Abstract
Background
Glioblastoma multiforme (GBM) is a highly aggressive tumor of the central nervous system with a dismal prognosis for affected patients. Aberrant protein kinase C (PKC) signaling has been implicated in gliomagenesis, and a member of the PKC-activated protein kinase D (PRKD) family, PRKD2, was identified as mediator of GBM growth in vitro and in vivo.
Methods
The outcome of PRKD2 silencing and pharmacological inhibition on glioma cell proliferation was established with different glioma cell lines. Western blotting, senescence assays, co-immunoprecipitation, fluorescence activated cell sorting, quantitative PCR, and immunofluorescence microscopy were utilized to analyze downstream signaling.
Results
RNA-interference (21-mer siRNA) and pharmacological inhibition (CRT0066101) of PRKD2 profoundly inhibited proliferation of p53wt (U87MG, A172, and primary GBM2), and p53mut (GM133, T98G, U251, and primary Gli25) glioma cells. In a xenograft experiment, PRKD2 silencing significantly delayed tumor growth of U87MG cells. PRKD2 silencing in p53wt and p53mut cells was associated with typical hallmarks of senescence and cell cycle arrest in G1. Attenuated AKT/PKB phosphorylation in response to PRKD2 silencing was a common observation made in p53wt and p53mut GBM cells. PRKD2 knockdown in p53wt cells induced upregulation of p53, p21, and p27 expression, decreased phosphorylation of CDK2 and/or CDK4, hypophosphorylation of retinoblastoma protein (pRb), and reduced transcription of E2F1. In p53mut GM133 and primary Gli25 cells, PRKD2 silencing increased p27 and p15 and reduced E2F1 transcription but did not affect pRb phosphorylation.
Conclusions
PRKD2 silencing induces glioma cell senescence via p53-dependent and -independent pathways.
Keywords: glioblastoma multiforme, protein kinase D2, p53, senescence, xenograft
Glioblastoma multiforme (GBM), which is classified as grade IV astrocytoma by the WHO, is the most common and aggressive primary brain tumor in adults. Despite advances in treatment options combining surgical resection, radiotherapy, and concomitant alkylating chemotherapy, the prognosis for GBM patients still remains dismal with a median survival of 14.2 months.1 The disproportionate malignancy of GBM is due to its invasive growth pattern and high inter-and intratumoral genetic heterogeneity.2,3 Frequent genetic alterations in GBM affect 3 major cancer pathways: (i) the receptor tyrosine kinase (RTK) pathway and its downstream oncogenic signaling by PI3K/AKT and Ras/MAPK, (ii) the p53, and (iii) the retinoblastoma protein (pRb) signaling network.4
Cellular senescence refers to the state of irreversible cell cycle arrest that occurs at the end of cells’ replicative life span or as a physiological response to different types of cellular stress including aberrant oncogenic activation, DNA damage, and oxidative stress. Depending on the type of senescence-inducing stimulus and their genetic background, different cell types display differential senescence responses. In addition to long-term exit from cell cycle, common characteristic features of the senescent phenotype are the induction of senescence-associated β-galactosidase (SA-β-Gal) activity, chromatin remodeling accompanied by the formation of senescence-associated heterochromatin foci, and morphological transformation, including vacuolization and change in morphology to a flat and enlarged cell shape. At molecular level, the senescence response can be triggered through several genetic effectors converging on the activation of tumor suppressor networks p53 and pRb.5,6 During early stages of tumorigenesis, senescence acts as an important anticancer mechanism. Aberrant activation of oncogenes, such as Ras, or the loss of tumor suppressors, as described for PTEN, can trigger senescence in vitro and in vivo. For further progression, tumor cells have to bypass the senescent arrest by additional mutations of tumor suppressor genes, including p53 and p16, or reactivation of telomerase. Thus, re-induction of the senescence program in tumor cells could represent an additional therapeutic approach. Potential intervention targets for prosenescence therapies are telomerase inhibition, modulation of cyclin-dependent kinase (CDK) activities, the reactivation of tumor suppressor genes (eg, p53) and the inactivation of oncogenes, as demonstrated for c-Myc addicted tumors.6–8
The protein kinase D (PRKD) family belongs to a subgroup of serine/threonine kinases within the calcium/calmodulin-dependent protein kinase superfamily and comprises 3 abundantly expressed mammalian isoforms. The 3 isoforms, PRKD1, PRKD2, and PRKD3, contain highly conserved structural motifs within their regulatory domains and are directly activated by phorbolesters, diacylglycerol (DAG), or DAG-activated protein kinase C (PKC) isoforms. By integrating downstream signaling of tyrosine kinase and G protein-coupled receptors, PRKDs have emerged as central players in tumor-promoting processes including proliferation, migration, invasion, and angiogenesis.9 PRKDs have been linked mechanistically to targets that are implicated in the regulation of cell proliferation, survival, and apoptosis including the Ras/MAPK pathway, HDAC, CREB, JNK, and AKT.10,11 Within a large-scale RNAi screen of human kinases, PRKD2, but not PRKD1 and PRKD3, was identified as an essential regulator of cell survival.12 In human cancers, PRKD2 is positively involved in dedifferentiation,13 survival,14 angiogenesis,15 and invasion.16 In astrocytoma, a positive correlation between PRKD2 expression and tumor grading was shown.17 This study further demonstrated that inhibition of PRKD2 results in an apoptosis-independent reduction of glioma cell proliferation in vitro and prevents tumor formation in a chicken chorioallantoic membrane assay. Just recently, we have identified PRKD2 as a potential kinase target to reduce glioblastoma cell motility and invasion.18
The present study aimed to investigate molecular pathways underlying PRKD2-silencing induced cell-cycle inhibition in glioma. We elucidated the effects of PRKD2 depletion on senescence-associated and RTK-mediated signaling in vitro and confirmed the role of PRKD2 in glioma growth in a xenograft model. Here we show for the first time that PRKD2 silencing induces a senescence-like program in p53wt and p53mut glioma cells and may represent a promising approach for prosenescence therapy in glioma.
Methods
Detailed information on chemicals, antibodies, siRNAs, adenoviruses, and primers is provided in Supplementary Materials and Methods.
Cell Culture
The human glioblastoma cell lines U87MG, A172, U251, and T98G were purchased from CLS Cell Line Service or ATTC, LGC Standards and maintained in Dulbecco's modified Eagle's medium /high glucose supplemented with 10% fetal calf serum (FCS) and 2% penicillin/streptomycin at 37°C under 5% CO2. For platelet-derived growth factor (PDGF) treatment, cells were pre-incubated in serum-free medium for 3 hours. Establishment and culture of GM133 cells has been previously described in detail.19
Glioma Primary Cell Cultures
Primary cell cultures (GBM2, Gli25) were established from glioblastoma multiforme tissue obtained during surgery and diagnosed according to the WHO classification. Cells were cultured in Dulbecco's modified Eagle's medium /high glucose supplemented with 10% FCS and 2% penicillin/streptomycin at 37°C under 5% CO2 and used up to passage 10. p53 status was sequenced as described in Supplementary Materials and Methods. The protocol was approved by the local ethical review boards.
RNA Interference
Transfections with siRNAs (20 nM) were performed using Oligofectamine transfection reagent (Invitrogen) according to the manufacturer’s instructions. Before seeding for transfection, cells were synchronized by serum-starvation for 36 hours. Untreated cells (control) and cells transfected with Oligofectamine alone (mock) or nontargeting siRNA (scrambled siRNA) were used as controls. Validated 21-bp siRNA duplexes specifically targeting PRKD2 and AKT were from Qiagen. The nontargeting siRNA (siScr) was purchased from Dharmacon.
Cell Proliferation
Glioma cells were seeded at 4 × 104 cells per well into 12-well or 6-well plates, grown for 24 hours, and treated with the indicated siRNAs, adenovirus constructs, or inhibitors. At the specified time points, cells were counted with a Casy®Cell Counter (Innovatis AG).
Flow Cytometric Analysis of Cell Cycle, DNA Synthesis, and Apoptosis
For flow cytometric analyses, untreated cells and cells transfected with nontargeting siRNA (siSCR) and targeting siRNA (siP5) were harvested by trypsinization on day 2 post transfection, washed, and resuspended in phosphate-buffered saline (PBS). For cell cycle analyses, cells were fixed in 70% ethanol for 10 minutes at 4°C, and propidiumiodide (PI) staining was performed using PI-staining buffer from Beckman Coulter. DNA synthesis was followed using the FITC BrdU Flow Kit (BD Biosciences) according to the manufacturer's instructions. Samples were run on a LSR II flow cytometer using FACSDiva software (Becton Dickinson) for acquisition and analysis. Cell cycle distribution of cells was calculated using ModFit (Verity Software House). For analysis of apoptosis, staining of cells was performed with APC Annexin V in a staining buffer containing PI (BioLegend) as recommended by the manufacturer. Samples were analyzed on LSR II using FACSDiva software for acquisition and FCS Express software (De Novo Software) for analysis.
