Abstract
Organofluorines represent a rapidly expanding proportion of molecules used in pharmaceuticals, diagnostics, agrochemicals, and materials. Despite the prevalence of fluorine in synthetic compounds, the known biological scope is limited to a single pathway that produces fluoroacetate. Here, we demonstrate that this pathway can be exploited as a source of fluorinated building blocks for introduction of fluorine into natural product scaffolds. Specifically, we have constructed pathways involving two polyketide synthase systems and show that fluoroacetate can be used to incorporate fluorine into the polyketide backbone in vitro. We further show that fluorine can be introduced site-selectively and introduced into polyketide products in vivo. These results highlight the prospects for the production of complex fluorinated natural products using synthetic biology.
The catalytic diversity of biological systems provides enormous potential for application of living cells to the scalable production of pharmaceuticals, fuels, and materials (1–4). However, the scope of innovation of living organisms is typically limited to functions that confer a direct advantage for cell growth, thereby maximizing biomass as the end product rather than a distinct molecule or reaction of interest. In contrast, synthetic biology approaches allow us to disconnect some of these remarkable biochemical transformations from cell survival and reconnect them differently for the targeted synthesis of alternative classes of compounds. One particularly interesting area of opportunity is the development of methods to introduce fluorine into complex small molecule scaffolds, which has become a powerful strategy for the design of synthetic pharmaceuticals. Indeed, it is estimated that 20–30% of drugs, including many of the top sellers, contain at least one fluorine atom (5–7). Recent innovations have expanded the scope of synthetic C–F bond forming methodologies, but the unusual elemental properties of fluorine that serve as the basis for its success also continue to restrict the range of molecular structures that can be accessed (8–11). As such, the invention of alternative routes for the site-selective introduction of fluorine into structurally diverse molecules, particularly under mild conditions, remains an outstanding challenge.
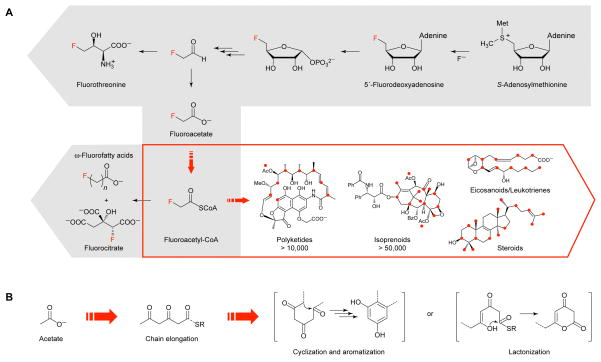
In comparison to synthetic small molecules, fluorine has limited distribution in naturally occurring organic compounds; the only organofluorine natural products characterized to date consist of a small set of simple molecules associated with the fluoroacetate pathway of Streptomyces cattleya, a soil bacterium that houses the remarkable ability to catalyze the formation of C–F bonds from aqueous fluoride (Figure 1A) (12, 13). Although these compounds lack the intricacy typically expected of secondary metabolites, they represent a potentially rich source of modular organofluorine building blocks for the production of complex fluorinated natural products. In this regard, the backbones of several large classes of medicinally-relevant natural products – including polyketides, isoprenoids, steroids, alkaloids, eicosanoids, leukotrienes, and others – are biosynthesized directly from the assembly and tailoring of simple acetate units (Figure 1A). Introduction of the fluoroacetate monomer in place of acetate would allow us to incorporate fluorine into the backbone of these targets and create new molecular function by combining the medicinal chemistry advantages of fluorine with the structural complexity and bioactivity of natural products. For example, the introduction of fluorine via synthetic or semisynthetic routes has enabled the improvement of the clinical properties of several natural products but remains challenging to achieve (14–17). While previous studies have shown that distal fluorine substituents can be accommodated in natural product biosynthetic pathways (18, 19), access to fluoromalonyl-CoA, a fluorinated analog of one of nature’s most powerful carbon nucleophiles, as an extender unit would enable a general method for direct incorporation of fluorine into any polyketide structure.
Fig. 1.
Synthetic biology of fluorine. (A) The fluoroacetate pathway and its metabolites represent the known scope of biological fluorine chemistry, starting with fluoride and S-adenosylmethionine, to produce fluoroacetate and fluorothreonine as the end products (right to left, grey box). This scope could be greatly expanded by engineering downstream pathways to use fluoroacetate as a building block for introduction of fluorine site-selectively into large families of natural products constructed from acetate backbones (left to right, red box). Red dots represent positions that could in principle be fluorinated by incorporation of a fluoroacetate monomer without altering the carbon skeleton, including locations where fluorine would replace a methyl group derived from propionate or where downstream tailoring steps have occurred on the final structure. (B) Assembly of acetate units in the biosynthesis of polyketide natural products.
