Abstract
Cohesins are evolutionarily conserved essential multi-protein complexes important for higher-order chromatin organization. They play pivotal roles in the maintenance of genome integrity through mitotic chromosome regulation, DNA repair and replication, as well as gene regulation critical for proper development and cellular differentiation. In this review, we will discuss the multifaceted functions of mammalian cohesins and their apparent functional hierarchy in the cell, with particular focus on their actions in gene regulation and their relevance to human developmental disorders.
Keywords: cohesin, NIPBL, CTCF, HP1, chromatin, CdLS
Cohesins are essential complexes for sister chromatid cohesion
Cohesins are required for sister chromatid cohesion and equal chromosome segregation in mitosis (Hirano, 2006, Nasmyth and Haering, 2009, Wood et al., 2010). While many fundamental functional and mechanistic discoveries were made in lower eukaryotes, the basic cohesin functions appear to be conserved in mammalian cells (Hoque and Ishikawa, 2002, Schmiesing et al., 1998, Hauf et al., 2001, Losada et al., 2005) with some differences in their regulation (see below). Cohesins contain a stable heterodimer of two Structural Maintenance of Chromosomes (SMC) family proteins (SMC1 and SMC3) and the associated non-SMC components Rad21 (Scc1/Mcd1) and SA1/SA2 (Scc3, stromalin, or STAG1/STAG2) (Fig. 1A) (Hirano, 2000, Nasmyth and Haering, 2009). The SMC1 and SMC3 subunits are bound at their hinge domains in a V-shaped configuration, while their ATPase heads are linked by the kleisin protein Rad21. Since biochemical and high-resolution microscopic analyses revealed that cohesins form a ring structure, it was hypothesized that cohesins trap two sister chromatids within the ring to hold them together (Fig. 1B) (Haering et al., 2002, Anderson et al., 2002). This is supported by evidence that cleavage of a cohesin subunit leads to loss of sister chromatid cohesion in vitro and in vivo, while covalent closure of the cohesin ring resulted in denaturation-resistant minichromosome concatenation (Gruber et al., 2003, Uhlmann et al., 2000, Haering et al., 2008, Ivanov and Nasmyth, 2007). SMC proteins have conserved ATPase motifs (Fig. 1C), and ATP binding and hydrolysis by SMC proteins was shown to be important for the complexes’ functions (Arumugam et al., 2003, Weitzer et al., 2003, Nasmyth and Haering, 2009). While a single Scc3 is present in yeast, two SA proteins, SA1 and SA2, are found in higher eukaryotes to form two distinct cohesin complexes (Losada et al., 2000, Sumara et al., 2000). Although both contribute to genome-wide sister chromatid cohesion (Losada et al., 2000), recent studies provided evidence that cohesinSA1 is particularly important for telomeric sister chromatid cohesion by its selective recruitment through the interaction of SA1 with telomere proteins (Canudas et al., 2007). For the remainder of this review, we will refer to both complexes as “cohesin”.
Fig. 1. Human cohesin.
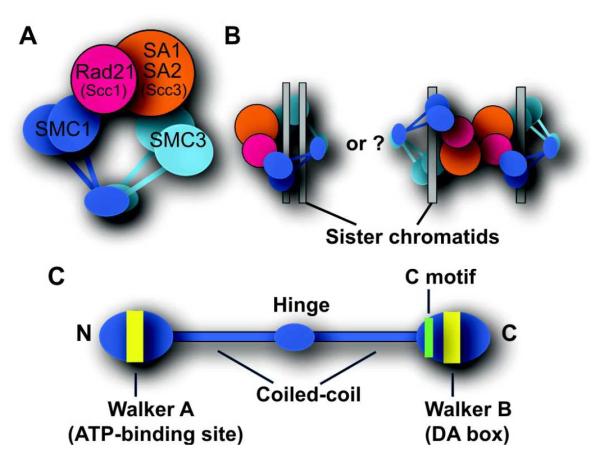
A. Human cohesin complex. B. Proposed models for chromatin trapping (cohesion) by cohesin. C. Structure of an SMC family protein.
Cell cycle-specific regulation of cohesin
Cohesin binding to chromatin and its function in sister chromatid cohesion are regulated in a cell cycle-specific manner (for detailed reviews, see Carretero et al., 2010, Nasmyth and Haering, 2009, Shintomi and Hirano, 2010). The loading of cohesin onto chromatin occurs in telophase in mammalian cells (as compared to the end of G1 phase in yeast (Ciosk et al., 2000)). This is mediated by the cohesin loading factor NIPBL (Scc2 or delangin) with its binding partner MAU-2 (Scc4), though the underlying mechanism remains undefined (Seitan et al., 2006, Watrin et al., 2006). The establishment of sister chromatid cohesion occurs during S phase, which requires additional factors including the yeast Eco1-related acetyltransferases ESCO1/ESCO2, sororin, and Pds5. ESCOs and sororin antagonize the cohesin destabilizing Wapl which interacts with Pds5 (Nishiyama et al., 2010, Terret et al., 2009). The cohesin subunit SMC3 is the target of ESCO acetyltransferases in both yeast and humans (Rolef Ben-Shahar et al., 2008, Ünal et al., 2008, Zhang et al., 2008). SMC3 acetylation appears to weaken the interaction of the Wapl-Pds5 complex with cohesin (Sutani et al., 2009) as well as promoting the binding of sororin to cohesin (Nishiyama et al., 2010). However, the exact mechanism of cohesion establishment is still under active investigation. In vertebrates, cohesin is removed from chromosomes in a two-step process in mitosis (Losada et al., 2000, Sumara et al., 2000, Waizenegger et al., 2000). The majority of cohesin dissociates from chromosomes in the first step (i.e., the “prophase pathway”), which is at least in part regulated by polo-like kinase through phosphorylation of SA2 (Waizenegger et al., 2000, Sumara et al., 2002, Hauf et al., 2005). Shugoshin (Sgo) associating with serine/threonine protein phosphatase 2A (PP2A) protects a small population of cohesin from phosphorylation at the centromeres and maintains centromeric cohesion during metaphase, which is important for proper spindle attachment and subsequent segregation of chromosomes (Kitajima et al., 2006, Riedel et al., 2006). At the end of metaphase, the protease separase, activated indirectly by the anaphase promoting complex, cleaves the cohesin subunit Rad21, which triggers separation of sister chromatids in anaphase (Uhlmann et al., 1999, Uhlmann et al., 2000, Hauf et al., 2001, Waizenegger et al., 2000). This constitutes the second step of cohesin removal from mitotic chromosomes. Cohesin is then reloaded onto chromosomes in telophase (in mammalian cells) for the next cell cycle.
Cohesin’s role in gene regulation
Evidence from multiple organisms indicates that cohesin has additional roles in gene regulation, in particular, cell type-specific developmental gene regulation (see (Dorsett, 2011). Studies now indicate that cohesin functions in gene regulation in at least three different contexts, which are summarized below.
(1) Insulator function of cohesin
In mammalian cells, more than 70% of cohesin sites were found to colocalize with CCCTC-binding factor (CTCF) binding sites genome-wide (Wendt et al., 2008, Stedman et al., 2008, Parelho et al., 2008, Rubio et al., 2008). CTCF is a zinc finger DNA-binding protein that acts as a transcriptional activator/repressor as well as an insulator binding protein blocking enhancer-promoter interaction (Zlatanova and Caiafa, 2009, Wallace and Felsenfeld, 2007). Depletion of cohesin compromised CTCF-mediated insulator function at the apolipoprotein gene cluster and at the c-myc, Igf2/H19, and interferon-γ (IFNγ) loci (Wendt et al., 2008, Parelho et al., 2008, Hadjur et al., 2009, Mishiro et al., 2009, Nativio et al., 2009). In addition, cohesin and CTCF at intragenic chromatin boundaries dictate the expression of the PUMA (p53 up-regulated modulator of apoptosis) gene by promoting the transcription of non-coding RNA (ncRNA) corresponding to the first half of the gene locus, which suppresses the transcription of the full-length PUMA transcript (Gomes and Espinosa, 2010). Cohesin loading onto chromatin does not require CTCF, but CTCF is necessary for the specific binding of cohesin to CTCF motifs (Wendt et al., 2008, Parelho et al., 2008). These breakthrough studies indicated that cohesin functions at insulator/boundary elements together with CTCF in mammalian cells.
(2) Cohesin in gene activation
In addition to the insulator function, cohesin appears to also function in gene activation. In Drosophila, both Nipped-B and cohesin preferentially localize to transcribed regions and coincide with RNA polymerase II (pol II) (Misulovin et al., 2008), and the expression of many cohesin-bound genes decreases following cohesin depletion (Schaaf et al., 2009). Similarly, in mouse embryonic stem cells (mESCs), there is significant overlap between the binding of cohesin, Nipbl and the Mediator complex, which associates with pol II, in the enhancer and promoter regions of active genes (Kagey et al., 2010). Cohesin or Nipbl depletion downregulates the expression of many of these genes. Thus, on a genome-wide level, there is a significant association of cohesin with gene activation.
In terms of individual genes, genetic analysis suggested that both the cohesin subunit Rad21 and Nipped-B promote hedgehog (hh) gene expression in Drosophila (Hallson et al., 2008). Cohesin also binds to and promotes several ecdysone-response genes in Drosophila salivary glands (Pauli et al., 2010). In zebrafish, cohesin is required for the hematopoietic expression of runx genes, though whether the effect is direct has not been established (Horsfield et al., 2007). Zebrafish embryos null for Rad21 displayed misregulation of multiple gene networks that include myca (c-myc), p53, and mdm2 where cohesin binding was detected (Rhodes et al., 2010). Importantly, Rad21 depletion reduced myca expression while inducing p53 and mdm2 expression (Rhodes et al., 2010). A similar correlation between cohesin binding and c-myc expression was observed in Drosophila and in mammalian cells (Kawauchi et al., 2009, Liu et al., 2009, Rubio et al., 2008, Schaaf et al., 2009, Stedman et al., 2008). Thus, c-myc appears to be a prominent target gene that requires cohesin for its expression in higher eukaryotes.
Interestingly, although cohesin was found at CTCF consensus sites at the c-myc locus, CTCF depletion did not have the same effect as cohesin on c-myc expression, suggesting a distinct function of cohesin in gene expression (Rhodes et al., 2010). In mESCs, Mediator, cohesin and Nipbl, but not CTCF, bind to and promote the expression of ESC transcription factor genes (Oct4, Nanog, and Sox2) (Kagey et al., 2010), further indicating the role of cohesin in gene activation in a CTCF-independent context.
(3) Cohesin in gene silencing and heterochromatin organization
In addition to gene activation, cohesin may play a role in gene silencing. A recent study reported the interaction of NIPBL with histone deacetylases (HDACs), suggesting its role in deacetylation of histones and transcriptional silencing (Jahnke et al., 2008). In Drosophila neural cells, both cohesin and Nipped-B, along with histone H3 lysine 27 tri-methylation (H3K27me3), simultaneously coat the entire Enhancer of split and invected-engrailed gene complexes, where cohesin appears to contribute to “restrained” transcription as depletion of cohesin led to upregulation of these genes (Schaaf et al., 2009). H3K27me3 is mediated by polycomb repressor complex 2 (PRC2), which in turn recruits PRC1 (Cao et al., 2005, Cao et al., 2002). Thus, the study suggests some functional interaction between cohesin and the polycomb silencing pathway. Indeed, cohesin associates with a subset of the polycomb group (PcG) complexes in Drosophila embryos (Strübbe et al., 2011).