Heterotopic Glioma Xenograft Model
Animal experiments were performed in accordance with animal care ethics approval and guidelines, as per Animal Care certificate BMWF-66.010/0089-11/3b/2011 of the Austrian Federal Ministry of Science and Research. Forty eight hours after transfection control (untreated, nontargeting siRNA) and treated (siP5) U87MG cells (1 × 106 in 100 µL media) were subcutaneously injected into the flank of Fox Chase SCID mutant (C.B-17/lcrHanTMHsd-Prkdcscid, Harlan Laboratories) 12-week old female mice (n = 5 per group). To follow tumor growth, tumor size was measured with a caliper 3 times a week, and tumor volume was calculated using the formula: volume = length x width2 × π/6. When tumors reached a volume of maximal 4000 mm3, animals were euthanized by cervical dislocation. For histological analyses, half of the tumor was fixed in formalin and embedded in paraffin using a tissue processor. Tumor tissue sections were deparaffinized, rehydrated, and subjected to hematoxylin-and-eosin and Ki-67 staining using an automated staining system (Dako-Autostainer). Quantification of Ki-67 positive cells was performed in tumor areas with dense tumor cell mass using ImageJ software.
Senescence-associated β-galactosidase Staining and Cell Size Calculation
For detection of senescence-associated β-galactosidase (SA-β-Gal) activity, we followed the protocol described by Dimri et al.20 For determination of cell size, the morphology of control (untreated and siScr transfected) and PRKD2-silenced (siP5) cells was recorded by phase-contrast microscopy at the times indicated. Four micrograph fields were randomly chosen for each condition. The total area occupied by the cells and the cell number were estimated using ImageJ, and cell size was calculated as total cell area/cell number. Measurements were done in triplicate.
Immunoblotting
For immunoblotting, whole cell extracts or nuclear and cytoplasmic protein fractions prepared with radio immunoprecipitation assay (RIPA) buffer or the NE-PER Nuclear and Cytoplasmic Kit (Pierce) were subjected to SDS-PAGE. Protein expression was normalized to appropriate loading controls (lamin A/C, glyceraldehyde 3-phosphate dehydrogenase, β-actin), and phosphorylation of proteins was normalized to the corresponding total protein.
Co-immunoprecipitation
Whole cell lysates (1 mg total protein) were incubated with 2 µg of anti-PRKD2 or anti-AKT IgG in RIPA buffer at 4°C overnight. Preclearing of cell lysates, using the appropriate preclearing matrix, and formation of the IP antibody-IP matrix complex (ExactaCruz) was performed at 4°C for 4 hours in PBS. Beads were washed with PBS, resuspended in reducing electrophoresis buffer, boiled for 3 minutes, and immunoblotted as described above using the horseradish peroxidase-conjugated reagent of the ExactaCruz detection system.
Quantitative Polymerase Chain Reaction
After transfection with the indicated siRNAs, total RNA was extracted and reverse transcribed. Quantitative PCRs (qPCRs) were performed using the Applied Biosystems 7900HT Fast Real Time PCR System, the QuantiFast SYBR Green PCR Kit, and Quantitect Primer Assays (Qiagen). Relative changes in gene expression were normalized to hypoxanthine phosphribosyltransferase 1 (HPRT1).
Statistical Analysis
Data are presented as mean ± SD. One-way ANOVA, followed by Bonferroni's post hoc comparison test, was used for analysis of statistical significance. *** P < .001, ** P < .01, * P < .05.
Statistical significance of differences in mRNA expression was analyzed with the relative expression software tool (REST©, http://www.gene-quantification.de/rest.html) using a pairwise fixed reallocation test.
Results
RNA Interference and Pharmacological Inhibition of PRKD2 Inhibits Glioma Cell Proliferation
In a first round of experiments, we established the efficacy and duration of PRKD2 silencing in U87MG cells using 3 different 21mer siRNA constructs. As shown in Fig. S1A, all siRNAs (siP4-P6) efficiently silenced PRKD2 protein expression up to day 6 post transfection, with siP5 showing highest efficacy. Mock transfection and a nontargeting siRNA were without effects. To investigate potential off-target effects of PRKD2 silencing within the PRKD family, PRKD1 and PRKD3 expression was analyzed by immunoblotting (Fig. S1B). Two of the siRNAs (siP4 and siP6) slightly increased PRKD1 expression compared with cells transfected with nontargeting siRNA, whereas PRKD3 was unaffected. In line with another report,17 silencing of PRKD2 resulted in a significant decrease of U87MG cell proliferation by a maximum of 83% (siP5; 6 days post silencing; Fig. S1C). Also in p53mut GM133 cells, siP4-6 efficiently decreased PRKD2 levels (Fig. S2A) and inhibited cell proliferation (Fig. S2B; maximum inhibition 61%).
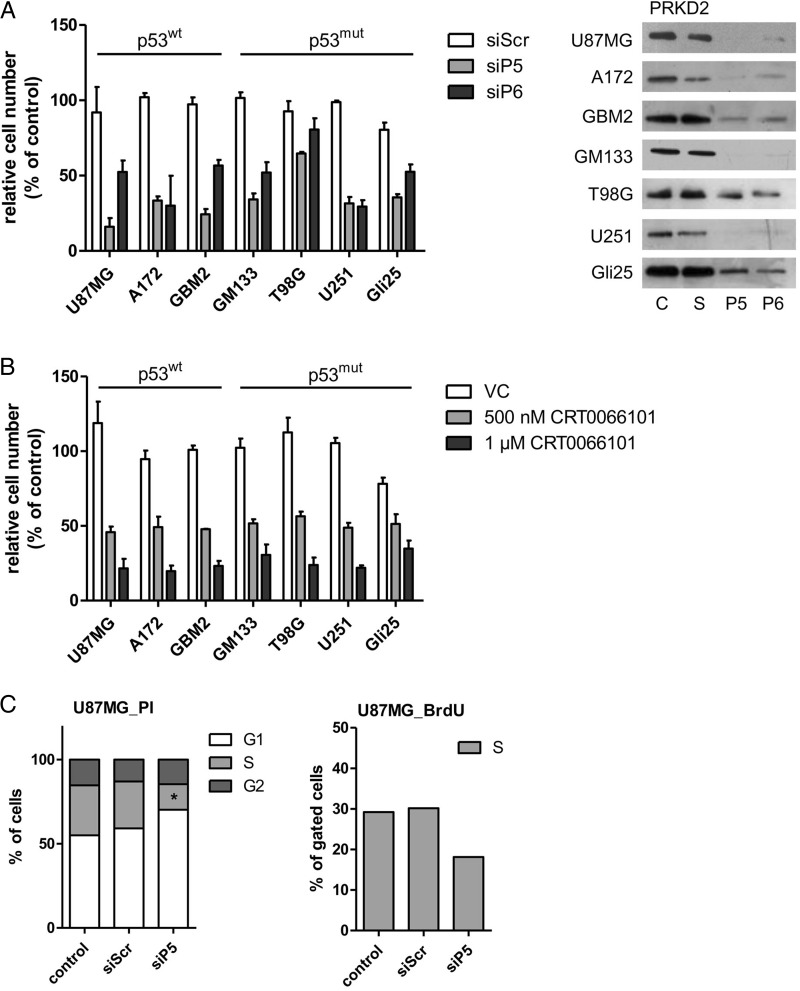
To get an indication whether the p53 status determined the outcome of PRKD2 silencing on cell proliferation, we used a panel of 5 established glioma cell lines (U87MG, GM133, A172, T98G, U251) and 2 primary cell cultures (GBM2, Gli25) established by our groups. Of these, U87MG and A172 expressed p53wt, while GM133, T98G, and U251 were p53mut. Sanger sequencing revealed that primary GBM2 cells are p53wt, and Gli25 are p53mut (Fig. S3). As observed with U87MG cells (Fig. S1C,) siP5 efficiently inhibited cell proliferation in all cells tested (lowest efficacy T98G, 30% reduction; highest efficacy U87MG, 83% reduction compared with siScr transfected cells; Fig. 1A), while siP6 was less potent in terms of inhibition of cell proliferation. Silencing efficacy is shown in the right panel of Fig. 1A. In the cell lines studied during these experiments, growth arrest in response to PRKD2 silencing occurred independently of the cellular p53 status.
Fig. 1.
Silencing and pharmacological inhibition of PRKD2 inhibits GBM cell growth. (A) Effect of PRKD2 RNAi on glioma cell proliferation. Two different siRNA constructs (siP5; siP6) were used to silence PRKD2 expression. Untreated cells and cells transfected with nontargeting siRNA (siScr) were used as controls. Cells were harvested and counted on day 6 post silencing. Results represent mean ± SD of relative cell numbers normalized to untreated cells from one representative experiment done in triplicate. The p53 status of the cells is indicated. Silencing efficacy is shown in the right panel. (B) Effects of pharmacological PRKD inhibition on GBM cell proliferation. The indicated p53wt and p53mut GBM cells were incubated in the presence of vehicle dimethyl sulfoxide (DMSO) or 0.5 and 1 µM CRT0066101 (added in DMSO). Cells were harvested and counted 3 days post CRT0066101/DMSO addition. Results represent mean ± SD of relative cell numbers normalized to untreated cells. Data from one representative experiment performed in triplicate are shown. (C) Analysis of cell cycle distribution and DNA synthesis of U87MG (upper panels) and GM133 (lower panels) cells. Two days post transfection control (untreated and siScr transfected) and PRKD2-silenced (siP5) cells were stained with propidium iodide (PI) (left panel) or bromodeoxyuridine (BrdU) (right panel) and analyzed by flow cytometry. Results are expressed as percentage of total cells and represent the mean values from 3 (PI) and 2 (BrdU) independent experiments. (**P < .01, *P < .05, one-way ANOVA). (D) Detection of Annexin V positive U87MG and GM133 cells. Four days post transfection cells (untreated, siScr, and siP5 transfected) were stained with APC Annexin V (‘A’) and PI and analyzed by fluorescence activated cell sorting. The percentages of A-/PI− (viable), A+/PI− (early apoptotic), A+/PI+ (late apoptotic) and A−/PI+ are shown. Mean values from 2 experiments are presented. (E) PRKD2 silencing in glioma xenografts. Forty-eight hours after transfection, 1 × 106 control or treated U87MG cells (siScr and siP5) were subcutaneously injected into the flank of SCID mice (n = 5 per group). Xenograft size was measured 3 times a week, and tumor volume was calculated. Results are expressed as mean ± SEM. Bar indicates delayed growth onset of siPRKD2 xenografts compared with siSCR xenografts (one-way ANOVA, **P < .01). Two animals (shown in grey circles) receiving silenced cells did not develop tumors until day 76 post inoculation.