Many acetate-based natural products, polyketides in particular, are generated through the iterative condensation of activated thioesters, resulting in reactive β-keto units that condense further to produce a wide range of structures (20, 21) (Figure 1B). The structural diversity of polyketides is especially striking given that the majority of polyketides draw on only two monomers, acetate and propionate, as the extender units that form their carbon skeletons (3, 20, 22). Although polyketide synthases (PKSs) have been observed to be promiscuous with regard to their starter units (23), the encoding of extender units has been found to be quite selective and many cellular acyl-CoAs are excluded from the backbone (22). However, progress in engineering extender unit incorporation has been made by domain engineering (23–25) or incorporation via a domain that encodes a rare extender unit (17, 26). Although fluoroacetate serves as a starter unit in nature to produce highly toxic ω-fluorofatty acids (Figure 1A) (13), fluorine has never been observed to date within the backbone, implying that chain extension reactions with the fluorinated acyl-CoA do not occur in these systems. The apparent inability of living systems to utilize fluoroacetate for the biosynthesis of complex small molecules likely results in part from the extreme properties of fluorine that affect biological as well as chemical synthesis. For example, the pKa of the α-proton, electrophilicity of the carbonyl group, and the stability of the acyl-CoA and its corresponding carbanion are all highly impacted by fluorine substitution. Furthermore, the fluoroacetyl group bears a clear similarity to the fluoromethylketone motif used for the design of covalent inhibitors, suggesting that the irreversible alkylation of active-site nucleophiles could also create problems (27). Thus, the development of a system to incorporate fluorinated extender units could dramatically increase the range of complex structures that can be accessed but must also address the challenges involved in activating the fluoroacetate monomer for the downstream C–C bond forming chemistry involved in chain extension reactions.
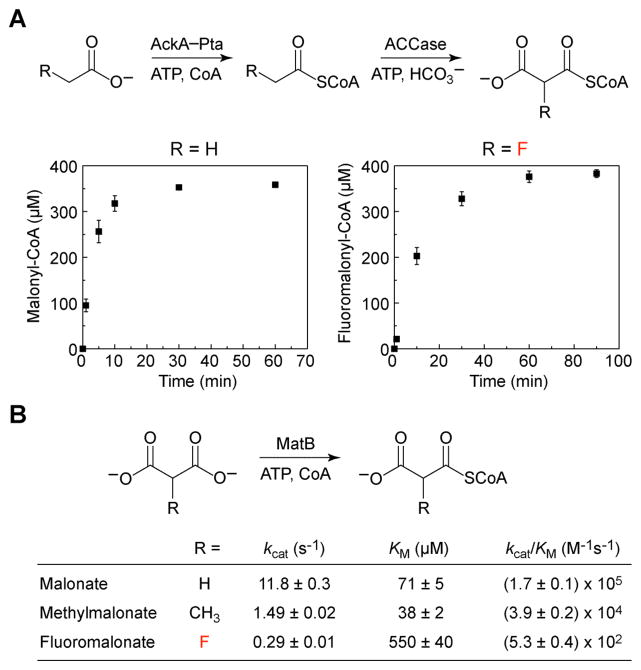
Chain elongation in polyketides and related fatty acid-based natural products relies on a separate pool of extender units formed by carboxylation of acyl-CoAs at the α-position. These malonyl-CoA derivatives are then used as masked enolates for C–C bond formation following decarboxylation. The fluorinated extender, fluoromalonyl-CoA, can be made through two routes: either a two-step activation of the biogenic fluoroacetate or a direct ligation of CoA to fluoromalonate (Figure 2). We reasoned that the acetate kinase (AckA)–phosphotransacetylase (Pta) pair would be effective at fluoroacetate activation, as mutations in this gene locus have been shown to lead to fluoroacetate resistance in Escherichia coli (28). The enzymes from E. coli were thus overexpressed and characterized biochemically, confirming that AckA and Pta serve as an effective activation system to rapidly produce both acetyl- and fluoroacetyl-CoA in nearly quantitative yield (Figures S1–S2). Analysis of the kinetic parameters for these enzymes with respect to fluorinated substrates indicated that neither appears to be affected by the fluorine substituent beyond inductive effects that alter the nucleophilicity of the carboxylic acid (AckA) or electrophilicity of the carbonyl (Pta) (29). Next, we purified the individual AccABCD subunits that make up the acetyl-CoA carboxylase (ACCase) from E. coli and added these enzymes to the AckA–Pta system in order to carry out the carboxylation of fluoroacetate in a one-pot reaction to generate the fluoromalonyl-CoA extender unit (Figure 2A, Figure S1). Under these conditions, the ligation of CoA with AckA–Pta to produce the acyl-CoA is rapid and production of the carboxylated product is limited by the ACCase. Although the rate of conversion is 4.5-fold slower for fluoroacetate compared to acetate, the overall extent of reaction is similar for both congeners and suggests that covalent inactivation of the ACCase by fluoroacetyl-CoA is not significant. In addition to the route from fluoroacetate, we also tested a malonyl-CoA synthetase (MatB) (30) for coupling CoA directly to fluoromalonate. Although MatB exhibits a 103-fold selectivity for malonate over fluoromalonate, fluoromalonyl-CoA is still produced at reasonable efficiency (Figure 2B, Figures S3–S4). Both of these systems also provide in situ regeneration capacity that can amplify product yields from polyketide synthases and we found that either system increased polyketide production by tetrahydroxynaphthalene synthase (31) compared to simple addition of malonyl-CoA (Figure S5).
Fig. 2.
