Abstract
Background
Primitive neuroectodermal tumors of the central nervous system (CNS-PNETs) are a rare group of neoplasms occurring in the CNS that includes supratentorial CNS-PNETs, medulloepitheliomas, and ependymoblastomas. While ependymoblastomas frequently carry chromosome 19q13.41 amplification and show aggressive clinical behavior, the biological mechanisms and molecular alterations contributing to the pathogenesis of supratentorial CNS-PNETs remain poorly understood. Moreover, genetic alterations suitable for molecular risk stratification are undefined to date.
Methods
In order to identify possible molecular markers, we performed multiplex ligation-dependent probe amplification (MLPA) and molecular inversion probe (MIP) analysis on DNA samples of 25 supratentorial CNS-PNETs (median age, 5.35 years; range, 2.41–17.28 years). Tumors with ependymoblastic rosettes (ependymoblastoma/ETANTR) and LIN28A positivity were excluded.
Results
MLPA and MIP analysis revealed large losses of genomic material of chromosomes 3, 4, 5, and 13, while frequent gains affected chromosomes 1, 17, 19, 20, and 22. High copy number gains (amplifications) were found in particular at chromosomes 2p24.3 (MYCN, n = 6 cases) and 4q12 (n = 2 cases). Patients with tumors harboring 2p gain or MYCN amplification showed unfavorable overall survival (P = .003 and P = .001, respectively).These markers were independent of the presence of metastases, which was indeed a clinical factor associated with poor overall survival (P = .01) in this series.
Conclusions
In the era of the personalized neuro-oncology, the identification of these molecular prognostic markers associated with patient outcome may represent a significant step towards improved patient stratification and risk-adapted therapeutic strategies for patients suffering from supratentorial CNS-PNETs.
Keywords: CNS-PNET, LIN28, molecular inversion probe analysis, MYCN amplification, multiplex ligation-dependent probe amplification, primitive neuroectodermal tumors of central nervous system, 2p gain
Primitive neuroectodermal tumors of the central nervous system (CNS-PNETs), which represent 3%–7% of all pediatric brain tumors, are a heterogeneous group of neoplasms occurring in the CNS composed of undifferentiated or poorly differentiated neuroepithelial cells that may display divergent differentiation along neuronal, astrocytic, and ependymal lines.1 According to the revised WHO classification (2007),1 this group of tumors includes supratentorial CNS-PNETs affecting the cerebral hemispheres, ependymoblastomas (EPBLs), and (the exceptionally rare) medulloepitheliomas.1 They are still considered to be a nosological entity with distinct biological behavior: they primarily affect infants and children and often present with cerebrospinal fluid dissemination. Standard treatment for older children and adolescents includes craniospinal radiotherapy and chemotherapy, whereas postoperative chemotherapy has been added to most treatment recommendations for young children in order to delay craniospinal radiotherapy. Although the development of high-resolution molecular analysis techniques and the increasing number of published collaborative international studies have led to better understanding of the biology of medulloblastomas (MBs) and atypical teratoid/rhabdoid tumors, the molecular alterations underlying supratentorial CNS-PNET pathogenesis remain poorly understood so far and are limited by their overall very low incidence.2,3 Expression analyses and a handful of comparative genomic hybridization studies (CGH)4,5 have indicated that supratentorial CNS-PNETs are genetically heterogeneous and may show a broad spectrum of copy number aberrations.2,5,6 To date, amplification of MYCN, PDGFRA and PDGFRB, as well as deletions of CDKN2A/2B and a few other sporadic alterations of different pathways (including RASSIF1A promoter methylation, p14ARF methylation and, transcriptional silencing of DLC-1) have been also reported.5–12 More recently, the identification of chr19q13.41 microRNA (miRNA) cluster (C19MC) amplification13–15 has permitted to better define among CNS-PNETs the ependymoblastoma/ETANTR (embryonal tumor with abundant neuropil and true rosettes) subgroup, which also shows aggressive clinical behavior, specific histopathological features and expression of stem cell marker LIN28A.15,16
In the era of personalized oncology, the identification of prognostic molecular markers may represent a significant step towards improved patient stratification and risk-adapted therapy for children with supratentorial CNS-PNET. In fact, despite multimodal therapy, less than half of affected patients currently survive 5 years after diagnosis, and severe late effects associated with current treatment protocols are a significant complication for survivors.17,18
In order to identify suitable genetic markers with predictive value for outcome, we performed a multiplex ligation-dependent probe amplification (MLPA) and molecular inversion probe (MIP)-based analysis of 25 supratentorial CNS-PNET cases enrolled in the German multicenter GPOH HIT study.
Materials and Methods
Patients and Study Design
The study included 25 patients with the histological diagnosis of supratentorial CNS-PNET according to the revised WHO classification of CNS tumors.1 The histology of tumors was reviewed by an expert neuropathologist (T.P.). Cases with histological features of ependymoblastoma/ETANTR (including presence of multilayered ependymoblastic rosettes, neuropil-rich areas, and LIN28A positivity) or medulloepithelioma features were excluded from this study (Supplementary Fig. 1). Twenty patients who underwent surgery between January 2001 and December 2011 in German, Austrian, and Swiss medical centers were enrolled in the prospective HIT 2000 study. Five patients diagnosed before the start of the HIT 2000 study were treated similarly and were also included in this study. Children aged 4–21 years were treated by postoperative chemotherapy (2 cycles of HIT-SKK chemotherapy)19 followed by radiotherapy and maintenance chemotherapy. Young children (aged <4 years) were treated in a risk-adapted manner with systemic chemotherapy (intraventricular methotrexate) followed by high-dose chemotherapy and administration of radiotherapy in case of residual tumor. Clinical data including age, metastatic disease status at presentation, sex, extent of surgical resection, event-free survival (EFS), and overall survival (OS) were collected. The HIT2000 trial and the HIT2000 registry were approved by institutional review boards, and informed consent was obtained from legal representatives of all patients.
