Abstract
The recent innovation of collecting X-ray scattering from solutions containing purified macromolecules in high-throughput has yet to be truly exploited by the biological community. Yet, this capability is becoming critical given that the growth of sequence and genomics data is significantly outpacing structural biology results. Given the huge mismatch in information growth rates between sequence and structural methods, their combined high-throughput and high success rate make high-throughput small angle X-ray scattering (HT-SAXS) analyses increasingly valuable. HT-SAXS connects sequence as well as NMR and crystallographic results to biological outcomes by defining the flexible and dynamic complexes controlling cell biology. Commonly falling under the umbrella of bio-SAXS, HT-SAXS data collection pipelines have or are being developed at most synchrotrons. How investigators practically get their biomolecules of interest into these pipelines, balance sample requirements and manage HT-SAXS data output format varies from facility to facility. While these features are unlikely to be standardized across synchrotron beamlines, a detailed description of HT-SAXS issues for one pipeline provides investigators with a practical guide to the general procedures they will encounter. One of the longest running and generally accessible HT-SAXS endstations is the SIBYLS beamline at the Advanced Light Source in Berkeley CA. Here we describe the current state of the SIBYLS HT-SAXS pipeline, what is necessary for investigators to integrate into it, the output format and a summary of results from 2 years of operation. Assessment of accumulated data informs issues of concentration, background, buffers, sample handling, sample shipping, homogeneity requirements, error sources, aggregation, radiation sensitivity, interpretation, and flags for concern. By quantitatively examining success and failures as a function of sample and data characteristics, we define practical concerns, considerations, and concepts for optimally applying HT-SAXS techniques to biological samples.
Keywords: High-throughput, SAXS, Conformation, Structure, Structural genomics, Macromolecules
1 Introduction
Small angle X-ray scattering (SAXS) has reemerged in its application to the study of biological macromolecules. SAXS from biomolecules was an early application of synchrotron radiation [1] in part because of its simplicity in terms of sample preparation. However with the realization of degree to which biomolecules could be crystallized yielding atomic resolution structures, macromolecular crystallography (MX) quickly became a focus of structural biologists. Relatively speaking, the application of SAXS and the development of analytical tools languished. Over the course of the last 10 years, SAXS has reemerged as a powerful complimentary tool to MX.
Three factors have contributed to the emerging power of SAXS. First, not all macromolecules of interest are amenable to crystallization. Even when a macromolecule has been crystallized and modeled to atomic resolution, biologically relevant alternate conformations can, at best, be inferred. Through a genomic analysis, 35–48 % of human gene products are predicted to have significant flexible regions when isolated [2]. SAXS provides an avenue to capture critical structural information from biomolecules even after an atomic resolution model is available. SAXS results suggest conformational variation is a general functional feature of macromolecules, so biologically relevant structural analyses will require a comprehensive approach that assesses both flexibility, as seen by SAXS, and detail, as determined by X-ray crystallography and NMR [3]. Indeed, SAXS also provides three-dimensional arrangements and oligomeric state for full-length proteins in solution, which is typically the functional assembly state, as seen for DNA break response framework proteins [4, 5], thermophilic superoxide dismutase [6], ATPase motors [7], and abscisic acid receptor [8]. Second, analysis tools have been developed and made accessible for the extraction of structural information. Shapes of macromolecule may be determined to ~15 Å resolution. Higher resolution information may be probed by complimenting SAXS with information from an atomic resolution model. Building upon the promise of early tools [9], the EMBL ATSAS [10] package has been transformative. Others have further contributed to the expanding suite of software available for analysis [11–14]. Additionally, the practical implementation of the Porod-Debye law in SAXS experiments of biopolymers provides a tool for assessing flexibility and for validation of SAXS models [15]. Flexible regions of macromolecules are often involved in interactions, as seen for antibody–protein binding [16, 17], and SAXS provides a means to define solution conformations with flexible regions. As generally appreciated, crystal contacts and constructs with missing regions may cause structural changes in the crystal structure relative to the SAXS solution results [18]. SAXS has recently been used to provide similarity maps of the functional conformational states of macromolecules independent of shape reconstructions [19]. Third, high signal to noise SAXS profiles are routinely collected from small quantities of sample with short exposure times. High-quality SAXS profiles are the result of advances in X-ray detectors and high brilliance synchrotron light with beam dimensions that match sample dimensions. Thus the motivation to move beyond the limits of MX, improved analysis tools and collection capabilities have all contributed to the increase in structural reports utilizing SAXS.
