Abstract
Background
We assessed the diagnostic accuracy of diffusion kurtosis imaging (DKI), dynamic susceptibility-weighted contrast-enhanced (DSC) MRI, and short echo time chemical shift imaging (CSI) for grading gliomas.
Methods
In this prospective study, 35 patients with cerebral gliomas underwent DKI, DSC, and CSI on a 3 T MR scanner. Diffusion parameters were mean diffusivity (MD), fractional anisotropy, and mean kurtosis (MK). Perfusion parameters were mean relative regional cerebral blood volume (rrCBV), mean relative regional cerebral blood flow (rrCBF), mean transit time, and relative decrease ratio (rDR). The diffusion and perfusion parameters along with 12 CSI metabolite ratios were compared among 22 high-grade gliomas and 14 low-grade gliomas (Mann–Whitney U-test, P < .05). Classification accuracy was determined with a linear discriminant analysis for each MR modality independently. Furthermore, the performance of a multimodal analysis is reported, using a decision-tree rule combining the statistically significant DKI, DSC-MRI, and CSI parameters with the lowest P-value. The proposed classifiers were validated on a set of subsequently acquired data from 19 clinical patients.
Results
Statistically significant differences among tumor grades were shown for MK, MD, mean rrCBV, mean rrCBF, rDR, lipids over total choline, lipids over creatine, sum of myo-inositol, and sum of creatine. DSC-MRI proved to be the modality with the best performance when comparing modalities individually, while the multimodal decision tree proved to be most accurate in predicting tumor grade, with a performance of 86%.
Conclusions
Combining information from DKI, DSC-MRI, and CSI increases diagnostic accuracy to differentiate low- from high-grade gliomas, possibly providing diagnosis for the individual patient.
Keywords: chemical shift imaging, diffusion kurtosis imaging, gliomas, perfusion, tumor grading
Adequate grading of gliomas presents many difficulties in clinical practice but is of capital importance because treatment regimens and prognosis depend on malignancy grade. The international classification published by the World Health Organization (WHO) based on histopathology of the resection specimen after neurosurgical intervention is currently recognized as the gold standard for the classification of gliomas.1 Although this procedure is invasive and subject to interreader variability and ∼10% sampling error, surgical intervention with subsequent histopathological examination is still considered the most adequate approach to determine the aggressiveness of gliomas.2
An imaging-based method for determining glioma grade is appealing due to its noninvasiveness and the possibility to cover larger areas of tissue in these often heterogeneous tumors, decreasing the chance of sampling errors. In contrast to conventional anatomical MRI, regarded as insufficient in determining glioma grade,3 more advanced and specialized MR techniques are of considerable interest, as various aspects of tumoral pathophysiology can be demonstrated.4–6 The WHO classification to grade gliomas is based on 5 histopathological criteria related to the degree of cellular aggressiveness: cellular density, nuclear atypia, mitosis, endothelial proliferation, and necrosis.1 These features find their counterparts in different advanced MR techniques. Endothelial proliferation and neovascularization can be mapped with perfusion-weighted MRI. Diffusion MRI reflects tissue organization and thereby can provide information on cellular density and necrosis.4,6,7
Several studies of advanced MR techniques to grade gliomas have been published with different setups and mixed results, indicating a widespread interest in the topic, with most of the reported results demonstrated on a group level.8–15
In order to find acceptance in clinical practice, prospective grading of gliomas should be performed on an individual patient level with sufficient accuracy. Moreover, combining different modalities has the potential to increase diagnostic accuracy, as the different advanced MR techniques yield complementary information.4
In this study, it was our aim to assess the separate diagnostic performances of diffusion kurtosis imaging (DKI), perfusion MRI using dynamic susceptibility-weighted contrast-enhanced (DSC) MRI, and short echo time chemical shift imaging (CSI) for grading gliomas and to examine whether a multimodal approach could be used to improve diagnostic power of the individual methods, leading to a diagnosis of glioma grade on an individual patient level.
Materials and Methods
Study Patient Population
The institutional human ethics review board approved this prospective study. Written informed consent was obtained from every patient before participation. Patients were recruited and scanned between October 2010 and August 2011. We examined 35 consecutive patients with suspicion of gliomas on conventional radiological imaging, prior to any treatment (12 females/23 males; age range: 22–78 y, median age: 55). None of the included patients had neurological disorders other than primary neoplasm. We enrolled 21 patients with high-grade glioma, of whom 18 had grade IV glioma (glioblastoma multiforme [GBM]) and 3 had grade III glioma. Thirteen patients with low-grade gliomas were recruited. Of these 13 patients, 3 were diagnosed with a diffuse fibrillary glioma grade II, 3 with a grade II pilocytic astrocytoma, 3 with a grade II oligodendroglioma, 1 with a grade II oligoastrocytoma, 1 with a grade II lesion with mixed findings of pilocytic astrocytoma, angiocentric glioma, and oligodendroglioma, and 2 with grade I oligodendroglioma. One patient had a grade II astrocytoma with focal progression to a grade III glioma. This resulted in 22 high-grade lesions and 14 low-grade lesions included in the study sample, with histopathological confirmation in all cases (2 biopsies, 33 total or near total resections in 35 patients). The lesions were classified according to grade using the 2007 WHO classification.1 The time between the MR examinations and surgery was maximally 19 days.
Validation Population
The results and the conclusions drawn from the study patient population were afterward evaluated through subsequent cases. To this purpose, a validation group of 19 new patients with histopathologically confirmed diagnoses (6 females/13 males; age range: 20–67 y, median age: 41) obtained in routine clinical settings was considered as a separate validation set. Ten patients presented with high-grade gliomas. Of these, 6 had GBM, 2 had diagnoses of grade III oligodendroglioma, and 2 had grade III astrocytoma. Nine patients were diagnosed with a low-grade glioma (3 oligodendrogliomas grade II, 3 oligoastrocytomas grade II, and 3 diffuse infiltrating astrocytomas).
Data Acquisition and Analysis
MRI was performed on a 3 T unit (Philips Achieva), using a body coil for transmission and an 8-channel head coil for signal reception.
Anatomical imaging
Acquired as high-resolution anatomical reference images were an axial spin echo T2-weighted MRI (repetition time [TR]/echo time [TE]: 3000/80 ms; slice/gap: 4/1 mm; turbo factor: 10; field of view [FOV]: 230 × 184 mm²; acquisition matrix: 400 × 300) and a T1-weighted 3D spoiled gradient echo scan (fast field echo, TR/TE: 9.7/4.6 ms; flip angle: 8°; turbo field echo factor: 180; acquisition voxel size: 0.98 × 0.98 × 1 mm³; 118 contiguous partitions; inversion time: 900 ms) after contrast administration.
Regions of interest (ROIs) were manually drawn around the solid part of the tumor, avoiding areas of necrosis or cystic components and the contralateral normal-appearing white matter (NAWM), using the medical image viewer MRIcro (http://www.sph.sc.edu/comd/rorden/mricro.html). ROIs comprising NAWM and non-contrast-enhancing tumor were delineated on the transverse T2-weighted images. Solid contrast-enhancing tumor was delineated on transversal 3-mm reconstructions from the 3D fast field echo sequences. This ROI delineation was copied to the DKI and DSC parameter maps after coregistration to the anatomical images.