Fig. 1.
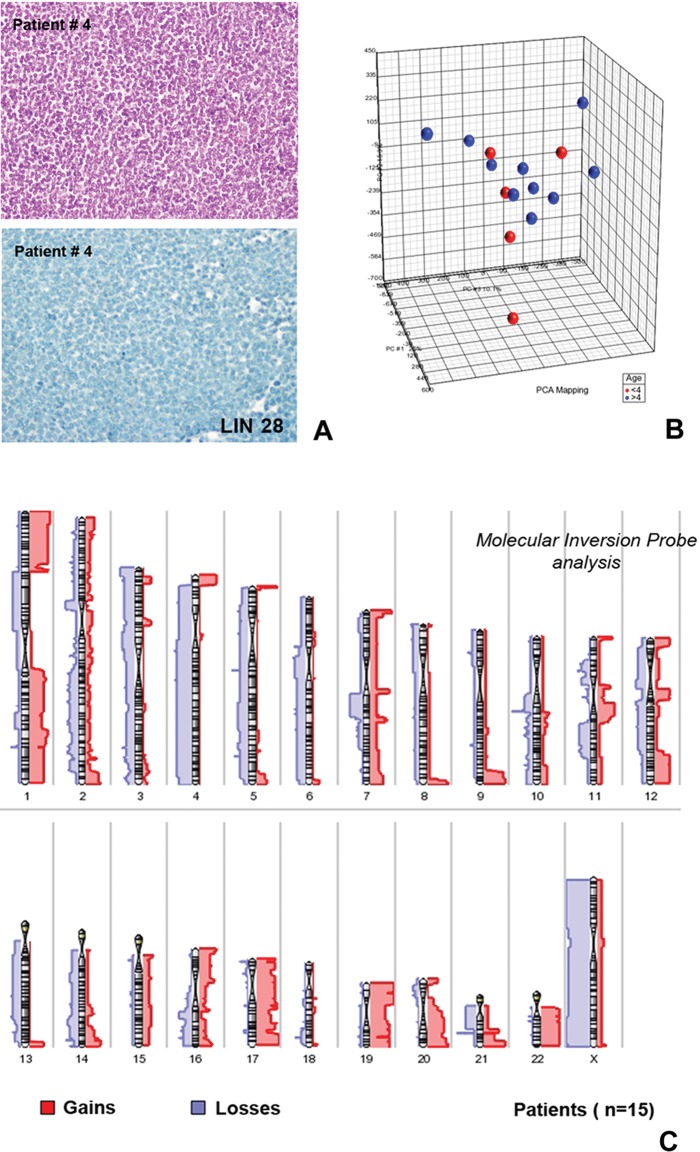
Principal component analysis and virtual karyotype results from the molecular inversion probe analysis of supratentorial CNS-PNET cases. The cases analyzed in this study commonly showed CNS-PNET histology with undifferentiated, small-cell cytology. (A, upper part: hematoxylin and eosin). All tumors included in the study were LIN28A negative (A, lower part: immunostaining with LIN28A antibody). The principal component analysis showed molecular heterogeneity of the 15 supratentorial CNS-PNETs examined with MIP. (B) No molecular subgroups were identifiable according to patient age (red points represent patients <4 years of age; blue points represent patients >4 years). The virtual karyotype (C) showed frequent gains of 1q, 17, 19, 20, and 22 as well as losses of 1p (in particular 1p33-1p21), 3, 4, 5, and 13. No case presented focal chromosome 19q13.41 amplification. Gains were indicated by the bars (red) on the right side of each chromosome, and losses (blue) are shown on the left side. Thicker bars indicate areas of recurrent change.
Tumor Samples, Immunohistochemistry and DNA Extraction
Formalin-fixed, paraffin-embedded (FFPE) tissue specimens were retrieved from the archives of the Institute of Neuropathology at the University of Bonn Medical Center and the German Brain Tumor Reference Center in Bonn. Immunohistochemical analyses (IHC) were performed on a semi-automated IHC Stainer (Tecan) or a Ventana Immunostainer (Roche-Ventana) with antibodies against Map-2 (Sigma), S-100 protein (Dako), epithelial membrane antigen (Dako), glial fibrillary acidic protein (GFAP, Dako), neurofilament protein (Dako), synaptophysin (Dako), INI-1 (Becton Dickinson), Vimentin (Dako), NeuN (Chemicon), OLIG-2 (R&D Systems), LIN28A (R&D Systems), and p53 (clone DO-7, Dako). Genomic DNA from FFPE tumor tissue was extracted using the QIAamp DNA Mini Tissue Kit (Qiagen) according to the manufacturer's instructions. Histological assessment of tissue fragments chosen for this study confirmed that all specimens consisted of at least 80% tumor cells.
Molecular Inversion Probe Assay
To identify copy number gains and losses, we performed a custom-designed OncoScan FFPE Express 330K MIP assay (Affymetrix) on 15 tumors, as described.20 Data were analyzed using Genomic Suite 6.6 Software (Partek). The segmentation analysis was performed according the software manufacturer's instructions. High-copy-gain (amplification) was defined as average copy gain higher than 10.
