Abstract
Background
Glial brain tumors cause considerable mortality and morbidity in children and adults. Innovative targets for therapy are needed to improve survival and reduce long-term sequelae. The aim of this study was to find a candidate tumor-promoting protein, abundantly expressed in tumor cells but not in normal brain tissues, as a potential target for therapy.
Methods
In silico proteomics and genomics, immunohistochemistry, and immunofluorescence microscopy validation were performed. RNA interference was used to ascertain the functional role of the overexpressed candidate target protein.
Results
In silico proteomics and genomics revealed pre-B-cell leukemia homeobox (PBX) interacting protein 1 (PBXIP1) overexpression in adult and childhood high-grade glioma and ependymoma compared with normal brain. PBXIP1 is a PBX-family interacting microtubule-binding protein with a putative role in migration and proliferation of cancer cells. Immunohistochemical studies in glial tumors validated PBXIP1 expression in astrocytoma and ependymoma but not in oligodendroglioma. RNAi-mediated PBXIP1-knockdown in glioblastoma cell lines strongly reduced proliferation and migration and induced morphological changes, indicating that PBXIP1 knockdown decreases glioma cell viability and motility through rearrangements of the actin cytoskeleton. Furthermore, expression of PBXIP1 was observed in radial glia and astrocytic progenitor cells in human fetal tissues, suggesting that PBXIP1 is an astroglial progenitor cell marker during human embryonic development.
Conclusion
PBXIP1 is a novel protein overexpressed in astrocytoma and ependymoma, involved in tumor cell proliferation and migration, that warrants further exploration as a novel therapeutic target in these tumors.
Keywords: astrocyte, astrocytoma, ependymoma, glioma, PBXIP1, progenitor
Malignant glial tumors are highly invasive brain tumors that carry a poor prognosis despite aggressive multimodal treatment strategies. The most frequently observed malignant glial tumors are high-grade gliomas (HGGs): WHO grade III anaplastic astrocytoma (AA), anaplastic oligodendroglioma, and WHO grade IV glioblastoma (GBM).1 Intracranial WHO grades II and III ependymomas are very rare in adults, but ependymomas are observed in up to 10% of all brain tumors in children.2,3 In adults, HGG is the most common group of primary malignant glial brain tumors, mainly comprising GBM that arises either as a primary (in 91% of cases) or as a secondary GBM after malignant transformation of a lower grade diffuse glioma (in 9% of cases). These secondary GBMs are more often observed in younger patients and usually harbor IDH1/2 mutations.4 In children, HGG is much more rare, with AA and GBM occurring in 2.8% and 1.9% of all childhood brain tumor cases, respectively.5 Standard therapy consists of surgery, aimed at maximal resection, followed by chemo-radiotherapy with temozolomide and maintenance temozolomide. Maximal surgery, eventually followed by focal radiotherapy, is the standard of care in both adult and childhood ependymoma. Chemotherapy has not proven to be of benefit in the treatment of ependymoma, warranting the need for the identification of new drug targets.2,3 Even with these multimodal regimens, prognosis still remains poor with 3-year overall survival (OS) rates of 16% (±4) in adult GBM, and 22% (±5) in childhood AA/GBM.6–8 Because of the poor outcome, especially in patients who are resistant to temozolomide, new therapies are warranted.
Gene expression profiling has significantly increased insight into glioma and ependymoma tumor biology and has already resulted in the identification of several putative therapeutic targets.9,10 Cancer Genome Atlas Research Network11,12 information on deregulated protein expression in tumor tissues might provide new clues on tumor cell behavior in its microenvironment, and this might allow for the identification of proteins overexpressed in tumor cells as compared with the normal microenvironment. To this purpose, we investigated which proteins are differentially overexpressed in malignant glial tumors compared with normal brain tissues using the Human Protein Atlas (HPA), a publicly available antibody-based proteomics platform.13,14 We identified pre-B-cell leukemia homeobox (PBX) interacting protein 1 (PBXIP1), also known as hematopoietic PBX-interacting protein (HPIP), as a protein overexpressed in astrocytoma, GBM, and ependymoma tissues, showed it to be a novel astrocyte progenitor cell-specific marker during human brain development, and provided evidence that it drives GBM cell proliferation and migration.
Materials and Methods
In Silico Proteomics and Genomics
The HPA,13,14 version 9.0 (November 11, 2011) was used to identify proteins that are differentially expressed between malignant glioma samples and normal human cortex. The following query was employed: ‘cancer_expression: malignant glioma: >80% strong, AND, normal_expression: cerebral cortex - glial cells: weak, OR, normal_expression: cerebral cortex - glial cells: negative, AND, normal_expression: cerebral cortex - neuronal cells: weak, OR, normal_expression: cerebral cortex - neuronal cells: negative.’ A total of 11 proteins were identified that fulfilled these criteria. The results of this search are depicted in alphabetical order in Table 1.
Table 1.
Differentially expressed proteins in malignant glioma (in alphabetical order)
| Protein | NCBI Gene Number | Description |
|---|---|---|
| C17orf66 | 256957 | chromosome 17 open reading frame 66 |
| FBXO18 | 84893 | F-box protein, helicase, 18 |
| JAM3 | 83700 | junctional adhesion molecule 3 |
| OCRL | 4952 | oculocerebrorenal syndrome of Lowe |
| OLIG2 | 10215 | oligodendrocyte lineage transcription factor 2 |
| PBXIP1 | 57326 | pre-B-cell leukemia homeobox interacting protein 1 |
| SQSTM1 | 8878 | sequestosome 1 |
| SYF2 | 25949 | SYF2 homolog, RNA splicing factor (S. cerevisiae) |
| SYT5 | 6861 | synaptotagmin V |
| ZCCHC16 | 340595 | zinc finger, CCHC domain containing 16 |
| ZFPM2 | 23414 | zinc finger protein, multitype 2 |
In silico proteomic analysis using The Human Protein Atlas reveals proteins differentially overexpressed (strong staining) in > 80% of malignant glioma cases compared with weak – negative staining in glial and neuronal cells in nonmalignant cerebral cortex.
R2, a microarray analysis and visualization platform provided by the Department of Oncogenomics of the Academic Medical Center, Amsterdam (http://r2.amc.nl), was used to obtain an overview of PBXIP1 mRNA expression in HGG. Affymetrix U133 Plus 2.0 genome-wide gene expression profiles in the public domain were downloaded from the NCBI GEO website and normalized for comparison using the MAS5.0 algorithm (Affymetrix). The datasets used were adult WHO grades II, III, and IV gliomas (n = 153; GSE4290),15 childhood WHO grades III and IV HGGs (n = 53; GSE19578)12 and diffuse intrinsic pontine gliomas (DIPGs) (n = 37; GSE26576),16 adult/pediatric WHO grades II and III ependymomas (n = 83; GSE21687),17 non-neoplastic prefrontal cortex (n = 44; GSE13564),18 and various normal tissues (GSE7307). Annotations and clinical data for these series are available from http://www.ncbi.nlm.nih.gov/geo/query/. The PBXIP1 214177_s_at the probe set was used for expression analyses. The R2 Transcript View Genomic Analysis and Visualization Tool was used to check if the probe set selected had an antisense position in an exon of the gene. The 214177_s_at the probe set used in this study fulfilled these criteria and showed the highest expression in all samples containing a present call for PBXIP1.
Tumor and Developmental Tissues
PBXIP1 immunohistochemistry and immunofluorescence microscopy were performed on 2 tissue microarrays (TMAs), with a total of 7 cases of WHO grade I pilocytic astrocytoma, 19 cases of WHO grade II diffuse astrocytoma, 19 cases of WHO grade III AA, 5 cases of WHO grade II oligodendroglioma, 13 cases of WHO grade III oligodendroglioma, 39 cases of WHO grade IV GBM, and 12 cases of WHO grade II/III ependymoma. A third TMA, consisting of paired diagnosis and relapse samples of 18 cases that initially presented with a WHO grade II or III glioma (astrocytoma/oligodendroglioma/oligoastrocytoma) and ultimately relapsed with a higher grade (WHO grade III or IV) glioma, was used to compare PBXIP1 expression at diagnosis and relapse. The TMAs contained 2–3 representative cores per patient tumor sample with a diameter of 0.6 mm from paraffin-embedded tissue. Postmortem paraffin-embedded tissue of normal neocortex was used as positive controls for immunohistochemistry. All cases were reviewed independently by 2 experienced neuropathologists, and the diagnosis of GBM was confirmed according to the revised 2007 WHO classification of tumors of the nervous system.1 The cases included in this study were obtained from the databases of the Departments of Pathology at VU University Medical Center in Amsterdam and Erasmus University Medical Center in Rotterdam (courtesy of Prof. J.M. Kros). General written informed consent was obtained previously from all patients for the use of tumor material for research. The VUmc Cancer Center Amsterdam Scientific Research Committee approved this study. Tissues were obtained and used in a manner compliant with the Declaration of Helsinki.
