Abstract
Four chemotypes of the Rough lipopolysaccharides (LPS) membrane from Pseudomonas aeruginosa were investigated by a combined approach of explicit water molecular dynamics (MD) simulations and Poisson-Boltzmann continuum electrostatics with the goal to deliver the distribution of the electrostatic potential across the membrane. For the purpose of this investigation, a new tool for modeling the electrostatic potential profile along the axis normal to the membrane, MEMPOT, was developed and implemented in DelPhi. Applying MEMPOT on the snapshots obtained by MD simulations, two observations were made: (a) the average electrostatic potential has a complex profile, but is mostly positive inside the membrane due to the presence of Ca2+ ions which overcompensate for the negative potential created by lipid phosphate groups; and (b) correct modeling of the electrostatic potential profile across the membrane requires taking into account the water phase, while neglecting it (vacuum calculations) results in dramatic changes including a reversal of the sign of the potential inside the membrane. Furthermore, using DelPhi to assign different dielectric constants for different regions of the LPS membranes, it was investigated whether a single frame structure before MD simulations with appropriate dielectric constants for the lipid tails, inner, and the external leaflet regions, can deliver the same average electrostatic potential distribution as obtained from the MD-generated ensemble of structures. Indeed, this can be attained by using smaller dielectric constant for the tail and inner leaflet regions (mostly hydrophobic) than for the external leaflet region (hydrophilic) and the optimal dielectric constant values are chemotype-specific.
Keywords: Glycolipids, Phospholipid Bilayers, Poisson-Boltzmann Equation, Multiple Dielectric Constants, Transmembrane Potential, Outer Membrane Remodeling, LPS Phenotype Variation
Introduction
The bacterial outer membrane is an asymmetric bilayer composed of lipopolysaccharides (LPS) in the external leaflet and phospholipids (DPPE) in the inner leaflet. The asymmetry resulting from the spatial separation of phospholipids and LPS generates an electrical potential across the membrane that is crucial for many membrane-related processes such as membrane current, membrane capacitance and transport. 1-3 The electrostatic asymmetry of bacterial outer membranes is of particular relevance for the development of novel antibacterial drugs since it affects the incorporation of membrane proteins into the membrane 4,5, and acts as the driving force for antimicrobial peptide association with the membrane. 6-8 The binding of various compounds to the LPS membrane was shown to activate the host immune system 9-12 and therefore understanding the effects governing the association are of great importance for drug development. 13 In addition to the immune system activation, the bacteria can be attacked directly via disruption of the LPS membrane by various cationic antimicrobial peptides 6,14 or synthetic derivatives. 15 In designing effective compounds, the knowledge of the electrostatic potential profile of bacterial outer membranes is of essential importance, especially for charged and polar substances. Therefore, the accurate modeling of the LPS electrostatic potential is crucial for understanding the forces and the effects behind membrane transport phenomena.
However, LPS membranes, like all other biological macromolecules, exist in an aqueous environment, which makes the calculation of electrostatic potential distribution not trivial. In principle it can be done with explicit models by calculating direct Coulombic potential created by all charged atoms of the LPS, and the potential generated by explicit ions and water molecules and averaging over long trajectories and over 3D space (or a 3D grid). However, the local fluctuations of water molecules and ions may introduce too much noise, especially in close proximity to their positions, which may not be reducible by extending the length of the simulation trajectory. Specifically for LPS membranes, the structural complexity renders the lateral diffusion of LPS monomers in outer membranes much slower (10−10 cm2 s−1) as compared to phospholipids (10−7-10−8 cm2 s−1). 16,17 A direct consequence of this is the requirement of simulations within the μs timescale in order to attain convergence of structural properties. 18,19 Alternatively, one may choose to neglect the water phase and to calculate the electrostatic potential profile in vacuum 20-22, but this may be a too crude approximation for many biologically relevant phenomena. At the other side of the spectrum are continuum electrostatic approaches 23-25, specifically Poisson-Boltzmann Equation (PBE) based approaches 26-28, which treat the membrane and embedded ions at the atomic level while the water phase is modeled as a continuum medium. The advantage is that the water phase is modeled at equilibrium while the effects of structural fluctuations of membrane atoms and embedded ions can be accessed via an ensemble of structures.
One particular implementation of a PBE solver is the DelPhi package 29. The DelPhi program utilizes Finite Difference (FD) method to solve PBE and to output the electrostatic potential distribution over the grid points of a Cartesian grid (such an output is typically referred as “potential map”). The potential map was demonstrated to be a very powerful tool for revealing the role of electrostatics in many reactions, including the guidance of the electrostatics of the substrate to the active site 30, electron pathways 31, protein-membrane binding 32, peptide-membrane binding 33 and many other phenomena 34,35. However, in membrane biophysics one needs to obtain the electrostatic potential profile across the membrane to be able to make comparisons with experimentally measurable quantities. This prompted us to develop a new feature for DelPhi program, termed MEMPOT (MEMbrane POTential), which uses the potential map to generate the electrostatic potential profile while avoiding artificial contributions from grid points being too close to charged atoms or ions.
Armed with this tool and assisted by MD simulations, in this work we analyze the electrostatic potential profiles of four LPS chemotypes. The goals are to reveal the similarities and dissimilarities of their profiles and how they are related to chemical composition of LPS. The potential is calculated with DelPhi using MD-derived structural ensembles and applying a uniform dielectric constant of 2 (electronic polarizablity only) for the entire membrane, while dielectric constant of water is kept at 80. The average profile is then compared with the profile calculated with a single structure (single frame, taken before MD simulations), and by using the potential map calculated with DelPhi by assigning different dielectric constants for lipid tails, inner leaflet regions, and the external leaflet region. It is demonstrated that the external leaflet region corresponding to the LPS polysaccharide chains should always be modeled with dielectric constant larger than the dielectric constant of the acyl tail region; however, the optimal values differ for different LPS chemotypes. It was found that for all LPS chemotypes the MEMPOT modeling of a single frame requires the use of a dielectric constant smaller than 2 for the hydrophobic region of the acyl chains. Furthermore, this dielectric constant was found to be lower than the dielectric constant used in multiple snapshot calculations, and this observation is conferred in the discussion section. For the external leaflet region, larger values of dielectric constant are needed. These findings indicate that different relaxation phenomena take place across the membrane, including charged groups and ions redistribution.
Another important component of this investigation is to reveal the effect of water phase on the calculations of the electrostatic potential profile. It is demonstrated that without taking into account the water phase (solving vacuum Poisson equation), the electrostatic potential profile is very different and even has opposite polarity from the profile obtained with Poisson or Poisson-Boltzmann equations in water. Because of that, correct modeling of the membrane potential, including the potential inside the membrane, requires solving Poisson or Poisson-Boltzmann equations in presence of a high dielectric water phase. These findings are examined in details in the discussion section.