Multiplex Ligation-dependent Probe Amplification
For MLPA analysis, the SALSA MLPA p302-A1, SALSA MLPA p303-A1, and SALSA p175 (MRC-Holland) assays were used. MLPA was performed in accordance with the manufacturer's instructions.21 In brief, 100 ng DNA was denatured for 5 minutes and cooled down to 25°C. Following addition of the probe mix, the sample was hybridized for 16 hours at 60°C. After ligation, PCR was performed in a total volume of 50 µl containing 10 µl of the ligation mix on a thermocycler (Biometra). Subsequently, a LIZ-labeled internal size standard was added to the tumor samples, and fragments were separated and quantified on an ABI 3730 capillary sequencer after denaturation (Applied Biosystems) and analysed using the Gene Mapper software (Applied Biosystems). Differences of ±3-fold SD from the mean were considered as significant gains or losses, respectively, after normalization of the assay using FFPE cerebellar tissue. A >5-fold mean value of the mean of controls was considered as genomic amplification.
Fluorescence in Situ Hybridization Analysis for MYCN
FFPE tumor slides were hybridized overnight with the Zyto-Light SPEC MYCN/2q11 Dual Color Probe (ZytoVision). Briefly, deparaffinization, protease treatment, and washes were performed on the half-automated VP2000 processor system (Abbott Molecular). After pretreatment, the slides were denatured in the presence of 10 μL probe for 5 minutes at 75°C and hybridized at 37°C overnight. Posthybridization saline-sodium citrate (SSC) washes were performed at 72°C, and the slides were stained with 4’,6-diamidino-2-phenylindole before analysis. Normal tissue served as internal positive control. Cases were further evaluated only if control tissue nuclei displayed 1 or 2 clearly distinct signals of each color. Tumor tissue was scanned for amplification hot spots by using × 40 or × 63 objectives (DM5500 fluorescent microscope; Leica). As the MYCN signals were homogeneously distributed, random areas were used for counting the signals. Twenty contiguous tumor cell nuclei were individually evaluated with the × 63 objective by counting green MYCN and orange 2q11 signals, and the MYCN/2q11 ratio was calculated. According to the diagnostic guidelines for neuroblastoma, cases were considered as amplified in case of a >4-fold MYCN copy number in relation to the copy number of chromosome 2, and a gain was defined as a 1.5-4-fold MYCN copy number in relation to the copy number of chromosome 2.
Statistical Analysis
OS was defined as the time period between date of diagnosis and date of death from any cause or to the date of last visit, while EFS was defined as the time period between date of diagnosis to date of first progression, relapse, death from any cause, or last contact; For statistical analysis, pairwise comparisons were made using Fisher' exact test (for categorical variables) and t tests (for continuous variables). Survival analysis was performed using the Kaplan–Meier method and log-rank test.22 Two-tailed P values were reported and were considered significant when <.05. The study was not powered for the multitude of statistical tests we performed; analyses should be understood as exploratory. Multivariable analysis Cox regression models were not assessable due to the low number of patients and events/deaths, respectively.23
Statistical analyses were performed using SAS 9.3 (SAS).
Results
The study included 15 male and 10 female patients with a median age at diagnosis of 5.35 years (range, 2.41–17.28 years). Presenting symptoms included headaches, seizures, nausea/vomiting, cranial nerve palsies, somnolence, weakness, and sensory abnormalities. No patient presented clinical features or familiar history suspicious for Li-Fraumeni syndrome or other genetic tumor-predisposing syndromes. The tumors arose in the cerebral hemispheres and in some cases involved multiple lobes, the basal ganglia, or the thalamus. Detailed neuroradiological data were available for 19 cases. Most tumors (8/19; 42%) were localized in the frontal lobe, while 6 cases affected the temporal or parietal lobe. One case affected the third ventricle region. The large majority of cases presented in homogeneous contrast enhancement (11/19; 57%) and did not appear to be sharply demarcated from the surrounding brain tissue. Intratumoral cysts were also observed frequently.
The median follow-up of surviving patients was 3.7 years (range, 0.66–11.43 years). Residual tumor was present in 15 patients, and 2 patients had metastatic disease. Among the clinical parameters examined, only the presence of metastatic disease was significantly associated with unfavorable OS (P = .011).
Histologically, all tumors were composed of undifferentiated cells with hyperchromatic nuclei (Fig. 1A) showing no definitive evidence of glial differentiation. The tumor cells did not show expression of glial markers (such GFAP) and presented variable expression of synaptophysin and constant and diffuse positivity of MAP-2. All cases were INI-1 positive. The proliferation index (MIB-1) showed a median value of 25%. All cases were LIN28A negative (Fig. 1A). The expression of OLIG-2 and p53 showed a large degree of inter- and intratumoral variability: 8 cases (32%) showed a significant expression (>50% of nuclei) for OLIG-2, and 8 cases (32%) strongly expressed p53 (> 25% of nuclei). However, both markers were found not to be statistically associated to shorter EFS or OS or related to other clinical parameters (such as age or residual tumor).