Cohesin was also found to associate with heterochromatin marked with H3 lysine 9 methylation (H3K9me). In S. pombe, Swi6 (a heterochromatin binding protein 1 (HP1) homolog) was found to be required for cohesin binding to pericentromeric heterochromatin (Bernard et al., 2001, Nonaka et al., 2002).Swi6 recognizes the methylated lysine 9 on the histone H3 tail and is important for the maintenance of heterochromatin structure and gene silencing (Nakayama et al., 2001). However, in this context, cohesin is important for proper centromeric sister chromatid cohesion, and does not appear to play any role in either Swi6 recruitment or gene silencing (Nonaka et al., 2002). Also, whether this pathway is conserved at mammalian centromeres (H3K9me-HP1-cohesin) has been controversial (Koch et al., 2008, Serrano et al., 2009). Interestingly, however, we found that cohesin specifically binds to a subtelomeric macrosatellite repeat sequence termed D4Z4 on chromosome 4q in human cells in a H3K9me3-dependent and CTCF-independent manner (Zeng et al., 2009). We demonstrated that one of the HP1 variants, HP1γ, and cohesin require each other to bind to D4Z4. This provided the first example that cohesin is actively involved in heterochromatin organization (by recruiting HP1) to H3K9me-marked heterochromatin. Interestingly, H3K9me3, HP1γ, and cohesin are specifically lost from D4Z4 in a muscular dystrophy called facioscapulohumeral dystrophy (FSHD) (Zeng et al., 2009). We proposed that cohesin-containing heterochromatin contributes to gene silencing, and its disruption results in abnormal gene expression in muscle cells that precipitates the clinical features of FSHD (Zeng et al., 2009). Indeed, a recent study indicated that a gene encoded in the D4Z4 repeat is upregulated in FSHD, which may be triggered by this heterochromatin loss (Lemmers et al., 2010). A similar role of cohesin in subtelomeric heterochromatin was recently reported in S. pombe where mutation of Rad21 phenocopied the Swi6 loss of function in heterochromatin organization and gene silencing (Dheur et al., 2011). Taken together, cohesin appears to be actively involved in heterochromatin organization and gene silencing in the context of the H3K9me-HP1 pathway.
Mechanism of cohesin recruitment to different genomic regions
How does cohesin mediate the distinct transcriptional functions described above and how does it differ from cohesin binding for sister chromatid cohesion? Cohesin appears to have little DNA sequence binding specificity on its own in vitro (Losada and Hirano, 2001) and the mechanism of its association with chromosomes in vivo is still under active investigation.
(1) Relationship with RNA transcription
Using chromatin immunoprecipitation (ChIP) assays, cohesin in S. cerevisiae was shown to bind to chromosome arms at 9-13 kb intervals with some preference for intergenic AT-rich regions, which appear to be at sites of transcriptional convergence (Laloraya et al., 2000, Blat and Kleckner, 1999, Lengronne et al., 2004, Tanaka et al., 1999, Glynn et al., 2004). It was postulated that cohesin is literally pushed by the transcription machinery to allow RNA transcription (Bausch et al., 2007, Lengronne et al., 2004, Glynn et al., 2004). This, however, does not appear to be conserved in higher eukaryotes. Cohesin was found to bind to transcribed regions co-localizing with RNA polymerase II in Drosophila and mESCs (Misulovin et al., 2008, Kagey et al., 2010) (Fig. 2A).
Fig. 2. Factors involved in cohesin recruitment to specific genomic regions.
A. Binding of RNA polymerase II, Mediator, and sequence-specific transcription factors (TFs), such as ERα, correlates with cohesin binding. B. Pre-replication complex (pre-RC), including the MCM complex, and cohesin binding to replication origins. C. ATP-dependent chromatin remodeling complex hSNF2h binds to cohesin and recruits it to a type of Alu repeat sequence in human cells. D. CTCF-dependent recruitment of cohesin to the insulator/boundary elements containing CTCF consensus binding sites. Additional factors, such as p68/SRA and ATRX, may dictate their binding selectivity. E. CTCF-independent cohesin recruitment to heterochromatic repeat sequences carrying histone H3K9me3 and HP1. At the pericentromeric heterochromatin in S. pombe, HP1 recruits cohesin. At the D4Z4 subtelomeric repeat regions in human cells, cohesin and HP1γ are co-recruited.
(2) Relationship with DNA replication
In Xenopus, the cohesin loading factor Scc2 (Nipbl) and cohesin require DNA pre-replication complex (pre-RC) assembly, suggesting that DNA replication origins may be the initial loading sites for Scc2 and cohesin in higher eukaryotes (Gillespie and Hirano, 2004, Takahashi et al., 2004) (Fig. 2B). In Drosophila, ChIP on chip analysis revealed that ORC2 preferentially binds to open chromatin regions and exhibited a high concordance with cohesin binding (MacAlpine et al., 2010). However, the bulk loading of cohesin does not appear to require the loading of the minichromosome maintenance (MCM) complex, an essential component for pre-RC assembly. A high concordance of cohesin and active replication origins was also found in human cells and cohesin interaction with the MCM complex was detected (Guillou et al., 2010). However, similar to Drosophila, the pre-RC is not required for bulk cohesin association with chromatin. Whether distribution of cohesin changes in the absence of the pre-RC remains to be determined. Furthermore, since the overlap with origins was observed with cohesin ChIP data in asynchronous cells, it would be interesting to determine whether the enrichment is cell cycle-regulated. Finally, it is unclear whether any of the identified binding sites affect expression of nearby genes and/or function as sister chromatid cohesion sites.
(3) Interactions with transcription factors
Several factors critical for cohesin recruitment were shown to physically interact with cohesin or Nipbl in mammalian cells. Originally, human cohesin was found to interact with the chromatin remodeling factor hSNF2h and to bind together to a type of Alu repeat sequence, which requires the ATPase activity of hSNF2h (Hakimi et al., 2002) (Fig. 2C). However, the function of this binding was not addressed.
As discussed above, more than 70% of cohesin binding sites in the mammalian genome contain CTCF binding sites where cohesin is recruited by CTCF. A weak but specific interaction of cohesin and CTCF has been observed (Stedman et al., 2008, Zeng et al., 2009, Rubio et al., 2008) (Fig. 2D). A recent study suggested that CTCF interacts with the SA2 subunit of cohesin (Xiao et al., 2011). More recently, it was shown that the DEAD-box RNA binding protein p68, together with its associated ncRNA called steroid receptor RNA activator (SRA), is co-recruited with cohesin to some of the CTCF binding sites. This appears to stabilize the interaction between CTCF and cohesin and promotes insulator function, for example, at the Igf2/H19 locus (Yao et al., 2010). Furthermore, ATRX, mutated in the Alpha-Thalassemia mental Retardation, X linked (ATR-X) syndrome, together with methyl-CpG binding protein 2 (MeCP2), was found to interact with cohesin and CTCF in the brain, affecting their binding and post-natal imprinting function at the Igf2/H19 and Gtl2/Dlk1 loci (Kernohan et al., 2010). It is conceivable that there are additional cell type- or tissue-specific factor(s) that modulate the interaction and chromatin binding of cohesin and CTCF to mediate cell type-specific insulator functions.
As described earlier, cohesin binding can also occur in a CTCF-independent context. For example, Mediator, whose binding sites significantly overlap with CTCF-free cohesin binding sites in mESCs, was shown to interact with Nipbl though the interaction appears to be weak (Kagey et al., 2010) (Fig. 2A). Whether Mediator is required for Nipbl and cohesin recruitment has not been examined. More recently, CTCF-free cohesin sites in mESCs were found to significantly overlap with the binding sites for pluripotency transcription factors, such as Oct4, Nanog, and Sox2 (Nitzsche et al., 2011). Although an interaction between cohesin and Nanog was detected, whether Nanog directly recruits cohesin to these sites is unknown.
A subset of cohesin binding sites that are CTCF-free tend to be cell type-specific and colocalize with estrogen receptor (ER)α binding sites in MCF-7 breast cancer cells and with liver-specific transcription factor Cebp binding sites in HepG2 hepatocellular carcinoma cells, suggesting a role for cohesin in cell type-specific gene regulation independent of CTCF (Schmidt et al., 2010) (Fig. 2A). However, the physical interaction of cohesin or NIPBL with these factors and/or the recruitment of cohesin by these factors have not been demonstrated. Interestingly, p68, mentioned above, was previously found to interact with ERα (Endoh et al., 1999), which may also contribute to the CTCF-free binding of cohesin at some of the ER target sites (Yao et al., 2010).
(4) Recruitment to heterochromatin
Cohesin recruitment to H3K9me3-marked D4Z4 heterochromatin is CTCF-independent and involves HP1 (Zeng et al., 2009) (Fig. 2E). Although it was originally suggested that cohesin directly interacts with Swi6/HP1 in S. pombe (Nonaka et al., 2002), more recent studies in both S. pombe and human cells demonstrated that Swi6/HP1 binds to NIPBL (Mis4 in S. pombe) rather than to cohesin (Fischer et al., 2009, Lechner et al., 2005, Serrano et al., 2009, Zeng et al., 2009). Interestingly, however, only HP1γ is co-recruited to the D4Z4 repeat together with cohesin (Zeng et al., 2009), despite the comparable binding of NIPBL to HP1α and HP1γ (Lechner et al., 2005) (Fig. 2E). Furthermore, despite the ubiquitous presence of H3K9me3 at D4Z4 in different cell types, cohesin and HP1γ binding was observed only in certain cell types, suggesting their roles in cell type-specific chromatin organization and gene regulation (Zeng et al., 2009). An additional cell type-specific factor(s) that may dictate their recruitment to D4Z4 remains to be determined (Fig. 2E).
(5) Relationship with Nipbl
Nipbl was suggested to play no role in cohesin recruitment to CTCF sites in mESCs based on the apparent lack of Nipbl peaks at cohesin-bound CTCF sites as indicated by ChIP sequencing (Kagey et al., 2010). However, in mouse embryonic liver and erythroleukemia cells, we found that Nipbl binding can be detected at CTCF insulator sites bound by cohesin in the β-globin locus, though it tends to be less prominent than at CTCF-free cohesin binding sites (Chien et al., 2011). Importantly, despite the weaker Nipbl binding, cohesin association with the CTCF site is sensitive to a decrease of Nipbl, strongly suggesting that Nipbl is universally required for cohesin binding to any genomic location (Chien et al., 2011). This is consistent with the major global loss of cohesin binding to chromatin in the HeLa cell nucleus following NIPBL depletion (Watrin et al., 2006). Taken together, the studies discussed above indicate that both cohesin and NIPBL, with other factors, appear to contribute to the binding specificity of cohesin at different chromosomal regions. It is possible that if the primary interaction of the recruiter is through cohesin, cohesin relies less on NIPBL in terms of binding site specificity, and NIPBL may be recruited by cohesin to these sites for further cohesin loading.