The advent and wide spread availability of high-throughput SAXS is relatively new. Pipelines for high-throughput SAXS have been reported at SSRL [20], SOLEIL [21], PETRA3 [22], and CHESS [23]. Several additional beamlines have developed these capabilities and are yet to be reported. SAXS at SIBYLS has been dedicated to HT-SAXS for the last 3 years with the initial application to structural genomics pipelines [24]. SIBYLS has leveraged tools developed for crystallography such as data control software and optimized features for SAXS [25].
A distinction of the SAXS at SIBYLS is that a significant fraction of samples are collected via mail-in/hand-in. Once an investigator’s samples have been delivered to the beamline their samples are placed into a queue and collected by beamline staff. The data output is a SAXS profile which tabulates the q value (X-ray momentum transfer) versus X-ray intensity with an error bar. This three-column format is electronically delivered post collection. One advantage to the mail-in/hand-in approach is an increase in flexibly arranging data collection times. Optimal sample preparation is often challenging and difficult to coordinate for a specific time. A second advantage is that “beamtime” is spent collecting data rather than training; thus increasing throughput. The disadvantage is that the investigators themselves are not there to guarantee every sample. Thus the guiding principle for development of the mail-in/hand-in program has been to enable data collection at as high qualities as if the investigator was present themselves. Over 160 laboratories have since taken advantage of this opportunity. Several results have been included in high profile reports [8, 26–29]. Our goal here is not to review post-processing analysis tools used to determine structural details. We suggest other sources for this purpose [10, 30–32]. We’ve also recently described more technical aspects of the control system and hardware elsewhere [25, 33]. Here we focus on optimal input and a detailed description of the output to improve coordination between investigators and synchrotron beamlines as required for true high-throughput. HT-SAXS appears rigid given the reduced interaction between the beamline and the investigator. In reality both data collection and data processing are flexible. Investigators are empowered to reprocess data by varying from the automated processing steps. By optimally taking advantage of HT-SAXS, new opportunities continue to be developed for the investigation of bio-molecules, such as comprehensive mapping of conformational states without requiring shape reconstructions [19].
2 Materials
HT-SAXS opportunities extend beyond experiments preformed at lower throughput. Optimal samples and procedures depend on the type of experiment being performed. Here we will provide general requirements for low signal samples acknowledging that at high concentrations, requirements may be relaxed.
2.1 Concentration
Concentration is an important parameter that impacts signal, problems from aggregation, and data collection requirements.
For organic macromolecules in an aqueous solvent, a useful rule of thumb for determining the required concentration for high-quality signal is concentration in mg/ml multiplied by molecular weight in kDa must be greater than 100 (mg/ml × kDa > 100).
With HT-SAXS the required concentration can be experimentally evaluated, as the desired signal to noise will vary from facility to facility and by the scattering power of the solvent.
2.2 Isolating the Solute Signal
The proper subtraction of background signal is often critical. Background includes the halo of the primary X-ray beam, scattering from windows in the beam path and scattering from solvent. To focus analysis on a solute (the macromolecule of interest), the SAXS from a solution containing all but the macromolecule of interest (referred to from here forward as the buffer) may be subtracted from the SAXS profile of the solution containing the macromolecule. This subtraction removes all three background components mentioned above.
2.3 Matching Buffers
Everything in solution scatters X-rays so having the appropriate matching buffers is critical.
Adequately matched buffers can be prepared by dialysis, size exclusion chromatography (SEC) or from a spin concentrator. However, these procedures must be carefully attended to, for example, filters in concentrators are typically covered in preservatives which must be washed at least three times before the flow through can be used as a proper buffer. Dialysis requires more time with viscous solvents. Some SEC fractions contain small amounts of column matrix so are not appropriate for use as a buffer.
Pipetting of cofactors into both the buffer and the sample, as a modification, is also possible provided the added volumes are equal to high accuracy (usually requires a minimum of 4 μL).
Added signal from improper buffer subtraction will typically reduce the apparent rate of intensity decay as a function of angle; giving the appearance of an unfolded polymer. Over subtracted signal often results in negative intensities at high values of q.