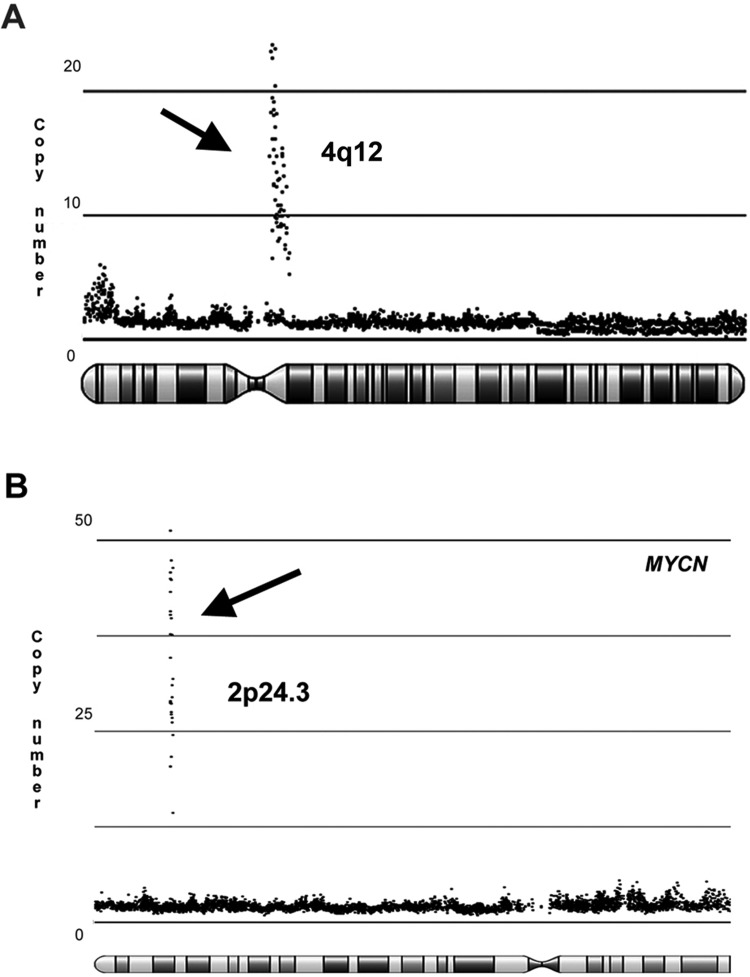
In order to define chromosomal alterations in the tumors of this series, we performed molecular inversion probe array (MIP) analysis in a representative group of 15 cases. MIP showed the frequent presence of large chromosomal aberrations, and a principal component analysis (PCA) (Fig. 1B) confirmed the marked genetic heterogeneity of these tumors. Most of the losses of chromosomal material involved chromosomes 1p, 3, 4, 5, and 13, while frequent gains were observed at chromosomes 1q, 17, 19, 20, and 22 (Fig. 1C). In some of these regions, homozygous deletions (ie, chromosome 3) and chromosome duplications were also observed. High copy number gains (amplifications) were found at 4q12 (including KIT, KDR, and PDGFRα genes) (Fig. 2A) in 2 cases, at chromosomes 2p24.3 (MYCN) in 4 cases (Fig. 2B), and at chromosome 12q13 in one case. None of such alterations was statistically associated with the age of the patient. In one case, loss of 9p21.3 (CDKN2A, CDKN2B) was identified. Recurrent cytogenetic alterations included gains at chromosomes 7q11.2, 7q22.1, and 12q13.11-2 and losses of material of chromosomes 16q23.1, 6p21.1 and 11q11. We found no evidence of MYCC amplification or high copy gains in regions of RTK genes (such as EGFR or MET) or affecting members of the PI3K pathway (such as KRAS, AKT1). Moreover, no evidence of CDK6 amplification was found. Focal chromosome 19q13.41 amplifications were absent in this LIN28A-negative cohort.
Fig. 2.
High copy gains at 4q12 and at 2p24.3 (MYCN locus) observed in the molecular inversion probe analysis. The plots represent (A) the 2 supratentorial CNS-PNET cases with 4q12 amplification and (B) 2 representative cases (of 4 identified) with high copy gain at 2p24.3 (MYCN locus).
In order to validate these data and expand the molecular profiling to the remaining supratentorial CNS-PNETs for MIP analysis, we performed a molecular analysis using MLPA on the whole cohort of patients (25 cases).
