Summary
Thiamine pyrophosphate (TPP) riboswitches regulate essential genes in bacteria by changing conformation upon binding intracellular TPP. Previous studies using fragment-based approaches identified small molecule “fragments” that bind this gene-regulatory mRNA domain. Crystallographic studies now show that, despite having micromolar Kds, four different fragments bind the TPP riboswitch site-specifically, occupying the pocket that recognizes the aminopyrimidine of TPP. Unexpectedly, the unoccupied site that would recognize the pyrophosphate of TPP rearranges into a structure distinct from that of the cognate complex. This idiosyncratic fragment-induced conformation, also characterized by small-angle X-ray scattering (SAXS) and chemical probing (SHAPE), represents a possible mechanism for adventitious ligand discrimination by the riboswitch, and suggests that off-pathway conformations of RNAs can be targeted for drug development. Our structures, together with previous screening studies, demonstrate the feasibility of fragment-based drug discovery against RNA targets.
Introduction
Riboswitches are cis-acting mRNA elements that specifically bind metabolites and modulate expression of genes involved in the metabolism of their cognate ligand (Roth and Breaker, 2009). Riboswitches are promising antimicrobial targets because of their highly specific small molecule recognition, prevalence in bacteria, and association with genes essential for pathogen survival or virulence. Moreover, retrospective studies on several antimicrobials for which the mechanism of action was previously unknown have revealed that they target riboswitches (Deigan and Ferré-D’Amaré, 2011).
Fragment-based approaches, in which weakly-binding small molecule “fragments” (Mr < 300 Da) are identified in modestly-sized screens and then elaborated into more potent ligands, have emerged as a promising method for drug discovery (Scott et al., 2012). Although extensively used against protein targets, only recently has this approach been applied to RNA, leading to the discovery of several small molecules that bind the Escherichia coli thiM TPP riboswitch aptamer with Kd between 20–700 μM (Cressina et al., 2010). Although even the most tightly-binding fragments did not influence expression of a downstream reporter gene in an in vitro assay (Cressina et al., 2010), a heterocyclic thiamine precursor has been shown to regulate expression of an endogenous gene via its TPP riboswitch (Moulin et al., 2013). Thus, even though substantial further development is necessary, fragments have the potential to be elaborated into ligands specific for TPP riboswitches. A critical step is to understand the exact mode by which the fragments bind RNA. Indeed, there are no reports showing that such weakly binding fragments can be crystallographically visualized bound to an RNA target. Further elaboration of the fragments is contingent on direct structural analysis that pinpoints their interaction with the riboswitch.
Results and Discussion
Structures of a TPP riboswitch in complex with fragments
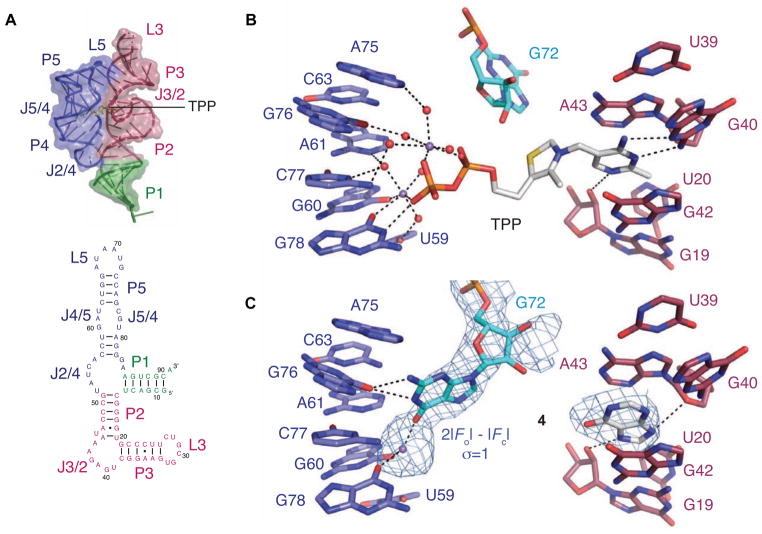
Riboswitch-TPP cocrystal structures demonstrate that this RNA forms a three-helix junction; two of the helical arms are bridged by the bound TPP (Figure 1A,B) (Edwards and Ferré-D’Amaré, 2006; Kulshina et al., 2010; Serganov et al., 2006; Thore et al., 2006). The aminopyrimidine ring of TPP is recognized by stacking and base pairing-like interactions with the J3/2 joining region of the “pyrimidine sensor helix”. The thiazole ring of TPP is in van der Waals contact with the sugar of G72 and, in some TPP-bound structures, the nucleobase of G72. The pyrophosphate moiety of TPP is coordinated by two partially hydrated divalent cations (Ba2+, Mg2+, or Mn2+ in different crystal structures), and makes direct, and water- and cation-mediated, contacts with residues in J4/5 and J5/4 in the “pyrophosphate sensor helix”. The RNA recognizes TPP as a divalent metal-ion chelate, thereby overcoming its unfavorable electrostatic character.
