk GB, Pinto J. Inhibition of riboflavin metabolism in cardiac and skeletal-muscles of rats by quinacrine and tetracycline. J Clin Biochem Nutr. 1988;4:203–8. [Google Scholar]
- 24.Beutler E. Effect of flavin compounds on glutathione reductase activity: in vivo and in vitro studies. J Clin Invest. 1969;48:1957–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beutler E. Glutathione reductase: stimulation in normal subjects by riboflavin supplementation. Science. 1969;165:613–5. [DOI] [PubMed] [Google Scholar]
- 26.National Research Council. Dietary reference intakes for thiamin, riboflavin, niacin, vitamin B6, folate, vitamin B12, pantothenic acid, biotin, and choline. Washington: National Academy Press; 1998. [PubMed] [Google Scholar]
- 27.Manthey KC, Chew YC, Zempleni J. Riboflavin deficiency impairs oxidative folding and secretion of apolipoprotein B-100 in HepG2 cells, triggering stress-response systems. J Nutr. 2005;135:978–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Werner R, Manthey KC, Griffin JB, Zempleni J. HepG2 cells develop signs of riboflavin deficiency within four days of culture in riboflavin-deficient medium. J Nutr Biochem. 2005;16:617–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manthey KC, Rodriguez-Melendez R, Hoi JT, Zempleni J. Riboflavin deficiency causes protein and DNA damage in HepG2 cells, triggering arrest in G1 phase of the cell cycle. J Nutr Biochem. 2006;17:250–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bernardi M, Maggioli C, Zaccherini G. Human albumin in the management of complications of liver cirrhosis. Crit Care. 2012;16:211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Camporeale G, Zempleni J. Oxidative folding of interleukin-2 is impaired in flavin-deficient Jurkat cells, causing intracellular accumulation of interleukin-2 and increased expression of stress response genes. J Nutr. 2003;133:668–72. [DOI] [PubMed] [Google Scholar]
- 32.Sauberlich HE, Judd JH, Nichoalds GE, Broquist HP, Darby WJ. Application of the erythrocyte glutathione reductase assay in evaluating riboflavin nutritional status in a high school student population. Am J Clin Nutr. 1972;25:756–62. [DOI] [PubMed] [Google Scholar]
- 33.Tietze F. Enzymic method for quantitative determination of nanogram amounts of total and oxidized glutathione: applications to mammalian blood and other tissues. Anal Biochem. 1969;27:502–22. [DOI] [PubMed] [Google Scholar]
- 34.Fang R, Barbera AJ, Xu Y, Rutenberg M, Leonor T, Bi Q, Lan F, Mei P, Yuan GC, Lian C, et al. Human LSD2/KDM1b/AOF1 regulates gene transcription by modulating intragenic H3K4me2 methylation. Mol Cell. 2010;39:222–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dahl JA, Collas P. MicroChIP—a rapid micro chromatin immunoprecipitation assay for small cell samples and biopsies. Nucleic Acids Res. 2008;36:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gralla M, Camporeale G, Zempleni J. Holocarboxylase synthetase regulates expression of biotin transporters by chromatin remodeling events at the SMVT locus. J Nutr Biochem. 2008;19:400–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pestinger V, Wijeratne SSK, Rodriguez-Melendez R, Zempleni J. Novel histone biotinylation marks are enriched in repeat regions and participate in repression of transcriptionally competent genes. J Nutr Biochem. 2011;22:328–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wei CL, Wu Q, Vega VB, Chiu KP, Ng P, Zhang T, Shahab A, Yong HC, Fu Y, Weng Z, et al. A global map of p53 transcription-factor binding sites in the human genome. Cell. 2006;124:207–19. [DOI] [PubMed] [Google Scholar]
- 39.SAS Institute. StatView reference. 3rd ed. Cary (NC): SAS Publishing; 1998. [Google Scholar]
- 40.Wissmann M, Yin N, Muller JM, Greschik H, Fodor BD, Jenuwein T, Vogler C, Schneider R, Gunther T, Buettner R, et al. Cooperative demethylation by JMJD2C and LSD1 promotes androgen receptor-dependent gene expression. Nat Cell Biol. 2007;9:347–53. [DOI] [PubMed] [Google Scholar]
- 41.Quinlan GJ, Martin GS, Evans TW. Albumin: biochemical properties and therapeutic potential. Hepatology. 2005;41:1211–9. [DOI] [PubMed] [Google Scholar]
- 42.Zempleni J, Link G, Bitsch I. Intrauterine vitamin B2 uptake of preterm and full-term infants. Pediatr Res. 1995;38:585–91. [DOI] [PubMed] [Google Scholar]
- 43.Zempleni J, Galloway JR, McCormick DB. Pharmacokinetics and utilization of orally and intravenously administered riboflavin in healthy humans. Am J Clin Nutr. 1996;63:54–66. [DOI] [PubMed] [Google Scholar]
- 44.Karytinos A, Forneris F, Profumo A, Ciossani G, Battaglioli E, Binda C, Mattevi A. A novel mammalian flavin-dependent histone demethylase. J Biol Chem. 2009;284:17775–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang Z, Jiang J, Stewart DM, Qi S, Yamane K, Li J, Zhang Y, Wong J. AOF1 is a histone H3K4 demethylase possessing demethylase activity-independent repression function. Cell Res. 2010;20:276–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Christensen J, Agger K, Cloos PA, Pasini D, Rose S, Sennels L, Rappsilber J, Hansen KH, Salcini AE, Helin K. RBP2 belongs to a family of demethylases, specific for tri-and dimethylated lysine 4 on histone 3. Cell. 2007;128:1063–76. [DOI] [PubMed] [Google Scholar]
- 47.Iwase S, Lan F, Bayliss P, de la Torre-Ubieta L, Huarte M, Qi HH, Whetstine JR, Bonni A, Roberts TM, Shi Y. The X-linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine 4 demethylases. Cell. 2007;128:1077–88. [DOI] [PubMed] [Google Scholar]
- 48.Klose RJ, Yan Q, Tothova Z, Yamane K, Erdjument-Bromage H, Tempst P, Gilliland DG, Zhang Y, Kaelin WG., Jr The retinoblastoma binding protein RBP2 is an H3K4 demethylase. Cell. 2007;128:889–900. [DOI] [PubMed] [Google Scholar]
- 49.Lee MG, Norman J, Shilatifard A, Shiekhattar R. Physical and functional association of a trimethyl H3K4 demethylase and Ring6a/MBLR, a polycomb-like protein. Cell. 2007;128:877–87. [DOI] [PubMed] [Google Scholar]
- 50.Tahiliani M, Mei P, Fang R, Leonor T, Rutenberg M, Shimizu F, Li J, Rao A, Shi Y. The histone H3K4 demethylase SMCX links REST target genes to X-linked mental retardation. Nature. 2007;447:601–5. [DOI] [PubMed] [Google Scholar]
- 51.Yamane K, Tateishi K, Klose RJ, Fang J, Fabrizio LA, Erdjument-Bromage H, Taylor-Papadimitriou J, Tempst P, Zhang Y. PLU-1 is an H3K4 demethylase involved in transcriptional repression and breast cancer cell proliferation. Mol Cell. 2007;25:801–12. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.