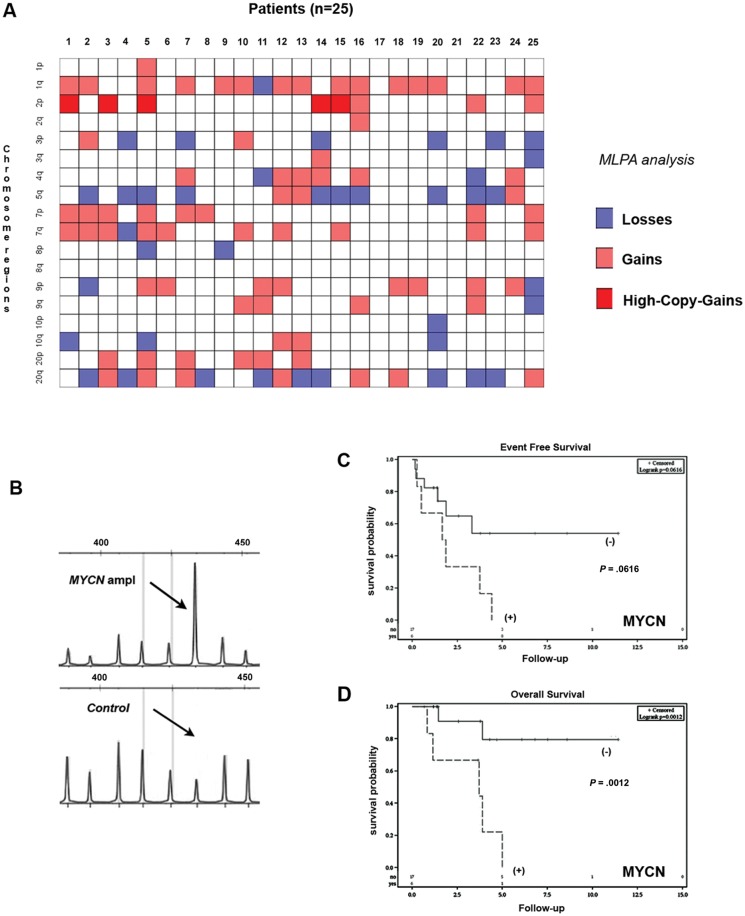
MLPA validated the results of MIP analysis and confirmed frequent losses of chromosomes 3p and 5q as well as gains of chromosomes 1q, 2p, and 7p (Fig. 3A) in the tumors. The analysis confirmed the presence of 4q21 amplifications and the absence of EGFR amplifications. Three cases showed heterozygous loss of CDKN2A. In addition to the 4 cases already identified in the MIP analysis, we found 2 additional cases harboring MYCN amplification (Fig. 3B). One tumor showed gain at MYCC locus, but no amplification. The fluorescence in situ hybridization (FISH) analysis was informative in 19 of the 25 cases investigated and revealed 4 cases (Supplementary Fig. 2) harboring MYCN amplification. The FISH results were concordant with MLPA data in all informative cases.
Fig. 3.
Multiplex ligation-dependent probe amplification (MLPA) analysis and Kaplan–Meier analysis according to MYCN status in 25 patients with supratentorial CNS-PNETs. (A) Intensity plot shows the results of MLPA (SALSA MLPA p302 and p303) analysis, which further revealed the frequent occurrence of chromosomal gains and losses (gains depicted in orange, losses in blue, amplifications/high-copy gains in red and unchanged region in white; on the x-axis, patients number; on the y-axis, chromosomal regions. (B) Representative elecropherograms of MLPA analysis of a tumor showing MYCN) amplification (SALSA MLPA p302-A1, MYCN probe 033327-L02466, length 436 bp. Normal cerebellar tissue was used as control). (C) In regard to event-free survival, the Kaplan–Meier analysis showed a clear trend to shorter survival, but significance was not reached (P= .062). (D) On the other hand, the Kaplan–Meier analysis revealed that patients with tumors harboring MYCN amplification showed shorter overall survival (P = .001).
Kaplan–Meier analysis revealed that patients with tumors harboring 2p gain had a less favorable OS (P = .003) compared with the remaining patients. Moreover, the patients with tumors presenting MYCN amplification revealed a poor OS (P = 0.001) (Fig. 3D). None of these parameters was significantly associated with shorter EFS. A trend toward significance was observed for MYCN amplification (Fig. 3C) (P = .062).
Discussion
Clinical and biological parameters have rarely been evaluated as prognostic markers in patients with supratentorial CNS-PNET, which were often analyzed together with medulloblastoma series in the past.2,23 Their molecular heterogeneity2,6 and absence of recurrent alterations have further hindered the identification of possible molecular markers suitable for clinical risk stratification and outcome prediction. Only a few recent studies attempted to integrate clinical and molecular markers in order to identify specific prognostic subgroups.15 Along with these experiences, we investigated the potential predictive value of molecular markers in supratentorial CNS-PNET patients enrolled in the GPOH HIT clinical study. This study is, to our knowledge, the first to investigate a homogenously treated series of supratentorial CNS-PNETs. EPBL patients were excluded (Supplementary Fig. 1) because they now represent a distinct molecularly defined subgroup of CNS-PNET that is characterized clinically by worse prognosis and poor response to therapy.
While extent of surgery, age at diagnosis, and residual tumor had no prognostic relevance in our cohort of supratentorial CNS-PNET patients,24–27 we found that patients with metastatic disease indeed had an unfavorable OS (P = .011). These data are similar to those observed in MB patients, where the M+ status is a main clinical adverse prognostic factor and the benchmark for all risk stratification and therapy.28 Recently, a molecularly homogeneous subgroup of supratentorial CNS-PNET (defined as “oligo-neural” group),15 which showed a low incidence of metastases clinically, has been identified and characterized by an increased expression of OLIG-2, a transcription factor involved in early development of the CNS.29 Due to the small number of cases, we could not demonstrate significant correlations between OLIG-2 expression and the M- status of our patients, but OLIG-2 expression was not statistically associated to OS or EFS. Also p53, whose increased expression has been associated with metastatic tumor potential and worst prognosis in MB,30 was statistically not associated with shorter OS or EFS in patients with supratentorial CNS-PNETs.
As M+ status, 2p gain was also found to be associated to a shorter OS. This alteration, as revealed previously, is not uncommon in supratentorial CNS-PNETs.5,8 Interestingly, 2p gain seems to be one of the most frequent aberrations maintained in primary and recurrent CNS-PNET pairs.5 While poorer OS in patients with tumor with 2p gain has been found to be associated to concomitant presence of chr.19q13.41 amplification in other studies,13 we did not include EPBL/ETANTR cases in our study. The association between the presence of 2p gain and unfavorable OS appeared to be related to the presence of MYCN amplification.