Although CTCF was shown to also colocalize with cohesin at centromeres in mitosis (Rubio et al., 2008), CTCF depletion failed to cause any significant sister chromatid cohesion defect (Wendt et al., 2008, Parelho et al., 2008). It is currently unclear whether some of these binding sites are also important for sister chromatid cohesion. Rather, accumulating evidence indicates that cohesin at these sites modulates gene expression at least partly by long-distance chromatin interaction or looping (see below).
Cohesin mediates long-distance chromatin interactions
An emerging aspect of gene regulation is the three-dimensional (3D) organization of chromosomes in the nucleus determined by long-distance chromatin: chromatin interactions, which can be considered a type of epigenetic chromatin regulation. Although a drastic change in chromatin structure is most easily observed during mitosis (i.e., condensation), evidence suggests that higher-order chromatin structure is also important during interphase, particularly for developmental and cell type-specific gene regulation in mammalian cells. Direct visualization of chromatin fibers using the lac operator/repressor system revealed that chromatin fibers undergo dynamic structural changes correlating with their transcriptional activity (Tumbar et al., 1999). Furthermore, studies indicate that gene regulation can be mediated by physical interactions between distant regulatory and promoter regions in cis or in trans (Tolhuis et al., 2002, Horike et al., 2005, Spilianakis and Flavell, 2004, Spilianakis et al., 2005, Cai et al., 2006). For example, the β-globin gene locus is known to form a chromatin loop termed the “active chromatin hub (ACH)” over an ~80 kb region through the interactions of the 5′-locus control region (LCR) with the 3′ insulator region and with promoters of the globin genes in a differentiation stage-specific fashion (Patrinos et al., 2004). MeCP2, which is mutated in Rett Syndrome, mediates chromatin looping of a ~70 kb region that is critical for silencing of the DLX5 gene in mice (Horike et al., 2005). The expression of IFNγ on chromosome 10 is coordinately regulated by an inter-chromosomal interaction with the LCR of the TH2 cytokine locus on chromosome 11 (Spilianakis et al., 2005). Thus, intra- and inter-chromosomal interactions of specific gene regions critically impact their expression patterns during cellular differentiation and development.
Currently, only a handful of proteins have been found to mediate chromatin interactions (Sexton et al., 2009). CTCF is one such protein and exerts at least some of its transcriptional effects by mediating long-distance chromatin interactions and loop formation in, for example, imprinting and X inactivation (Kurukuti et al., 2006, Xu et al., 2007, Ling et al., 2006). CTCF is also involved in chromatin tethering to a subnuclear site important for insulator function (Yusufzai et al., 2004). Thus, the discovery that cohesin is an important mediator of some, if not all, of CTCF’s transcriptional function suggested that cohesin also dictates gene expression by facilitating such higher-order chromatin organization. It is postulated that in a manner potentially similar to cohesin’s proposed embrace of two sister chromatids in its ring (Gruber et al., 2003), cohesin may trap two distant chromatin fibers to mediate chromatin interactions or chromatin looping. Recent studies indeed revealed the involvement of cohesin in chromatin interactions in several different contexts (see below).
(1) CTCF-dependent chromatin looping
Cohesin’s functions in long-distance chromatin interactions were demonstrated at CTCF insulator sites at specific loci such as IFNγ, Igf2/H19, the apolipoprotein cluster, and β-globin (Hadjur et al., 2009, Mishiro et al., 2009, Nativio et al., 2009, Hou et al., 2010, Chien et al., 2011). Cohesin’s involvement in chromatin interactions at CTCF sites in the MHC-II locus was also suggested although the effect of cohesin depletion has not been examined (Majumder and Boss, 2010). Although CTCF is important for the recruitment of cohesin, it is cohesin that appears to play a primary role in long-distance chromatin interaction (Hadjur et al., 2009). These studies suggested the architectural role of cohesin in chromatin domain organization important for developmental gene regulation in the context of the CTCF binding sites (Fig. 3A).
Fig. 3.
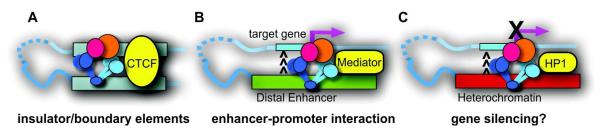
Different types of chromatin looping mediated by cohesin. A. Between insulator/boundary elements. B. Between distal enhancer and promoter regions. C. Between heterochromatic regions (C is hypothetical).
(2) CTCF-independent chromatin looping
CTCF-independent chromatin looping was observed at active gene promoters and at upstream regulatory regions co-occupied by cohesin, Nipbl, and Mediator in mESCs (Kagey et al., 2010) (Fig. 3B). Depletion of cohesin resulted in reduced loop formation correlating with decreased gene expression at pluripotency genes such as Nanog and Oct4 (Kagey et al., 2010). Consistent with this, neither cohesin binding nor enhancer-promoter interaction was observed in mouse embryonic fibroblasts in which these genes were silenced (Kagey et al., 2010). These findings suggest that cohesin mediates gene activation by bridging the enhancer and promoter regions. Similarly, our recent study of both the mouse and human β-globin loci revealed that while cohesin together with CTCF mediates the 5′ and 3′ insulator interaction, cohesin specifically mediates the enhancer-promoter interaction important for β-globin expression in a CTCF-independent manner (Chien et al., 2011).
As described above, some of the CTCF-free cohesin binding sites overlap with ERα binding sites in MCF7 cells (Schmidt et al., 2010). Interestingly, these sites appear to correspond to the ERα binding sites that anchor chromatin loops (Schmidt et al., 2010). Previously, a genome-wide analysis of ERα binding revealed that the majority of its binding sites are located in intergenic regions far from promoters (Lin et al., 2007). Using the novel high-throughput genome-wide chromatin interaction assay called “Chromatin Interaction Analysis with Paired-End Tag sequencing” (ChIA-PET), it was found that ERα mediates extensive chromatin looping at these binding sites for coordinated target gene regulation in estrogen-treated MCF7 cells (Fullwood et al., 2009). Whether cohesin is required for ERα-dependent chromatin loop formation has not been determined.
Although it has not been experimentally proven, it is conceivable that cohesin bound to heterochromatin may also mediate long-distance chromatin interactions to silence distant genes (e.g., at D4Z4 heterochromatin)(Zeng et al., 2009) (Fig. 3C). Repositioning and physical association of heterochromatic regions were suggested to be important for coordinated gene silencing in Drosophila and in mammalian cells (Li et al., 2003, Su et al., 2004). Indeed, the association of cohesin with the PcG complexes in Drosophila mentioned earlier may be important for pairing-sensitive gene silencing (Strübbe et al., 2011). It is possible that cohesin may be recruited to polycomb response elements (PREs) by the PcG complexes and mediate silencing by facilitating the pairing of PREs on different chromosomes.
Based on high-throughput ChIP analysis, cohesin binds to ~9,000 to 44,000 sites in mammalian cells depending on the analysis and cell types (Kagey et al., 2010, Wendt et al., 2008). Accumulating evidence summarized above suggests that cohesin may be a genome-wide mediator of both constitutive and cell type-specific long-distance chromatin interactions critical for gene expression. Cohesin may serve as an important molecular glue or clamp that bonds different genomic regions. It is currently unclear if all the cohesin binding sites are involved in chromatin interactions, if these interactions take place along some type of anchoring scaffold structure, and how this relates to sister chromatid cohesion.
Cohesinopathies
Human syndromes resulting from cohesin dysfunction are called “cohesinopathies” (Bose and Gerton, 2010, Liu and Krantz, 2008). This class of diagnoses not only highlights the significance of the cohesin pathway in development, but also provides important clues as to how cohesin and cohesin-associated factors function in the cell.
(1) Cornelia de Lange Syndrome (CdLS)
CdLS (OMIM 122470, 300590, 610759) is a dominantly inherited disorder characterized by growth and cognitive retardation, abnormalities of the upper limbs, gastroesophageal dysfunction, cardiac, ophthalmologic and genitourinary anomalies, and characteristic facial features. More than 50% of CdLS cases are associated with NIPBL mutations (Krantz et al., 2004, Tonkin et al., 2004). Since NIPBL homologs are essential in yeast, flies, and mice, it is likely that homozygous mutation of the NIPBL gene causes lethality in humans. It has been widely reported that NIPBL alleles containing truncating mutations lead to possible haploinsufficiency of the NIPBL protein, resulting in the most severe clinical phenotypes (Dorsett and Krantz, 2009, Liu and Krantz, 2009, Selicorni et al., 2007). Some CdLS cases are associated with genomic rearrangements involving NIPBL (Ratajska et al., 2010). NIPBL missense mutations and somatic mosaicism were also found, which have a milder clinical presentation (Liu and Krantz, 2009, Castronovo et al., 2010). Mutations in the cohesin subunit genes SMC1A (hSMC1) and hSMC3, found in a subset of often phenotypically milder CdLS cases (Musio et al., 2006, Deardorff et al., 2007), suggested that both NIPBL and cohesin are in the same pathway impaired in the disorder. Evidence suggests that the point mutations of NIPBL and SMC proteins result in altered protein function (Gard et al., 2009, Jahnke et al., 2008, Revenkova et al., 2009).
Importantly, cohesin/NIPBL mutations analyzed so far do not seem to cause significant sister chromatid cohesion defects (Kaur et al., 2005, Castronovo et al., 2009, Vrouwe et al., 2007), suggesting the deficiency may lie in non-mitotic function(s), such as gene regulation. Indeed, Nipbl heterozygous mutant (Nipbl+/-) mutant mice, in which Nipbl is reduced by only 30% (similar to CdLS patient levels), faithfully reproduced many CdLS features, supporting the notion that haploinsufficiency causes CdLS (Kawauchi et al., 2009). Mutant mouse tissues and cells exhibited modest changes in the expression of many genes without any significant sister chromatid cohesion defects, suggesting the CdLS phenotype is caused by relatively subtle gene regulation changes. Consistent with this notion, cohesin and NIPBL binding was reduced at some of the misregulated gene loci in human CdLS lymphoblastoid cells, relative to unchanged or silent genes (Liu et al., 2009). We found that cohesin and Nipbl binding was partially reduced at the β-globin locus in Nipbl+/− mutant embryonic mouse liver (Chien et al., 2011). Importantly, this partial reduction was sufficient to decrease both the long-distance chromatin interactions at this locus and β-globin gene expression (Chien et al., 2011). These results suggest that the impairment of proper chromatin interactions due to reduced cohesin binding may be the cause of the global gene expression changes in CdLS.
Finally, it is also possible that NIPBL may potentially have other functions besides cohesin loading, perhaps loading other SMC complexes as was demonstrated in yeast (D’Ambrosio et al., 2008, Gard et al., 2009).