In order to confirm the importance of PRKD2 for glioma proliferation, we used an adenoviral system to overexpress the kinase in U87MG cells. Although virus transduction induced elevated PRKD2 expression (Fig. S4A), there was no pro-proliferative effect on untreated (Fig. S4B) and silenced (not shown) U87MG cells. This might (at least in part) result from the formation of multinucleated giant cells (reminiscent of syncytia with massive PRKD2 overexpression) that eventually detached (Fig. S4C) and reduced cell counts (Fig. S4B; MOI 5–50).
As an alternative approach to RNAi,21 we used the small molecule PRKD family inhibitor CRT0066101 (a pyrazine-benzamide compound).22 This compound potently inhibited glioma cell proliferation between 45% and 81% (Fig. 1B). Dose response curves for CRT0066101 (shown in Fig. S5) revealed IC50 values of 270 and 470 nM CRT0066101 for U87MG (A) and GM133 (B) cells, respectively.
Next, we analyzed the outcome of PRKD2 silencing on cell cycle progression in U87MG and GM133 cells. In these experiments, we observed an accumulation of silenced cells in the G1 phase and a decrease in the S phase of the cell cycle (PI staining and bromodeoxyuridine incorporation; Fig. 1C), suggesting G0/G1 arrest in response to PRKD2 depletion. In addition, cell cycle arrest occurred in both diploid and aneuploid GM133 populations (Fig. 1B, lower panel). Cell cycle arrest in PRKD2-silenced cells was not accompanied by apoptosis as evaluated by Annexin V/PI staining, since the percentage of apoptotic cells (Annexin V+/PI+) was always < 5% (Fig. 1D).
In a xenograft model, all animals in the control groups developed tumors (mean onset of tumor development was 27 ± 0.9 days and 29 ± 4.6 days, untreated and nontargeting siRNA transfected cells, respectively), whereas a significant delay in tumor onset was observed in 3 animals receiving siPRKD2-transfected glioma cells (mean onset 42.3 ± 5.1 days; Fig. 1E, open squares). Two animals (represented by open circles) in the siRNA group remained tumor free until termination of the study (day 76 post inoculation). Linear regression analysis revealed that the rate of tumor growth was also lower (though statistically not significant) in PRKD2-targeted xenografts (mean slope of untreated, nontargeting siRNA- and siPRKD2-transfected xenografts was 216 ± 13.9, 177 ± 33.4, and 154 ± 23.7 mm3/day, respectively). Xenograft staining for the proliferation marker Ki-67 revealed no differences in the proliferation index, since tumor volume was defined as experimental endpoint (data not shown).
Interference with PRKD2 Expression Induces Cellular Senescence
To get an indication about potential alterations of the transcriptional profile of PRKD2-silenced cells, we performed qPCR array analyses of U87MG cells and primary GBM2 cultures (Supplementary Tables S1 and S2). In U87MG cells, these analyses revealed significant alterations in RNA expression of BRCA1, CDC25A, CDK2, CDKN1A, E2F1, and MYC. In primary GBM2 cells CDC25A, CDK2, CDKN1A, and E2F expression was also significantly altered in silenced cells.
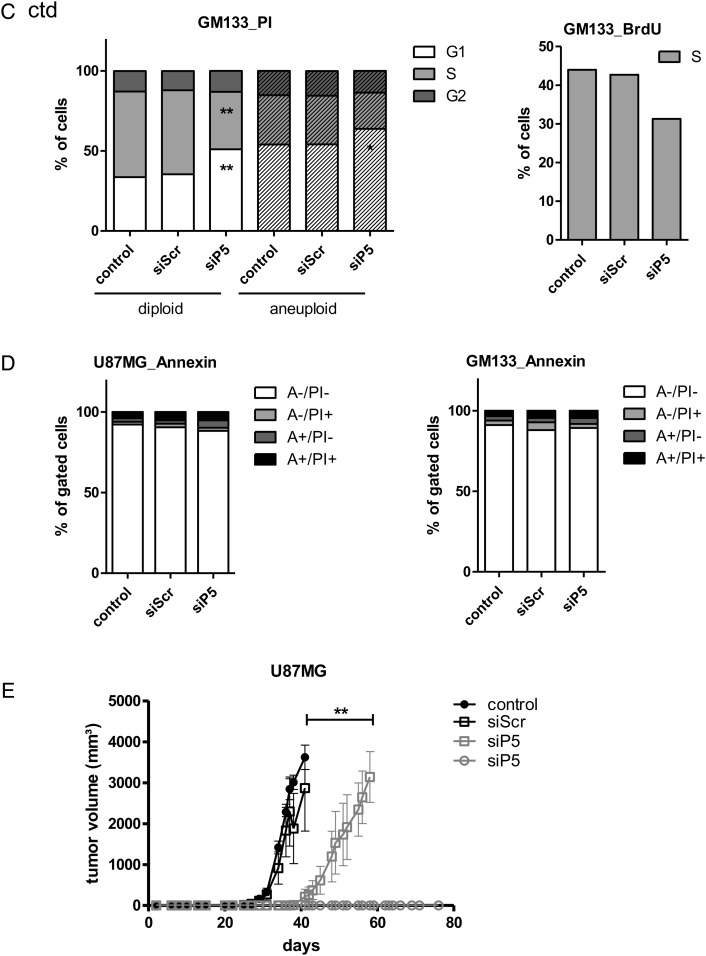
To investigate whether PRKD2 silencing induced senescence-like growth arrest, we examined characteristic features that define the senescent state. As shown in Fig. 2A, silencing of PRKD2 induced acidic senescence-associated β-galactosidase activity (SA-β-gal) in U87MG (64 ± 10.4% positive) and GM133 (41 ± 5.4% positive) cells. SA-β-gal activity was also observed in both primary glioma cultures (GBM2, Gli25) in response to PRKD2 silencing (Fig. S6). In addition, the distinctive flattened and enlarged morphology of senescent cells was observed in PRKD2 deficient cells. Analysis of the cell surface area (Fig. 2B) revealed a significant increase in cell area of PRKD2-silenced U87MG (2 200, 1 625, 1 450 µm2; siP5, controls, and siScr) and GM133 (6 648, 3550, and 3 702 µm2; siP5, controls, and siScr) cells. In line with these indications for cellular senescence, qPCR analyses revealed reduced hTERT mRNA levels in silenced p53wt U87MG by 70%.
Fig. 2.
Interference with PRKD2 expression induces cellular senescence. (A) Untreated U87MG (left panels) and GM133 (right panels) cells and cells transfected with siScr or siP5 were stained for SA-β-Gal activity (blue) on day 3 (U87MG) and day 5 (GM133) post transfection. (B) Bar graphs represent cell surface area of control and transfected (siScr or siP5) cells. Three (U87MG) and 5 (GM133) days post transfection, 4 randomly chosen micrograph fields (in triplicates) were recorded. The total area occupied by the cells and the total cell number was calculated with ImageJ. Mean surface area/cell was calculated as total cell area/total cell number (mean ± SEM, one-way ANOVA, ***P < .001, **P < .01, *P < .05).
