Abstract
Isolated mitochondria display a wide range of sizes plausibly resulting from the coexistence of subpopulations, some of which may be associated with disease or aging. Strategies to separate subpopulations are needed to study the importance of these organelles in cellular functions. Here, insulator-based dielectrophoresis (iDEP) was exploited to provide a new dimension of organelle separation. The dielectrophoretic properties of isolated Fischer 344 (F344) rat semimembranosus muscle mitochondria and C57BL/6 mouse hepatic mitochondria in low conductivity buffer (0.025–0.030 S/m) at physiological pH (7.2–7.4) were studied using polydimethylsiloxane (PDMS) microfluidic devices. First, direct current (DC) and alternating current (AC) of 0–50 kHz with potentials of 0–3000 V applied over a channel length of 1 cm were separately employed to generate inhomogeneous electric fields and establish that mitochondria exhibit negative DEP (nDEP). DEP trapping potential thresholds at 0–50 kHz were also determined to be weakly dependent on applied frequency and were generally above 200 V. Second, we demonstrated a separation scheme using DC potentials <100 V to perform the first size-based iDEP sorting of mitochondria. Samples of isolated mitochondria with heterogeneous sizes (150 nm–2 μm diameters) were successfully separated into sub-micron fractions, indicating the ability to isolate mitochondria into populations based on their size.
INTRODUCTION
The study of mitochondria is important due to the organelle's significant contribution to many cellular functions such as energy production, metabolism, cellular signaling, and apoptosis.1, 2, 3 Mitochondria have intrinsic heterogeneous sizes and morphologies suggesting the existence of mitochondrial subpopulations dividing into normal mitochondria of about 0.1–1 μm in diameter4, 5 and atypically sized giant mitochondria (>1 μm) which have been observed in various pathological states and models of aging.5 Therefore, a proper subcellular separation method to distinguish and harvest various sizes of mitochondria is needed to elucidate the role of this organelle in cellular function. Here, we propose a novel method to manipulate mitochondria, namely by their dielectrophoretic properties, in a microfluidic device. To accomplish this, we studied the dielectrophoretic migration of isolated mitochondria under DC and low frequency AC conditions and applied dielectrophoretic migration to fractionate mitochondria into various size groups using a microfluidic sorter.
Conventional methods to separate and prepare mitochondrial fractions are based on either size and density such as differential centrifugation,6, 7 density gradient centrifugation,7, 8, 9, 10 and flow field-flow fractionation,11 or charge-dependent methods such as free flow electrophoresis,10, 12, 13 isoelectric focusing,14 and capillary electrophoresis.15 In addition, immunomagnetic isolation of mitochondria has also been reported.16 However, these methods are mainly focused on isolating mitochondria from cell cytoplasm and few have considered the separation of mitochondrial subpopulations.17
Dielectrophoresis (DEP) is an electric field gradient-based technique allowing the manipulation of polarizable particles in an inhomogeneous electric field.18 In the case where DEP is the dominating force, particles can be trapped in regions with electric field gradients. In conjunction with other transport phenomena, concentration of particles into streams or separation can also occur.19, 20 DEP is governed by the polarization of a particle and the surrounding medium in which it is suspended. The arising DEP force, , on a spherical particle can be expressed as:21, 22
| (1) |
where R is particle radius, is medium permittivity, is particle permittivity, and is electric field. The component in Eq. 1 represents the Clausius-Mossotti factor (fCM) which governs the dielectrophoretic properties of suspended particles. When is less than , fCM is negative, thus negative DEP (nDEP) behavior is exhibited in which particles are repelled from regions of high . Conversely, when is greater than rendering fCM positive, an attraction to regions of high occurs and the corresponding migration and trapping behavior is termed positive DEP (pDEP).
Most commonly, traditional DEP applications focus on the use of electrodes to evoke electric field gradients,23, 24 as utilized with a variety of biological samples, namely cells25, 26 and tissue.27, 28, 29 Observed variations in the DEP response of such samples have led to techniques that can separate and distinguish cell lines such as various malignant or cancerous cells30, 31, 32 or detect and concentrate bacteria.33, 34 While the use of DEP for the characterization and analysis of organelles was originally suggested by Pohl18 a few decades ago, only one DEP-based application toward organelles was found in the literature in which mitochondria were purified from cell homogenates in an electrode-based DEP microfluidic device.35
While with electrode-based DEP the frequency dependence of the DEP response can be tested, the DC and low frequency DEP behavior of biological entities can also be examined with insulator-based DEP (iDEP).26, 36, 37, 38, 39, 40 In iDEP, electric field gradients are generated by integrating insulating structures within a microfluidic device. Upon application of a potential difference between the ends of such a device, the insulating structures (in our case posts) deviate electric field lines into an inhomogeneous distribution, creating electric field gradients at these locations. This method has the advantage of providing uniform electric field gradients spanning the entire microfluidic cross section, in addition to avoiding involved fabrication of embedded electrodes and reactions at electrodes within a microchannel.41
Here, we describe the manipulation of mitochondria by iDEP in two microfluidic devices. We investigated the iDEP migration and trapping of mitochondria under both DC and low frequency AC conditions, and the trapping potential thresholds under a range of AC fields and frequencies (0–50 kHz). Also, we demonstrate the first realization of iDEP size-based sorting of mitochondria as a potential precursor to isolating biologically relevant mitochondria of different sizes.