(2) Roberts’ Syndrome
Roberts’ Syndrome (RBS; OMIM 268300) (more recently, Roberts’ Syndrome/SC phocomelia) is a recessive developmental disorder caused by mutation of both alleles of ESCO2 (Vega et al., 2005). RBS/SC phocomelia spectrum patients also have a wide range of clinical phenotypes that include upper and lower limb defects, growth retardation, craniofacial anomalies, and tetraphocomelia and mental retardation with some similarity to the CdLS phenotype (Vega et al., 2005, Dorsett, 2007). Unlike CdLS, which exhibits a minimal cohesion defect, RBS chromosomes exhibit premature centromere separation and heterochromatin puffing, indicative of a sister chromatid cohesion defect (Tomkins et al., 1979). Chromosome segregation abnormalities have also been reported (Vega et al., 2005). In addition, patient cells show hypersensitivity to specific DNA damaging agents, suggesting that chromosomal stability is compromised (Gordillo et al., 2008, van der Lelij et al., 2009). Interestingly, inhibition of chromosome segregation genes such as INCENP, ZWINT-1, and ZW10 resulted in RBS-like cellular phenotypes (Musio et al., 2004). Taken together, while CdLS may be caused by NIPBL deficiency affecting gene expression, sister chromatid cohesion and segregation defects may play an important role in the pathogenesis of RBS.
Cohesin as a multi-faceted regulator of genome functions
In addition to cell cycle-regulated sister chromatid cohesion and cell type-specific chromatin architectural organization and gene regulation, cohesin has additional functions in both mitosis and interphase including mitotic spindle organization, DNA repair, and more recently, DNA replication (Fig. 4). In order to properly analyze the different cohesin defect phenotypes, it is important to take these different functions of cohesin into consideration.
Fig.4.
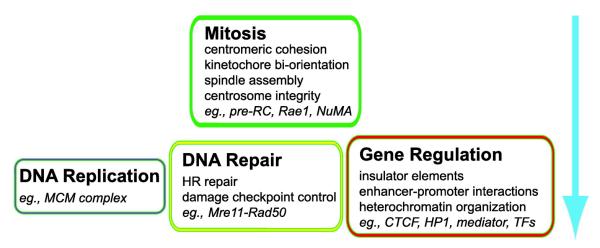
Multiple functions of cohesin in the cell and functional hierarchy. Four major areas of cohesin functions and examples of factors that recruit cohesin to specific sites are listed. Mitotic functions appear to be the least dose-sensitive and most essential, while other interphase functions, such as DNA repair and gene regulation, are more dose-sensitive. Further analysis is needed to determine whether there are significant hierarchical differences among the three non-mitotic functions. TFs: transcription factors.
(1) Mitotic spindle regulation at centrosomes/spindle poles
In mitosis, cohesin not only localizes to centromere-kinetochore regions, but also clusters to the centrosomes/spindle poles (Gregson et al., 2001, Kong et al., 2009). This mitosis-specific recruitment of cohesin to spindle poles appears to be modulated by its interaction with the pericentrosomal factor NuMA and RNA export factor 1 (Rae1) (Kong et al., 2009, Wong and Blobel, 2008). Centrosome/spindle pole association of cohesin is mitosis-specific, and while cohesin depletion has no effect on interphase spindle organization, its depletion compromised pole-originated spindle reassembly in mitosis, centrosome integrity, and caused multipolar spindles (Díaz-Martínez et al., 2010, Kong et al., 2009, Losada et al., 2005, Wong and Blobel, 2008, Wong, 2010). Thus, in mitosis, cohesin has dual functions on chromosomes and at spindle poles to ensure proper chromosome organization and segregation. Although the exact mechanism of cohesin function in spindle assembly regulation is currently unclear, one possibility is that cohesin may bundle spindle microtubules in a way similar to how it is proposed to embrace sister chromatids (Kong et al., 2009, Wong and Blobel, 2008).
(2) DNA double-strand break (DSB) repair
Cohesin was also shown to be important for DSB repair and damage-induced cell cycle checkpoint control. In human cells, cohesin accumulates at the damage sites only in S/G2 phase when sister chromatids are present, which is dependent on the DSB repair factor Mre11-Rad50 complex (Kim et al., 2002a). Consistent with this cell cycle specificity, cohesin is involved in homologous recombination (HR) repair between sister chromatids, but not in non-homologous endjoining (NHEJ) or other types of HR in human cells (Potts et al., 2006). As predicted from the elegant studies in yeast demonstrating the damage-induced activation of cohesin to establish de novo sister chromatid cohesion (Ström et al., 2004, Ünal et al., 2004, Ström et al., 2007, Ünal et al., 2007, Heidinger-Pauli et al., 2009), mammalian cohesin also promotes sister chromatid HR possibly by establishing local cohesion and facilitating homologous pairing of the damaged and intact sister chromatids (Ball and Yokomori, 2008).
Cohesin also plays a role in the S phase and G2/M DNA damage checkpoint responses (Kim et al., 2002b, Kitagawa et al., 2004, Luo et al., 2008, Yazdi et al., 2002, Watrin and Peters, 2009). SMC1 and SMC3 are targets of the DNA damage checkpoint kinase ATM for the intra-S checkpoint (inhibition of DNA synthesis) in mammalian cells (Kim et al., 2002b, Luo et al., 2008, Yazdi et al., 2002). Although the exact mechanism is unclear, a recent study indicated that DSB damage induces the enhancement of cohesin binding to its binding sites in a genome-wide fashion (Kim et al., 2010). Thus, it is possible that the enhanced genome-wide cohesin binding may provide a roadblock to halt DNA replication for the S phase checkpoint. How this relates to the regulation of the cohesion function of cohesin at the damage sites is currently unclear.
(3) DNA replication
A recent study demonstrated that depletion of cohesin slows down S phase progression, decreases the number of active DNA replication origins, and increases the size of the chromatin loops tethered to the nuclear matrix (Guillou et al., 2010). It remains to be evaluated whether the increase of chromatin loop length is the cause of the DNA replication defect.
(4) Functional hierarchy of cohesin
The discoveries of these different cohesin functions raise the question: how is the functional specificity of cohesin determined and regulated? Based on the studies described above, the functional specificity of cohesin appears to be mediated by interactions with other factors that recruit cohesin to distinct subcellular and genomic sites (Figs. 2, 3 and 4). Given that cohesin has such diverse roles, it is important to ask whether a functional hierarchy exists when the cohesin level is limited in the cell (Fig. 4). In human cells, transient depletion of cohesin by small interfering RNA (siRNA) that removes more than 80% of the total cohesin did not cause any significant defect in sister chromatid cohesion and failed to completely displace cohesin from spindle poles in metaphase cells (Kong et al., 2009). In contrast, similar siRNA treatment effectively changed the expression of cohesin target genes (Wendt et al., 2008) and HP1 recruitment to D4Z4 heterochromatin in the same cell line (Zeng et al., 2009). Similarly, in S. cerevisiae, a greater than 80% reduction of Rad21 affected chromosome condensation, DNA repair, and DNA repeat stability, but not sister chromatid cohesion and chromosome segregation (Heidinger-Pauli et al., 2010). In Drosophila cells, 80% depletion of cohesin or Nipped-B affected gene expression but did not cause sister chromatid cohesion or chromosome segregation defects (Schaaf et al., 2009). These studies indicate that there is a functional hierarchy of cohesin in the cell ensuring the most fundamental function of cohesin in mitotic chromosome organization and segregation (Fig. 4). These results are particularly important for understanding the pathogenesis of CdLS. As described earlier, the majority of CdLS is caused by partial impairment of cohesin function without any significant cohesion defect. In addition to the altered gene expression, CdLS patient cells also exhibit increased DNA damage sensitivity, similar to the observation in yeast (Vrouwe et al., 2007). Furthermore, although DNA replication defects have not been reported in CdLS patient cells, the fact that replication defects can be observed following conventional cohesin siRNA depletion (Guillou et al., 2010) raises the possibility that this activity of cohesin may also be affected. How the repair and replication functions of cohesin contribute to CdLS pathogenesis is currently unclear. In Drosophila, a 50% decrease of Nipped-B reduced viability during pupal development (Rollins et al., 2004), and in mice, a Nipbl heterozygous mutation significantly increased perinatal mortality (Kawauchi et al., 2009). In both cases, there was no significant sister chromatid cohesion defect, indicating that the non-mitotic functions of cohesin are indeed critical in vivo.
The realization that cohesin has many dose-sensitive functions poses some challenges for evaluating cohesin function. Even within cohesin’s role in gene regulation, there may be further differential dose sensitivities. At the Enhancer of split and invected-engrailed loci in Drosophila, a modest degree of cohesin or Nipped-B depletion led to silencing while a higher degree, and longer term, of depletion led to activation of the same genes (Schaaf et al., 2009). Thus, different degrees of cohesin reduction may have distinct effects on gene expression. Furthermore, it is possible that individual cohesin binding sites on the genome have different sensitivities to cohesin reduction depending on how cohesin is recruited to those sites. Although the transcriptional role of cohesin was not tested, partial loss of cohesin in yeast led to a significant reduction of cohesin binding to chromosome arm regions, but not to pericentromeric domains (Heidinger-Pauli et al., 2010). Thus, it is possible that drastic experimental cohesin depletion may result in a gene expression phenotype different from the more subtle partial impairment that occurs in CdLS. Finally, it is formally possible that any phenotype observed following cohesin depletion is indirect, possibly caused by expression changes of some of the cohesin target genes. Understanding the multifaceted role of cohesin in the cell, and carefully considering the differential effects of cohesin depletion, will be critical for further characterization of this fascinating chromatin regulator.
Acknowledgment
The work in the Yokomori laboratory was supported in part by NIH AR058548, HD062951, MDA4026, and the David and Helen Younger Research Fellowship from the FSH Society (FSHS-DHY-001) to KY, and the FSH Society Helen Younger and David Younger Fellowship Research Grant FSHS-DHY-002 to WZ. RC is supported by NIH T32-CA113265. We apologize to the researchers whose work we could not cite in this chapter due to space limitations.