Silencing of PRKD2 Impairs RTK Downstream Signaling in Glioma Cells
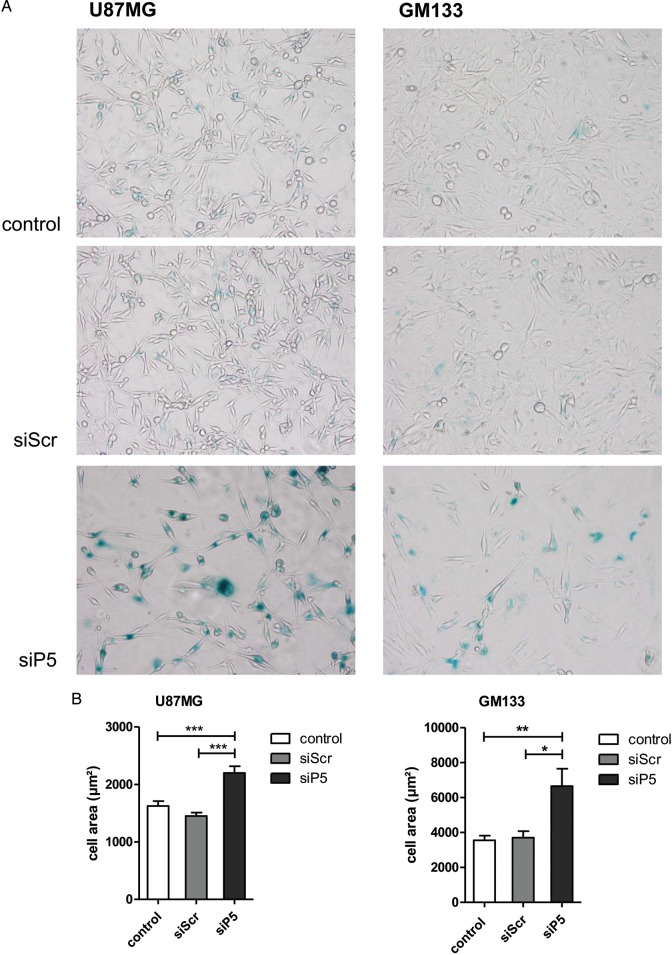
Hyperactive RTK signaling through the PI3K/AKT and RAS/MAPK pathways represents fundamental events in gliomagenesis.23 To investigate whether PRKD2 communicated to AKT in glioma cells (probably in a similar manner as shown for PRKD3 in prostate cancer cells11), basal and PDGF-mediated activation of PRKD2 and AKT (the antibody used recognizes all 3 isoforms) was studied. During these experiments, we used U87MG and GM133 cells (Fig. 3A) as well as primary GBM2 and Gli25 cultures (Fig. S7). As shown in Fig. 3A, stimulation with PDGF resulted in activation of PRKD2 (increased autophosphorylation of Ser876). The phosphorylation status of AKT in response to PDGF was cell-line dependent: in U87MG, PDGF was without effect on pAKT levels, most probably due to high basal activation caused by an in-frame deletion of exon 3 of the PTEN gene.24 In GM133 cells, PDGF treatment increased pAKT levels. Importantly, PRKD2 knockdown reduced AKT phosphorylation under basal and PDGF-stimulated conditions in both cell lines, while total AKT levels remained unchanged. In addition, diminished AKT phosphorylation was observed in the cytoplasmic and nuclear compartment where AKT isoforms execute their functions (Fig. 3B). Also during these experiments, total AKT levels were unaffected by PRKD2 silencing. Additionally, depletion of PRKD2 resulted in decreased AKT phosphorylation in primary GBM2 (Fig. S7A) and Gli25 (Fig. S7B) cultures. These results suggest that PRKD2 silencing attenuates activity of the classical proto-oncogenic AKT pathway under both basal and growth-factor stimulated conditions.
Fig. 3.
PRKD2 modulates AKT activity in glioma cells. (A) Three days post silencing, U87MG and GM133 were stimulated with 10 ng/ml PDGF for 5 and 30 minutes. Total cell lysates were immunoblotted, and activation of indicated proteins was monitored using phospho-specific antibodies. To ensure comparable loading of lysates, the corresponding pan-proteins or actin were used as controls. One representative immunoblot (of at least 3 independent experiments) is shown. AKT antibodies used recognize AKT isoforms 1, 2, and 3. (B) The effect of PRKD2 silencing on AKT phosphorylation was studied in cytosolic and nuclear fractions by Western blotting. C = untreated cells, S = siScr, P5 = siP5. (C) Co-immunoprecipitation experiments indicating physical interaction of PRKD2 and AKT. Protein extracts of U87MG cells (3 independent dishes, ‘1, 2, 3′) were immunoprecipitated with the indicated antibodies, followed by immunoblotting. The association of proteins was confirmed by reverse immunoprecipitation. Input levels were detected to ensure comparable loading. Sham precipitation (“-”) was performed to exclude unspecific immunoreactions. (D) The indicated cell lines were incubated in the presence of 5 and 10 µM triciribine. Ethanol was used as vehicle control. Five days after addition of triciribine/ethanol, cells were harvested and counted. Results represent mean ± SD (n = 4) of relative cell numbers normalized to untreated cells. (E) Two different siRNA constructs (si1; si2) were used to silence AKT1 and AKT3 expression. Untreated cells and cells transfected with nontargeting siRNA (siScr) were used as controls. Cells were harvested and counted on day 6 post silencing. Results represent mean ± SD of relative cell numbers normalized to untreated cells from one representative experiment done in triplicates.
To test whether the observed decrease of AKT phosphorylation could be a result of physical interaction between AKT and PRKD2, we performed co-immunoprecipitation experiments in U87MG cells. As shown in Fig. 3C, reciprocal pull-down indicated protein-protein interaction between PRKD2 and AKT. These results make it reasonable to assume that PRKD2 directly mediates AKT phosphorylation in glioma cells and potentially accounts for the growth-promoting effects of PRKD2 in GBM cells.
To get pharmacological evidence that AKT drives proliferation of the 7 glioma cell lines, triciribine was used as antagonist. Triciribine inhibited growth of all cell lines at both concentrations (5 and 10 µM) used. The extent of inhibition was, however, cell-line dependent and ranged between 29% and 62% (Fig. 3D), observations in line with another report.25 Silencing of AKT1 (Fig. 3E) decreased U87MG proliferation (57%–63%), while AKT3 knockdown was less efficient (18%–34%). We did not target AKT2, since relative RNA expression in U87MG cells is low (http://biogps.org; AKT1:AKT2:AKT3 = 653:7:115).
We have also analyzed the effect of PRKD2 knockdown on activation of MAPK family members; however, the outcome of these experiments was (in contrast to AKT) not uniform among the 2 cell lines (Fig. S8). Treatment with PDGF induced phosphorylation of ERK, JNK, and p38 in U87MG and GM133 cells. Silencing of PRKD2 attenuated p44/42 MAPK, p54, and p46 JNK phosphorylation in U87MG cells (Fig. S8A) but not in GM133 cells (Fig. S8B).
PRKD2 Silencing Induces the Expression of Tumor-suppressor Proteins
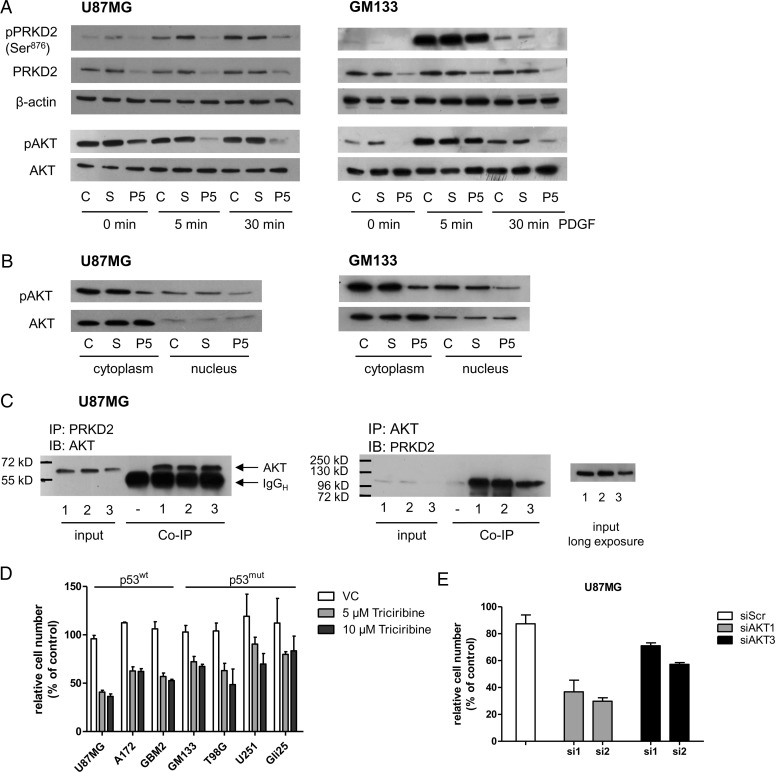
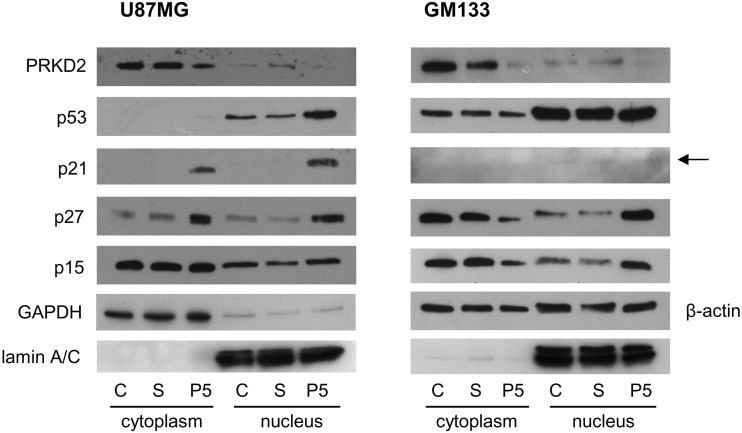
On the molecular level, activation of the senescence program is mediated by the p53 and CDKN2A/p16-pRb pathways.5 Accumulation of CDKN2A/p16, p53, and other pathway-related proteins, notably CDKN1A/p21, CDKN2A/p27, and CDKN2B/p15, indicated activation of these signaling cascades. Therefore, we analyzed the expression and subcellular localization (cytoplasm vs nucleus) of p53 and cyclin-dependent kinase inhibitor proteins in response to PRKD2 silencing. As shown in Fig. 4, p21 and p27 were upregulated in the cytoplasm of PRKD2-silenced U87MG cells. Moreover, knockdown of PRKD2 induced an increase of p53, p21, and p27 protein in the nucleus as compared with untreated and nontargeting siRNA-transfected controls. CDKN2A/p16 was undetectable because of endogenous gene deletion in U87MG cells.26 Although the reported complete homozygous deletion on chromosome 9 contains both CDKN2A/p16 and CDKN2B/p15, p15 was detected, although no regulation was observed in response to PRKD2 silencing. We have also confirmed nuclear accumulation of p53, p27, and p21 in U87MG cells by immunofluorescence microscopy. For better visualization of cell borders, overexposed micrographs are presented in Fig. S9.