Because of the importance of proper buffer subtraction and because buffer is typically inexpensive, we recommend preparing larger buffer volumes than required for samples and collecting identical buffers both before and after the sample.
2.4 Sample Format
Robotic sample loading from 96-well plates requires decisions regarding shipping, seal against evaporation, and safe volumes for loading the sample cell. If frozen, the plate should be transported in sub-freezing conditions. If unfrozen, care must be taken so that samples do not slow freeze during transport but remain cool. A kilogram of Blue Ice at 5° packed on both sides of the sample plate in a well-sealed (taped) Styrofoam box is a reasonable option.
HT-SAXS facilities have specific sample formats as precise sample locations in three dimensions are required for robotic loading. The sample format at SIBYLS is a specific, commercially available, full-skirt 96 conical well plate. Samples sent in alternate plate types cause delay as samples must be transferred to the proper plate type.
A safe volume for filling the sample cell above the incident beam path is 24 μL.
Plates must also be covered with an appropriate seal for transport to prevent mixing between wells, evaporation and contamination from the sealing material. Plates are typically covered with a commercially available silicone mat.
Once samples are sealed they are ready for shipment or delivery. Flash freezing of samples is possible but usually unnecessary with 24 h shipping times and a maximum of two additional days between delivery and collection. Flash freezing may be accomplished by placing the plate over a shallow bath of liquid nitrogen. Practice with plates containing water is recommended.
2.5 Homogeneity Requirements for Shape Determination
Shape reconstruction requires homogeneous samples and removal of concentration-dependent signals.
A significant fraction of investigators use SAXS data for shape determination. Strategies for data collection for this purpose have been reported [34]. Important procedures include collecting a concentration series to identify and possibly remove concentration- dependent signals contaminating the signal characterizing macro-molecular shape.
SAXS by itself cannot determine heterogeneity so supporting data such as elution profiles from chromatographic purification, native gels or multi-angle light scattering are required for quality assessment of homogeneity. Many problems with SAXS experiments on RNA samples derive from heterogeneity of the folded RNA so separation by sizing chromatography or other means is important [35]. The reporting of a single shape representing an entire population of macromolecules that contribute to the SAXS signal assumes homogeneity.
2.6 Organizing Data Collection
An organized plan for sample and washing steps impacts efficiency.
The SIBYLS HT-SAXS pipeline utilizes formatted spreadsheets, filled out by investigators, for organizing data collection. The spreadsheet describes the order of data collection, the desired naming of output experimental files from each sample, which wells contain buffers and at which points in the data collection washes are necessary.
Washing is not required between every well, if sample collection order is strategically chosen. For example a concentration series collected in the order of lowest to highest does not need washing steps. Washing is a significant bottleneck in data collection so the fewer washes the higher the throughput.
3 Methods
3.1 Instrument Calibration
Significant calibration of the SAXS instrumentation is applied prior to data collection. Investigators should be aware of four important calibration procedures which will affect all data sets.
The incident beam orientation, sample position, and detector orientation must all be accurately defined in order to calculate scattering plots of Intensity versus q. This is typically done through the collection and analysis of a crystalline powder pattern. Inaccuracy in this calibration will result in blurred SAXS curves where sharp peaks are broadened and the small q scattering may have larger variation.
The incident X-ray wavelength is calibrated typically by measuring absorbance from metal filters with fluorescence near an electron orbital edge. Inaccuracy in wavelength leads to shifted and stretched SAXS profiles with peaks occurring at an alternate apparent q value.
The beamstop and other shadows blocking scattering from the beamline to the detector are masked out. Inaccuracy in defining these regions will lead to large drops in intensity at small q near the beamstop. If the mask is too large, valuable low q data may be obscured.
A solute of known molecular weight and concentration is collected to enable plotting data on an absolute scale. This calibration can be valuable for calculating molecular weight when the concentration of the macromolecule is known. However the scattering contrast between buffer and solute must be considered relative to the calibrant. Including a calibrant on the sample plate is an alternative. These calibration files are readily available if desired.
3.2 Sample Handling
Communicating sample handling procedures is important as the assumption is that samples are to be stored in cool conditions and centrifuged prior to data collection.
Once samples have been delivered to the facility they are stored at an appropriate temperature (−80 °C for frozen and 4 °C for unfrozen).