MYCN is a member of the MYC family of proto-oncogenes and encodes a 60–63 kDa protein. MYC transcription factors (MYCC, MYCN, and MYCL) have a conserved structure that includes a transcriptional activation domain in the N-terminus and a C-terminus basic helix-loop-helix leucine zipper (HLH-zip) domain that plays a role in protein dimerization, sequence-specific DNA binding, and regulation of transcription.31
Normal MYCN expression is restricted to the central and peripheral nervous systems, kidneys, lungs, and spleen during embryonic development. MYCN is a transcription factor that controls expression of many target genes implicated in basic cellular functions including proliferation, cell growth, protein synthesis, metabolism, apoptosis, and differentiation. While genetically engineered mouse models harboring amplification of MYCN develop peripheral neuroblastoma,32 recent evidences also show that orthotopic transplantation of MYCN-transduced neural stem cells generates different tumor subtypes in which their histology is dependent on the site of origin of precursor cells.33
In addition to peripheral neuroblastoma, where amplification is described in 40% of cases, MYCN and MYCC amplifications have been reported in supratentorial CNS-PNETs5,8,15 and MB.34,35 While amplifications of MYCC (8q24) and MYCN (2p24.3) have been detected in 5%–10% of MB, often with large cell/anaplastic features, and are associated with poor prognosis,34,35 the number of cases of CNS-PNET showing MYCN amplifications have varied largely among different studies (ranging from 5% to 50%) and have not been described to date as being clinically associated with shorter OS.5,8,15 However, we found a significant association between shorter OS and MYCN amplification. Patients with MYCN-amplified tumors appear to have slightly less favorable EFS. The presence of MYCN amplification seems to be unrelated to the age of the patient. Notably, 4 cases with MYCN amplification were localized in the temporal or parietal lobes, while only one case arose in the frontal lobe. On the other hand, we did not find MYCC amplification in tumors in our study, which seems to be rare among supratentorial CNS-PNETs8: the single patient showing 8q24 gain presented a rapidly progressive clinical course with extremely short survival.
Because MYCN is already one of the most important prognostic parameters in the existing panels of molecular MB markers and it can be routinely evaluated by quantitative PCR, FISH, or MLPA-based approaches on FFPE or frozen material, routine evaluation of its status could be also performed for supratentorial CNS-PNET. Its determination may provide a very useful tool for reliable identification of high-risk patients with supratentorial CNS-PNETs.
We also found some other recurrent cytogenetic alterations besides MYCN amplification. Recurrent gains in two regions of chromosome 7 and in a larger region of chromosome 12 were observed in the MIP analysis. This region (12q13.11-2) contains several genes, including PGA2A4 and ERBB3, that are potentially implicated in tumor biology. On the contrary, only a few high-copy gains were seen. One tumor in particular showed amplification of 12q13, a region that harbors GLI-1, whose alteration/upregulation has been frequently observed in MB.34,35
Large chromosomal losses and homozygous deletions were frequently revealed by MIP analysis, but focal deletions, on the contrary, were rarely observed. We identified only a few cases showing loss of the cyclin-dependent kinase inhibitors CDKN2A and CDKN2B. Although CDKN2A and CDKN2B (9p21.3) are frequently lost in ∼10%–15% of primary supratentorial CNS-PNET cases,5 especially those affecting older patients, their possible role in predicting survival in patients with CNS-PNET is undetermined. In our study, the univariate analysis did not show an association between CDKN2A and CDKN2B losses and shorter OS or EFS.
As previously highlighted by other studies, we also noted some genetic alterations that have been described in pediatric glioblastoma. Although an insight of molecular relationships between pediatric glioblastoma and supratentorial CNS-PNET exceeds the goals of our study, it cannot be excluded that CNS-PNET and glioblastomas may share a defined grade of homology. Pediatric glioblastoma presents significantly fewer DNA copy number alterations compared with supratentorial CNS-PNET, but some alterations (such as gains at 1p, 2q, and 21q as well as losses at 6q, 4q, 11q, and 16q) have been described for both tumor entities.36,37 To further support this evidence, we found two supratentorial CNS-PNET cases with amplification of the 4q12 region harboring, among others, PDGFRα. PDGFRα amplification is, by far, the most common genomic event identified in pediatric glioblastomas.36,37 On the other hand, MYCN amplifications have also been observed in pediatric glioblastomas.36 In this regard, further studies are needed to definitively explore the possible molecular relationship and overlapping features between these two tumor entities. From a diagnostic point of view, supratentorial CNS-PNETs may also be difficult to distinguish from diffuse high-grade gliomas and often represent an “exclusion-diagnosis” based on the identification of a supratentorial embryonal neoplasm with medulloblastoma-like histology that is frequently associated with expression of neuronal markers.1 However, definitive histological distinction between a supratentorial CNS-PNET and a glioblastoma sometimes remains impossible; in such cases, the final diagnosis depends on the expertise of the neuropathologist.
In conclusion, the identification of MYCN amplification as a molecular marker associated with patient outcome may represent a further significant step towards the risk stratification of patients suffering from supratentorial CNS-PNET.
Supplementary Material
Funding
We acknowledge the following sources of funding: German Children's Cancer Foundation/Deutsche Kinderkrebsstiftung (MG, AOVB, MWM, SR, TP).
Supplementary Material
Aknowledgments
Special thanks to the participating centers for their valuable cooperation. We thank Wiebke Treulieb and Christine Lindow (HIT data center) for their data management and Katharina Petrasch for central review of cerebrospimal fluid assessments.
