Abstract
Background
Glioblastoma multiforme (GBM) is one of the most aggressive human tumors, and the establishment of an effective therapeutic reagent is a pressing priority. Recently, it has been shown that the tumor tissue consists of heterogeneous components and that a highly aggressive population should be the therapeutic target.
Methods
Through a single subcutaneous passage of GBM cell lines LN443 and U373 in mice, we have developed highly aggressive variants of these cells named LN443X, U373X1, and U373X2, which showed increased tumor growth, colony-forming potential, sphere-forming potential, and invasion ability. We further investigated using microarray analysis comparing malignant cells with their parental cells and mRNA expression analysis in grades II to IV glioma samples.
Results
Adipocyte enhancer binding protein 1, epiregulin (EREG), and microfibrillar associated protein 5 were identified as candidate genes associated with higher tumor grade and poor prognosis. Immunohistochemical analysis also indicated a correlation of a strong expression of EREG with short overall survival. Furthermore, both EREG stimulation and EREG introduction of GBM cell lines were found to increase phosphorylation of epidermal growth factor receptor (EGFR) and extracellular signal-regulated kinase and resulted in the promotion of colony formation, sphere formation, and in vivo tumor formation. Gefitinib treatment inhibited phosphorylation of EGFR and extracellular signal-regulated kinase and led to tumor regression in U373-overexpressed EREG.
Conclusion
These results suggested that EREG is one of the molecules involved in glioma malignancy, and EGFR inhibitors may be a candidate therapeutic agent for EREG-overexpressing GBM patients.
Keywords: brain tumor, epiregulin, ERK/MAPK, GBM
Glioma is the most common primary tumor of the central nervous system, accounting for ∼30%, and is classified by the World Health Organization (WHO) into 4 clinical grades, from I to IV. The most aggressive form of glioma is glioblastoma multiforme (GBM), with a 5-year survival rate of ∼8%.1,2 Surgical resectability is the most important prognostic factor, as effects of additional chemotherapy and radiotherapy are limited. Temozolomide (TMZ) is an alkylating agent used in the treatment of malignant gliomas, including GBM.3 In a study of 573 patients with newly diagnosed, histologically confirmed GBM randomly assigned to receive radiotherapy alone or radiotherapy plus continuous daily TMZ, statistically significant survival benefits were shown in patients given TMZ.4 However, the prognosis for most patients with GBM remains dismal, with a median survival of only 14.6 months.4
A greater understanding of the biological mechanisms for GBM oncogenesis will contribute to the development of targeted therapies that can improve patient outcome. The genome-wide analysis performed by The Cancer Genome Atlas has shown that the most frequent genetic abbreviations were identified in the signaling pathways involving receptor tyrosine kinase, phosphatidylinositol-3 kinase (PI3K), p53, and retinoblastoma protein.5 In addition, it has been suggested that GBM can be classified into 3 subgroups: proneuronal, proliferative, and mesenchymal.6 Although various targeted molecular agents have been used either as single agents or in combination therapy for GBM, few have been reported to be effective in phase II trials so far.7 Therefore, identification of new molecular targets is still of paramount importance.
The cancer stem cell hypothesis proposes that tumors are driven by subpopulations of tumor cells with stem cell–like properties.8 Cancer stem cells have been isolated in multiple tumor types, including GBM. Several molecules, such as cluster of differentiation (CD )133, sex determining region Y–box 2 (Sox2), CD15, integrin-α6, and the L1 cell adhesion molecule, have been proposed as markers for cancer-initiating cells.9–16 Especially, by promoter analysis for CD133, the pathway of epidermal growth factor receptor (EGFR)/extracellular signal-regulated kinase (ERK)/mitogen-activated protein kinase (MAPK)–ERK has been shown to be involved in CD133 gene expression through Ets-family transcription factors.17 Considering the evaluation for their prognostic value, none of them was proven to be clinically useful in large-scale studies. Given the heterogeneity of GBM, further investigations are necessary to identify the treatment-resistant cell population, as these occasionally overlap with cancer-initiating cell properties. It is essential to develop tailored treatments to target this population with increased tumorigenic potential.18
In this study, through single subcutaneous passage in mice, we have developed highly aggressive variants of human GBM cell lines LN443 and U373, which showed increased tumor growth, colony-forming potential, sphere-forming potential, and invasiveness compared with the parental cell lines. Using DNA microarray analysis, we identified a novel molecular mechanism for the pathogenicity of GBM and explored new therapeutic agents that can be used for this disease.
Materials and Methods
Cells
The human GBM cell lines LN443 and U373 were kindly provided by Dr Erwin G. Van Meir (Emory University School of Medicine, Atlanta, Georgia). LN443 and U373 were cultured in Dulbecco's modified minimal essential medium (DMEM; Wako) supplemented with 10% fetal bovine serum. Cell line authentication was not carried out by the authors within the last 6 months.
Reagents
Human epiregulin (EREG) was purchased from Cell Signaling Technology. Gefitinib was purchased from Cayman Chemical Company. All reagents were used following the manufacturers' instructions.
Preparation of Retrovirus and Establishment of Stable Cell Line
For retrovirus production, the pCX4 vector system was used.47,48 The complete sequences of pCX4pur (puromycin) are available from the GenBank database (AB086386). Full-length cDNAs for human EREG were subcloned into pCX4pur. Retroviruses were obtained by using 293T cells as packaging cells, infected to the KMG4 glioma cell line, and selected with puromycin (2 μg/mL).
Xenograft
For xenograft preparation, the indicated number of cells was s.c. injected into 6- to 8-week-old female athymic nude mice (BALB/cA Jcl-nu/nu; Clea Japan). For evaluation of gefitinib treatment, gefitinib (200 mg/kg) or control dimethyl sulfoxide was i.p. administered on days 25–29 and 32–36 (for U373-EREG) or days 3–7 and 10–14 (for U373X1). Tumor volume (in cubic millimeters) was calculated by the following formula: (length × width2)/2. For the in vivo orthotopic tumor model, tumor cells (3 × 105) suspended in 10 µL PBS were i.c. injected into the BALB/cA Jcl-nu/nu mice. Mice were maintained under specific pathogen-free conditions, and all animal procedures were carried out according to the protocol approved by the Institutional Animal Care and Use Committee at Hokkaido University Graduate School of Medicine. Kaplan–Meier curves were constructed, and the brains were dissected and snap frozen immediately after mice died. The sections (10 μm) were stained with hematoxylin and eosin using standard protocols.
