Abstract
BMP signaling plays an essential role in second heart field-derived heart and arterial trunk development, including myocardial differentiation, right ventricular growth, and interventricular, outflow tract and aortico-pulmonary septation. It is mediated by a number of different BMP ligands, and receptors, many of which are present simultaneously. The mechanisms by which they regulate morphogenetic events and degree of redundancy amongst them have still to be elucidated. We therefore assessed the role of BMP Type I receptor AcvR1 in anterior second heart field-derived cell development, and compared it with that of BmpR1a.
By removing Acvr1 using the driver Mef2c[AHF]-Cre, we show that AcvR1 plays an essential role in arterial pole morphogenesis, identifying defects in outflow tract wall and cushion morphology that preceded a spectrum of septation defects from double outlet right ventricle to common arterial trunk in mutants. Its absence caused dysregulation in gene expression important for myocardial differentiation (Isl1, Fgf8) and regional identity (Tbx2, Tbx3, Tbx20, Tgfb2). Although these defects resemble to some degree those in the equivalent Bmpr1a mutant, a novel gene knock-in model in which Bmpr1a was expressed in the Acvr1 locus only partially restored septation in Acvr1 mutants. These data show that both BmpR1a and AcvR1 are needed for normal heart development, in which they play some non-redundant roles, and refine our understanding of the genetic and morphogenetic processes underlying Bmp-mediated heart development important in human congenital heart disease.
Keywords: cardiac development, second heart field, conotruncal defects, BMP signaling, receptors
Introduction
Heart morphogenesis is a complex process involving many secreted growth factors and their receptors (reviewed by (Bruneau, 2013)). Bone morphogenetic protein (BMP)-mediated signaling has been strongly implicated in the process by which separate ventricular outlets, valves and arterial trunks are formed from a single tube by septation requiring precise shape and positioning of contributory structures (reviewed by (Dyer and Kirby, 2009)). Failure in this process results in formation of outflow tract (OFT) septation defects, one of the common cardiac birth defects in humans.
The arterial pole of the developing heart is largely composed of mesodermally-derived second heart field (SHF), which forms myocardium of the interventricular septum and right ventricle (RV), and myocardium and future smooth muscle (arterial trunk) of the OFT wall. It also contributes to the adjacent endocardium, and cushion mesenchyme, especially proximally, by endocardial-to-mesenchymal transformation (EMT). The mesenchyme of OFT cushions is largely composed of cardiac neural crest-derived cells (CNCC), which also contribute to arterial trunk smooth muscle (Dyer and Kirby, 2009; Snarr et al., 2008; Verzi et al., 2005).
BMPs have been shown to exert complex, dose-dependent and even opposing effects on myocardial differentiation, functioning both as promoters and inhibitors of cardiac growth(de Pater et al., 2012). It has been suggested that a delicate balance between FGF and BMP signaling controls proliferation and differentiation of SHF progenitors(Hutson et al., 2010) and that BMPs can block FGF signaling (Tirosh-Finkel et al., 2010). In mouse embryos, inactivation of genes encoding BMPs 2,4 and 7 or BmpR1a (Bone morphogenetic protein receptor, type Ia) specifically in the second heart field has been shown to result in formation of common arterial trunk and OFT cushion defects (Bai et al., 2013; Briggs et al., 2013; Wang et al., 2010; Yang et al., 2006). BMPs have also been shown to regulate myocardial differentiation through a microRNA-mediated mechanism, and Bmpr1a needed for expression of T-box transcription factors Tbx2 and Tbx3. These are important for myocardial differentiation and arterial pole morphogenesis(Mesbah et al., 2012; Singh et al., 2012).
BMPs signal via tetrameric transmembrane receptor complexes composed of two Type II (BmpRII, AcvRIIa or AcvRIIb) and two Type I (BmpRIa [Alk3], BmpRIb [Alk6] or AcvRI [Alk2; Activin A receptor, type Is]) receptors(Massague and Chen, 2000). Ligand binding results in activation of Type I receptor kinase activity, which subsequently phosphorylates receptorregulated R-Smads 1,5 or 8 (Derynck and Zhang, 2003). Phosphorylated R-Smads and Smad4 form a complex which accumulates in the nucleus where it functions as a transcriptional coregulator. Evidence suggests there is a limited degree of redundancy between BMP receptors underlying regulation of developmental events (Kishigami and Mishina, 2005). The situation is complex; heteromeric as well as homomeric ligand dimers can bind with different binding affinities to oligomeric receptor complexes composed of different type II and type I and coreceptors(Ehrlich et al., 2012), at least in model systems.
Although an essential role for BmpRIa has been demonstrated, it is not the only type I receptor expressed in the heart. AcvRI has been shown to play critical roles in endocardial cushion and OFT valve development in mice (Kaartinen et al., 2004; Thomas et al., 2012; Wang et al., 2005) and mutations in ACVR1 have been implicated in congenital cardiac defects in humans (Smith et al., 2009). We therefore chose to investigate the role of AcvRI specifically in the second heart field and its derivatives.
Materials and Methods Mice
Mice
Mice carrying the conditional Acvr1-flox allele (Acvr1F), Bmpr1a-flox allele (Bmpr1aF), ‘constitutively active’ Acvr1 conditional allele (ACVR1 carrying Q207D mutation, ‘caAcvr1’) and Mef2c[AHF]-Cre transgenic mice (kindly provided by BL Black) were used and genotyped as previously described(Ahn et al., 2001; Fukuda et al., 2006; Kaartinen and Nagy, 2001; Verzi et al., 2005). Timed matings between Acvr1KO/WTMef2c[AHF]-Cre+ males and Acvr1F/F female mice were used to obtain Alk2F/KOMef2c[AHF]-Cre+ tissue-specific mutant embryos (‘Acvr1-cKO’). Some females were also R26RYFP/YFP or R26RlacZ/lacZ to enable Mef2c[AHF]-Cre lineage tracing, and FACS-based isolation of recombined second heart field cells (see below). Controls were either Cre0, or Acvr1F/WT, Mef2c[AHF]-Cre+ littermates (‘Acvr1-cHet’). To generate other experimental genotypes, females were generated carrying various combinations of floxed and wild type alleles of both Acvr1 and Bmpr1a, or with Acvr1Bmpr1a-KI, or Acvr1ACVR1-KI (see below, and main text), and Bmpr1F/WT Mef2c[AHF]-Cre+ and Acvr1F/WT,Bmpr1F/WT,Mef2c[AHF]-Cre+ males.
Ethics Statement
This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The experiments described in this study were specifically approved by the University Committee on Use and Care of Animals of the University of Michigan-Ann Arbor (Protocol Number: #09944).
Generation of Acvr1nlsLacZ bac transgenic mice
A nlsLacZ-pA cassette was inserted inframe into the first coding exon of Acvr1 (exon 3) in the bac RP23-366D15 (Children’s Hospital Oakland Research Institute) by using standard recombineering techniques(Warming et al., 2005) (see Figure 4). Bac transgenic mice were generated in the University of Michigan Transgenic Animal Model Core. Three of five founders displayed similar staining patterns consistent with authentic Acvr1 expression (data not show). The line #934 was used for further studies.
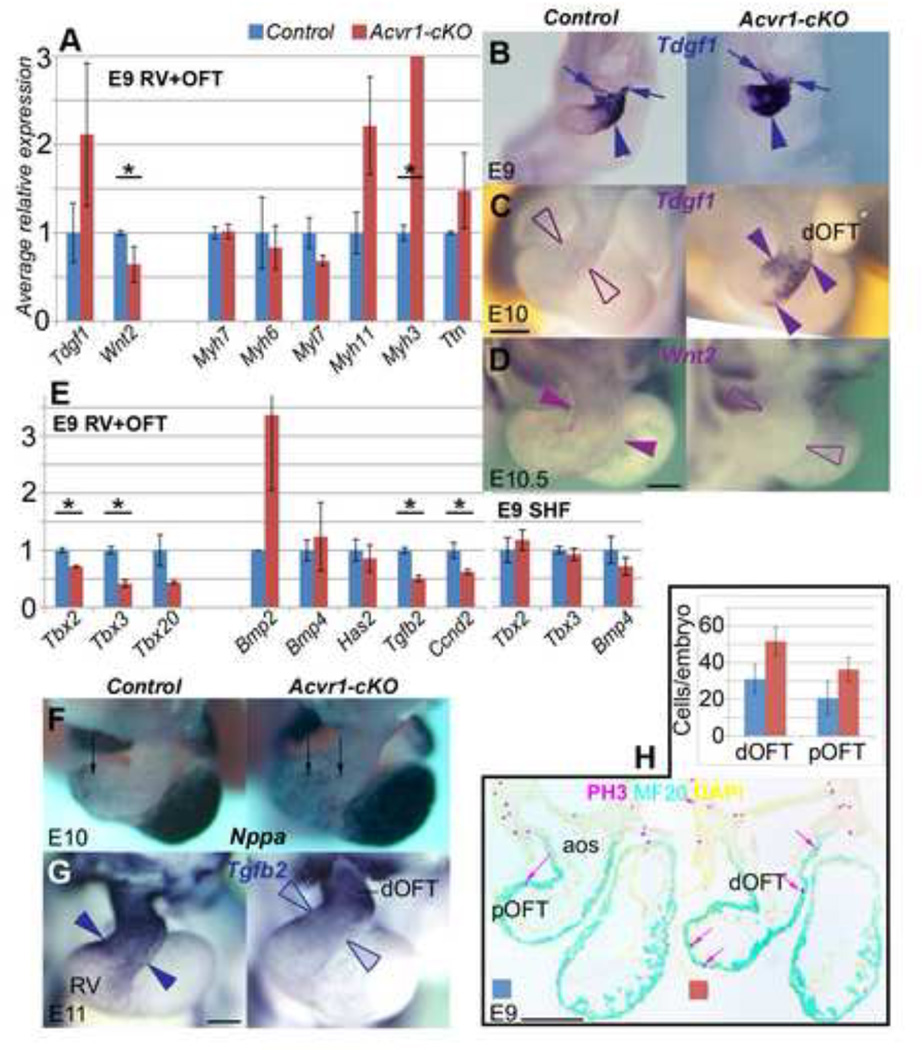
Figure 4. Gene expression differences between Acvr1-cKO and controls (II).
A, qRT-PCR on E9 RV+OFT for muscle-related gene expression differentiation (control, blue; mutant, red): Though average expression of sarcomeric protein genes Myh7, Myh6 not affected, Myl7 lower, Myh11 and Myh3 higher (expression levels too low for WMT ISH). Tdgf1 (Cripto) variably higher, Wnt2 lower in mutants. Results are average of n>=3 each control, Acvr1 cKO samples, +/− SEM, * p≤0.05.
B-D E9, E10 WMT ISH for gene expression reported in A, control on the left, mutant on the right: B, Tdgf1 expressed in OFT distally (arrows) to mid/proximally (arrowhead) more strongly in mutant than control, still detectable E10 (arrowheads) only in mutants. L, Wnt2 detectable throughout OFT (arrowheads) but not in mutant.
E, qRT-PCR on E9 RV+OFT, SHF for average Bmp/Tbx regional gene expression, control blue, mutant red: Tbx2, Tbx3, Tbx20, Tgfb2, Ccnd2 all reduced in mutant RV+OFT; Tbx2, 3 not affected in mutant SHF; Has2, Bmp4 no difference; Bmp2 higher in mutant RV+OFT. Results are average of n>=3 each control, Acvr1 cKO samples, +/− SEM. * p≤0.05
F-H, WMT ISH and proliferation related to gene expression reported in E, control on the left, mutant on the right: F, Nppa expressed more broadly mid-OFT in mutant (arrows); G, Tgfb2 much lower mid-proximal OFT (arrowheads) in mutant. H, average proliferation higher in mutant than control MF20 (cyan)-positive OFT wall (a-PH3-labelled nucleus: pink arrow): bar chart (n=3 control, mutant) on left, representative sections of control (middle) and mutant (right).
aos, aortic sac; dOFT, pOFT distal, proximal outflow tract; PA pharyngeal arch; PH3, phospho-histone 3. * p≤0.05 Scale bar, 200µm.
Generation of Acvr1Bmpr1a-Flag (Bmpr1a-KI) and Acvr1ACVR1-His6Myc (ACVR1-KI) knockin mice
The coding region of the Bmpr1a cDNA (from ATCC) was subcloned into pcDNA3.1 vector (Invitrogen), and sequences encoding the Flag-tag inserted in frame into the 3’end of the cDNA by site-directed mutagenesis (QuikChange II XL; Agilent). A similar strategy was used to modify human ACVR1A cDNA (kindly provided by R. Derynck) by inserting sequences encoding the His6/Myc-tag into its 3’end. Targeting vectors were generated as follows: a long 5’ homology arm covering a 5-kb segment of the mouse genomic DNA upstream of the Acvr1 translational start in exon 3 and a short 3’ homology arm covering a 2.6-kb segment of DNA downstream of the exon 3/intron 3 boundary were PCR-amplified using the mouse bac DNA as a template, and subcloned into a cloning vector containing the Flag-tagged Bmpr1a–bGHpolyA (or Myc-His6-tagged ACVR1-bGHpolyA) and the loxP-PGK-Neo-polyA-loxP cassettes (DTA was used a negative selection marker) (see Figure 7). Electroporation, screening, validation, removal of the selection marker, ES cell expansion, blastocyst injections and production of chimeric and heterozygote mice were performed according to standard procedures by genOway (France).
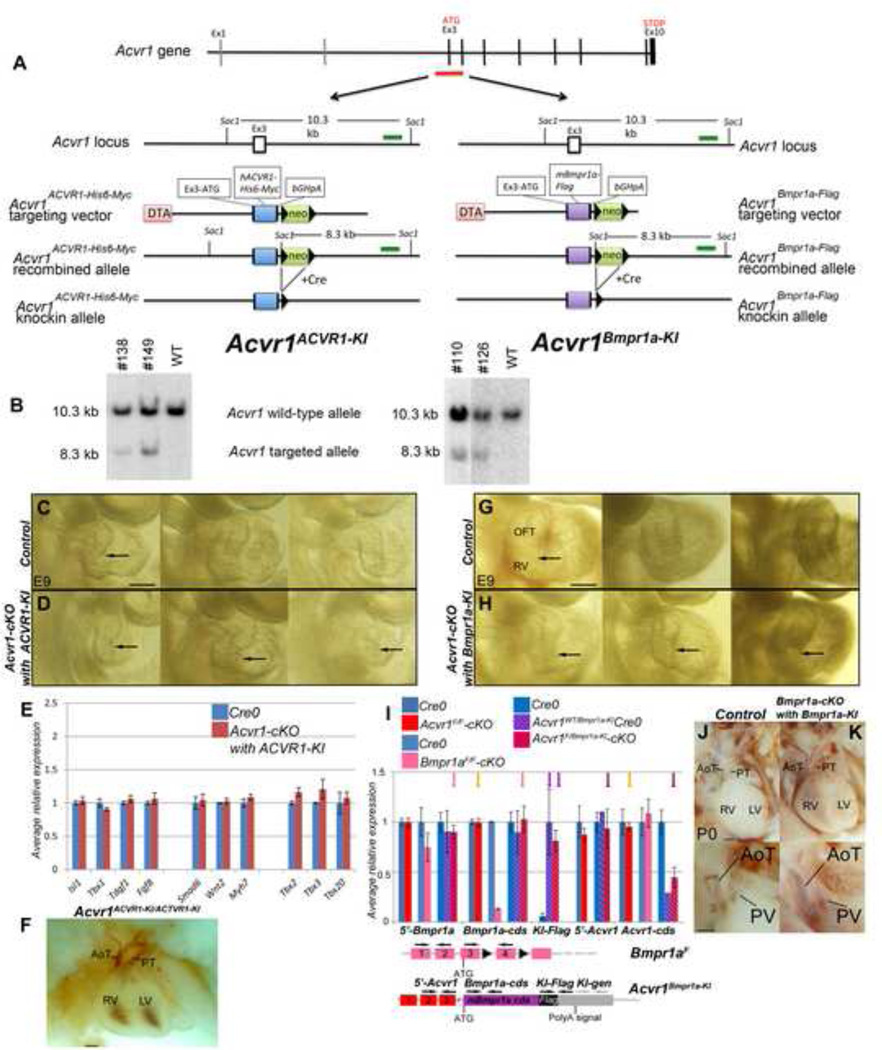
Figure 7. Generation and analysis of mice carrying the Acvr1ACVR1- and AcvrBmpr1a- knock-in alleles.