Methods
Preparation and simulation of the LPS chemotype membranes
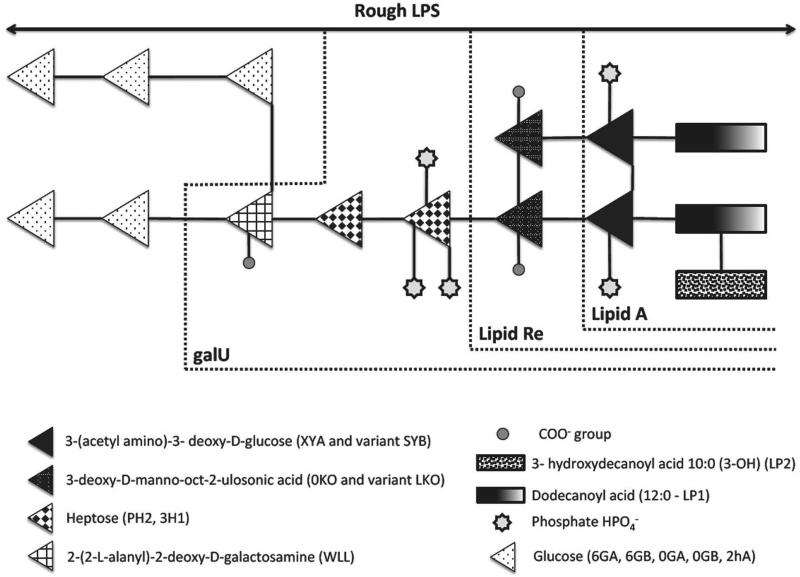
The outer membrane models consisted of bilayers made of DPPE (inner leaflet) and four distinct LPS chemotypes (outer leaflet), namely rough LPS, galU, Re and Lipid-A (Table I, Figure 1). The Glycam-compatible atomic parameters for the LPS chemotypes and DPPE molecules were previously published.18,36 Atomic coordinates for the initial configurations of the galU, Re and lipid-A chemotypes were built by removing the respective monosaccharide units from a rough LPS membrane pre-equilibrated for 1 μs.18 Counter-ions were added to neutralize the charged functional groups on the saccharide units at pH 7. A total of 288 Ca2+ cations were added to the galU LPS membranes, 144 Ca2+ cations to the Re LPS membrane and 72 Ca2+ cations to the lipid-A LPS membrane. The placement of the cations in the inner and outer core of the molecule was carried out via a multiple-step protocol previously developed for this purpose.19 Periodic boundary conditions were applied to the simulations based on a rectangular box containing the bilayer (LPS chemotype and DPPE) and explicit solvent. The TIP3P water model 37 was used as in our previous simulations of LPS membranes. 18,19,38,39 All bond lengths in the solute were kept constant using the LINCS 40,41 algorithm, and the water geometry was maintained with the SETTLE 42 algorithm. Simulations were performed in the isothermal-isobaric ensemble (NPT) with a time step of 2 fs, and in the absence of any artificial external surface tension parameter. The center of mass motion was removed at every 5 steps. The temperatures of the solute and solvent degrees of freedom were separately coupled to a Berendsen thermostat 43 at 328 K with a relaxation time of 0.4 ps. The pressure was maintained by weakly coupling 43 the particle coordinates and box dimensions in the xy plane and separately along the z axis, to a pressure bath at 1.0 bar by means of semi-isotropic coordinate scaling with a relaxation time of 0.4 ps and a compressibility of 4.5 × 10−5 bar−1 as appropriate for water. Bond lengths between hydrogen and heavy atoms, and the geometry of the water molecules were constrained using the linear constraint solver algorithm with a tolerance of 10−4.40 The reaction field correction and a cutoff of 1.4 nm were used for both vdW and long-range electrostatic interactions with a permittivity dielectric constant of 66. 44 In all cases, the pair list for short-range nonbonded and long-range electrostatic interactions were updated with a frequency of 5 timesteps. Configurations of the trajectory were recorded every 100 ps. To investigate the time-dependent behavior of LPS membranes, two time-windows for outputting snapshots were used, namely the first 10ns and (set 1) and last 10ns (set 2). The first time-window represents the early stages of the equilibration, while the second one indicates more equilibrated system. The software package Gromacs v.4.04 was used for the analysis of the simulations in conjunction with in-house developed tools.45-47
Table 1.
Number of Atoms and Charged Groups in the Simulated Systems.
| Chemotype | Number of Atoms |
||||
|---|---|---|---|---|---|
| Solute | Solvent | Ca2+ cations | PO4− groups | COO− groups | |
| Rough LPS | 47376 | 48024 | 288 | 360 | 216 |
| galU | 39888 | 56193 | 288 | 360 | 216 |
| LPS Re | 32832 | 62244 | 144 | 144 | 144 |
| Lipid-A | 29016 | 66021 | 72 | 144 | 0 |
Figure 1.
Schematic representation of the chemical structure of the LPS chemotypes from Pseudomonas aeruginosa.
The MEMPOT algorithm
In order to analyze the electrostatic potential distribution in membranes, a new tool has been implemented in Delphi. 29 The MEMbrane POTential (MEMPOT) algorithm is outlined below. The first step is to calculate the electrostatic potential by solving the Poisson-Boltzmann equation (PBE) 26:
| (2) |
where is the electrostatic potential as a function of , is the dielectric constant which has different values in different regions of space, ε0 is the dielectric constant of vacuum, is the distribution of the permanent charges within biological macromolecules, membranes and/or geometric objects, e is the electron charge, Cib is the bulk concentration of the i-th ion type, zi is ion's valence, “k” is Boltzmann constant and T is the absolute temperature. The distribution of the potential is obtained by solving the equation with the finite difference method after the application of appropriate boundary conditions. Note that in modeling membrane-water system, the dielectric constant(s) of membrane (εmem=2) is distinctive different from the dielectric constant of water (εwat=80).
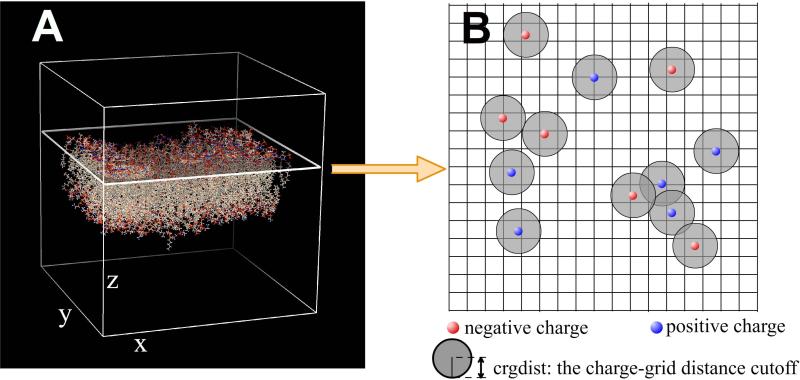
In this step, the structure of the membrane is placed in a grid box defined by N*N*N periodic grids (Figure 2a). The surface of the membrane is set along the plane defined by the x and y axes whereas the z axis is parallel to the membrane normal. The quantity of interest is the average potential value along z-axis, which is a macroscopic quantity independent of the position along the x-y plane. Therefore, the potential must be integrated over the x-y plane and subsequently normalized. Following existing approaches in MD packages, the system is sliced into tiny slabs along the z-axis and the following procedure is carried out for each of the slabs.