Enzymatic production of activated extender units for C–C bond formation reactions. (A) Formation of malonyl-CoA (left) and fluoromalonyl-CoA (right) from 500 μM CoA and either acetate or fluoroacetate, respectively (AckA, acetate kinase; Pta, phosphotransacetylase; ACCase, acetyl-CoA carboxylase). Values are reported as the mean ± s.d. (n = 3). (B) Kinetic parameters for malonate activation (MatB, malonyl-CoA synthetase). Kinetic parameters are reported as mean ± s.e. (n = 3) as determined from non-linear curve-fitting. Error in the kcat/KM parameter was obtained from propagation of error from the individual kinetic terms.
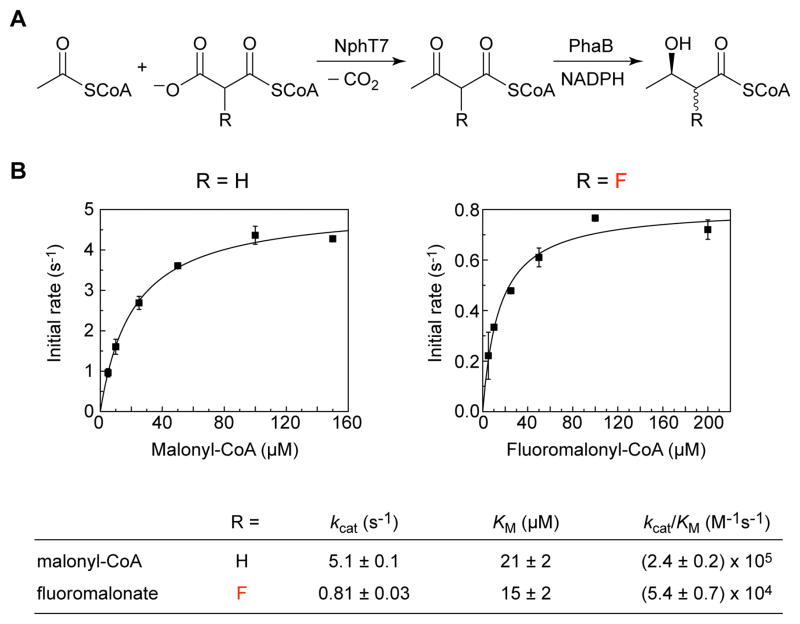
We next turned our attention to utilizing the fluoromalonyl-CoA monomer for downstream chain elongation reactions. To start, we examined the behavior of a simple polyketide synthase system with regard to one cycle of chain extension and ketoreduction, which is a key functionality of larger multimodular systems for controlling downstream cyclization and rearrangements within the polyketide backbone (Figure 3A) (3, 20). We constructed a synthetic gene encoding NphT7 from Streptomyces sp. CL190 (32), which appears to be a free-standing ketosynthase that is related at the structural level to the ketosynthase domain of more complex polyketide synthases (Figure S6), and isolated the heterologously-expressed enzyme for biochemical characterization (Figure S1). Using a coupled assay with an R-hydroxyl forming acetoacetyl-CoA reductase (PhaB), we found that NphT7 is competent to catalyze the formation of acetofluoroacetyl-CoA using an acetyl-CoA starter and fluoromalonyl-CoA extender with only a five-fold defect in catalytic efficiency (kcat/KM) derived from a drop in kcat with the fluorinated substrate (Figure 3). This lower turnover rate observed with the fluorinated substrate is possibly related to the reduced reactivity of the enolate species, which would be stabilized by the fluorine substituent. However, the overall yield was comparable for both fluorinated and nonfluorinated substrates, which shows that a decarboxylative Claisen condensation with fluoromalonyl-CoA can take place at a similar extent of conversion compared to malonyl-CoA. Furthermore, these experiments also show that the 2-fluoro-3-keto motif produced with the fluoromalonyl-CoA extender can be accepted by ketoreductases, as PhaB is capable of efficiently reducing the acetofluoroacetyl-CoA substrate (Figure S7). The 1H and 19F NMR spectra of the reduced product indicate that both diastereomers are produced in this reaction (Figure S7), which may result either from lack of stereochemical preference of NphT7 with respect to the fluorine substituent or from racemization of the product prior to reduction by PhaB. Although PhaB does not appear to show diastereoselectivity with respect to the fluorine group, the polyketide synthase ketoreductases are known to be selective with regard to their native α-substituent and could potentially carry out the stereochemical resolution of the fluorine modification upon reduction (33).
Fig. 3.
A chain extension and ketoreduction cycle with a fluorinated extender using a simple polyketide synthase, NphT7. (A) Reactions catalyzed by NphT7 and PhaB. (B) Steady-state kinetic parameters for NphT7-catalyzed C–C bond formation measured using a coupled assay with PhaB. Data points are reported as the mean ± s.d. (n = 3). Kinetic parameters are reported as mean ± s.e. (n = 3) as determined from non-linear curve-fitting. Error in the kcat/KM parameter was obtained from propagation of error from the individual kinetic terms.