Figure 1. TPP- and fragment-bound riboswitch structures.
(A) Three-dimensional structure of the TPP riboswitch bound to TPP (Edwards and Ferré-D’Amaré, 2006) and secondary structure of the TPP riboswitch.
(B) Interactions between the TPP riboswitch and bound cations with TPP. Metal ions and water molecules are in purple and red, respectively.
(C) Interactions between the TPP riboswitch and bound cation with fragment 4. A portion of a composite, simulated annealing-omit 2|Fo| - |Fc| Fourier synthesis, contoured around the final refined fragment 4, G72, and the bound metal ion at 1.0 sd, is in cyan.
See also Table S1, Figures S1, S2, S3 and S5.
To understand how compounds considerably smaller than TPP can bind the TPP riboswitch specifically, and as a starting point for elaborating these into more potent ligands, we determined crystal structures of the E. coli thiM TPP riboswitch in complex with compounds 1–4, identified by previous fragment-based discovery experiments (Cressina et al., 2010) (Table 1) at resolutions between 2.65 Å and 3.1 Å (Table S1). These four fragments selectively bind a TPP riboswitch over an unrelated lysine riboswitch, and are representative of the range of Kd values and structural diversity of the set of 17 fragments identified by Cressina et al. (2010). In each case, the RNA adopts a global structure that superimposes closely (rmsd = 0.6–0.8 Å for 78 or 79 C1’ atom pairs) on that of its TPP complex (Edwards and Ferré-D’Amaré, 2006; Serganov et al., 2006). Well-defined |Fo|-|Fc| electron density features (peak heights > 5 sd) appropriate for each of the fragments were present in J3/2 of the RNA (Figure S1). Even hypoxanthine (4), which binds the riboswitch with Kd = 670 μM, exhibited clear unbiased electron density.
Table 1.
Structure and Kd values for TPP, thiamine, and fragments 1–4 (Cressina et al., 2010). See also Table S3.
| compound | structure | Kd (μM) |
|---|---|---|
| TPP |
|
0.01 – 0.1 |
| thiamine |
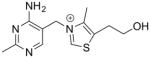
|
1.5 |
| 1 |
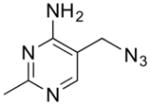
|
49 ± 4 |
| 2 |
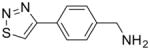
|
103 ± 6 |
| 3 |
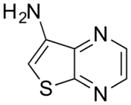
|
280 ± 30 |
| 4 |
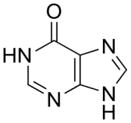
|
670 ± 50 |
Consistent with their conjugated heterocyclic character, fragments 1–4 all occupy the aminopyrimidine-binding site of the riboswitch. Fragment 4 is representative (Figure 1C). It stacks between the nucleobases of G42 and A43 and forms hydrogen bonds with nucleotides in J3/2 and in P2, reminiscent of the recognition of the aminopyrimidine moiety of TPP.
Idiosyncratic rearrangement of an unoccupied site
In addition to exhibiting features in the aminopyrimidine-binding site corresponding to the fragments, our experimental electron density maps unexpectedly indicate that G72 from the pyrophosphate sensor helix has rearranged. Alignment of TPP-bound E. coli thiM TPP riboswitch structures shows G72 in three distinct conformations (Figure S3). In our structures of the riboswitch bound to 1–4, G72 adopts an unprecedented conformation, with its nucleobase pointing into the pyrophosphate binding pocket, where it makes contacts with J4/5 or J5/4 through N1 and N2 (Figure 1C, Figure S1). In addition, in the structures bound to 2–4, unambiguous anomalous difference electron density corresponding to one Mn2+ is observed (a Mg2+ is observed in the complex with 1, and a second cation is observed in the complex with 3). The metal ion is located in the same position as one of the pyrophosphate-bound divalent cations in the TPP complex. Rather than being coordinated by the pyrophosphate of TPP, however, in the complexes with 1–4, the cation is coordinated by the Watson-Crick face of the rearranged G72.