Figure 2.
Scheme of the MEMPOT algorithm. a) The membrane is placed inside the grid box where a single slab is shown as an example. b) Charge distribution on the surface of a given slab of the grid box. Areas in gray represent charge-neighbor grids.
Step two involves the integration, or summation in Delphi, of the potentials in each slab. The resulting potential is a normalized collection of potentials outputted by the DelPhi algorithm at each grid point in the lab. However, it is plausible that some grid points will be located quite close to a real charge yielding an unrealistically high potential (due to the inverse distance term in the Coulombic formula). These grid points must be removed from the summation, which is done in DelPhi by introducing a charge-grid distance cutoff, and a grid point is flagged as “charged neighbor” if the distance between this grid and any point charge is less than the cutoff “crgdist” (Figure 2b). This option is activated with the keyword “crgdist” which will call the MEMPOT routine in the DelPhi package. Thus the calculated grid potentials pi,j,k at each grid point (i,j,k) are pruned as:
| (3) |
Step three calculates the average potential distribution along the membrane-crossing direction (z direction). Potential on each grid pi,j,k are taken from eq. (3). Then, the average potential at each layer along the z-axis Pkaverage is calculated as
| (4) |
where the nremoved,k is the total number of “charged neighbor” grids in layer k.
Parameters used in DelPhi calculations
A key issue in modeling electrostatic potential in membranes, which require specific considerations, is how to account for the fact that the average membrane properties are periodic along x,y axes (infinite membrane) (for more comprehensive analysis see recent work of Luo and co-workers 48). In addressing this, one should apply periodic boundary conditions along x,y axes. However, in terms of finite-difference (FD) algorithm, another question that should be addressed is how to position the membrane into the FD computational box. Requiring 100% filling will place the edges of the membrane on the edges of the FD box and some atoms, charged atoms, may be positioned at the FD box walls. Such a scenario would have large impact on setting the boundary conditions and will result in large non-realistic potential values being assigned to the boundary points and will be computationally very expensive. To avoid this problem, the calculations were done with perfil of 95%, leaving a tiny water layer between membrane edges and the edges of the FD box while keeping the membrane charges away from the FD walls. While this deforms the potential at the membrane edges, it has a negligible effect on the average potential and the protocol was adopted for computational efficiency. Thus, the initial boundary conditions were set up with the dipole option, the perfil was 95%, and the salt concentration was varied from I=0M to I= 0.02M. The grid spacing, or scale, was 2.5 Grids/A. This scale was selected after extensive testing of the sensitivity of the results for Lipid-A as a function of the scale. The internal dielectric constant was 2 and the external (water phase) dielectric constant was kept at 80. The snapshot structures taken from MD simulations were processed into PB calculations by simply removing all water molecules, while keeping Ca2+ ions within the membrane and treating them explicitly.
Some of the modeling with MEMPOT was done utilizing the DelPhi feature of assigning multi-dielectric regions. 49,50 For this purpose, lipid tails and DPPE regions were modeled with a dielectric constant ε1, while the external leaflet with different dielectric constant ε2 (see Fig. 1 for details). The values of these dielectric constants were systematically varied and the effect on the calculated electrostatic potential profile was analyzed. Such a treatment was motivated by the expectation that dielectric properties of the lipid tails and DPPE region are different from the properties of the highly charged polysaccharide region and effective dielectric constants applied on single frame structure can mimic the conformational changes occurring in MD simulations. It should be mentioned that more sophisticated approaches were recently reported considering the macromolecular systems, including membranes, as made of domains and each domain has its own biophysical properties, as dielectric function, charge distribution, surface tension and pressure 51.
Results and Discussion
Density profiles of functional groups and their time-evolution
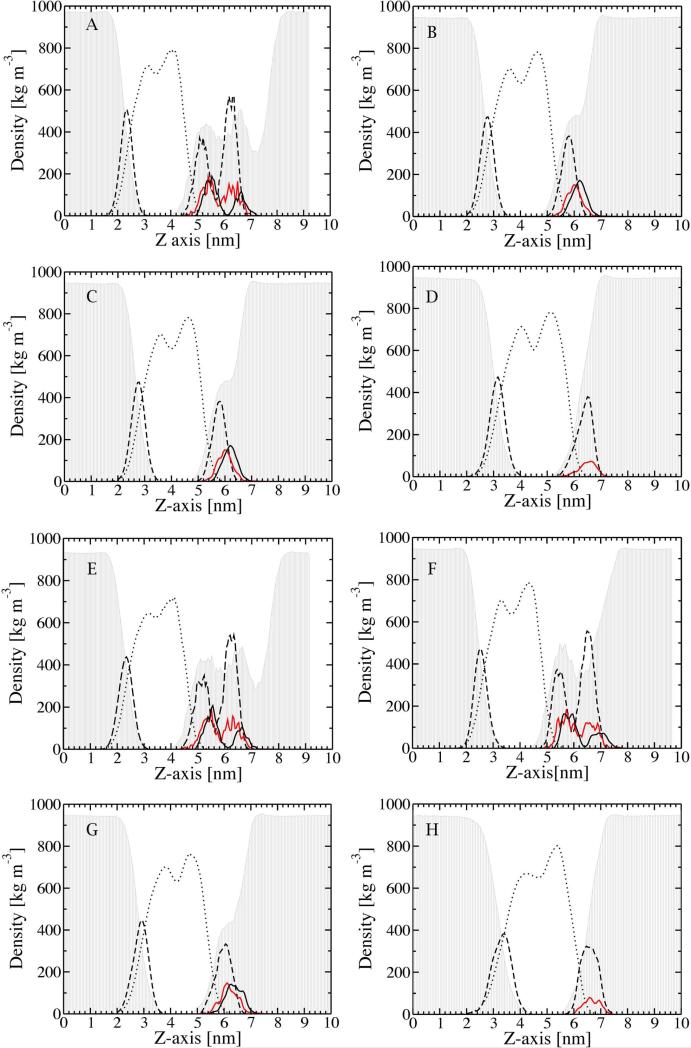
Previous MD simulations have shown that the rough LPS membrane maintains a lamellar aggregate structure in presence of Ca2+ ions and water molecules without a significant conformational change in the course of 1μs simulation time 18,52. The Rough LPS chemotypes also maintain a stable bilayer arrangement in presence of Ca2+ ions within the simulated time of 100 ns.36 Divalent cations bind to negatively charged groups in the LPS molecule stabilize lamellar aggregates via cross-bridge of neighboring molecules.18,53 A comparison of the density profiles of chemical groups obtained from the early (set 1) and late stages (set 2) of MD simulations of the Rough LPS and chemotypes show minor changes over the simulation time (Figure 3). These changes are most noticeable in the chemotypes with short polysaccharide chains, i.e. Lipid-A and LPS Re, and can be summarized as the broadening of phosphate peaks in both LPS and DPPE leaflets, and increase of the bilayer thickness for Lipid-A. Counter-ion peaks are nearly invariable between the two time windows, and overlap with the phosphate and carboxylate peaks (Figure 3). It should also be noted that the polysaccharide region is highly hydrated and each counter-ion is coordinated on average by four water molecules 18,52, an effect found by other researchers as well. 54 These observations may suggest that the electrostatic potential profile of the LPS membranes should not vary significantly over the time, however, small changes of charge distributions in such highly charged systems can have a huge impact on the resulting electrostatic potential. Below we summarize these changes for each chemotype separately, and the effect of these structural rearrangements on the electrostatic potential profiles for each chemotype will be discussed in the next section.