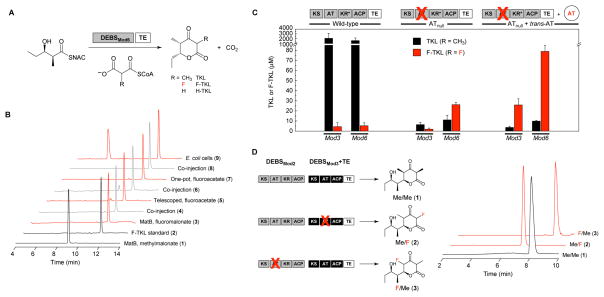
With this information in hand, we sought to extend our biosynthetic method for fluorine introduction to more complex polyketide synthase systems, which use the chain elongation reaction for the biosynthesis of many bioactive and clinically important natural products, such as erythromycin and rapamycin (3, 20). Of the multimodular polyketide systems, 6-deoxyerythronolide B synthase (DEBS) is likely the best understood and also responsible for production of the erythromycin precursor (34). We therefore focused our studies on the sixth module of DEBS, including the terminal thioesterase (DEBSMod6+TE) (35). Using a diketide substrate (NDK-SNAC), DEBSMod6+TE can catalyze a single round of chain elongation with its native methylmalonyl-CoA extender unit and then cyclize the tethered product to form a methyltriketide lactone (TKL) (Figure 4A, R = CH3; Figure 4B, 1; Figure S8) (36). We found that DEBSMod6+TE is also able to accept the fluorinated monomer in chain extension catalysis to form the 2-fluoro-2-desmethyltriketide lactone (F-TKL) and incorporate fluorine into the polyketide backbone (Figure 4B, 2–4; Figure S9). The identity of the F-TKL was established by comparison to an authentic synthetic standard by reverse-phase HPLC monitored by ESI-MS and further confirmed by characterization of the isolated compound by high resolution MS, GC-MS, and 19F NMR spectroscopy (Figures S10–S13). Although the 2S keto tautomer is generated in ≥94% diastereomeric excess (d.e.) (Figure S12), this ratio appears to be set by the compound’s stereoelectronic factors rather than the stereochemical preference of DEBSMod6+TE, as the F-TKL is fully enolized in aqueous solution. The F-TKL can also be produced directly from fluoroacetate using the AckA–Pta/ACCase activation system in either a multi-stage (Figure 4B, 5–6) or single-pot reaction (Figure 4B, 7–8) with DEBSMod6+TE in a similar yield to the MatB reaction, which allows us to connect fluorinated polyketide production directly to the biosynthetically available fluorinated building block (Figure 1A, Scheme S1).
Fig. 4.
Production of fluorinated polyketides in vitro and in vivo by DEBSMod6+TE. (A) Reaction catalyzed by DEBSMod6+TE using the NDK-SNAC substrate with various extender units (NDK-SNAC, native diketide N-acetylcysteamine thioester, (2S,3R)-2-methyl-3-hydroxypentanoyl-N-acetylcysteamine thioester). (B) Chain extension by DEBSMod6+TE to form triketide lactones monitored by LC-MS (TKL, m/z = 169; F-TKL, m/z = 173). CoA, ATP, and ATP regeneration system are included in all in vitro reactions. Data are normalized with respect to the TKL peak. (C) Selectivity of DEBSMod6+TE and DEBSMod3+TE for methylmalonyl-CoA vs. fluoromalonyl-CoA extender unit as monitored by TKL (m/z = 169) and F-TKL (m/z = 173) formation. Conditions include wild-type modules, AT0 modules, and AT0 modules in conjunction with the trans-AT from the disorazole PKS (DszsAT). Values are reported as the mean ± s.d. (n = 3). (KR*, the KR domain of Mod3 is inactive). (D) LC-MS traces showing regioselective tetraketide lactone formation using the DEBS mini-PKS consisting of DEBSMod2 and DEBSMod3+TE (Me/Me, 2-methyl-4-methyl-tetraketide lactone, m/z = 227; Me/F, 2-fluoro-4-methyl-tetraketide lactone, m/z = 231; F/Me, 2-methyl-4-fluoro-tetraketide lactone, m/z = 231). Me/Me was produced using DEBSMod2/DEBSMod3+TE and methylmalonate (1). Me/F was produced using DEBSMod2/DEBSMod3AT0+TE, DszsAT, methylmalonyl-CoA, and fluoromalonate (2). F/Me was produced using DEBSMod2AT0/DEBSMod3+TE, methylmalonyl-CoA, and fluoromalonate (3). Data are normalized with respect to the Me/Me peak. All reactions contained MatB and the ATP regeneration system.
In contrast to the chain extension reaction catalyzed by NphT7, DEBSMod6+TE does not incorporate fluorinated extender units into the triketide lactone product as efficiently as its native methylmalonyl-CoA extender. Preliminary studies indicate that the reduced efficiency of DEBSMod6+TE with the fluorinated extender is not due to covalent inactivation of the enzyme (Figure S14), but rather to the more complex biochemistry of polyketide synthases with regard to monomer selection (37). Extender unit hydrolysis, which occurs even for the native substrate (Table S2), appears to limit fluoromalonyl-CoA incorporation based on the observations that MatB and ATP are needed for fluoromalonyl-CoA regeneration and that fluoromalonate remains the major organofluorine species even in their presence (Figure S15). The fluoromalonyl-CoA extender is however incorporated at higher efficiency by DEBSMod6+TE than malonyl-CoA (R = H), which is reported to be naturally excluded by DEBS (38). In fact, DEBSMod6+TE produces at least 10-fold more F-TKL than H-TKL in a direct competition experiment with equimolar amounts (1 mM) of fluoromalonyl-CoA and malonyl-CoA (Table S3).