MATERIALS AND METHODS
Chemicals
For muscle isolation buffer, sucrose was purchased from MP Biomedicals (Solon, OH, USA), potassium chloride (KCl) and potassium phosphate (K2HPO4) were from Mallinckrodt (Paris, KY, USA), Tris-HCl was from Teknova (Hollister, CA, USA), ethylene glycol tetraacetic acid (EGTA) was from Amresco (Solon, OH, USA), and bovine serum albumin (BSA) was from Roche (Indianapolis, IN, USA). For liver isolation buffer, sucrose and EGTA were purchased from Sigma-Aldrich (St. Louis, MO, USA), 3-(N-morpholino)propanesulfonic acid (MOPS) was from Acros (Geel, Belgium), and Tris was from Fisher (Fair Lawn, NJ, USA). Nagarse and dimethyl sulfoxide (DMSO) were from Sigma-Aldrich. For buffers used in DEP experiments, 4-(2-hydroxyethyl)piperazine-1-ethanesulfonic acid (HEPES), poly(ethylene glycol)-block-poly(propylene glycol)-block-poly(ethylene glycol) (brand name Pluronic® F108), potassium hydroxide (KOH), and sucrose were purchased from Sigma-Aldrich. Deionized (DI) water was from a Synergy purification system (Millipore, USA). SYLGARD® 184 silicone elastomer kit for polydimethylsiloxane (PDMS) was obtained from Dow Corning Corporation (Midland, MI, USA). Glass slides of 0.15 mm thickness were purchased from Electron Microscopy Sciences (Gold-Seal coverslip; Hatfield, PA, USA) and platinum wire was purchased from Alfa Aesar (Ward Hill, MA, USA).
Microchip fabrication
A silicon master wafer patterned with the microfluidic structures was fabricated by photolithography as reported previously,42 followed by elastomer molding via soft lithography. Briefly, the PDMS silicon elastomer base and curing agent were mixed at a 10:1 ratio (w/w), poured onto the master wafer, degassed under vacuum, and cured in an oven for at least 4 h at 80 °C. The PDMS mold was then peeled off of the master wafer and 2 mm diameter reservoirs were manually punched at the channel ends. The PDMS mold and a glass slide were cleaned with isopropanol and distilled water in an ultrasonic bath, dried in a stream of nitrogen, and treated with oxygen plasma (PDC-001; Harrick Plasma, Ithaca, New York, USA) at high RF for 1 min. After treatment, the PDMS mold was irreversibly bonded to the glass slide to form a sealed microchannel system for all DC and AC experiments.
Mitochondria preparation and labeling
Mitochondria were prepared at the University of Minnesota according to procedures based on mechanical homogenization and differential centrifugation from the semimembranosus muscle of a Fischer 344 (F344) rat43, 44 (hereafter called muscle mitochondria) and from the liver of a C57BL/6 mouse45 (hereafter called hepatic or liver mitochondria). For all procedures, animals were housed in a central specific pathogen-free facility and were treated in an optimally ethical and humane fashion using protocols approved by the Institutional Animal Care and Use Committee.
For mitochondria prepared from rat semimembranosus muscle, the muscle was excised from an anesthetized animal and placed in ice-cold muscle isolation buffer (100 mM sucrose, 100 mM KCl, 50 mM Tris-HCl, 1 mM K2HPO4, 0.1 mM EGTA, 0.2% (w/v) BSA, adjusted to pH 7.4). All subsequent procedures for muscle and liver mitochondria isolation were performed on ice or at 4 °C unless otherwise noted. The sample was minced into small (∼1 mm) pieces and rinsed with muscle isolation buffer to remove blood. The pieces (∼1 g total) were transferred to 5 ml of muscle isolation buffer containing 0.2 mg/ml Nagarse (bacterial proteinase type XXIV) and incubated on ice for 1 min. The sample was then blended with an electric homogenizer (Tissue-Tearor, Biospec, Bartlesville, OK, USA) for three 20 s intervals. Muscle isolation buffer was added to a total volume of 9 ml and the sample was centrifuged at 700×g for 10 min. The pellet was discarded and this step was repeated. The supernatant was centrifuged at 10000×g for 10 min. The resulting pellet was washed twice (10000×g for 10 min) and resuspended in 1.2 ml of muscle isolation buffer containing 10% DMSO. Aliquots (50 μl each) were flash-frozen in a liquid nitrogen dewar.
For mitochondria prepared from mouse liver, the animal was anesthetized and the liver was excised. The liver was immersed in ice-cold liver isolation buffer (200 mM sucrose, 10 mM MOPS, 10 mM Tris, 1.0 mM EGTA, adjusted to pH 7.4). The liver was rinsed with liver isolation buffer and minced into small (∼1 mm) pieces. The pieces were rinsed, suspended in 5 ml of buffer, and transferred to a glass 15 ml Potter-Elvehjem homogenizer (Wheaton, Millville, NJ). The liver was homogenized with 3–5 strokes of a motor-driven Teflon pestle operated at 1600 RPM (Wheaton). The homogenate was centrifuged at 600×g for 10 min and the resulting pellet was discarded. The supernatant was centrifuged at 7000×g for 10 min, then the pellet was washed once with liver isolation buffer and centrifuged at 7000×g for 10 min. Mitochondria were resuspended in 4 ml of liver isolation buffer containing 10% DMSO, divided into four aliquots, and flash-frozen in a liquid nitrogen dewar. When needed, mitochondrial samples were transported on dry ice and otherwise stored in a nitrogen dewer. Samples were transferred to a −80 °C freezer one day prior to the experiment. Isolation buffer was stored at 4 °C and subjected to a 0.2 μm sterile filter before use.
Mitochondria were labeled at the time of experimentation with MitoTracker Green (Life Technologies, USA). 1 mM MitoTracker Green stock solution in DMSO was thawed to room temperature, diluted by isolation buffer, and added to the mitochondrial sample to reach a final concentration of 800 nM MitoTracker Green. The mitochondrial sample was incubated at 37 °C with gentle shaking (300 RPM) for 15 min, then centrifuged (10000×g) at 4 °C for 10 min followed by removal of the supernatant. The resulting mitochondria-containing pellet was then resuspended in Buffer B (250 mM sucrose, 250 μM or 1 mM F108, 10 mM HEPES, pH adjusted to 7.2–7.4 with KOH, σm = 0.025–0.030 S/m).