Conflict of interest statement. None declared.
References
- 1.McLendon RE, Judkins AR, et al. Central nervous system primitive neuroectodermal tumours. In: Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, editors. WHO Classification of Tumours of the Central Nervous System. Lyon: IARC press; 2007. pp. 141–146. [Google Scholar]
- 2.Pomeroy SL, Tamayo P, Gaasenbeek M, et al. Prediction of central nervous system embryonal tumour outcome based on gene expression. Nature. 2002;415:436–442. doi: 10.1038/415436a. [DOI] [PubMed] [Google Scholar]
- 3.Li MH, Bouffet E, Hawkins CE, et al. Molecular genetics of supratentorial primitive neuroectodermal tumors and pineoblastoma. Neurosurg Focus. 2005;19:E3. doi: 10.3171/foc.2005.19.5.4. [DOI] [PubMed] [Google Scholar]
- 4.McCabe MG, Ichimura K, Liu L, et al. High-resolution array-based comparative genomic hybridization of medulloblastomas and supratentorial primitive neuroectodermal tumors. J Neuropathol Exp Neurol. 2006;65:549–561. doi: 10.1097/00005072-200606000-00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miller S, Rogers HA, Lyon P, et al. Genome-wide molecular characterization of central nervous system primitive neuroectodermal tumor and pineoblastoma. Neuro Oncol. 2011;13:866–879. doi: 10.1093/neuonc/nor070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.von Bueren AO, Gerss J, Hagel C, et al. DNA copy number alterations in central primitive neuroectodermal tumors and tumors of the pineal region: an international individual patient data meta-analysis. J Neurooncol. 2012;109:415–423. doi: 10.1007/s11060-012-0911-7. [DOI] [PubMed] [Google Scholar]
- 7.Kraus JA, Felsberg J, Tonn JC, et al. Molecular genetic analysis of the TP53, PTEN, CDKN2A, EGFR, CDK4 and MDM2 tumour-associated genes in supratentorial primitive neuroectodermal tumours and glioblastomas of childhood. Neuropathol Appl Neurobiol. 2002;28:325–333. doi: 10.1046/j.1365-2990.2002.00413.x. [DOI] [PubMed] [Google Scholar]
- 8.Behdad A, Perry A. Central nervous system primitive neuroectodermal tumors: a clinicopathologic and genetic study of 33 cases. Brain Pathol. 2010;20:441–450. doi: 10.1111/j.1750-3639.2009.00314.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pfister S, Remke M, Toedt G, et al. Supratentorial primitive neuroectodermal tumors of the central nervous system frequently harbor deletions of the CDKN2A locus and other genomic aberrations distinct from medulloblastomas. Genes Chromosomes Cancer. 2007;46:839–851. doi: 10.1002/gcc.20471. [DOI] [PubMed] [Google Scholar]
- 10.Inda MM, Castresana JS. RASSF1A promoter is highly methylated in primitive neuroectodermal tumors of the central nervous system. Neuropathology. 2007;27:341–346. doi: 10.1111/j.1440-1789.2007.00788.x. [DOI] [PubMed] [Google Scholar]
- 11.Inda MM, Munoz J, Coullin P, et al. High promoter hypermethylation frequency of p14/ARF in supratentorial PNET but not in medulloblastoma. Histopathology. 2006;48:579–587. doi: 10.1111/j.1365-2559.2006.02374.x. [DOI] [PubMed] [Google Scholar]
- 12.Pang JC, Chang Q, Chung YF, et al. Epigenetic inactivation of DLC-1 in supratentorial primitive neuroectodermal tumor. Hum Pathol. 1995;36:36–43. doi: 10.1016/j.humpath.2004.09.021. [DOI] [PubMed] [Google Scholar]
- 13.Li M, Lee KF, Lu Y, et al. Frequent amplification of a chr19q13.41 microRNA polycistron in aggressive primitive neuroectodermal brain tumors. Cancer Cell. 2009;16:533–546. doi: 10.1016/j.ccr.2009.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Korshunov A, Remke M, Gessi M, et al. Focal genomic amplification at 19q13.42 comprises a powerful diagnostic marker for embryonal tumors with ependymoblastic rosettes. Acta Neuropathol. 2010;120:253–260. doi: 10.1007/s00401-010-0688-8. [DOI] [PubMed] [Google Scholar]
- 15.Picard D, Miller S, Hawkins CE, et al. Markers of survival and metastatic potential in childhood CNS primitive neuro-ectodermal brain tumours: an integrative genomic analysis. Lancet Oncol. 2012;3:838–848. doi: 10.1016/S1470-2045(12)70257-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.von Bueren AO. CNS PNET molecular subgroups with distinct clinical features. Lancet Oncol. 2012;13:753–754. doi: 10.1016/S1470-2045(12)70260-7. [DOI] [PubMed] [Google Scholar]
- 17.Cohen BH, Zeltzer PM, Boyett JM, et al. Prognostic factors and treatment results for supratentorial primitive neuroectodermal tumors in children using radiation and chemotherapy: a Children's Cancer Group randomized trial. J Clin Oncol. 1995;13:1687–1696. doi: 10.1200/JCO.1995.13.7.1687. [DOI] [PubMed] [Google Scholar]
