Abstract
There are many mechanisms of lifespan extension, including the disruption of insulin/IGF-1 signaling, metabolism, translation, or feeding. Despite the disparate functions of these pathways, inhibition of each induces responses that buffer stress and damage. Here, emphasizing data from genetic analyses in C. elegans, we explore the effectors and upstream regulatory components of numerous cytoprotective mechanisms activated as major elements of longevity programs, including detoxification, innate immunity, proteostasis, and oxidative stress response. We show that their induction underpins longevity extension across functionally diverse triggers and across species. Intertwined with the evolution of longevity, cytoprotective pathways are coupled to the surveillance of core cellular components, with important implications in normal and aberrant responses to drugs, chemicals, and pathogens.
Keywords: Cytoprotection, Hormesis, Detoxification, Stress, Longevity, Aging
The Biology of Aging
Aging in many organisms is accompanied by diverse pathologies, suggesting that it may be the product of many physiological mechanisms of decay. The pace of aging varies greatly across species and manifests distinct pathologies amongst the tissues of individual organisms. Beneath the variation, however, lies a common deterioration of function over time. The universality of the decay of biological integrity suggests that a single phenomenon may underlie the process: a balance of damage accumulation, defense, and repair that determines the rate of cellular deterioration. Analysis of the mechanisms by which this balance is regulated may reveal the regulatory axes of the aging process and allow the eventual development of anti-aging therapeutics. Already, genetic analyses have revealed a network of homeostatic mechanisms that couple cytoprotection and lifespan. Here we review the evidence for the involvement of cytoprotective functions, including detoxification, innate immunity, oxidative stress response, proteostasis, and others, in aging and how variation in such pathways might inform our understanding of the aging process itself. While many studies have addressed these mechanisms in isolation, we present evidence that they are evolutionarily, genetically, and functionally integrated.
Insulin/IGF-1 Signaling
Many organisms can enter states of quiescence during which aging may decrease to a negligible rate. Yeast enter a state of diapause in response to nutrient deprivation, fruit flies enter a state of diapause in response to cold, some mammals, such as bats, are able to hibernate, and the nematode C. elegans enters the semi-dormant dauer state [1–9]. These states are characterized by decreased metabolism, increased cellular maintenance, and reduced exposure to predation [10–12].
Studies of the C. elegans dauer arrest have been particularly informative to aging. Dauer formation is an alternative developmental path induced by adverse environmental conditions, such as starvation and, most potently, crowding [5, 13]. An impermeable cuticle, cessation of feeding, and the activation of somatic maintenance pathways protect the animal from aging. Animals recovered after 5 to 60 days in dauer retain the same adult lifespan, egg production, and egg viability as animals that proceeded directly through normal reproductive development [14]. The animals, by this measure, do not age at all during a ~3-lifetime suspension in the dauer state.
Dauer arrest is regulated by insulin/IGF-1 and TGF-β endocrine signaling pathways [15–22]. The genes encoding the insulin/IGF-1 receptor (daf-2), the signaling PI-3 kinase (age-1), and the downstream FoxO transcription factor (daf-16) comprise the major elements of the pathway [21, 23–36]. Under conditions that support growth and reproduction, signaling through DAF-2 suppresses emergency protection- and maintenance-promoting functions activated by the DAF-16 transcription factor. Disruption of this insulin/IGF-1 signaling cascade drives the nuclear translocation and activation of DAF-16. In developing animals, the result is dauer formation. However, when the cascade is disrupted in adult C. elegans, the animals continue to feed and reproduce, albeit at a reduced level, but live two-fold longer than wild-type [37]. In a sense, the genetic pathway that allows dauer-arrested animals to suspend the aging process can be activated in non-dauer animals, if insulin/IGF-1 signaling is attenuated.