Microfluidic chip preparation
The following general preparation was followed for both microfluidic devices shown in Figures 14. After assembly, all microfluidic channels were immediately filled with Buffer A (500 μM or 1 mM F108, 10 mM HEPES, pH adjusted to 7.2–7.4 with KOH) by capillarity and the chip was placed in a humid environment overnight to ensure complete coating of F108. Buffer A was then removed by vacuum suction, and the channels were washed with Buffer B three times and refilled by adding Buffer B to the outlet reservoirs.
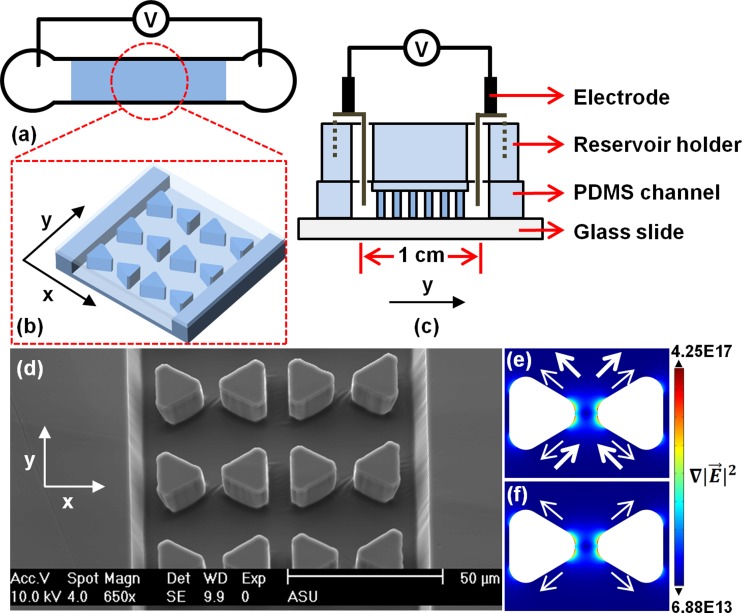
Figure 1.
(a)–(d) Schematics of the microchip (not to scale). (a) Channel top view with shaded area indicating the post region. (b) Zoom-in of the post region. (c) Channel side cross section with PDMS reservoir holder and integrated electrodes. (d) SEM image of the post region. (e) and (f) show numerical simulations of for a post pair based on a DC experiment where 3000 V is applied across a 1 cm long channel. Under DC conditions (e), small arrows indicate the nDEP force direction and large arrows indicate the electrokinetic forces resulting from electroosmosis and electrophoresis. Under AC conditions (f), only nDEP forces are prevalent. Note that arrow size does not represent the magnitude of the forces. For all experiments, the electric field is applied along the y axis.
Figure 4.
(a) Schematic of the microfluidic device employed to sort hepatic mitochondria. The device consists of one inlet channel (I) and five outlet channels (O: outer, MO: mid-outer, C: center) with reservoirs at all channel ends (not shown). The mitochondrial sample is injected into the inlet reservoir and transported through the device via electroosmosis. The zoomed in region shows the constriction area where inhomogeneous electric fields are generated, creating areas of high to induce DEP. Scale bar is 20 μm. (b) Fluorescence microscopy image of the constriction region during a mitochondria sorting experiment. Applied potentials are +60 V in I, −60 V in C, and 0 V in MO and O. As shown, large particles focus into the center outlet channel (C) due to greater nDEP forces repelling them from areas of high (Multimedia view) .
DEP characterization and trapping experiments were performed using the microfluidic device shown in Figure 1. For this device, a PDMS holder was employed to increase reservoir volume and provide stability for the electrodes. The holder was ∼ 0.5 cm thick, consisting of 5 mm diameter reservoirs that matched the channel design allowing it to be reversibly placed onto the assembled chip. The prepared mitochondrial sample was added to an inlet reservoir and mineral oil was added on top of both reservoirs to prevent evaporation. Platinum electrodes attached to the reservoirs were connected via micro-clamps (LabSmith, Livermore, CA, USA) to either a DC power supply (HVS448-6000D High Voltage Sequencer, LabSmith) commanded by Sequence software (version 1.150, LabSmith), or an AC power supply from a high voltage amplifier (AMT-3B20, Matsusada Precision Inc.) amplified from a Multifunction DAQ card (USB X Series, National Instruments, TX, USA) programmed by LabVIEW 2010 (version 10.0.1, National Instruments).
The microfluidic device shown in Figure 4 was employed for sorting experiments. Outlet reservoirs were filled with 5 μl of Buffer B prior to the experiment and at the beginning of the experiment, 5 μl of mitochondrial sample was injected into the inlet reservoir. Positive DC potentials ranging from +20 to +60 V were applied to the inlet and negative DC potentials ranging from −20 to −60 V were applied to the outlets. Sorting was performed over a duration of ∼1 h and solutions were extracted from all reservoirs for size analysis.
Detection and data analysis
Fluorescence images were acquired with an inverted microscope (IX71, Olympus, Center Valley, PA, USA) equipped with a 100 W mercury burner (U-RFL-T, Olympus) and fluorescence filter set (exciter ET470/40, dichroic T495LP, emitter ET525/50, Semrock, USA). A 60× (UPLSAPO60 × W, water immersion, NA = 1.20) or 100× (UPLSAPO100xO, oil immersion, NA = 1.40) objective was used to visualize mitochondrial migration in the microchannel. Images were captured at an interval of 100 or 150 ms using a CCD camera (for DC experiments: iXon X3, Andor Technology, Belfast, Northern Ireland; for AC experiments: QuantEM:512SC, Photometrics, Tucson, AZ, USA) and Micro-Manager software (version 1.4.7, Vale Lab, UCSF, CA, USA). The data were then analyzed with ImageJ software (version 1.47d, NIH). For sorting experiments, dynamic light scattering (DLS) (Spectro Size 302, Molecular Dimensions, UK) was used for size characterization of particle distributions in the inlet and each outlet. A 3 μl hanging droplet was set up in a 24-well crystallization plate and aligned to the DLS laser until a response signal was obtained. Each sample was subjected to 10 consecutive measurements lasting 20 s each. The results were combined into histograms of particle size distribution as well as signal intensity heat maps.