Fig. 4.
PRKD2 silencing upregulates expression of tumor suppressor proteins. U87MG and GM133 cells were transfected with nontargeting (S) and PRKD2 targeting siRNA (P5), and expression analysis of tumor suppressor proteins was performed by immunoblotting in cytosolic and nuclear fractions 3 days post transfection. Untreated cells (“C”) were used as controls. Nuclear and cytoplasmic lysates were analyzed for expression of PRKD2, p53, p21, p27, and p15. Lamin A/C, and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) or β-actin was used as loading controls.
The p53mut GM133 cell line showed no response on p53 level, but nuclear p27 and p15 levels were upregulated in silenced cells (Fig. 4, right panel). p21 was barely detectable in GM133 cells, and p16 was undetectable, in line with an earlier report.19
In primary p53wt GBM2 cells, PRKD2 silencing induced comparable changes as observed in U87MG cells, namely upregulation of cytoplasmic p21 and p27 (and to a lesser extent p15) as well as nuclear p53, p21, p27, and p15 content (Fig. S10A). In primary p53mut Gli25 cells, PRKD2 silencing (using siRNA P5 and P6) induced pronounced upregulation of nuclear p27 and a slight increase of p15 (but not p53 or p21; Fig. S10B), which was comparable to that observed in the GM133 cell line.
PRKD2 Knockdown Interferes with G1/S Transition
Altered signaling events observed in response to PRKD2 silencing are likely to affect regulation of the G1/S restriction point machinery. G1 to S progression is tightly regulated by pRb protein phosphorylation. Phosphorylation at several critical sites by G1-specific CDKs inactivates pRb protein and leads to enhanced transcriptional activity of the E2F family and cell cycle advancement.27 In turn, CDKs and their regulatory cyclin partners are subject to negative regulation by the Ink4 and Cip/Kip families of CDK inhibitors.
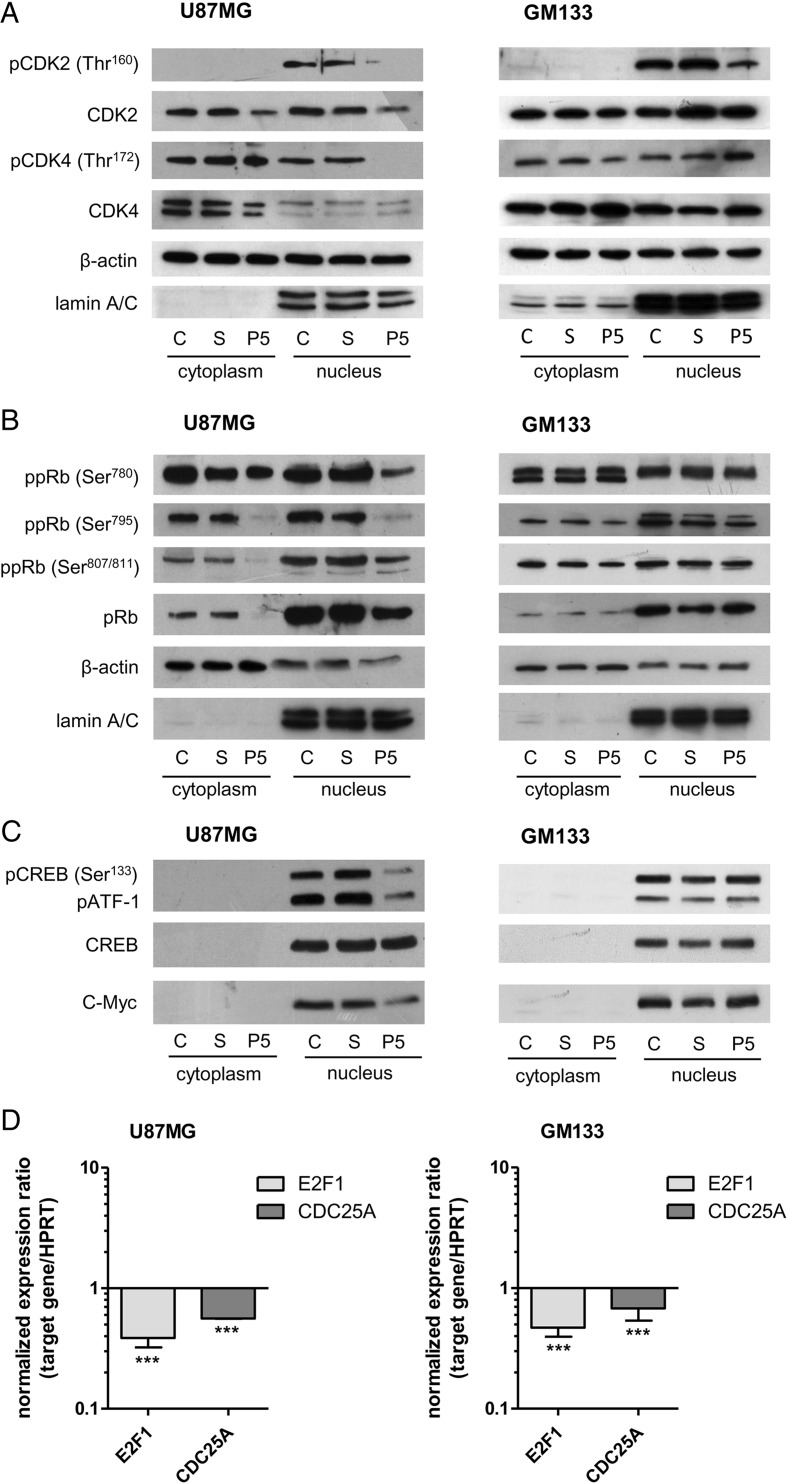
Thus, we examined the impact of PRKD2 silencing on the activation state of the CDK/pRb/E2F regulatory complex in U87MG and GM133 cells. As shown in Fig. 5A for U87MG cells, phosphorylation of G1-associated kinases CDK2 at Thr160 and CDK4 at Thr172 (both are activating phosphorylation sites) was reduced in nuclear fractions of silenced cells. Total CDK2 was reduced in nuclear and cytoplasmic fractions, whereas total CDK4 remained unchanged. This is in line with downregulated CDK2 mRNA (reduction by 61%) in response to PRKD2 silencing (Supplementary Table S1), while CDK4 mRNA levels were almost unaffected (Table S1). In line with Azoitei et al.17, protein expression of CDK4-associated cyclin D1 was reduced in PRKD2-silenced cells. In contrast, cyclin E1 mRNA and nuclear protein levels remained unchanged in response to PRKD2 knockdown, whereas cytoplasmic cyclin E protein levels were slightly elevated (Table S1 and Fig. S11). In PRKD2-silenced primary GBM2 cells, pCDK2 was slightly reduced in the cytoplasm, while total protein was reduced in nuclear fractions of silenced cells (Fig. S12A). In p53mut GM133 cells only CDK2 phosphorylation at Thr160 was affected by PRKD2 knockdown (Fig. 5A, right panel).
Fig. 5.
PRKD2 silencing interferes with the G1/S transition complex. U87MG (left panels) and GM133 (right panels) cells were transfected with nontargeting (S) and PRKD2 targeting (P5) siRNA. C = control, untreated cells. One representative immunoblot out of 3 independent experiments is shown. (A) Analysis of phosphorylated and total CDK2 and CDK4 proteins was performed by immunoblotting in cytosolic and nuclear fractions 3 days post transfection. Lamin A/C and β-actin were used as loading controls. (B) Phosphorylation of pRb on Ser780, Ser795, and Ser807/811 and expression of total pRb was determined by immunoblotting in nuclear and cytoplasmic fractions. Lamin A/C and β-actin were used as loading controls. (C) CREB was analyzed for phosphorylation on Ser133 by immunoblotting, and total CREB was used as loading control. The phospho-antibody used also detects the phosphorylated form of CREB-related protein ATF-1. The lower line shows c-Myc expression in control and silenced cells. (D) On day 4, (U87MG) and 6 (GM133) post transfection target gene expression was analyzed by qPCR using validated primer pairs. HPRT1 was used as housekeeping gene. Relative gene expression of target genes is presented in relation to scrambled RNAi. Results represent mean ± SD from 3 biological replicates (***P < .001). Gene expression ratios were calculated by REST as described in Materials and Methods.
In p53wt U87MG cells, immunoblot analyses of phosphorylated and nonphosphorylated pRb species revealed decreased pRb-phosphorylation at CDK recognition sites (RbSer780, Ser795, and Ser807/811) in response to PRKD2 silencing (Fig. 5B). Expression of total pRb protein was also significantly reduced, whereas mRNA levels were unaffected (Table S1). In p53mut GM133 cells, neither Rb phosphorylation nor total Rb levels were influenced by PRKD2 silencing (Fig. 5B). PRKD2-silenced primary GBM2 cells displayed similar alterations in phosphorylated and total pRb levels as seen in U87MG cells (Fig. S12B).