References
- Anderson DE, Losada A, Erickson HP, Hirano T. Condensin and cohesin display different arm conformations with characteristic hinge angles. J. Cell Biol. 2002;156:419–424. doi: 10.1083/jcb.200111002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arumugam P, Gruber S, Tanaka K, Haering CH, Mechtler K, Nasmyth K. ATP hydrolysis is required for cohesin’s association with chromosomes. Curr. Biol. 2003;13:1941–1953. doi: 10.1016/j.cub.2003.10.036. [DOI] [PubMed] [Google Scholar]
- Ball AR, Yokomori K. Damage-induced reactivation of cohesin in postreplicative DNA repair. Bioessay. 2008;30:5–9. doi: 10.1002/bies.20691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bausch C, Noone S, Henry JM, Gaudenz K, Sanderson B, Seidel C, Gerton JL. Transcription alters chromosomal locations of cohesin in Saccharomyces cerevisiae. Mol. Cell Biol. 2007;27:8522–8532. doi: 10.1128/MCB.01007-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard P, Maure J-F, Partridge JF, Genier S, Javerzat J-P, Allshire RC. Requirement of heterochromatin for cohesion at centromeres. Science. 2001;294:2539–2542. doi: 10.1126/science.1064027. [DOI] [PubMed] [Google Scholar]
- Blat Y, Kleckner N. Cohesins bind to preferential sites along yeast chromosome III, with differential regulation along arms versus the centric region. Cell. 1999;98:249–259. doi: 10.1016/s0092-8674(00)81019-3. [DOI] [PubMed] [Google Scholar]
- Bose T, Gerton JL. Cohesinopathies, gene expression, and chromatin organization. J. Cell Biol. 2010;189:201–210. doi: 10.1083/jcb.200912129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai S, Lee CC, Kohwi-Shigematsu T. SATB1 packages densely looped, transcriptionally active chromatin for coordinated expression of cytokine genes. Nat. Genet. 2006;38:1278–1288. doi: 10.1038/ng1913. [DOI] [PubMed] [Google Scholar]
- Canudas S, Houghtaling BR, Kim JY, Dynek JN, Chang WG, Smith S. Protein requirements for sister telomere association in human cells. EMBO J. 2007;26:4867–4878. doi: 10.1038/sj.emboj.7601903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Tsukada Y, Zhang Y. Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol. Cell. 2005;20:845–854. doi: 10.1016/j.molcel.2005.12.002. [DOI] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- Carretero M, Remeseiro S, Losada A. Cohesin ties up the genome. Curr. Opin. Cell Biol. 2010;22:781–787. doi: 10.1016/j.ceb.2010.07.004. [DOI] [PubMed] [Google Scholar]
- Castronovo P, Delahaye-Duriez A, Gervasini C, Azzollini J, Minier F, Russo S, Masciadri M, Selicorni A, Verloes A, Larizza L. Somatic mosaicism in Cornelia de Lange syndrome: a further contributor to the wide clinical expressivity? Clin. Genet. 2010;78:560–564. doi: 10.1111/j.1399-0004.2010.01408.x. [DOI] [PubMed] [Google Scholar]
- Castronovo P, Gervasini C, Cereda A, Masciadri M, Milani D, Russo S, Selicorni A, Larizza L. Premature chromatid separation is not a useful diagnostic marker for Cornelia de Lange syndrome. Chromosome Res. 2009;17:763–771. doi: 10.1007/s10577-009-9066-6. [DOI] [PubMed] [Google Scholar]
- Chien R, Zeng W, Kawauchi S, Bender MA, Santos R, Gregson HC, Schmiesing JA, Newkirk D, Kong X, Ball ARJ, Calof AL, Lander AD, Groudine MT, Yokomori K. Cohesin mediates chromatin interactions that regulate mammalian {beta}-globin expression. J. Biol. Chem. 2011 doi: 10.1074/jbc.M110.207365. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciosk R, Shirayama M, Shevchenko A, Tanaka T, Toth A, Shevchenko A, Nasmyth K. Cohesin’s binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol. Cell. 2000;5:243–254. doi: 10.1016/s1097-2765(00)80420-7. [DOI] [PubMed] [Google Scholar]
- D’Ambrosio C, Schmidt CK, Katou Y, Kelly G, Itoh T, Shirahige K, Uhlmann F. Identification of cis-acting sites for condensin loading onto budding yeast chromosomes. Genes Dev. 2008;22:2215–2227. doi: 10.1101/gad.1675708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deardorff MA, Kaur M, Yaeger D, Rampuria A, Korolev S, Pie J, Gil-Rodríguez C, Arnedo M, Loeys B, Kline AD, Wilson M, Lillquist K, Siu V, Ramos FJ, Musio A, Jackson LS, Dorsett D, Krantz ID. Mutations in cohesin complex members SMC3 and SMC1A cause a mild variant of cornelia de Lange syndrome with predominant mental retardation. Am. J. Hum. Genet. 2007;80:485–494. doi: 10.1086/511888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dheur S, Saupe SJ, Genier S, Vazquez S, Javerzat JP. Role for cohesin in the formation of a heterochromatic domain at fission yeast subtelomeres. Mol. Cell Biol. 2011;31:1088–1097. doi: 10.1128/MCB.01290-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz-Martínez LA, Beauchene NA, Furniss K, Esponda P, Giménez-Abián JF, Clarke DJ. Cohesin is needed for bipolar mitosis in human cells. Cell Cycle. 2010;9:1764–1773. doi: 10.4161/cc.9.9.11525. [DOI] [PubMed] [Google Scholar]
- Dorsett D. Roles of the sister chromatid cohesion apparatus in gene expression, development, and human syndromes. Chromosoma. 2007;116:1–13. doi: 10.1007/s00412-006-0072-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett D. Cohesin: genomic insights into controlling gene transcription and development. Curr. Opin. Genet. Dev. 2011;21:199–206. doi: 10.1016/j.gde.2011.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett D, Krantz ID. On the molecular etiology of Cornelia de Lange syndrome. Ann. N. Y. Acad. Sci. 2009;1151:22–37. doi: 10.1111/j.1749-6632.2008.03450.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endoh H, Maruyama K, Masuhiro Y, Kobayashi Y, Goto M, Tai H, Yanagisawa J, Metzger D, Hashimoto S, Kato S. Purification and identification of p68 RNA helicase acting as a transcriptional coactivator specific for the activation function 1 of human estrogen receptor alpha. Mol Cell Biol. 1999;19:5363–5372. doi: 10.1128/mcb.19.8.5363. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Fischer T, Cui B, Dhakshnamoorthy J, Zhou M, Rubin C, Zofall M, Veenstra TD, Grewal SI. Diverse roles of HP1 proteins in heterochromatin assembly and functions in fission yeast. Proc. Natl. Acad. Sci. 2009;106:8998–9003. doi: 10.1073/pnas.0813063106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullwood MJ, Liu MH, Pan YF, Liu J, Xu H, Mohamed YB, Orlov YL, Velkov S, Ho A, Mei PH, Chew EG, Huang PY, Welboren WJ, Han Y, Ooi HS, Ariyaratne PN, Vega VB, Luo Y, Tan PY, Choy PY, Wansa KD, Zhao B, Lim KS, Leow SC, Yow JS, Joseph R, Li H, Desai KV, Thomsen JS, Lee YK, Karuturi RK, Herve T, Bourque G, Stunnenberg HG, Ruan X, Cacheux-Rataboul V, Sung WK, Liu ET, Wei CL, Cheung E, Ruan Y. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature. 2009;462:58–64. doi: 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gard S, Light W, Xiong B, Bose T, McNairn AJ, Harris B, Fleharty B, Seidel C, Brickner JH, Gerton JL. Cohesinopathy mutations disrupt the subnuclear organization of chromatin. J. Cell Biol. 2009;187:455–462. doi: 10.1083/jcb.200906075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie PJ, Hirano T. Scc2 couples replication licensing to sister chromatid cohesion in Xenopus egg extracts. Curr. Biol. 2004;14:1598–1603. doi: 10.1016/j.cub.2004.07.053. [DOI] [PubMed] [Google Scholar]
- Glynn EF, Megee PC, Yu HG, Mistrot C, Ünal E, Koshland DE, DeRisi JL, Gerton JL. Genome-wide mapping of the cohesin complex in the yeast Saccharomyces cerevisiae. PLoS Biol. 2004;2:E259. doi: 10.1371/journal.pbio.0020259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes NP, Espinosa JM. Gene-specific repression of the p53 target gene PUMA via intragenic CTCF-Cohesin binding. Genes Dev. 2010;24:1022–1034. doi: 10.1101/gad.1881010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordillo M, Vega H, Trainer AH, Hou F, Sakai N, Luque R, Kayserili H, Basaran S, Skovby F, Hennekam RC, Uzielli ML, Schnur RE, Manouvrier S, Chang S, Blair E, Hurst JA, Forzano F, Meins M, Simola KO, Raas-Rothschild A, Schultz RA, McDaniel LD, Ozono K, Inui K, Zou H, Jabs EW. The molecular mechanism underlying Roberts syndrome involves loss of ESCO2 acetyltransferase activity. Hum Mol Genet. 2008;17:2172–2180. doi: 10.1093/hmg/ddn116. [DOI] [PubMed] [Google Scholar]
- Gregson HC, Schmiesing JA, Kim J-S, Kobayashi T, Zhou S, Yokomori K. A potential role for human cohesin in mitotic spindle aster assembly. J. Biol. Chem. 2001;276:47575–47582. doi: 10.1074/jbc.M103364200. [DOI] [PubMed] [Google Scholar]
- Gruber S, Haering CH, Nasmyth K. Chromosomal cohesin forms a ring. Cell. 2003;112:765–777. doi: 10.1016/s0092-8674(03)00162-4. [DOI] [PubMed] [Google Scholar]
- Guillou E, Ibarra A, Coulon V, Casado-Vela J, Rico D, Casal I, Schwob E, Losada A, Méndez J. Cohesin organizes chromatin loops at DNA replication factories. Genes Dev. 2010;24:2812–2822. doi: 10.1101/gad.608210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjur S, Williams LM, Ryan NK, Cobb BS, Sexton T, Fraser P, Fisher AG, Merkenschlager M. Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature. 2009;460:410–413. doi: 10.1038/nature08079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haering CH, Farcas AM, Arumugam P, Metson J, Nasmyth K. The cohesin ring concatenates sister DNA molecules. Nature. 2008;454:297–301. doi: 10.1038/nature07098. [DOI] [PubMed] [Google Scholar]
- Haering CH, Lowe J, Hochwagen A, Nasmyth K. Molecular architecture of SMC proteins and the yeast cohesin complex. Mol. Cell. 2002;9:773–788. doi: 10.1016/s1097-2765(02)00515-4. [DOI] [PubMed] [Google Scholar]
- Hakimi MA, Bochar DA, Schmiesing JA, Dong Y, Barak OG, Speicher DW, Yokomori K, Shiekhattar R. A chromatin remodeling complex that loads cohesin onto human chromosomes. Nature. 2002;418:994–998. doi: 10.1038/nature01024. [DOI] [PubMed] [Google Scholar]