SAXS and SHAPE characterization of fragment-induced riboswitch folding
Previous SAXS experiments have shown that the thiM TPP riboswitch compacts in the presence of physiological Mg2+ concentrations, but achieves complete folding only when it has bound TPP (Baird and Ferré-D’Amaré, 2010). Although the conformation of the riboswitch bound to 1–4 in our cocrystals is very similar to that of the TPP-bound RNA, it is possible that crystallization selected a subset of molecules that had achieved complete folding. To examine the average global conformation of the fragment-bound riboswitch, we performed SAXS experiments using 2 as a representative ligand. As judged by the radius of gyration (Rg) and the distance distribution function P(r), binding to this fragment results in a degree of compaction intermediate between those of the free and TPP-bound riboswitch (Figure 2A and Table S2).
Figure 2. SAXS analysis of fragment-induced RNA folding.
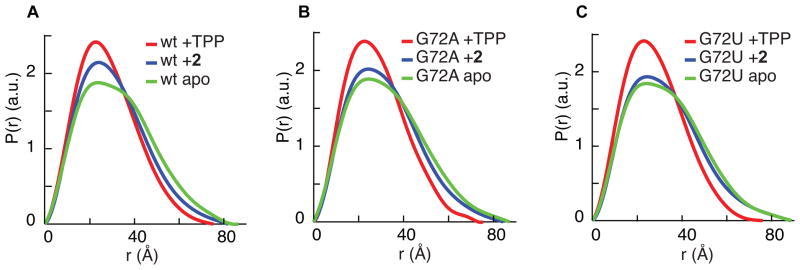
(A) SAXS analysis of the wild-type TPP riboswitch. P(r) plots reflect differences in shape and maximum dimension between the ligand-free, TPP-bound and fragment 2-bound states of the riboswitch. SAXS behavior of the wild-type RNA, free or bound to TPP, is consistent with a previous report (Baird and Ferré-D’Amaré, 2010).
(B) SAXS analysis of the G72A TPP riboswitch.
(C) SAXS analysis of the G72U TPP riboswitch.
See also Table S2.
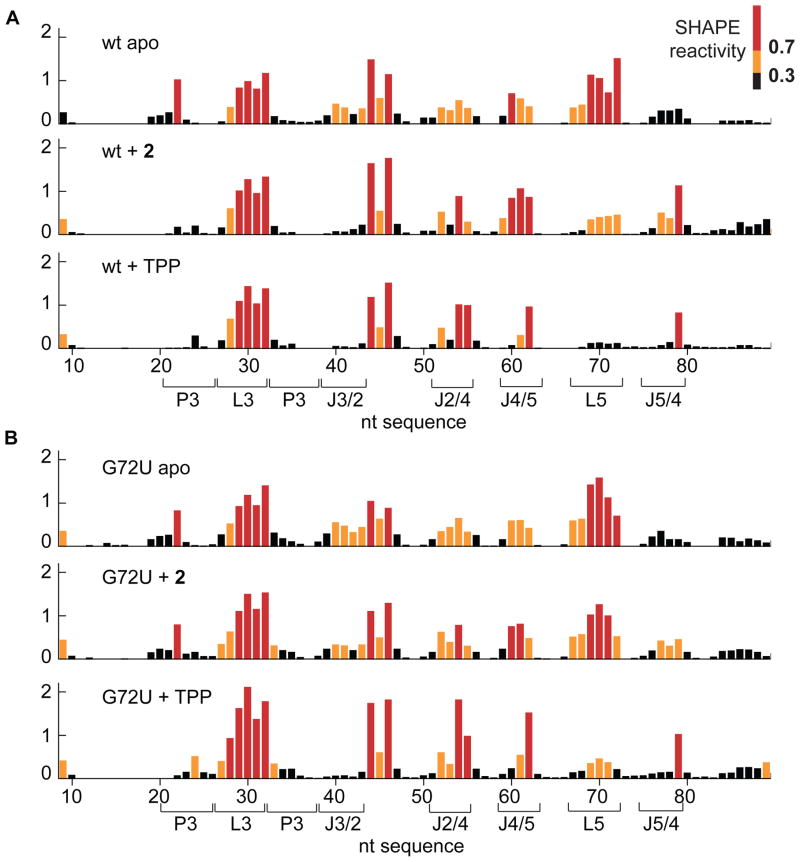
We probed the effect of fragment binding to the RNA at single-nucleotide resolution using SHAPE (Wilkinson et al., 2006) (Figure 3A). The riboswitch bound to 2 exhibits regions that have folded to approximately the TPP-bound structure, while other regions are only partially folded. At J3/2 and P3, which together form the pyrimidine-binding pocket, the SHAPE reactivity is low in the fragment-bound structure, in agreement with the crystallographic observation that the fragment is bound similarly to the aminopyrimidine of TPP. At J2/4, J4/5 and J5/4, which together form the pyrophosphate-binding pocket, as well as in L5, the SHAPE reactivity of the fragment-bound RNA is intermediate between those of the free and TPP-bound states.