Diffusion kurtosis imaging
DKI data were acquired according to a previously described protocol.16 An echo planar imaging (EPI) diffusion-weighted (DW) imaging sequence with a spin echo read-out was used to acquire the DKI data. Implemented b-values were 700, 1000, and 2800 s/mm², applied in 25, 40, and 75, respectively, uniformly distributed directions. Additionally, 10 images without diffusion sensitization were obtained. Other imaging parameters were kept constant throughout the DKI data acquisition sequences: TR/TE: 3200/90 ms; δ/Δ: 20/48.3 ms; FOV: 240 × 240 mm²; matrix: 96 × 96; 1 signal average acquired; section thickness/gap: 2.5/0 mm. The scan time was 17 min 29 s.
The DKI data were corrected for head motion and eddy current distortions by means of global affine transformations.17 The correction was followed by b-matrix rotation and DW signal intensity modulation, which are needed to correct for the severe voxel volume changes due to large eddy current distortions at high b-values.18 In each voxel, the diffusion tensor and the diffusion kurtosis tensor were estimated with a constrained nonlinear least squares estimator, which accounted for the Rician data statistics. A more detailed overview of the tensor estimation is given by Veraart et al.19 From the tensors were derived the diffusion and kurtosis parameters of fractional anisotropy, mean diffusivity (MD), and mean kurtosis (MK).16,20 A nonlinear registration of the parameter maps to the anatomical imaging data was performed to minimize the local misalignment between the EPI-distorted DKI data and the anatomical data on which the ROIs were manually positioned.21
Dynamic susceptibility-weighted contrast-enhanced imaging
Perfusion images were obtained using the DSC technique with a gradient echo-EPI sequence (TR/TE: 1350/30 ms; section thickness/gap: 3/0 mm; dynamic scans: 60; FOV: 200 × 200 mm²; matrix: 112 × 109; scan time: 1 min 26 s). EPI data were acquired during the first pass following a rapid injection of a 0.1 mmol/kg body weight bolus of meglumine gadoterate (Dotarem, Guerbet) via a mechanical pump at a rate of 4 mL/s, followed by a 20-mL bolus of saline.
DSC data were analyzed using DPTools (www.fmritools.org). Firstly, spatial realignment and temporal slice time correction were performed. The arterial input function was defined in the tissue near the middle cerebral artery. Regional cerebral blood flow (rCBF) was calculated according to the indicator dilution theory and singular value decomposition.22 After baseline correction, which corrects for the leakage of contrast in the extravascular extracellular space in case of a disrupted blood–brain barrier,23–26 concentration-time curves of the first pass of the bolus were calculated. Subsequently, these curves were fitted with a gamma variate function in order to derive regional cerebral blood volume (rCBV). By using the central volume theorem, the mean transit time (MTT) was calculated as the ratio of rCBV to rCBF.27 The signal decrease ratio (DR) was calculated as described by Ducreux et al,28 with DR = S(TTP)/S0, where S(TTP) is the signal value at the peak time, and S0 is the baseline signal value. The obtained parameter maps were coregistered with the anatomical data using in-house software based on mutual information rigid coregistration, and the ROIs of tumor and NAWM were afterward copied to the parameter maps. The mean values of the considered perfusion parameter were retrieved in tumoral tissue and NAWM. We report relative rCBV (rrCBV), relative rCBF (rrCBF), and relative DR (rDR) of tumoral tissue by using the corresponding parameter values in the contralateral NAWM as internal reference.
Chemical shift imaging
A 2D-CSI protocol was used as previously described.29 In brief, a point-resolved spectroscopy sequence was used as the volume selection technique with a bandwidth of 1.3 kHz for the conventional slice selective pulses; TR/TE: 2000/35 ms; FOV: 160 × 160 mm²; volume of interest (VOI): 80 × 80 mm²; section thickness: 10 mm; acquisition voxel size: 10 × 10 mm²; reconstruction voxel size: 5 × 5 mm²; receiver bandwidth: 2000 Hz; samples: 2048; number of signal averages: 1; water suppression method: MOIST (multiple optimizations insensitive suppression train)30; first- and second-order pencil beam shimming; parallel imaging: sensitivity encoding with reduction factors of 2 (left-right) and 1.8 (anterior-posterior); scan time: 3 min 30 s. Automated prescanning optimized the shim in order to yield a peak width consistently <20 Hz full-width half-maximum (FWHM). The slice was positioned in the center of the tumor. The VOI was adjusted to contain pathological tissue and NAWM from the contralateral hemisphere.
Spectra were processed using the MatLab 2010b environment (MathWorks) with SPID (Simulation Package based on In vitro Databases) graphical user interface (http://homes.esat.kuleuven.be/~biomed/software.php#SpidGUI) as described in detail by Van Cauter et al.29 As the quantification method, we used AQSES-MRSI (Accurate Quantification of Short-echo Time Magnetic Resonance Spectroscopic Signals–MR Spectroscopic Imaging)31 for the following metabolites: N-acetyl aspartate (NAA), glutamine (Gln), glutamate (Glu), total creatine (Cre), phosphorylcholine (PCh), glycerophosphorylcholine (GPCh), myo-inositol (Myo), and macromolecules/lipids (Lips) at 0.9 and 1.3 ppm, referred to as Lip 1 and Lip 2, respectively. Glu + Gln and PCh + GPCh are reported as Glx and tCho (total choline), respectively. For each metabolite, AQSES-MRSI reports the error estimates as Cramèr-Rao lower bounds (CRLBs). After quantification, 10 representative voxels were selected in the solid tumoral area and the NAWM, based on the CRLBs and spectral quality assessment as recommended by Kreis (FWHM of metabolites <0.07–0.1 ppm, no unexplained features in the residuals, no doubled peaks or evidence for movement artifacts, symmetric lineshape, no outer volume ghosts or other artifacts present).32 Considered sufficient were CRLB <20% for tCho, NAA, Glx, Cre, and Lips and CRLB <50% for Myo.
From these representative voxels, the mean metabolite ratios as proposed by Kounelakis et al8 were calculated: NAA/tCho, NAA/sum, tCho/sum, NAA/Cre, Lips/tCho, tCho/Cre, Myo/sum, Cre/sum, Lips/Cre, and Glx/sum (10 parameters). The sum represents the sum of the peak areas of all quantified metabolites. Also, normalized tCho/NAA and tCho/Cre were calculated, leading thus to a total of 12 CSI parameters. Normalized metabolite ratios were calculated by the metabolite amplitude ratio between tumor tissue and NAWM for each metabolite.
Statistical Analysis
Parameter values obtained from the study population (35 patients) were compared among high- and low-grade gliomas using the Mann-Whitney-Wilcoxon test with Bonferroni correction, taking into account the number of the considered parameters (DKI: 3, DSC: 4, CSI: 12). The median values and interquartile ranges were calculated for the group of low- and high-grade gliomas for each parameter. The parameters with P < .05 were selected as statistically significant for differentiating between high- and low-grade gliomas and were considered for further analysis.
We applied a linear discriminant analysis separately on the selected (ie, statistically significant) DKI, DSC-MRI, and CSI parameters. The classifier and its performance, estimated on the study patient population (35 cases), were estimated using 10-fold cross-validation and averaged over 100 different random partitions of the patient population (later referred to as “runs”). Then, the generalization ability of the classifier was determined by building a linear discriminant analysis classifier using all 35 cases within the study patient population and testing its performance on the subsequent validation population set (19 cases).