To address the issue of site- or regioselective fluorine incorporation, we turned our attention to exploiting the greater reactivity of the fluorinated extender unit towards acylation reactions. In this regard, we hypothesized that it would be possible for a fluorinated substrate to selectively acylate either the AT or ACP domains of individual DEBS modules in the presence of a catalytically compromised or inactive AT domain, an approach that has been shown to facilitate malonyl incorporation by DEBS (39). Experiments with DEBSMod6+TE showed that not only does F-TKL yield increase as expected but fluorine selectivity also improves upon introduction of a key S2107A mutation, reversing the selectivity of the wild-type module (Figure 4C). Indeed, when the NDK-SNAC substrate is used with its native module, DEBSMod2, in conjunction with the analogous S2652A mutation, extension with fluoromalonyl-CoA to form F-TKL reaches 30% efficiency compared to methylmalonyl-CoA (Figure S16). Furthermore, we found that the standalone trans-AT from the disorazole polyketide synthase (40, 41) accepts fluoromalonyl-CoA and can further enhance F-TKL formation by the AT-null mutant (Figure 4C). Using this approach, we began to explore the possibility of site-selective fluorine incorporation with a mini-PKS model system, consisting of DEBSMod2 and DEBSMod3+TE, that was designed to carry out two chain extension reactions from the NDK-SNAC substrate (42). Using the appropriate AT-null constructs, we were able to observe exclusive production of either regioisomer of the fluoro-methyl tetraketide lactone (tetraKL). The identity of the 2-fluoro-4-methyl tetraKL and 2-methyl-4-fluoro tetraKL were established by both HR ESI-MS and LC-MS based on their different retention times, as well as their mass fragmentation patterns which are consistent with the incorporation of fluorine at the expected sites (Figure 4D, Figure S17). These studies also indicate that further chain extension after fluorine insertion can be achieved and that downstream reactions of fluorinated intermediates could potentially be tolerated. This observation is consistent with previous work that has shown that intermediates with non-native substituents, including fluorine, can be extended and tailored to the final structure (3, 17–20, 23) and gives promise that larger fluorinated polyketide targets may be accessible through this approach.
The observed selectivity for fluoromalonyl- over malonyl-CoA extender units suggested that polyketide chain extension reactions with fluoromalonyl-CoA could possibly be catalyzed in vivo in E. coli, which contains a significant malonyl-CoA pool (~35 μM) (43) but almost no methylmalonyl-CoA (44, 45). We carried out preliminary 19F-NMR studies of cells expressing MatB, NphT7 and PhaB and fed with non-toxic levels of fluoromalonate. Analysis of the media and cell extracts indicated that flux through fluoromalonyl-CoA could reach 100 μM to 1 mM, which is sufficient for use by PKSs in live cells (Table S4). Next, we tested the ability of DEBSMod6+TE to catalyze chain elongation in cell lysates prepared from E. coli BAP1 co-expressing DEBSMod6+TE and MatB. Under these conditions, F-TKL is produced with no observable H-TKL upon addition of only NDK-SNAC, fluoromalonate, CoA, ATP, and the ATP regeneration system (Figure S18A). Negative controls with either no DEBSMod6+TE/MatB expressed or no NDK-SNAC substrate show no production of F-TKL (Figure S18A). These results demonstrate that the intracellular level of expression of the DEBSMod6+TE and MatB enzymes is sufficient for the incorporation of the fluorinated extender unit. They also further imply that fluorine could be introduced into the polyketide backbone inside living cells, which are capable of generating ATP through normal metabolic processes. We therefore cultured E. coli BAP1 co-expressing DEBSMod6+TE and MatB and harvested the cells after induction. These cells were then fed with the fluoromalonate precursor, which resulted in the production of F-TKL upon addition of NDK-SNAC (Figure 4B, 9; Figure S18B). The identity of the F-TKL under these conditions were established by LC-MS, co-injection with an authentic standard, as well as high resolution MS. Moreover, F-TKL can also be produced directly in cell culture with the simple addition of a mixture of both substrates to the media after induction of DEBSMod6+TE and MatB (Figure S18C). Taken together, these studies show that the natural selectivity of the polyketide synthase allows for the site-selective introduction of fluorine over hydrogen into the polyketide backbone inside living cells.
To close, we have demonstrated that we can expand the fluorine chemistry of living systems using engineered pathways to link simple biogenic organofluorine building blocks into more complex fluorinated small molecule targets. Because of the modular nature of the biosynthetic pathways used to produce polyketides and related acetate-derived natural products, these findings open the door to general strategies for exploring the fluorine synthetic biology of complex natural products.