Figure 3.
Density of selected chemical groups along the axis normal to the LPS bilayers. The chemotypes are Rough LPS (A, E), galU (B, F), LPS Re (C, G) and Lipid-A (D, H). Chemical groups are acyl chains (dotted line), water molecules (grey shaded area), phosphate groups (dashed line), carboxylate group (black line) and Ca2+ ions (red line). Values are averaged over 100 snapshots over the first 10 ns (set 1) (A-D) and the last 10 ns (set 2) of the simulations (E-H).
(a) Rough LPS. The main effect of the simulation time is seen for the distribution of phosphate groups on both sides of the membrane. The corresponding peaks in the last 10ns window are smaller but broader than seen in the first 10ns window. As result, the water penetration is significantly decreased at the end of the simulation time (second time window). The density of the acyl chains also decreases with simulation time (Fig. 3).
(b) galU LPS. In this case, the only significant change is associated with the distribution of the phosphate groups at the external leaflet. Their density is significantly lower at the end of simulation time, but the water penetration does not change much.
(c) LPS Re. The major changes include a decrease of the density of phospholipid head groups on both sides of the membrane, but simultaneously the peaks shift toward the membrane interior. Water penetration is less on the outer side of the membrane at the last 10ns of the simulations.
(d) Lipid A. Within the simulation time several prominent changes are seen. The left peak of the density of the acyl chains decreases, and both peaks of the density of phosphate groups decrease as well. In addition, the position of the phosphate density peak at the inner side of the membrane moves slightly toward the membrane interior at the last 10ns of the simulations.
Potential distribution and its time evolution
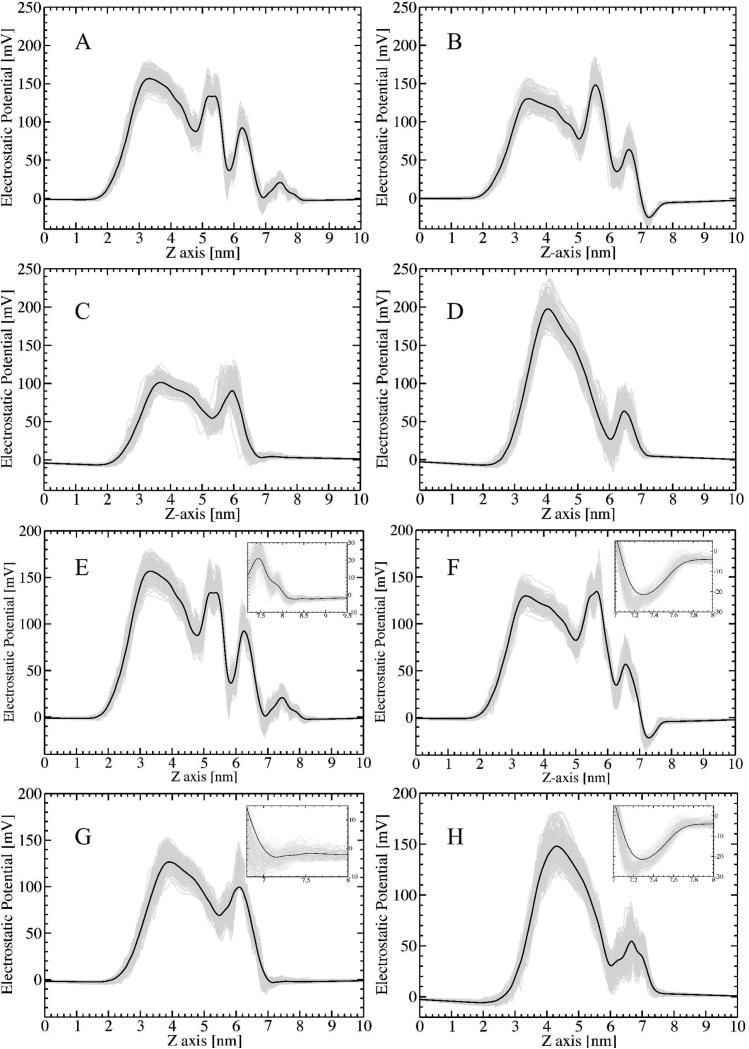
The membrane potential originates from charged atoms in the LPS molecule and counter-ions inside of the membrane. These atoms are not homogenously distributed, and the distinct chemical composition of LPS chemotypes generates different potential profiles (Figure 4). For this reason, the atomic structure of LPS membranes, including embedded Ca2+ ions, should be taken into account when calculating the electrostatic potential. The MEMPOT tool was applied to MD-derived structures of LPS membranes to obtain the electrostatic potential profile along the axis normal to the membrane. To reveal the time evolution of the electrostatic potential profile, two sets of snapshot structures (set 1 and set 2) were used as explained in the method section. The prominent features and changes of the electrostatic potential profile for each chemotype are summarized below.
Figure 4.
Electrostatic potential calculated for the chemotype membranes: A. Rough LPS, B. galU, C. LPS Re and D. Lipid-A. Calculations were performed for an ensemble of 100 snapshots from the first 10 ns (set 1) and the last 10 ns (set 2) of the simulations. The potentials were calculated under conditions of ionic strength of 0 mM and dielectric constants of εmem=2 (ε1=ε2=2) and εwat=80. Insets show details of the potential profile at the LPS surface for the respective chemotypes.
(a) Rough LPS. The resulting profile has 4 prominent peaks, the largest one being at the middle of the acyl chains. However, the other three peaks are larger at the last 10ns of the simulations compared with the first 10ns. The changes are modest, but detectable. These changes can be explained with the structural changes discussed above and the observation that the snapshots at the last 10ns are less hydrated in the external leaflet of the membrane. The decreased density of water molecules in the external leaflet region reduces the screening and the potential is greater as compared with the potential calculated with the snapshots taken from the first 10ns. The effect is further facilitated by the slight decrease of the phosphate group densities.
(b) galU LPS. Three prominent peaks of the electrostatic potential are seen inside the LPS membrane. The peak at the acyl groups is not the largest in this case, but the largest peak is associated with the Ca2+ ions inside the membrane. The amplitude of the peak at the location of acyl group is unaffected by the simulation time, while the other two peaks slightly decrease with the time. This is attributed to the structural relaxation discussed above; indicating that the density of the phospholipid groups decreases while the water penetration does not change with the simulation time. Reduced density of phospholipid groups causes reduced density of the Ca2+ ions, which are the primary source of positive potential. As result, the potential slightly decreases in the last 10ns compared to the initial 10ns simulations.