To combine the DKI, DSC-MRI, and CSI information, we proposed a decision-tree rule. At each decision level, a different modality was considered by exploiting for each modality only the statistically significant parameter with the lowest P-value. The order in which the modalities were used was determined by the results of the linear discriminant analysis. The modality with the highest diagnostic accuracy was used in the first step, the modality with the second highest accuracy in the second step, and the modality with the lowest accuracy in the last step. The receiver operating characteristic (ROC) curve was used to determine a low-confidence interval for the considered parameter. The low-confidence interval, where poor separation between high- and low-grade gliomas was achieved, was defined by the 2 cutoff values with maximum sensitivity and specificity, respectively. If at any of these decision-tree levels the considered parameter value was within the low-confidence intervals, no prediction was archived and the next level was performed. If the specific parameter in a considered case had a value outside the low-confidence interval, the tumor grade was predicted and a further step was not needed. The performance of the proposed decision tree was estimated both on the study patient population (35 cases) (using leave-one-out cross-validation techniques and reporting the results over 100 runs) and on the subsequent validation population (19 cases).
Finally, in order to illustrate possible correlations among the parameters considered in the decision tree, we performed linear regression analyses between pairs of parameters (DKI parameter vs DSC-MRI parameter, CSI parameter vs DSC-MRI parameter, and CSI parameter vs DKI parameter) for all patients where both parameters were available.
Statistical analysis was performed in SPSS 19.0 for Windows (IBM) and MatLab Statistics Toolbox (MathWorks).
Results
Data Quality Assessment in the Study Patient Population
One of the 35 DKI datasets showed extensive movement artifacts and was therefore discarded. DSC-MRI failed in 3 patients because of problems of insufficient pressure buildup of the mechanical pump. Two additional DSC-MRI datasets were rejected due to extensive movement artifacts, not adequately corrected for as mentioned in the Material and Methods section. Four CSI datasets did not meet the quality criteria as recommended by Kreis.32 One dataset showed doubled peaks suggestive of movement artifacts, and in 3 datasets the FWHM of the metabolites was higher than 0.1 ppm. These data were therefore not used for further analysis. At least 2 of the 3 advanced MR modalities were of sufficient quality in each patient.
Data Quality Assessment in the Validation Population
The data of 19 patients in the validation group were obtained in a routine clinical setting. In 17 of 19 patients, DSC-MRI and CSI data were of good quality. DKI data were acquired in only 3 patients, as this sequence has been only recently implemented in the clinical setting in our hospital.
Comparison of DKI, DSC-MRI, and CSI for Grading of Gliomas
Univariate analysis
Median values and interquartile ranges for the 3 diffusion parameters, the 4 DSC parameters (Table 1), and the 12 CSI parameters (Table 2) are reported for the group of low- and high-grade gliomas.
Table 1.
Median diffusion and perfusion parameter values (interquartile ranges) in the differentiation of low-grade glioma (LGG) and high-grade glioma (HGG)
| Parameter | LGG | HGG | P | Optimal Cutoff |
|---|---|---|---|---|
| DKI | ||||
| n = 13 | n = 22 | |||
| MK | 0.48 (0.42–0.49) | 0.58 (0.50–0.58) | .001* | 0.5 |
| FA | 0.11 (0.09–0.13) | 0.13 (0.10–0.16) | .195 | |
| MD (10−3 mm²/s) | 1.64 (1.62–1.82) | 1.40 (1.28–1.62) | .003* | 1.56 |
| DSC imaging | ||||
| n = 12 | n = 19 | |||
| Mean rrCBV | 0.95 (0.76–1.24) | 2.44 (1.88–2.78) | .001* | 1.49 |
| Mean rrCBF | 0.94 (0.70–1.19) | 2.44 (1.89–3.05) | .001* | 1.45 |
| MTT, s | 75.37 (70–87.98) | 92.10 (66.38–104.37) | 1 | |
| rDR | 1.08 (0.88–1.17) | 1.51 (1.31–2.18) | .001* | 1.2 |
Abbreviation: FA, fractional anisotropy.
Optimal cutoff values to separate high- from low-grade gliomas in this study population are provided for the statistically significant parameters.
MD values in 10−3 mm²/s. MTT in sec. The remaining parameters are dimensionless.
*Indicates statistical significance (P < .05 Bonferroni corrected).
Table 2.
Median CSI parameter values (interquartile ranges) in the differentiation of low-grade glioma (LGG) and high-grade glioma (HGG)
| Parameter | LGG | HGG | P | Optimal Cutoff |
|---|---|---|---|---|
| CSI | ||||
| n = 14 | n = 18 | |||
| Norm tCho/Cre | 1.35 (1.14–1.40) | 1.49 (1.16–1.74) | .33 | |
| Norm tCho/NAA | 1.95 (1.55–2.27) | 1.95 (1.66–2.31) | 1 | |
| NAA/tCho | 1.29 (1.06–1.55) | 1.25 (1.14–1.63) | 1.58 | |
| NAA/sum | 0.13 (0.11–0.15) | 0.11 (0,10–0.12) | .16 | |
| tCho/sum | 0.11 (0.10–0.12) | 0.09 (0.08–0.10) | .13 | |
| NAA/Cre | 0.72 (0.58–0.64) | 0.83 (0.67–1.01) | .37 | |
| Lips/tCho | 1.62 (1.15–2.41) | 4.49 (2.84–5.96) | .002* | 2.16 |
| tCho/Cre | 0.58 (0.48–0.64) | 0.66 (0.56–0.76) | .15 | |
| Myo/sum | 0.11 (0.08–0.13) | 0.07 (0.05–0.08) | .02* | 0.10 |
| Cre/sum | 0.19 (0.17–0.20) | 0.14 (0.12–0.17) | .004* | 0.18 |
| Lips/Cre | 0.15 (0.12–0.22) | 0.35 (0.25–0.39) | .004* | 0.19 |
| Glx/sum | 0.29 (0.25–0.32) | 0.25 (0.22–0.29) | .42 | |
Optimal cutoff values to separate high- from low-grade gliomas in this study population are provided for the statistically significant parameters. All parameters are dimensionless.
*Indicates statistical significance (P < .05 Bonferroni corrected).
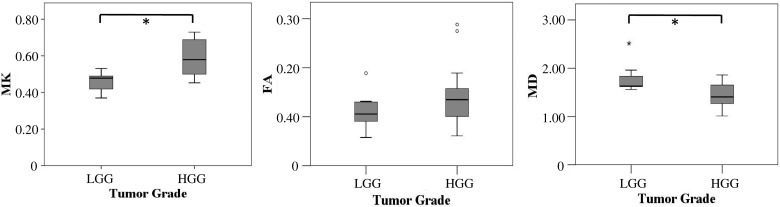
While MK was significantly higher, MD was significantly lower in high-grade compared with low-grade gliomas (P < .001 and P = .003, respectively). Fractional anisotropy did not significantly differ between high- and low-grade glioma (P = .195).
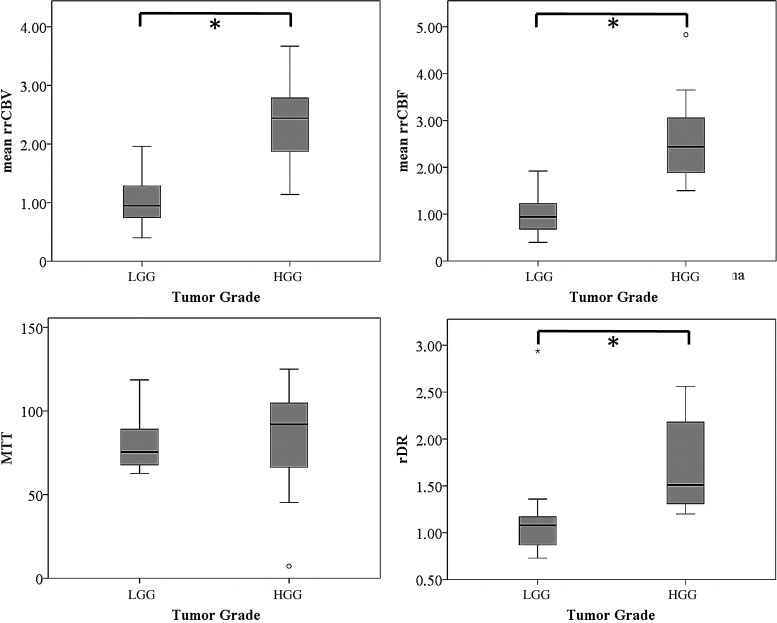
Mean rrCBV, mean rrCBF, and rDR were significantly higher in high-grade glioma than in low-grade glioma (P < .001 for all 3 parameters). MTT did not show statistically significant differences between tumor grades (P = 1).