Just prior to data collection they are spun in a centrifuge to condense the sample and sediment large aggregates. Once centrifuged, the sealing mat is replaced with a thinner pierceable seal for better sample delivery by the sample loading needle.
3.3 Sample Temperature Control
Temperature is an important and underutilized parameter.
The plate deck and the sample cell are cooled to 15 °C during data collection using a water chiller. The temperature can be decreased, but the dew point must be considered as condensation on the sample cell windows can negatively affect buffer subtractions.
Helium can be added to the sample cell environment to minimize the surrounding humidity, effectively lowering the dew point. The sample cell can also be heated up to 70 °C using a Peltier; however, the temperature is typically kept at 15 °C.
3.4 Data Collection
Strategic data collection and guarding against interfering bubbles is key for efficiency and data quality.
Three plates may be held on the SAXS instrument at one time. At a rate of 4 h/plate this conveniently enables unsupervised overnight collection.
Procedures are in place to automatically stop data collection and alert the beamline scientists when problems occur. If the X-ray source is shutdown for example, the system stops and sends a text message alert. Sample loading and data collection can be monitored by beamline staff remotely.
A snapshot of every loaded sample is taken so that samples with bubbles can be diagnosed after data collection. Often, sufficient volume remains in the plate to recollect these samples.
Samples are pipetted one at a time from the plate into the sample cell, exposed, then pipetted back into the plate.
Typically, the aspiration rate for sample delivery is set at 4 μl/s but can be decreased for viscous, low volume, or bubble-prone samples.
Samples are exposed with a 1011 photon/s, 12 keV monochromatic beam in a series of exposures: 0.5, 1.0, 2.0, and 4.0 s in that order. A range of exposure times are collected to identify radiation damage and overcome the limited dynamic range of the detector.
Images from the sample are named using a prefix designated in the investigator prepared spreadsheet followed by the well location, followed by the exposure number. Results from these images are later merged together by the investigator to maximize quality.
3.5 From Images to SAXS Profiles
Once the images are collected from each sample, data processing begins.
Automated scripts subtract the images of the closest collected buffer before the sample and the closest collected buffer collected after the sample. The two profiles are averaged creating a total of three scattering profiles for each sample exposure.
The subtraction process requires normalization for the number of X-rays during the exposure of the buffer and the sample. X-ray flux is monitored by a diode within the beamstop. Extracting an accurate value for the flux during the exposure to the high accuracy required is not a trivial procedure and is a source of error.
Once a subtracted image is created a mask is applied blocking out unwanted pixels for integration.
Subtracted and masked images are then integrated utilizing geometric and wavelength parameters determined from pre- collection calibration.
3.6 Sources of Error and Error Bars
The calculation of error bars and examination of the buffer subtraction impacts quality of data analysis.
Since SAXS images contain many observations at equivalent q, an error bar may be calculated using the standard deviation and average intensity.
A second error of the subtraction process involves slight but random variations in detector background between sample and buffer. In some cases these can be significant.
Mechanisms are in place to enable investigators to repeat the subtraction and integration process using alternate pairings of sample and buffer.
Raw images are rarely desired, thus investigators typically receive the one dimensional SAXS profile of X-ray intensity as a function of q with error bars.
4 Preliminary Visualization and Interpretation of Results
4.1 Sample Report
A sample report and assessment of scattering profiles provides the basis for appropriate data processing.
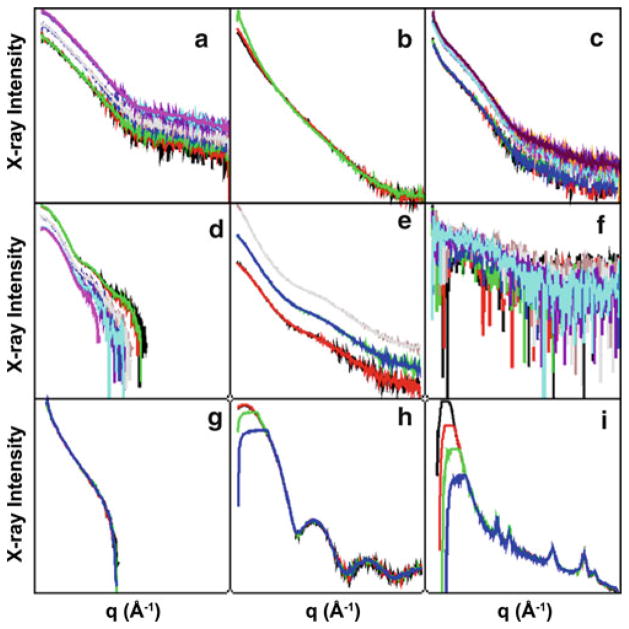
Besides receiving scatting data files, investigators also receive an html formatted sample report. The report is viewable utilizing web browser software and enables mouse click based zooming for visualization of individual profiles. A partial example is shown in Fig. 1.