A-B, Construction of Acvr1ACVR1- and Acvr1Bmpr1a- knock-in alleles. A: A segment of the first coding exon (3) and next donor splice site of Acvr1 was replaced by Myc-tagged ACVR1 cDNA (left panel) or Flag-tagged Bmpr1a cDNA (right panel). B: Targeted ES-cell clones were first identified by PCR (data not shown) and the correct targeting was confirmed by Southern blotting. DNAs isolated from control (WT) ES cells and putative correctly targeted ES cell clones (#138 and #149 for Acvr1ACVR1-KI, left panel; #110 and #126 for Acvr1Bmpr1aKI, right panel) were digested with SacI and detected with an external probe (green bar in A). The wild-type and targeted alleles show 10.3-kb and 8.3-kb SacI fragments, respectively.
Note that, for ease of reading, these knock-in alleles are sometimes referred to as ACVR1-KI (rather than Acvr1ACVR1-KI) and Bmpr1a-KI (rather than Acvr1Bmpr1a-KI) in Figures 7 and 8.
C-D, E9 hearts from right: Comparison of RV, OFT morphology of three representative control (C: no Cre) and Acvr1-cKO with ACVR1-KI mutant (D: Acvr1F/ACVR1-KI, Mef2c[AHF]-Cre+) embryos, showing similar range in size of right ventricle in mutants as controls (arrows). RV+OFT of these samples used for qRT-PCR shown in E.
E, qRT-PCR on E9 RV+OFT Cre0, Acvr1ACVR1-KI-negative controls (blue), Acvr1 f/acvr1-ki-cKO mutants (red columns). In contrast to Acvr1F/F-cKO (Figure 3) and Acvr1F/Bmpr1-KI-cKO (Figure 8A), average gene expression in mutants is the same or very similar to that in controls. Each bar average of 3 separate samples, +/− SEM.
F, E15 4-chamber view of heart in Acvr1ACVR1-KI/ACVR1KI embryo showing normal gross morphology, including complete AP septation.
G-H, E9 hearts from right: Comparison of RV, OFT morphology of three representative control (G: no Cre) and Acvr1-cKO with Bmpr1a-KI mutant (H: Acvr1F/Bmpr1-KI Mef2c[AHF]-Cre+) embryos, showing variable reduction in size of right ventricle in mutants (arrows). RV+OFT of these samples used for qRT-PCR, Figure 6A,B.
I, qRT-PCR on E9 RV+OFT of various controls and mutant genotype samples. We assessed relative levels of Acvr1Bmpr1a-KI expression in three ways. Transcripts from Acvr1Bmpr1a-KI were specifically detectable (using ‘KI-Flag’ primers) in Acvr1Bmpr1aKI control and mutant samples (purple arrows). The presence of one Acvr1Bmpr1a-KI allele in mutants increased the average level of Bmpr1a by only 14% (compare the results (pink arrows) of ‘Bmpr1a-cds’, which detects both Bmpr1a and Acvr1 Bmpr1a-KI, and ‘5’-Bmpr1a’, which only detects Bmpr1a)This suggests that only a subset of cells would have experienced supra-normal levels of Bmpr1a. Loss of signaling via Acvr1 did not alter the average amount of either Acvr1 or Bmpr1a RNA (orange arrows). Acvr1Bmpr1a-KIallele was expressed at about the same level asAcvr1WTorAcvr1Fl: It includes 5’UTR exons from Acvr1 but not the Acvr1 cds. The relative level of Acvr1 5’UTR remained the same in samples with or without an Acvr1Bmpr1a-KI allele, whereas the level of ‘ Acvr1 cds’ detected was halved when one of the Acvr1 alleles was Acvr1Bmpr1a-KI (brown arrows). These results also suggest that the levels of expression driven by the Bmpr1a locus and Acvr1 locus may differ (and vary with cell type: see Figure 5). Genomic DNA contamination (assessed using ‘KI-gen’ primers) did not contribute (data not shown).
J-K P0 4-chamber, and oblique right view below: Gross morphology and trunk (AoT, PT) septation in Bmpr1a-cKO also carrying one Acvr1Bmpr1aKI allele (K) very similar to Cre0 control (J).
AoT, aortic trunk; cds, coding sequence; OFT, outflow tract; PV, pulmonary valve; RV, right ventricle. Scale bar 200µm (C,D,G,H) or 500µm (F,J,K).
LiCl treatment
Pregnant female mice were injected intraperitoneally with LiCl (200 mg/kg in saline) once a day at E7-E10 (NaCl [200 mg/kg] was used as a control) as described(Tian et al., 2010).
Histology, immunohistochemistry and in situ hybridization
Embryos for assessment were collected in sterile DBPS and fixed overnight in commercial formalin or fresh 4% paraformaldehyde in PBS at 4oC overnight. Those for wax embedding were then washed, dehydrated, and oriented and embedded in fresh Blue Ribbon Tissue Embedding/Infiltration Medium (Leica Surgipath) after three changes. 7µm sections were cut and mounted on Superfrost plus slides (Fisher) and stored at room temperature or 4°C. Haematoxylin and eosin staining was performed using a standard protocol, taking care to ensure complete, but not over-, staining to allow maximum discrimination of cell types. Immunohistochemistry performed on rehydrated wax sections: Antigen retrieval (15 mins at 95–100°C) in citrate buffer pH6 was used in some cases (see Supplemental Table 1 for details), tissues blocked with 2–3% BSA, 0.1% Triton-X100 in PBS and TBS prior to incubation with primary antibodies overnight at 4°C. Binding was visualized with Alexafluor-594 or −418-conjugated secondary antibodies (Life Technologies) on slides mounted with Vectamount/DAPI (Vector Labs).
Some fixed embryos were processed for cryo-embedding: washed in PBS, allowed to sink in sterile 10% sucrose in PBS, then 7% gelatin (Sigma G6650, 75 bloom), 15% sucrose in PBS, oriented and embedded in fresh 7% gelatin, 15% sucrose in PBS on ice then dry ice, and stored at −20 or −80 °C. 10µm cryo sections were cut and stored at −20 or −80°C.
Immunohistochemistry performed on cryo sections
Antigen retrieval was not used unless stronger/more complete MF20 or α-SMαA signal was needed. α-GFP detected YFP lineagetracing but any heat-based antigen retrieval prevented its detection.
For quantification of proportion of pSmad1/5/8-positive cells in OFT column cells, immunofluorescence was performed using α-pSmad1/5/8 and either α-Isl1 or MF20 on 10µm cryostat sections of E11 control and mutant samples, cut transverse to the OFT. Using single channel gray scale images and combined color images, column cells (positive for Isl1 but negative to MF20) were quantified in each section, and the proportion showing positive α-pSmad1/5/8 staining as assessed by comparison with positive non-recombined cells determined.
For cell proliferation analysis, immunofluorescence was used on 10µm cryostat sections cut 4-chamber, parallel to OFT, with α-phosphohistone3 (PH3) and either MF20, α-GFP or α-Isl1. Every α-PH3-positive wall cell was counted that was also within MF20/α-GFP-positive region within the morphologically definable OFT. MF20-negative second heart field cells were not included. Proximal and distal OFT were distinguished by direction of heart tube. Three separate control and mutant samples paired by size were counted (19–22 pair somite stage) were assessed. Apoptotic cells were detected using Dead End Fluorometric TUNEL system (Promega) following manufacturer's instruction.
RNA In situ hybidization
Embryos for ISH were fixed in freshly thawed 4% paraformaldehyde in PBS over night at 4°C. Those for section ISH were washed and wax-embedded, sectioned at 10µm and mounted as described above. DIG-labelled RNA probes (see Supplemental Table 2 for details) were made using a DIG-labeled NTP mix (Roche Applied Sciences) according to manufacturer’s instructions, stored at −80, or −20°C diluted in hybridization buffer. Section ISH was performed as described(Moorman et al., 2001) with probes diluted to 1ng µl−1 or less. After staining, sections were washed, fixed and mounted in Immumount (Thermo Scientific).
Embryos for wholemount ISH were fixed as above, washed in PBST, dehydrated and stored in 95% methanol, 5% PBST at −20°C. After rehydration they were trimmed to enable access of reagents, visualization of OFT or other tissues in cross section without sectioning in some cases, and easy discrimination of controls and mutants (which were then processed together for all steps), proteinase K-treated, post-fixed and pre-hybridized rotating for at least 1 hour in standard non-SDS-based hybridization buffer, pH5, at 70°C. Probe (for final concentration of 0.1–1ngµl−1) was added, or buffer replaced with pre-heated previously used probe in hyb buffer, and left rolling overnight. Used probe was recovered and stored at −20°C, but discarded when staining level reduced. Embryos were then washed (rolling) for 20 mins three times in preheated hybridization buffer at 70°C, once in hybridization buffer/TBST, then in at least 6 changes TBST at room temperature over 1 hour, blocked in 0.05% Roche block (one hour) and rocked O/N at 4°C with α-DIG Fab fragments (Roche Applied Sciences,1:2000–5000); washed with ten changes of TBST the next day (RT), rolling, over several hours, and O/N at 4°C, stationary if E9 or 10; rolled for 10 mins (RT) twice in NTMT pH9.5, and stained with BM Purple AP substrate or 1× NBT/BCIP (Roche Applied Sciences) at RT in the dark, overnight if necessary. Stained embryos were washed several times in PBS, fixed in formalin and stored at 4°C. Some were dehydrated through graded ethanols and Histoclear (National Diagnostics), wax-embedded, sectioned at 7–10µm and mounted in Immumount (Thermo Scientific).
β-galactosidase staining
Embryos for wholemount staining (for Mef2c[AHF]-Cre, R26RlacZ lineage tracing, or Acvr1nlsLacZ detection (see below), were fixed for 15 mins in 0.5% glutaraldehyde in DPBS, washed at least three times in detergent rinse, then stained using standard procedures(Hogan et al., 1994). After sufficient staining had occurred, embryos were washed in detergent rinse and PBS and fixed in formalin. Some were processed to wax using Histoclear (National Diagnostics), sectioned at 7 or 10 µm, and mounted in Immumount (Thermo Scientific).
Microscopy and photography
Results were viewed and recorded on an Olympus BX51 (sections, bright field and fluorescence) with Olympus DP71 and software, Leica MZ95 microscope (wholemount or sections, bright field) with Olympus DP72 and software, or Leica M165FC (wholemount, brightfield) with DP73 and software. A dish of agarose was used to obtain consistent orientations of small tissues. Adobe Photoshop 4 and 6 were used to invert immunofluorescence images and prepare figures for publication according to editors’ requirements.
Sample recovery for quantitative RT-PCR
Second heart field cells were recovered by FACS from Mef2c-Cre+, R26RYFP/WT E9 and E10 torsos (from which the heart tube had been completely removed), cut between first and second pharyngeal arches distally and immediately anterior to the atrial position posteriorly. Each torso piece was digested in 1% Collagenase I (Worthington) diluted 1:9 with Hanks BSS for 15–20 mins at 37°C, pipetted gently to disperse cells, cell-strained and stored on ice before sorting using the GFP channel on a Beckman Coulter MoFlo Astria Cell Sorter. Cells from each sample were collected directly into 500µl of RLT (Qiagen) to which 20µl of 1M DTT was added, transferred to 1.5ml tubes and stored at −80°C. RNA was purified using RNeasy (Qiagen) and Omniscript (Qiagen) used to make cDNA. RT-PCR was used to confirm the genotype of each sample (data not shown).
‘RV+OFT’ tissue was dissected from E9 embryonic hearts by cutting across the OFT immediately adjacent to the second heart field distally and through the interventricular groove/inner curvature proximally. Each sample was immediately placed in a separate labeled 1.5ml tube, excess DPBS removed, and 100µl RLT buffer (Qiagen) added. Samples were stored at −80°C until RNA isolation. RNA was isolated using RNeasy micro kit (Qiagen) following manufacturer’s instructions. All 12µl of RNA solution was immediately used to make 20µl cDNA using Omniscript (Qiagen) and random primers (Life Technology), and stored at −20°C. To increase the number of different gene expressions that could be assessed on the Acvr1,Mef2c-Cre samples, pre-amplification was performed using the TaqMan Pre Amp master mix (Applied Biosystems) according to manufacturer’s instructions: sense and antisense primers used in the Roche Universal system (see below) for qRT-PCR were diluted to the correct concentration and used in place of TaqMan assay reagents. Ten cycles of pre-amplification were performed on 1µl of cDNA per 20µl reaction, and the resulting solutions diluted 1:5 with TE, and stored at −20°C. Assays on cDNA and pre-amplified DNA were used to confirm that relative quantification was unaltered following pre-amplification (data not shown).
Real time quantitative PCR
Most experiments were carried out using Universal Probe Library-based assays (Roche Applied Science) with gene-specific primer sequences generated by the manufacturer’s online algorithm and TaqMan Universal PCR master mix (Applied Biosystems) (see Supplemental Table 3 for sequences). Some used Taqman Assay reagents (see Supplemental Table 3). 30µl assays were quantified using Applied Biosystems ABI7300 PCR and ViiA7 detection systems and software. All Ct values were checked by eye. Actb levels were used to normalize other expression levels. cDNA was diluted to avoid Cts lower than 18. At least three separate control (Cre negative or conditional heterozygote) and three separate mutant samples were assessed for each genotype/condition, where possible from the same litter, or at least stage-matched.
miRNA assays
Individual E9 RV+OFT pieces were dissected manually as described above and stored in QIAzol lysis reagent (Qiagen) at −80°C until RNA isolation using miRNeasy mini kit (Qiagen). Individual E9 second heart field-enriched torso pieces without heart were dissected manually and stored at −80°C until RNA isolation using miRNA purification kit from Norgen Biotek. After quantification, samples were shipped overnight on dry ice to LC Sciences (Houston Texas) who assayed the samples miR-17 and miR-20a (using Taqman miRNA assays Ids 2308, 0580) and control RNAs snoRNA202 and noRNA234 (assay Ids 1232, 1234). Three stage-matched pairs of controls and mutants were used for each genotype assayed; each of these assayed in triplicate for each assay, along with a no template control.