PBXIP1 expression during human cortical development was examined on brain material from 4 fetuses at 9, 15, 20, and 23 gestational weeks obtained from spontaneous or medically induced abortions. These tissues were obtained from the Department of Pathology at the Academic Medical Center in Amsterdam. Neuropathological examination of these fetal cases did not show any significant brain pathology. In all cases, formalin-fixed, paraffin-embedded tissue sections (temporal cortical areas) were analyzed. Additionally, we obtained control temporal cortex/white matter at autopsy from 2 children (aged 8 weeks and 8 months) and 1 adult female control (aged 39 years) without a history of neurological disease. All autopsies were performed with appropriate consent within 12 hours after death. Appropriate parental written consent for brain autopsy was given in accordance with the Declaration of Helsinki.
Cell Culture
Human Embryonic Kidney 293 cells (HEK293), human glioma cell lines U-251 MG, U-373 MG, and U-87 MG (ATCC-LGC Standards GmbH), and early passage primary glioblastoma cells E98 and VU148 were maintained in Dulbecco's modified Eagle's medium (DMEM; PAA) supplemented with 10% [v/v] fetal calf serum (PAA), penicillin 100 U/mL (Sigma-Aldrich Chemie B.V.) and streptomycin 100 μg/mL (Sigma-Aldrich) in a humidified atmosphere at 37°C and 5% CO2.
Immunohistochemistry
Tissue was fixed in 4% buffered formalin and embedded in paraffin. 5-µm thick tissue sections were mounted on organosilane-coated slides (Sigma-Aldrich) and used for immunohistochemical staining, as described below.
For single-label immunohistochemistry, tissue sections were deparaffinized, rehydrated, and incubated for 20 min in 0.3% H2O2 diluted in methanol to quench the endogenous peroxidase activity. After retrieving the antigen by incubation for 10 min at 121°C in citrate buffer (0.01 M, pH 6.0), sections were washed with phosphate-buffered saline (PBS) and incubated for 30 min in 10% normal goat serum (Harlan Sera-Lab). Sections were then incubated with a primary polyclonal rabbit anti-human PBXIP1 antibody (HPA006949; dilution 1:400; Sigma-Aldrich) overnight at 4°C. Hereafter, sections were washed in PBS and developed using PowerVision Peroxidase Poly-HRP-anti-mouse/rabbit/rat IgG (Immunologic) and 3,3′-diaminobenzidine (DAB; Sigma-Aldrich) as chromogen. Sections were counterstained with hematoxylin, dehydrated, and coverslipped. Sections incubated without the primary antibody were always completely blank.
For double-labeling immunofluorescence studies, PBXIP1 antibody was combined with antibodies specific for GFAP, vimentin, or Ki67. These were GFAP (monoclonal mouse; dilution 1:50; Dako Netherlands BV), vimentin (monoclonal mouse V9; 1:1,000; Dako), and Ki67 (monoclonal mouse MIB-1; dilution 1:200; Dako). After incubation with the primary antibodies overnight at 4°C, sections were incubated for 2 h at room temperature (RT) with Alexa Fluor 568-conjugated anti-rabbit and Alexa Fluor 488 anti-mouse IgG or anti-goat IgG (1:100; Molecular Probes). Sections were then analyzed using a laser scanning confocal microscope (Leica TCS Sp2).
For the double-labeling of PBXIP1 and GFAPδ, we employed a rabbit polyclonal GFAPδ antibody (dilution 1:500; kindly provided by Dr. E. Hol) as previously described.19–21 PBXIP1 was visualized with a polymer-alkaline phosphatase (AP)-labeled anti-rabbit antibody (BrightVision #DPVR55AP; Immunologic) and liquid permanent red (K0640, Dako) as chromogen. To remove the first primary antibody, sections were incubated at 121°C in a citrate buffer for 10 min, as previously described.22 Sections were then incubated for 1 h at RT with the GFAPδ antibody. The second primary antibody was then visualized with poly-AP anti-rabbit antibody (BrightVision #DPVR55AP; Immunologic) and Vector Blue (AP substrate kit III, #SK-5300; Vector Laboratories) as chromogen.
For double-labeling of PBX1P1 with mutant IDH1, the first primary antibody PBX1P1, was visualized as described above, with Vector Red (AP substrate kit III, #SK-5100, Vector Laboratories) as chromogen. Primary antibody was then removed by incubation at 121°C in Tris/EDTA buffer (10 mM Tris and 1 mM EDTA; pH 9,0) for 10 minSections were incubated for 1 h at room temperature with the second primary antibody (IDH1 R132H, cloneH09, 1:75; Dianova), which was visualized with Vector Blue (Vector Laboratories).
Knockdown Studies Using Lentiviral shRNA Constructs
A nontargeted control and 2 different puromycin selectable pLKO.1-based shRNA constructs of the TRC/MISSION Library (Sigma-Aldrich), numbers TRCN0000015073 and -4, were used with the PBXIP1 mRNA targeting sequences 5′- CAGGCATTAAGGCAAGAGTTA – 3′ and 5′- GATGATGAAGTAGATGACTTT – 3′. A third generation split-genome lentiviral packaging system in the presence of the designated shRNA was used to transfect HEK293 cells and produce lentivirus, as described previously.23 The glioma cells were transduced with the shRNA lentivirus. The medium was refreshed after 24 h and 48 h post-transduction selection medium containing 1.0 μg/mL puromycin (Life Technologies Europe BV) was added for 48 h before using the cells for the assays described below.
Immunofluorescence Microscopy
Cells were seeded on glass coverslips, fixed 24 h later using 3.7% formaldehyde (Sigma-Aldrich), and immunostained. To stain the F-actin fibers, cells were incubated for 15 min with 2.0 U/mL rhodamine-phalloidin (Molecular Probes). Subsequently, cells were counterstained with 0.1 μg/mL 4′,6-diamidino-2-phenylindole (DAPI; Sigma-Aldrich). Images were captured using a fluorescence microscope (Leica) with a 20x objective and Leica Application Software.
Cell Viability Assay and Live-cell Imaging
To study the effect of loss of PBXIP1 on cell growth, the shRNA-transduced glioblastoma cell lines were seeded in a 96-well plate. After 4 days, the cells were fixed with 3.7% formaldehyde for 15 min at RT and washed with PBS, and the nuclei were stained using 0,1 μg/mL DAPI (Sigma-Aldrich) for 15 min at RT. The number of cells was quantified by determining the number of nuclei using the Acumen eX3 Laser-Scanning Fluorescence Microplate Cytometer (TTP LabTech) (GBM cell lines). For the primary GBMs, cells were seeded in a 6-well plate and counted manually after 4 days.
To study cellular motility using live-cell imaging, cells were seeded in an 8-well chamber slide (Ibidi) for 24 h prior to imaging. Time-lapse images were taken using an Olympus IX81 inverted, motorized, fluorescent microscope (Olympus Europe) placed in a humidified 37°C incubator with 5% CO2. Images were taken with a 20x objective every 10 min using Olympus Cell^R software. Quantitative analysis of cellular motility was performed using ImageJ software and the chemotaxis plugin (National Institute of Health, USA).
Flow Cytometric Analysis
Four days after seeding in a 6-well plate, the shRNA-transduced cells were harvested, including the floating cells in the supernatant, pelleted at 300 × g for 5 min at room temperature, washed with ice-cold PBS, and fixated overnight in ice-cold 70% ethanol. Half an hour before flow cytometric analysis using the FACScalibur (BD Biosciences), cells were treated with 125 µg/mL RNase A (Sigma-Aldrich) and 50 µg/mL propidium iodide (Sigma-Aldrich).
Apoptosis Assay
To determine the percentage of apoptotic cells based on the expression of activated caspases, 4 days after plating in a 96-well plate the shRNA-transduced cells were incubated with 10 µM CaspACE™ FITC-VAD-FMK In Situ Marker (Promega) for 90 min at 37°C, washed with PBS, and fixed with 3.7% formaldehyde for 15 min at RT. Nuclei were stained with 0.1 μg/mL DAPI for 15 min at RT. The percentage of FITC-VAD-FMK-positive cells relative to the total number of DAPI-positive cells was determined using the Acumen eX3 (TTP LabTech).
5-bromo-2-deoxyuridine Assay
Four days after plating in a 96-well plate, shRNA-transduced cells were incubated for 1 h at 37°C with 1x 5-bromo-2-deoxyuridine (BrdU)-labeling buffer (Invitrogen), fixed with 75% ethanol for 20 min, and washed twice with PBS. Next, BrdU was visualized using an Alexa Fluor 488 conjugated BrdU mouse monoclonal antibody (Invitrogen). Hereto, cells were permeabilized with 0.1% Triton X-100 for 1 h at RT, followed by DNA denaturation with 2N HCl for 30 min at 37°C. Cells were washed twice for 5 min with 0.1 M sodium borate (pH 8.5) and once with 0.1% Triton X-100. Cells were incubated with BrdU antibody (1:100) at 4°C overnight. The next day, nuclei were counterstained with 0.1 μg/mL DAPI (Sigma-Aldrich) for 15 min at RT. The percentage of BrdU-positive cells relative to the total number of DAPI-positive cells was determined using the Acumen eX3 (TTP LabTech).