(c) LPS Re. There are two prominent positive peaks of the electrostatic potential inside the LPS. The largest peak is at the middle of the acyl chains and the second one at the location of Ca2+ ions. The simulation time causes both peaks to increase slightly. The increase of the positive potential at the middle of the acyl chains can be attributed to the observed decrease of the phosphate group density at the inner membrane side, thus reducing the negative potential contribution to the total potential profile. The slight increase of the second positive peak can be explained by the observation that the external leaflet is less hydrated, an effect that overcompensates for the interplay between the phosphate groups and Ca2+ ions density charges.
(d) Lipid A. Two positive peaks are seen in the membrane interior (Fig. 4). The largest is located at the middle of acyl chains, and the second one at the Ca2+ ions positions. The magnitude of the first peak is greatly affected by the simulation time, resulting in almost 25% reduction of the potential at the last 10ns of the simulations. The second peak is practically unaffected by the time of the simulations. The calculated drastic change of the magnitude of the potential at the acyl group positions can be explained by the observation that the density of acyl group decreases significantly at the last 10ns of the calculations allowing water penetration at the inner leaflet of the membrane. The penetrated water further screens the potential and reduces its amplitude.
The above results indicate that small structural changes and small redistributions of the charged groups and structured ions can have significant impact on the electrostatic potential profile. The reason for this is that the distances between charges within membrane are very short, on the order of Angstroms, resulting in strong potentials generated by positive and negative changed entities. The resulting potential is a product of the cancellation of such large contributions and any small change of the position of the individual charges or ions may cause significant effect on the electrostatic potential profile. However, since the changes are found not to change the shape of the electrostatic potential profile, the rest of the manuscript focuses on the results obtained at the last 10ns of the simulations (set 2).
Membrane potential
The chemical composition of the LPS chemotypes leads to rather complex electrostatic potential profiles at the LPS surface of the bilayers. The decrease in the total negative charge per LPS molecule, as monosaccharides are stripped from the longer to the shorter chemotypes, leads to less negative potentials at the LPS surface. Ultimately the potential at the LPS surface of the Lipid-A membrane reaches a positive value due to the clustering of the Ca2+ ions at the membrane surface where they are attracted by the phosphate groups of this chemotype (Figure 1). The average potentials at the LPS surface of the chemotypes are: −2.8 mV for the Rough LPS, −21.3 mV for the galU, −3.6 mV for the LPS Re and 3.5 mV for the Lipid-A. The potential at the DPPE surface is slightly negative, and varies from −1.07 mV (Rough LPS) to −5.79 mV (Lipid-A). It should be cautioned that the magnitude of these potentials depends on how the surface of the membrane is determined and may change dramatically if molecular surface definition is altered.
Mimicking the relaxation effects with optimized dielectric constants
MD simulations of LPS membranes require sampling of large systems across a fairly long time scale.18,19 These calculations remain computationally demanding even nowadays with increasing processing power and faster MD codes. The DelPhi program has been shown to successfully mimic the conformational relaxation of proteins with appropriate effective dielectric constants assigned to different regions of protein. 49,50 In principle, the same approach can be applied to LPS membranes by defining two distinctive dielectric space regions (Fig. 1): the hydrophobic region corresponding to acyl chains (including the hydrophilic DPPE leaflet) and the polysaccharide region of the LPS in the external leaflet. For simplicity, the phosphate and amine groups in the DPPE molecule are treated as dielectric medium similar to acyl chains. These two dielectric regions were assigned different dielectric constants, ε1 and ε2, which were systematically varied to assess the sensitivity of the potential to such approach.
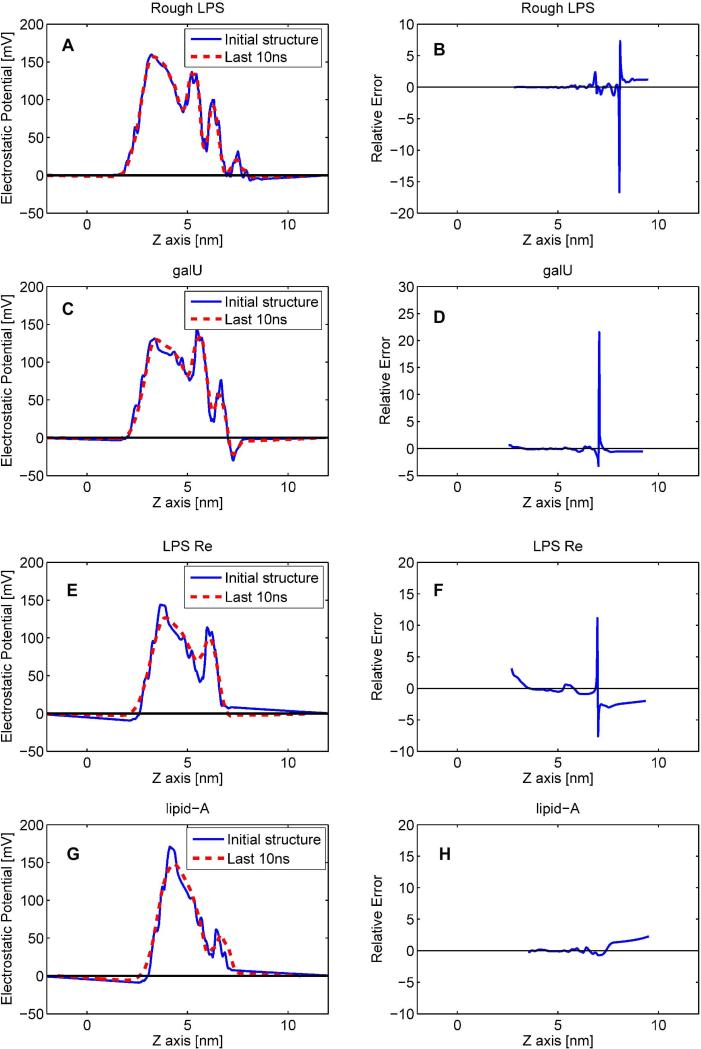
The structures before the phase production of MD simulations (BFMD) (one structure per chemotype), and the average electrostatic potential profile obtained from modeling set 2 (one average profile per chemotype), were used for the electrostatic potential calculations with MEMPOT. Each potential profile was simplified into 240 values along the z-axis of the membrane in the interval z = 1 – 9 nm. The ε1 and ε2 were varied to obtain the best fit (linear regression) between calculations done with BFMD structures and average electrostatic potential profiles generated from the last 10 ns snapshots. The best results in terms of smallest RMSD between BFMD delivered potential and the average potential from set 2 are shown in Figure 5. The profiles derived from the single BFMD chemotype structures match very closely the average electrostatic potential profiles calculated from the set 2. The resemblance between the profiles derived from the two sets of structures is particularly noticeable for the Rough LPS and galU membranes. The increase in the membrane (hydrophobic) thickness observed for LPS Re and Lipid-A causes a small drift of the membrane position upon the MD simulations. Because of that, the positions of profiles generated from BFMD structures were adjusted to minimize the corresponding RMSD. At the end, the overall similarity between the profiles obtained from a single structure and the average potential from multiple structures collected during the last 10 ns (out of 100 ns) of simulations are either almost identical or very similar. These findings indicate that the electrostatic potential profiles obtained from a single pre-equilibrated LPS membrane structure can be representative of the profile obtained from a MD-derived ensemble of structures by using the appropriate dielectric constants for the hydrophobic and hydrophilic regions of the LPS membrane. The optimal values obtained for ε1 and ε2 along with the RMSD between the electrostatic potential profiles are presented in Table 2. It can be seen that optimal dielectric constants are chemotype specific and that ε1 is always smaller than ε2. The finding that the acyl groups should be modeled with lower dielectric constant than the external leaflet is not surprising since acyl tails are apolar. Even when acyl chains are flexible, they are not expected to affect the dielectric properties of the membrane (excluding water penetration or depletion). In contrast, the membrane external leaflet is composed of highly charged polysaccharide chains and bounded counter-ions for which any structural reorganization is expected to affect the dielectric properties of the region. For this reason the modeling of the membrane external leaflet requires larger dielectric constant than the membrane acyl chains. For comparison, the results of calculations using dielectric constant ε1 = ε2 =2.0 (homogeneous membrane) are also shown in Table 2. One can see the significant improvement by treating acyl groups and external leaflet as media with different dielectric constants.