Immunoblotting
Immunoblotting was performed by the method described elsewhere. Cells were lysed with buffer containing 0.5% NP40 (nonyl phenoxypolyethoxylethanol), 10 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1 mM EDTA, 50 mM NaF, 1 mM phenylmethylsulfonyl fluoride, and 1 mmol/L Na3VO4. Proteins were subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis, and separated proteins were transferred to a polyvinylidene difluoride filter (Immobilon-P; Millipore). Filters were probed with antibodies obtained from the following sources: anti-EREG (D405I) monoclonal antibody (mAb), p44/42 MAPK (Erk1/2) polyclonal antibody, anti–phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204) polyclonal antibody, anti–signal transducers and activators of transcription (STAT)3 mAb, anti–phospho-STAT3 (Tyr705) polyclonal antibody, anti–phospho-EGFR (Tyr1068) (D7A5) rabbit mAb (Cell Signaling Technology), anti-actin mAb (Chemicon), and anti-EGFR antibody (D-20) (Santa Cruz Biotechnology). Bound antibodies were detected with peroxidase-labeled goat antibody to mouse IgG, goat antibody to rabbit IgG, or rabbit antibody to goat IgG and visualized by enhanced chemiluminescence reagents (Amersham Pharmacia Biotech).
Immunohistochemical analysis
Formalin-fixed paraffin-embedded tissues were sectioned and stained using anti–adipocyte enhancer binding protein 1 (AEBP1) mouse mAb (1D2) (MT3.1) (Abnova) and anti-EREG polyclonal antibody (Lifespan Biosciences). The intensity scores were 0 = negative or weakly positive and 1 = strongly positive; the proportional scores were: 0 = 0%; 1 = 1%–10%; 2 = 11%–50%; 3 = 51%–100%. By total score (intensity score + proportional score), immunohistochemical (IHC) positivity was classified as negative (total score = 0), weakly positive (total score = 1, 2), or strongly positive (total score = 3, 4).
Matrigel Invasion Assay
The invasive potential of GBM cells was assessed in vitro in Matrigel-coated invasion chambers (Becton Dickinson Biosciences) in accordance with the manufacturer's instructions. Briefly, cells in log phase of growth were serum starved for 24 h prior to seeding, detached by brief trypsinization, and resuspended in medium containing the appropriate treatment. The Matrigel invasion inserts were rehydrated and prepared as described in the manufacturer's instructions. Cells (5 × 104 /mL in 0.5 mL serum-free medium) were added in suspension to the upper chamber, and medium (0.75 mL, supplemented with 10% fetal bovine serum as a chemoattractant) containing the same treatment was added to the bottom well. After incubation for 24 h, the noninvasive cells were removed from the upper surface of the membrane, and the invasive cells on the lower surface of the membrane were stained with 0.04% crystal violet and counted microscopically. Experiments were done in triplicate.
Immunocytofluorescence and Confocal Microscopy
Glioblastoma multiforme cells grown on Lab-Tek chamber slides (Nalge Nunc International) were fixed with 3% paraformaldehyde in phosphate buffered saline (PBS) for 15 min, permeabilized with 0.1% Triton X-100 in PBS for 5 min, and incubated with 1% bovine serum albumin in PBS for 20 min at room temperature. They were then incubated overnight at 4°C with mouse mAb specific for paxillin (1:5000; BD Transduction Laboratories), then for 1 h at room temperature with Alexa Fluor 488–conjugated secondary antibodies (Molecular Probes), and finally for 30 min at 37°C with Alexa Fluor 594–conjugated phalloidin (Molecular Probes). Fluorescent images were obtained using a fluorescence microscope (Olympus).
Neurosphere Culture
GBM cells were plated as single cells in ultralow attachment plates at a low density (2500 cells/mL) and were grown in neurosphere culture medium with or without gefitinib (Cayman Chemical Company) at the indicated concentrations for 4–7 days. Neurosphere culture medium was serum-free DMEM containing 10 μg/mL bovine insulin, 100 μg/mL human transferrin, 100 μg/mL bovine serum albumin, 60 ng/mL progesterone, 16 μg/mL putrescine, 40 ng/mL sodium selenite, 63 μg/mL N-acetylcysteine, 5 μM forskolin, 50 units/mL penicillin, and 50 μg/mL streptomycin (Gibco), as well as 10 ng/mL basic fibroblast growth factor or 10 ng/mL platelet-derived growth factor, or both.
Reverse Transcription PCR Analysis
Total RNA was isolated from cells using TRI Reagent (Sigma) and resuspended in RNA secure resuspension solution (Ambion). Reverse transcription was carried out with Superscript II RT (Invitrogen). The resulting first-strand cDNA was used as a template and amplified by PCR using KOD-Plus DNA polymerase (Toyobo).
Microarray Analysis
Total RNA was extracted using TRI Reagent (Sigma) and qualified using an Agilent 2100 Bioanalyzer. All samples showed RNA integrity numbers >9.5 and were subjected to microarray experiments according to the manufacturer's instructions. In brief, RNA samples were labeled using the Low Input Quick Amp Labeling Kit (Agilent Technologies). Labeling of 100 ng total RNA was performed using cyanine 3-CTP. Hybridization was carried out using the Gene Expression Hybridization Kit (Agilent Technologies). Samples of 1.65 μg cRNA were subjected to fragmentation (30 min at 60°C) and then hybridized on a 4 × 180K Agilent Whole Human Genome Oligo DNA Custom Microarray (G4862A) in a rotary oven (10 rpm at 65°C for 17 h). The array included 180K probes designed to identify transcripts of coding and predicted noncoding genes, including long intergenic noncoding RNAs. The annotation file about the probes on the array is shown in Supplementary Table 1. Slides were washed in Agilent Gene Expression Wash Buffers 1 and 2 and scanned with an Agilent DNA Microarray Scanner. To adjust for differences in the probe intensity distribution across different chips, gene expression values measured with the microarrays for U373, U373X1, LN443, and LN443X were normalized with GeneSpring software (Agilent Technologies) by using the 75th percentile value.
Statistical analysis was performed by unpaired Student t-test; as a multiple testing correction, Benjamini Hochberg false discovery rate was exploited. P < .05 was considered statistically significant. Fold changes were calculated by comparing normalized expression values.