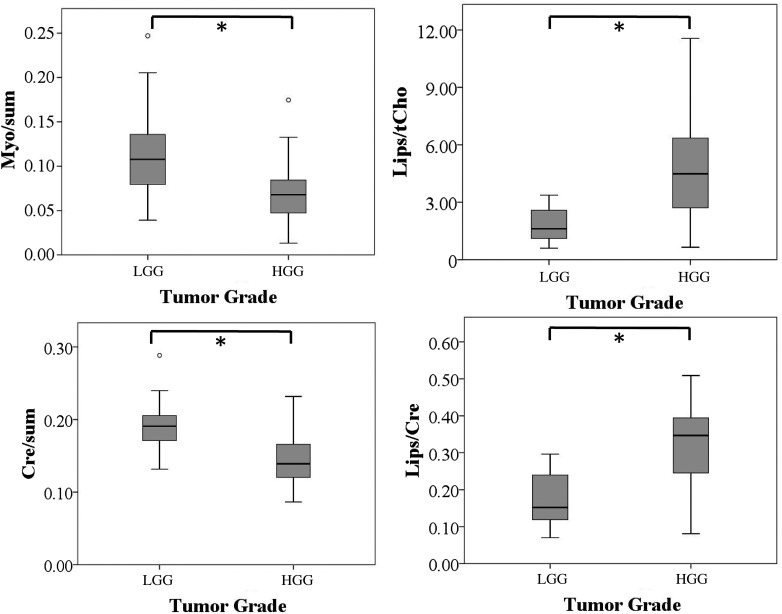
Lips/tCho, Lips/Cre, Myo/sum, and Cre/sum showed statistically significant differences between high- and low-grade glioma (P = .002, P = .004, P = .02, and P = .004, respectively). Lips/tCho and Lips/Cre increased with higher tumor grade, whereas Myo/sum and Cre/sum were lower in high-grade compared with low-grade gliomas. Normalized tCho/NAA, tCho/Cre, NAA/tCho, NAA/sum, tCho/sum, NAA/Cre, tCho/Cre, and Glx/sum did not significantly differ between tumor grades (P = .328, P = 1, P = 1, P = .16, P = .13, P = .37, P = .15, and P = .42, respectively).
Box plots with values for perfusion and diffusion parameters are shown in Figs. 1 and 2, respectively. For the sake of clarity, box plots of only the statistically significant CSI parameters are shown in Fig 3. Box plots with values for all CSI parameters are provided as a Supplementary Figure.
Fig. 1.
Box plots of MK, fractional anisotropy (FA), and MD against tumor grade. Asterisk (*) indicates statistically significant differences (P < .05, Bonferroni corrected) of the respective diffusion parameters with tumor grade. MD values in 10−3 mm²/s. The remaining parameters are dimensionless. Error bars indicate interquartile ranges.
Fig. 2.
Box plots of mean rrCBV, mean rrCBF, MTT, and rDR against tumor grade. Asterisk (*) indicates statistically significant differences (P < .05, Bonferroni corrected) of the respective perfusion parameter with tumor grade. MTT values in seconds. The remaining parameters are dimensionless. Error bars indicate interquartile ranges.
Fig. 3.
Box plots of the statistically significant CSI parameters against tumor grade. For the box plots of all considered CSI parameters, refer to Supplementary Fig. 1. Asterisk (*) indicates statistically significant differences (P < .05, Bonferroni corrected) of the respective CSI parameters with tumor grade. All metabolite ratios are dimensionless. Error bars indicate interquartile ranges.
Linear discriminant analysis
The classification accuracy, sensitivity, specificity, negative predictive value, and positive predictive value for differentiating low- from high-grade glioma are reported for the DKI, DSC, and CSI datasets.33,34 The cross-validation results on the study population are presented in Table 3. The mean/SD of these classification parameters over 100 runs were computed. When considering the statistically significant DSC parameters (mean rrCBV, mean rrCBF, and rDR), the performance reached 83%. Based on the DKI and CSI data, the classification performance for the considered parameters is lower.
Table 3.
Linear discriminant analysis performance for the separation between high- and low-grade gliomas
| Detection Value | DKI | DSC | CSI |
|---|---|---|---|
| MK and MD | Mean rrCBV, Mean rrCBF, and rDR | Lips/tCho, Myo/sum, Cre/sum, and Lips/Cre | |
| Diagnostic accuracy | 77% (±5%) | 83% (±6%) | 75% (±4%) |
| Sensitivity | 68% (±9%) | 78% (±9%) | 72% (±7%) |
| Specificity | 92% (±4%) | 91% (±5%) | 78% (±8%) |
| Positive predictive value | 93% (±4%) | 93% (±3%) | 81% (±8%) |
| Negative predictive value | 63% (±5%) | 73% (±11%) | 68% (±6%) |
Reported are the mean values and standard deviation of the accuracy, sensitivity, specificity, negative predictive value, and positive predictive value over 100 runs.
Linear discriminant analysis classification performance on the independent, subsequently acquired, validation population is in agreement with the cross-validation results. Again, DSC parameters show to be most discriminative in separating among different glioma grades. All the 17 patients for which DSC-MRI was acquired were correctly classified. For the CSI modality, 12 of 17 cases for which CSI was acquired were correctly classified. For the 3 cases in which DKI was acquired, 2 were correctly classified.
Combining DKI, DSC-MRI, and CSI to Grade Gliomas
DKI, DSC-MRI, or CSI data might be inconclusive or suffer from artifacts, patient movement, or failures of the protocol (eg, contrast agent injection). Therefore, on an individual patient basis, a combination of parameters would potentially benefit, in order to always provide a conclusive diagnosis.