Supplementary Material
Acknowledgments
We thank Brooks Bond-Watts and Ioana Aanei for assembly and cloning of the synthetic NphT7 gene, Xingye Yu for providing plasmids for the E. coli ACCase, Colin Harvey for synthetic chemistry assistance, Fong Tian Wong for helpful discussions, and Andreas Lingel (Novartis) for QCI CryoProbe use. The College of Chemistry NMR Facility at U.C. Berkeley is supported in part by the National Institutes of Health (1S10RR023679-01 and S10 RR16634-01). M.C.W. and B.W.T acknowledge the support of a National Institutes of Health NRSA Training Grant (1 T32 GMO66698) and the Gerald K. Branch Predoctoral Fellowship and UC Cancer Research Council Committee Predoctoral Fellowship (to B.W.T). L.K.C. acknowledges support from the National Cancer Institute (F32 CA137994). This work was funded by generous support from U.C. Berkeley and the National Institutes of Health (1 DP2 OD008696 to M.C.Y.C. and R01 GM087934 to C.K.).
Footnotes
Materials and Methods
References and Notes
- 1.Ro DK, et al. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature. 2006;440:940–943. doi: 10.1038/nature04640. [DOI] [PubMed] [Google Scholar]
- 2.Atsumi S, Hanai T, Liao JC. Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature. 2008;451:86–89. doi: 10.1038/nature06450. [DOI] [PubMed] [Google Scholar]
- 3.Cane DE, Walsh CT, Khosla C. Harnessing the biosynthetic code: Combinations, permutations, and mutations. Science. 1998;282:63–68. doi: 10.1126/science.282.5386.63. [DOI] [PubMed] [Google Scholar]
- 4.Weeks AM, Chang MCY. Constructing de novo biosynthetic pathways for chemical synthesis inside living cells. Biochemistry. 2011;50:5404–5418. doi: 10.1021/bi200416g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Müller K, Faeh C, Diederich F. Fluorine in pharmaceuticals: Looking beyond intuition. Science. 2007;317:1881–1886. doi: 10.1126/science.1131943. [DOI] [PubMed] [Google Scholar]
- 6.O’Hagan D. Understanding organofluorine chemistry. An introduction to the C-F bond. Chem Soc Rev. 2008;37:308–319. doi: 10.1039/b711844a. [DOI] [PubMed] [Google Scholar]
- 7.Furuya T, Kamlet AS, Ritter T. Catalysis for fluorination and trifluoromethylation. Nature. 2011;473:470–477. doi: 10.1038/nature10108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ball ND, Sanford MS. Synthesis and reactivity of a mono-σ-aryl palladium(IV) fluoride complex. J Am Chem Soc. 2009;131:3796–3797. doi: 10.1021/ja8054595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Watson DA, et al. Formation of ArF from LPdAr(F): Catalytic conversion of aryl triflates to aryl fluorides. Science. 2009;325:1661–1664. doi: 10.1126/science.1178239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rauniyar V, Lackner AD, Hamilton GL, Toste FD. Asymmetric electrophilic fluorination using an anionic chiral phase-transfer catalyst. Science. 2011;334:1681–1684. doi: 10.1126/science.1213918. [DOI] [PubMed] [Google Scholar]
- 11.Lee E, et al. A fluoride-derived electrophilic late-stage fluorination reagent for PET imaging. Science. 2011;334:639–642. doi: 10.1126/science.1212625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dong C, et al. Crystal structure and mechanism of a bacterial fluorinating enzyme. Nature. 2004;427:561–565. doi: 10.1038/nature02280. [DOI] [PubMed] [Google Scholar]
- 13.O’Hagan D. Recent developments on the fluorinase from Streptomyces cattleya. J Fluorine Chem. 2006;127:1479–1483. [Google Scholar]
- 14.Rivkin A, Biswas K, Chou TC, Danishefsky SJ. On the introduction of a trifluoromethyl substituent in the epothilone setting: Chemical issues related to ring forming olefin metathesis and earliest biological findings. Org Lett. 2002;4:4081–4084. doi: 10.1021/ol0268283. [DOI] [PubMed] [Google Scholar]
- 15.Bégué JP, Bonnet-Delpon D. Recent advances (1995–2005) in fluorinated pharmaceuticals based on natural products. J Fluorine Chem. 2006;127:992–1012. [Google Scholar]
- 16.Llano-Sotelo B, et al. Binding and action of CEM-101, a new fluoroketolide antibiotic that inhibits protein synthesis. Antimicrob Agents Chemother. 2010;54:4961–4970. doi: 10.1128/AAC.00860-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mo S, et al. Biosynthesis of the allylmalonyl-CoA extender unit for the FK506 polyketide synthase proceeds through a dedicated polyketide synthase and facilitates the mutasynthesis of analogues. J Am Chem Soc. 2010;133:976–985. doi: 10.1021/ja108399b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Runguphan W, Maresh JJ, O’Connor SE. Silencing of tryptamine biosynthesis for production of nonnatural alkaloids in plant culture. Proc Natl Acad Sci USA. 2009;106:13673–13678. doi: 10.1073/pnas.0903393106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goss RJM, et al. An expeditious route to fluorinated rapamycin analogues by utilising mutasynthesis. ChemBioChem. 2010;11:698–702. doi: 10.1002/cbic.200900723. [DOI] [PubMed] [Google Scholar]