Results
Establishment of Highly Malignant Glioma by Single Subcutaneous Passage in Mice
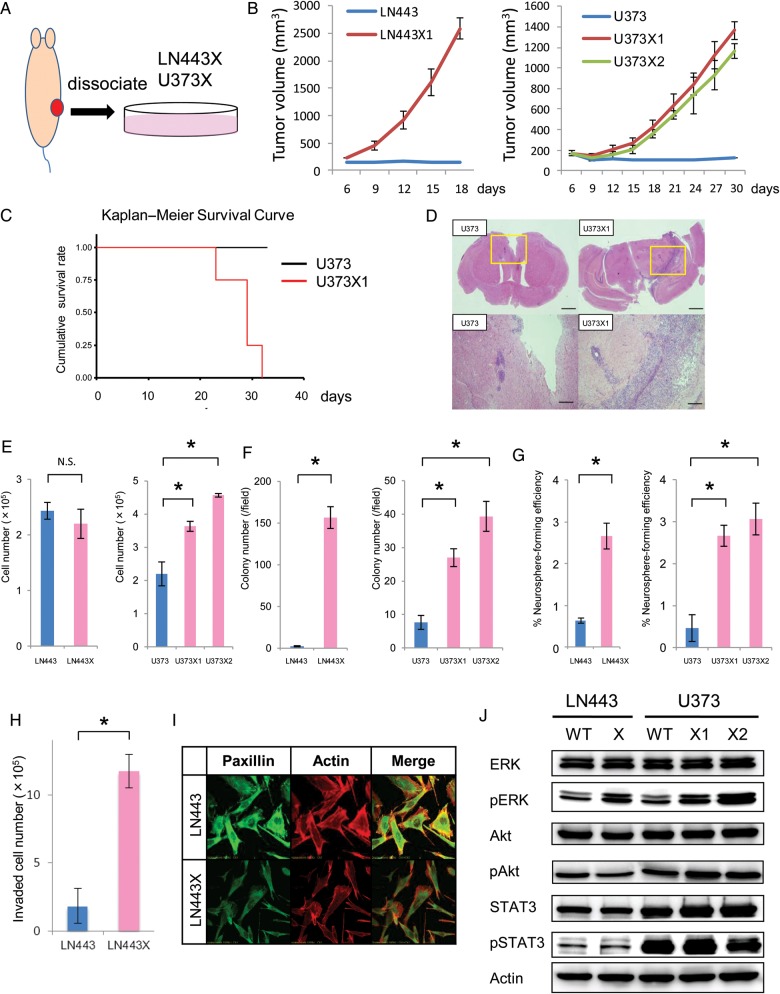
Tumor environment has been shown to affect tumor growth, and an aggressive population may be enriched in vivo. To examine whether single subcutaneous passage in mice promotes tumor progression in GBM, we injected GBM cell lines LN443 and U373 into nude mice subcutaneously and formed tumor masses in vivo. GBM cells were then isolated from these subcutaneous tumors and expanded in vitro to create 3 cell lines, designated LN443X, U373X1, and U373X2 (Fig. 1A).
Fig. 1.
Establishment of highly malignant glioma by (A) the scheme of a single subcutaneous passage in mice. In the tissue of mice were s.c. injected 5 × 106 cells of LN443 and U373. The xenografts at 3 months were removed, dissociated, and cultured in the tissue culture dishes. The established cell line from LN443 was designated LN443X and those from 2 subcutaneous tumors of U373 were designated U373X1 and U373X2. (B) Evaluation of the growth in vivo. In the subcutaneous tissue of mice were injected 5 × 106 cells of LN443, LN443X, U373, U373X1, and U373X2. The tumor volumes were calculated as described in Materials and Methods. (C) Kaplan–Meier curves were constructed for mice i.c. implanted with U373 or U373X1. Analysis was performed in 4 mice from each group. Overall survival of mice implanted with U373X1 is significantly worse (P = .012). (D) Representative photographs of intracranial tumors with hematoxylin and eosin staining, consisting of U373 or U373X1. The upper and lower panels show the whole brain sections and magnifications of the insets in the upper panels, respectively. Scale bar, 1 mm (upper panels) and 200 μm (lower panels). (E) Evaluation of the growth in vitro. Cultured in DMEM were 5 × 104 cells of LN443, LN443X, U373, U373X1, and U373X2. The cell numbers of each group were counted at day 5. *P < .01. N.S., not significant. (F) Soft-agar colony formation assay. The number of colonies was counted at day 21 in each group. Error bars represent SD of 3 independent experiments. *P < .01. (G) Neurosphere formation assay. GBM cells were plated as single cells in ultralow attachment plates at a low density (2500 cells/mL) and were grown in neurosphere culture medium for 4–7 days. The spheres were counted and the percentage of nuerosphere-forming cells was determined in each group. Error bars represent SD of 3 independent experiments. *P < .01. (H) Matrigel invasion assay. The invasive cells of LN443 and LN443X on the lower surface of the membrane were counted microscopically. Error bars represent SD of 3 independent experiments. *P < .01. (I) Immunocytofluorescence. LN443 and LN443X were fixed, permeabilized, stained with rhodamine-phalloidin to visualize filamentous actin (red) and with anti-paxillin antibodies to visualize focal adhesions (green), and examined by confocal microscopy. Magnification is ×600. (J) Immunoblot analysis of ERK/MAPK, Akt/PI3K, and Janus kinase/STAT pathways. The levels of ERK, pERK, Akt, pAkt, STAT3, and pSTAT3 were evaluated (top panel to the 6th panel from the top). Actin is shown as a loading control (bottom panel). WT, wild type.
First, we evaluated in vivo tumor growth of these established cells by re-injecting them into nude mice. Tumors formed from LN443X, U373X1, and U373X2 were observed within 10 days, while those derived from the original cell lines LN443 and U373 were observed only after 2 months (Fig. 1B, Supplementary Fig. 1A). Furthermore, in an orthotopic xenograft model, we found that the overall survival of mice implanted with U373X1 was significantly poorer than those implanted with U373 (P = .012; Fig. 1C). The intracranial tumor formed by U373X also exhibited a more invasive appearance compared with the U373-derived tumor (Fig. 1D). In culture, morphology of LN443X and U373X1 appeared more spindled, with piled-up growth compared with LN443 and U373 (Supplementary Fig. 1B). To confirm the growth advantage of LN443X, U373X1, and U373X2 seen in vivo, in vitro growth rates were measured, soft-agar colony formation assays were performed to assess the anchorage-independent growth, and neurosphere assays were performed to evaluate the neural stem cell component. Although in vitro growth of LN443X was not significantly different from that of LN443, the growth of U373X was faster than that of U373 (Fig. 1E). In soft-agar colony formation assays and neurosphere formation assays, the number of colonies and neurospheres were increased in LN443X, U373X1, and U373X2 compared with their parental cell lines (Fig. 1F and G, Supplementary Fig. 1C and 1D). In invasion assay, LN443X showed higher invasiveness compared with LN443, while U373X1 did not (Fig. 1H and Supplementary Fig. 1E). Furthermore, immunofluorescence analysis showed an irregular and disordered distribution of actin filaments and paxillin in LN443X but not LN443, in which regular focal adhesions were observed (Fig. 1I and Supplementary Fig. 2A). A too bizarre cellular morphology of U373X1 was exhibited, precluding evaluation of the distribution of actin or paxillin (Supplementary Fig. 2B). As highly malignant phenotypes could be observed in LN443X, U373X1, and U373X2, we hypothesized that GBM-related signaling pathways, including ERK/MAPK, Akt/PI3K, and Janus kinase/STAT, were activated in these cells. The phosphorylated form of ERK was increased in LN443X, U373X1, and U373X2, while phosphorylation levels of Akt and STAT3 were unchanged (Fig 1J).