Figure 3. SHAPE analysis of fragment-induced RNA folding.
(A) SHAPE analysis of the ligand-free, fragment 2-bound and TPP-bound native sequence riboswitch. Black, yellow and red in SHAPE profiles denote low, medium and high reactivities, respectively. SHAPE data on the wild-type RNA, free or bound to TPP, are consistent with a previous report (Steen et al., 2010).
(B) SHAPE analysis of ligand-free, fragment 2-bound and TPP-bound G72U mutant riboswitches.
See also Figure S4.
Importance of G72 for fragment-induced folding
We examined the role of G72 in fragment-induced partial folding by synthesizing G72A and G72U mutant RNAs, and examining them free or bound to either 2 or TPP. SAXS shows that while the mutations have little impact on the overall conformations of TPP-bound or free RNA, the mutants are less compact in the presence of the fragment (Figure 2B,C and Table S2), indicating that the G72 mutations inhibit folding, G72U more so than G72A. In SHAPE experiments, the ligand-free mutants also show little change from the wild-type, while the TPP-bound mutants are slightly less ordered than the native sequence (Figure 3B, Figure S4). In agreement with the SAXS experiments, SHAPE experiments on the fragment-bound mutants show that mutations at G72 (particularly G72U) inhibit the ability of the RNA to fold in the presence of 2. Throughout the entire RNA, but particularly in L5 and in the TPP-binding site (J3/2, J5/4), the SHAPE reactivity of the 2-bound mutant RNAs is closer to that of the free RNA than to the TPP-bound RNA.
Our SHAPE and SAXS data show that a point mutation at G72, which does not contact fragments 1–4, destabilizes the fragment-bound TPP riboswitch. G72 is thus critical for fragment-induced partial folding of the TPP riboswitch, presumably due to its metal-mediated and direct interactions with the pyrophosphate sensor helix. Nucleotides that form the pyrophosphate binding pocket have previously been shown to be critical for gene regulation in vivo, and are strongly conserved (Kulshina et al., 2010; Ontiveros-Palacios et al., 2008). G72 mutants, especially G72U, are unable to recapitulate these contacts to stabilize the pyrophosphate-binding site, which impairs the ability of the RNA to fold in the presence of the fragments.
We investigated further the function of G72 in TPP riboswitch stabilization through isothermal titration calorimetry (ITC) (Table S3). The G72U mutant binds 2 with Kd ~ 100 μM, essentially the same as wild-type, suggesting that disruption of fragment-induced folding in a G72U mutant is not due to weaker fragment binding. Surprisingly, ITC revealed that the G72U mutant binds TPP with Kd ~ 2 μM, approximately two orders of magnitude weaker than wild-type, even though G72U does not significantly impact the overall shape and local conformation of the unfolded free RNA or the folded TPP-bound RNA (SAXS and SHAPE data, respectively). This may be due to the role G72 plays in stabilizing the L5-P3 tertiary interactions, which have been shown previously to be critical for TPP binding and TPP-induced compaction (Kulshina et al., 2010).