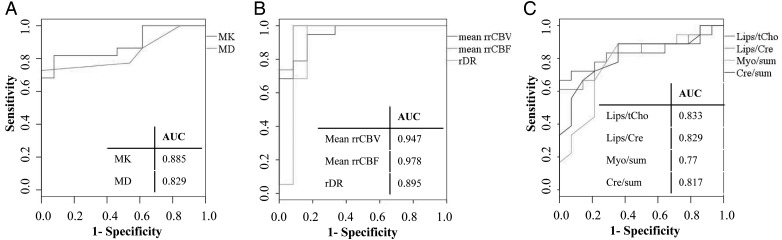
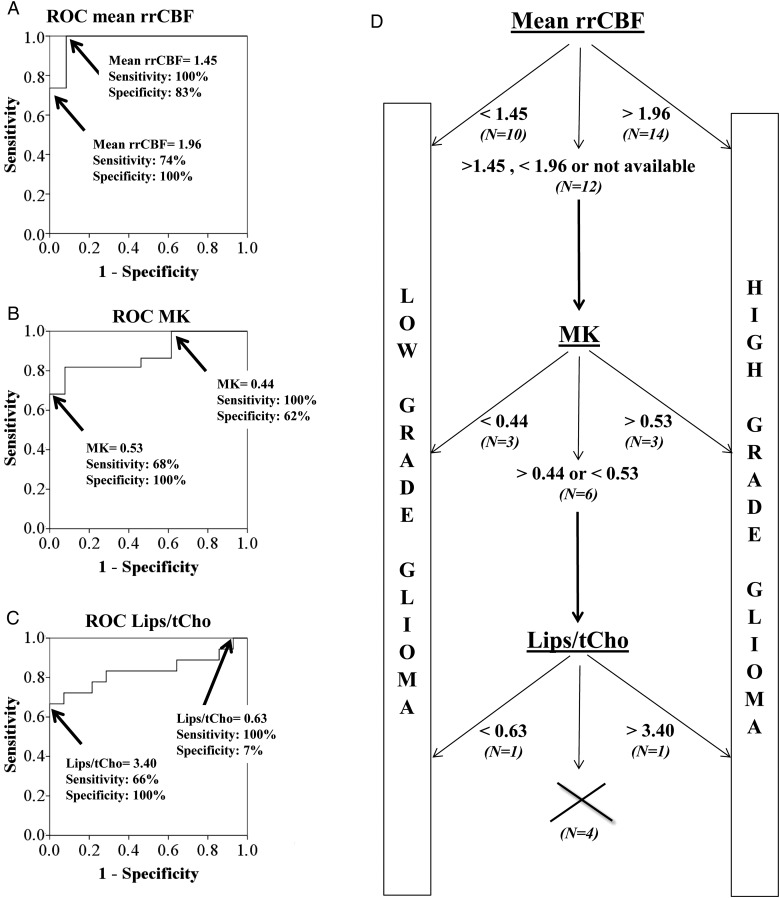
We proposed a decision-tree rule based on the ROC curves, determined for the statistically significant diffusion, perfusion, and MR spectroscopic parameters. These ROC curves with the corresponding area under the curve values are shown in Fig 4. We consider the parameter with the lowest P-value in each modality—mean rrCBF for DSC-MRI, MK for DKI, and Lips/tCho for CSI.
Fig. 4.
ROC curves and area under the curve values for MK and MD (panel A); mean rrCBV, mean rrCBF, and rDR (panel B); and Lips/tCho, Lips/Cre, Myo/sum, and Cre/sum (panel C) in solid tumor in order to differentiate between low- and high-grade glioma.
The performance averaged over 100 runs of the proposed decision tree was 86%; that is, 86% of the cases were correctly classified, 5% of the cases were misclassified, and for 9% of the cases diagnosis was not reached, since all the considered parameters were within the low-confidence interval. We also observed that 75% of the cases were classified at the first decision-tree level (based on mean rrCBF), out of which 74% of cases were correctly classified; 11% of the cases were classified in the second decision-tree level based on MK, out of which 9% were correctly classified; and 5% of the cases were classified in the third step based on Lips/tCho, out of which 3% were correctly classified. In Fig. 5 we demonstrate the decision tree by presenting the rules extracted from a randomly selected run within the cross-validation step. Based on the ROC analysis, the mean rrCBF low-confidence interval for the current dataset was 1.45–1.96. The low-confidence interval for MK was 0.44 to 0.53 and for Lips/tCho 0.63 to 3.40. The combination of a high mean rrCBF, a high MK, and a high Lips/tCho was indicative for high-grade glioma (Fig 6).
Fig. 5.
(A–C) ROC curves indicating the sensitivities and specificities of mean rrCBF-based, MK-based, and Lips/tCho-based differentiation between low- and high-grade gliomas, respectively. The 2 indicated points show the range where misclassifications can occur in this specific study population. The cutoff values indicated on the ROCs are an example obtained during a validation run randomly selected in the leave-one-out cross-validation. (D) Decision tree to distinguish low- from high-grade glioma in our study population based on the ranges of possible misclassification of mean rrCBF, MK, and Lips/tCho obtained for the leave-one-out cross-validation. The percentage of undecided cases after each decision step is indicated at each level. Nine percent of cases could not be classified using the proposed decision algorithm.
Fig. 6.
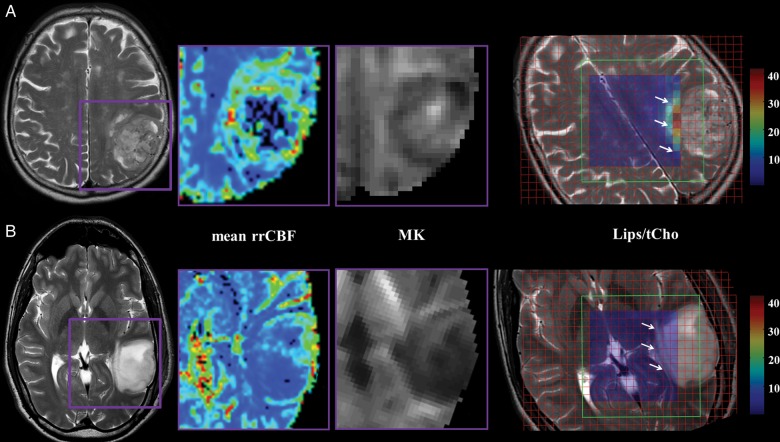
T2-weighted MRI, mean rrCBF, MK, and Lips/tCho maps of a 71-year-old female patient with GBM in the left parietal lobe (panel A) and a 35-year-old male patient with a grade II pilocytic astrocytoma in the left temporal lobe (panel B). The mean rrCBF and MK maps display the tumoral area in detail as indicated on the T2-weighted MRI with the purple box. Notice the high mean rrCBF and MK values in the high-grade glioma (top row) compared with the low-grade glioma (bottom row). The VOI of the CSI (green box) is superimposed on the T2-weighted images showing Lips/tCho ratios per voxel. The tumoral area is indicated with white arrows. Lips/tCho ratios are higher in high-grade glioma compared with low-grade glioma. Notice that only the center voxels are displayed in the color map for the sake of clarity, as the outer rows are affected by the chemical shift displacement error (CSDE). This CSDE can be defined as the difference in location of the center of the excitation or refocusing slices of 2 resonances with a different chemical shift, ie, Lips (0.9 and 1.3 ppm) and the carrier frequency of the water suppressed spectrum (2.2 ppm). Intensities of Lips/tCho are equally scaled in the parameter maps of the low- and high-grade glioma patient.
Furthermore, the proposed combination and cutoff values of imaging parameters of the decision-tree rule were validated on subsequent acquired data coming from 19 new patients. Sixteen of the 19 cases were correctly classified. In 3/19 cases, no final diagnosis could be made, and therefore the cases were classified as undecided. None of the cases was misclassified.
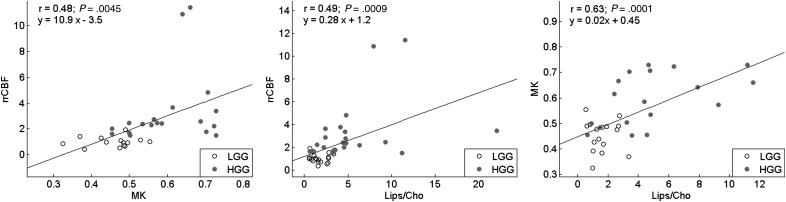
The linear regression analysis among the most significant parameters for each modality illustrates positive correlations among rrCBF, MK, and Lips/tCho (Fig 7). It can be noted that the low-grade glioma cases are consistently more tightly clustered toward lower values of the considered biomarkers, while the high-grade cases have larger and more scattered values for all parameters.