RESULTS AND DISCUSSION
The DEP behavior of muscle mitochondria and hepatic mitochondria was tested in the device depicted in Figure 1. The iDEP device consists of a 1 cm long linear channel, in which an array of triangular insulating posts is integrated. A dynamic coating with Pluronic® F108 was utilized to significantly reduce adsorption to channel walls as well as sample aggregation.46, 47, 48 Figures 1a, 1b, 1c show the device schematically and Figure 1d shows a scanning electron microscopy image of the post array in the PDMS channel. The PDMS mold was attached to a reservoir holder which also defined the volume of the reservoirs accessing the inlet and outlet. Mitochondrial iDEP was tested under both DC and AC conditions (up to 50 kHz). Figures 1e, 1f show the arising around the inward facing tips of two triangular posts representative of all post pairs in the channel. Additionally, arrows indicate the direction of electrokinesis and nDEP for DC (Figure 1e) and nDEP only in the case of AC (Figure 1f). From this simulation considering an fCM of −0.5, the acting on mitochondria with diameters of 150 nm to 2 μm ranges from −9.4 × 10−14 to −2.2 × 10−10 N at the post region (max= 1017 V2/m3) and −9.4 × 10−16 to −2.2 × 10−12 N away from the posts (max = 1015 V2/m3), a two orders of magnitude difference. The applied fCM of −0.5 was obtained according to a shell model21—derived mitochondrial conductivity of 6 × 10−5 S/m.
DC experiments
Mitochondrial DEP was studied under DC conditions at various electric field strengths (0–3000 V applied across a channel length of 1 cm). We chose a physiological pH and a low conductivity buffer similar to previous capillary electrophoresis studies on mitochondria.49 Generally, mitochondria were observed moving away from the inward facing tips of the posts where the highest electric field gradient regions are present, indicative of nDEP. An example of this behavior can be seen in Figure 2a which shows a fluorescence snapshot of muscle mitochondria under a DC field with a voltage-to-distance ratio of 3000 V/cm. Three typical mitochondria migration behaviors were observed and classified as “wiggling,” “trapping,” and “trap hopping.” We define wiggling as mitochondria moving in-between two adjacent post pairs along the flow direction. This behavior could result from a combination of several factors: (1) electroosmotic flow (EOF), which drives the mitochondria toward the cathode, (2) migration of mitochondria to the anode due to their negative electrophoretic mobility at physiological pH,49 and/or (3) nDEP forces pushing the mitochondria away from the inward facing post tips. Because the magnitude of the DEP force is directly proportional to organelle size and the electric field gradient, the mitochondria experience unbalanced forces at varying locations within the post array resulting in the wiggling behavior observed in the DC experiments.
Figure 2.
Behaviors of (a) muscle mitochondria under DC conditions at 3000 V and (b) hepatic mitochondria under AC conditions at 180 V and 50 kHz across the 1 cm long channel. Scale bar is 30 μm. One row of posts and the channel edges are outlined in each figure; the other rows of posts are indicated by stars. In (a), the thinnest arrows point at mitochondria wiggling in-between adjacent rows of posts, the medium thickness arrows point at those trapped at the edges of posts or away from the inward facing tips of the posts (nDEP trapping), and the thickest arrows point at those migrating in-between several rows of posts quickly (trap hopping). In (b), AC iDEP trapping is shown. Since electroosmotic flow is suppressed under AC conditions, mitochondria were attracted toward the edges of the posts immediately when voltage was applied (Multimedia view) .
The second phenomenon is trapping. When mitochondria were approaching a post pair, the nDEP force was large enough to repel them away to areas of lower along the edges of posts, effectively trapping them. The third phenomenon, trap hopping, is in principle similar to wiggling; however, the mitochondria also migrate perpendicularly to the flow direction. In this way, the mitochondria could escape high regions before being trapped and continue migrating longitudinally until they were randomly trapped by another post pair downstream. In real time, this appeared as though the mitochondria were hopping between post pair sections, hence the designated term. Figure 2a provides a snapshot of each of the three mitochondrial migration phenomena with arrows indicating the distinct migration mechanisms occurring.
AC experiments
AC frequencies ranging from 0 to 50 kHz were tested in the same chip under varying applied potentials. As expected, the DEP trapping behavior was pronounced since the electrokinetic component supporting the wiggling and trap hopping phenomena was suppressed. Upon the application of AC signal, the mitochondria were attracted to the edges of the posts where is low (Figure 2b), again indicating nDEP trapping in accordance with the DC experiments.