Furthermore, PRKD2 silencing considerably impaired phosphorylation of CREB at Ser133 and CREB-related protein ATF-1 in U87MG when compared with controls. Moreover, PRKD2-deficient cells showed a distinct reduction in the expression of c-Myc on mRNA (reduction by 60%; Table S1) and protein level (Fig. 5C). None of these alterations were observed in p53mut GM133 cells (Fig. 5C; right panel). mRNA expression of transcription factor E2F1 was significantly attenuated in p53wt U87MG and p53mut GM133 cells (Fig. 5D; reduction by 62 and 52%, respectively). Active E2F1 induces the expression of CDC25A phosphatase that plays an essential role during G1/S transition.28 qPCR of U87MG and GM133 cells (Fig. 5D) analysis revealed downregulated mRNA of CDC25A (reduction by 44 and 33%). Primary GBM2 cultures (p53wt) responded in a similar manner as U87MG cells (Table S2).
Discussion
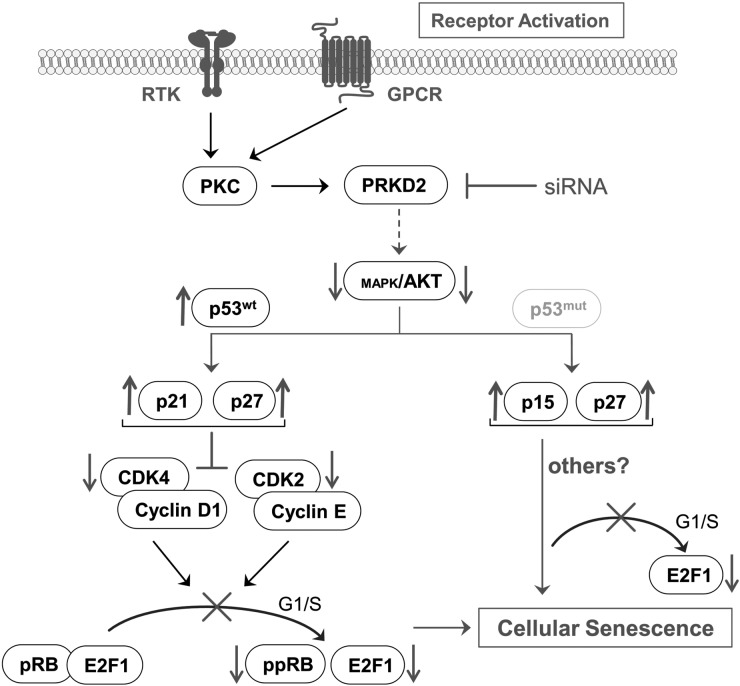
We here show that RNA interference and pharmacological inhibition of PRKD2 blocks malignant proliferation of glioma cells in vitro and in vivo. Moreover, multiple p53wt and p53mut cell lines and primary cultures responded similarly (ie, induction of senescence and reduced proliferation), underscoring the therapeutic relevance of our findings. We found that PRKD2 silencing induced cellular senescence via p53-dependent and p53-independent pathways, upregulated specific sets of tumor suppressor proteins, attenuated RTK-mediated mitogenic AKT signaling, and inhibited the G1/S transition machinery in glioma cells. A schematic summary of our findings is displayed in Fig. 6.
Fig. 6.
Summary of signaling events leading to PRKD2-dependent induction of senescence in p53wt and p53mut GBM cells. RTK and/or GPCR signaling induces PKCs and downstream PRKD2 activation. Silencing of PRKD2 attenuates AKT activation (and MAPK activation in U87MG cells). In p53wt U87MG and primary GBM2 cells, this leads to increased p53, p21, and p27 expression. As a consequence, diminished cyclin-CDK activation, hypophosphorylation of pRb, and reduced transcription of E2F target genes induce senescence. In p53mut GM133 and Gli25 primary cells, PRKD2 silencing results in decreased AKT activation, upregulation of p15 and p27, G1/S arrest, and downregulated E2F1 transcription. Also this pathway induces senescence; however, the signaling modules downstream of p15 and p27 that are ultimately responsible for the induction of senescence remain to be elucidated.
Data obtained here demonstrate a high antiproliferative potential of PRKD2 silencing for glioma cells in vitro and in vivo, findings in line with another report.17 There, Azoitei and colleagues have shown that PRKD2 silencing inhibits glioma cell proliferation in vitro and in a chicken allantoic membrane model. Data of the present study support and extend these findings to a xenograft mouse model and the pharmacological PRKD family inhibitor CRT0066101 (Fig. 1). This is in line with a report demonstrating high antiproliferative properties and good oral bioavailability of this inhibitor in an orthotopic pancreatic tumor model.22 In addition, the present study demonstrates that growth arrest in response to PRKD2 silencing in p53wt and p53mut glioma cells is characterized by 2 hallmarks of senescence (Fig. 2): namely, positive SA-β-gal staining and enlarged, flattened cell morphology.5 Downstream-signaling events that induce senescence in response to PRKD2 silencing are relatively clear-cut in p53wt cells, while the responsible mechanisms remain partially elusive in p53mut cells.
In p53wt cells, silencing of PRKD2 resulted in pronounced upregulation of p53, p21, and p27 (Fig. 4), which are known mediators of senescence.6 Corroborating our results, p53 and p21 were shown to be indispensable for the induction of premature senescence in response to ionizing radiation in PTEN-deficient glioma cells.29 The mechanisms that increase p53 protein (but not mRNA, Tables S1 and S2) levels in response to PRKD2 silencing are currently unclear. However, the ubiquitin ligase MDM2 facilitates proteasomal p53 degradation in an AKT-dependent manner.30 Therefore, it is reasonable to assume that decreased AKT activation in PRKD2-silenced cells (Fig. 3 and S7) could inactivate Mdm2 and thereby stabilize p53. During the present study, p21 upregulation was observed only in p53wt but not in p53mut cells (Fig. 4 and S10), indicating that the increase in p21 expression depends on intact p53. Expression of p27 was identified as a central checkpoint for senescence in an AKT1-driven prostate cancer model,31 and reduced p27 expression in PTEN deficient cells correlates with enhanced expression of the ubiquitin ligase Skp2.32 Proteolysis of p27 by nuclear Skp2 and the cytoplasmic ligase KPC is triggered by oncogenic signaling through PI3K/AKT, Ras/MAPK, and CDK2 activity.32,33 Therefore, reduced AKT (Fig. 3 and S7A) and CDK2 (Fig. 5 and S12; Tables S1 and 2) transcription/activation in PRKD2-silenced U87MG and GBM2 cells is compatible with increased p27 levels.
In a variety of cancers including glioma, restoration of the telomerase complex represents a common mechanism to bypass telomere shortening and replicative senescence. Thus, targeting telomerase activity or hTERT expression was proposed as a powerful prosenescence approach in cancer therapy.34 During the present study, U87MG cells displayed the most pronounced senescent phenotype. This might be due to a reduction of hTERT expression in response to PRKD2 silencing. Among others, c-Myc, Ras/MAPK, and JNK are inducers of hTERT transcription, whereas p53 represses hTERT expression.35 In addition, AKT acts as a posttranslational modulator of the catalytic telomerase subunit.35 Thus, downregulated c-Myc, JNK, p44/42 MAPK, and AKT expression/activation and upregulated p53 could contribute to reduced hTERT transcription. However, these observations were restricted to the U87MG cell line during the present study.
Hyperactive RTK signaling mediated by PI3K/AKT and Ras/MAPK networks occurs in 88% of gliomas.36 Co-immunoprecipitation of PRKD2 and AKT (Fig. 3C) suggests the possibility of PRKD2-mediated AKT phosphorylation. Notably, reduced AKT activation was a common observation independent of the cellular p53 status in response to PRKD2 silencing (Fig. 3 and S7). As observed here for PRKD2, depletion of PRKD3 attenuates basal activation of AKT in prostate cancer cells.11 Activated AKT represents a nodal point in the regulation of G1/S transition through inhibitory phosphorylation of downstream proteins including GSK3β, p27, p21, FOXO, and pRb as well as activating phosphorylation of mTOR and MDM2.37 In line with results obtained during the present study (Fig. 3D), pharmacological antagonism of AKT was shown to inhibit tumor cell proliferation in vitro25 and in vivo.38 Some small molecule inhibitors of PI3K/AKT are in the early phases of clinical trials for targeting glioma.23 Lee and colleagues have demonstrated that silencing of AKT decreased pRb and increased p53 protein expression in U87MG cells29 in a similar manner as observed here (Fig. 4 and 5B). In contrast to the present study, these events were accompanied by U87MG apoptosis.