Transwell Migration Assay
Four days after plating, shRNA-transduced cells were harvested, washed with PBS, and resuspended in DMEM without serum. 50 000 cells were seeded in the upper chamber of a 24-well transwell with an 8.0 µm pore membrane (Greiner Bio-one). Sixteen hours later, cells were fixed with 3.7% formaldehyde for 15 min at RT, and nuclei were stained with 0.1 μg/mL DAPI (Sigma-Aldrich) for 15 min at RT. The nonmigrated cells at the upper side of the membrane were wiped off using a cotton swap. TIFF images were taken of the migrated cells at the bottom of the membrane with an Olympus microscope using a 10x objective. The number of migrated cells was quantified using ImageJ software (National Institute of Health, USA).
Western Blot Analysis and Quantitative Reverse Transcriptase Polymerase Chain Reaction
Western blot analysis was performed according to standard procedures. In brief, RIPA solution (150 mM NaCl, 50 mM Tris-HCl (pH 8.0), 1% NP-40, 0.5% deoxycholate, 0.1% SDS) supplemented with Protease Inhibitor Cocktail (Roche Applied Sciences) was used to lyse the cells. Proteins were separated using NuPAGE® Novex 4%–12% Bis-Tris 1.0 mm gel (Life Technologies) and the NuPAGE® system from Life Technologies and were transferred onto an Immobilon-P PVDF Membrane 0.45 μm pore (Millipore). For immunoblotting, the membranes were blocked in 5% milk (Campina) in Tris-Buffered Saline Tween-20 [TBS-T]) and consecutively incubated for 1 h with the PBXIP1 rabbit polyclonal antibody (1:1000) or a mouse monoclonal antibody against actin (MAB1501R; Millipore; dilution 1:10 000) and with the secondary antibodies HRP-labeled goat-anti-rabbit (P0448) and goat-anti-mouse antibody (P0447, both DAKO), respectively.
For quantitative reverse transcriptase polymerase chain reaction (qRT-PCR) total RNA was isolated using Trizol reagent (Life Technologies). Single-stranded cDNA was synthesized from 1 μg total RNA using a mixture of random hexamer primers and oligonucleotide dT primers (Life Technologies) and the Omniscript RT kit (Qiagen). QRT-PCR was performed with the FastStart Universal SYBR Green Master mix (Roche) and validated gene-specific primers (Qiagen) on a ABI7500 real-time thermal cycler (Life Technologies) according to the instructions provided. The delta-delta CT method was applied to obtain relative PBXIP1 mRNA levels.
Statistics
For in silico mRNA analysis, ANOVA was used to compare PBXIP1 mRNA expression between a dataset of normal brain tissue and each dataset of glial tumors. A P value <.001 was considered statistically significant. Statistical analyses and visualization of graphs were performed using GraphPad Prism version 5.03 (GraphPad). For all functional assays, significance was determined using the 2-tailed Student t test analysis.
Results
In Silico Proteomics and Transcriptomics Reveal Differential Overexpression of PBXIP1 in Astrocytomas and Ependymomas
To identify novel proteins involved in HGG tumors, an in silico proteomics meta-analysis was initiated using the HPA,13,14 which is based on immunohistochemistry and contains validated immunostainings for more than 10 000 genes on tissue microarrays of 46 different normal human tissues types and 20 different cancer types. We searched for proteins that were highly expressed in >80% of malignant glioma tissues and showed only weak or negative expression in normal cortex, the micro-environment of these tumors. The results of this search are depicted in Table 1. Results included Olig2, a protein described in glioma and tumor stem cells,24,25 which we understood to be a validation of our search. We next focused on PBXIP1 because it showed strong staining in 91% of the malignant gliomas, the highest percentage of strong expression in all tumor types examined by HPA. Moreover, PBXIP1 was previously described as being required for proliferation and migration in breast cancer.26,27
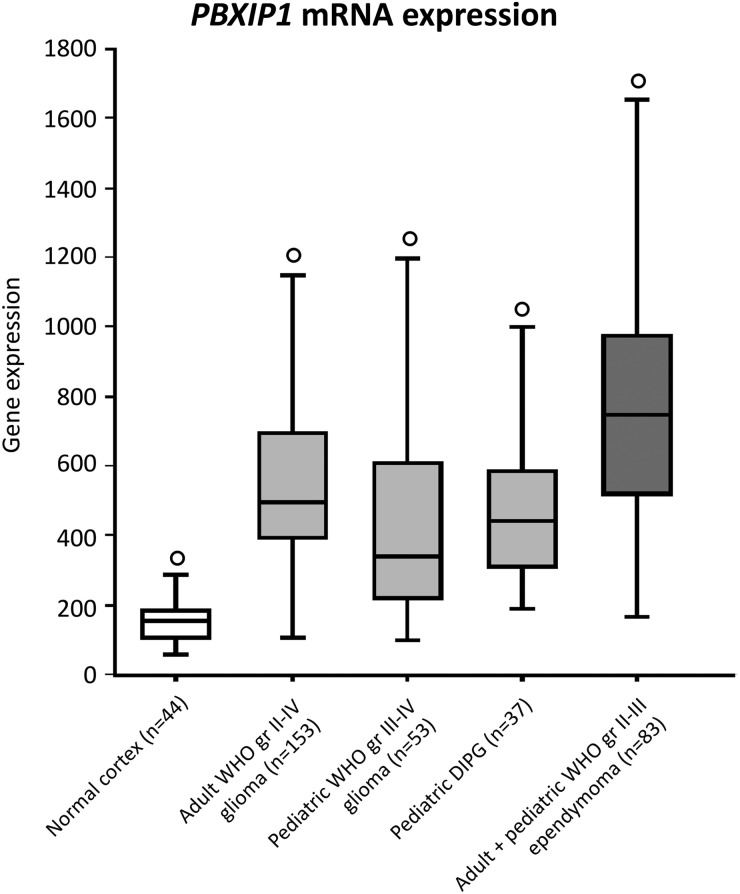
To assess PBXIP1 overexpression in glial tumors with an independent technique, PBXIP1 mRNA expression in adult and pediatric HGG (including DIPG), ependymoma, and normal cortex was determined by in silico analysis of publicly available microarray data using R2 analysis software.12,15–18 PBXIP1 expression was significantly (P < .001) higher in all high-grade glioma and ependymoma datasets, compared with normal cortex tissue (Fig. 1). Furthermore, to ascertain how PBXIP1 expression in these tumor datasets was related in silico to expression in other normal tissues, a normal tissue dataset was used (Supplementary Fig. 1). Significant overexpression was observed between ependymomas and normal tissues (P < .00005), and adult WHO grade II-IV gliomas showed significant overexpression relative to normal male/female reproductive tissues (P < .001). No significant differences were observed in PBXIP1 expression levels in childhood HGGs and DIPGs versus normal tissues.
Fig. 1.
PBXIP1 mRNA expression. In silico analysis of PBXIP1 mRNA expression using R2 analysis software on public datasets of non-neoplastic brain tissues of cerebral cortex (white) versus datasets of adult high-grade glioma, pediatric high-grade glioma, pediatric diffuse intrinsic pontine glioma (DIPG) (grey) and adult/pediatric ependymoma (dark grey). Outliers are indicated by “o”. All cancers showed significant differences in mRNA expression relative to the normal cortex (P < .00001).
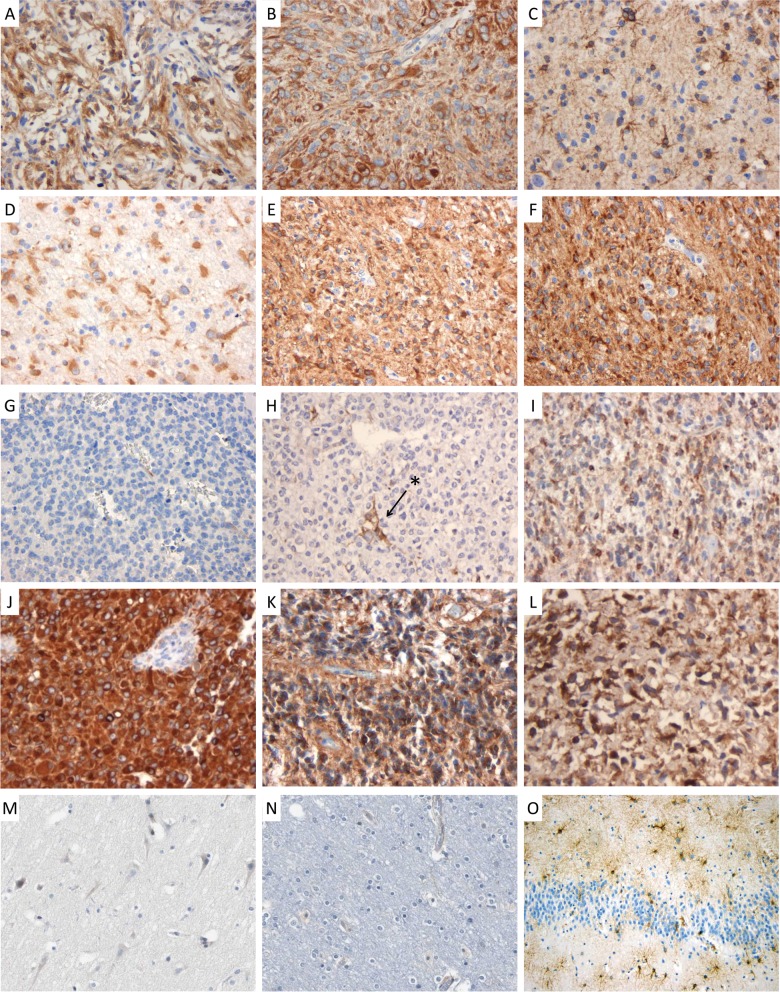
We next validated these results using immunohistochemical analysis on a wide panel of glial tumors. To this purpose, we used a TMA that included WHO grade I pilocytic astrocytoma (n = 7), WHO grade II diffuse astrocytoma (n = 19) and oligodendroglioma (n = 5), WHO grade III AA (n = 19) and oligodendroglioma (n = 13), WHO grade IV GBM (n = 39), and WHO grade II/III ependymoma (n = 12). Normal cortex was used as a control. Representative pictures of the staining are shown in Fig. 2, and results of semiquantitative scoring of these tissues are shown in Table 2. Conforming to the HPA data, we found that PBXIP1 was not expressed in normal cortex (Fig. 2M and/N). No clear correlation could be observed with tumor grade: PBXIP1 was highly expressed in all grades of astrocytoma, with strong staining in more than 50% of the tumor cells in 71%, 37%, 47%, and 41% of the WHO grades I, II, and III astrocytoma and WHO grade IV GBM cases, respectively (Table 2). GBM samples were also observed with moderate staining in a lower percentage of tumor cells. Surprisingly, WHO grades II and III oligodendroglioma cells were immunonegative for PBXIP1 (Fig. 2G and H), while positive reactive astrocytes were detected in some sections in these tumors (Fig. 2H, arrow and asterisk).