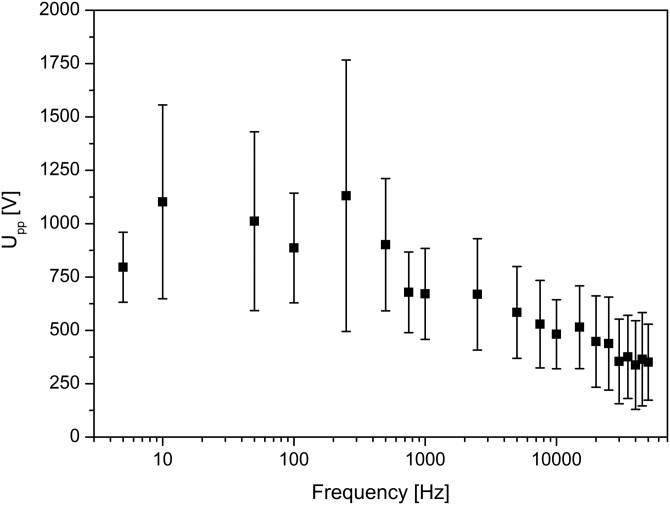
In addition, we studied the potential threshold at which the mitochondria began to exhibit significant trapping behavior, as demonstrated in Figure 3 for AC frequencies ranging from 0 to 50 kHz using seven devices with about 100 mitochondria per potential tested. The large variation observed at each frequency, shown as error bars, is expected from the natural size distribution of mitochondria in the sample. Consequently, Figure 3 shows that the trapping potential threshold for heterogeneous mitochondria is weakly dependent on frequency. Furthermore, the data were collected from highest to lowest frequency over the duration of each single experiment; therefore, a possible reason for the increasing error at low frequencies may be the longer exposure of the mitochondria to the electric field (while residing in the chip). This exposure could cause changes in their DEP properties over the course of the experiment and could also lead to mitochondria aggregation. Additionally, at low frequencies, EOF is not completely suppressed since a considerable DC component is acting in each half period of the periodic potential (as tested with an oscilloscope). This could also explain why the potential required to trap the mitochondria was somewhat higher at low frequencies compared to high frequencies.
Figure 3.
The dependence of trapping potential threshold on frequency for muscle mitochondria and hepatic mitochondria (n = 7). In general, mitochondria can be trapped at applied peak-to-peak potentials above 200 V across the 1 cm long channel and at frequencies between 0 and 50 kHz.
To elucidate the observed behavior for mitochondria, the DEP properties of 0.87 μm polystyrene beads were also studied as they have been demonstrated to be a good model particle for characterizing iDEP devices.36, 38, 40 With an estimated conductivity of 0.001 S/m,50 polystyrene beads are expected to show nDEP behavior40 under the same experimental conditions (σm = 0.025–0.030 S/m) used for the mitochondria. As such, these beads provided a reference for nDEP behavior. The results of these experiments, shown in the supplementary material (Figures S1 and S2),51 support the conclusion that mitochondria exhibit nDEP since the polystyrene beads resemble the migration behavior of the mitochondria using the same conditions and microfluidic device.
Additionally, we note that for both DC and AC experiments, currents never exceeded 10 μA, even at the largest potentials tested, and power dissipation is on the order of a few tens of mW. A considerable heating and temperature rise above 37 °C within the device are thus only expected for larger potentials applied (>100 V), such as examined by Chaurey et al.,52 and over long durations (several minutes to hours). If potentials are kept small, effects on mitochondria viability are expected to be marginal and other assays on viable mitochondria can be performed after iDEP experiments. Consequently, low applied potentials on the order of 100 V were used to test iDEP mitochondrial sorting into subpopulations. We also note that considerable pH gradients can occur in iDEP experiments; however, when the applied potentials are small and durations on the order of minutes are not exceeded, considerable pH variations are not expected.53
Mitochondria sorting by nDEP
Based on our study of mitochondrial iDEP, we investigated iDEP sorting of hepatic mitochondria using a microfluidic sorter previously developed for sorting nanobeads and protein nanocrystals by nDEP.40 A schematic of this device can be seen in Figure 4a in which six different microchannels (I: inlet, 2 × O: outer, 2 × MO: midouter, C: center) are employed for particle fractionation. Mitochondria of various sizes were introduced in the inlet reservoir (I) and DC potentials were applied to all channels to transport the organelle downstream by EOF. The zoomed in region shows a numerical simulation of at the constriction area connecting the inlet to the outlets where electric field lines converge to establish an inhomogeneous electric field inducing nDEP. Areas of high (red), three orders of magnitude larger than the wide inlet channel (blue), form and repel larger mitochondria with a greater from constriction walls, per Eq. 1. Consequently, the sorting principle is based on the varying magnitude of experienced by differently sized mitochondria flowing through the device.
As a proof of principle that this sorting mechanism could be applied to mitochondria, the sorter was set up with DC voltages of +20 to +60 V in I, −20 to −60 V in C, and 0 V in MO and O (low potentials were used to reduce Joule heating effects that could potentially damage the sample). The optimal potential scheme was determined to be −60 V I, +60 V C, and 0 V MO and O in which upon entering the constriction, large mitochondria experiencing a greater nDEP force focus into the center outlet channel (C), as shown in Figure 4b. Conversely, small mitochondria experiencing lesser nDEP forces are only marginally influenced by nDEP at the constriction and therefore can deflect into the O and MO outlet channels. Because small mitochondria can have a sub-micron size range, they are difficult to detect with optical microscopy as they are either below the resolution limit or have weak fluorescence emission indistinguishable from the background. Therefore, DLS was used to detect and quantify the size distribution of each fraction collected after ∼1 h of sorting at the optimized potentials (Figure 5).
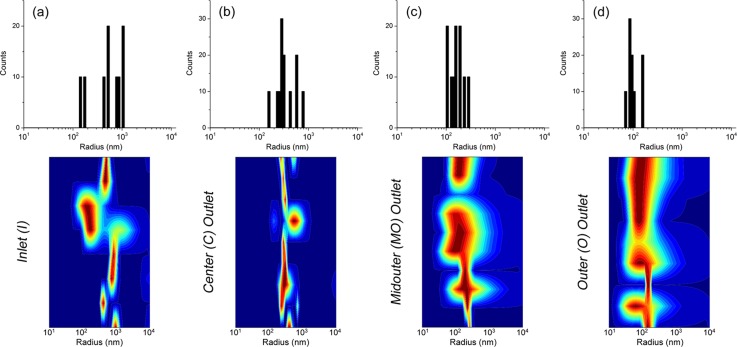
Figure 5.