- Hallson G, Syrzycka M, Beck SA, Kennison JA, Dorsett D, Page SL, Hunter SM, Keall R, Warren WD, Brock HW, Sinclair DA, Honda BM. The Drosophila cohesin subunit Rad21 is a trithorax group (trxG) protein. Proc. Natl. Acad. Sci. 2008;105:12405–12410. doi: 10.1073/pnas.0801698105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauf S, Roitinger E, Koch B, Dittrich CM, Mechtler K, Peters JM. Dissociation of cohesin from chromosome arms and loss of arm cohesion during early mitosis depends on phosphorylation of SA2. PLoS Biol. 2005;3:e69. doi: 10.1371/journal.pbio.0030069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauf S, Waizenegger IC, Peters J-M. Cohesin cleavage by separase required for anaphase and cytokinesis in human cells. Science. 2001;293:1320–1323. doi: 10.1126/science.1061376. [DOI] [PubMed] [Google Scholar]
- Heidinger-Pauli JM, Mert O, Davenport C, Guacci V, Koshland D. Systematic reduction of cohesin differentially affects chromosome segregation, condensation, and DNA repair. Curr Biol. 2010;20:957–963. doi: 10.1016/j.cub.2010.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidinger-Pauli JM, Unal E, Koshland D. Distinct targets of the Eco1 acetyltransferase modulate cohesion in S phase and in response to DNA damage. Mol. Cell. 2009;34:311–321. doi: 10.1016/j.molcel.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T. Chromosome cohesion, condensation, and separation. Annu. Rev. Biochem. 2000;69:115–144. doi: 10.1146/annurev.biochem.69.1.115. [DOI] [PubMed] [Google Scholar]
- Hirano T. At the heart of the chromosome: SMC proteins in action. Nat. REv. Mol. Cell. Biol. 2006;7:311–322. doi: 10.1038/nrm1909. [DOI] [PubMed] [Google Scholar]
- Hoque MD, Ishikawa F. Cohesin defects lead to premature sister chromatid separation, kinetochore dysfunction and spindle-assembly checkpoint activation. J. Biol. Chem. 2002;277:42306–42314. doi: 10.1074/jbc.M206836200. [DOI] [PubMed] [Google Scholar]
- Horike SI, Cai S, Miyano M, Cheng JF, Kohwi-Shigematsu T. Loss of silent-chromatin looping and impaired imprinting of DLX5 in Rett syndrome. Nat. Genet. 2005;37:31–40. doi: 10.1038/ng1491. [DOI] [PubMed] [Google Scholar]
- Horsfield JA, Anagnostou SH, Hu JK, Cho KH, Geisler R, Lieschke G, Crosier KE, Crosier PS. Cohesin-dependent regulation of Runx genes. Development. 2007;134:2639–2649. doi: 10.1242/dev.002485. [DOI] [PubMed] [Google Scholar]
- Hou C, Dale R, Dean A. Cell type specificity of chromatin organization mediated by CTCF and cohesin. Proc. Natl. Acad. Sci. 2010;107:3651–3656. doi: 10.1073/pnas.0912087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov D, Nasmyth K. A physical assay for sister chromatid cohesion in vitro. Mol. Cell. 2007;27:300–310. doi: 10.1016/j.molcel.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Jahnke P, Xu W, Wülling M, Albrecht M, Gabriel H, Gillessen-Kaesbach G, Kaiser FJ. The Cohesin loading factor NIPBL recruits histone deacetylases to mediate local chromatin modifications. Nuc. Acids Res. 2008;36:6450–6458. doi: 10.1093/nar/gkn688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, Taatjes DJ, Dekker J, Young RA. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur M, Descipio C, McCallum J, Yaeger D, Devoto M, Jackson LG, Spinner NB, Krantz ID. Precocious sister chromatid separation (PSCS) in Cornelia de Lange syndrome. Am. J. Med. Genet. 2005;138:27–31. doi: 10.1002/ajmg.a.30919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawauchi S, Calof AL, Santos R, Lopez-Burks ME, Young CM, Hoang MP, Chua A, Lao T, Lechner MS, Daniel JA, Nussenzweig A, Kitzes L, Yokomori K, Hallgrimsson B, Lander AD. Multiple organ system defects and transcriptional dysregulation in the nipbl mouse, a model of cornelia de lange syndrome. PLoS Genet. 2009;5:e1000650. doi: 10.1371/journal.pgen.1000650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kernohan KD, Jiang Y, Tremblay DC, Bonvissuto AC, Eubanks JH, Mann MR, Bérubé NG. ATRX partners with cohesin and MeCP2 and contributes to developmental silencing of imprinted genes in the brain. Dev. Cell. 2010;18:191–202. doi: 10.1016/j.devcel.2009.12.017. [DOI] [PubMed] [Google Scholar]
- Kim BJ, Li Y, Zhang J, Xi Y, Li Y, Yang T, Jung SY, Pan X, Chen R, Li W, Wang Y, Qin J. Genome-wide reinforcement of cohesin binding at pre-existing cohesin sites in response to ionizing radiation in human cells. J. Biol. Chem. 2010 doi: 10.1074/jbc.M110.134577. Epub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J-S, Krasieva TB, LaMorte VJ, Taylor AMR, Yokomori K. Specific recruitment of human cohesin to laser-induced DNA damage. J. Biol. Chem. 2002a;277:45149–45153. doi: 10.1074/jbc.M209123200. [DOI] [PubMed] [Google Scholar]
- Kim S-T, Xu B, Kastan MB. Involvement of the cohesin protein, Smc1, in Atm-dependent and independent responses to DNA damage. Genes & Dev. 2002b;16:560–570. doi: 10.1101/gad.970602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa R, Bakkenist CJ, McKinnon PJ, Kastan MB. Phosphorylation of SMC1 is a critical downstream event in the ATM–NBS1–BRCA1 pathway. Genes Dev. 2004;18:1423–1438. doi: 10.1101/gad.1200304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima TS, Sakuno T, Ishiguro K, Iemura S, Natsume T, Kawashima SA, Watanabe Y. Shugoshin collaborates with protein phosphatase 2A to protect cohesin. Nature. 2006;441:46–52. doi: 10.1038/nature04663. [DOI] [PubMed] [Google Scholar]
- Koch B, Kueng S, Ruckenbauer C, Wendt KS, Peters JM. The Suv39h-HP1 histone methylation pathway is dispensable for enrichment and protection of cohesin at centromeres in mammalian cells. Chromosoma. 2008;117:199–210. doi: 10.1007/s00412-007-0139-z. [DOI] [PubMed] [Google Scholar]
- Kong X, Ball ARJ, Sonoda E, Feng J, Takeda S, Fukagawa T, Yen TJ, Yokomori K. Cohesin associates with spindle poles in a mitosis-specific manner and functions in spindle assembly in vertebrate cells. Mol. Biol. Cell. 2009;20:1289–1301. doi: 10.1091/mbc.E08-04-0419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krantz ID, McCallum J, DeScipio C, Kaur M, Gillis LA, Yaeger D, Jukofsky L, Wasserman N, Bottani A, Morris CA, Nowaczyk MJ, Toriello H, Bamshad MJ, Carey JC, Rappaport E, Kawauchi S, Lander AD, Calof AL, Li HH, Devoto M, Jackson LG. Cornelia de Lange syndrome is caused by mutations in NIPBL, the human homolog of Drosophila melanogaster Nipped-B. Nat. Genet. 2004;36:631–635. doi: 10.1038/ng1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurukuti S, Tiwari VK, Tavoosidana G, Pugacheva E, Murrell A, Zhao Z, Lobanenkov V, Reik W, Ohlsson R. CTCF binding at the H19 imprinting control region mediates maternally inherited higher-order chromatin conformation to restrict enhancer access to Igf2. Proc. Natl. Acad. Sci. 2006;103:10684–10689. doi: 10.1073/pnas.0600326103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laloraya S, Guacci V, Koshland D. Chromosomal addresses of the cohesin component Mcd1p. J. Cell Biol. 2000;151:1047–1056. doi: 10.1083/jcb.151.5.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner MS, Schultz DC, Negorev D, Maul GG, Rauscher F. J. r. The mammalian heterochromatin protein 1 binds diverse nuclear proteins through a common motif that targets the chromoshadow domain. Biochem. Biophys. Res. Commun. 2005;331:929–937. doi: 10.1016/j.bbrc.2005.04.016. [DOI] [PubMed] [Google Scholar]
- Lemmers RJ, van der Vliet PJ, Klooster R, Sacconi S, Camaño P, Dauwerse JG, Snider L, Straasheijm KR, van Ommen GJ, Padberg GW, Miller DG, Tapscott SJ, Tawil R, Frants RR, van der Maarel SM. A unifying genetic model for facioscapulohumeral muscular dystrophy. Science. 2010;329:1650–1653. doi: 10.1126/science.1189044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengronne A, Katou Y, Mori S, Yokobayashi S, Kelly GP, Itoh T, Watanabe Y, Shirahige K, Uhlmann F. Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature. 2004;430:573–578. doi: 10.1038/nature02742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Danzer JR, Alvarez P, Belmont AS, Wallrath LL. Effects of tethering HP1 to euchromatic regions of the Drosophila genome. Development. 2003;130:1817–1824. doi: 10.1242/dev.00405. [DOI] [PubMed] [Google Scholar]
- Lin CY, Vega VB, Thomsen JS, Zhang T, Kong SL, Xie M, Chiu KP, Lipovich L, Barnett DH, Stossi F, Yeo A, George J, Kuznetsov VA, Lee YK, Charn TH, Palanisamy N, Miller LD, Cheung E, Katzenellenbogen BS, Ruan Y, Bourque G, Wei CL, Liu ET. Whole-genome cartography of estrogen receptor alpha binding sites. PLoS Genet. 2007;3:e87. doi: 10.1371/journal.pgen.0030087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling JQ, Li T, Hu JF, Vu TH, Chen HL, Qiu XW, Cherry AM, Hoffman AR. CTCF mediates interchromosomal colocalization between Igf2/H19 and Wsb1/Nf1. Science. 2006;312:269–272. doi: 10.1126/science.1123191. [DOI] [PubMed] [Google Scholar]
- Liu J, Krantz ID. Cohesin and human disease. Annu. Rev. Genomics Hum. Genet. 2008;9:303–320. doi: 10.1146/annurev.genom.9.081307.164211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Krantz ID. Cornelia de Lange syndrome, cohesin, and beyond. Clin. Genet. 2009;76:303–314. doi: 10.1111/j.1399-0004.2009.01271.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Zhang Z, Bando M, Itoh T, Deardorff MA, Clark D, Kaur M, Tandy S, Kondoh T, Rappaport E, Spinner NB, Vega H, Jackson LG, Shirahige K, Krantz ID. Transcriptional dysregulation in NIPBL and cohesin mutant human cells. PLoS Biol. 2009;7:e1000119. doi: 10.1371/journal.pbio.1000119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losada A, Hirano T. Intermolecular DNA interactions stimulated by the cohesin complex in vitro: Implications for sister chromatid cohesion. Curr. Biol. 2001;11:268–272. doi: 10.1016/s0960-9822(01)00066-5. [DOI] [PubMed] [Google Scholar]
- Losada A, Yokochi T, Hirano T. Functional contribution of Pds5 to cohesin-mediated cohesion in human cells and Xenopus egg extracts. J. Cell Sci. 2005;118:2133–2141. doi: 10.1242/jcs.02355. [DOI] [PubMed] [Google Scholar]