Statistical analyses
For histological and expression (qRT-PCR) analyses three or more samples were analyzed unless otherwise stated. Averages, standard error and probability (Student’s t-test, 1-tailed) were calculated. Probability (P) equal or less than 0.05 was marked as significant.
Acvr1 in the AHF is required for OFT septation and RV development
To address the role of the BMP type I receptor AcvRI in the SHF, Acvr1 was deleted using Mef2c[AHF]-Cre+ driver mice(Verzi et al., 2005). The resulting Acvr1F/F/Mef2c[AHF]-Cre+embryos (Acvr1-cKO) lacked most of the functional Acvr1 in the Mef2c[AHF]-Cre recombination domain (Online Figure 1).
Defects in post-septation stage Acvr1 mutants
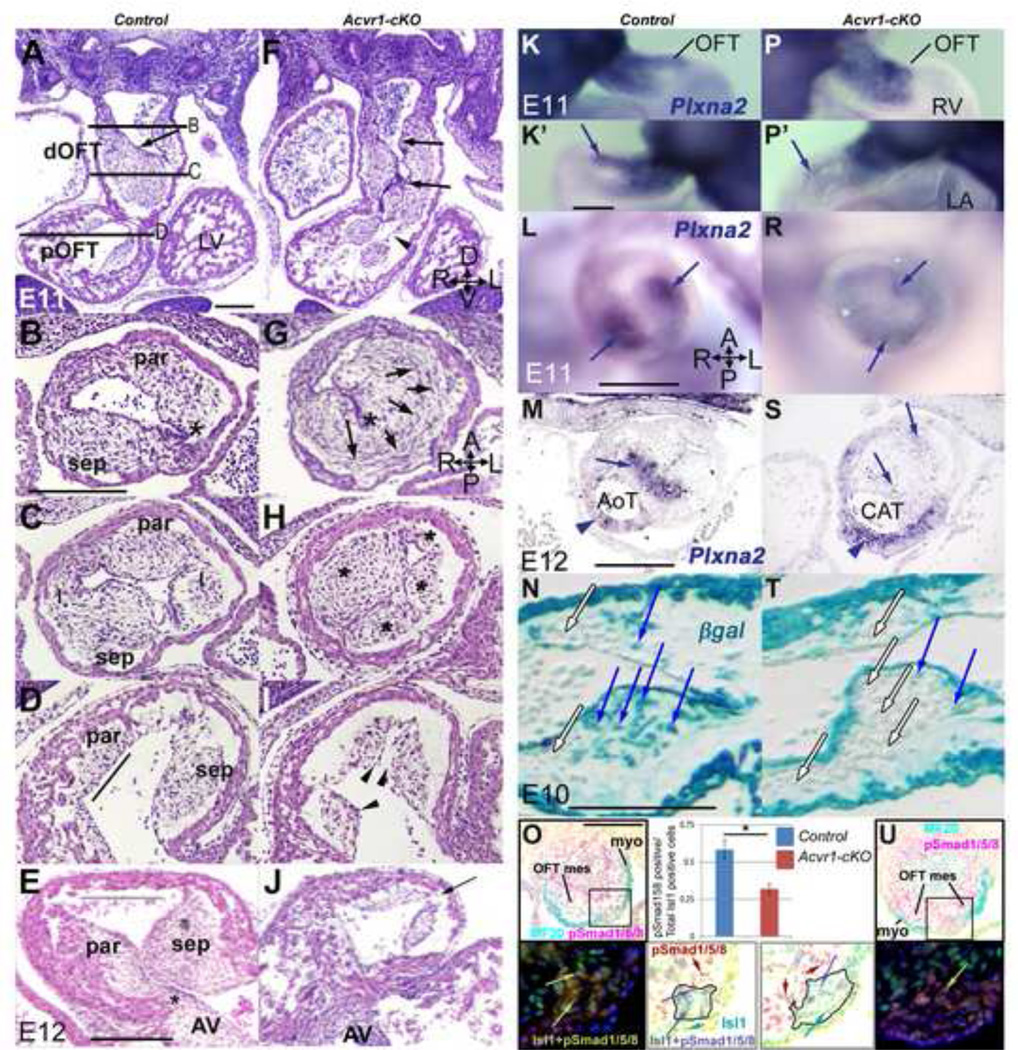
Acvr1-cKO embryos consistently had morphological defects in ventricular and OFT septation (Figure 1A–Fb). Membranous ventricular septal defects (VSD; Figure 1Fa, Fb) were accompanied by OFT, valve and aortico-pulmonary (AP) trunk septation defects that formed a spectrum in severity, from the mildest, DORV (double outlet right ventricle, Figure 1Da, Ea, Fa) to CAT (common arterial trunk, from which pulmonary arteries arose directly posteriorly). In some examples of CAT, septation of the arterial valves had not occurred, but two trunks of similar size (Online Figure 2), or one rudimentary (Figure 1Eb, Online Figure 2Lb), were present distally. Despite these malformations, six (or more) OFT valve cushions/leaflets could often be identified in partially or distally unseptated CAT, and coronary ostia were grossly normal (Online Figure 2). Defects in arterial tree downstream, and atrial/AV septation upstream were sometimes found but were not further examined in this study.
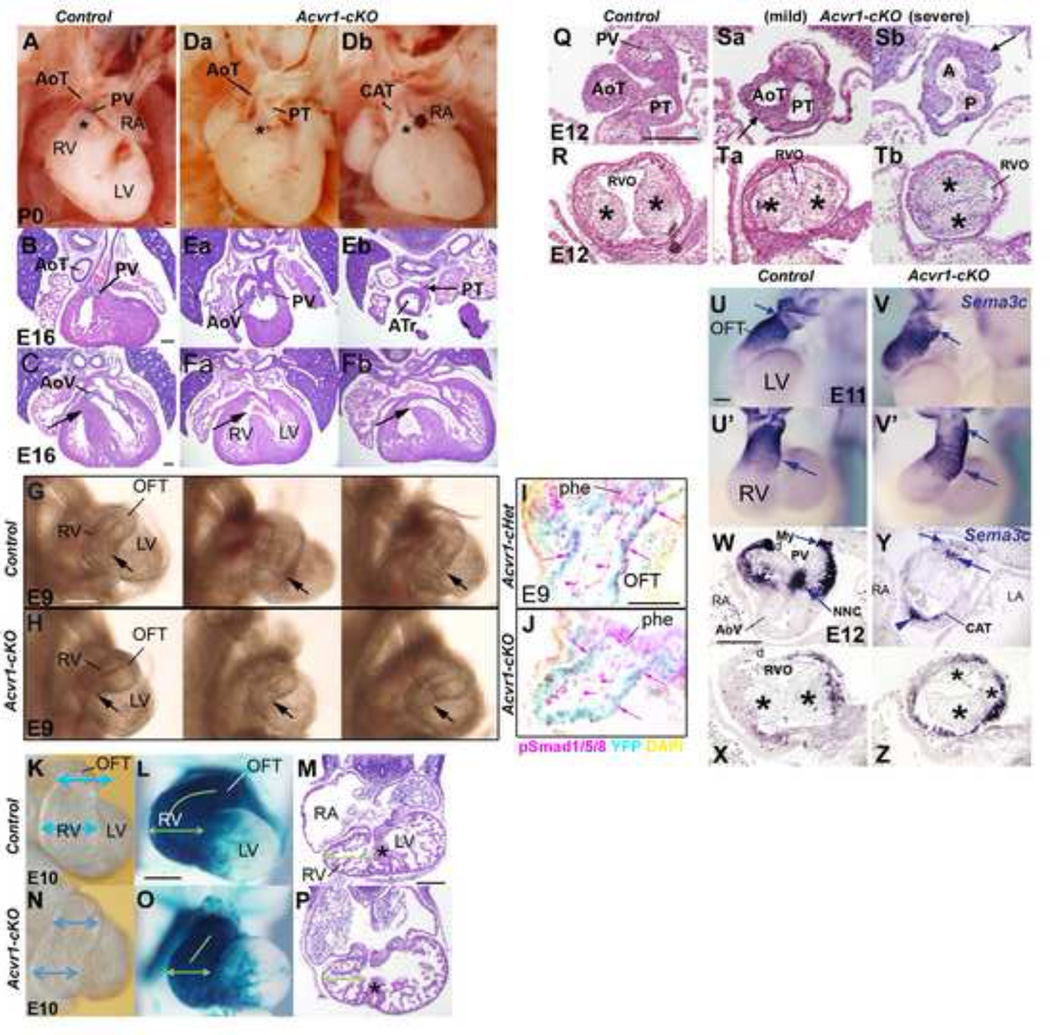
Figure 1. Abnormal morphology in Acvr1-cKO hearts and trunks.
A,Da,Db, P0 hearts; B,C,Ea-Fb, E16 4-chamber H&E: Post-septation stage controls (A,B) have aortic and pulmonary trunks at right angles to one another, proximal AoT behind sub-pulmonary sleeve (asterisk in A). In ‘mildest’ Acvr1 cKO (Da,Ea,Fa), DORV, trunks parallel to one another, valves on same level as one another, proximal AoT visible (Da) as subpulmonary sleeve inadequate, insufficient displacement of AoT to the left posterior/PT to the right anterior. In ‘severe’ Acvr1 cKO (Db,Eb,Fb) single trunk close to valve, though often distally septated to form separate full size (not shown) or rudimentary second trunk (PT in Eb connected to pulmonary arteries). DORV (Fa), CAT (Fb) accompanied by subvalvular VSD (black arrow).
G,H,E9; I,L, E10; hearts in situ from right: Three representative E9 mutant hearts (H) distinguishable from three controls (G) by variably narrower RV apex (arrow) versus OFT (RV+OFT of these samples used for qRT-PCR, Figures 3,4). I, J, E9 4-chamber cryosections showing less nuclear phosho-Smad1/5/8 immunostaining (pink) in OFT wall (pink arrows) in Mef2c[AHF]-Cre-recombined cells (YFP, cyan) in Acvr1-cKO (J) than in Acvr1-cHet control (I). Endocardium also positive (arrowheads). Unrecombined endoderm same in both (phe).
E10 control RV broader (blue arrows) than OFT (K) unlike mutant (N). L,O, Mef2c[AHF]-Cre lineage pgal stain (blue), apical view: control (L) RV is broader laterally (green arrows), proximal OFT curves to reach midline (green line) unlike mutant (O). N,P, E11, 4-chamber,H&E: mutant (P) RV apex grows but remains narrower laterally than in control (N), though wall thickness and trabeculation very similar and muscular septum (asterisk) present (contacts AV cushion in different section from N). Scale bar, 200µm.
‘Rotation’ of OFT less in mutant hearts.
Q-Tb, E12 transverse to OFT, H&E: In control (Q), AP septation complete, distal trunks separate, lying in correct relation to one another; proximal to valve cushions, parietal and septal cushions (*) lie parallel to A-P axis (R) so RVO anterior-most. In mild mutant, some partial distal trunk septation (Sa) present only, but main OFT cushions (*) near correct orientation (Ta); in severe mutant, no distal trunk septation, aortic (A) and pulmonary (P) lumens unseparated (Sb). More proximally (Tb), cushion faces 60 degrees short of normal orientation. Note irregular trunk wall (arrow, Sa,Sb).
U-Z, Sema3C expression, E11, WMT ISH; E12, section ISH: In control E11 (U from left, U’ from anterior) strongest (small arrows) on distal anterior/left face; in mutant V from left, V from anterior) strongest distal left to posterior, less cleanly defined. Expression extends more proximally in mutant (large arrow). Control E12, valve level (W), strongest myocardial (My) expression on anterior face (RVO) (tissue split anteriorly, d) but in mutant (Y in which distal septation failed), very weak on anterior face (small arrow), strong posteriorly (arrowhead); positive NC cells abnormally located (large arrow). Proximally, expression weak in control (X), more extensive in mutant (Z: note abnormal cushion* morphology).
d artifactual damage. Scale bar, 200 µm.
Defects in pre-septation stage Acvr1 mutants
Acvr1-cKO hearts could usually be distinguished morphologically from controls between E9 and E11 especially by a smaller RV apex (Figure 1G–N): The inner curvature could also appear narrower (Online Figure 3A, B). A reduction in Bmp signaling could be detected in recombined Acvr1-cKO heart cells at E9 (Figure 1I,J) and the myocardium of these phenotypically abnormal structures was all recombined by Mef2c[AHF]-Cre (Figure 1L,O). Unrecombined regions in control and Acvr1-cKO hearts appeared morphologically indistinguishable (Figure 1 and data not shown).
It has been suggested that the appropriate degree of OFT ‘rotation’ is required to position aortic and pulmonary trunks with respect to the ventricular outlets and ventricular septum appropriately(Bajolle et al., 2006), which can be defective when BMP signaling is compromised(Beppu et al., 2009). At E12, mutant OFTs were found to be (variably) less ‘rotated’ as judged by the angle of the opposing faces of the parietal and septal cushions to the embryonic anterior-posterior (A-P) axis (Figure 1Q–Tb). Sema3c expression, strongest in sub-pulmonary myocardium which should be anterior-by E12(Theveniau-Ruissy et al., 2008), was also abnormal at E11 and 12 in mutants (Figure 1U–Z).
Other regional differences in wall morphology were also present in mutants. By E10, mutant OFT wall was more varied in thickness (Online Figure 3C–H): the anterior face of the mid-proximal OFT wall appeared thicker, and an area of much thinner wall on the posterior/left side of mutant OFTs surrounded part of the septal cushion (Online Figure 3I–W). The wall cells in the thin region appeared to contain much less cytoplasm and less, poorly organized, striated muscle myosin than those in other parts of the wall, or control cells, (Online Figure 3O,P). Despite this, the cells were typical of wall, not mesenchymal cell phenotype, as they were Isl1-and SMαA-positive, and Sox9-negative (Online Figure 3I–L). Some apoptosis was present nearby but not localized within the thin wall itself (Online Figure 3R–S). This area was barely overlain by epicardium by E12 (Figure 3T,U). The right ventricular wall did not appear to differ in thickness between mutants and controls, and its trabeculation was not grossly abnormal (data not shown).