Gene Expression Profiling in LN443X and U373X1
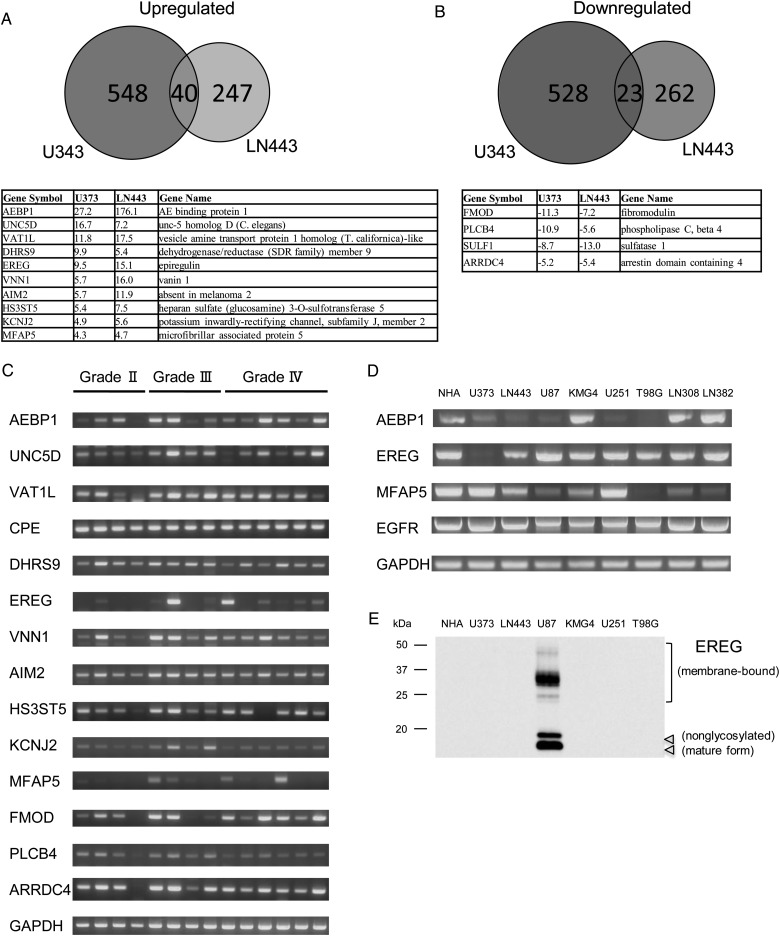
We employed DNA microarray analysis to examine alteration in gene expression during the tumor progression of LN443 and U373 to LN443X and U373X1. The number of upregulated genes with a >2-fold increase in LN443 compared with LN443 was 287, and that in U373X1 compared with U373 was 588. Forty and 10 genes showed a 2- and 4-fold upregulation in both cell lines, respectively (Fig. 2A). On the other hand, the expression levels of 285 genes were downregulated in LN443X by >2-fold compared with LN443, while the levels of 551 genes were decreased in U373X1 compared with U373. Twenty-three and 4 genes showed a 2- and 4-fold downregulation in both cell lines, respectively (Fig. 2B).
Fig. 2.
Gene expression profiling in LN443X and U373X1. (A) Diagram of the number of genes that were specifically or commonly upregulated in LN443X and U373X1 compared with LN443 and U373, respectively. Commonly upregulated genes by more than 4-fold are indicated in the list. (B) Diagram of the number of genes that were specifically or commonly downregulated in LN443X and U373X1 compared with LN443 and U373, respectively. Commonly downregulated genes by more than 4-fold are indicated in the list. (C) Expression analysis of commonly up- or downregulated genes in 14 glioma samples (WHO grades II to IV) by reverse transcription PCR. Expression glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as an internal control. (D) Expression analysis of AEBP1, EREG, MFAP5, EGFR, and GAPDH in GBM cell lines by reverse transcription PCR. GAPDH expression was used as an internal control. NHA, normal human astrocytes. (E) Immunoblot analysis of EREG was evaluated in NHA and GBM cell lines.
We further investigated the expression levels of the 14 genes with 4-fold alterations in 14 human glioma samples (ranging from WHO grades II to IV) by reverse transcription PCR (Fig. 2C). The expression levels of AEBP1, EREG, and microfibrillar associated protein 5 (MFAP5) were different among the samples. Especially, the levels of EREG were not clearly observed in low-grade glioma as grade II but were relatively high in high-grade glioma as grades III and IV (Fig. 2C). The mRNA levels of AEBP1, EREG, and MFAP5 were evaluated in 10 human GBM cell lines using normal human astrocytes as control, and higher expression of these genes was observed in 5 to 6 cell lines with different expression profiles (Fig. 2D). Immunoblotting analysis revealed that expression levels of EREG protein were remarkably high in U87 cells, corresponding with its mRNA level (Fig. 2E). For further investigation, we utilized the public database obtained from the Repository for Molecular Brain Neoplasia Data (REMBRANDT) of the National Cancer Institute (http://rembrandt.nci.nih.gov). According to REMBRANDT, AEBP1 upregulation is observed in 71.7% of GBM, and this is significantly associated with shorter median survival compared with tumors with intermediate expression (P < 1 × 10−7; Supplementary Fig. 3).
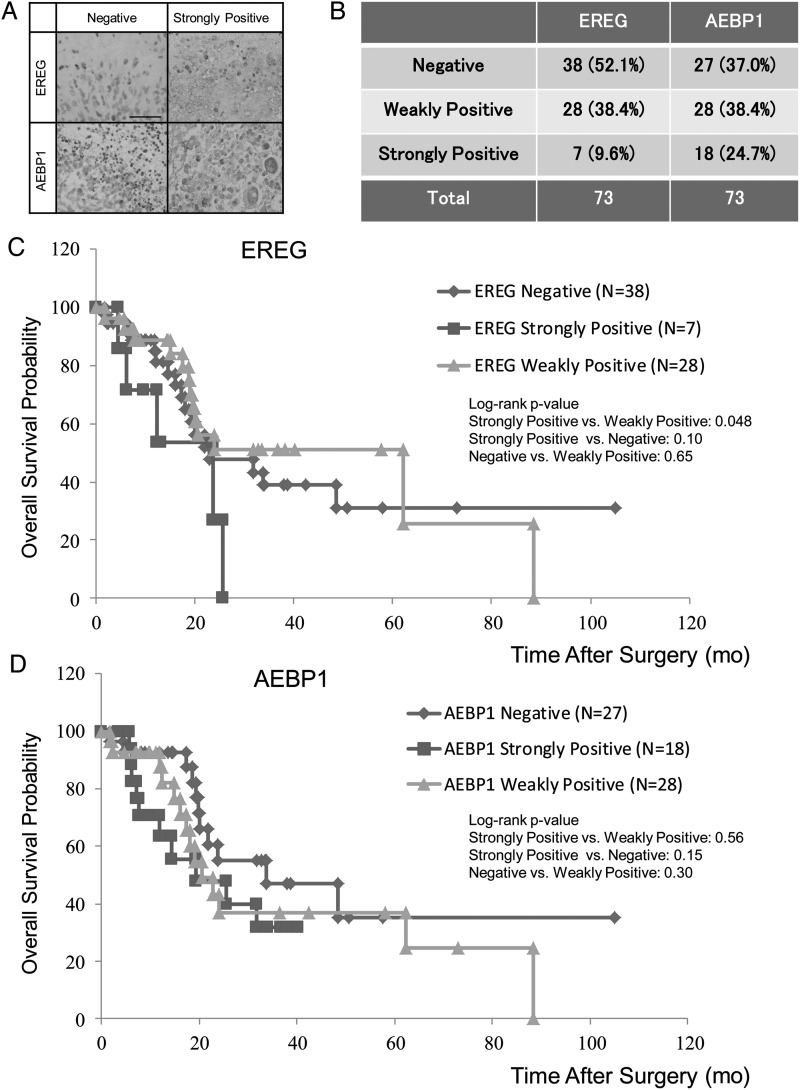
Immunohistochemical Analysis of EREG and AEBP1 in GBM
As we had observed a correlation of AEBP1 with poor prognosis and a higher expression of EREG in malignant gliomas, we further analyzed the expression levels of AEBP1 and EREG by IHC in 73 GBM cases with known clinical follow-up. Immunohistochemical stainings of AEBP1 and EREG were scored as negative, weakly positive, or strongly positive according to the criteria described in Materials and Methods (Fig. 3A). Seven (9.6%) and 18 cases (24.7%) showed strong positive staining for EREG and AEBP1, respectively (Fig. 3B). Patients with GBM who exhibited positive staining for EREG had significantly shorter overall survival compared with those with weak immunopositivity (P < .05; Fig. 3C). However, the immunophenotypic status of AEBP1 did not show any correlation with patients' overall survival (Fig. 3D).