Figure 5.
The left panels show the best fit between the electrostatic potential profiles obtained from BFMD structures and the average potential profile obtained from the last 10ns (set 2) snapshots via linear regression optimizing the ε1 and ε2 in the calculations using BFMD structures; the right panels show the relative error between the potential profiles obtained from BFMD structures and the average potential profile obtained from the last 10ns.
Table 2.
Optimal dielectric constants and the corresponding RMSD for using single structures (set I) to mimic the average electrostatic potential profile of the structural ensemble (set II) from the last 10 ns of the MD simulations.
| Chemotypes | ε1 (DPPE leaflet) and (lipid tails) | ε2 (LPS leaflet) | RMSD (mV) with ε1 and ε2 | RMSD(mV) with ε1 = ε2=2.0 |
|---|---|---|---|---|
| Rough LPS | 1.9 | 2.0 | 5.62 | 6.23 |
| galU | 1.5 | 1.9 | 7.66 | 13.05 |
| LPS Re | 1.2 | 1.8 | 10.98 | 22.77 |
| Lipid-A | 1.0 | 2.0 | 10.28 | 31.20 |
However, it was unexpected that the optimal dielectric constants ε1 and ε2 were found to be generally lower than the dielectric constant of 2 used in the calculations of the electrostatic potential for structures in set 2. As it was previously outlined, the LPS membranes undergo structural rearrangements that cause the drift of charged groups and structured ions and alter the water penetration in the polysaccharide region. 36 We conjecture that the slight charged groups and ions redistributions are the primary reasons for the need of lowering the dielectric constant in the modeling using BFMD structures. The plausible physical explanation is offered in the conclusion section.
Role of water phase
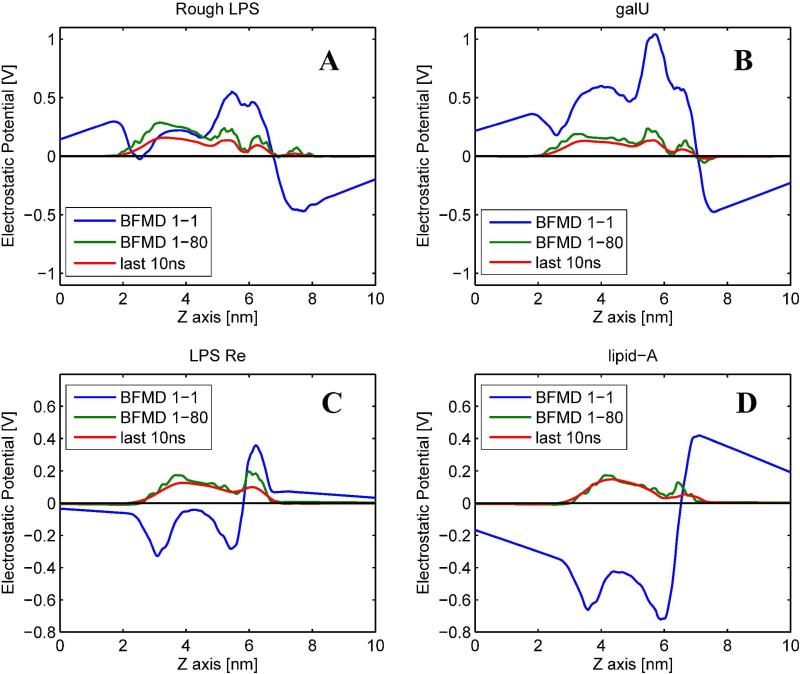
All biological macromolecules exist in water and computing the electrostatic potential, presumably, should include the water phase into the computational protocol. To test the role of the water for modeling the electrostatic potential distributions, the MEMPOT protocol was applied forcing dielectric constant of membrane and the water phase to be the same and equal to unity (εmem=εwat=1). The results are shown in Fig. 6 along with the potential profiles obtained solving Poisson equation in two media (εmem=1 and εwat=80). It can be seen that the calculated profiles with vacuum mimicking MEMPOT are very different from the profiles obtained in presence of water. Not only is the magnitude of the potential calculated in vacuum much larger than the potential calculated with system containing water, but the polarity changes as well. The most striking case is Lipid-A, for which the electrostatic potential profile is calculated to be almost entirely negative in vacuum environment, but is almost entirely positive in the presence of water. Similar, but not so dramatic, differences are observed for three other chemotypes, indicating that the water phase must be taken into account in modeling electrostatic potential. To indicate that structural rearrangement is not as important as taking into account the water phase, Fig. 6 shows the average potential profile obtained from set 2. It can be seen that it very closely resembles the profile obtained with BFMD structure and solving Poisson equation in presence of water, and it is very different from vacuum calculations.
Figure 6.
The electrostatic potential profiles for each chemotype calculated with vacuum Poisson equation (εmembrane=εwat=1) and with Poisson equation in two dielectric media ((εmembrane=1 and εwat=80). Modeling was done using BFMD structures. For comparison, the corresponding average potential profiles of last 10 ns are also show for all the chmotypes.
Role of salt concentration in the water phase
The effect of ionic strength was evaluated through the use of salt concentration values of 0M and 0.02M. Practically no differences were observed (Fig. SI1-SI4 in supplementary material) indicating that the presence of non-structured (mobile) ions in the water phase is not important for the membrane potential, in contrast with structured ions inside the membrane. Of course, if one increases the ionic strength above the used value of 0.02M, then the effect of the salt concentration will become observable. However, even at 0.1M salt concentration, the Debye length is about 10Å, which indicates that the potential at distance 10Å away from the membrane surface will be only “e=2.72” times smaller. Still the dominant effect of the water phase will be the high dielectric constant of water, reducing the magnitude of the potential 80 times.