Structure of a thiamine-bound TPP riboswitch
To understand the effect of an intermediate-size ligand on the stability of the pyrophosphate-binding pocket, we solved the cocrystal structure of the thiM TPP riboswitch with thiamine (Figure S5). We observe no direct contacts between thiamine and the pyrophosphate sensor helix. There is no electron density indicative of either metal ions or a reoriented G72 in the pyrophosphate-binding pocket. Furthermore, several nucleotides in the pyrophosphate-binding pocket are disordered. Relative to TPP or 1–4, thiamine is shifted out of the pyrimidine-binding pocket such that stacking of the aminopyrimidine is sub-optimal. The destabilization of the pyrophosphate-binding pocket and the position of thiamine is reminiscent of a structure of the RNA bound to the thiamine antimetabolite pyrithiamine (Edwards and Ferré-D’Amaré, 2006) (Figure S5).
Significance
Our crystallographic, SAXS, SHAPE and ITC analyses suggest that the pyrophosphate-binding site of the TPP riboswitch can be stabilized either through contacts with a metal ion-bound pyrophosphate moiety, or via contacts with a reoriented G72. Thiamine and pyrithiamine cannot make direct contacts with the pyrophosphate-binding site, but appear to be large enough to sterically block G72 reorientation. The inability of thiamine to stabilize the pyrophosphate helix may be a mechanism to facilitate discrimination in vivo between TPP and thiamine by the riboswitch.
Despite its polyanionic nature, RNA has been observed to bind anions and electronegative groups (Auffinger et al., 2004). An Rfam (Burge et al., 2013) analysis of 115 TPP riboswitches indicates that a purine is 99% conserved at residue 72. In addition to facilitating ligand discrimination, a purine at this position may aid in recognition of the pyrophosphate moiety of TPP, possibly through direct contacts via the nucleobase, similar to the interaction between a guanine in the glmS riboswitch-ribozyme and the phosphate of its cognate ligand, glucosamine-6-phosphate (Klein et al., 2007).
To our knowledge, our cocrystal structures provide the first direct structural demonstration of site-specific binding of fragments derived from a screen to a target biological RNA. Observation of specific binding provides proof of concept for RNA-targeted fragment-based methods, and insight into strategies for structure-based fragment elaboration against an RNA target. Our results reveal that the fragments studied here induce a novel, alternate conformation of the TPP riboswitch, distinct from that induced by the cognate ligand. This unexpected structural rearrangement may be exploited to develop small molecule chemical tools with a mechanism of action reminiscent of the anticancer drug Gleevec, which binds to an off-pathway conformation of the Abl kinase (Schindler et al., 2000), or of oligonucleotides that promote misfolding of a group I intron (Childs et al., 2002).
Experimental Procedures
Crystallization
For structures bound to 2, 3, 4 and thiamine, TPP riboswitch RNA transcribed from plasmid pTB13 (150 μM) was incubated with ligand (1 mM fragment 2, 1 mM fragment 3, 0.75 mM fragment 4, or 1 mM thiamine) in 5 mM Tris-HCl (pH 8.1), 3 mM MgCl2, 10 mM NaCl, 100 mM KCl, 0.5 mM spermine and 0–3% (v/v) DMSO at 37 °C for 30 min. Cocrystals were grown by vapor diffusion at 21 °C of hanging drops prepared by mixing 2 μL of an RNA-fragment complex solution with 1 μL of reservoir solution [2: 30% (w/v) polyethyleneglycol 2000 (PEG-2000), 0.35 M NH4Cl, 50 mM succinate-KOH (pH 7.0), 10 mM MnCl2; 3: 30% (w/v) PEG-2000, 0.5 M NH4Cl, 50 mM BisTris-HCl (pH 6.5), 10 mM MnCl2; 4: 30% (w/v) PEG-2000, 0.5 M NH4Cl, 50 mM succinate-KOH (pH 7.0), 10 mM MnCl2; thiamine: 30% (w/v) PEG-2000, 0.35 M NH4Cl, 50 mM succinate-KOH (pH 7.0), 10 mM ZnCl2]. Prior to data collection, crystals were transferred to drops containing reservoir solution supplemented with 1 mM fragment and cryoprotectant [15% (v/v) glycerol, 10–20% (v/v) ethylene glycol or 10–20% (w/v) sucrose].