Fig. 1.
Exemplary SIBYLS output format of data sets collected from a sample plate. Scattering profiles are grouped by concentration series and graphed on log plots. In the web-enabled version, individual plots can be enlarged for easier viewing. (a) A concentration series of a well-behaved sample. (b) A sample flagged as radiation sensitive. Aggregation induced through damage has occurred during the highest exposure shown in green. (c) The extrapolation of X-ray intensity at q = 0 is impossible for the curves shown assuming a particle size smaller than 600 Å. Particles of larger size are considered aggregates at SIBYLS. (d) Profiles are over subtracted indicating an error in buffer subtraction (either an inappropriate buffer or instrumental error). (e) A slight concentration dependence can be observed as the low q region that increases with concentration (SAXS curves from higher intensity plots). This effect can also be seen in plot. (f) The low signal to noise indicates low concentration or insufficient exposure times. (g) A sharp drop to negative intensity at low q is characteristic of bubbles or insufficient volume in the sample cell. Images of the sample cell during these exposures may be referenced for further diagnosis. (h) The red and black curves show a smooth downturn in intensity approaching Izero, indicating the presence of inter-particle repulsive forces. The effect is masked by detector saturation in the long exposures (green and blue curves). (i) Aside from major detector saturation, the curve shows the rare presence of micro-crystals as indicated by sharp peaks of intensity
Using this comprehensive view of the data, beamline staff provides guidance on which of the three profiles from each sample to use for further processing.
4.2 Judging Buffer Subtraction
Data redundancy and consistency of buffer subtraction guide further data processing.
If the SAXS profile from the sample analyzed with a buffer collected before the sample agrees to within noise to that analyzed with a buffer collected after then the average is used. If the two do not agree then a judgment is made.
Above we described errors that may occur during data collection and may cause this disagreement between buffer subtraction (improperly matched buffer, incorrect measure of the incident X-ray flux, and detector background oscillations). These errors create obvious features in the data.
Significant redundancy often exists in collected data. For example, in concentration series, the q dependent intensity decay rate of high q data is nearly always consistent. Thus outliers can often be identified and eliminated.
When an obvious choice is not possible, the average is taken.
4.3 Red Flags for Further Analysis
Once all scattering profiles are selected and plotted, further comments are added. Comments are based on a visual inspection of the data. These comments are meant to serve as flags of concern rather than a definitive judgment on further processing of data. The following lists typical comments and examples are shown in Fig. 1.
4.3.1 Aggregation or Undefined Guineir Region
The intensity at zero scattering angle (I(0)) cannot be extrapolated from aggregated data. Similarly particles of size greater than 600 Å cannot be fully characterized with the available q range at SIBYLS. The scattering angles required for Guineir analysis are smaller than can be measured. Further analysis of data without a Guineir region is limited from a shape restoration perspective as the Guineir region is valued for quality control.
4.3.2 Radiation Sensitivity
X-ray radiation damages samples, but the damage rate cannot be determined a priori. Some samples show no noticeable differences in SAXS for all exposure lengths. Others are damaged by the first exposure. Radiation damage is identified as increase in I(0) with exposure toward features of aggregation. Use of the low exposure data in this q region is thus critical for further analysis.
4.3.3 Detector Saturation
Extremely high concentration samples will scatter with intensities that saturate the detector in some regions of q. Data in these regions cannot be analyzed and must be compensated by utilizing shorter exposures or more dilute concentrations.
4.3.4 Low Concentration
At low concentration the difference between sample and buffer approaches zero. The small q region may have sufficient intensity to identify the radius of gyration Rg. However scattering features quickly blend in to flat, near zero values.