Until now, phosphorylation targets of PRKD family members were reported only for PRKD1. One of them, the Ras and Rab interactor 1 (RIN1) inhibits Ras/Raf interaction in its unphosphorylated form, thereby preventing MAPK/ERK activation. PRKD1-mediated phosphorylation of RIN1 reverses this block and leads to activation of MAPK pathways.10 Data obtained during the present study suggest that PRKD2 regulates MAPK activation in U87MG but not in GM133 cells (Fig. S8); therefore, these findings appear to be cell-context specific. MAPK/ERK downstream targets involve, among others, c-Myc,39 cyclin D1,40 and CREB,10 a known modulator of proliferation through regulation of cyclin D1 transcription.41 In U87MG cells, reduced c-Myc (Fig. 5C and Table S1), CREB (Fig. 5C), and cyclin D1 (Fig. S11) expression/activation might be a result of attenuated MAPK signaling. Expression levels of c-Myc are positively linked to astrocytoma malignancy and stemness.42 In RAS-driven tumors, c-Myc counteracts oncogene-induced senescence through downregulation of p16 and p21 and upregulation of hTERT.43 Though experimentally not validated during the present study, this raises the possibility that reduced CDK2 activation results in decreased c-Myc phosphorylation and thereby promotes cellular senescence.44
Here we show that senescence of PRKD2-silenced p53wt U87MG and GBM2 cells is accompanied by hypophosphorylation of CDK2, CDK4, and pRb (Fig. 5 and S12), inducing an arrest in G1. Entry and progression through G1 depends on the sequential activation/phosphorylation of cyclin D-CDK4/6 and cyclin E-CDK2 complexes leading to pRb phosphorylation and expression of E2F family target genes.27 In line with our data demonstrating reduced phosphorylation of pRb in PRKD2-silenced U87MG and GBM2 cells, pharmacological inhibition of CDK4/6 and subsequent hypophosphorylation of pRb arrest proliferation of glioma xenografts.45
In addition to altered phosphorylation patterns, PRKD2 silencing decreased mRNA and/or protein expression of G1/S cyclins A2 and D1, CDK2, pRb, and E2F1 in p53wt cells. Transcription and stability of cyclin D1 protein depends on Ras/MAPK and PI3K/AKT signaling.40 AKT-mediated phosphorylation of GSK3β was shown to inhibit nuclear export and proteasomal degradation of cyclin D1.40 Along this line, it is important to note that PRKD1 is a regulator of GSK3β activity in endothelial cells,46 indicating that PRKDs could be directly involved in cyclin stabilization. Dynamic control of CDK2 levels during cell cycle is mediated by the ubiquitin-proteasome.47 The transcription of E2F genes is under control of AP-1 (which is a MAPK target) in breast cancer cells.48 In turn, E2F is a transcriptional regulator of pRb expression, as indicated by direct binding of E2F1 to the pRb promotor in T98G glioma cells.49 In PRKD2-silenced U87MG and GBM2 cells, we observed decreased CDC25A mRNA levels. CDC25A phosphatase promotes entry into the S phase by dephosphorylating CDK2 at Thr14/15. In line with our findings, p53-dependent downregulation of CDC25 on mRNA and protein level was demonstrated.50
The signaling events inducing cellular senescence in PRKD2-silenced p53mut cells (with the exception of reduced AKT activation) are less clear-cut. Nevertheless a growing body of evidence indicates that cancer cells lacking functional p53 and pRb retain the capacity for therapy-induced senescence,8 as observed here. GM133 cells are characterized by a loss of function mutation in p53 and defective p16 protein expression,19 while the primary Gli25 culture is characterized by a 30 bp deletion in exon 5 (Fig. S3). PRKD2 silencing in these cells induced a senescence-like phenotype (though less pronounced as that observed for the p53wt cells) independent of p53 and p16/pRb but with concomitant upregulation of p15 and p27 (Fig. 4 and S10). Downregulation of Skp2, the regulator ubiquitin ligase of p27, was described as eliciting p53-independent cellular senescence.51 In addition to pRb, pRb family member p107 has been reported to interact with Skp252 and induce pRb-independent cellular senescence, albeit in association with p53-mediated stabilization of p27.53 Hence, the exact pathways that facilitate p53-independent senescence in PRKD2-silenced GM133 cells remain elusive.
In conclusion, data obtained during the present study (summarized in Fig. 6) demonstrate that silencing of PRKD2 induces glioma cell senescence via p53-dependent and p53-independent pathways, indicating that PRKD2 could represent a pharmacological target for therapy-induced senescence. Our finding that PRKD2 silencing reduces AKT phosphorylation independently of the cellular p53 status makes it reasonable to assume that this is the unifying principle of these observations.
Supplementary Material
Funding
Financial support was provided by the Austrian Research Promotion Agency (FFG; grant No. Bridge P820107), the Austrian Science Fund (FWF; SFB LIPOTOX-F3007 and DK MOLIN-W1241), the Austrian Nationalbank (Anniversary Fund, project number 14534) and the National Institutes of Neurological Disease and Stroke (NS73831 to PSM).
Supplementary Material
Acknowledgments
We thank Helga Reicher, Doris Treier, Stephanie Müller, Andrea Randig, and Marina Koch for their expert technical assistance.
Conflict of interest statement. None declared.
References
- 1.Johnson DR, O'Neill BP. Glioblastoma survival in the United States before and during the temozolomide era. J Neurooncol. 2012;107(2):359–364. doi: 10.1007/s11060-011-0749-4. [DOI] [PubMed] [Google Scholar]
- 2.Harada K, Nishizaki T, Ozaki S, Kubota H, Ito H, Sasaki K. Intratumoral cytogenetic heterogeneity detected by comparative genomic hybridization and laser scanning cytometry in human gliomas. Cancer Res. 1998;58(20):4694–4700. [PubMed] [Google Scholar]
- 3.Kim Y, Kim E, Wu Q, et al. Platelet-derived growth factor receptors differentially inform intertumoral and intratumoral heterogeneity. Genes Dev. 2012;26(11):1247–1262. doi: 10.1101/gad.193565.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dunn GP, Rinne ML, Wykosky J, et al. Emerging insights into the molecular and cellular basis of glioblastoma. Genes Dev. 2012;26(8):756–784. doi: 10.1101/gad.187922.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuilman T, Michaloglou C, Mooi WJ, Peeper DS. The essence of senescence. Genes Dev. 2010;24(22):2463–2479. doi: 10.1101/gad.1971610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nardella C, Clohessy JG, Alimonti A, Pandolfi PP. Pro-senescence therapy for cancer treatment. Nat Rev Cancer. 2011;11(7):503–511. doi: 10.1038/nrc3057. [DOI] [PubMed] [Google Scholar]
- 7.Acosta JC, Gil J. Senescence: a new weapon for cancer therapy. Trends Cell Biol. 2012;22(4):211–219. doi: 10.1016/j.tcb.2011.11.006. [DOI] [PubMed] [Google Scholar]
- 8.Ewald JA, Desotelle JA, Wilding G, Jarrard DF. Therapy-induced senescence in cancer. J Natl Cancer Inst. 2010;102(20):1536–1546. doi: 10.1093/jnci/djq364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.LaValle CR, George KM, Sharlow ER, Lazo JS, Wipf P, Wang QJ. Protein kinase D as a potential new target for cancer therapy. Biochim Biophys Acta. 2010;1806(2):183–192. doi: 10.1016/j.bbcan.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fu Y, Rubin CS. Protein kinase D: coupling extracellular stimuli to the regulation of cell physiology. EMBO Rep. 2011;12(8):785–796. doi: 10.1038/embor.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen J, Deng F, Singh SV, Wang QJ. Protein kinase D3 (PKD3) contributes to prostate cancer cell growth and survival through a PKCepsilon/PKD3 pathway downstream of Akt and ERK 1/2. Cancer Res. 2008;68(10):3844–3853. doi: 10.1158/0008-5472.CAN-07-5156. [DOI] [PubMed] [Google Scholar]
- 12.MacKeigan JP, Murphy LO, Blenis J. Sensitized RNAi screen of human kinases and phosphatases identifies new regulators of apoptosis and chemoresistance. Nat Cell Biol. 2005;7(6):591–600. doi: 10.1038/ncb1258. [DOI] [PubMed] [Google Scholar]
- 13.Shabelnik MY, Kovalevska LM, Yurchenko MY, Shlapatska LM, Rzepetsky Y, Sidorenko SP. Differential expression of PKD1 and PKD2 in gastric cancer and analysis of PKD1 and PKD2 function in the model system. Exp Oncol. 2011;33(4):206–211. [PubMed] [Google Scholar]
- 14.Giroux V, Iovanna J, Dagorn JC. Probing the human kinome for kinases involved in pancreatic cancer cell survival and gemcitabine resistance. FASEB J. 2006;20(12):1982–1991. doi: 10.1096/fj.06-6239com. [DOI] [PubMed] [Google Scholar]
- 15.Azoitei N, Pusapati GV, Kleger A, et al. Protein kinase D2 is a crucial regulator of tumour cell-endothelial cell communication in gastrointestinal tumours. Gut. 2010;59(10):1316–1330. doi: 10.1136/gut.2009.206813. [DOI] [PubMed] [Google Scholar]