Fig. 7.
Relationships between MK and rrCBF (left), Lips/tCho and rrCBF (center), and Lips/tCho and MK (right). The fitted regression lines, Pearson correlation coefficients (r), P-values, and equation of the fitted line are shown. Low-grade glioma (LGG) cases are plotted as grey circles, while high-grade glioma (HGG) cases are plotted as black dots.
Discussion
Noninvasive preoperative grading of gliomas is important for treatment planning and prediction of prognosis. In this study, we proposed a combination of mean rrCBF, MK, and Lips/tCho presented in a decision tree, which showed a high diagnostic performance in this study for differentiating between low- and high-grade gliomas. With this decision tree, we provide an alternative when perfusion, commonly considered as the most accurate technique to grade gliomas, does not give a decisive answer or technically fails or when contrast administration is not possible. Moreover, new techniques such as DKI and less commonly used perfusion and spectroscopy parameters were further explored.
One of the risks of incorrect grading is inappropriate therapy. Diagnostic accuracy derived from biopsy and surgical procedures can be misleading, because these procedures provide information on only a fraction of the neoplastic tissue.14 With an overall complication rate of 6% after biopsy in patients with gliomas,35 the need for a more reliable noninvasive grading method is considerable. Moreover, prospective noninvasive grading is important for lesions that cannot be treated surgically, for lesions for which the functional risks of biopsy are high, or during follow-up of low-grade gliomas. With this study, we show that advanced MR techniques have the potential of differentiating between glioma grades. As each individual MR method has its merits and drawbacks, we have examined the possible synergy between the different MR-derived parameters. Moreover, we propose a decision tree where DSC, CSI, and DKI are combined, and we demonstrate the complementarity of different advanced MR techniques for grading gliomas, potentially providing a noninvasive alternative for glioma grading on an individual patient level.
However, in clinical practice, acquiring all sequences and performing all postprocessing steps is currently time-consuming and limited to specialized research centers. Therefore, we propose a decision-tree structure of combining information in such a way that the most discriminatory parameters are used first. If these are already conclusive, the remaining parameters do not even have to be acquired. In case of uncertainty, the other modalities can then be acquired in a second imaging session. This is a clinically much more intuitive diagnostic approach, as not all imaging and postprocessing need to be performed for each patient. In case there is no classification possible with the proposed decision-tree rule, a biopsy is still warranted.
Mean rrCBV and mean rrCBF were found to be very accurate predictors of tumor grade. This is in concordance with previously published results.11,36,37 Mean rrCBF has rarely been studied to determine tumor grade. However, some studies have demonstrated the usefulness of this parameter.11,38 Although the results of DSC-MR are promising, DSC-MR acquisition was not always satisfactorily performed because of inconsistent performance of the automated pressure injector and substantial movement artifacts related to bolus injections.
Regional hypoxia and hypoglycemia in glial metabolism stimulate the production of vascular endothelial growth factor (VEGF) and other angiogenesis promotors, which in turn increase local capillary permeability and promote endothelial proliferation, new vessel formation, and ingrowth of new vessels into the interstitial stroma.39 The greatest expression in VEGF among different brain tumors is seen in gliomas where levels are directly related to tumor grade. Moreover, endothelial proliferation is one of the 5 pathological hallmarks of the WHO classification of glioma grades.1 It is therefore not surprising that MR perfusion metrics are well related with tumor grade and histological findings of increased tumor vascularity.
In previous studies,13,15 diffusion kurtosis parameters were assigned as potential biomarkers for grading gliomas because better separation between high- and low-grade gliomas could be demonstrated with kurtosis parameters than with conventional diffusion MRI parameters. Similarly in this study, MK was the most accurate diffusion parameter to grade gliomas. DKI describes the deviation from unrestricted diffusion, which is determined by the complexity of the cytoarchitectonic environment.40,41 In that view, kurtosis can be regarded as a biomarker to map properties of microstructural tissue. Increased values of kurtosis parameters in high-grade glioma might reflect a higher degree of tissue complexity.42,43 Invasion in anatomical structures, increased cellularity, heterogeneity due to necrosis, hemorrhage, and endothelial proliferation are histopathological hallmarks of malignant gliomas, whereas low-grade gliomas are more homogeneous, with a lower cell density.1 Several studies have suggested that diffusion MR does not make fundamental contributions to the characterization of gliomas when multiple advanced MR techniques are available.4,14,44 However, in this study, we demonstrated that MK can be used as a valid contributing parameter, when for example perfusion MRI fails or when MR perfusion parameter values are inconclusive.
The diagnostic value of MR spectroscopy in grading gliomas has been widely studied, and the literature reports variable results using different MR spectroscopic methods.4,8,10,14,44 Most studies assigned a pivotal role to choline-containing metabolites in determining glioma grade.4,14,44 In addition, the presence of high amounts of lipids is indicative of necrosis, another hallmark for high-grade gliomas.45 Our results are only partly in concordance with previous results, as solely Lips/tCho showed a significant difference between low- and high-grade gliomas. We demonstrated a trend of increasing tCho with tumor grade. Apart from the grade of malignancy, the large spatial heterogeneities within the tumor tissue have a crucial role in the tCho signal. Increase of tCho in glial tumors is associated with cell proliferation and cell density and hence tumor grade. Due to the presence of necrosis in GBM, a decreased tCho can be expected in those regions.44 Therefore, in high-grade gliomas, the increase in tCho related to the hypercellular nature of more aggressive tumors is counterbalanced by the decrease of tCho induced by necrosis. The statistical significance of Lips/tCho most likely relies mainly on the contribution of lipids, associated with necrosis.
Furthermore, we demonstrated a potential role of Myo in grading gliomas, in concordance with Castillo et al46 and Kounelakis et al8 but contradicting the results of McKnight et al.10 It has been reported that Myo is a precursor of phosphatidylinositol, which is involved in a metabolic pathway to activate proteolytic enzymes such as matrix metalloproteases—key players in the events that underlie tumor dissemination. Tumor infiltration is a well-documented feature of high-grade glioma.46
We present a decision tree based on a study population of 35 patients. To validate the proposed decision-tree performance, cross-validation techniques were considered. Previous studies have shown that a good generalization is an important property of a classification algorithm because it provides information about the quality of the developed classifier or algorithm.47 Thus, in this study, the conclusions and results drawn on the study population were further validated on a new population set of 19 cases, which were subsequently acquired. We believe that such an evaluation over time provides the clinical community with robust results and can consolidate the confidence of clinicians in the potential applicability of the classifiers. Moreover, these results should be further explored in a larger, preferably multicenter study population, where advanced and automated algorithms can be tested for combining multiparametric data.
We did not administer a preloading dose of contrast agent before acquiring DSC-MRI, in order to minimize relaxivity effects in case of contrast leakage with a disrupted blood–brain barrier, which typically results in underestimation of rCBV values. We did account for this effect in postprocessing, where we performed a baseline correction. Hu et al25 stated that an optimal experimental approach for measuring rCBV in case of blood–brain barrier rupture consists of combining baseline correction methods with administration of a preloading dose. We only performed a correction in the postprocessing of DSC-MRI data, and therefore this can be regarded as a technical limitation of the study.23–25
In conclusion, this study demonstrates the complementarity of DSC-MRI, DKI, and CSI for grading gliomas. The most accurate parameters for determining glioma grade were mean rrCBF, MK, and Lips/tCho. The best performing MR modality, when considering the techniques separately, was DSC-MRI. However, a combination of the parameters of mean rrCBF, MK, and Lips/tCho could still provide a better differentiation between high- and low-grade gliomas, possibly providing diagnosis on the individual patient level.