Conclusions
Four LPS chemotypes, Rough LPS, galU, LPS Re and Lipid-A were investigated in this work and it was shown that charged groups, counter-ions and water molecules change their distributions within the simulations time. The changes are small from structural point of view, but since the membrane leaflets are highly charged, these small structural changes result, in some cases, in a significant perturbation of the electrostatic potential profile along the z-axis. For some chemotypes, the MD simulations cause an increase of the local magnitude of the electrostatic potential profile, while for others the simulations result in a decrease. The different behavior, in most of cases, was attributed to the concerted fluctuations of the phosphate groups and Ca2+ ions. Such a complex and chemotype-dependent outcome indicates the interplay between structural flexibility and the electrostatic interactions between charged groups and associated counter-ions.
Another important finding of this work is that the average electrostatic potential profile obtained from MD generated snapshots can easily be calculated using the BFMD structure and assigning appropriate dielectric constants for the acyl tails and DPPE regions and the external leaflet region. This allows for tremendous speedup of the electrostatic calculations for such types of membranes. While the value of the dielectric constant of the LPS leaflet was found to be larger than the dielectric constant of the acyl tails for all chemotypes, the observation that both are either smaller or equal to the dielectric constant used for the calculations of the snapshots is surprising, if one neglects the interplay between structural changes and ions distribution. In terms of classical physics, the dielectric constant reflects the magnitude, density and ability of existing and induced dipoles to oppose external electrostatic field (see also a recent work by Wei and co-worker on hyperpolarization 55). However, in membrane biophysics, even more important is the rearrangement of charged groups and structured ions and their ability to change the local field. The arrangement can result from various interactions including interactions which may not be primarily electrostatic in origin. As mentioned above, these structural changes, mainly associated with phosphate groups and Ca2+ ions distributions, make the corresponding distributions sharper. As a result, the potential obtained via the snapshots from the last 10ns of the simulation is larger than the potential calculated with BFMD structures (using identical dielectric constants for the membrane). This is the reason why the optimal dielectric constant values for BFMD calculations are lower than the dielectric constant used for MD snapshot calculations, since they have to mimic the increase of the electrostatic potential due to sharper charge distributions inside the membranes caused by MD simulations.
The third important outcome of this work is the observation of the importance of taking into account the water phase in the calculations of the electrostatic potential profile. Neglecting the water phase results in (a) overestimation of the magnitude of the potential, (b) incorrect profile of the electrostatic potential and (c) incorrect polarity of the membrane potential. The developed routine, the MEMPOT DelPhi routine, overcomes this limitation and allows the potential to be calculated in the presence of water and mobile ions in the water phase. The simple physical interpretation of this effect stems from the well-known result of high dielectric region “repelling” electrostatic field out towards the low dielectric region, the membrane. This is specifically important for charges and ions close to the interface membrane-water as Ca2+ and the presence of water increases their contribution to the internal membrane potential.
Supplementary Material
Acknowledgment
This work was supported by the Brazilian funding agencies FACEPE, CNPq and BioNanotec/CAPES, the Swedish funding agency STINT and the National Institutes of Health, National Institute of General Medical Science, grant number R01 GM093937. Computational resources were partially provided by the High Performance Computing Center North (HPC2N), by the Argonne Leadership Computer Facility, a scientific user facility sponsored by the U.S. Department of Energy and Clemson Palmetto Supercomputer. RPD acknowledges a Junior Post-Doctoral Fellowship from CNPq. The Delphi program with newly developed MEMPOT routine is freely available for download from http://www.compbio.clemson.edu/delphi.php. We thank Shannon Stefl for proofreading the manuscript.
References
- 1.Hagge SO, Wiese A, Seydel U, Gutsmann T. Biophysical journal. 2004;86(2):913–922. doi: 10.1016/S0006-3495(04)74167-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gutsmann T, Riekens B, Bruhn H, Wiese A, Seydel U, Leippe M. Biochemistry. 2003;42(32):9804–9812. doi: 10.1021/bi034686u. [DOI] [PubMed] [Google Scholar]
- 3.Gurtovenko AA, Vattulainen I. Journal of the American Chemical Society. 2005;127(50):17570–17571. doi: 10.1021/ja053129n. [DOI] [PubMed] [Google Scholar]
- 4.Wiese A, Brandenburg K, Carroll SF, Rietschel ET, Seydel U. Biochemistry. 1997;36(33):10311–10319. doi: 10.1021/bi970177e. [DOI] [PubMed] [Google Scholar]
- 5.Gutsmann T, Haberer N, Carroll SF, Seydel U, Wiese A. Biological chemistry. 2001;382(3):425–434. doi: 10.1515/BC.2001.052. [DOI] [PubMed] [Google Scholar]
- 6.Ravi HK, Stach M, Soares TA, Darbre T, Reymond JL, Cascella M. Chemical communications. 2013;49(78):8821–8823. doi: 10.1039/c3cc44912b. [DOI] [PubMed] [Google Scholar]
- 7.Domingues MM, Inacio RG, Raimundo JM, Martins M, Castanho MA, Santos NC. Biopolymers. 2012;98(4):338–344. doi: 10.1002/bip.22095. [DOI] [PubMed] [Google Scholar]
- 8.Wiese A, Gutsmann T, Seydel U. Journal of endotoxin research. 2003;9(2):67–84. doi: 10.1179/096805103125001441. [DOI] [PubMed] [Google Scholar]
- 9.Mueller M, Lindner B, Kusumoto S, Fukase K, Schromm AB, Seydel U. The Journal of biological chemistry. 2004;279(25):26307–26313. doi: 10.1074/jbc.M401231200. [DOI] [PubMed] [Google Scholar]
- 10.Zhu W, Ma H, Miao J, Huang G, Tong M, Zou S. International immunopharmacology. 2013;15(2):457–465. doi: 10.1016/j.intimp.2012.12.007. [DOI] [PubMed] [Google Scholar]
- 11.Lee DK, Park EJ, Kim EK, Jin J, Kim JS, Shin IJ, Kim BY, Lee H, Kim DE. Cellular physiology and biochemistry : international journal of experimental cellular physiology, biochemistry, and pharmacology. 2012;30(3):499–511. doi: 10.1159/000341433. [DOI] [PubMed] [Google Scholar]