For the structure bound to 1, TPP riboswitch RNA derived from the pIDT-TPP plasmid (500 μM) was mixed with fragment 1 (2 mM) in 50 mM KOAc (pH 6.9), 5 mM MgCl2 and 2% (v/v) DMSO. Cocrystals were grown by vapor diffusion at 21 °C of hanging drops prepared by mixing 1.5 μL of an RNA-fragment complex solution with 0.75 μL of reservoir solution [30% (w/v) polyethyleneglycol 4000 (PEG-4000), 0.1 M NaOAc (pH 5.1), 0.3 M NH4Cl]. Prior to data collection, crystals were transferred to a drop containing reservoir solution supplemented with an additional 5% (w/v) PEG-4000, 50 mM KOAc (pH 6.9), 5 mM MgCl2, and 2 mM fragment 1. Crystals were mounted in nylon loops and flash frozen by plunging into liquid nitrogen.
Diffraction data collection, structure determination, and refinement
Diffraction data were collected either at beamlines 5.0.1 or 5.0.2 of the Advanced Light Source (ALS) (2, 3, 4 and thiamine) and were collected, indexed, integrated, and scaled with the program HKL2000 (Otwinowski and Minor, 1997) or at beamline 24-ID-C of the Advanced Photon Source (APS) (1) and collected with the NE-CAT RAPD pipeline and indexed, integrated, and scaled with HKL2000 (Table S1). The structures were solved by molecular replacement, in which the E. coli thiM TPP riboswitch (1: PDB 2GDI; 2, 3, 4, thiamine: PDB 2HOJ) lacking TPP, metal ions or water molecules was employed as a search model in the program PHASER (McCoy et al., 2007). Rounds of manual rebuilding (Jones et al., 1991), interspersed with simulated-annealing, positional, and restrained individual B-factor refinement against a maximum likelihood target (Brünger et al., 1998), yielded the current crystallographic models (Table S1). Small-molecule coordinates, parameter files, and topology files were obtained from the PRODRG server (Schüttelkopf and van Aalten, 2004) for the fragments, and from the HIC-UP server (Kleywegt and Jones, 1998) for thiamine.
Supplementary Material
Highlights for.
Despite micromolar Kds, fragments bind a TPP riboswitch site-specifically
Fragment binding induces an off-pathway rearrangement of the ligand binding site
RNA rearrangement may suppress adventitious ligand binding in vivo
Structural studies prove feasibility of RNA-targeted fragment-based drug discovery
Acknowledgments
We thank the staff at beamlines 24-ID-C of APS and 5.0.1 and 5.0.2 of ALS for crystallographic data collection support, N. Baird, Y.-X. Wang and X. Fang for assistance with SAXS, G. Piszczek for assistance with ITC, and N. Baird, C. Jones, M. Lau, J. Posakony, S. Ryder, A. Serganov, M. Warner, and J. Zhang for discussions. This work is partly based on research conducted at ALS on the BCSB beamlines, and at APS on the NE-CAT beamlines, all of which are supported by the National Institute for General Medical Sciences, NIH. Use of ALS and APS was supported by the U.S. Department of Energy. This work was supported in part by the NIH (grant GM098662 to K.M.W.), the BBSRC of the UK, and the NIH-Oxford/Cambridge Research Scholars program. This work was supported in part by the intramural program of the National Heart, Lung and Blood Institute, NIH.
Footnotes
Accession Codes
Coordinates and structure factors have been deposited with the Protein Data Bank with accession codes 4NYA, 4NYB, 4NYC, 4NYD, and 4NYG, respectively, for the fragment 1-, 2-, 3-, 4-, and thiamine-bound structures.