- 20.Staunton J, Weissman KJ. Polyketide biosynthesis: A millennium review. Nat Prod Rep. 2001;18:380–416. doi: 10.1039/a909079g. [DOI] [PubMed] [Google Scholar]
- 21.Croteau R, Kutchan TM, Lewis NG. In: Biochemistry and molecular biology of plants. Buchanan RB, Gruissem W, Jones R, editors. ASPB; Rockville, MD: 2000. pp. 1250–1318. [Google Scholar]
- 22.Chan YA, Podevels AM, Kevany BM, Thomas MG. Biosynthesis of polyketide synthase extender units. Nat Prod Rep. 2009;26:90–114. doi: 10.1039/b801658p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McDaniel R, et al. Multiple genetic modifications of the erythromycin polyketide synthase to produce a library of novel “unnatural” natural products. Proc Natl Acad Sci USA. 1999;96:1846–1851. doi: 10.1073/pnas.96.5.1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sundermann U, et al. Enzyme-directed mutasynthesis: A combined experimental and theoretical approach to substrate recognition of a polyketide synthase. ACS Chem Biol. 2012;8:443–450. doi: 10.1021/cb300505w. [DOI] [PubMed] [Google Scholar]
- 25.Koryakina I, McArthur JB, Draelos MM, Williams GJ. Promiscuity of a modular polyketide synthase towards natural and non-natural extender units. Org Biomol Chem. 2013:4449–4458. doi: 10.1039/c3ob40633d. [DOI] [PubMed] [Google Scholar]
- 26.Eustaquio A, O’Hagan D, Moore B. Engineering fluorometabolite production: Fluorinase expression in Salinispora tropica yields fluorosalinosporamide. J Nat Prod. 2010;73:378–382. doi: 10.1021/np900719u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Powers JC, Asgian JL, Ekici ÖD, James KE. Irreversible inhibitors of serine, cysteine, and threonine proteases. Chem Rev. 2002;102:4639–4750. doi: 10.1021/cr010182v. [DOI] [PubMed] [Google Scholar]
- 28.Brown TDK, Jones-Mortimer MC, Kornberg HL. The enzymic interconversion of acetate and acetyl-coenzyme A in Escherichia coli. J Gen Microbiol. 1977;102:327–336. doi: 10.1099/00221287-102-2-327. [DOI] [PubMed] [Google Scholar]
- 29.Walker MC, Wen M, Weeks AM, Chang MCY. Temporal and fluoride control of secondary metabolism regulates cellular organofluorine biosynthesis. ACS Chem Biol. 2012:1576–1585. doi: 10.1021/cb3002057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hughes AJ, Keatinge-Clay A. Enzymatic extender unit generation for in vitro polyketide synthase reactions: Structural and functional showcasing of Streptomyces coelicolor MatB. Chem Biol. 2011;18:165–176. doi: 10.1016/j.chembiol.2010.12.014. [DOI] [PubMed] [Google Scholar]
- 31.Izumikawa M, et al. Expression and characterization of the type III polyketide synthase 1,3,6,8-tetrahydroxynaphthalene synthase from Streptomyces coelicolor A3(2) J Ind Microbiol Biot. 2003;30:510–515. doi: 10.1007/s10295-003-0075-8. [DOI] [PubMed] [Google Scholar]
- 32.Okamura E, Tomita T, Sawa R, Nishiyama M, Kuzuyama T. Unprecedented acetoacetyl-coenzyme A synthesizing enzyme of the thiolase superfamily involved in the mevalonate pathway. Proc Natl Acad Sci USA. 2010;107:11265. doi: 10.1073/pnas.1000532107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Siskos AP, et al. Molecular basis of Celmer’s rules: Stereochemistry of catalysis by isolated ketoreductase domains from modular polyketide synthases. Chem Biol. 2005;12:1145–1153. doi: 10.1016/j.chembiol.2005.08.017. [DOI] [PubMed] [Google Scholar]
- 34.Khosla C, Tang Y, Chen AY, Schnarr NA, Cane DE. Structure and mechanism of the 6-deoxyerythronolide B synthase. Annu Rev Biochem. 2007;76:195–221. doi: 10.1146/annurev.biochem.76.053105.093515. [DOI] [PubMed] [Google Scholar]
- 35.Gokhale RS, Tsuji SY, Cane DE, Khosla C. Dissecting and exploiting intermodular communication in polyketide synthases. Science. 1999;284:482–485. doi: 10.1126/science.284.5413.482. [DOI] [PubMed] [Google Scholar]
- 36.Wu N, Kudo F, Cane DE, Khosla C. Analysis of the molecular recognition features of individual modules derived from the erythromycin polyketide synthase. J Am Chem Soc. 2000;122:4847–4852. [Google Scholar]
- 37.Bonnett SA, et al. Acyl-CoA subunit selectivity in the pikromycin polyketide synthase PikAIV: Steady-state kinetics and active-site occupancy analysis by FTICR-MS. Chem Biol. 2011;18:1075–1081. doi: 10.1016/j.chembiol.2011.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liou GF, Lau J, Cane DE, Khosla C. Quantitative analysis of loading and extender acyltransferases of modular polyketide synthases. Biochemistry. 2002;42:200–207. doi: 10.1021/bi0268100. [DOI] [PubMed] [Google Scholar]