- 18.Friedrich C, von Bueren AO, von Hoff K, et al. Treatment of young children with CNS-primitive neuroectodermal tumors/pineoblastomas in the prospective multicenter trial HIT 2000 using different chemotherapy regimens and radiotherapy. Neuro Oncol. 2013;15:224–234. doi: 10.1093/neuonc/nos292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rutkowski S, Bode U, Deinlein F, et al. Treatment of early childhood medulloblastoma by postoperative chemotherapy alone. N Engl J Med. 2005;352:978–979. doi: 10.1056/NEJMoa042176. [DOI] [PubMed] [Google Scholar]
- 20.Wang Y, Cottman M, Schiffman JD. Molecular inversion probes: a novel microarray technology and its application in cancer research. Cancer Genetics. 2012;205:341–355. doi: 10.1016/j.cancergen.2012.06.005. [DOI] [PubMed] [Google Scholar]
- 21.Hömig-Hölzel C, Savola S. Multiplex Ligation-dependent Probe Amplification (MLPA) in Tumor Diagnostics and Prognostics. Diagn Mol Pathol. 2012;21:189–206. doi: 10.1097/PDM.0b013e3182595516. [DOI] [PubMed] [Google Scholar]
- 22.Kaplan EL, Meier P. Nonparametric estimation from incomplete observations. J Am Stat Assoc. 1958;53:457–481. [Google Scholar]
- 23.Peduzzi P, Concato J, Feinstein AR, Holford TR. Importance of events per independent variable in proportional hazards regression analysis. II. Accuracy and precision of regression estimates. J Clin Epidemiol. 1995;48:1503–1510. doi: 10.1016/0895-4356(95)00048-8. [DOI] [PubMed] [Google Scholar]
- 24.Johnston DL, Keene DL, Lafay-Cousin L, et al. Supratentorial primitive neuroectodermal tumors: a Canadian pediatric brain tumor consortium report. J Neurooncol. 2008;86:101–108. doi: 10.1007/s11060-007-9440-1. [DOI] [PubMed] [Google Scholar]
- 25.Pizer BL, Weston CL, Robinson KJ, et al. Analysis of patients with supratentorial primitive neuro-ectodermal tumours entered into the SIOP/UKCCSG PNET 3 study. Eur J Cancer. 2006;42:1120–1128. doi: 10.1016/j.ejca.2006.01.039. [DOI] [PubMed] [Google Scholar]
- 26.Albright AL, Wisoff JH, Zeltzer P, et al. Prognostic factors in children with supratentorial (nonpineal) primitive neuroectodermal tumors. A neurosurgical perspective from the Children's Cancer Group. Pediatr Neurosurg. 1995;22:1–7. doi: 10.1159/000121292. [DOI] [PubMed] [Google Scholar]
- 27.Mikaeloff Y, Raquin MA, Lellouch-Tubiana A, et al. Primitive cerebral neuroectodermal tumors excluding medulloblastomas: a retrospective study of 30 cases. Pediatr Neurosurg. 1998;29:170–177. doi: 10.1159/000028717. [DOI] [PubMed] [Google Scholar]
- 28.Zeltzer PM, Boyett JM, Finlay JL, et al. Metastasis stage, adjuvant treatment, and residual tumor are prognostic factors for medulloblastoma in children: conclusions from the Children's Cancer Group 921 randomized phase III study. J Clin Oncol. 1999;17:832–845. doi: 10.1200/JCO.1999.17.3.832. [DOI] [PubMed] [Google Scholar]
- 29.Ligon KL, Alberta JA, Kho AT, et al. The oligodendroglial lineage marker OLIG2 is universally expressed in diffuse gliomas. J Neuropathol Exp Neurol. 2004;63:499–509. doi: 10.1093/jnen/63.5.499. [DOI] [PubMed] [Google Scholar]
- 30.Gessi M, von Bueren AO, Rutkowski S, et al. p53 expression predicts dismal outcome for medulloblastoma patients with metastatic disease. J Neurooncol. 2012;106:135–141. doi: 10.1007/s11060-011-0648-8. [DOI] [PubMed] [Google Scholar]
- 31.Gustafson WC, Weiss WA. Myc proteins as therapeutic targets. Oncogene. 2010;29:1249–1259. doi: 10.1038/onc.2009.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chesler L, Weiss WA. Genetically engineered murine models-contribution to our understanding of the genetics, molecular pathology and therapeutic targeting of neuroblastoma. Semin Cancer Biol. 2011;21:245–255. doi: 10.1016/j.semcancer.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Swartling FJ, Savov V, Persson AI, et al. Distinct neural stem cell populations give rise to disparate brain tumors in response to N-MYC. Cancer Cell. 2012;21:601–613. doi: 10.1016/j.ccr.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ellison DW. Childhood medulloblastoma: novel approaches to the classification of a heterogeneous disease. Acta Neuropathol. 2010;120:305–316. doi: 10.1007/s00401-010-0726-6. [DOI] [PubMed] [Google Scholar]
- 35.Giangaspero F, Eberhart CG, Haapasalo HH, et al. In: WHO Classification of Tumours of the Central Nervous System. Louis DN, Ohgaki H, Wiestler OD, editors. Lyon: IARC press; 2007. pp. 132–140. [Google Scholar]
- 36.Fangusaro J. Pediatric high grade glioma: a review and update on tumor clinical characteristics and biology. Front Oncol. 2012;2:105–116. doi: 10.3389/fonc.2012.00105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paugh BS, Qu C, Jones C, et al. Molecular genetic profiling of pediatric high-grade gliomas reveals key differences with the adult disease. J Clin Oncol. 2010;28:3061–3068. doi: 10.1200/JCO.2009.26.7252. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.