- Losada A, Yokochi T, Kobayashi R, Hirano T. Identification and characterization of SA/Scc3p subunits in the Xenopus and human cohesin complexes. J. Cell Biol. 2000;150:405–416. doi: 10.1083/jcb.150.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo H, Li Y, Mu JJ, Zhang J, Tonaka T, Hamamori Y, Jung SY, Wang Y, Qin J. Regulation of intra-S phase checkpoint by IR-dependent and IR-independent phosphorylation of SMC3. J. Biol. Chem. 2008 doi: 10.1074/jbc.M802299200. Epub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAlpine HK, Gordân R, Powell SK, Hartemink AJ, MacAlpine DM. Drosophila ORC localizes to open chromatin and marks sites of cohesin complex loading. Genome Res. 2010;20:201–211. doi: 10.1101/gr.097873.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumder P, Boss JM. CTCF controls expression and chromatin architecture of the human major histocompatibility complex class II locus. Mol. Cell. Biol. 2010;30:4211–4223. doi: 10.1128/MCB.00327-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishiro T, Ishihara K, Hino S, Tsutsumi S, Aburatani H, Shirahige K, Kinoshita Y, Nakao M. Architectural roles of multiple chromatin insulators at the human apolipoprotein gene cluster. EMBO J. 2009;28:1234–1245. doi: 10.1038/emboj.2009.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misulovin Z, Schwartz YB, Li XY, Kahn TG, Gause M, Macarthur S, Fay JC, Eisen MB, Pirrotta V, Biggin MD, Dorsett D. Association of cohesin and Nipped-B with transcriptionally active regions of the Drosophila melanogaster genome. Chromosoma. 2008;117:89–102. doi: 10.1007/s00412-007-0129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musio A, Mariani T, Montagna C, Zambroni D, Ascoli C, Ried T, Vezzoni P. Recapitulation of the Roberts syndrome cellular phenotype by inhibition of INCENP, ZWINT-1 and ZW10 genes. Gene. 2004;331:33–40. doi: 10.1016/j.gene.2004.01.028. [DOI] [PubMed] [Google Scholar]
- Musio A, Selicorni A, Focarelli ML, Gervasini C, Milani D, Russo S, Vezzoni P, Larizza L. X-linked Cornelia de Lange syndrome owing to SMC1L1 mutations. Nat. Genet. 2006;38:528–530. doi: 10.1038/ng1779. [DOI] [PubMed] [Google Scholar]
- Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SIS. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- Nasmyth K, Haering CH. Cohesin: its roles and mechanisms. Annu. Rev. Genet. 2009;43:525–528. doi: 10.1146/annurev-genet-102108-134233. [DOI] [PubMed] [Google Scholar]
- Nativio R, Wendt KS, Ito Y, Huddleston JE, Uribe-Lewis S, Woodfine K, Krueger C, Reik W, Peters JM, Murrell A. Cohesin is required for higher-order chromatin conformation at the imprinted IGF2-H19 locus. PLoS Genet. 2009;5:e1000739. doi: 10.1371/journal.pgen.1000739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama T, Ladurner R, Schmitz J, Kreidl E, Schleiffer A, Bhaskara V, Bando M, Shirahige K, Hyman AA, Mechtler K, Peters JM. Sororin mediates sister chromatid cohesion by antagonizing Wapl. Cell. 2010;143:737–749. doi: 10.1016/j.cell.2010.10.031. [DOI] [PubMed] [Google Scholar]
- Nitzsche A, Paszkowski-Rogacz M, Matarese F, Janssen-Megens EM, Hubner NC, Schulz H, de Vries I, Ding L, Huebner N, Mann M, Stunnenberg HG, Buchholz F. RAD21 Cooperates with Pluripotency Transcription Factors in the Maintenance of Embryonic Stem Cell Identity. PLos ONE. 2011;6:e19470. doi: 10.1371/journal.pone.0019470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonaka N, Kitajima T, Yokobayashi S, Xiao G, Yamamoto M, Grewal SS, Watanabe Y. Recruitment of cohesin to heterochromatic regions by Swi6/HP1 in fission yeast. Nat. Cell Biol. 2002;4:89–93. doi: 10.1038/ncb739. [DOI] [PubMed] [Google Scholar]
- Parelho V, Hadjur S, Spivakov M, Leleu M, Sauer S, Gregson HC, Jarmuz A, Canzonetta C, Webster Z, Nesterova T, Cobb BS, Yokomori K, Dillon N, Aragon L, Fisher AG, Merkenschlager M. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell. 2008;132:422–433. doi: 10.1016/j.cell.2008.01.011. [DOI] [PubMed] [Google Scholar]
- Patrinos GP, de Krom M, de Boer E, Langeveld A, Imam AMA, Strouboulis J, de Laat W, Grosveld FG. Multiple interactions between regulatory regions are required to stabilize an active chromatin hub. Genes Dev. 2004;18:1495–1509. doi: 10.1101/gad.289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli A, van Bemmel JG, Oliveira RA, Itoh T, Shirahige K, van Steensel B, Nasmyth K. A direct role for cohesin in gene regulation and ecdysone response in Drosophila salivary glands. Curr. Biol. 2010;20:1787–1798. doi: 10.1016/j.cub.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potts PR, Porteus MH, Yu H. Human SMC5/6 complex promotes sister chromatid homologous recombination by recruiting the SMC1/3 cohesin complex to double-strand breaks. EMBO J. 2006;25:3377–3388. doi: 10.1038/sj.emboj.7601218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratajska M, Wierzba J, Pehlivan D, Xia Z, Brundage EK, Cheung SW, Stankiewicz P, Lupski JR, Limon J. Cornelia de Lange syndrome case due to genomic rearrangements including NIPBL. Eur. J. Med. Genet. 2010;53:378–382. doi: 10.1016/j.ejmg.2010.08.002. [DOI] [PubMed] [Google Scholar]
- Revenkova E, Focarelli ML, Susani L, Paulis M, Bassi MT, Mannini L, Frattini A, Delia D, Krantz I, Vezzoni P, Jessberger R, Musio A. Cornelia de Lange syndrome mutations in SMC1A or SMC3 affect binding to DNA. Hum. Mol. Genet. 2009;18:418–427. doi: 10.1093/hmg/ddn369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes JM, Bentley FK, Print CG, Dorsett D, Misulovin Z, Dickinson EJ, Crosier KE, Crosier PS, Horsfield JA. Positive regulation of c-Myc by cohesin is direct, and evolutionarily conserved. Dev Biol. 2010;344:637–649. doi: 10.1016/j.ydbio.2010.05.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedel CG, Katis VL, Katou Y, Mori S, Itoh T, Helmhart W, Gálová M, Petronczki M, Gregan J, Cetin B, Mudrak I, Ogris E, Mechtler K, Pelletier L, Buchholz F, Shirahige K, Nasmyth K. Protein phosphatase 2A protects centromeric sister chromatid cohesion during meiosis I. Nature. 2006;441:53–61. doi: 10.1038/nature04664. [DOI] [PubMed] [Google Scholar]
- Rolef Ben-Shahar T, Heeger S, Lehane C, East P, Flynn H, Skehel M, Uhlmann F. Eco1-dependent cohesin acetylation during establishment of sister chromatid cohesion. Science. 2008;321:563–566. doi: 10.1126/science.1157774. [DOI] [PubMed] [Google Scholar]
- Rollins RA, Korom M, Aulner N, Martens A, Dorsett D. Drosophila Nipped-B protein supports sister chromatid cohesion and opposes the stromalin/Scc3 cohesion factor to facilitate long-range activation of the cut gene. Mol. Cell. Biol. 2004;24:3100–3111. doi: 10.1128/MCB.24.8.3100-3111.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio ED, Reiss DJ, Welcsh PL, Disteche CM, Filippova GN, Baliga NS, Aebersold R, Ranish JA, Krumm A. CTCF physically links cohesin to chromatin. Proc. Natl. Acad. Sci. 2008;105:8309–8314. doi: 10.1073/pnas.0801273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaaf CA, Misulovin Z, Sahota G, Siddiqui AM, Schwartz YB, Kahn TG, Pirrotta V, Gause M, Dorsett D. Regulation of the Drosophila Enhancer of split and invected-engrailed gene complexes by sister chromatid cohesion proteins. PLos ONE. 2009;4:e6202. doi: 10.1371/journal.pone.0006202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D, Schwalie P, Ross-Innes CS, Hurtado A, Brown G, Carroll J, Flicek P, Odom D. A CTCF-independent role for cohesin in tissue-specific transcription. Genome Res. 2010;20:578–588. doi: 10.1101/gr.100479.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmiesing JA, Ball AR, Gregson HC, Alderton J, Zhou S, Yokomori K. Identification of two distinct human SMC protein complexes involved in mitotic chromosome dynamics. Proc. Natl. Acad. Sci. USA. 1998;95:12906–12911. doi: 10.1073/pnas.95.22.12906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitan VC, Banks P, Laval S, Majid NA, Dorsett D, Rana A, Smith J, Bateman A, Krpic S, Hostert A, Rollins RA, Erdjument-Bromage H, Tempst P, Benard CY, Hekimi S, Newbury SF, Strachan T. Metazoan Scc4 Homologs Link Sister Chromatid Cohesion to Cell and Axon Migration Guidance. PLoS Biol. 2006;4 doi: 10.1371/journal.pbio.0040242. Epub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selicorni A, Russo S, Gervasini C, Castronovo P, Milani D, Cavalleri F, Bentivegna A, Masciadri M, Domi A, Divizia MT, Sforzini C, Tarantino E, Memo L, Scarano G, Larizza L. Clinical score of 62 Italian patients with Cornelia de Lange syndrome and correlations with the presence and type of NIPBL mutation. Clin. Genet. 2007;72:98–108. doi: 10.1111/j.1399-0004.2007.00832.x. [DOI] [PubMed] [Google Scholar]
- Serrano A, Rodríguez-Corsino M, Losada A. Heterochromatin protein 1 (HP1) proteins do not drive pericentromeric cohesin enrichment in human cells. PLos ONE. 2009;4:e5118. doi: 10.1371/journal.pone.0005118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton T, Bantignies F, Cavalli G. Genomic interactions: chromatin loops and gene meeting points in transcriptional regulation. Semin Cell Dev Biol. 2009;20:849–855. doi: 10.1016/j.semcdb.2009.06.004. [DOI] [PubMed] [Google Scholar]
- Shintomi K, Hirano T. Sister chromatid resolution: a cohesin releasing network and beyond. Chromosoma. 2010;119:459–467. doi: 10.1007/s00412-010-0271-z. [DOI] [PubMed] [Google Scholar]
- Spilianakis CG, Flavell RA. Long-range intrachromosomal interactions in the T helper type 2 cytokine locus. Nat. Immunol. 2004;5:1017–1027. doi: 10.1038/ni1115. [DOI] [PubMed] [Google Scholar]
- Spilianakis CG, Lalioti MD, Town T, Lee GR, Flavell RA. Interchromosomal associations between alternatively expressed loci. Nature. 2005;435:637–645. doi: 10.1038/nature03574. [DOI] [PubMed] [Google Scholar]