Fig. 2.
PBXIP1 immunohistochemistry on paraffin-embedded sections. Representative images of cytoplasmic PBXIP1 staining in WHO grade I pilocytic astrocytoma (A and B); WHO grade II diffuse astrocytoma (C and D); WHO grade III anaplastic astrocytoma (E and F); WHO grade III anaplastic oligodendroglioma (G and H; arrow and asterisk indicate reactive astrocytes); WHO grade IV glioblastoma multiforme (I and J); WHO grade III anaplastic ependymoma (K and L); non-malignant cerebral cortex (M and N) and reactive astrogliosis (O).
Table 2.
Overview of semiquantitative scoring of immunohistochemical PBXIP1 staining in glioma (astrocytoma, oligodendroglioma, glioblastoma multiforme), and ependymoma patient material. The intensity of cytoplasmic PBXIP1 staining (negative/moderate/strong) and the percentage of positive tumor cells was determined by 2 independent reviewers (EA/DVV)
| Staining | ||||
|---|---|---|---|---|
| Positive cells (%) | Negative | Moderate | Strong | |
| WHO gr I pilocytic astrocytoma (n = 7) | <10% | 0 | 0 | 0 |
| 10%–50% | N/A | 0 | 2 | |
| >50% | N/A | 0 | 5 | |
| WHO gr II astrocytoma (n = 19) | <10% | 0 | 1 | 1 |
| 10%–50% | N/A | 4 | 5 | |
| >50% | N/A | 1 | 7 | |
| WHO gr III astrocytoma (n = 19) | <10% | 0 | 1 | 0 |
| 10%–50% | N/A | 1 | 7 | |
| >50% | N/A | 1 | 9 | |
| WHO gr II oligodendroglioma (n = 5)a | <10% | 5 | 0 | 0 |
| 10%–50% | N/A | 0 | 0 | |
| 50%–90% | N/A | 0 | 0 | |
| WHO gr III oligodendroglioma (n = 13)b | <10% | 12 | 1 | 0 |
| 10%–50% | N/A | 0 | 0 | |
| >50% | N/A | 0 | 0 | |
| WHO gr IV glioblastoma multiforme (n = 39) | <10% | 0 | 8 | 1 |
| 10%–50% | N/A | 8 | 4 | |
| >50% | N/A | 2 | 16 | |
| Ependymoma (n = 12) | <10% | 0 | 4 | 2 |
| 10%–50% | N/A | 0 | 2 | |
| >50% | N/A | 0 | 4 | |
aIPBXIP1-positive reactive astrocytes were observed in some oligodendroglioma sections (example Fig. 2D), while tumor cells were consistently negative.
Among ependymomas, 33% of the cases showed strong PBXIP immunopositivity in the majority of tumor cells. To further investigate a role for PBXIP1 in reactive astrocytes, we analyzed PBXIP1 expression in non-neoplastic astrogliotic brain tissue. Interestingly, we found strong PBXIP1-immunoreactivity in activated astrocytes (Fig. 2O) in striking contrast with resting astrocytes in the normal cortex, which were consistently negative (Fig. 2M and N).
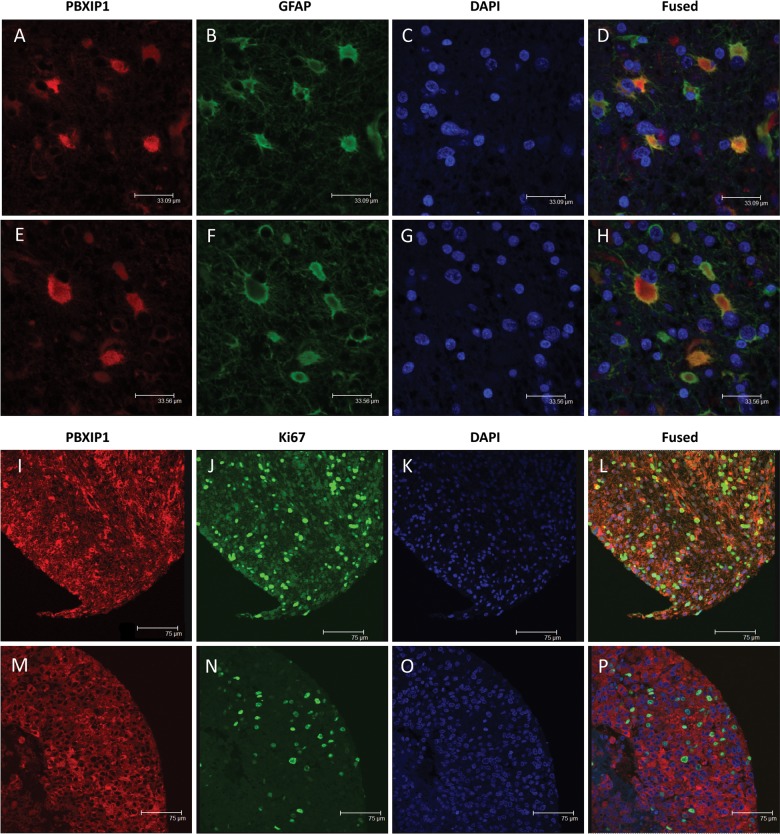
To determine the subcellular localization of PBXIP1 and to ascertain whether PBXIP1 is expressed in astrocytic tumor cell areas, double stainings were performed for PBXIP1/GFAP, PBXIP1/Ki67 in GBM (Fig. 3) and for PBXIP1/IDH1 in IDH1-mutated WHO grade III astrocytoma (Supplementary Fig. 2). PBXIP1 was predominantly confined to the cytoplasm of GBM cells, largely overlapping the GFAP expression pattern (Fig. 3D and H). Furthermore, PBXIP1 was expressed in both Ki67-expressing and Ki67-negative tumor cells (Fig. 3L and P). Since IDH1 mutation is an early tumorigenic mutation and is therefore considered to be present in all tumor cells,28 mutant IDH1 immunostaining can distinguish between tumor cells and reactive astrocytes that could be present in the astrocytoma tumor area, as was observed for the PBXIP1-negative oligodendroglial tumors. Indeed, within IDH1-mutated GBM tumors, both PBXIP1/IDH1 double-positive tumor cells were observed, but PBXIP1-positive/IDH1-negative cells were also seen, presumably reflecting reactive astrocytes. Together, these data suggest that PBXIP1 is expressed in reactive and malignant glial cells of the astrocytic lineage.
Fig. 3.
Immunofluorescence microscopy on paraffin-embedded sections of WHO grade IV glioblastoma multiforme. Two representative images of costaining with PBXIP1 (A and E; red), GFAP (B and F; green), and DAPI (C and G; blue) and the merged images (D and H) showing partial colocalization of PBXIP1 and GFAP expression. Two representative images of costaining with PBXIP1 (I and M; red), Ki67 (J and N; green), and DAPI (K and O; blue) and the merged images (L and P) showing PBXIP1 expression in Ki67-positive tumor area.