- 16.Zou Z, Zeng F, Xu W, et al. PKD2 and PKD3 promote prostate cancer cell invasion by modulating NF-kappaB- and HDAC1-mediated expression and activation of uPA. J Cell Sci. 2012;125(Pt 20):4800–4811. doi: 10.1242/jcs.106542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Azoitei N, Kleger A, Schoo N, et al. Protein kinase D2 is a novel regulator of glioblastoma growth and tumor formation. Neuro Oncol. 2011;13(7):710–724. doi: 10.1093/neuonc/nor084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bernhart E, Damm S, Wintersperger A, et al. Protein kinase D2 regulates migration and invasion of U87MG glioblastoma cells in vitro. Exp Cell Res. 2013 doi: 10.1016/j.yexcr.2013.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y, Zhu S, Cloughesy TF, Liau LM, Mischel PS. p53 disruption profoundly alters the response of human glioblastoma cells to DNA topoisomerase I inhibition. Oncogene. 2004;23(6):1283–1290. doi: 10.1038/sj.onc.1207244. [DOI] [PubMed] [Google Scholar]
- 20.Dimri GP, Lee X, Basile G, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci U S A. 1995;92(20):9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sigoillot FD, King RW. Vigilance and validation: Keys to success in RNAi screening. ACS Chem Biol. 2011;6(1):47–60. doi: 10.1021/cb100358f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harikumar KB, Kunnumakkara AB, Ochi N, et al. A novel small-molecule inhibitor of protein kinase D blocks pancreatic cancer growth in vitro and in vivo. Mol Cancer Ther. 2010;9(5):1136–1146. doi: 10.1158/1535-7163.MCT-09-1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krakstad C, Chekenya M. Survival signalling and apoptosis resistance in glioblastomas: opportunities for targeted therapeutics. Mol Cancer. 2010;9:135. doi: 10.1186/1476-4598-9-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Furnari FB, Lin H, Huang HS, Cavenee WK. Growth suppression of glioma cells by PTEN requires a functional phosphatase catalytic domain. Proc Natl Acad Sci U S A. 1997;94(23):12479–12484. doi: 10.1073/pnas.94.23.12479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gursel DB, Connell-Albert YS, Tuskan RG, et al. Control of proliferation in astrocytoma cells by the receptor tyrosine kinase/PI3 K/AKT signaling axis and the use of PI-103 and TCN as potential anti-astrocytoma therapies. Neuro Oncol. 2011;13(6):610–621. doi: 10.1093/neuonc/nor035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clark MJ, Homer N, O'Connor BD, et al. U87MG decoded: the genomic sequence of a cytogenetically aberrant human cancer cell line. PLoS Genet. 2010;6(1):e1000832. doi: 10.1371/journal.pgen.1000832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Henley SA, Dick FA. The retinoblastoma family of proteins and their regulatory functions in the mammalian cell division cycle. Cell Div. 2012;7(1):10. doi: 10.1186/1747-1028-7-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vigo E, Muller H, Prosperini E, et al. CDC25A phosphatase is a target of E2F and is required for efficient E2F-induced S phase. Mol Cell Biol. 1999;19(9):6379–6395. doi: 10.1128/mcb.19.9.6379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee JJ, Kim BC, Park MJ, et al. PTEN status switches cell fate between premature senescence and apoptosis in glioma exposed to ionizing radiation. Cell Death Differ. 2011;18(4):666–677. doi: 10.1038/cdd.2010.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou BP, Liao Y, Xia W, Zou Y, Spohn B, Hung MC. HER-2/neu induces p53 ubiquitination via Akt-mediated MDM2 phosphorylation. Nat Cell Biol. 2001;3(11):973–982. doi: 10.1038/ncb1101-973. [DOI] [PubMed] [Google Scholar]
- 31.Majumder PK, Grisanzio C, O'Connell F, et al. A prostatic intraepithelial neoplasia-dependent p27 Kip1 checkpoint induces senescence and inhibits cell proliferation and cancer progression. Cancer Cell. 2008;14(2):146–155. doi: 10.1016/j.ccr.2008.06.00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chu IM, Hengst L, Slingerland JM. The Cdk inhibitor p27 in human cancer: prognostic potential and relevance to anticancer therapy. Nature reviews. Cancer. 2008;8(4):253–267. doi: 10.1038/nrc2347. [DOI] [PubMed] [Google Scholar]
- 33.Wang H, Xu Y, Fang Z, Chen S, Balk SP, Yuan X. Doxycycline regulated induction of AKT in murine prostate drives proliferation independently of p27 cyclin dependent kinase inhibitor downregulation. PLoS One. 2012;7(7):e41330. doi: 10.1371/journal.pone.0041330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harley CB. Telomerase and cancer therapeutics. Nat Rev Cancer. 2008;8(3):167–179. doi: 10.1038/nrc2275. [DOI] [PubMed] [Google Scholar]
- 35.Gladych M, Wojtyla A, Rubis B. Human telomerase expression regulation. Biochem Cell Biol. 2011;89(4):359–376. doi: 10.1139/o11-037. [DOI] [PubMed] [Google Scholar]
- 36.Cancer Genome Atlas Research N. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu N, Lao Y, Zhang Y, Gillespie DA. Akt: a double-edged sword in cell proliferation and genome stability. J Oncol. 2012;2012:951724. doi: 10.1155/2012/951724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang L, Dan HC, Sun M, et al. Akt/protein kinase B signaling inhibitor-2, a selective small molecule inhibitor of Akt signaling with antitumor activity in cancer cells overexpressing Akt. Cancer Res. 2004;64(13):4394–4399. doi: 10.1158/0008-5472.CAN-04-0343. [DOI] [PubMed] [Google Scholar]
- 39.Meyer N, Penn LZ. Reflecting on 25 years with MYC. Nat Rev Cancer. 2008;8(12):976–990. doi: 10.1038/nrc2231. [DOI] [PubMed] [Google Scholar]
- 40.Kim JK, Diehl JA. Nuclear cyclin D1: an oncogenic driver in human cancer. J Cell Physiol. 2009;220(2):292–296. doi: 10.1002/jcp.21791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee RJ, Albanese C, Stenger RJ, et al. pp60(v-src) induction of cyclin D1 requires collaborative interactions between the extracellular signal-regulated kinase, p38, and Jun kinase pathways. A role for cAMP response element-binding protein and activating transcription factor-2 in pp60(v-src) signaling in breast cancer cells. J Biol Chem. 1999;274(11):7341–7350. doi: 10.1074/jbc.274.11.7341. [DOI] [PubMed] [Google Scholar]
- 42.Wang J, Wang H, Li Z, et al. c-Myc is required for maintenance of glioma cancer stem cells. PLoS One. 2008;3(11):e3769. doi: 10.1371/journal.pone.0003769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hydbring P, Bahram F, Su Y, et al. Phosphorylation by Cdk2 is required for Myc to repress Ras-induced senescence in cotransformation. Proc Natl Acad Sci U S A. 2010;107(1):58–63. doi: 10.1073/pnas.0900121106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hydbring P, Larsson LG. Tipping the balance: Cdk2 enables Myc to suppress senescence. Cancer Res. 2010;70(17):6687–6691. doi: 10.1158/0008-5472.CAN-10-1383. [DOI] [PubMed] [Google Scholar]
- 45.Michaud K, Solomon DA, Oermann E, et al. Pharmacologic inhibition of cyclin-dependent kinases 4 and 6 arrests the growth of glioblastoma multiforme intracranial xenografts. Cancer Res. 2010;70(8):3228–3238. doi: 10.1158/0008-5472.CAN-09-4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shin S, Wolgamott L, Yoon SO. Regulation of endothelial cell morphogenesis by the protein kinase D (PKD)/glycogen synthase kinase 3 (GSK3)beta pathway. Am J Physiol Cell Physiol. 2012;303(7):C743–756. doi: 10.1152/ajpcell.00442.2011. [DOI] [PubMed] [Google Scholar]
- 47.Fang Y, Zhou X, Lin M, et al. The ubiquitin-proteasome pathway plays essential roles in ATRA-induced leukemia cells G0/G1 phase arrest and transition into granulocytic differentiation. Cancer Biol Ther. 2010;10(11):1157–1167. doi: 10.4161/cbt.10.11.13556. [DOI] [PubMed] [Google Scholar]
- 48.Shen Q, Uray IP, Li Y, et al. The AP-1 transcription factor regulates breast cancer cell growth via cyclins and E2F factors. Oncogene. 2008;27(3):366–377. doi: 10.1038/sj.onc.1210643. [DOI] [PubMed] [Google Scholar]
- 49.Burkhart DL, Ngai LK, Roake CM, et al. Regulation of RB transcription in vivo by RB family members. Mol Cell Biol. 2010;30(7):1729–1745. doi: 10.1128/MCB.00952-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rother K, Kirschner R, Sanger K, Bohlig L, Mossner J, Engeland K. p53 downregulates expression of the G1/S cell cycle phosphatase Cdc25A. Oncogene. 2007;26(13):1949–1953. doi: 10.1038/sj.onc.1209989. [DOI] [PubMed] [Google Scholar]
- 51.Lin HK, Chen Z, Wang G, et al. Skp2 targeting suppresses tumorigenesis by Arf-p53-independent cellular senescence. Nature. 2010;464(7287):374–379. doi: 10.1038/nature08815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rodier G, Makris C, Coulombe P, et al. p107 inhibits G1 to S phase progression by down-regulating expression of the F-box protein Skp2. J Cell Biol. 2005;168(1):55–66. doi: 10.1083/jcb.200404146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lehmann BD, Brooks AM, Paine MS, Chappell WH, McCubrey JA, Terrian DM. Distinct roles for p107 and p130 in Rb-independent cellular senescence. Cell Cycle. 2008;7(9):1262–1268. doi: 10.4161/cc.7.9.5945. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.