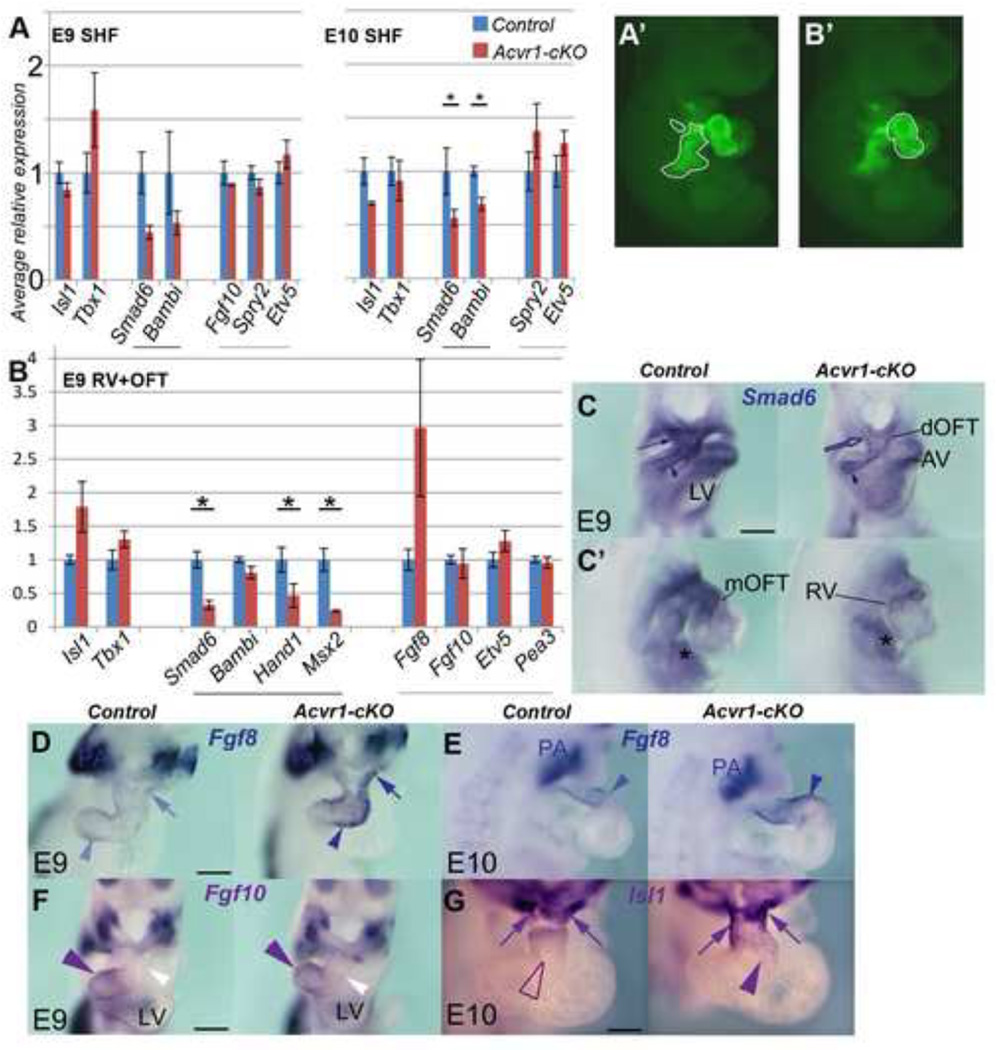
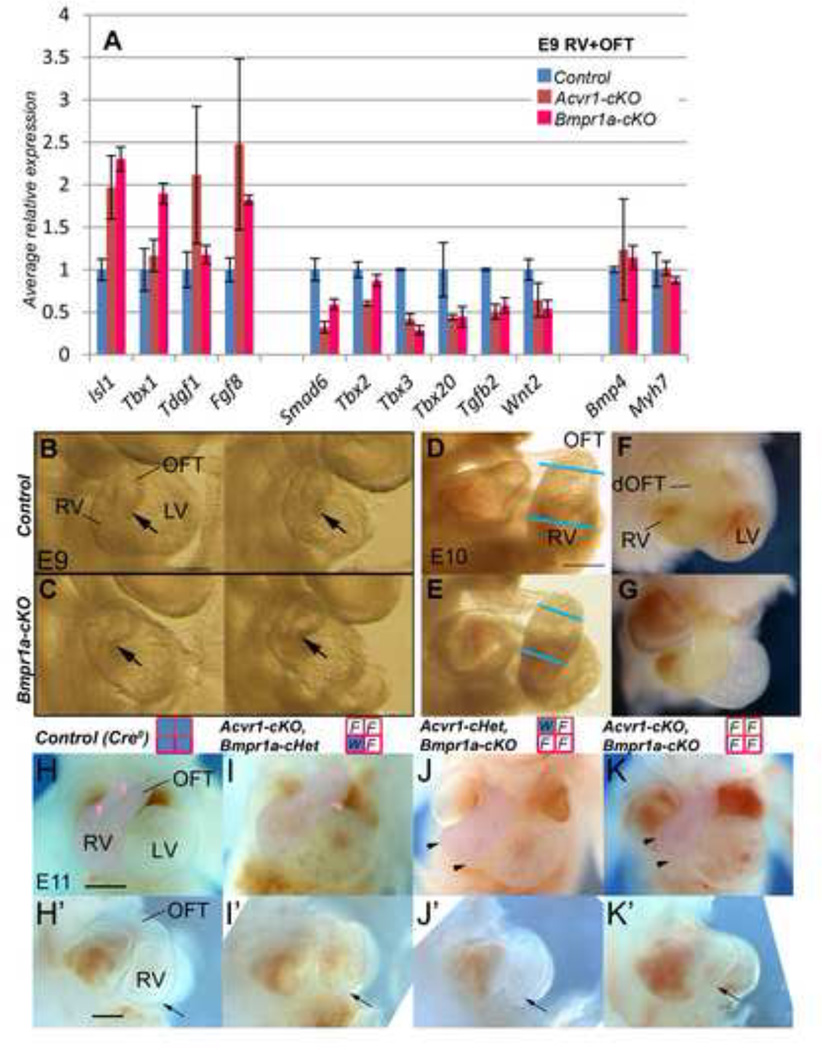
Figure 3. Gene expression differences betweenAcvr1-cKOand controls (I).
A, qRT-PCR, E9 and E10, SHF cells (YFP+ve Mef2c[AHF]-Cre-recombined population, outlined in A’ at E9, from right) FACS-sorted after removing the OFT/heart tube. control, blue bars; Acvr1-cKO, red bars. Average Bmp synexpression gene expression (black underline) reduced in mutants at both time points. Fgf synexpression target gene expression (grey underline) slightly elevated at E10. Tbx1 expression higher at E9, and Isl1 less at E10 in mutants. Results are average of n>=3 each control, Acvr1 cKO samples, +/− SEM, * p≤0.05.
B, qRT-PCR on E9 RV+OFT (manually dissected region, outlined in B’, in YFP+ve Mef2c[AHF]-Cre-recombined population). Average BMP target gene expression (black underline) reduced in mutants, Fgf8 expression much higher, but not Fgf10 or FGF targets. Isl1 and Tbx1 also elevated.
C-G, E9, E10 WMT ISH for gene expression reported in B, control on left, mutant on right. C 4-chamber, C from right, Smad6 strong in distal (arrow)-mid OFT and adjacent SHF in control, greatly reduced in mutant (also inner curvature/RAV). Endocardium (arrowhead), LAV and liverbud (asterisk) positive in both. D, Fgf8 strong distal (arrow) to proximal OFT (arrowhead) in mutant but weak in control; E, Fgf8 stronger, extends more proximally (arrowhead) in mutant. Endoderm expression strong in all (PA). F, Fgf10 expression similar in both control and mutant, stronger in proximal/mid OFT (purple arrowhead) than distal (white). G, Isl1 strong distally (arrows) but also extends to mid-OFT (arrowhead) in mutant.
dOFT, mOFT, distal, mid, outflow tract * p≤0.05 Scale bar, 200µm.
Mesenchymal cells are added to the initially cell-free ECM cushions along the length of the OFT both distally (by migration of CNCC via the pharyngeal region) and locally, especially proximally, through EMT from overlying endocardium. At E11, instead of the usual 2(+2) cushion arrangement, Acvr1-cKO mutants had highly populated cushions of differing sizes mid-OFT, and a single mesenchymal cushion usually packed to one side distally (Figure 2A–C,F–H) in which mesenchyme underwent more cell death at E12 (Online Figure 4Q–T). The lumen(s) formed by these cushions were tortuous, and the endocardium itself was irregular in contour (Figure 2F–I). Proximally, mutant cushions were abnormally shaped, and cushion continuity to AV cushion absent (Figure 2D,E,I,J). Cushion mesenchyme organization was abnormal and Plexina2 expression (expressed by CNCC) reduced in mutants (Figure 2K–S). Recombined endocardium appeared to make little contribution to cushion mesenchyme (Figure 2N,T), so CNCC likely contributed most of the mesenchyme as endocardium appeared increasingly recombined during this period. Consistent with this, Smad1/5/8 phosphorylation was clearly detectable in both control and mutant OFT mesenchyme at E11 (Figure 2O,U). OFT cushion morphology was frequently so abnormal in Acvr1-cKO that two separate lumens were not maintained through them to the aortic sac (Figure 2G, Online Figure 4F), so they would not have aligned with the nascent aortic and pulmonary trunk flow routes distally if formed. Distal SHF-derived OFT and aortic sac wall morphology was abnormal. A recognizable column of Isl1-positive, MF20-negative SHF cells on the posterior/left of the distal OFT showed variably reduced phospho- Smad1/5/8 immuno staining (Figure2O,U) and abnormal morphology in mutants, and lay adjacent to the area of thin myocardium (Online Figure 4A–L).
Figure 2. Abnormalities in OFT cushions in Acvr1-cKO.
A-J, E11, E12 H&E; B-E,G-J transverse to OFT: Parallel to the distal OFT, endocardial contour (arrow) smooth in control (A) but irregular (arrows) and cushion continuity poor (arrowhead) in mutant (F). Distally, two cushions (par, sep) present in control (B) leave endocardium fairly close to OFT wall (asterisk), but in mutant (G) one large dysmorphic cushion, with circumferentially oriented cells on left side (arrows), create a greater distance to between wall and endocardium (asterisk) and functional lumen remains only in right anterior position. Mid-OFT, four cushions (par, sep, I, I) in control (C), abnormal cushion size and location (asterisks) in mutant (H). Proximal cushions faces are parallel to flow (line) in control (D), but an abnormal pyramidal cushion contour (arrowheads) in mutants (I). By E12, parietal cushion continuity with AV cushion present (asterisk), wide cushion-free RV outlet (line) anteriorly in control (E) but undersized proximal cushions, no continuity with AV cushion, no cushion-free outlet space (arrow) in mutant (J).
K-S, E11–12, Plxna2 ISH: From both right (K,P) and left (K’,P’) Plxna2-positive cells present in both control (K,K’) and mutant (P,P’) OFT, though staining mid-OFT in mutant (arrows) weaker. Mid-OFT, strong expression in central cells of each main OFT cushion in control (L), faint misplaced expression in mutant (R). By E12, strong expression in condensed central mesenchyme (arrow) at level of valve septation in control (M), faint in aortic trunk (arrowhead); faint in mutant cushion (S, arrows) but strong expression in irregular thickness arterial wall (arrowhead).
N,T, E10, pgal stain, sagittal sections: in Acvr1-cHet Mef2c[AHF]-Cre control, numerous recombined mesenchymal cells (blue arrows) underlie recombined endocardium (N) in proximal cushion, but very few in Acvr1-cKO (T). Unrecombined mesenchyme (white arrows) and endocardium also present.
O,U, E11, transverse distal OFT sections, immunostaining: Upper panels show MF20, a-pSmad1/5/8 immunostaining on sections in which the region including a particular column of SHF-derived cells is indicated by a box (see also Online Figure 4). Lower panels: darkfield and color-inverted images show an enlargement of the equivalent areas in sister sections to those in the upper panels, demonstrating that the population of Isl1-positive, MF20-negative cells there (outlined in color-inverted images) contained more pSmad1/5/8-positive cells in the control than in the mutant sample (quantification shown in bar chart; n=4, +/− SEM, * p<0.01). Examples of Isl1-positive, pSmad1/5/8-negative cells (green arrows); Isl1-negative, pSmad1/5/8-positive cells (red arrows); and Isl1-positive/pSmad1/5/8-positive cells (yellow arrows on darkfield, blue arrows on color-inverted image) identified. Note many pSmad1/5/8-positive mesenchymal cells in both control and mutant sections (red cells in color-inverted images).
AV, atrioventricular cushion; dOFT, distal OFT; I, intercalated cushion; pOFT, proximal OFT; par, parietal cushion; sep, septal cushion. Embryonic axes in F,G,M: A, anterior; D, dorsal; L, left; P, posterior; R, right; V, ventral. Scale bar, 200µm.
These results show that AcvR1 in SHF-derived cells made an essential contribution to the development of structures important for ventricular, OFT and AP septation, that its removal affected the behavior of both SHF-derived cells themselves (myocardium and endocardium) and those genetically normal with which they interact (CNCC).
Loss of Acvr1 in the SHF results in altered gene expression, increased cell proliferation and altered regional identity in OFT myocardium
To investigate molecular pathways underlying the effects of removing AcvR1 function, we chose to study E9, as later events may be secondary to earlier regulatory defects, and the more complex anatomy make it harder to discriminate different processes.
Chick anterior SHF explant studies suggest that FGFs and BMPs co-ordinate the balance between proliferation and differentiation of anterior SHF progenitors, in part by inhibiting one another via distinctive groups of ‘synexpressed’ genes(Hutson et al., 2010; Tirosh-Finkel et al., 2010). Cardiomyocyte and smooth muscle precursors continue to be added to the mouse heart tube from the anterior SHF between days 9 and 12 (reviewed by Vincent and Buckingham(Vincent and Buckingham, 2010)) so to determine if AcvRI-mediated BMP signaling mediates FGF repression in anterior SHF progenitor cells themselves, we assessed their gene expression by qRT-PCR at E9 and E10 in FACS-sorted populations purified from individual Mef2c[AHF]-Cre-positive Acvr1F/WT (control) and Acvr1F/F (cKO, mutant) embryos heterozygous for R26R-YFP. The results (Figure 3A) were consistent with BMP signaling being reduced in mutants but FGF synexpression only slightly affected. We also assessed other genes implicated in SHF-derived heart tube development, Isl1, Tbx1, and Fgf10(Cai et al., 2003; Watanabe et al., 2012; Xu et al., 2004)(Figure 3A). Average Tbx1 expression was higher at E9 and Isl1 expression less by E10, in mutants.