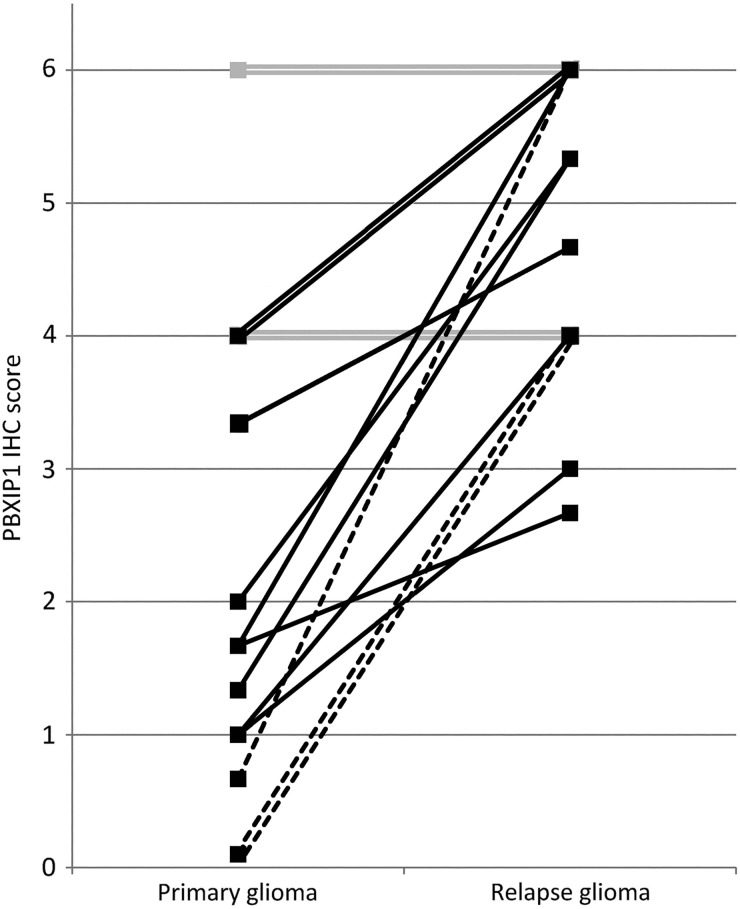
PBXIP1 Protein Upregulation Relates to Malignant Transformation in Glioma
To investigate whether PBXIP1 correlates with malignant progression of astrocytoma and oligodendroglioma, we compared tissue samples obtained from patients who were initially diagnosed with WHO grade II or III astrocytoma or oligodendroglioma and later presented a higher grade relapse (eg, WHO grade III astrocytoma or oligodendroglioma or WHO grade IV GBM). Semiquantitative immunohistochemistry scoring showed a marked upregulation of PBXIP1 expression in 12 of 16 (75%) patients upon malignant transformation (Fig. 4). Interestingly, 3 patients with a PBXIP1-negative oligodendroglioma at the time of first diagnosis showed strong PBXIP1immunoreactivity in the progressed GBM that was diagnosed at relapse (Fig. 4, dashed lines). The remaining 4 of 16 patients, who had moderate to high expression at first diagnosis, showed equal PBXIP1 expression at relapse (Fig. 4, grey lines); in no instance was PBXIP1 expression found to decrease during malignant transformation. These data suggest that PBXIP1 expression is associated with glioma progression.
Fig. 4.
PBXIP1 expression relates to tumor grade. PBXIP1 immunohistochemistry score (staining intensity × tumor cell positivity) in patients with a WHO grade II or III astrocytoma (solid lines) or oligodendroglioma (dashed lines), who had a higher grade tumor (WHO grade III astrocytoma or oligodendroglioma, or WHO grade IV glioblastoma multiforme) at relapse. Twelve of 18 patients showed increased PBXIP1 staining upon malignant transformation. Grey lines represent 4 of 18 patients with equal immunohistochemistry score at diagnosis and relapse.
PBXIP1 Depletion Strongly Reduces Glioma Cell Growth
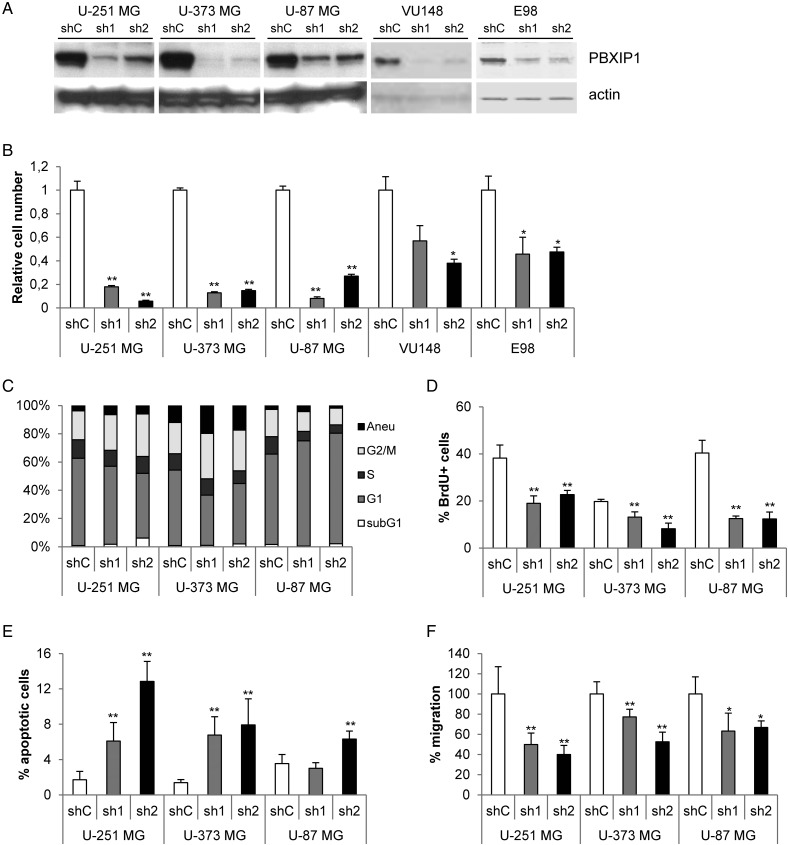
To address the functional role of PBXIP1 in glioma, we investigated the effects of loss of PBXIP1 expression using RNAi technology. Two distinct shRNAs were used to knockdown PBXIP1 mRNA levels in the PBXIP1-expressing HGG cell lines U-251 MG, U-373 MG, and U-87 MG and 2 primary GBM cell cultures E98 and VU148. Western blot analysis showed that these shRNAs efficiently reduced PBXIP1 expression levels in the glioma cells (Fig. 5A). Loss of PBXIP1 expression clearly reduced cell growth in all 3 glioma cell lines and 2 primary GBM cell cultures tested (Fig. 5B). To better understand how loss of PBXIP1 expression affects cell growth, we performed several cell cycle analyses on the 3 glioma cell lines (Fig. 5C). Flow cytometric analysis showed that the cell cycle profile was affected in all 3 glioma cell lines. In U-251 MG and U-373 MG, an increase in the G2/M phase and concurrent decrease in G1 and S-phase were observed. In the U-87 MG glioma cells, we observed a decrease in the G1 cell cycle phase concurrent with a decrease in S phase. To better assess the effects on proliferation, BrdU incorporation assays were performed. Upon depletion of PBXIP1, DNA synthesis was reduced significantly in all GBM cell lines tested (Fig. 5D), suggesting that proliferation was tempered in these cells. To determine if the reduced growth rate also involved apoptosis, the number of apoptotic cells was determined by incubating the cells briefly with FITC-VAD-FMK to visualize the cells that contained activated caspases. Loss of PBXIP1 significantly induced apoptosis in all glioma cell lines tested, which was the most clear in U-251 MG cells, where the percentage of apoptotic cells increased from 1.7% in the shControl cells to 12.8% in the sh2-mediated PBXIP1 depleted cells (Fig. 5E). Together these data suggest that loss of PBXIP1 reduces cellular growth by reducing proliferation and increasing apoptosis.
Fig. 5.
Loss of PBXIP1 expression affects glioma cell survival. Effective knockdown of PBXIP1 expression on protein (A) by 2 independent shRNA constructs (sh1 and sh2, respectively) compared with a nontargeting control shRNA (shC). (B) PBXIP1-knockdown resulted in reduced cell growth in U-251 MG, U-373 MG, and U-87 MG glioma cells as well as in the primary glioblastoma multiforme cell cultures VU148 and E98. The graph shows the cell number of the shRNA transduced glioma cells relative to nontargeting control shRNA after 4 days growth. (C) The graph shows the percentages of each cell cycle phase (subG1, G1, S, and G2/M. “aneu” is the fraction of aneuploidic cells with a DNA content larger than 4N) as determined by flow cytometry for the glioma cell lines that were transduced with the shRNAs indicated (D) The bar graph shows the percentage of BrdU-positive cells relative to the total number of (DAPI-positive) cells. (E) The bar graph shows the percentage of apoptotic cells that were visualized and counted after incubation with the CaspACE™ FITC-VAD-FMK In Situ Marker relative to the total number of (DAPI-stained) cells. (F) The bar graph shows the results of transwell migration assay. The number of migrated cells in the shPBXIP1-transduced glioma cells was quantified and depicted relative to the number of migrated cells of the nontargeting shRNA control-transduced glioma cells, which were set to 100%. (* indicates a statistical significance of P < .01 and ** a significance of P < .001 as determined by 2-tailed student t test analysis).