Fig. 3.
Immunohistochemical analysis of EREG and AEBP1 in GBM. (A) Representative photographs of IHC analysis for EREG and AEBP1 in surgical specimens of malignant glioma whose IHC expression was classified as negative, weakly positive, or strongly positive, as determined by a combination of both intensity score and proportion score described in Materials and Methods. Magnification is ×400. Scale bar, 100 μm. (B) The proportion of IHC positivity for EREG and AEBP1 are indicated. (C and D) Kaplan–Meier survival curves of 73 GBM patients grouped by IHC expression of EREG (C) and AEBP1 (D). Blue line: negative. Red line: strongly positive. Green line: weakly positive. Log-rank P values are indicated.
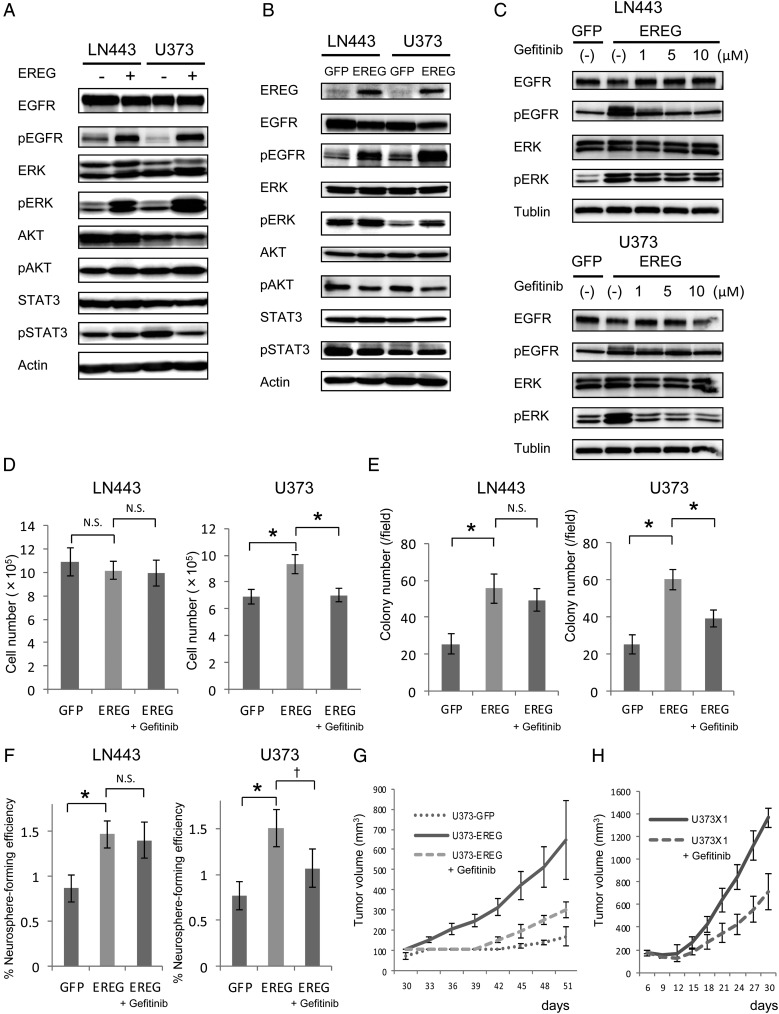
EREG Activates MAPK Pathway and Promotes Colony Formation, Sphere Formation, and Tumor Formation
As EREG is known to activate EGFR (ErbB1 and ErbB4) as its ligand, we examined whether EREG phosphorylates EGFR and promotes subsequent activation of its downstream signaling pathway, including phosphorylation of ERK, Akt, and STAT3 in GBM cell lines. Twenty-four hours after EREG stimulation, phosphorylation levels of EGFR and ERK were elevated in both LN443 and U373, while those of Akt and STAT3 remained unchanged (Fig. 4A). We then evaluated the effect of EREG on primary GBM cells and found that EREG stimulation was also able to promote the phosphorylations of EGFR and ERK (Supplementary Fig. 4).
Fig. 4.
EREG activates MAPK pathway and promotes colony formation, sphere formation, and tumor formation. (A) Immunoblot analyses of EGFR, pEGFR, ERK, pERK, Akt, pAkt, STAT3, and pSTAT3 were evaluated in LN443 and U373 treated with 10 μM of EREG for 24 h (top panel to the 8th panel from the top). Actin is shown as a loading control (bottom panel). (B) Immunoblot analyses of EREG, EGFR, pEGFR, ERK, pERK, Akt, pAkt, STAT3, and pSTAT3 were evaluated in LN443- and U373-introduced GFP or EREG by retroviral vector (top panel to the 9th panel from the top). Actin is shown as a loading control (bottom panel). (C) Immunoblot analyses of EGFR, pEGFR, ERK, and pERK, were evaluated in LN443-GFP, LN443-EREG, U373-GFP, and U373-EREG treated with gefitinib or control dimethyl sulfoxide (DMSO; top panel to the 5th panel from the top). Tubulin is shown as a loading control (bottom panel). (D) Evaluation of the growth in vitro. Cultured were 5 × 104 cells of LN443-GFP, LN443-EREG, U373-GFP, and U373-EREG in DMEM treated with gefitinib (10 μM) or control DMSO. The cell number of each group was counted at day 6. Error bars represent SD of 3 independent experiments. *P < .01. N.S., not significant. (E) Soft-agar colony formation assay. Seeded were 5 × 104 cells of LN443-GFP, LN443-EREG, U373-GFP, and U373-EREG into 6-cm tissue culture dishes and treated with gefitinib (10 μM) or control DMSO. The number of colonies was counted at 30 days in each group. Error bars represent SD of 3 independent experiments. *P < .01. N.S., not significant. (F) Neurosphere formation assay. GBM cells were plated as single cells in ultralow attachment plates at a low density (2500 cells/mL) and were grown in neurosphere culture medium with or without gefitinib (10 μM) for 4–7 days. The spheres were counted and the percentage of nuerosphere-forming cells was determined in each group. Error bars represent SD of 3 independent experiments. *P < .01, †P < .05. N.S., not significant. (G) Evaluation of the growth in vivo. Injected were 5 × 106 cells of U373-GFP and U373-EREG, and gefitinib (200 mg/kg) or control DMSO was i.p. administered at days 25–29 and day 32–36. The tumor volumes were calculated as described in Materials and Methods. (H) Evaluation of the growth in vivo. Injected were 5 × 106 cells of U373X1, and gefitinib (200 mg/kg) or control DMSO was i.p. administered at days 3–7 and days 10–14. The tumor volumes were calculated as described in Materials and Methods.