AcvR1 might still have mediated Fgf repression in anterior SHF-derived cells that have joined the heart tube itself, so we compared the expression of these and other genes implicated in regulating proliferation and differentiation in the RV/OFT segment of individual control and mutant heart tubes at E9 (Figure 3B–H, 4A–G). BMP signaling was reduced in recombined mutant RV/OFT relative to controls (Figure 1I,J). Only Fgf8 expression was much higher in mutants, (not Fgf10, or other synexpression genes, Figure 3B, E–G), but the abnormal expression of a number of other markers in mutants was consistent with a continued proliferative/less-differentiated phenotype: Isl1, Tdgf1, Tbx1, Wnt2 (Figure 3B,H; Figure 4A–D). A delay in differentiation did not extend to average sarcomeric muscle gene expression which was little altered for those encoding functionally important quantities (Figure 4A). Although Nppa levels were on average lower in mutants (data not shown), WMT ISH showed not only that Nppa expression was still present in RV trabeculations, but ectopic patches were present distally beyond the RV chamber and more widely around the circumference of the OFT at E9, 10 and 11 (Figure 4F, Online Figure 4A–C, data not shown). Nppa expression is normally restricted to the ‘chamber’ myocardium of the ventricles and atrial appendages and does not occur in AV junction or OFT myocardium. The boundary between phenotypic chamber and non-chamber in AV region heart myocardium is tightly regulated by Tbx2, Tbx3, and Tbx20(Cai et al., 2005). These may play a similar role in OFT(Mesbah et al., 2012; Ribeiro et al., 2007; Sakabe et al., 2012). Expression of these (BMP-target) genes was reduced in mutants, as was another regional marker, Tgfb2 (Figure 4E,G). Proliferation is normally low in non-chamber AV region myocardium, but was higher on average in mutant versus control OFT (Figure 4H). Note that average expression of Bmp4 and Bmp2 at E9 was not reduced (Figure 4H), so their signaling through other receptors, and in non-recombined cells (such as neural crest cells) would not be expected to be reduced.
Stabilization of β-catenin did not substantially rescue Acvr1-cKO phenotype
Embryos lacking Ctnnb1 in SHF have very small RV, CAT and dysmorphic OFT cushions(Ai et al., 2007). We therefore wondered whether any relevant Wnt/Ctnnb1 signaling lay downstream of Bmp signaling and tested this by stabilizing canonical Wnt signaling by maternal LiCl injection(Tian et al., 2010). A number of genes showed some normalization in average levels in treated embryos, suggesting β-catenin signaling might normally act downstream of AcvR1-mediated BMP signaling to regulate Fgf8, Tgfb2, and Ccnd2 expression. Tbx3, Tbx20, Isl1, and Wnt2 expression remained abnormal, and LiCl-treated Acvr1-cKO hearts also remained macroscopically morphologically abnormal at post-septation stages (Online Figure 5).
Do AcvR1 and BmpR1a regulate the same pathways and processes in anterior SHF-derived cells?
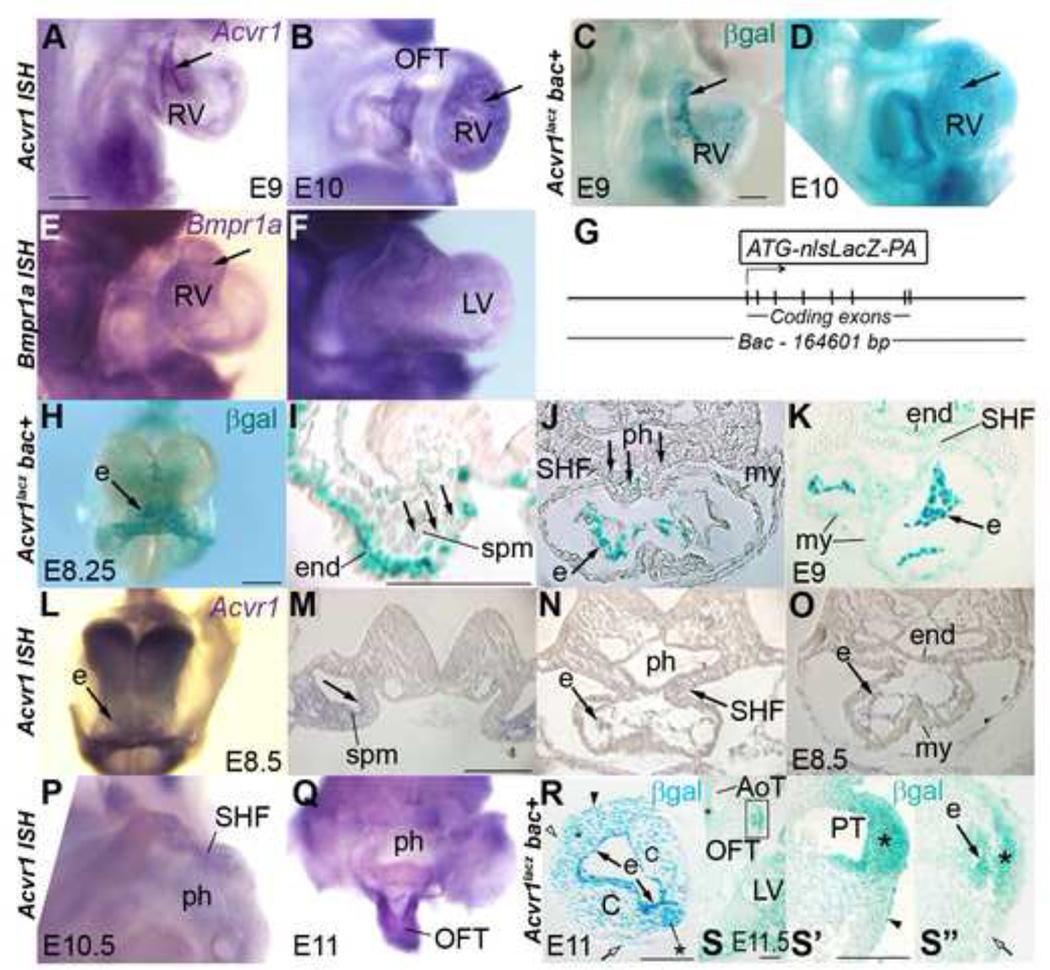
Both AcvR1 and BmpR1 are BMP Type I receptors, yet abnormal development occurs if the expression of one of these genes is deleted from the anterior SHF. This may be because they are expressed in different physical or chronological domains, and/or that each controls distinctive regulatory events. Using RNA WMT ISH and a novel lacZ-bac line to report Acvr1 expression, we found expression in SHF and heart tube cells, as reported for Bmpr1a(Klaus et al., 2007; Song et al., 2007) but whilst Bmpr1a was broadly expressed at similar levels (Figure 5E,F), Acvr1 was most strongly expressed in endocardium, and cushion mesenchyme (Figure 5A–D, H, K–O, R). Weaker expression was also found in splanchnic lateral plate mesenchyme, variably in SHF cells and gradually increasing in myocardial cells after tube formation. It was detectable in SHF cells at E10 and in non-myocardial OFT cells at E11 (Figure 5H–S”).
Figure 5. Acvr1 is expressed in SHF and endocardium.
A-F, E9, E10 from right except F, apical: Strongest expression of Acvr1 in endocardium (arrows) but Bmpr1a similar level in all cardiac tissues. G, Transgenic bac reporter line construct, nls-lacz-polyA inserted in Acvr1 exon 3.
H-O Acvr1 expression in early anterior (I) and later posterior (M) splanchnic mesoderm (spm, arrows, I,M). Strongest expression in endocardium (e) throughout heart tube from E8.25 (H,J,K,L,N,O). Weaker, intermittant expression in SHF (J, K, N), myocardium (my) and pharyngeal endoderm (end) by E9 (K). J: section from H, M-O: sections from L.
P-S” Acvr1 expression in mid SHF (P, cut above atrium, viewed from posterior) and distal OFT (rest of heart removed, Q), including morphologically distinct populations of SHF-derived cells (*), some that contribute to aortic (AoT) and pulmonary trunk (PT) (S-S”). Strong expression in endocardium (e); in neural crest cushion cells (C), some epicardial cells (arrowheads R, S’), less expression in myocardial wall (open arrows). R,transverse, S oblique, sections of distal OFT of WMT pgal-stained E11–11.5. S” is detail from S, S’ from more distal section.
C, cushion; e, endothelium; end, endoderm; my, myocardium; ph, pharynx; spm, splanchnic mesoderm. Scale bar 200µm, except R’,S, 100µm
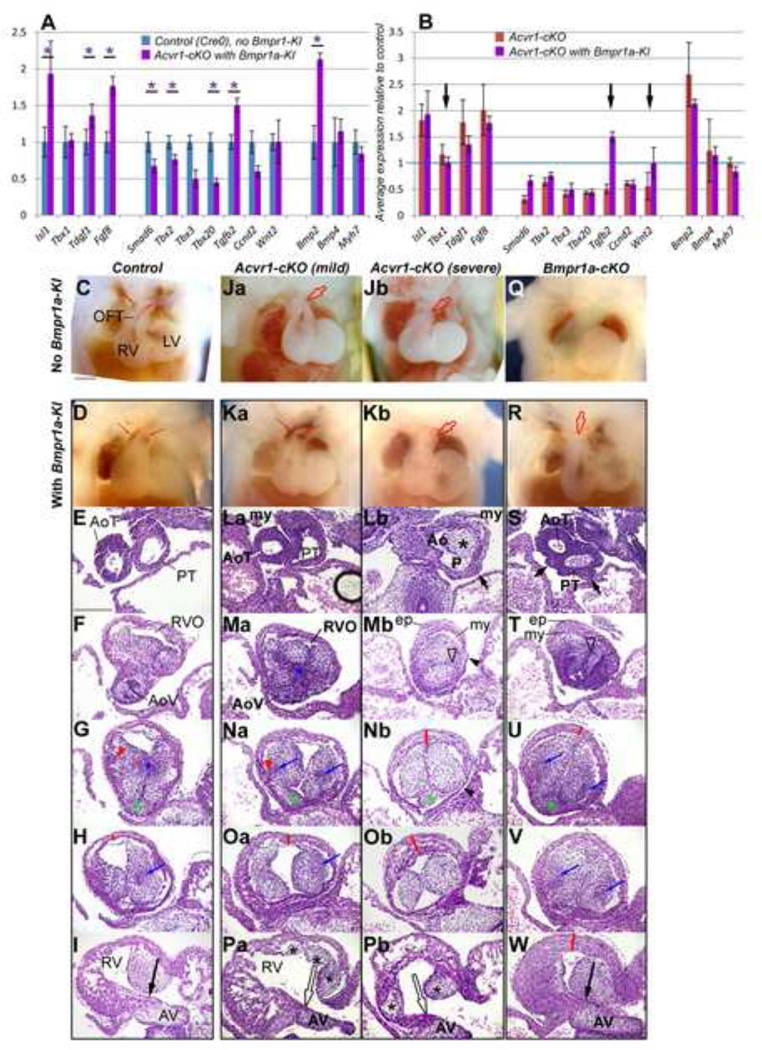
We also found that most of the genes showing altered expression in Acvr1-cKO RV+OFT at E9 were similarly altered in Bmpr1-cKO (Figure 6A). The morphological phenotype of Bmpr1-cKO was also similar, but more extreme (compare Figure 1H vs 6C, and Online Figures 3,4 vs 6) including small RV (E9–10), abnormal OFT cushion development, abnormal OFT wall and CAT-consistent morphology at E12. All were dead by E13. Hearts containing only one or no Acvr1/Bmpr1a alleles also showed progressively more abnormal RV and OFT phenotypes (Figure 6H–K’), so mal-development might be the result of a lack of sufficient BMP Type I receptor capacity remaining for normal development if only either Acvr1 or Bmpr1a were absent. To test this, we added Bmpr1a back in place of Acvr1 genetically in Acvr1-cKO mice by making two novel knock-in mouse lines (Figure 7A,B) carrying an Acvr1 allele in which either Bmpr1a cDNA (to make Acvr1Bmpr1a-KI) or ACVR1 cDNA (to make Acvr1ACVR1-KI) coding sequence under Acvr1 locus control replaced the endogenous Acvr1 sequence (Figures 7 and 8). Acvr1ACVR1-KIallele (‘ACVR1A–KI’) functioned similarly to Acvr1WT, and the presence of Acvr1Bmpr1a–KI (‘Bmpr1a–KI’) was not lethal, as Acvr1ACVR1-KI/ ACVR1-KI homozygote knock-in, Acvr1Bmpr1a-KI/ ACVR1-KI, Acvr1WT/Bmpr1a-KI and Cre-negative Acvr1F/Bmpr1a-KI animals were viable, fertile and grossly normal (data not shown), and Acvr1ACVR1-KI/F-cKO resembled Cre-negative controls in morphology and E9 RV+OFT gene expression (Figure 7C–F). However, only limited normalization of average gene expression was found in E9 RV+OFT from Acvr1Bmpr1a-KI/F-cKO at E9: Tbx1 and Wnt2 to control levels, Smad6 partially restored, and Tgfb2 from under-expression to over-expression (Figure 8A,B). From E12, these embryos still displayed a spectrum of septation defect phenotypes ranging from CAT to successful AP septation (Figure 8, Table 1), but with variable normalization of mid-OFT cushion orientation, organization and number, and restoration of OFT wall thickness (Figure 8La–Pb). This was despite the fact that expression of Acvr1Bmpr1a-KI RNA could be detected at E9 (Figure 7I), and produced functional protein as there was some improvement in average Smad6 expression at E9, and Bmpr1a-cKO, Acvr1WT/Bmpr1a-KImice survived beyond E13 and showed a substantial improvement in morphology (Figure 7J,K, Figure 8Q–W, Table 1).
Figure 6. Effect of Bmpr1a-cKO on gene expression in OFT/RV and on OFT/RV development.
A, qRT-PCR on E9 RV+OFT: Bmpr1a-cKO RVOFT tissues (pink bars) showed similar differences in selected gene expression to those of Acvr1-cKO (red bars) relative to controls (blue). However, Tbx1 was more highly expressed in Bmpr1a-cKO mutants. Tdgf1 and Tbx2 in Bmpr1a-cKOs differed very little from controls. * p≤0.05 Bmpr1a control vs cKO.
B-G, E9, E10 hearts, B-E from right, F-G 4-chamber. At E9, RV (arrow) in two representative Bmpr1a-cKO hearts (B) appeared narrower vs OFT than those of controls (C)(RV+OFT of these samples used for qRT-PCR). At E10, RV appeared wider in control (D,F) than mutant (E,G) though OFT of similar width (blue lines in D,E; RV and OFT colored yellow in F,G).
H-K’, Progressive loss of BMP Type I receptor alleles increased abnormality in size and shape of RV, and OFT at E11. Functional (blue) alleles remaining following Mef2c[AHF]-Cre recombination indicated in grid by title: top row, Acvr1; lower row Bmpr1a; W, wild type; F, floxed. Arrow (H’-K’) marks position of anterior face of posterior RV which appeared progressively narrower, and flattened apically (arrowheads, J,K). One Bmpr1a allele appeared more effective at maintaining RV than one Acvr1 allele. OFT also became shorter (J’,K’) and narrower (K). Straight RV outlet route visible (pink arrowheads) in control (H) but twisting in mutant (I). H, K 4 chamber view, RV+OFT colored pink; H’-K’ view from right, RV+OFT outlined. dOFT, distal OFT. Scale bar 200µm.
Figure 8. Effect of Bmpr1a knock-in into Acvr1 locus (Bmpr1a-KI) on gene expression and OFT morphology.