Size analysis by dynamic light scattering of the sorted hepatic mitochondrial sample. The upper frame shows the histogram size distributions of detected particles and the lower frame shows the heat maps of scattering intensities with respect to particle size (blue = lowest, red = highest, y-axis spans the measurement time). (a) Size distribution of the injected inlet solution. A wide distribution (∼300 nm–2 μm diameters) is observed, qualifying the need for sorting. The smallest particles detected in other fractions are not seen due to their significantly lower scattering intensities. (b) Size distribution of the fraction collected from the C outlet which has the same wide distribution as the inlet since all sample particles flow here. (c) Size distribution of the MO fraction which is shifted towards a smaller particle size range of ∼200 –600 nm. (d) Size distribution of the O fraction showing a further shift towards an even smaller size range of ∼150–350 nm. As observed, the mitochondrial sample was fractionated into three size groups, decreasing from the C to O outlet channels.
In the upper half of the figure, histograms are shown illustrating the detected particle size distributions in each reservoir and in the lower half, scattering intensity heat maps with respect to particle size in each reservoir are shown. Figure 5a illustrates the expected large size distribution of the injected bulk mitochondrial sample ranging from ∼300 nm to 2 μm diameters. Because the entire sample, namely the larger mitochondria, focuses into the center outlet, a similar distribution is seen in Figure 5b. The smaller mitochondria ideally fractionate into the side outlet channels MO and O, which is shown in Figures 5c, 5d as both having sub-micron distributions. Surprisingly, an even more desirable effect is seen here as the MO fraction has a measured size range of ∼200–600 nm, whereas further fractionation occurs in O which was measured at ∼150–350 nm. In total, three fractions of decreasing size from C to O were obtained, indicating that the nDEP properties of mitochondria can be exploited for size-based sorting. Furthermore, from a typical loading of 5 μl of sample containing 106 mitochondria, analysis of the obtained fractions show that from a bulk mitochondrial preparation, large mitochondria can be separated from small mitochondria. For an outlet reservoir, the estimated recovery is ∼105 mitochondria per reservoir. This amount is sufficient for further analysis such as the measurement of membrane potentials,54 mitochondrial respiration,55 or proteomics studies.56
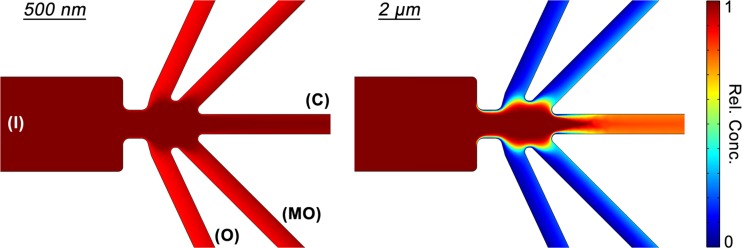
To help establish and support this optimized potential scheme, we also simulated the sorting experiments using Comsol Multiphysics with a numerical model considering DEP,38 EOF,38 and diffusion acting within an exact replica of the microfluidic device geometry (further detail on the physics and parameters considered in the simulation can be found in the supplementary material).51 This numerical method was successfully applied to the previously mentioned sorter developed to fractionate nanocrystals,40, 57 thus we employed the same physical model adapted to mitochondrial DEP parameters. The Transport of Diluted Species module was utilized to obtain relative concentration profiles (Figure 6, zoomed in to the constriction region) of two representative particle sizes, 500 nm and 2 μm diameters, with nDEP characteristics (fCM = −0.5). The resulting profile of the smaller 500 nm particles illustrates complete deflection (>90% rel. conc.) into all outlet reservoirs due to a weak influence of DEP. Under the same conditions, the corresponding profile of the larger 2 μm particles illustrates DEP driven focusing into the C outlet channel (>90% focused, <10% deflected). Consequently, these simulation results agree with expected migration trends.
Figure 6.
Simulated relative concentration profiles of the nDEP sorting mechanism supporting the optimized potential scheme for mitochondria sorting shown experimentally in Figure 4b (+60 V in I, −60 V in C, and 0 V in MO and O). Two representative particle sizes were studied (500 nm and 2 μm diameters). As expected, the larger particles focus into the C outlet, whereas the smaller particles deflect into the MO and O outlets, effectively sorting and isolating them.
CONCLUSION
In summary, we studied the DEP properties of isolated mitochondria from Fischer 344 (F344) rat semimembranosus muscle and C57BL/6 mouse liver and elucidated that they exhibit nDEP behavior. This was performed using a surface treated PDMS microfluidic device with DC (0–3000 V) and AC (0–50 kHz) electric fields applied across a 1 cm long channel containing media with a conductivity of 0.025–0.030 S/m at physiological pH. The trapping potential thresholds for mitochondria at the applied frequency range were generally above 200 V and were found to be weakly dependent on frequency. These conclusions were confirmed by a model study utilizing 0.87 μm polystyrene beads known to exhibit nDEP, since their behavior within the post array was in agreement with that of the mitochondria under the same conditions. Moreover, the DEP-based sorting experiment conducted at low DC potentials successfully sorted mitochondria into various size fractions. The size range investigated is highly compatible with separations of “giant” mitochondria observed under conditions of disease and aging.5 To the best of our knowledge, this is the first time that iDEP sorting of an organelle type, specifically mitochondria, has been demonstrated. Additionally, we emphasize that the amount of sorted mitochondria is suitable for most ensuing studies and negative effects due to iDEP manipulation are greatly suppressed due to the low potentials applied in the sorting device. Overall, this study provides important information about mitochondria under both DC and AC conditions, contributing to a foundation for future development of mitochondrial subpopulation separation and applications to investigate their role in aging and disease.
ACKNOWLEDGMENTS
We thank Dr. Fernanda Camacho-Alanis in the Chemistry and Biochemistry Department at Arizona State University for her help with SEM imaging. G.G.W. and E.A.A. acknowledge funding from the National Institutes of Health (Grant No. ROI-AG020866). G.G.W. was supported in part by NIH Training Grant No. T32-AG029796. B.G.A. and A.R. acknowledge funding from the National Institutes of Health (Grant No. R01-GM095583) and fruitful discussions with Dr. Petra Fromme at the Chemistry and Biochemistry Department at Arizona State University.