Loss of PBXIP1 Expression Changes the Cytoskeleton and Affects Glioma Migration
Since diffuse migration is an important feature of infiltrative glioma cells, we investigated the effect of loss of PBXIP1 on glioma cell migration by performing a transwell migration assay. This showed that loss of PBXIP1 significantly decreased migration of all glioblastoma cell lines tested, ranging from 23% in U-373 MG shPBXIP1-1 cells to 60% in U-251 MG cells shPBXIP1-2 cells relative to the corresponding shControl cells (Fig. 5F).
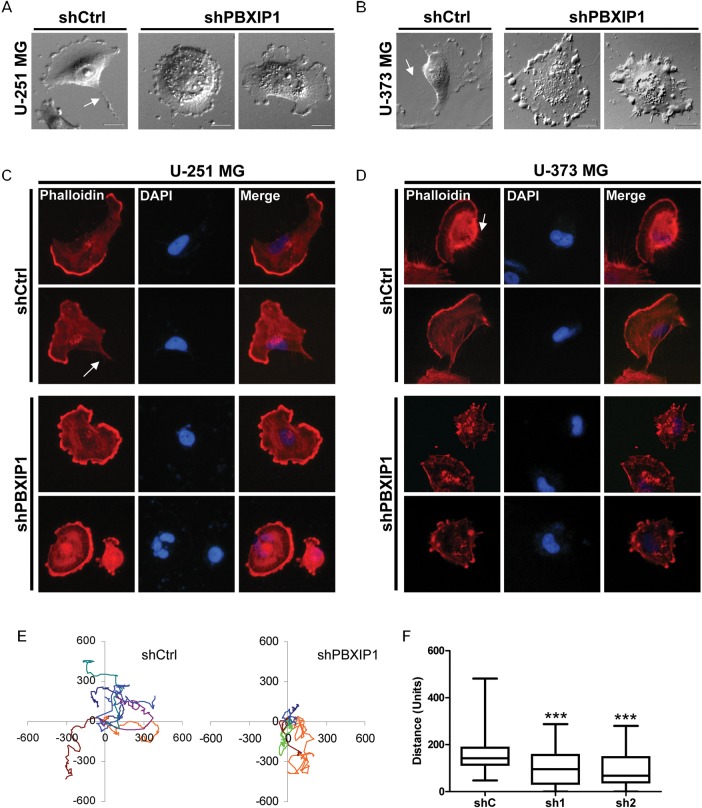
Close microscopic observations showed marked changes in cellular morphology. Bright field microscopy showed that many of the PBXIP1-knockdown cells were more rounded in cellular shape and had dramatic changes in membrane ruffling and cellular protrusions (Fig. 6A and B). The cells had lost the fan-shaped ruffles typical of moving cells and showed circular membrane ruffles or spiky protrusions at the cellular edges. Staining of the filamentous actin fibers of U-251 MG and U-373 MG glioma cells, using rhodamine-labeled phalloidin, showed that the PBXIP1-knockdown cells had rounded up and mostly lost their ventral stress fibers that normally mark the rear of a moving cell (Fig. 6C and D). In parallel, PBXIP1-knockdown cells had more dorsal stress fibers than the control cells.
Fig. 6.
PBXIP1 depletion results in changes in cellular morphology and actin stress fiber formation and reduces motility of glioma cells. Representative bright-field images show the effect of loss of PBXIP1 expression in U-251 MG (A, right 2 panels) and U-373 MG HGG cells (B, right 2 panels) compared with the nontargeting shRNA control cells (A and B, left panels). Representative images of rhodamine-phalloidin staining in U-251 MG (C) and U-373 MG cells (D) with PBXIP1-knockdown (lower panels) or the nontargeting shRNA control cells (upper panels). Arrows indicate ventral actin stress fibers. PBXIP1-knockdown (E, right panel) impaired motility compared with the nontargeting shRNA control U-251 MG cells (E, left panel). Panel F shows the quantification of the distance over 24 h of the nontargeting shRNA control and the 2 independent shRNAs targeting PBXIP1 in U-251 MG cells. At least 50 cells were tracked. Distance is measured in arbitrary units. Plot F shows the mean, min, and max whiskers. The difference in motility with the control was significant for both shRNAs (P < .001).
We next investigated glioma migration capacities by determining the cellular motility using live-cell imaging and tracking cellular moves of U-251 MG glioma cells that were transduced with shControl or shPBXIP1. Quantification of the live-cell imaging data showed that motility of U-251 MG glioma cells was impaired in the absence of PBXIP1 (Fig. 6E and F). While the control-cell population had a median traveled distance of 144 units per followed track, the motility in the PBXIP1-knockdown cells was reduced to 96 units and 70 units, respectively, per track for the 2 independent shRNAs. The impact of the cellular changes deriving from PBXIP1-depletion varied, although the decrease in traveled distance was highly significant (respective P values using the 2-tailed t test were .0008 and .0001; Fig. 6E and F). We postulated that this was caused in part by the varying levels of PBXIP1 mRNA depletion between cells in the PBXIP1-knockdown cell population. In part, this might also reflect the nature of glioma itself: it is a highly heterogeneous tumor, and some cells can be highly mobile within a glioma cell population, while others move slowly. Indeed, this could be observed in the variability in cellular motility in the control cells (Fig. 6F). Together, these data suggest a role for PBXIP1 in glioma cell migration.
PBXIP1 Is Expressed in Astrocyte Progenitor Cells During Human Cortical Development
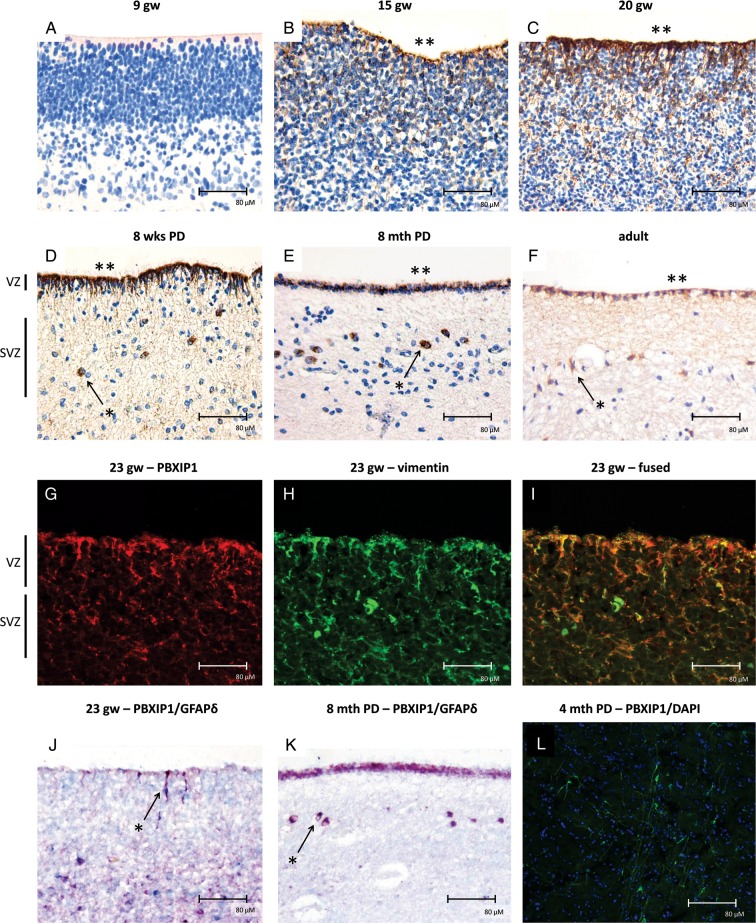
Considering the role of PBXIP1 in hematopoietic and embryonic stem cells,29 and since high grade gliomas are thought to either arise from multipotent neural stem cells or lineage-restricted neural progenitor cells,30 or to result from dedifferentiation of astrocyte- or oligodendrocyte-lineage committed cells, we investigated PBXIP1 expression in the fetal human brain (Fig. 7). Tissue sections were obtained from brain areas that included the ventricular zone (VZ) and the subventricular zone (SVZ), since these are well known to harbor high numbers of glial progenitor cells.20,31 At 9 weeks of gestational age (GA), we observed no expression of PBXIP1 in the VZ (Fig. 7A), but strong PBXIP1 staining in radial glia neural stem cells could be observed in the VZ from 15 weeks GA onwards (Fig. 7B and C). Expression of PBXIP1 was also observed in the SVZ between 20 and 40 gestational weeks (data not shown). Coexpression of vimentin (Fig. 7I) and GFAP-δ (Fig. 7J and K) suggests that PBXIP1 is specifically expressed in radial glia (Fig. 7J) and astrocyte progenitor cells (Fig. 7K). PBXIP1-positive astrocyte progenitor cells with bipolar morphology were detected up to 4 months post natally (Fig. 7L). Albeit, strongly reduced in number, a ribbon of PBXIP1-positive SVZ progenitor cells could still be detected in the postnatal SVZ and even into adulthood (Fig. 7D–F). PBXIP1-positive cells consistently coexpressed GFAP-δ, a marker for progenitor cells in the SVZ.32 Except for 9 weeks GA, the progenitor cells in the ependymal layer stained positive for PBXIP1 during all stages of development.
Fig. 7.