4.3.5 Bubble, Low Volume, or Empty Sample Cell
Bubbles, low volume, and empty sample cells often resemble profiles with over subtracted buffers. Radial streaks near the detector beamstop indicate that the incident X-ray beam is hitting a liquid/air surface. High q is the most clearly affected region.
4.3.6 Bad Buffer Subtraction
See Subheadings 3.5, 3.6, and 4.2 above for identification and causes of this error.
4.3.7 Repulsion
Repulsion is indicated by a gradual dip at low q and is caused by inter-particle interference. This effect most often occurs at high concentration. Unless the additional structure factor is of experimental interest, an extrapolation to zero concentration using a concentration series is often necessary.
4.3.8 Concentration- Dependent Effects
Concentration dependence includes multimerization, aggregation, or inter-particle interference, all of which contribute to characteristic changes in the scattering profiles from different concentrations.
4.3.9 Micro Crystals
Sharp peaks along the scattering curve indicate micro-crystal formation in the sample solution (Fig. 1d).
5 Conclusions and Perspectives
By compiling statistics over the course of 2 years (2011 and 2012), below we provide a picture of data collection using the mail-in/hand-in system. SIBYLS collected 267 plates from 106 different labs. Of these labs, 73 % requested subsequent data collection. While most plates were shipped at 4 C, 10 % were shipped frozen. Figure 2 also breaks down the frequency at which each comment was made. The scattering from the samples was sufficient to cause detector saturation in 39 % of samples, usually during the longest exposure. 45 % of samples were sensitive to radiation after 8 s of exposure, while 16 % showed significant radiation damage after only 3 s. 10 % of samples had an undefined Guineir region due to aggregation or molecular dimensions too large for our SAXS configuration. 7 % of samples had poorly matching buffer blanks. Concentration dependence affected 6 % of samples. Another 6 % were below the required concentration. Approximately 1 % of samples were lost by bubbles in the beam path or because of insufficient volume. Repulsion and micro-crystal formation were observed in less than 1 % of samples. Through visual inspection of each scattering curve by the SIBYLS staff, it was estimated that 78 % of all data could be used for further processing after a merging of different exposures and concentrations.
Fig. 2.

SIBYLS SAXS sample quality statistics for 2 years of data collection. Each SAXS profile generated through the mail-in/hand-in system is visually inspected by beamline staff and commented upon for sample quality. Though many samples receive comments, when further merged and processed with other exposures and concentrations 78 % are estimated to be suitable for further analysis (pie chart inset)
HT-SAXS systems enable wide spread use of SAXS for structural characterization. The introduction of HT-SAXS data collection has been accompanied with criticism for being metric driven rather than science driven. Looking forward, we’d like to connect HT-SAXS efforts with problems being addressed in biology. Biological macromolecules are increasingly appreciated as parts of larger networks. Frequently, even components of these networks are challenging to work with and require specific laboratory expertise. Few single laboratories can successfully purify, characterize, and study many interacting components within a network. HT-SAXS facilities complement efforts to compose more comprehensive pictures of networks by drawing upon samples from many laboratories and enabling facile structural characterization.
SAXS is a solution-based technique so components may be examined individually, in the presence of partners or under a host of chemical conditions. Besides providing access to SAXS, HT-SAXS facilities continue to develop tools to aid in the analysis and integration of information collected; the staff at these facilities thus play a key part of the broader effort of post-genomic science. Further, new opportunities have been enabled with HT-SAXS [19] and by analysis of HT-SAXS data [36]. We anticipate more high impact results in the near future from HT-SAXS as well as from the combination of HT-SAXS with crystallography, NMR, and other biophysical methods.
Acknowledgments
This work and the operation of the SIBYLS beamline has been supported by the Integrated Diffraction Analysis Technologies (IDAT) program, the DOE Office of Biological and Environmental Research plus the National Institutes of Health grant MINOS (Macromolecular Insights on Nucleic Acids Optimized by Scattering) GM105404.