To confirm the significance of EREG on EGFR-dependent signaling, we generated EREG-overexpressed cells (LN443-EREG and U373-EREG) by using retroviral gene transfer into LN443 and U373 (Fig. 4B). Western blotting analysis provided evidence of elevated phosphorylation levels of EGFR and ERK both in LN443-EREG and U373-EREG (Fig. 4B).
To further validate our findings, we examined whether a selective EGFR–tyrosine kinase inhibitor such as gefitinib could suppress the phosphorylation levels of EGFR and ERK. The suppression of phosphorylation of both ERK and EGFR was observed in U373-EREG, and a similar, though more subtle suppression was also observed in LN443-EREG (Fig. 4C). The in vitro growth of U373-EREG was faster than control U373 (U373–green fluorescent protein [GFP]), and this growth enhancement was suppressed with gefitinib treatment (Fig. 4D). It should be noted that the growth of LN443-EREG was not significantly different from GFP-introduced control cells (LN443-GFP). Anchorage-independent growth and serum-independent growth were measured by soft-agar colony formation assays and sphere formation assays, and the numbers of colonies and spheres were increased in both LN443-EREG and U373-EREG cells compared with LN443-GFP and U373-GFP cells, respectively (Fig. 4E and F). Gefitinib treatment suppressed these enhanced growths in U373-EREG (Fig. 4E and F). Small interfering RNA knockdown of EREG also inhibited Matrigel invasion ability (Supplementary Fig. 5A–5C) and neurosphere-forming activity (Supplementary Fig. 6A and 6B) of U373X1 and LN443X cells. Although we performed neurosphere formation assays using several primary GBM cells, none of these cells formed neurospheres even with EREG stimulation (data not shown).
To examine the growth potential in vivo, the cells were injected subcutaneously into the flanks of nude mice and the size of subsequent tumors were measured. It was found that the growth of tumors derived from U373-EREG was faster than that derived from U373-GFP and that gefitinib treatment inhibited this growth enhancement. Similar findings were observed with U373X1 cells (Fig. 4G and H).
Discussion
By using single subcutaneous passage in mice, we have established highly aggressive variants of human GBM cell lines LN443 and U373. These showed enhanced growth, colony-forming and sphere-forming potential, invasive ability, and tumor-forming potential in vivo compared with the parental cell lines. It has been shown recently that malignant tumors exhibit genetic heterogeneity, and in this study we found that the subcutaneous microenvironment may affect clonal expansion of aggressive subpopulations of GBM cells. Data from expression profiling analysis have suggested that GBM can be classified into 3 subtypes: proneuronal, proliferative, and mesenchymal, and recurrent tumors are thought to exhibit a mesenchymal signature. However, we were unable to classify the aggressive cells established in this study into any of these subtypes (Supplementary Fig. 7). A previous paper also could not detect comparable transcriptional subtypes in immortalized cell lines.19 The discrepancy between cell lines and patients' samples may be due to loss of specific gene expression or selection during the culture process. Therefore, we validated the results from our in vivo experiments by performing IHC analysis of patients' samples.
Using DNA microarray analysis, EREG was found to be a candidate gene for a malignant phenotype. EREG is a member of the epidermal growth factor (EGF) family, which includes heparin-binding EGF-like growth factor, transforming growth factor (TGF)–α, epigen, amphiregulin, betacellulin, and neuregulins; EREG functions as a ligand of EGFR and human EGFR 4.20,21 Not only has EREG been shown to enhance cell growth, but it also plays a role in differentiation, migration, adhesion, and tumor vessel assembly.22,23 Overexpression of EREG has been observed in various cancers arising in the bladder, lung, kidney, and colon24,25; this study is the first to demonstrate its involvement in brain tumors.
Considering the increased sphere-forming potentials of LN443X, U373X1, and U373X2, these cell populations include cancer stem–like/cancer-initiating cells. Various pathways, such as those of EGFR dependency,26,27 PI3K/Akt,28,29 Wnt/β-catenin,30 c-Met signaling,31 STAT3 signaling,32,33 TGF-β signaling,34 and Notch signaling,35,36 have been implicated in maintaining cancer stemness in GBM. Previously, we reported that under the EGFR/ERK-dependent signaling pathway, the Ets family of transcription factors were involved in the expression of CD133, which is thought to be one of the cancer stem cell markers for GBM.17 Considering that the EGFR/ERK pathway regulates cancer stemness, the identification of EREG in our experimental system for tumor aggressiveness may explain the reason to enrich tumor aggressiveness. In fact, another growth-factor family of proteins known as heregulin has been reported to regulate mammosphere formation in breast cancer.37 We tried to evaluate an association between expression of EREG and the stem cell characters in U373 and LN443 by comparing the expression of Sox2 in U373, U373X1, LN443, and LN443X. Immunofluorescence analysis revealed that Sox2 was highly expressed in all U373 cells, but no expression was observed in LN443 (Supplementary Fig. 8A). No significant changes in Sox2 protein expression were detected after subcutaneous passage in mice (Supplementary Fig. 8B), suggesting that Sox2 is not a marker for stemness in U373 and LN443. CD133, another stemness marker, was also hardly detectable in these cells (data not shown).
Inhibition of the EGFR-dependent signaling pathway may be a candidate for targeted molecular therapy because our data indicated that EREG immunopositivity found in 9.6% of tumors correlated with poor prognosis, and we also confirmed the contribution of EREG to the malignant phenotype by establishing EREG overexpressing cell lines. However, phase II trials have so far shown limited clinical benefit of the small-molecule EGFR inhibitor erlotinib in patients with either recurrent or newly diagnosed GBM, either in combination regimens38–41 or as monotherapy.42,43 While gefitinib inhibited the phosphorylation of EGFR in both LN443 and U373, inhibition of ERK phosphorylation was observed in only U373, suggesting the presence of an alternative mechanism of phosphorylation of ERK other than by EGFR signaling in LN443. Besides, although gefitinib treatment was performed for the subcutaneous tumors of LN443X in nude mice, we observed no effect on tumor growth inhibition (data not shown). Further studies to elucidate the pathogenesis of GBM is essential to identify additional molecular targets for treatment of patients.