PBXIP1 immunoreactivity (IR) during human brain development. (A) At gestational week 9 (9 gw), PBXIP1 was not detectable in the ventricular zone (VZ). At 15 (B) and 20 gw (C) PBXIP1 expression was found in the VZ. PBXIP1 expression in the SVZ (arrows and asterisks) was still detected at 8 weeks (D) and at 8 months (E) post delivery (PD) and even until adulthood (F). The ependymal layer (**) was positive during all phases of development, except at the early stage of 9 weeks. Hematoxylin counterstain shows nuclei in blue. Panels G-I show confocal images of VZ/subventricular zone (SVZ) at 23 gw stained with PBXIP1 (red; G) and vimentin (green; H), and the merged image (I) showing colocalization in radial glial cells. Panel J shows colocalization (purple; arrow and asterisk) of PBXIP1 (red) with GFAP-delta (blue) in radial glial neural stem cells in VZ at 23 gw. Panel K (8 months PD) shows PBXIP1 IR in ependymal layer and in GFAP-delta positive progenitors of SVZ astrocytes (arrow and asterisk). Panel L shows PBXIP1 staining (green; nuclear staining with DAPI, blue) in astrocyte progenitor cells in neonatal cortex (age 4 months). Scale bar in A-L: 80 µm.
Discussion
The mortality in both adult and pediatric patients with high-grade glial cancers is very high, and the limited treatment possibilities call for new therapeutic strategies. To identify putative novel targets for therapy with a potentially high therapeutic index, we performed in silico proteomics and transcriptomics using existing data platforms to search for genes that are differentially expressed in malignant glioma but not in normal brain. These analyses revealed PBXIP1 as a candidate tumor-specific gene with overexpression to normal brain (Fig. 1). Correspondingly, PBXIP1 was previously shown to be overexpressed in pancreatic cancer as compared with normal pancreatic tissues.27,33,34 Interestingly, we found that PBXIP1 is overexpressed in malignant astrocytoma, GBM, and ependymoma, whereas oligodendroglioma did not show any positive PBXIP1 staining. No apparent differences in PBXIP1 expression were observed between different astrocytic tumor grades (Table 2), although larger numbers of patient samples are needed to correlate tumor grade with PBXIP1 expression. Still, increased intrapatient expression of PBXIP1 was observed upon malignant transformation of both astrocytoma and oligodendroglioma to GBM, although we cannot exclude that the observed increase in PBXIP1 expression upon malignant progression is due to infiltration of reactive astrocytes in progressive glial tumors, as was observed in IDH1-mutated anaplastic astrocytoma (Supplementary Fig. 2). This warrants further research. Notably, discrimination of reactive astrocytes from (non-IDH1 mutated) tumor astrocytes is still difficult. In addition, while increased PBXIP1 expression levels within patient grading suggest that PBXIP1 can be a biomarker to ascertain tumor grade, the observed variation of PBXIP1 expression levels between patients with the same WHO grading abrogates this suggestion. Another explanation of the increased PBXIP1 expression levels could be the increased proliferation and/or migration of tumor cells usually observed in glioma recurrences. In strong support of this, shRNA-mediated knockdown of PBXIP1 resulted in significantly reduced migration and changed cell cycle profiles, including reduced proliferation and increase in apoptosis. In addition, PBXIP1 depletion induced progressive changes in stress fiber formation and cellular protrusions with loss of cellular motility and therefore possibly compromised the migration potential of glioma cells. As PBXIP1 was significantly overexpressed in adult and pediatric HGG and ependymomas relative to normal brain, expression in some other normal tissues was comparable to that observed in most of these tumors, except for ependymomas (Supplementary Fig. 1). Systemic therapy against PBXIP1 therefore necessitates close observation of possible toxicity to these normal tissues in future studies. Together, these data suggest PBXIP1 to be an interesting, potential therapeutic target that warrants further investigations.
We found that PBXIP1 is important for glioma cell growth, which might be explained in several ways. One possible mode of action might be that PBXIP1 acts through the PI3K-Akt pathway, of which PBXIP1 is an activator that is frequently altered in glioma.11,34 This may explain the observed effect of PBXIP1 depletion on actin remodeling, since PI3K-Akt signaling regulates cancer cell migration and growth, actin remodeling, and microtubule organization.35–37 A second scenario might be that PBXIP1 acts through the regulation of PBX homeobox proteins. We found that PBXIP1 is predomiantly expressed in the cytoplasm of glioma cells. Cytoplasmic expression of PBXIP1 was previously shown to control localization and functioning of transcription factors such as the PBX homeobox proteins, as well as estrogen receptor alpha and beta.27,34,38,39 Interestingly, PBX1 is highly expressed in SVZ progenitor cells during brain development, and other PBX proteins are broadly expressed in the telencephalon.40,41,39,40 An attractive hypothesis is that self-renewing stem cell-like tumor cells are able to retain an immature phenotype because of sequestration of PBX proteins in the cytosol. A third scenario involves CK2, a potential regulator of PBXIP1.38 CK2 is involved in migration and tumor growth, and its inhibition affects cellular morphology of cultured primary astrocytes and retinal endothelial cells, thereby inducing rounding of astrocytes due to collapse and disorganization of the cytoskeleton similar to that observed after depletion of PBXIP1 in HGG cells.42 By acting via multiple pathways, inhibiting PBXIP1 might allow for a one hit-multiple targets principle, while maintaining its tumor specificity. Clearly, more research is needed to delineate the pathways through which PBXIP1 acts in glial cancers.
High-grade gliomas and ependymomas often share stem-cell like characteristics with radial and early progenitor cells, either being ontologically derived from these cells or resulting from dedifferentiation of mature cells.43,44 We therefore investigated the expression of PBXIP1 in the developing human brain. Indeed, PBXIP1 was strongly coexpressed with the astrocytic progenitor marker GFAP-δ in the SVZ of the developing human brain, indicating that PBXIP1 plays a role in astrocyte progenitor cells. Interestingly, we also found PBXIP1 to be overexpressed in radial glia, which are the neural stem cells that give rise to GFAP-δ positive SVZ astrocyte progenitor cells during human brain development.31,32,30,31 Radial glia cells ultimately differentiate into astrocytes and ependymal cells,31,45 whereas oligodendrocytes are derived from oligodendrocyte precursor cells.30,44 Interestingly, in analogy to this expression pattern, we showed that PBXIP1 is expressed in malignant astrocytoma and ependymoma but not in oligodendroglioma.
In conclusion, we discovered PBXIP1 to be a protein strongly expressed in astrocytic and ependymal tumors. Depletion of PBXIP1 strongly affects migration and proliferation of glioma cells in vitro, suggesting that PBXIP1 is a driving factor in glioma tumorigenesis and thus a possible candidate target for gene-specific antitumor therapy. These data warrant further exploration of PBXIP1 as a novel therapeutic target for astrocytomas and ependymomas.
Supplementary Material
Funding
This work was financially supported by VUmc Cancer Center Amsterdam (VUmc CCA). EH and DB are supported by Stichting Semmy. EA is supported by the Stichting Kinderen Kankervrij (KiKa). PS is supported by Stichting STOPhersentumoren.nl.
Supplementary Material
Acknowledgements
We are grateful to J.J. Anink for his technical contribution and thank Dr. P. Wesseling for critically reading this manuscript.
Conflict of interest statement. None declared.