Supplementary Material
Funding
S.V.H., D.M.S., and A.C.S. are supported by the Research Council of the KU Leuven (GOA MaNet, PFV/10/002 [OPTEC]), the Flemish government (FWO projects G.0427.10N [Integrated EEG-fMRI], G.0108.11 [Compressed Sensing], G.0869.12N [Tumor imaging]; IWT [innovation mandate 120213]; iMinds; and the Belgian Federal Science Policy Office IUAP P6/04 [DYSCO, 2007–2011]), and EU MC ITN TRANSACT 2012 (n°16679). This work was further supported by the Olivia Hendrickx Research Fund (www.olivia.be). Support was also obtained from the Herman Memorial Research Fund (www.hmrf.be), the James E. Kearney Memorial Fund (www.jekfoundation.org), and gifts from private families and service clubs. Additionally, grants were obtained from “Stichting tegen Kanker”, IWT (TBM projects), the Stem Cell Institute Leuven, the Emmanuel van der Schueren Fund, the International Union against Cancer, the Klinisch Onderzoeksfonds UZ Leuven, and the Fund for Scientific Research–Flanders (FWO-V). S.V.G. is senior clinical investigator at the FWO-V. S.S. is supported by grant no. G.048010N from Fonds Wetenschappelijk Onderzoek–Vlaanderen; by Stichting tegen Kanker; by grant no. IUAP EMF-B6772- p6/29 from Inter-University-Attraction- Pole; and by grant no. EF/05/014 from Excellentie-Financiering. U.H. is supported by the Flemish government (FWO projects G.0869.12N [Tumor imaging]) and by the Research Council of the KU Leuven (PF IMIR). For the remaining authors, no funding was declared.
Conflict of interest statement. None declared.
Supplementary Material
References
- 1.Louis DN, Ohgaki H, Wiestler OD, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114(2):97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McGirt MJ, Woodworth GF, Coon AL, et al. Independent predictors of morbidity after image-guided stereotactic brain biopsy: a risk assessment of 270 cases. J Neurosurg. 2005;102(5):897–901. doi: 10.3171/jns.2005.102.5.0897. [DOI] [PubMed] [Google Scholar]
- 3.Scott JN, Brasher PM, Sevick RJ, Rewcastle NB, Forsyth PA. How often are nonenhancing supratentorial gliomas malignant? A population study. Neurology. 2002;59(6):947–949. doi: 10.1212/wnl.59.6.947. [DOI] [PubMed] [Google Scholar]
- 4.Heiss WD, Raab P, Lanfermann H. Multimodality assessment of brain tumors and tumor recurrence. J Nucl Med. 2011;52(10):1585–1600. doi: 10.2967/jnumed.110.084210. [DOI] [PubMed] [Google Scholar]
- 5.Morita N, Harada M, Otsuka H, Melhem ER, Nishitani H. Clinical application of MR spectroscopy and imaging of brain tumor. Magn Reson Med Sci. 2010;9(4):167–175. doi: 10.2463/mrms.9.167. [DOI] [PubMed] [Google Scholar]
- 6.Lee SK. Diffusion tensor and perfusion imaging of brain tumors in high-field MR imaging. Neuroimaging Clin N Am. 2012;22(2):123–134, ix. doi: 10.1016/j.nic.2012.02.001. [DOI] [PubMed] [Google Scholar]
- 7.Cha S. Update on brain tumor imaging: from anatomy to physiology. AJNR Am J Neuroradiol. 2006;27(3):475–487. [PMC free article] [PubMed] [Google Scholar]
- 8.Kounelakis MG, Dimou IN, Zervakis ME, et al. Strengths and weaknesses of 1.5T and 3T MRS data in brain glioma classification. IEEE Trans Inf Technol Biomed. 2011;15(4):647–654. doi: 10.1109/TITB.2011.2131146. [DOI] [PubMed] [Google Scholar]
- 9.Slotboom J, Schaer R, Ozdoba C, et al. A novel method for analyzing DSCE-images with an application to tumor grading. Invest Radiol. 2008;43(12):843–853. doi: 10.1097/RLI.0b013e3181893605. [DOI] [PubMed] [Google Scholar]
- 10.McKnight TR, Smith KJ, Chu PW, et al. Choline metabolism, proliferation, and angiogenesis in nonenhancing grades 2 and 3 astrocytoma. J Magn Reson Imaging. 2011;33(4):808–816. doi: 10.1002/jmri.22517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Server A, Graff BA, Orheim TE, et al. Measurements of diagnostic examination performance and correlation analysis using microvascular leakage, cerebral blood volume, and blood flow derived from 3T dynamic susceptibility-weighted contrast-enhanced perfusion MR imaging in glial tumor grading. Neuroradiology. 2011;53(6):435–447. doi: 10.1007/s00234-010-0770-x. [DOI] [PubMed] [Google Scholar]
- 12.Stadlbauer A, Gruber S, Nimsky C, et al. Preoperative grading of gliomas by using metabolite quantification with high-spatial-resolution proton MR spectroscopic imaging. Radiology. 2006;238(3):958–969. doi: 10.1148/radiol.2382041896. [DOI] [PubMed] [Google Scholar]
- 13.Van Cauter S, Veraart J, Sijbers J, et al. Gliomas: diffusion kurtosis MR imaging in grading. Radiology. 2012;263(2):492–501. doi: 10.1148/radiol.12110927. [DOI] [PubMed] [Google Scholar]
- 14.Zonari P, Baraldi P, Crisi G. Multimodal MRI in the characterization of glial neoplasms: the combined role of single-voxel MR spectroscopy, diffusion imaging and echo-planar perfusion imaging. Neuro-radiology. 2007;49(10):795–803. doi: 10.1007/s00234-007-0253-x. [DOI] [PubMed] [Google Scholar]
- 15.Raab P, Hattingen E, Franz K, Zanella FE, Lanfermann H. Cerebral gliomas: diffusional kurtosis imaging analysis of microstructural differences. Radiology. 2010;254(3):876–881. doi: 10.1148/radiol.09090819. [DOI] [PubMed] [Google Scholar]
- 16.Poot DH, den Dekker AJ, Achten E, Verhoye M, Sijbers J. Optimal experimental design for diffusion kurtosis imaging. IEEE Trans Med Imaging. 2010;29(3):819–829. doi: 10.1109/TMI.2009.2037915. [DOI] [PubMed] [Google Scholar]
- 17.Tournier JD, Mori S, Leemans A. Diffusion tensor imaging and beyond. Magn Reson Med. 2011;65(6):1532–1556. doi: 10.1002/mrm.22924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leemans A, Jones DK. The B-matrix must be rotated when correcting for subject motion in DTI data. Magn Reson Med. 2009;61(6):1336–1349. doi: 10.1002/mrm.21890. [DOI] [PubMed] [Google Scholar]
- 19.Veraart J, Rajan J, Peeters RR, Leemans A, Sunaert S, Sijbers J. Comprehensive framework for accurate diffusion MRI parameter estimation. Magn Reson Med. 2012 doi: 10.1002/mrm.24529. [Published online ahead of print November 06, 2012]. doi:10.1002/mrm.24529. [DOI] [PubMed] [Google Scholar]
- 20.Le Bihan D, Mangin JF, Poupon C, et al. Diffusion tensor imaging: concepts and applications. J Magn Reson Imaging. 2001;13(4):534–546. doi: 10.1002/jmri.1076. [DOI] [PubMed] [Google Scholar]