- Stedman W, Kang H, Lin S, Kissil JL, Bartolomei MS, Lieberman PM. Cohesins localize with CTCF at the KSHV latency control region and at cellular c-myc and H19/Igf2 insulators. EMBO J. 2008;27:654–666. doi: 10.1038/emboj.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ström L, Karlsson C, Lindroos HB, Wedahl S, Katou Y, Shirahige K, Sjögren C. Postreplicative formation of cohesion is required for repair and induced by a single DNA break. Science. 2007;317:242–245. doi: 10.1126/science.1140649. [DOI] [PubMed] [Google Scholar]
- Ström L, Lindroos HB, Shirahige K, Sjögren C. Postreplicative recruitment of cohesin to double-strand breaks is required for DNA repair. Mol. Cell. 2004;16:1003–1015. doi: 10.1016/j.molcel.2004.11.026. [DOI] [PubMed] [Google Scholar]
- Strübbe G, Popp C, Schmidt A, Pauli A, Ringrose L, Beisel C, Paro R. Polycomb purification by in vivo biotinylation tagging reveals cohesin and Trithorax group proteins as interaction partners. Proc. Natl. Acad. Sci. 2011;108:5572–5577. doi: 10.1073/pnas.1007916108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su RC, Brown KE, Saaber S, Fisher AG, Merkenschlager M, Smale ST. Dynamic assembly of silent chromatin during thymocyte maturation. Nat. Genet. 2004;36:502–506. doi: 10.1038/ng1351. [DOI] [PubMed] [Google Scholar]
- Sumara I, Vorlaufer E, Gieffers C, Peters BH, Peters J-M. Characterization of vertebrate cohesin complexes and their regulation in prophase. J. Cell Biol. 2000;151:749–761. doi: 10.1083/jcb.151.4.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumara I, Vorlaufer E, Stukenberg PT, Kelm O, Redemann N, Nigg EA, Peters J-M. The dissociation of cohesin from chromosomes in prophase is regulated by Polo-like kinase. Mol. Cell. 2002;9:515–525. doi: 10.1016/s1097-2765(02)00473-2. [DOI] [PubMed] [Google Scholar]
- Sutani T, Kawaguchi T, Kanno R, Itoh T, Shirahige K. Budding yeast Wpl1(Rad61)-Pds5 complex counteracts sister chromatid cohesion-establishing reaction. Curr. Biol. 2009;19:492–497. doi: 10.1016/j.cub.2009.01.062. [DOI] [PubMed] [Google Scholar]
- Takahashi TS, Yiu P, Chou MF, Gygi S, Walter JC. Recruitment of Xenopus Scc2 and cohesin to chromatin requires the pre-replication complex. Nat. Cell Biol. 2004;6:991–996. doi: 10.1038/ncb1177. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Cosma MP, Wirth K, Nasmyth K. Identification of cohesin association sites at centromeres and along chromosome arms. Cell. 1999;98:847–858. doi: 10.1016/s0092-8674(00)81518-4. [DOI] [PubMed] [Google Scholar]
- Terret ME, Sherwood R, Rahman S, Qin J, Jallepalli PV. Cohesin acetylation speeds the replication fork. Nature. 2009;462:231–234. doi: 10.1038/nature08550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol. Cell. 2002;10:1453–1465. doi: 10.1016/s1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- Tomkins D, Hunter A, Roberts M. Cytogenetic findings in Roberts-SC phocomelia syndrome(s) Am J Med Genet. 1979;4:17–26. doi: 10.1002/ajmg.1320040104. [DOI] [PubMed] [Google Scholar]
- Tonkin ET, Wang TJ, Lisgo S, Bamshad MJ, Strachan T. NIPBL, encoding a homolog of fungal Scc2-type sister chromatid cohesion proteins and fly Nipped-B, is mutated in Cornelia de Lange syndrome. Nat. Genet. 2004;36:636–641. doi: 10.1038/ng1363. [DOI] [PubMed] [Google Scholar]
- Tumbar T, Sudlow S, Belmont AS. Large-scale chromatin unfolding and remodeling induced by VP16 acidic activation domain. J. Cell Biol. 1999;145:1341–1354. doi: 10.1083/jcb.145.7.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlmann F, Lottspeich F, Nasmyth K. Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature. 1999;400:37–42. doi: 10.1038/21831. [DOI] [PubMed] [Google Scholar]
- Uhlmann F, Wernic D, Poupart M-A, Koonin EV, Nasmyth K. Cleavage of cohesin by the CD clan protease separin triggers anaphase in yeast. Cell. 2000;103:375–386. doi: 10.1016/s0092-8674(00)00130-6. [DOI] [PubMed] [Google Scholar]
- Ünal E, Arbel-Eden A, Sattler U, Shroff R, Lichten M, Haber JE, Koshland D. DNA damage response pathway uses histone modification to assemble a double-strand break-specific cohesin domain. Mol. Cell. 2004;16:991–1002. doi: 10.1016/j.molcel.2004.11.027. [DOI] [PubMed] [Google Scholar]
- Ünal E, Heidinger-Pauli JM, Kim W, Guacci V, Onn I, Gygi SP, Koshland DE. A molecular determinant for the establishment of sister chromatid cohesion. Science. 2008;321:566–569. doi: 10.1126/science.1157880. [DOI] [PubMed] [Google Scholar]
- Ünal E, Heidinger-Pauli JM, Koshland D. DNA double-strand breaks trigger genome-wide sister-chromatid cohesion through Eco1 (Ctf7) Science. 2007;317:245–248. doi: 10.1126/science.1140637. [DOI] [PubMed] [Google Scholar]
- van der Lelij P, Godthelp BC, van Zon W, van Gosliga D, Oostra AB, Steltenpool J, de Groot J, Scheper RJ, Wolthuis RM, Waisfisz Q, Darroudi F, Joenje H, de Winter JP. The cellular phenotype of Roberts syndrome fibroblasts as revealed by ectopic expression of ESCO2. PLos ONE. 2009;4:e6936. doi: 10.1371/journal.pone.0006936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega H, Waisfisz Q, Gordillo M, Sakai N, Yanagihara I, Yamada M, van Gosliga D, Kayserili H, Xu C, Ozono K, Jabs EW, Inui K, Joenje H. Roberts syndrome is caused by mutations in ESCO2, a human homolog of yeast ECO1 that is essential for the establishment of sister chromatid cohesion. Nat. Genet. 2005;37:468–470. doi: 10.1038/ng1548. [DOI] [PubMed] [Google Scholar]
- Vrouwe MG, Elghalbzouri-Maghrani E, Meijers M, Schouten P, Godthelp BC, Bhuiyan ZA, Redeker EJ, Mannens MM, Mullenders LH, Pastink A, Darroudi F. Increased DNA damage sensitivity of Cornelia de Lange syndrome cells: evidence for impaired recombinational repair. Hum. Mol. Genet. 2007;16:1478–1487. doi: 10.1093/hmg/ddm098. [DOI] [PubMed] [Google Scholar]
- Waizenegger IC, Hauf S, Meinke A, Peters J-M. Two distinct pathways remove mammalian cohesin from chromosome arms in prophase and from centromeres in anaphase. Cell. 2000;103:399–410. doi: 10.1016/s0092-8674(00)00132-x. [DOI] [PubMed] [Google Scholar]
- Wallace JA, Felsenfeld G. We gather together: insulators and genome organization. Curr Opin Genet Dev. 2007;17:400–407. doi: 10.1016/j.gde.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watrin E, Peters JM. The cohesin complex is required for the DNA damage-induced G2/M checkpoint in mammalian cells. EMBO J. 2009;28:2625–2635. doi: 10.1038/emboj.2009.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watrin E, Schleiffer A, Tanaka K, Eisenhaber F, Nasmyth K, Peters JM. Human Scc4 is required for cohesin binding to chromatin, sister-chromatid cohesion, and mitotic progression. Curr. Biol. 2006;16:863–874. doi: 10.1016/j.cub.2006.03.049. [DOI] [PubMed] [Google Scholar]
- Weitzer S, Lehane C, Uhlmann F. A model for ATP hydrolysis-dependent binding of cohesin to DNA. Curr. Biol. 2003;13:1930–1940. doi: 10.1016/j.cub.2003.10.030. [DOI] [PubMed] [Google Scholar]
- Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, Tsutsumi S, Nagae G, Ishihara K, Mishiro T, Yahata K, Imamoto F, Aburatani H, Nakao M, Imamoto N, Maeshima K, Shirahige K, Peters JM. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- Wong RW. Interaction between Rae1 and cohesin subunit SMC1 is required for proper spindle formation. Cell Cycle. 2010;9:198–200. doi: 10.4161/cc.9.1.10431. [DOI] [PubMed] [Google Scholar]
- Wong RW, Blobel G. Cohesin subunit SMC1 associates with mitotic microtubules at the spindle pole. Proc. Natl. Acad. Sci. 2008;105:15441–15445. doi: 10.1073/pnas.0807660105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood AJ, Severson AF, Meyer BJ. Condensin and cohesin complexity: the expanding repertoire of functions. Nat. Rev. Genet. 2010;11:391–404. doi: 10.1038/nrg2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao T, Wallace J, Felsenfeld G. Specific Sites in the C Terminus of CTCF Interact with the SA2 Subunit of the Cohesin Complex and Are Required for Cohesin-Dependent Insulation Activity. Mol. Cell Biol. 2011;31:2174–2183. doi: 10.1128/MCB.05093-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N, Donohoe ME, Silva SS, Lee JT. Evidence that homologous X-chromosome pairing requires transcription and Ctcf protein. Nat. Genet. 2007;39:1390–1396. doi: 10.1038/ng.2007.5. [DOI] [PubMed] [Google Scholar]
- Yao H, Brick K, Evrard Y, Xiao T, Camerini-Otero RD, Felsenfeld G. Mediation of CTCF transcriptional insulation by DEAD-box RNA-binding protein p68 and steroid receptor RNA activator SRA. Genes Dev. 2010;24:2543–2555. doi: 10.1101/gad.1967810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazdi PT, Wang Y, Zhao S, Patel N, Lee EY-HP, Qin J. SMC1 is a downstream effector in the ATM/NBS1 branch of the human S-phase checkpoint. Genes & Dev. 2002;16:571–582. doi: 10.1101/gad.970702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusufzai TM, Tagami H, Nakatani Y, Felsenfeld G. CTCF tethers an insulator to subnuclear sites, suggesting shared insulator mechanisms across species. Mol. Cell. 2004;13:291–298. doi: 10.1016/s1097-2765(04)00029-2. [DOI] [PubMed] [Google Scholar]
- Zeng W, de Greef JC, Chen Y-Y, Chien R, Kong X, Gregson HC, Winokur ST, Pyle A, Robertson KD, Schmiesing JA, Kimonis VE, Balog J, R. FR, Ball J,AR, Lock LF, Donovan PJ, van der Maarel S, Yokomori K. Specific loss of histone H3 lysine 9 trimethylation and HP1γ/cohesin binding at D4Z4 repeats is associated with facioscapulohumeral dystrophy (FSHD) PLoS Genet. 2009;5:e1000559. doi: 10.1371/journal.pgen.1000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Shi X, Li Y, Kim BJ, Jia J, Huang Z, Yang T, Fu X, Jung SY, Wang Y, Zhang P, Kim ST, Pan X, Qin J. Acetylation of Smc3 by Eco1 is required for S phase sister chromatid cohesion in both human and yeast. Mol. Cell. 2008;31:143–151. doi: 10.1016/j.molcel.2008.06.006. [DOI] [PubMed] [Google Scholar]
- Zlatanova J, Caiafa P. CTCF and its protein partners: divide and rule? J. Cell Sci. 2009;122:1275–1284. doi: 10.1242/jcs.039990. [DOI] [PubMed] [Google Scholar]