- 39.Kumar P, Koppisch AT, Cane DE, Khosla C. Enhancing the modularity of the modular polyketide synthases: Transacylation in modular polyketide synthases catalyzed by malonyl-CoA:ACP transacylase. J Am Chem Soc. 2003;125:14307–14312. doi: 10.1021/ja037429l. [DOI] [PubMed] [Google Scholar]
- 40.Wong FT, Chen AY, Cane DE, Khosla C. Protein-protein recognition between acyltransferases and acyl carrier proteins in multimodular polyketide synthases. Biochemistry. 2009;49:95–102. doi: 10.1021/bi901826g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wong FT, Jin X, Mathews II, Cane DE, Khosla C. Structure and mechanism of the trans-acting acyltransferase from the disorazole synthase. Biochemistry. 2011;50:6539–6548. doi: 10.1021/bi200632j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsuji SY, Cane DE, Khosla C. Selective protein-protein interactions direct channeling of intermediates between polyketide synthase modules. Biochemistry. 2001;40:2326–2331. doi: 10.1021/bi002463n. [DOI] [PubMed] [Google Scholar]
- 43.Bennett BD, et al. Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat Chem Biol. 2009;5:593–599. doi: 10.1038/nchembio.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haller T, Buckel T, Rétey J, Gerlt JA. Discovering new enzymes and metabolic pathways: Conversion of succinate to propionate by Escherichia coli. Biochemistry. 2000;39:4622–4629. doi: 10.1021/bi992888d. [DOI] [PubMed] [Google Scholar]
- 45.Pfeifer BA, Admiraal SJ, Gramajo H, Cane DE, Khosla C. Biosynthesis of complex polyketides in a metabolically engineered strain of E. coli. Science. 2001;291:1790–1792. doi: 10.1126/science.1058092. [DOI] [PubMed] [Google Scholar]
- 46.Rouillard JM, et al. Gene2Oligo: Oligonucleotide design for in vitro gene synthesis. Nucleic Acids Res. 2004;32:W176–W180. doi: 10.1093/nar/gkh401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gibson DG, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 48.Bond-Watts BB, Bellerose RJ, Chang MCY. Enzyme mechanism as a kinetic control element for designing synthetic biofuel pathways. Nat Chem Biol. 2011;7:222–227. doi: 10.1038/nchembio.537. [DOI] [PubMed] [Google Scholar]
- 49.Kumar P, Khosla C, Tang Y. Manipulation and analysis of polyketide synthases. Method Enzymol. 2004:269–293. doi: 10.1016/S0076-6879(04)88023-6. [DOI] [PubMed] [Google Scholar]
- 50.Theodorou V, Skobridis K, Tzakos AG, Ragoussis V. A simple method for the alkaline hydrolysis of esters. Tetraherdon Lett. 2007;48:8230–8233. [Google Scholar]
- 51.Williamson JR, Corkey BE. Assays of intermediates of the citric acid cycle and related compounds by fluorometric enzyme methods. Method Enzymol. 1969:434–513. [Google Scholar]
- 52.Huang F, et al. The gene cluster for fluorometabolite biosynthesis in Streptomyces cattleya: A thioesterase confers resistance to fluoroacetyl-coenzyme A. Chem Biol. 2006;13:475–484. doi: 10.1016/j.chembiol.2006.02.014. [DOI] [PubMed] [Google Scholar]
- 53.Funa N, et al. A new pathway for polyketide synthesis in microorganisms. Nature. 1999;400:897–899. doi: 10.1038/23748. [DOI] [PubMed] [Google Scholar]
- 54.Cane DE, Tan W, Ott WR. Nargenicin biosynthesis. Incorporation of polyketide chain elongation intermediates and support for a proposed intramolecular Diels-Alder cyclization. J Am Chem Soc. 1993;115:527–535. [Google Scholar]
- 55.Akoka S, Barantin L, Trierweiler M. Concentration measurement by proton NMR using the ERETIC method. Anal Chem. 1999;71:2554–2557. doi: 10.1021/ac981422i. [DOI] [PubMed] [Google Scholar]
- 56.Hinterding K, Singhanat S, Oberer L. Stereoselective synthesis of polyketide fragments using a novel intramolecular Claisen-like condensation/reduction sequence. Tetrahedron lett. 2001;42:8463–8465. [Google Scholar]
- 57.Luo G, Pieper R, Rosa A, Khosla C, Cane DE. Erythromycin biosynthesis: Exploiting the catalytic versatility of the modular polyketide synthase. Bioorg Med Chem. 1996;4:995–999. doi: 10.1016/0968-0896(96)00096-x. [DOI] [PubMed] [Google Scholar]
- 58.Kelley LA, Sternberg MJE. Protein structure prediction on the Web: A case study using the Phyre server. Nat Prot. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 59.Tang Y, Kim CY, Mathews II, Cane DE, Khosla C. The 2.7 Å crystal structure of a 194-kDa homodimeric fragment of the 6-deoxyerythronolide B synthase. Proc Nat Ac Sc USA. 2006;103:11124–11129. doi: 10.1073/pnas.0601924103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mohanta PK, Davis TA, Gooch JR, Flowers RA. Chelation-controlled diastereoselective reduction of α-fluoroketones. J Am Chem Soc. 2005;127:11896–11897. doi: 10.1021/ja052546x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.