References
- 1.Louis DN, Ohgaki H, Wiestler OD, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gilbert MR, Ruda R, Soffietti R. Ependymomas in adults. Curr Neurol Neurosci Rep. 2010;10:240–247. doi: 10.1007/s11910-010-0109-3. [DOI] [PubMed] [Google Scholar]
- 3.Zacharoulis S, Moreno L. Ependymoma: an update. J Child Neurol. 2009;24:1431–1438. doi: 10.1177/0883073809339212. [DOI] [PubMed] [Google Scholar]
- 4.Ohgaki H, Kleihues P. Genetic profile of astrocytic and oligodendroglial gliomas. Brain Tumor Pathol. 2011;28:177–183. doi: 10.1007/s10014-011-0029-1. [DOI] [PubMed] [Google Scholar]
- 5.Kaatsch P, Rickert CH, Kuhl J, et al. Population-based epidemiologic data on brain tumors in German children. Cancer. 2001;92:3155–3164. doi: 10.1002/1097-0142(20011215)92:12<3155::aid-cncr10158>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 6.Cohen KJ, Pollack IF, Zhou T, et al. Temozolomide in the treatment of high-grade gliomas in children: a report from the Children's Oncology Group. Neuro Oncol. 2010;13:317–323. doi: 10.1093/neuonc/noq191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stupp R, Hegi ME, Mason WP, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10:459–466. doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 8.MacDonald TJ, Aguilera D, Kramm CM. Treatment of high-grade glioma in children and adolescents. Neuro Oncol. 2011;13:1049–1058. doi: 10.1093/neuonc/nor092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grill J, Bergthold G, Ferreira C. Pediatric ependymomas: will molecular biology change patient management? Curr Opin Oncol. 2011;23:638–642. doi: 10.1097/CCO.0b013e32834b5310. [DOI] [PubMed] [Google Scholar]
- 10.Kilday JP, Rahman R, Dyer S, et al. Pediatric ependymoma: biological perspectives. Mol Cancer Res. 2009;7:765–786. doi: 10.1158/1541-7786.MCR-08-0584. [DOI] [PubMed] [Google Scholar]
- 11.The Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paugh BS, Qu C, Jones C, et al. Integrated molecular genetic profiling of pediatric high-grade gliomas reveals key differences with the adult disease. J Clin Oncol. 2010;28:3061–3068. doi: 10.1200/JCO.2009.26.7252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Uhlen M, Bjorling E, Agaton C, et al. A human protein atlas for normal and cancer tissues based on antibody proteomics. Mol Cell Proteomics. 2005;4:1920–1932. doi: 10.1074/mcp.M500279-MCP200. [DOI] [PubMed] [Google Scholar]
- 14.Uhlen M, Oksvold P, Fagerberg L, et al. Towards a knowledge-based Human Protein Atlas. Nat Biotechnol. 2010;28:1248–1250. doi: 10.1038/nbt1210-1248. [DOI] [PubMed] [Google Scholar]
- 15.Sun L, Hui AM, Su Q, et al. Neuronal and glioma-derived stem cell factor induces angiogenesis within the brain. Cancer Cell. 2006;9:287–300. doi: 10.1016/j.ccr.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 16.Paugh BS, Broniscer A, Qu C, et al. Genome-wide analyses identify recurrent amplifications of receptor tyrosine kinases and cell-cycle regulatory genes in diffuse intrinsic pontine glioma. J Clin Oncol. 2011;29:3999–4006. doi: 10.1200/JCO.2011.35.5677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson RA, Wright KD, Poppleton H, et al. Cross-species genomics matches driver mutations and cell compartments to model ependymoma. Nature. 2010;466:632–636. doi: 10.1038/nature09173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harris LW, Lockstone HE, Khaitovich P, et al. Gene expression in the prefrontal cortex during adolescence: implications for the onset of schizophrenia. BMC Med Genomics. 2009;2:28. doi: 10.1186/1755-8794-2-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van den Berge SA, Middeldorp J, Zhang CE, et al. Longterm quiescent cells in the aged human subventricular neurogenic system specifically express GFAP-delta. Aging Cell. 2010;9:313–326. doi: 10.1111/j.1474-9726.2010.00556.x. [DOI] [PubMed] [Google Scholar]
- 20.Roelofs RF, Fischer DF, Houtman SH, et al. Adult human subventricular, subgranular, and subpial zones contain astrocytes with a specialized intermediate filament cytoskeleton. Glia. 2005;52:289–300. doi: 10.1002/glia.20243. [DOI] [PubMed] [Google Scholar]
- 21.Martinian L, Boer K, Middeldorp J, et al. Expression patterns of glial fibrillary acidic protein (GFAP)-delta in epilepsy-associated lesional pathologies. Neuropathol Appl Neurobiol. 2009;35:394–405. doi: 10.1111/j.1365-2990.2009.00996.x. [DOI] [PubMed] [Google Scholar]
- 22.Aronica E, Boer K, Baybis M, et al. Co-expression of cyclin D1 and phosphorylated ribosomal S6 proteins in hemimegalencephaly. Acta Neuropathol. 2007;114:287–293. doi: 10.1007/s00401-007-0225-6. [DOI] [PubMed] [Google Scholar]
- 23.Mir SE, De Witt Hamer PC, Krawczyk PM, et al. In silico analysis of kinase expression identifies WEE1 as a gatekeeper against mitotic catastrophe in glioblastoma. Cancer Cell. 2010;18:244–257. doi: 10.1016/j.ccr.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rhee W, Ray S, Yokoo H, et al. Quantitative analysis of mitotic Olig2 cells in adult human brain and gliomas: implications for glioma histogenesis and biology. Glia. 2009;57:510–523. doi: 10.1002/glia.20780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ligon KL, Huillard E, Mehta S, et al. Olig2-regulated lineage-restricted pathway controls replication competence in neural stem cells and malignant glioma. Neuron. 2007;53:503–517. doi: 10.1016/j.neuron.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abramovich C, Chavez EA, Lansdorp PM, et al. Functional characterization of multiple domains involved in the subcellular localization of the hematopoietic Pbx interacting protein (HPIP) Oncogene. 2002;21:6766–6771. doi: 10.1038/sj.onc.1205784. [DOI] [PubMed] [Google Scholar]
- 27.Wang X, Yang Z, Zhang H, et al. The estrogen receptor-interacting protein HPIP increases estrogen-responsive gene expression through activation of MAPK and AKT. Biochim Biophys Acta. 2008;1783:1220–1228. doi: 10.1016/j.bbamcr.2008.01.026. [DOI] [PubMed] [Google Scholar]
- 28.Camelo-Piragua S, Jansen M, Ganguly A, et al. A sensitive and specific diagnostic panel to distinguish diffuse astrocytoma from astrocytosis: chromosome 7 gain with mutant isocitrate dehydrogenase 1 and p53. J Neuropathol Exp Neurol. 2011;70:110–115. doi: 10.1097/NEN.0b013e31820565f9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manavathi B, Lo D, Bugide S, et al. Functional regulation of pre-B-cell leukemia homeobox interacting protein 1 (PBXIP1/HPIP) in erythroid differentiation. J Biol Chem. 2012;287:5600–5614. doi: 10.1074/jbc.M111.289843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xie Z. Brain tumor stem cells. Neurochem Res. 2009;34:2055–2066. doi: 10.1007/s11064-009-0079-5. [DOI] [PubMed] [Google Scholar]
- 31.Merkle FT, Tramontin AD, Garcia-Verdugo JM, et al. Radial glia give rise to adult neural stem cells in the subventricular zone. Proc Natl Acad Sci U S A. 2004;101:17528–17532. doi: 10.1073/pnas.0407893101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Middeldorp J, van den Berge SA, Aronica E, et al. Specific human astrocyte subtype revealed by affinity purified GFAP antibody; unpurified serum cross-reacts with neurofilament-L in Alzheimer. PLoS One. 2009;4:e7663. doi: 10.1371/journal.pone.0007663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grutzmann R, Pilarsky C, Staub E, et al. Systematic isolation of genes differentially expressed in normal and cancerous tissue of the pancreas. Pancreatology. 2003;3:169–178. doi: 10.1159/000070087. [DOI] [PubMed] [Google Scholar]
- 34.Manavathi B, Acconcia F, Rayala SK, et al. An inherent role of microtubule network in the action of nuclear receptor. Proc Natl Acad Sci U S A. 2006;103:15981–15986. doi: 10.1073/pnas.0607445103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cain RJ, Ridley AJ. Phosphoinositide 3-kinases in cell migration. Biol Cell. 2009;101:13–29. doi: 10.1042/BC20080079. [DOI] [PubMed] [Google Scholar]
- 36.Vivanco I, Sawyers CL. The phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nat Rev Cancer. 2002;2:489–501. doi: 10.1038/nrc839. [DOI] [PubMed] [Google Scholar]
- 37.Buttrick GJ, Wakefield JG. PI3-K and GSK-3: Akt-ing together with microtubules. Cell Cycle. 2008;7:2621–2625. doi: 10.4161/cc.7.17.6514. [DOI] [PubMed] [Google Scholar]
- 38.Abramovich C, Shen WF, Pineault N, et al. Functional cloning and characterization of a novel nonhomeodomain protein that inhibits the binding of PBX1-HOX complexes to DNA. J Biol Chem. 2000;275:26172–26177. doi: 10.1074/jbc.M001323200. [DOI] [PubMed] [Google Scholar]
- 39.Yu YH, Siao FP, Hsu LC, Yen PH. TEX11 modulates germ cell proliferation by competing with estrogen receptor beta for the binding to HPIP. Mol Endocrinol. 2012;26:630–642. doi: 10.1210/me.2011-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Redmond L, Hockfield S, Morabito MA. The divergent homeobox gene PBX1 is expressed in the postnatal subventricular zone and interneurons of the olfactory bulb. J Neurosci. 1996;16:2972–2982. doi: 10.1523/JNEUROSCI.16-09-02972.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Toresson H, Parmar M, Campbell K. Expression of Meis and Pbx genes and their protein products in the developing telencephalon: implications for regional differentiation. Mech Dev. 2000;94:183–187. doi: 10.1016/s0925-4773(00)00324-5. [DOI] [PubMed] [Google Scholar]
- 42.Kramerov AA, Ahmed K, Ljubimov AV. Cell rounding in cultured human astrocytes and vascular endothelial cells upon inhibition of CK2 is mediated by actomyosin cytoskeleton alterations. J Cell Biochem. 2012;113:2948–2956. doi: 10.1002/jcb.24171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zong H, Verhaak RG, Canolk P. The cellular origin for malignant glioma and prospects for clinical advancements. Expert Rev Mol Diagn. 2012;12:383–394. doi: 10.1586/erm.12.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu Q, Wang X. Neuronal stem cells in the central nervous system and in human diseases. Protein Cell. 2012;3:262–270. doi: 10.1007/s13238-012-2930-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spassky N, Merkle FT, Flames N, et al. Adult ependymal cells are postmitotic and are derived from radial glial cells during embryogenesis. J Neurosci. 2005;25:10–18. doi: 10.1523/JNEUROSCI.1108-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.