Another molecule associated with tumor aggressiveness found in this study was AEBP1, originally identified as a transcriptional repressor for the AE1 element located in the proximal promoter region of the adipose P2 gene, which codes adipocyte-specific fatty acid binding protein 4 regulating adipogenesis.44,45 Recently, it has been reported that AEBP1 affects tumor growth and survival in patients with gliomas.46 However, the correlation of protein expression by IHC has not been studied. It should be noted that while results from the REMBRANDT database suggest a correlation of AEBP1 upregulation with poor prognosis, our IHC analysis did not show any significant correlation. One of the reasons for this discrepancy may be the limited number of cases in our cohort. A larger-scale IHC analysis is needed to clarify the significance of AEBP1 positivity in GBM. REMBRANDT data of EREG expression analysis seem to be inconsistent with our IHC-derived data, due to possible differences of expression levels between EREG mRNA and its protein. Indeed, REMBRANDT evaluated the expression levels of mRNA, while we evaluated protein levels by IHC. Specifically, REMBRANDT categorized 93.9% of cases into an intermediate category in the expression analysis of EREG, whereas 52% of cases were negative by IHC in our study, suggesting posttranscriptional regulation of EREG. Therefore, we propose that evaluating EREG expression by IHC may be a more reliable way to predict patient survival.
In this study, we found that 2 molecules, EREG and AEBP1, are involved in the acquisition of the malignant potential of GBM cell lines in vivo, and these may be potential therapeutic targets for the treatment of GBM. However, the pathogenicity of GBM cannot be entirely accounted for by these 2 molecules; thus, additional studies using other GBM cell lines or pairing with human GBM primary cultures should be performed to identify other therapeutic targets for establishing tailor-made therapy to eradicate this highly aggressive tumor.
Supplementary Material
Funding
This work was supported in part by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan.
Supplementary Material
Acknowledgments
We would like to thank Dr Erwin G. Van Meir (Emory University School of Medicine, Atlanta, Georgia) for providing cell lines and Dr Tsuyoshi Akagi and Dr Ken Sasai (KAN Research Institute, Inc, Kobe, Japan) for providing the plasmid. We are also grateful to Shiori Akesaka for technical assistance.
Conflict of interest statement. None declared.
References
- 1.Louis DN, Ohgaki H, Wiestler OD, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114(2):97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Van Meir EG, Hadjipanayis CG, Norden AD, et al. Exciting new advances in neuro-oncology: the avenue to a cure for malignant glioma. CA Cancer J Clin. 2010;60(3):166–193. doi: 10.3322/caac.20069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mrugala MM, Chamberlain MC. Mechanisms of disease: temozolomide and glioblastoma—look to the future. Nat Clin Pract Oncol. 2008;5(8):476–486. doi: 10.1038/ncponc1155. [DOI] [PubMed] [Google Scholar]
- 4.Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 5.Network TCGAR. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Phillips HS, Kharbanda S, Chen R, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9(3):157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 7.Wick W, Weller M, Weiler M, et al. Pathway inhibition: emerging molecular targets for treating glioblastoma. Neuro Oncol. 2011;13(6):566–579. doi: 10.1093/neuonc/nor039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reya T, Morrison SJ, Clarke MF, et al. Stem cells, cancer, and cancer stem cells. Nature. 2001;414(6859):105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 9.Liu K, Lin B, Zhao M, et al. The multiple roles for Sox2 in stem cell maintenance and tumorigenesis. Cell Signal. 2013;25(5):1264–1271. doi: 10.1016/j.cellsig.2013.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lathia JD, Gallagher J, Heddleston JM, et al. Integrin alpha 6 regulates glioblastoma stem cells. Cell Stem Cell. 2010;6(5):421–432. doi: 10.1016/j.stem.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ogden AT, Waziri AE, Lochhead RA. Identification of A2B5+CD133- tumor-initiating cells in adult human gliomas. Neurosurgery. 2008;62(2):505–514. doi: 10.1227/01.neu.0000316019.28421.95. discussion 514–515. [DOI] [PubMed] [Google Scholar]
- 12.Read TA, Fogarty MP, Markant SL, et al. Identification of CD15 as a marker for tumor-propagating cells in a mouse model of medulloblastoma. Cancer Cell. 2009;15(2):135–147. doi: 10.1016/j.ccr.2008.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Son MJ, Woolard K, Nam DH, et al. SSEA-1 is an enrichment marker for tumor-initiating cells in human glioblastoma. Cell Stem Cell. 2009;4(5):440–452. doi: 10.1016/j.stem.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bao S, Wu Q, Li Z, et al. Targeting cancer stem cells through L1CAM suppresses glioma growth. Cancer Res. 2008;68(15):6043–6048. doi: 10.1158/0008-5472.CAN-08-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Galli R, Binda E, Orfanelli U, et al. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res. 2004;64(19):7011–7021. doi: 10.1158/0008-5472.CAN-04-1364. [DOI] [PubMed] [Google Scholar]
- 16.Singh SK, Hawkins C, Clarke ID, et al. Identification of human brain tumour initiating cells. Nature. 2004;432(7015):396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 17.Tabu K, Kimura T, Sasai K, et al. Analysis of an alternative human CD133 promoter reveals the implication of Ras/ERK pathway in tumor stem-like hallmarks. Mol Cancer. 2010;9:39. doi: 10.1186/1476-4598-9-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brescia P, Richichi C, Pelicci G. Current strategies for identification of glioma stem cells: adequate or unsatisfactory? J Oncol. 2012;2012:376894. doi: 10.1155/2012/376894. Article ID. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Verhaak RG, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eltarhouny SA, Elsawy WH, Radpour R, et al. Genes controlling spread of breast cancer to lung “gang of 4.”. Exp Oncol. 2008;30(2):91–95. [PubMed] [Google Scholar]
- 21.Komurasaki T, Toyoda H, Uchida D, et al. Epiregulin binds to epidermal growth factor receptor and ErbB-4 and induces tyrosine phosphorylation of epidermal growth factor receptor, ErbB-2, ErbB-3 and ErbB-4. Oncogene. 1997;15(23):2841–2848. doi: 10.1038/sj.onc.1201458. [DOI] [PubMed] [Google Scholar]
- 22.Inatomi O, Andoh A, Yagi Y, et al. Regulation of amphiregulin and epiregulin expression in human colonic subepithelial myofibroblasts. Int J Mol Med. 2006;18(3):497–503. [PubMed] [Google Scholar]