A, qRT-PCR on E9 RV+OFT: controls (no Cre, no Bmpr1a-KI), blue columns; Acvr1-cKO with Bmpr1a-KI mutants (ie Acvr1F, Bmpr1a-KI, Mef2c[AHF]-Cre+), purple columns; and B, Acvr1-cKO (red columns) vs Acvr1-cKO with Bmpr1a-KI (purple columns) showing that when Acvr1 was replaced by expression from one allele of Bmpr1a cDNA in the Acvr1 locus average Tbx1 and Wnt2 expression became normal, and Tgfb2 expression was over rather than under expressed (arrows) but expression of other genes shown remained only partially normalized (Smad6) or abnormal in a similar way to Acvr1 cKO. Each bar average of 3, +/− SEM, * p≤0.05.
E12 4-chamber views: Gross morphology of Control, mild and severe Acvr1-cKO and Bmpr1a-cKO without (C, Ja, Jb, Q) and with Bmpr1a-KI (D, Ka, Kb, R) dissected to reveal trunks where possible. Presence of Bmpr1a-KI in Acvr1-cKO improved morphology of RV, OFT in some (Ka vs Ja) but not all (Kb vs Jb) embryos, enlarging RV size and improving OFT orientation (R vs Q, green area) in Bmpr1a-cKO. Blood in arterial trunks shows septation in controls and mild Acvr1-cKO (Ka).
E12 sections, transverse to OFT, H&E: Control (I-U), Acvr1-cKO mild (Ja-Pa) and severe (Jb-Pb) phenotype and Bmpr1a-cKO (Q-W), all withBmpr1a-KI. OFT, from arterial trunk level (top row, sections) to proximal OFT (bottom row). In presence of Bmpr1a-KI, all cKO showed some improved wall thickness and cushion organization; but uneven wall thickness and distal epicardium (ep) was still seen where distal septation failed. In mild Acvr1-cKO with Bmpr1a-KI, trunk septation and mid-OFT cushion formation including condensed areas (blue *, arrows), myocardial growth into cushion (red arrowhead) more similar to control, but proximal cushion morphology (black*) and continuity with AV cushion (open arrow, Pa) absent. In the severe mutant with Bmpr1-KI, trunk septation failed (Lb), uneven trunk wall (Lb, short arrow), RVO lumen displaced (open arrowhead), distal (Mb) and proximal cushions abnormal (Pb), mid- to proximal OFT anterior wall too thick (red lines), some wall very thin (arrowheads Mb, Nb) but mid-OFT orientation relatively normal. In Bmpr1-cko with Bmpr1a-KI, trunk septation only partial, uneven trunk wall (K, short arrows), separate RVO lumen lost (open arrow) but mid- and proximal orientation, condensation (blue arrows), intercalated leaflet (green*) and proximal cushion continuity with AV cushion (W, black arrow), wall thickness except proximally (red lines) similar to control. Compare with Acvr1- and Bmpr1a-cKO E12 morphology in Figures 1–2 and Online Figures 2–4. Spaces between cushions and walls are artifacts.
Ao, ‘aortic’ lumen; AV, atrioventricular cushion; ep, epicardium; my, myocardium; P, ‘pulmonary’ lumen. Scale bar, 200µm.
Table I. Incidence of different arterial trunk/OFT phenotypes.
Mutant genotype samples were categorized according to number of arterial trunks. ‘Two trunk’ phenotype includes DORV (where AP and OFT valve septation complete) but also CAT with AP septation variably distal to the level of the valve leaflets (no valve septation). ‘One trunk’ phenotypes showed no AP septation, such that the pulmonary arteries arose from the back of the common arterial trunk.
Range of stages of embryos include in totals is indicated, with percentage (in brackets) of samples of those stages genotyped as cKO showing that number of trunks. These data include dead embryos/newborns in which genotype and anatomy could be determined.
| Acvr1-cKO | Acvr1-cKO,Bmpr1a-KI | Bmpr1a-cKO | Bmpr1a-cKO, Bmpr1a-KI | |
|---|---|---|---|---|
| 2 trunksa | E12-E15: 8 (53%) E12-P0: 17 (40%) |
E12-E15: 5 (63%) | E12-E14: 0 (0%) | E12-P0: 4 (100%)c |
| 1 trunk | E12-E15: 7 (47%) E12-P0: 25 (60%) |
E12-E15: 3 (37%) | E12 (live): 6 (100%) E13–14 (deadb): 15 (100%) |
E12-P0: 0 (0%)c |
Complete AP and valve septation was present in two Acvr1-cKO embryos, one Acvr1-cKO with Bmpr1a-KI embryo and two Bmpr1a-cKO with Bmpr1a-KI embryos.
Although variably necrotic, Bmpr1a-cKO mutant anatomy could still be determined at E13–14, and limb and eye morphology resembled that of E12 at the latest. The cause of death was not obviously of cardiac origin. An extra-cardiac explanation for lethality has been suggested (1).
The marked improvement in cardiovascular morphology of Bmpr1a-cKO in the presence of Bmpr1a-KI contrasts with the more limited effects of the allele on Acvr1-cKO anatomy overall. A population of Mef2c[AHF]-Cre-recombined cells which expresses both Acvr1 and Bmpr1a normally but in which BmpR1A performs an essential role for embryonic survival that AcvRI cannot, is implied by the survival and development well beyond E12 of otherwise Bmpr1a-cKO mice.
1. Briggs, L. E., Phelps, A. L., Brown, E., Kakarla, J., Anderson, R. H., van den Hoff, M. J., and Wessels, A. (2013) Expression of the BMP receptor Alk3 in the second heart field is essential for development of the dorsal mesenchymal protrusion and atrioventricular septation. Circ Res 112, 1420–1432
We therefore conclude that AcvR1 and BmpR1 each mediate signaling required for normal development in the anterior SHF-derived tissues, and propose that the abnormalities that arise from the deletion of either gene may result in part from a Tbx2/3/20-mediated misregulation of the regional identity of the OFT myocardium. Despite the similarities in their location and in the effects of their absence, AcvR1 and BmpR1a play some non-redundant roles in the development of anterior SHF-derived cardiovascular tissues.
Discussion
Implications for BMP-dependent arterial pole cardiovascular morphogenesis
In this, the first report of the Acvr1 arterial pole cKO phenotype, we have shown that a spectrum of SHF-related cardiovascular septation defects occurred but survival was possible to birth. Analysis of morphology at different times and positions along the OFT suggests a model in which the degree of distal (aortico-pulmonary trunk) septation is in part dependent on the correct positioning of two nascent OFT lumens in relation to those formed by the most distal component of the septation complex, a neural crest-dependent protrusion in the dorsal wall of the aortic sac, which lies between the routes to pharyngeal arch III,IV vessels and arch VI vessels. Loss or mis-positioning of one lumen means that a permanently unseptated truncal region, even if more distal septation still occurs. The mechanism underlying the variability is still unclear but key morphological features of normal and abnormal development of this region, and similarity to human congenital malformations of the variety of abnormal phenotypes we saw in Acvr1-cKO are examined in detail in (Anderson et al., 2012).
We detected reduced Bmp signaling via Smad1/5/8 phosphorylation in recombined OFT wall cells at both E9 and E11, but it is not possible solely from this approach to determine which events are key to normal morphogenesis without considering morphological evidence. Preseptation, we found that OFT ‘rotation’ was less than normal, and that OFT wall morphology was regionally abnormal. This could be because specific SHF populations failed to make their normal contribution to the OFT. RV/OFT walls are formed from cells originating in different areas of SHF(Bertrand et al., 2011; Dominguez et al., 2012; Takahashi et al., 2012) and their regional contribution to the tube may act to displace others in relation to the axes of the rest of the heart, i.e., relative ‘rotation’(Bajolle et al., 2006; Scherptong et al., 2012), including forming the distinctive subpulmonary myocardial population essential for normal truncal origin position and proximal trunk wall cells. We observed consistent regions of abnormally thin and thick OFT walls, areas of thickened trunk wall, and an insufficiency of sub-pulmonary myocardium suggesting AcvR1 and BmpR1a-mediated BMP signaling is required for normal formation, location and differentiation of specific SHF cell populations. Both OFT wall thickness and ‘rotation’ were improved by adding Bmpr1a-KI expression to Acvr1-cKOs consistent with the rescue of a related ‘thin wall’ model(Wang et al., 2010).
OFT cushion morphology is important for OFT septation but also affected lumen position and number in both Acvr1- and Bmpr1a-cKOs particularly distally. The regulation of OFT cushion morphogenesis, size and location remains poorly understood at the molecular level. Certain SHF populations, including those identified by Sizarov et al (Sizarov et al., 2012) and abnormal in morphology and Smad1/5/8 phosphorylation in our models, might play a role in the maintenance of separate nascent truncal lumens close to the OFT wall, perhaps via effects on OFT cushion morphogenesis. As many endocardial /endothelial cells are also recombined by Mef2c[AHF]-Cre, their behavior and signaling may also contribute, especially as cushion shape regulation is sensitive to flow(Colvee and Hurle, 1983; Manner et al., 1993) which might be expected to be abnormal from at least E9 as the right ventricle is abnormally small. However, as AP and valve septation were normal in endothelium-specific Acvr1-cKOs(Wang et al., 2005) an isolated role in septation failure is unlikely. Abnormalities in OFT mesenchymal behavior including abnormal (circumferential) alignment, and elevated cell death distally, were found in Acvr1-cKOs from E11, but also some degree of normal organization and gene expression (Plxna2, Sox9). Although Bmp4 expression has been shown to be important in normal OFT cushion development (Jia et al., 2007), other factors may be abnormal in our model, as average expression of Bmp4 was normal in mutant tissues at E9, and mesenchyme (mostly genotypically normal CNCC) stained strongly for phospho-Smad1/5/8 at E11, and expressed BMP-sensitive Sox9 in both control and mutants. However, we did see abnormalities in regional expression of Sema3C (Bajolle et al., 2008) and elevated levels of OFT Fgf8 expression may have affected cushion shape and cell number proximally(Sugi et al., 2003). Contribution to OFT cushion mesenchymal cells by EMT from recombined cells in Acvr1-cKO appeared reduced at E10, consistent with previous results(Wang et al., 2005). Neural crest cells probably made a greater contribution to cushions including valve leaflets where non-CNCC normally contribute, including to OFT valve leaflets, but proximally, continuity between OFT and AV cushions still formed poorly, contributing to the VSD phenotype in Acvr1 mutants. The walls over which these usually form are totally recombined by Mef2c[AHF]-Cre, and were not well rescued by the addition of Bmpr1a-KI expression to Acvr1 mutants, unlike Bmpr1a-cKO, consistent with AcvR1 playing a specific role in their formation.
Role of AcvR1-mediated signaling in regulating gene expression in SHF-derived cells
We did not find strong evidence that an autocrine element of the ‘BMP/FGF balance’ model(Tirosh-Finkel et al., 2010) in recombined cells contributed to the Acvr1-cKO phenotype. However, altered expression levels of other genes implicated in precursor/proliferation (elevated) and differentiation (reduced) were found in E9 RV+OFT Acvr1 mutants: Fgf8 (promotes SHF proliferation(Ilagan et al., 2006; Park et al., 2006)), Isl1 (needed for SHF progenitors(Cai et al., 2003)), and Tdgf1 (Cripto, associated with muscle precursor development/proliferation(Guardiola et al., 2012)) elevated; and Wnt2 (required for myocardial differentiation(Onizuka et al., 2012; Tian et al., 2010) reduced. Fgf8, Isl1 and Wnt2 are all Tbx1 targets(Liao et al., 2008); average expression of Tbx1 itself was mildly elevated in E9 SHF, (where it maintains proliferation(Zhang et al., 2006)) so may contribute to their altered expression as they enter the heart tube. Elevated expression of Tbx1 detected in E9 RV+OFT in Acvr1- and Bmpr1a-cKO mutants is hard to detect in OFT wall by WMT ISH, but cells present distally by the aortic sac at E9(Chen et al., 2009) would be included in RV+OFT by dissection. Not only was average Fgf8, Isl1, and Tdgf1 expression elevated at E9, but their expression also extended along the OFT more proximally and for longer in time in Acvr1-cKO, also less differentiated’ phenotypes. FGF signaling has also been implicated in the differentiation of distal SHF into arterial smooth muscle of the trunk walls(Hutson et al., 2010). Some ‘smooth muscle’ genes are normally expressed in early embryonic myocardium (such as Acta2(Franco et al., 1999)) so were not assayed, but the higher expression of Myh11 (even though at a very low level), not normally expressed until E14, may reflect a subtle shift in transcriptional milieu to favor smooth muscle-related fate.
T-box transcription factors, Tbx2, 3 and 20 are good candidates to be key downstream targets for AcvR1-dependent BMP signaling and the lack of expression of which could be responsible for the phenotypes we report here in Acvr1-cKO. That levels of all three are reduced in our mutants could result in a phenotype where partial loss of any one alone would not. They can be directly up-regulated by BMP R-Smad1/5/8 signaling(Singh et al., 2009; Yang et al., 2006). Tbx2 and 3 have previously been identified as targets in OFT development in Bmpr1a/Isl1-Cre-cKO model(Yang et al., 2006) and Tbx20 as required for suppression of expression of Isl1(Cai et al., 2005) and expression of Tbx3 in the AV(Cai et al., 2011). Their individual and double KOs have smaller RV and abnormal OFT(Mesbah et al., 2012). Study of their roles in AV morphological development have shown that they repress the expression of ‘chamber’ (i.e., ventricular apex and atrial appendage) region genes, including Nppa, morphology (trabeculation) and higher proliferation (Ribeiro et al., 2007); and thereby maintain the presence of cushions and formation of cushion mesenchyme, through expression of genes such as Has2 and Tgfb2. Average Tgfb2 expression was also reduced in Acvr1- and Bmpr1a-cKO, and the Tgfb2-KO phenotype also resembles that of Acvr1-cKO, including a range of OFT/AP septation defects from DORV to CAT with variable degree of distal trunk septation and failure of cushion fusion(Bartram et al., 2001). Mis-expression of Nppa and Tgfb2, and thicker anterior proximal OFT myocardium in Acvr1-cKO suggest that at least the boundary between the right ventricle chamber and non-chamber mid-proximal areas of the OFT wall was less distinct and OFT non-chamber identity compromised. Higher expression of muscle genes in part of the OFT myocardium together with a reduced contribution by the smaller RV might contribute to the similarity of average muscle gene expression in Acvr1-cKO and control samples. Like Isl1 expression, Tbx2 and Tbx3 expression only became abnormal in Acvr1-cKO after SHF had joined the heart tube, suggesting an AcvR1-mediated process maintained their expression and Tbx20 from this point. Despite apparently normal levels of Bmp4 and higher than normal Bmp2 at E9, remaining Type I BMP receptors were insufficient to normalize levels of these three genes in Acvr1-cKO. There is feedback regulation between these T-box factors (Cai et al., 2011; Dupays et al., 2009; Gavrilov et al., 2013), so Acvr1-cKO is an interesting model where all three were reduced but none was absent. Again, the failure in Acvr1- and Bmpr1a-cKO to establish correct chamber and non-chamber-type regions would represent a failure of normal differentiation.