Supplemental information includes three tables, five figures, additional experimental procedures, and supplemental references.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Auffinger P, Bielecki L, Westhof E. Anion binding to nucleic acids. Structure. 2004;12:379–388. doi: 10.1016/j.str.2004.02.015. [DOI] [PubMed] [Google Scholar]
- Baird NJ, Ferré-D’Amaré AR. Idiosyncratically tuned switching behavior of riboswitch aptamer domains revealed by comparative small-angle X-ray scattering analysis. RNA. 2010;16:598–609. doi: 10.1261/rna.1852310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Burge SW, Daub J, Eberhardt R, Tate J, Barquist L, Nawrocki EP, Eddy SR, Gardner PP, Bateman A. Rfam 11.0: 10 years of RNA families. Nucleic Acids Res. 2013;41:D226–D232. doi: 10.1093/nar/gks1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs JL, Disney MD, Turner DH. Oligonucleotide directed misfolding of RNA inhibits Candida albicans group I intron splicing. Proc Natl Acad Sci USA. 2002;99:11091–11096. doi: 10.1073/pnas.172391199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cressina E, Chen L, Abell C, Leeper FJ, Smith AG. Fragment screening against the thiamine pyrophosphate riboswitch thiM. Chem Sci. 2010;2:157–165. [Google Scholar]
- Deigan KE, Ferré-D’Amaré AR. Riboswitches: discovery of drugs that target bacterial gene-regulatory RNAs. Acc Chem Res. 2011;44:1329–1338. doi: 10.1021/ar200039b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards TE, Ferré-D’Amaré AR. Crystal structures of the thi-box riboswitch bound to thiamine pyrophosphate analogs reveal adaptive RNA-small molecule recognition. Structure. 2006;14:1459–1468. doi: 10.1016/j.str.2006.07.008. [DOI] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Klein DJ, Wilkinson SR, Been MD, Ferré-D’Amaré AR. Requirement of helix P2.2 and nucleotide G1 for positioning the cleavage site and cofactor of the glmS ribozyme. J Mol Biol. 2007;373:178–189. doi: 10.1016/j.jmb.2007.07.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleywegt GJ, Jones TA. Databases in protein crystallography. Acta Crystallogr D. 1998;54:1119–1131. doi: 10.1107/s0907444998007100. [DOI] [PubMed] [Google Scholar]
- Kulshina N, Edwards TE, Ferré-D’Amaré AR. Thermodynamic analysis of ligand binding and ligand binding-induced tertiary structure formation by the thiamine pyrophosphate riboswitch. RNA. 2010;16:186–196. doi: 10.1261/rna.1847310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulin M, Nguyen GTDT, Scaife MA, Smith AG, Fitzpatrick TB. Analysis of Chlamydomonas thiamin metabolism in vivo reveals riboswitch plasticity. Proc Natl Acad Sci USA. 2013;110:14622–14627. doi: 10.1073/pnas.1307741110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ontiveros-Palacios N, Smith AM, Grundy FJ, Soberon M, Henkin TM, Miranda-Ríos J. Molecular basis of gene regulation by the THI-box riboswitch. Mol Microbiol. 2008;67:793–803. doi: 10.1111/j.1365-2958.2007.06088.x. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Meth Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Roth A, Breaker RR. The structural and functional diversity of metabolite-binding riboswitches. Annu Rev Biochem. 2009;78:305–334. doi: 10.1146/annurev.biochem.78.070507.135656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler T, Bornmann W, Pellicena P, Miller WT, Clarkson B, Kuriyan J. Structural mechanism for STI-571 inhibition of abelson tyrosine kinase. Science. 2000;289:1938–1942. doi: 10.1126/science.289.5486.1938. [DOI] [PubMed] [Google Scholar]
- Schüttelkopf AW, van Aalten D. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr D. 2004;60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- Scott DE, Coyne AG, Hudson SA, Abell C. Fragment-based approaches in drug discovery and chemical biology. Biochemistry. 2012;51:4990–5003. doi: 10.1021/bi3005126. [DOI] [PubMed] [Google Scholar]
- Serganov A, Polonskaia A, Phan AT, Breaker RR, Patel DJ. Structural basis for gene regulation by a thiamine pyrophosphate-sensing riboswitch. Nature. 2006;441:1167–1171. doi: 10.1038/nature04740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steen KA, Malhotra A, Weeks KM. Selective 2′-hydroxyl acylation analyzed by protection from exoribonuclease. J Am Chem Soc. 2010;132:9940–9943. doi: 10.1021/ja103781u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thore S, Leibundgut M, Ban N. Structure of the eukaryotic thiamine pyrophosphate riboswitch with its regulatory ligand. Science. 2006;312:1208–1211. doi: 10.1126/science.1128451. [DOI] [PubMed] [Google Scholar]
- Wilkinson KA, Merino EJ, Weeks KM. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat Protoc. 2006;1:1610–1616. doi: 10.1038/nprot.2006.249. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.