- 23.Gupta GP, Nguyen DX, Chiang AC, et al. Mediators of vascular remodelling co-opted for sequential steps in lung metastasis. Nature. 2007;446(7137):765–770. doi: 10.1038/nature05760. [DOI] [PubMed] [Google Scholar]
- 24.Lee D, Pearsall RS, Das S, et al. Epiregulin is not essential for development of intestinal tumors but is required for protection from intestinal damage. Mol Cell Biol. 2004;24(20):8907–8916. doi: 10.1128/MCB.24.20.8907-8916.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yun J, Song SH, Park J, et al. Gene silencing of EREG mediated by DNA methylation and histone modification in human gastric cancers. Lab Invest. 2012;92(7):1033–1044. doi: 10.1038/labinvest.2012.61. [DOI] [PubMed] [Google Scholar]
- 26.Jin X, Yin J, Kim SH, et al. EGFR-AKT-Smad signaling promotes formation of glioma stem-like cells and tumor angiogenesis by ID3-driven cytokine induction. Cancer Res. 2011;71(22):7125–7134. doi: 10.1158/0008-5472.CAN-11-1330. [DOI] [PubMed] [Google Scholar]
- 27.Mazzoleni S, Politi LS, Pala M, et al. Epidermal growth factor receptor expression identifies functionally and molecularly distinct tumor-initiating cells in human glioblastoma multiforme and is required for gliomagenesis. Cancer Res. 2010;70(19):7500–7513. doi: 10.1158/0008-5472.CAN-10-2353. [DOI] [PubMed] [Google Scholar]
- 28.Bleau AM, Hambardzumyan D, Ozawa T, et al. PTEN/PI3 K/Akt pathway regulates the side population phenotype and ABCG2 activity in glioma tumor stem-like cells. Cell Stem Cell. 2009;4(3):226–235. doi: 10.1016/j.stem.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hambardzumyan D, Becher OJ, Rosenblum MK, et al. PI3 K pathway regulates survival of cancer stem cells residing in the perivascular niche following radiation in medulloblastoma in vivo. Genes Dev. 2008;22(4):436–448. doi: 10.1101/gad.1627008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu X, Morales FC, Agarwal NK, et al. Moesin is a glioma progression marker that induces proliferation and Wnt/beta-catenin pathway activation via interaction with CD44. Cancer Res. 2013;73(3):1142–1155. doi: 10.1158/0008-5472.CAN-12-1040. [DOI] [PubMed] [Google Scholar]
- 31.De Bacco F, Casanova E, Medico E, et al. The MET oncogene is a functional marker of a glioblastoma stem cell subtype. Cancer Res. 2012;72(17):4537–4550. doi: 10.1158/0008-5472.CAN-11-3490. [DOI] [PubMed] [Google Scholar]
- 32.Guryanova OA, Wu Q, Cheng L, et al. Nonreceptor tyrosine kinase BMX maintains self-renewal and tumorigenic potential of glioblastoma stem cells by activating STAT3. Cancer Cell. 2011;19(4):498–511. doi: 10.1016/j.ccr.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sherry MM, Reeves A, Wu JK, et al. STAT3 is required for proliferation and maintenance of multipotency in glioblastoma stem cells. Stem Cells. 2009;27(10):2383–2392. doi: 10.1002/stem.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ikushima H, Todo T, Ino Y, et al. Autocrine TGF-beta signaling maintains tumorigenicity of glioma-initiating cells through Sry-related HMG-box factors. Cell Stem Cell. 2009;5(5):504–514. doi: 10.1016/j.stem.2009.08.018. [DOI] [PubMed] [Google Scholar]
- 35.Fassl A, Tagscherer KE, Richter J, et al. Notch1 signaling promotes survival of glioblastoma cells via EGFR-mediated induction of anti-apoptotic Mcl-1. Oncogene. 2012;31(44):4698–4708. doi: 10.1038/onc.2011.615. [DOI] [PubMed] [Google Scholar]
- 36.Fan X, Khaki L, Zhu TS, et al. NOTCH pathway blockade depletes CD133-positive glioblastoma cells and inhibits growth of tumor neurospheres and xenografts. Stem Cells. 2010;28(1):5–16. doi: 10.1002/stem.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hinohara K, Kobayashi S, Kanauchi H, et al. ErbB receptor tyrosine kinase/NF-kappaB signaling controls mammosphere formation in human breast cancer. Proc Natl Acad Sci U S A. 2012;109(17):6584–6589. doi: 10.1073/pnas.1113271109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Groot JF, Gilbert MR, Aldape K, et al. Phase II study of carboplatin and erlotinib (Tarceva, OSI-774) in patients with recurrent glioblastoma. J Neurooncol. 2008;90(1):89–97. doi: 10.1007/s11060-008-9637-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peereboom DM, Shepard DR, Ahluwalia MS, et al. Phase II trial of erlotinib with temozolomide and radiation in patients with newly diagnosed glioblastoma multiforme. J Neurooncol. 2010;98(1):93–99. doi: 10.1007/s11060-009-0067-2. [DOI] [PubMed] [Google Scholar]
- 40.Prados MD, Chang SM, Butowski N, et al. Phase II study of erlotinib plus temozolomide during and after radiation therapy in patients with newly diagnosed glioblastoma multiforme or gliosarcoma. J Clin Oncol. 2009;27(4):579–584. doi: 10.1200/JCO.2008.18.9639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reardon DA, Desjardins A, Vredenburgh JJ, et al. Phase 2 trial of erlotinib plus sirolimus in adults with recurrent glioblastoma. J Neurooncol. 2010;96(2):219–230. doi: 10.1007/s11060-009-9950-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van den Bent MJ, Brandes AA, Rampling R, et al. Randomized phase II trial of erlotinib versus temozolomide or carmustine in recurrent glioblastoma: EORTC Brain Tumor Group study 26034. J Clin Oncol. 2009;27(8):1268–1274. doi: 10.1200/JCO.2008.17.5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thiessen B, Stewart C, Tsao M, et al. A phase I/II trial of GW572016 (lapatinib) in recurrent glioblastoma multiforme: clinical outcomes, pharmacokinetics and molecular correlation. Cancer Chemother Pharmacol. 2010;65(2):353–361. doi: 10.1007/s00280-009-1041-6. [DOI] [PubMed] [Google Scholar]
- 44.He GP, Muise A, Li AW, et al. A eukaryotic transcriptional repressor with carboxypeptidase activity. Nature. 1995;378(6552):92–96. doi: 10.1038/378092a0. [DOI] [PubMed] [Google Scholar]
- 45.Ro HS, Roncari DA. The C/EBP-binding region and adjacent sites regulate expression of the adipose P2 gene in human preadipocytes. Mol Cell Biol. 1991;11(4):2303–2306. doi: 10.1128/mcb.11.4.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ladha J, Sinha S, Bhat V, et al. Identification of genomic targets of transcription factor Aebp1 and its role in survival of glioma cells. Mol Cancer Res. 2012;10(8):1039–1051. doi: 10.1158/1541-7786.MCR-11-0488. [DOI] [PubMed] [Google Scholar]
- 47.Akagi T, Sasai K, Hanafusa H. Refractory nature of normal human diploid fibroblasts with respect to oncogene-mediated transformation. Proc Natl Acad Sci U S A. 2003;100(23):13567–13572. doi: 10.1073/pnas.1834876100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kohsaka S, Sasai K, Takahashi K, et al. A population of BJ fibroblasts escaped from Ras-induced senescence susceptible to transformation. Biochem Biophys Res Commun. 2011;410(4):878–884. doi: 10.1016/j.bbrc.2011.06.082. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.