The effects of elevation of Wnt/beta-catenin signaling (by LiCl injection) or BMP signaling (using Acvr1 locus-regulated Bmpr1a-KI or ACVR1-KI) on average gene expression in E9 RV+OFT (Online Figure 7) suggest a variety of regulatory pathways controlled the expression of genes altered in Acvr1-cKO. Although Tbx2,3,20 expression was not normalized, the downstream target gene Tgfb2 became over-expressed by Bmpr1a-KI in Acvr1-cKO. This may have enabled the degree of context-dependent improvement in morphological development. Comparison of gene expression at E9 with morphological phenotypes suggests additional genes regulated by AcvR1-mediated signaling remain to be identified, and that more region-specific studies are needed to advance understanding of this complex environment.
AcvR1 as one component of BMP signaling in SHF development
AcvR1 and BmpR1a are the principal Type I BMP receptors expressed in the developing RV and OFT wall. Either one alone is insufficient to maintain normal development in arterial pole SHF-derived structures, or expression of known BMP-Smad target genes such as Smad6, Msx2, Id1 and Bambi at E9 even though Bmpr1a and Acvr1 are expressed at grossly similar times and places. Nevertheless, they appear to regulate many of the same processes (gene regulation, morphogenesis). The sequential loss of their functional alleles appeared to create a somewhat gradient-like of loss of normality of morphology (Figure 6), and of gene expression (though sensitivity/mechanism varied from gene to gene) (Online Figure 7). Yet even though Acvr1Bmpr1a-KI was expressed at E9 (Figure 7I) and substantially rescued Bmpr1a-cKO phenotype despite its expression being restricted to the Acvr1 expression domain, Bmpr1a-KI did not restore expression of all assayed genes to normal levels in Acvr1-cKO RV+OFT at E9. Surprising amongst these were Smad6, Tbx2, 3 and 20. One interpretation is that AcvR1 and BmpR1a have some non-redundant roles, consistent with their known difference in ligand preference(Macias-Silva et al., 1998), but the similarity of gene expression and phenotypes affected suggest that their signaling also needs to be co-ordinated or combined to be effective. It may also be restricted to a very specific region and time. There is evidence in other models for heteromeric receptor formation(Little and Mullins, 2009; Shimmi et al., 2005), but other mechanisms, such as interaction with as yet unidentified regulatory factors, may be responsible.
AcvR1 in congenital heart disease
Although AcvR1 itself is best known for its constitutively active mutant form that results in fibrodysplasia ossificans progressiva(Shore et al., 2006), the ACVR1 L343P allele was identified in a screen of atrioventricular septal defect patients and shows greatly reduced signaling capacity and transcriptional activity(Smith et al., 2009). Abnormalities in a number of genes, the normal expression of which we found dependent on AcvR1 have already been identified as causative of CHD (TBX20(Kirk et al., 2007) and TGFB2(Gao et al., 2012; Lindsay et al., 2012)), or as potential contributory (TBX2(Pang et al., 2013),TBX3(Chen et al., 2013), BMP4(Goracy et al., 2012), TDGF1(Roessler et al., 2008; Wang et al., 2011) and ISL1(Stevens et al., 2010)). Even a mildly defective allele of Acvr1 could influence the expression of many genes known to be required for normal heart development and when in combination with other such alleles of these genes account for cases of CHD. The results reported here illustrate the importance of considering the effect of abnormal, though apparently ‘redundant’, gene products in the understanding of human CHD.
Supplementary Material
Highlights.
Second heart field-specific deletion of Acvr1 results in cardiovascular defects.
AcvR1 is required for normal myocardial differentiation and regional development.
AcvR1 and BmpR1a play both overlapping and unique roles in the second heart field.
Acknowledgements
We thank Wanda Filipiak and Margaret van Keuren (University of Michigan Transgenic Animal Model Core) for preparation of transgenic mice, and Martin White and Mike Pihalja (University of Michigan Flow Cytometry Core).
Sources of Funding
This study was supported by the NIH RO1 grant HL HL074862 (VK).
Glossary
- AoT
Aortic trunk
- AoV
Aortic valve
- AP
Aortico-pulmonary
- AV
Atrioventricular (region)
- BMP
Bone morphogenetic protein
- CAT
Common arterial trunk
- cHet
conditional heterozygote
- Cko
conditional knock out
- CNCC
cardiac neural crest-derived cells
- CHD
Congenital heart disease
- DORV
Double outlet right ventricle
- E9
Embryonic day 9
- EMT
Epithelial-to-mesenchymal transformation
- FGF
Fibroblast growth factor
- ISH
In situ hybridization
- LA
Left atrium
- LV
Left ventricle
- OFT
Outflow tract
- P0
Postnatal, within first 24 hours
- PT
Pulmonary trunk
- PV
Pulmonary valve
- RA
Right atrium
- RV
Right ventricle
- RVO
Right ventricular outlet
- SHF
Second heart field
- SmαA
Smooth muscle alpha actin
- VSD
Ventricular septal defect
- WMT
Whole mount (pieces of tissue, not sections)
Footnotes
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures
None
References
- Ahn K, Mishina Y, Hanks MC, Behringer RR, Crenshaw EB., 3rd BMPR-IA signaling is required for the formation of the apical ectodermal ridge and dorsal-ventral patterning of the limb. Development. 2001;128:4449–4461. doi: 10.1242/dev.128.22.4449. [DOI] [PubMed] [Google Scholar]
- Ai D, Fu X, Wang J, Lu MF, Chen L, Baldini A, Klein WH, Martin JF. Canonical Wnt signaling functions in second heart field to promote right ventricular growth. Proc Natl Acad Sci U S A. 2007;104:9319–9324. doi: 10.1073/pnas.0701212104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson RH, Chaudhry B, Mohun TJ, Bamforth SD, Hoyland D, Phillips HM, Webb S, Moorman AF, Brown NA, Henderson DJ. Normal and abnormal development of the intrapericardial arterial trunks in humans and mice. Cardiovasc Res. 2012;95:108–115. doi: 10.1093/cvr/cvs147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y, Wang J, Morikawa Y, Bonilla-Claudio M, Klysik E, Martin JF. Bmp signaling represses Vegfa to promote outflow tract cushion development. Development. 2013;140:3395–3402. doi: 10.1242/dev.097360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajolle F, Zaffran S, Kelly RG, Hadchouel J, Bonnet D, Brown NA, Buckingham ME. Rotation of the myocardial wall of the outflow tract is implicated in the normal positioning of the great arteries. Circ Res. 2006;98:421–428. doi: 10.1161/01.RES.0000202800.85341.6e. [DOI] [PubMed] [Google Scholar]
- Bajolle F, Zaffran S, Meilhac SM, Dandonneau M, Chang T, Kelly RG, Buckingham ME. Myocardium at the base of the aorta and pulmonary trunk is prefigured in the outflow tract of the heart and in subdomains of the second heart field. Dev Biol. 2008;313:25–34. doi: 10.1016/j.ydbio.2007.09.023. [DOI] [PubMed] [Google Scholar]
- Bartram U, Molin DG, Wisse LJ, Mohamad A, Sanford LP, Doetschman T, Speer CP, Poelmann RE, Gittenberger-de Groot AC. Double-outlet right ventricle and overriding tricuspid valve reflect disturbances of looping, myocardialization, endocardial cushion differentiation, and apoptosis in TGF-beta(2)-knockout mice. Circulation. 2001;103:2745–2752. doi: 10.1161/01.cir.103.22.2745. [DOI] [PubMed] [Google Scholar]
- Beppu H, Malhotra R, Beppu Y, Lepore JJ, Parmacek MS, Bloch KD. BMP type II receptor regulates positioning of outflow tract and remodeling of atrioventricular cushion during cardiogenesis. Dev Biol. 2009;331:167–175. doi: 10.1016/j.ydbio.2009.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand N, Roux M, Ryckebusch L, Niederreither K, Dolle P, Moon A, Capecchi M, Zaffran S. Hox genes define distinct progenitor sub-domains within the second heart field. Dev Biol. 2011;353:266–274. doi: 10.1016/j.ydbio.2011.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs LE, Phelps AL, Brown E, Kakarla J, Anderson RH, van den Hoff MJ, Wessels A. Expression of the BMP receptor Alk3 in the second heart field is essential for development of the dorsal mesenchymal protrusion and atrioventricular septation. Circ Res. 2013;112:1420–1432. doi: 10.1161/CIRCRESAHA.112.300821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruneau BG. Signaling and transcriptional networks in heart development and regeneration. Cold Spring Harbor perspectives in biology. 2013;5:a008292. doi: 10.1101/cshperspect.a008292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai CL, Liang X, Shi Y, Chu PH, Pfaff SL, Chen J, Evans S. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev Cell. 2003;5:877–889. doi: 10.1016/s1534-5807(03)00363-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai CL, Zhou W, Yang L, Bu L, Qyang Y, Zhang X, Li X, Rosenfeld MG, Chen J, Evans S. T-box genes coordinate regional rates of proliferation and regional specification during cardiogenesis. Development. 2005;132:2475–2487. doi: 10.1242/dev.01832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Nomura-Kitabayashi A, Cai W, Yan J, Christoffels VM, Cai CL. Myocardial Tbx20 regulates early atrioventricular canal formation and endocardial epithelial-mesenchymal transition via Bmp2. Dev Biol. 2011;360:381–390. doi: 10.1016/j.ydbio.2011.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Qiao Y, Meng H, Pang S, Huang W, Zhang H, Yan B. Genetic analysis of the TBX3 gene promoter in ventricular septal defects. Gene. 2013;512:185–188. doi: 10.1016/j.gene.2012.10.066. [DOI] [PubMed] [Google Scholar]
- Chen L, Fulcoli FG, Tang S, Baldini A. Tbx1 regulates proliferation and differentiation of multipotent heart progenitors. Circ Res. 2009;105:842–851. doi: 10.1161/CIRCRESAHA.109.200295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colvee E, Hurle JM. Malformations of the semilunar valves produced in chick embryos by mechanical interference with cardiogenesis. An experimental approach to the role of hemodynamics in valvular development. Anat Embryol (Berl) 1983;168:59–71. doi: 10.1007/BF00305399. [DOI] [PubMed] [Google Scholar]
- de Pater E, Ciampricotti M, Priller F, Veerkamp J, Strate I, Smith K, Lagendijk AK, Schilling TF, Herzog W, Abdelilah-Seyfried S, Hammerschmidt M, Bakkers J. Bmp signaling exerts opposite effects on cardiac differentiation. Circ Res. 2012;110:578–587. doi: 10.1161/CIRCRESAHA.111.261172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature. 2003;425:577–584. doi: 10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
- Dominguez JN, Meilhac SM, Bland YS, Buckingham ME, Brown NA. Asymmetric fate of the posterior part of the second heart field results in unexpected left/right contributions to both poles of the heart. Circ Res. 2012;111:1323–1335. doi: 10.1161/CIRCRESAHA.112.271247. [DOI] [PubMed] [Google Scholar]
- Dupays L, Kotecha S, Angst B, Mohun TJ. Tbx2 misexpression impairs deployment of second heart field derived progenitor cells to the arterial pole of the embryonic heart. Dev Biol. 2009;333:121–131. doi: 10.1016/j.ydbio.2009.06.025. [DOI] [PubMed] [Google Scholar]
- Dyer LA, Kirby ML. The role of secondary heart field in cardiac development. Dev Biol. 2009;336:137–144. doi: 10.1016/j.ydbio.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich M, Gutman O, Knaus P, Henis YI. Oligomeric interactions of TGF-beta and BMP receptors. FEBS letters. 2012;586:1885–1896. doi: 10.1016/j.febslet.2012.01.040. [DOI] [PubMed] [Google Scholar]
- Franco D, Markman MM, Wagenaar GT, Ya J, Lamers WH, Moorman AF. Myosin light chain 2a and 2v identifies the embryonic outflow tract myocardium in the developing rodent heart. The Anatomical record. 1999;254:135–146. doi: 10.1002/(SICI)1097-0185(19990101)254:1<135::AID-AR17>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- Fukuda T, Scott G, Komatsu Y, Araya R, Kawano M, Ray MK, Yamada M, Mishina Y. Generation of a mouse with conditionally activated signaling through the BMP receptor, ALK2. Genesis. 2006;44:159–167. doi: 10.1002/dvg.20201. [DOI] [PubMed] [Google Scholar]
- Gao Y, Ma XJ, Huang GY, Zhang J, Wang HJ, Ma D, Wu Y. DNA sequencing of TGFbeta2 in sporadic patients with tetralogy of Fallot. Experimental and therapeutic medicine. 2012;3:878–880. doi: 10.3892/etm.2012.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavrilov S, Harvey RP, Papaioannou VE. Lack of Genetic Interaction between Tbx20 and Tbx3 in Early Mouse Heart Development. PLoS One. 2013;8:e70149. doi: 10.1371/journal.pone.0070149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goracy I, Safranow K, Dawid G, Skonieczna-Zydecka K, Kaczmarczyk M, Goracy J, Loniewska B, Ciechanowicz A. Common genetic variants of the BMP4, BMPR1A, BMPR1B, and ACVR1 genes, left ventricular mass, and other parameters of the heart in newborns. Genetic testing and molecular biomarkers. 2012;16:1309–1316. doi: 10.1089/gtmb.2012.0164. [DOI] [PubMed] [Google Scholar]
- Guardiola O, Lafuste P, Brunelli S, Iaconis S, Touvier T, Mourikis P, De Bock K, Lonardo E, Andolfi G, Bouche A, Liguori GL, Shen MM, Tajbakhsh S, Cossu G, Carmeliet P, Minchiotti G. Cripto regulates skeletal muscle regeneration and modulates satellite cell determination by antagonizing myostatin. Proc Natl Acad Sci U S A. 2012;109:E3231–E3240. doi: 10.1073/pnas.1204017109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan B, Beddington R, Costantini F, Lacy E. A laboratory manual. New York: Cold Spring Harbor Laboratory Press; 1994. Manipulating the mouse embryo. [Google Scholar]