References
- 1.Koch MHJ. SAXS instrumentation for synchrotron radiation then and now. J Phys Conf Ser. 2010;247:012001. [Google Scholar]
- 2.Fukuchi S, Hosoda K, Homma K, et al. Binary classification of protein molecules into intrinsically disordered and ordered segments. BMC Struct Biol. 2011;11:29. doi: 10.1186/1472-6807-11-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rambo RP, Tainer JA. Bridging the solution divide: comprehensive structural analyses of dynamic RNA, DNA, and protein assemblies by small-angle X-ray scattering. Curr Opin Struct Biol. 2010;20:128–137. doi: 10.1016/j.sbi.2009.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hammel M, Yu Y, Fang S, et al. XLF regulates filament architecture of the XRCC4. ligase IV complex. Structure. 2010;18:1431–1442. doi: 10.1016/j.str.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hammel M, Rey M, Mani RS, et al. XRCC4 protein interactions with XRCC4-like factor (XLF) create an extended grooved scaffold for DNA ligation and double strand break repair. J Biol Chem. 2011;286:32638–32650. doi: 10.1074/jbc.M111.272641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shin DS, Didonato M, Barondeau DP, et al. Superoxide dismutase from the eukaryotic thermophile Alvinella pompejana: structures, stability, mechanism, and insights into amyotrophic lateral sclerosis. J Mol Biol. 2009;385:1534–1555. doi: 10.1016/j.jmb.2008.11.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamagata A, Tainer JA. Hexameric structures of the archaeal secretion ATPase GspE and implications for a universal secretion mechanism. EMBO J. 2007;26:878–890. doi: 10.1038/sj.emboj.7601544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nishimura N, Hitomi K, Arvai AS, et al. Structural mechanism of abscisic acid binding and signaling by dimeric PYR1. Science. 2009;326:1373–1379. doi: 10.1126/science.1181829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chacon P, Moran F, Diaz JF, et al. Low-resolution structures of proteins in solution retrieved from X-ray scattering with a genetic algorithm. Biophys J. 1998;74:2760–2775. doi: 10.1016/S0006-3495(98)77984-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Petoukhov MV, Franke D, Shkumatov AV, et al. New developments in the ATSAS program package for small-angle scattering data analysis. J Appl Crystallogr. 2012;45:342–350. doi: 10.1107/S0021889812007662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pelikan M, Hura GL, Hammel M. Structure and flexibility within proteins as identified through small angle X-ray scattering. Gen Physiol Biophys. 2009;28:174–189. doi: 10.4149/gpb_2009_02_174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bernado P, Mylonas E, Petoukhov MV, et al. Structural characterization of flexible proteins using small-angle X-ray scattering. J Am Chem Soc. 2007;129:5656–5664. doi: 10.1021/ja069124n. [DOI] [PubMed] [Google Scholar]
- 13.Grishaev A, Wu J, Trewhella J, et al. Refinement of multidomain protein structures by combination of solution small-angle X-ray scattering and NMR data. J Am Chem Soc. 2005;127:16621–16628. doi: 10.1021/ja054342m. [DOI] [PubMed] [Google Scholar]
- 14.Schneidman-Duhovny D, Hammel M, Sali A. FoXS: a web server for rapid computation and fitting of SAXS profiles. Nucleic Acids Res. 2010;38:W540–W544. doi: 10.1093/nar/gkq461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rambo RP, Tainer JA. Characterizing flexible and intrinsically unstructured biological macromolecules by SAS using the Porod-Debye law. Biopolymers. 2011;95:559–571. doi: 10.1002/bip.21638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Getzoff ED, Tainer JA, Lerner RA, et al. The chemistry and mechanism of antibody binding to protein antigens. Adv Immunol. 1988;43:1–98. doi: 10.1016/s0065-2776(08)60363-6. [DOI] [PubMed] [Google Scholar]
- 17.Tainer JA, Getzoff ED, Alexander H, et al. The reactivity of anti-peptide antibodies is a function of the atomic mobility of sites in a protein. Nature. 1984;312:127–134. doi: 10.1038/312127a0. [DOI] [PubMed] [Google Scholar]