Paper submitted as part of a special collection covering contributions related to the American Electrophoresis Society's symposium at the SciX 2013 meeting (Guest Editors: A. Ros, E. D. Goluch) held in Milwaukee, Wisconsin, September 29–October 4, 2013.
References
- Newmeyer D. D. and Ferguson-Miller S., Cell 112, 481 (2003). 10.1016/S0092-8674(03)00116-8 [DOI] [PubMed] [Google Scholar]
- McBride H. M., Neuspiel M., and Wasiak S., Curr. Biol. 16, R551 (2006). 10.1016/j.cub.2006.06.054 [DOI] [PubMed] [Google Scholar]
- Abou-Sleiman P. M., Muqit M. M. K., and Wood N. W., Nat. Rev. Neurosci. 7, 207 (2006). 10.1038/nrn1868 [DOI] [PubMed] [Google Scholar]
- Brown T. A., Fetter R. D., Tkachuk A. N., and Clayton D. A., Methods 51, 458 (2010). 10.1016/j.ymeth.2010.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navratil M., Terman A., and Arriaga E. A., Exp. Cell Res. 314, 164 (2008). 10.1016/j.yexcr.2007.09.013 [DOI] [PubMed] [Google Scholar]
- Fernandez-Vizarra E., Lopez-Perez M. J., and Enriquez J. A., Methods 26, 292 (2002). 10.1016/S1046-2023(02)00034-8 [DOI] [PubMed] [Google Scholar]
- Michelsen U. and von Hagen J., Methods Enzymol. 463, 305 (2009). 10.1016/S0076-6879(09)63019-6 [DOI] [PubMed] [Google Scholar]
- Storrie B. and Madden E. A., Methods Enzymol. 182, 203 (1990). 10.1016/0076-6879(90)82018-W [DOI] [PubMed] [Google Scholar]
- Strack A., Duffy C. F., Malvey M., and Arriaga E. A., Anal. Biochem. 294, 141 (2001). 10.1006/abio.2001.5148 [DOI] [PubMed] [Google Scholar]
- Hartwig S., Feckler C., Lehr S., Wallbrecht K., Wolgast H., Mueller-Wieland D., and Kotzka J., Proteomics 9, 3209 (2009). 10.1002/pmic.200800344 [DOI] [PubMed] [Google Scholar]
- Kang D., Oh S., Reschiglian P., and Moon M. H., Analyst 133, 505 (2008). 10.1039/b716851a [DOI] [PubMed] [Google Scholar]
- Pasquali C., Fialka I., and Huber L. A., J. Chromatogr. B: Biomed. Sci. Appl. 722, 89 (1999). 10.1016/S0378-4347(98)00314-4 [DOI] [PubMed] [Google Scholar]
- Eubel H., Lee C. P., Kuo J., Meyer E. H., Taylor N. L., and Millar A. H., Plant J. 52, 583 (2007). 10.1111/j.1365-313X.2007.03253.x [DOI] [PubMed] [Google Scholar]
- Lu H., Gaudet S., Schmidt M. A., and Jensen K. F., Anal. Chem. 76, 5705 (2004). 10.1021/ac049794g [DOI] [PubMed] [Google Scholar]
- Johnson R. D., Navratil M., Poe B. G., Xiong G., Olson K. J., Ahmadzadeh H., Andreyev D., Duffy C. F., and Arriaga E. A., Anal. Bioanal. Chem. 387, 107 (2007). 10.1007/s00216-006-0689-6 [DOI] [PubMed] [Google Scholar]
- Hornig-Do H.-T., Guenther G., Bust M., Lehnartz P., Bosio A., and Wiesner R. J., Anal. Biochem. 389, 1 (2009). 10.1016/j.ab.2009.02.040 [DOI] [PubMed] [Google Scholar]
- Kostal V., Fonslow B. R., Arriaga E. A., and Bowser M. T., Anal. Chem. 81, 9267 (2009). 10.1021/ac901508x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohl H. A., Bull. Am. Phys. Soc. 15, 1362 (1970). [Google Scholar]
- Nakano A., Chao T.-C., Camacho-Alanis F., and Ros A., Electrophoresis 32, 2314 (2011). 10.1002/elps.201100037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho-Alanis F., Gan L., and Ros A., Sens. Actuators, B 173, 668 (2012). 10.1016/j.snb.2012.07.080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones T. B., Electromechanics of Particles (Cambridge University Press, New York, USA, 1995). [Google Scholar]
- Jones T. B., IEEE Eng. Med. Biol. Mag. 22, 33 (2003). 10.1109/MEMB.2003.1304999 [DOI] [PubMed] [Google Scholar]
- Asbury C. L., Diercks A. H., and van den Engh G., Electrophoresis 23, 2658 (2002). [DOI] [PubMed] [Google Scholar]
- Hughes M. P., Electrophoresis 23, 2569 (2002). [DOI] [PubMed] [Google Scholar]
- Pethig R., Menachery A., Pells S., and De Sousa P., J. Biomed. Biotechnol. 2010, 182581 (2010). 10.1155/2010/182581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnon Z. R., Electrophoresis 32, 2466 (2011). 10.1002/elps.201100060 [DOI] [PubMed] [Google Scholar]
- Ho C.-T., Lin R.-Z., Chang W.-Y., Chang H.-Y., and Liu C.-H., Lab Chip 6, 724 (2006). 10.1039/b602036d [DOI] [PubMed] [Google Scholar]
- Sebastian A., Buckle A., and Markx G. H., Biotechnol. Bioeng. 98, 694 (2007). 10.1002/bit.21416 [DOI] [PubMed] [Google Scholar]
- Yang M. and Zhang X., Sens. Actuators, A 135, 73 (2007). 10.1016/j.sna.2006.06.071 [DOI] [Google Scholar]