- Hutson MR, Zeng XL, Kim AJ, Antoon E, Harward S, Kirby ML. Arterial pole progenitors interpret opposing FGF/BMP signals to proliferate or differentiate. Development. 2010;137:3001–3011. doi: 10.1242/dev.051565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilagan R, Abu-Issa R, Brown D, Yang YP, Jiao K, Schwartz RJ, Klingensmith J, Meyers EN. Fgf8 is required for anterior heart field development. Development. 2006;133:2435–2445. doi: 10.1242/dev.02408. [DOI] [PubMed] [Google Scholar]
- Jia Q, McDill BW, Li SZ, Deng C, Chang CP, Chen F. Smad signaling in the neural crest regulates cardiac outflow tract remodeling through cell autonomous and non-cell autonomous effects. Dev Biol. 2007;311:172–184. doi: 10.1016/j.ydbio.2007.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaartinen V, Dudas M, Nagy A, Sridurongrit S, Lu MM, Epstein JA. Cardiac outflow tract defects in mice lacking ALK2 in neural crest cells. Development. 2004;131:3481–3490. doi: 10.1242/dev.01214. [DOI] [PubMed] [Google Scholar]
- Kaartinen V, Nagy A. Removal of the floxed neo gene from a conditional knockout allele by the adenoviral Cre recombinase in vivo. Genesis. 2001;31:126–129. doi: 10.1002/gene.10015. [DOI] [PubMed] [Google Scholar]
- Kirk EP, Sunde M, Costa MW, Rankin SA, Wolstein O, Castro ML, Butler TL, Hyun C, Guo G, Otway R, Mackay JP, Waddell LB, Cole AD, Hayward C, Keogh A, Macdonald P, Griffiths L, Fatkin D, Sholler GF, Zorn AM, Feneley MP, Winlaw DS, Harvey RP. Mutations in cardiac T-box factor gene TBX20 are associated with diverse cardiac pathologies, including defects of septation and valvulogenesis and cardiomyopathy. American journal of human genetics. 2007;81:280–291. doi: 10.1086/519530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishigami S, Mishina Y. BMP signaling and early embryonic patterning. Cytokine & growth factor reviews. 2005;16:265–278. doi: 10.1016/j.cytogfr.2005.04.002. [DOI] [PubMed] [Google Scholar]
- Klaus A, Saga Y, Taketo MM, Tzahor E, Birchmeier W. Distinct roles of Wnt/beta-catenin and Bmp signaling during early cardiogenesis. Proc Natl Acad Sci U S A. 2007;104:18531–18536. doi: 10.1073/pnas.0703113104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao J, Aggarwal VS, Nowotschin S, Bondarev A, Lipner S, Morrow BE. Identification of downstream genetic pathways of Tbx1 in the second heart field. Dev Biol. 2008;316:524–537. doi: 10.1016/j.ydbio.2008.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay ME, Schepers D, Bolar NA, Doyle JJ, Gallo E, Fert-Bober J, Kempers MJ, Fishman EK, Chen Y, Myers L, Bjeda D, Oswald G, Elias AF, Levy HP, Anderlid BM, Yang MH, Bongers EM, Timmermans J, Braverman AC, Canham N, Mortier GR, Brunner HG, Byers PH, Van Eyk J, Van Laer L, Dietz HC, Loeys BL. Loss-of-function mutations in TGFB2 cause a syndromic presentation of thoracic aortic aneurysm. Nat Genet. 2012;44:922–927. doi: 10.1038/ng.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little SC, Mullins MC. Bone morphogenetic protein heterodimers assemble heteromeric type I receptor complexes to pattern the dorsoventral axis. Nature cell biology. 2009;11:637–643. doi: 10.1038/ncb1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macias-Silva M, Hoodless PA, Tang SJ, Buchwald M, Wrana JL. Specific activation of Smad1 signaling pathways by the BMP7 type I receptor, ALK2. J.Biol.Chem. 1998;273:25628–25636. doi: 10.1074/jbc.273.40.25628. [DOI] [PubMed] [Google Scholar]
- Manner J, Seidl W, Steding G. Correlation between the embryonic head flexures and cardiac development. An experimental study in chick embryos. Anat Embryol (Berl) 1993;188:269–285. doi: 10.1007/BF00188218. [DOI] [PubMed] [Google Scholar]
- Massague J, Chen YG. Controlling TGF-beta signaling. Genes Dev. 2000;14:627–644. [PubMed] [Google Scholar]
- Mesbah K, Rana MS, Francou A, van Duijvenboden K, Papaioannou VE, Moorman AF, Kelly RG, Christoffels VM. Identification of a Tbx1/Tbx2/Tbx3 genetic pathway governing pharyngeal and arterial pole morphogenesis. Human molecular genetics. 2012;21:1217–1229. doi: 10.1093/hmg/ddr553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorman AF, Houweling AC, de Boer PA, Christoffels VM. Sensitive nonradioactive detection of mRNA in tissue sections: novel application of the whole-mount in situ hybridization protocol. J.Histochem.Cytochem. 2001;49:1–8. doi: 10.1177/002215540104900101. [DOI] [PubMed] [Google Scholar]
- Onizuka T, Yuasa S, Kusumoto D, Shimoji K, Egashira T, Ohno Y, Kageyama T, Tanaka T, Hattori F, Fujita J, Ieda M, Kimura K, Makino S, Sano M, Kudo A, Fukuda K. Wnt2 accelerates cardiac myocyte differentiation from ES-cell derived mesodermal cells via non-canonical pathway. J Mol Cell Cardiol. 2012;52:650–659. doi: 10.1016/j.yjmcc.2011.11.010. [DOI] [PubMed] [Google Scholar]
- Pang S, Liu Y, Zhao Z, Huang W, Chen D, Yan B. Novel and functional sequence variants within the TBX2 gene promoter in ventricular septal defects. Biochimie. 2013;95:1807–1809. doi: 10.1016/j.biochi.2013.05.007. [DOI] [PubMed] [Google Scholar]
- Park EJ, Ogden LA, Talbot A, Evans S, Cai CL, Black BL, Frank DU, Moon AM. Required, tissue-specific roles for Fgf8 in outflow tract formation and remodeling. Development. 2006;133:2419–2433. doi: 10.1242/dev.02367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro I, Kawakami Y, Buscher D, Raya A, Rodriguez-Leon J, Morita M, Rodriguez Esteban C, Izpisua Belmonte JC. Tbx2 and Tbx3 regulate the dynamics of cell proliferation during heart remodeling. PLoS One. 2007;2:e398. doi: 10.1371/journal.pone.0000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessler E, Ouspenskaia MV, Karkera JD, Velez JI, Kantipong A, Lacbawan F, Bowers P, Belmont JW, Towbin JA, Goldmuntz E, Feldman B, Muenke M. Reduced NODAL signaling strength via mutation of several pathway members including FOXH1 is linked to human heart defects and holoprosencephaly. American journal of human genetics. 2008;83:18–29. doi: 10.1016/j.ajhg.2008.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakabe M, Kokubo H, Nakajima Y, Saga Y. Ectopic retinoic acid signaling affects outflow tract cushion development through suppression of the myocardial Tbx2-Tgfbeta2 pathway. Development. 2012;139:385–395. doi: 10.1242/dev.067058. [DOI] [PubMed] [Google Scholar]
- Scherptong RW, Jongbloed MR, Wisse LJ, Vicente-Steijn R, Bartelings MM, Poelmann RE, Schalij MJ, Gittenberger-De Groot AC. Morphogenesis of outflow tract rotation during cardiac development: the pulmonary push concept. Dev Dyn. 2012;241:1413–1422. doi: 10.1002/dvdy.23833. [DOI] [PubMed] [Google Scholar]
- Shimmi O, Umulis D, Othmer H, O’Connor MB. Facilitated transport of a Dpp/Scw heterodimer by Sog/Tsg leads to robust patterning of the Drosophila blastoderm embryo. Cell. 2005;120:873–886. doi: 10.1016/j.cell.2005.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore EM, Xu M, Feldman GJ, Fenstermacher DA, Cho TJ, Choi IH, Connor JM, Delai P, Glaser DL, LeMerrer M, Morhart R, Rogers JG, Smith R, Triffitt JT, Urtizberea JA, Zasloff M, Brown MA, Kaplan FS. A recurrent mutation in the BMP type I receptor ACVR1 causes inherited and sporadic fibrodysplasia ossificans progressiva. Nat Genet. 2006;38:525–527. doi: 10.1038/ng1783. [DOI] [PubMed] [Google Scholar]
- Singh R, Hoogaars WM, Barnett P, Grieskamp T, Rana MS, Buermans H, Farin HF, Petry M, Heallen T, Martin JF, Moorman AF, t Hoen PA, Kispert A, Christoffels VM. Tbx2 and Tbx3 induce atrioventricular myocardial development and endocardial cushion formation. Cell Mol Life Sci. 2012;69:1377–1389. doi: 10.1007/s00018-011-0884-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Horsthuis T, Farin HF, Grieskamp T, Norden J, Petry M, Wakker V, Moorman AF, Christoffels VM, Kispert A. Tbx20 interacts with smads to confine tbx2 expression to the atrioventricular canal. Circ Res. 2009;105:442–452. doi: 10.1161/CIRCRESAHA.109.196063. [DOI] [PubMed] [Google Scholar]
- Sizarov A, Lamers WH, Mohun TJ, Brown NA, Anderson RH, Moorman AF. Three-dimensional and molecular analysis of the arterial pole of the developing human heart. J Anat. 2012;220:336–349. doi: 10.1111/j.1469-7580.2012.01474.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KA, Joziasse IC, Chocron S, van Dinther M, Guryev V, Verhoeven MC, Rehmann H, van der Smagt JJ, Doevendans PA, Cuppen E, Mulder BJ, Ten Dijke P, Bakkers J. Dominant-negative ALK2 allele associates with congenital heart defects. Circulation. 2009;119:3062–3069. doi: 10.1161/CIRCULATIONAHA.108.843714. [DOI] [PubMed] [Google Scholar]
- Snarr BS, Kern CB, Wessels A. Origin and fate of cardiac mesenchyme. Dev Dyn. 2008;237:2804–2819. doi: 10.1002/dvdy.21725. [DOI] [PubMed] [Google Scholar]
- Song L, Fassler R, Mishina Y, Jiao K, Baldwin HS. Essential functions of Alk3 during AV cushion morphogenesis in mouse embryonic hearts. Dev.Biol. 2007;301:276–286. doi: 10.1016/j.ydbio.2006.08.004. [DOI] [PubMed] [Google Scholar]
- Stevens KN, Hakonarson H, Kim CE, Doevendans PA, Koeleman BP, Mital S, Raue J, Glessner JT, Coles JG, Moreno V, Granger A, Gruber SB, Gruber PJ. Common variation in ISL1 confers genetic susceptibility for human congenital heart disease. PLoS One. 2010;5:e10855. doi: 10.1371/journal.pone.0010855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugi Y, Ito N, Szebenyi G, Myers K, Fallon JF, Mikawa T, Markwald RR. Fibroblast growth factor (FGF)-4 can induce proliferation of cardiac cushion mesenchymal cells during early valve leaflet formation. Dev.Biol. 2003;258:252–263. doi: 10.1016/s0012-1606(03)00099-x. [DOI] [PubMed] [Google Scholar]
- Takahashi M, Terasako Y, Yanagawa N, Kai M, Yamagishi T, Nakajima Y. Myocardial progenitors in the pharyngeal regions migrate to distinct conotruncal regions. Dev Dyn. 2012;241:284–293. doi: 10.1002/dvdy.23714. [DOI] [PubMed] [Google Scholar]
- Theveniau-Ruissy M, Dandonneau M, Mesbah K, Ghez O, Mattei MG, Miquerol L, Kelly RG. The del22q11.2 candidate gene Tbx1 controls regional outflow tract identity and coronary artery patterning. Circ Res. 2008;103:142–148. doi: 10.1161/CIRCRESAHA.108.172189. [DOI] [PubMed] [Google Scholar]
- Thomas PS, Sridurongrit S, Ruiz-Lozano P, Kaartinen V. Deficient signaling via Alk2 (Acvr1) leads to bicuspid aortic valve development. PLoS One. 2012;7:e35539. doi: 10.1371/journal.pone.0035539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y, Yuan L, Goss AM, Wang T, Yang J, Lepore JJ, Zhou D, Schwartz RJ, Patel V, Cohen ED, Morrisey EE. Characterization and in vivo pharmacological rescue of a Wnt2-Gata6 pathway required for cardiac inflow tract development. Dev Cell. 2010;18:275–287. doi: 10.1016/j.devcel.2010.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh-Finkel L, Zeisel A, Brodt-Ivenshitz M, Shamai A, Yao Z, Seger R, Domany E, Tzahor E. BMP-mediated inhibition of FGF signaling promotes cardiomyocyte differentiation of anterior heart field progenitors. Development. 2010;137:2989–3000. doi: 10.1242/dev.051649. [DOI] [PubMed] [Google Scholar]
- Verzi MP, McCulley DJ, De Val S, Dodou E, Black BL. The right ventricle, outflow tract, and ventricular septum comprise a restricted expression domain within the secondary/anterior heart field. Dev Biol. 2005;287:134–145. doi: 10.1016/j.ydbio.2005.08.041. [DOI] [PubMed] [Google Scholar]
- Vincent SD, Buckingham ME. How to make a heart: the origin and regulation of cardiac progenitor cells. Curr Top Dev Biol. 2010;90:1–41. doi: 10.1016/S0070-2153(10)90001-X. [DOI] [PubMed] [Google Scholar]
- Wang B, Yan J, Peng Z, Wang J, Liu S, Xie X, Ma X. Teratocarcinoma-derived growth factor 1 (TDGF1) sequence variants in patients with congenital heart defect. International journal of cardiology. 2011;146:225–227. doi: 10.1016/j.ijcard.2009.08.046. [DOI] [PubMed] [Google Scholar]
- Wang J, Greene SB, Bonilla-Claudio M, Tao Y, Zhang J, Bai Y, Huang Z, Black BL, Wang F, Martin JF. Bmp signaling regulates myocardial differentiation from cardiac progenitors through a MicroRNA-mediated mechanism. Dev Cell. 2010;19:903–912. doi: 10.1016/j.devcel.2010.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Sridurongrit S, Dudas M, Thomas P, Nagy A, Schneider MD, Epstein JA, Kaartinen V. Atrioventricular cushion transformation is mediated by ALK2 in the developing mouse heart. Dev.Biol. 2005;286:299–310. doi: 10.1016/j.ydbio.2005.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warming S, Costantino N, Court DL, Jenkins NA, Copeland NG. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 2005;33:e36. doi: 10.1093/nar/gni035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y, Zaffran S, Kuroiwa A, Higuchi H, Ogura T, Harvey RP, Kelly RG, Buckingham M. Fibroblast growth factor 10 gene regulation in the second heart field by Tbx1, Nkx2–5, and Islet1 reveals a genetic switch for down-regulation in the myocardium. Proc Natl Acad Sci U S A. 2012;109:18273–18280. doi: 10.1073/pnas.1215360109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Morishima M, Wylie JN, Schwartz RJ, Bruneau BG, Lindsay EA, Baldini A. Tbx1 has a dual role in the morphogenesis of the cardiac outflow tract. Development. 2004;131:3217–3227. doi: 10.1242/dev.01174. [DOI] [PubMed] [Google Scholar]
- Yang L, Cai CL, Lin L, Qyang Y, Chung C, Monteiro RM, Mummery CL, Fishman GI, Cogen A, Evans S. Isl1Cre reveals a common Bmp pathway in heart and limb development. Development. 2006;133:1575–1585. doi: 10.1242/dev.02322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Huynh T, Baldini A. Mesodermal expression of Tbx1 is necessary and sufficient for pharyngeal arch and cardiac outflow tract development. Development. 2006;133:3587–3595. doi: 10.1242/dev.02539. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.