- 12.Shimada M, Kadowaki T, Taniguchi Y, Inagawa H, Okazaki K, Soma G. Anticancer research. 2012;32(6):2337–2341. [PubMed] [Google Scholar]
- 13.Kim BW, Koppula S, Hong SS, Jeon SB, Kwon JH, Hwang BY, Park EJ, Choi DK. PloS one. 2013;8(2):e55792. doi: 10.1371/journal.pone.0055792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clausell A, Garcia-Subirats M, Pujol M, Busquets MA, Rabanal F, Cajal Y. The journal of physical chemistry B. 2007;111(3):551–563. doi: 10.1021/jp064757+. [DOI] [PubMed] [Google Scholar]
- 15.Katz M, Tsubery H, Kolusheva S, Shames A, Fridkin M, Jelinek R. The Biochemical journal. 2003;375(Pt 2):405–413. doi: 10.1042/BJ20030784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schindler M, Osborn MJ, Koppel DE. Nature. 1980;285(5762):261–263. doi: 10.1038/285261a0. [DOI] [PubMed] [Google Scholar]
- 17.Wu ES, Jacobson K, Papahadjopoulos D. Biochemistry. 1977;16(17):3936–3941. doi: 10.1021/bi00636a034. [DOI] [PubMed] [Google Scholar]
- 18.Kirschner K, Lins R, Maass A, Soares T. Journal of Chemical Theory and Computation. 2012;28:14849–14854. doi: 10.1021/ct300534j. [DOI] [PubMed] [Google Scholar]
- 19.Soares T, Straatsma T. Molecular Simulation. 2008;34:295–307. [Google Scholar]
- 20.Gurtovenko AA, Vattulainen I. Journal of the American Chemical Society. 2007;129(17):5358–5359. doi: 10.1021/ja070949m. [DOI] [PubMed] [Google Scholar]
- 21.Gurtovenko AA, Vattulainen I. The Journal of chemical physics. 2009;130(21):215107. doi: 10.1063/1.3148885. [DOI] [PubMed] [Google Scholar]
- 22.Gurtovenko AA, Vattulainen I. The journal of physical chemistry B. 2009;113(20):7194–7198. doi: 10.1021/jp902794q. [DOI] [PubMed] [Google Scholar]
- 23.Brannigan G, Lin LC, Brown FL. European biophysics journal : EBJ. 2006;35(2):104–124. doi: 10.1007/s00249-005-0013-y. [DOI] [PubMed] [Google Scholar]
- 24.Sigalov G, Fenley A, Onufriev A. The Journal of chemical physics. 2006;124(12):124902. doi: 10.1063/1.2177251. [DOI] [PubMed] [Google Scholar]
- 25.Mongan J, Simmerling C, McCammon JA, Case DA, Onufriev A. J Chem Theory Comput. 2007;3(1):156–169. doi: 10.1021/ct600085e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li C, Li L, Petukh M, Alexov E. Molecular based mathematical biology. 2013:1. doi: 10.2478/mlbmb-2013-0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baker NA. Methods in enzymology. 2004;383:94–118. doi: 10.1016/S0076-6879(04)83005-2. [DOI] [PubMed] [Google Scholar]
- 28.Zhou YC, Feig M, Wei GW. J Comput Chem. 2008;29(1):87–97. doi: 10.1002/jcc.20769. [DOI] [PubMed] [Google Scholar]
- 29.Li L, Li C, Sarkar S, Zhang J, Witham S, Zhang Z, Wang L, Smith N, Petukh M, Alexov E. BMC biophysics. 2012;5:9. doi: 10.1186/2046-1682-5-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Z, Zheng Y, Petukh M, Pegg A, Ikeguchi Y, Alexov E. PLoS computational biology. 2013;9(2):e1002924. doi: 10.1371/journal.pcbi.1002924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li C, Petukh M, Li L, Alexov E. J Comput Chem. 2013;34(22):1949–1960. doi: 10.1002/jcc.23340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murray D, Hermida-Matsumoto L, Buser CA, Tsang J, Sigal CT, Ben-Tal N, Honig B, Resh MD, McLaughlin S. Biochemistry. 1998;37(8):2145–2159. doi: 10.1021/bi972012b. [DOI] [PubMed] [Google Scholar]
- 33.Bechor D, Ben-Tal N. Biophysical journal. 2001;80(2):643–655. doi: 10.1016/S0006-3495(01)76045-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hirayama T, Okaniwa M, Imada T, Ohashi A, Ohori M, Iwai K, Mori K, Kawamoto T, Yokota A, Tanaka T, Ishikawa T. Bioorganic & medicinal chemistry. 2013;21(17):5488–5502. doi: 10.1016/j.bmc.2013.05.067. [DOI] [PubMed] [Google Scholar]
- 35.Bishop EP, Rohs R, Parker SC, West SM, Liu P, Mann RS, Honig B, Tullius TD. ACS chemical biology. 2011;6(12):1314–1320. doi: 10.1021/cb200155t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dias RP, Hora G. C. A. d., Ramstedt M, Soares TA. Accepted for publication in Journal of Chemical Theory and Computation. doi: 10.1021/ct500075h. [DOI] [PubMed] [Google Scholar]
- 37.Jorgensen W, Chandrasekhar J, Madura J, Impey R, Klein M. Journal of Chemical Physics. 1983;79:926–936. [Google Scholar]
- 38.Lins RD, Straatsma TP. Biophysical journal. 2001;81(2):1037–1046. doi: 10.1016/S0006-3495(01)75761-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Straatsma TP, Soares TA. Proteins. 2009;74(2):475–488. doi: 10.1002/prot.22165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hess B. Journal of Chemical Theory and Computation. 2007;4:116–122. doi: 10.1021/ct700200b. [DOI] [PubMed] [Google Scholar]
- 41.Hess B, Bekker H, Berendsen H, Fraaije J. J Comput Chem. 1997;18:1463–1472. [Google Scholar]
- 42.Miyamoto S, Kollman P. J Comput Chem. 1992;13:952–962. [Google Scholar]
- 43.Berendsen H, Postma J, DiNola A, Haak J. Journal of Chemical Physics. 1984;81:3684–3691. [Google Scholar]
- 44.Tironi I, Sperb R, Smith P, van Gunsteren W. Journal of Chemical Physics. 1995;102:5451–5459. [Google Scholar]
- 45.Hess B, Kutzner C, van der Spoel D, Lindahl E. Journal of Chemical Theory and Computation. 2008:4. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 46.Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJ. J Comput Chem. 2005;26(16):1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 47.Pontes F, Rusu V, Soares T, Lins R. Journal of Chemical Theory and Computation. 2012;8:3830–3838. doi: 10.1021/ct300084v. [DOI] [PubMed] [Google Scholar]
- 48.Botello-Smith WM, Liu X, Cai Q, Li Z, Zhao H, Luo R. Chem Phys Lett. 2013;555:274–281. doi: 10.1016/j.cplett.2012.10.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang L, Zhang Z, Roccia W, Alexov E. Commun Comput Phys. 2013;13:13. doi: 10.4208/cicp.300611.120911s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rocchia W, Sridharan S, Nicholls A, Alexov E, Chiabrera A, Honig B. J Comput Chem. 2002;23(1):128–137. doi: 10.1002/jcc.1161. [DOI] [PubMed] [Google Scholar]
- 51.Wei G-W. Journal of Theoretical and Computational Chemistry. 2013;12(08) doi: 10.1142/S021963361341006X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nascimento A, Jr., Pontes FJ, Lins RD, Soares TA. Chemical communications. 2014;50(2):231–233. doi: 10.1039/c3cc46918b. [DOI] [PubMed] [Google Scholar]
- 53.Jeworrek C, Evers F, Howe J, Brandenburg K, Tolan M, Winter R. Biophysical journal. 2011;100(9):2169–2177. doi: 10.1016/j.bpj.2011.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu EL, Engstrom O, Jo S, Stuhlsatz D, Yeom MS, Klauda JB, Widmalm G, Im W. Biophysical journal. 2013;105(6):1444–1455. doi: 10.1016/j.bpj.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hu L, Wei GW. Biophysical journal. 2012;103(4):758–766. doi: 10.1016/j.bpj.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.