- 18.Tsutakawa SE, Hura GL, Frankel KA, et al. Structural analysis of flexible proteins in solution by small angle X-ray scattering combined with crystallography. J Struct Biol. 2007;158:214–223. doi: 10.1016/j.jsb.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 19.Hura GL, Budworth H, Dyer KN, et al. Comprehensive macromolecular conformations mapped by quantitative SAXS analysis. Nat Methods. 2013;10:453–454. doi: 10.1038/nmeth.2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martel A, Liu P, Weiss TM, et al. An integrated high-throughput data acquisition system for biological solution X-ray scattering studies. J Synchrotron Radiat. 2012;19:431–434. doi: 10.1107/S0909049512008072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.David G, Perez J. Combined sampler robot and high-performance liquid chromatography: a fully automated system for biological small-angle X-ray scattering experiments at the Synchrotron SOLEIL SWING beamline. J Appl Crystallogr. 2009;42:892–900. [Google Scholar]
- 22.Franke D, Kikhney AG, Svergun DI. Automated acquisition and analysis of small angle X-ray scattering data. Nucl Instrum Methods A. 2012;689:52–59. [Google Scholar]
- 23.Nielsen SS, Moller M, Gillilan RE. High-throughput biological small-angle X-ray scattering with a robotically loaded capillary cell. J Appl Crystallogr. 2012;45:213–223. doi: 10.1107/S0021889812000957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hura GL, Menon AL, Hammel M, et al. Robust, high-throughput solution structural analyses by small angle X-ray scattering (SAXS) Nat Methods. 2009;6:606–U683. doi: 10.1038/nmeth.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Classen S, Rodic I, Holton J, et al. Software for the high-throughput collection of SAXS data using an enhanced Blu-Ice/DCS control system. J Synchrotron Radiat. 2010;17:774–781. doi: 10.1107/S0909049510028566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Williams RS, Dodson GE, Limbo O, et al. Nbs1 flexibly tethers Ctp1 and Mre11-Rad50 to coordinate DNA double-strand break processing and repair. Cell. 2009;139:87–99. doi: 10.1016/j.cell.2009.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Christie JM, Arvai AS, Baxter KJ, et al. Plant UVR8 photoreceptor senses UV-B by tryptophan-mediated disruption of cross-dimer salt bridges. Science. 2012;335:1492–1496. doi: 10.1126/science.1218091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chao LH, Stratton MM, Lee IH, et al. A mechanism for tunable autoinhibition in the structure of a human Ca2+/calmodulin-dependent kinase II holoenzyme. Cell. 2011;146:732–745. doi: 10.1016/j.cell.2011.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dueber EC, Schoeffler AJ, Lingel A, et al. Antagonists induce a conformational change in cIAP1 that promotes autoubiquitination. Science. 2011;334:376–380. doi: 10.1126/science.1207862. [DOI] [PubMed] [Google Scholar]
- 30.Putnam CD, Hammel M, Hura GL, et al. X-ray solution scattering (SAXS) combined with crystallography and computation: defining accurate macromolecular structures, conformations and assemblies in solution. Q Rev Biophys. 2007;40:191–285. doi: 10.1017/S0033583507004635. [DOI] [PubMed] [Google Scholar]
- 31.Petoukhov MV, Svergun DI. Analysis of X-ray and neutron scattering from biomacro-molecular solutions. Curr Opin Struc Biol. 2007;17:562–571. doi: 10.1016/j.sbi.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 32.Rambo RP, Tainer JA. Super-resolution in solution X-ray scattering and its applications to structural systems biology. Ann Rev Biophys. 2013;42:415–441. doi: 10.1146/annurev-biophys-083012-130301. [DOI] [PubMed] [Google Scholar]
- 33.Classen S, Hura GL, Holton JM, et al. Implementation and performance of SIBYLS: a dual endstation small-angle X-ray scattering and macromolecular crystallography beamline at the Advanced Light Source. J Appl Crystallogr. 2013;46:1–13. doi: 10.1107/S0021889812048698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jacques DA, Guss JM, Svergun DI, et al. Publication guidelines for structural modelling of small-angle scattering data from biomolecules in solution. Acta Crystallogr D. 2012;68:620–626. doi: 10.1107/S0907444912012073. [DOI] [PubMed] [Google Scholar]
- 35.Rambo RP, Tainer JA. Improving small-angle X-ray scattering data for structural analyses of the RNA world. RNA. 2010;16:638–646. doi: 10.1261/rna.1946310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rambo RP, Tainer JA. Accurate assessment of mass, models and resolution by small-angle scattering. Nature. 2013;496:477–481. doi: 10.1038/nature12070. [DOI] [PMC free article] [PubMed] [Google Scholar]