- Becker F. F., Wang X.-B., Huang Y., Pethig R., Vykoukal J., and Gascoyne P. R. C., Proc. Natl. Acad. Sci. U.S.A. 92, 860 (1995). 10.1073/pnas.92.3.860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascoyne P., Satayavivad J., and Ruchirawat M., Acta Trop. 89, 357 (2004). 10.1016/j.actatropica.2003.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascoyne P. R. C., Noshari J., Anderson T. J., and Becker F. F., Electrophoresis 30, 1388 (2009). 10.1002/elps.200800373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu K., Kaprelyants A. S., Salina E. G., and Markx G. H., Biomicrofluidics 4, 022809 (2010). 10.1063/1.3435335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Carmen Jaramillo M., Torrents E., Martinez-Duarte R., Madou M. J., and Juarez A., Electrophoresis 31, 2921 (2010). 10.1002/elps.201000082 [DOI] [PubMed] [Google Scholar]
- Moschallski M., Hausmann M., Posch A., Paulus A., Kunz N., Duong T. T., Angres B., Fuchsberger K., Steuer H., Stoll D., Werner S., Hagmeyer B., and Stelzle M., Electrophoresis 31, 2655 (2010). 10.1002/elps.201000097 [DOI] [PubMed] [Google Scholar]
- Lapizco-Encinas B. H., Simmons B. A., Cummings E. B., and Fintschenko Y., Anal. Chem. 76, 1571 (2004). 10.1021/ac034804j [DOI] [PubMed] [Google Scholar]
- Lapizco-Encinas B. H., Simmons B. A., Cummings E. B., and Fintschenko Y., Electrophoresis 25, 1695 (2004). 10.1002/elps.200405899 [DOI] [PubMed] [Google Scholar]
- Bhattacharya S., Chao T.-C., and Ros A., Electrophoresis 32, 2550 (2011). 10.1002/elps.201100066 [DOI] [PubMed] [Google Scholar]
- Braff W. A., Pignier A., and Buie C. R., Lab Chip 12, 1327 (2012). 10.1039/c2lc21212a [DOI] [PubMed] [Google Scholar]
- Abdallah B. G., Chao T.-C., Kupitz C., Fromme P., and Ros A., ACS Nano 7, 9129 (2013). 10.1021/nn403760q [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano A. and Ros A., Electrophoresis 34, 1085 (2013). 10.1002/elps.201200482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano A., Camacho-Alanis F., Chao T.-C., and Ros A., Biomicrofluidics 6, 034108 (2012). 10.1063/1.4742695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya S. K., Thakar J. H., Johnson P. L., and Shanklin D. R., Anal. Biochem. 192, 344 (1991). 10.1016/0003-2697(91)90546-6 [DOI] [PubMed] [Google Scholar]
- Madsen K., Ertbjerg P., and Pedersen P. K., Anal. Biochem. 237, 37 (1996). 10.1006/abio.1996.0197 [DOI] [PubMed] [Google Scholar]
- Frezza C., Cipolat S., and Scorrano L., Nat. Protoc. 2, 287 (2007). 10.1038/nprot.2006.478 [DOI] [PubMed] [Google Scholar]
- Hellmich W., Regtmeier J., Duong T. T., Ros R., Anselmetti D., and Ros A., Langmuir 21, 7551 (2005). 10.1021/la0510432 [DOI] [PubMed] [Google Scholar]
- Viefhues M., Manchanda S., Chao T.-C., Anselmetti D., Regtmeier J., and Ros A., Anal. Bioanal. Chem. 401, 2113 (2011). 10.1007/s00216-011-5301-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greif D., Galla L., Ros A., and Anselmetti D., J. Chromatogr. A 1206, 83 (2008). 10.1016/j.chroma.2008.07.013 [DOI] [PubMed] [Google Scholar]
- Fuller K. M. and Arriaga E. A., J. Chromatogr. B: Anal. Technol. Biomed. Life Sci. 806, 151 (2004). 10.1016/j.jchromb.2004.03.050 [DOI] [PubMed] [Google Scholar]
- Zhang C., Khoshmanesh K., Tovar-Lopez F. J., Mitchell A., Wlodarski W., and Klantar-zadeh K., Microfluid. Nanofluid. 7, 633 (2009). 10.1007/s10404-009-0419-4 [DOI] [Google Scholar]
- See supplementary material at http://dx.doi.org/10.1063/1.4866852 for further information on the polystyrene bead experiments using DC and AC conditions (Figures S1 and S2), and the physics and parameters considered in the simulation for iDEP sorting.
- Chaurey V., Rohani A., Su Y.-H., Liao K.-T., Chou C.-F., and Swami N. S., Electrophoresis 34, 1097 (2013). 10.1002/elps.201200456 [DOI] [PubMed] [Google Scholar]
- Gencoglu A., Camacho-Alanis F., Nguyen V. T., Nakano A., Ros A., and Minerick A. R., Electrophoresis 32, 2436 (2011). 10.1002/elps.201100090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim T.-S., Davila A., Wallace D. C., and Burke P., Lab Chip 10, 1683 (2010). 10.1039/c001818j [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers G. W., Brand M. D., Petrosyan S., Ashok D., Elorza A. A., Ferrick D. A., and Murphy A. N., PLoS One 6, e21746 (2011). 10.1371/journal.pone.0021746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Zhu G., Li Y., Yang P., and Dovichi N. J., Anal. Chem. 84, 8715 (2012). 10.1021/ac3019608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdallah B. G., Chao T.-C., Fromme P., and Ros A., in 16th μTAS Proceedings, Okinawa, Japan, 28 October-1 November 2012, pp. 458–460.