- 21.Irfanoglu MO, Walker L, Sarlls J, Marenco S, Pierpaoli C. Effects of image distortions originating from susceptibility variations and concomitant fields on diffusion MRI tractography results. Neuroimage. 2012;61(1):275–288. doi: 10.1016/j.neuroimage.2012.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu O, Ostergaard L, Sorensen AG. Technical aspects of perfusion-weighted imaging. Neuroimaging Clin N Am. 2005;15(3):623–637, xi. doi: 10.1016/j.nic.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 23.Boxerman JL, Schmainda KM, Weisskoff RM. Relative cerebral blood volume maps corrected for contrast agent extravasation significantly correlate with glioma tumor grade, whereas uncorrected maps do not. AJNR Am J Neuroradiol. 2006;27(4):859–867. [PMC free article] [PubMed] [Google Scholar]
- 24.Donahue KM, Krouwer HG, Rand SD, et al. Utility of simultaneously acquired gradient-echo and spin-echo cerebral blood volume and morphology maps in brain tumor patients. Magn Reson Med. 2000;43(6):845–853. doi: 10.1002/1522-2594(200006)43:6<845::aid-mrm10>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 25.Hu LS, Baxter LC, Pinnaduwage DS, et al. Optimized preload leakage-correction methods to improve the diagnostic accuracy of dynamic susceptibility-weighted contrast-enhanced perfusion MR imaging in posttreatment gliomas. AJNR Am J Neuroradiol. 2010;31(1):40–48. doi: 10.3174/ajnr.A1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmainda KM, Rand SD, Joseph AM, et al. Characterization of a first-pass gradient-echo spin-echo method to predict brain tumor grade and angiogenesis. AJNR Am J Neuroradiol. 2004;25(9):1524–1532. [PMC free article] [PubMed] [Google Scholar]
- 27.Jackson A, Buckley DL, Parker GJM. Dynamic Contrast-Enhanced Magnetic Resonance Imaging in Oncology. New York: Springer; 2005. [Google Scholar]
- 28.Ducreux D, Petit-Lacour MC, Marsot-Dupuch K, Bittoun J, Lasjaunias P. MR perfusion imaging in a case of cerebral proliferative angiopathy. Eur Radiol. 2002;12(11):2717–2722. doi: 10.1007/s00330-001-1293-y. [DOI] [PubMed] [Google Scholar]
- 29.Van Cauter S, Sima DM, Luts J, et al. Reproducibility of rapid short echo time CSI at 3 Tesla for clinical applications. J Magn Reson Imaging. 2013;37(2):445–456. doi: 10.1002/jmri.23820. [DOI] [PubMed] [Google Scholar]
- 30.Ogg RJ, Kingsley PB, Taylor JS. WET, a T1- and B1-insensitive water-suppression method for in vivo localized 1H NMR spectroscopy. J Magn Reson B. 1994;104(1):1–10. doi: 10.1006/jmrb.1994.1048. [DOI] [PubMed] [Google Scholar]
- 31.Croitor Sava AR, Sima DM, Poullet J-B, Wright AJ, Heerschap A, Van Huffel S. Exploiting spatial information to estimate metabolite levels in two-dimensional MRSI of heterogeneous brain lesions. NMR Biomed. 2013;26(3):307–319. doi: 10.1002/nbm.1628. [DOI] [PubMed] [Google Scholar]
- 32.Kreis R. Issues of spectral quality in clinical 1H-magnetic resonance spectroscopy and a gallery of artifacts. NMR Biomed. 2004;17(6):361–381. doi: 10.1002/nbm.891. [DOI] [PubMed] [Google Scholar]
- 33.Nicoll D, McPhee SJ, Pignone M, Detmer WM, Chou TM. Pocket Guide to Diagnostic Tests. 3rd edn. New York: McGraw-Hill; 2001. [Google Scholar]
- 34.Theodoridis S, Koutroumbas K. Pattern Recognition. 3rd edn. Academic Press; 1999. p 69–112. [Google Scholar]
- 35.Kulkarni AV, Guha A, Lozano A, Bernstein M. Incidence of silent hemorrhage and delayed deterioration after stereotactic brain biopsy. J Neurosurg. 1998;89(1):31–35. doi: 10.3171/jns.1998.89.1.0031. [DOI] [PubMed] [Google Scholar]
- 36.Knopp EA, Cha S, Johnson G, et al. Glial neoplasms: dynamic contrast-enhanced T2*-weighted MR imaging. Radiology. 1999;211(3):791–798. doi: 10.1148/radiology.211.3.r99jn46791. [DOI] [PubMed] [Google Scholar]
- 37.Law M, Yang S, Babb JS, et al. Comparison of cerebral blood volume and vascular permeability from dynamic susceptibility contrast-enhanced perfusion MR imaging with glioma grade. AJNR Am J Neuroradiol. 2004;25(5):746–755. [PMC free article] [PubMed] [Google Scholar]
- 38.Weber MA, Zoubaa S, Schlieter M, et al. Diagnostic performance of spectroscopic and perfusion MRI for distinction of brain tumors. Neurology. 2006;66(12):1899–1906. doi: 10.1212/01.wnl.0000219767.49705.9c. [DOI] [PubMed] [Google Scholar]
- 39.Carmeliet P, Jain RK. Angiogenesis in cancer and other diseases. Nature. 2000;407(6801):249–257. doi: 10.1038/35025220. [DOI] [PubMed] [Google Scholar]
- 40.Jensen JH, Helpern JA. MRI quantification of non-Gaussian water diffusion by kurtosis analysis. NMR Biomed. 2010;23(7):698–710. doi: 10.1002/nbm.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu EX, Cheung MM. MR diffusion kurtosis imaging for neural tissue characterization. NMR Biomed. 2010;23(7):836–848. doi: 10.1002/nbm.1506. [DOI] [PubMed] [Google Scholar]
- 42.Fieremans E, Jensen JH, Helpern JA. White matter characterization with diffusional kurtosis imaging. Neuroimage. 2011;58(1):177–188. doi: 10.1016/j.neuroimage.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cheung MM, Hui ES, Chan KC, Helpern JA, Qi L, Wu EX. Does diffusion kurtosis imaging lead to better neural tissue characterization? A rodent brain maturation study. Neuroimage. 2009;45(2):386–392. doi: 10.1016/j.neuroimage.2008.12.018. [DOI] [PubMed] [Google Scholar]
- 44.Wagner M, Nafe R, Jurcoane A, et al. Heterogeneity in malignant gliomas: a magnetic resonance analysis of spatial distribution of metabolite changes and regional blood volume. J Neurooncol. 2011;103(3):663–672. doi: 10.1007/s11060-010-0443-y. [DOI] [PubMed] [Google Scholar]
- 45.Delikatny EJ, Chawla S, Leung DJ, Poptani H. MR-visible lipids and the tumor microenvironment. NMR Biomed. 2011;24(6):592–611. doi: 10.1002/nbm.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Castillo M, Smith JK, Kwock L. Correlation of myo-inositol levels and grading of cerebral astrocytomas. AJNR Am J Neuroradiol. 2000;21(9):1645–1649. [PMC free article] [PubMed] [Google Scholar]
- 47.Garcia-Gomez JM, Luts J, Julia-Sape M, et al. Multiproject-multicenter evaluation of automatic brain tumor classification by magnetic resonance spectroscopy. Magma. 2009;22(1):5–18. doi: 10.1007/s10334-008-0146-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.