In wild-type adults, daf-16 serves important stress-responsive functions, lying upstream of numerous mechanisms responsible for cytoprotection (the protection of cells from damage). It is required for tolerance of heat, radiation, osmotic stress, anoxia, oxidative damage, and heavy metals, as well as pathogens [38–43]. Long-lived daf-2 mutants that activate DAF-16/FOXO nuclear translocation and transcriptional activation are resistant to these challenges [38–44]. The mechanisms that promote stress tolerance in dauers may be the same as those that, when activated in healthy adult animals, cause lifespan extension, with cytoprotection as the critical link upon which the two phenomena converge, as detailed below.
daf-16-Regulated Cytoprotective Mechanisms of Lifespan Extension
What target genes are regulated by DAF-16 to extend lifespan and modulate resistance to diverse stressors? Transcriptional analyses comparing wild-type, dauer, and long-lived insulin/IGF-1 signaling mutant C. elegans first identified heat shock proteins and the antioxidant enzyme sod-3 as the most DAF-16-responsive downstream genes, suggesting roles for proteostasis and ROS detoxification [44–46]. More in depth analysis comparing daf-2 and WT adult animals confirmed that heat shock proteins were upregulated by as much as 140-fold and also found that neuropeptide-like proteins associated with pathogen defense were upregulated 2-fold [47].
Microarray studies identified still more DAF-16 targets. Microarray comparisons of daf-2 and age-1 mutants, or of daf-2 gene inactivation with and without daf-16 mutation or inactivation, over 8 days of adulthood identified 254 genes upregulated and 243 downregulated by daf-16. Activated stress response mechanisms were found to include heat shock proteins (hsp-16.1, hsp-16.2, hsp-12.3, and hsp-12.6), catalase and superoxide dismutase antioxidant enzymes (ctl-1, ctl-2, and sod-3), and putative antimicrobial effectors (such as LYS, SPP, CLEC, and NLP class genes) [48]. The analysis also revealed upregulation of detoxification mechanisms (such as, cytochrome, UDP, and glutathione transferase genes, etc.). Several analyses of dauer and daf-2 mutant transcription focused upon the detoxification response and, prophetically, suggested it may play a critical role in protective functions [49, 50].
The number and diversity of cytoprotective genes and gene classes upregulated downstream of DAF-16 suggests that the daf-2 aging phenotype is not mediated by a single effector mechanism. Because of the free radical theory of aging [51], oxidative stress resistance mechanisms were the first to be explored in detail. Oxidative damage of nuclear and mitochondrial DNA, proteins, and lipids increases with age in organisms ranging from invertebrates to humans, and correlates with species longevity [52–60]. Intraspecies observations of the correlation between ROS tolerance and strain lifespan have yielded mixed results, often contradicting the suggestion of a simple linear relationship between ROS and aging [61–65]. Many long-lived mutants are resistant to treatment with ROS, and mutants selected for resistance to ROS are also long-lived. In every case however, ROS levels could not be isolated from innumerable covariants and causality was not established [66].
Numerous antioxidative enzymes (catalases, superoxide dismutases, glutathione enzymes, metal-binding proteins) provided promising avenues for mechanistic analyses of ROS defenses [67–74]. The long-lived age-1 mutant is resistant to chemically induced oxidative damage and induces expression of the antioxidant enzymes Cu/Zn superoxide dismutase and catalase [68–74]. Microarray data demonstrates that daf-16 positively regulates the expression of sod-3, ctl-1, ctl-2, and mtl-1; inactivation of each of these genes subtly abrogates daf-2 lifespan extension [48]. Other studies have identified glutathione s-transferase 4 (gst-4) as a longevity-linked ROS response enzyme [72, 75].
Despite a well-developed set of candidates, mutant studies of ROS response genes have failed to reveal a causal role in the regulation of lifespan. In C. elegans, antioxidant enzymes include 5 superoxide dismutase genes and 3 catalases. Individual deletion of the superoxide dismutases did not affect lifespan, with the exception of sod-2 deletion, which increased lifespan [76, 77]. In mice, knockouts of five antioxidant enzymes (superoxide dismutase 2, glutathione peroxidases 1 and 4, methionine sulfoxide reductase A, and thioredoxin 2) increase sensitivity to ROS but do not decrease lifespan. Antioxidant enzyme overexpression (superoxide dismutase 1 and 2, catalase, glutathione peroxidase 4, or binary combinations of superoxide dismutase and catalase) fails to extend lifespan, with the exceptions of sod-1 and, from separate experiments, mitochondria-targeted catalase [66, 78, 79]. Although some studies have conflicted with these negative results, the study of anti-oxidant enzymes has imparted little clarity to the ROS hypothesis, and instead raised serious doubts regarding its validity [53, 66, 76]. Reports of the response to exogenous treatment with chemical antioxidants conflict as well [53, 80–85]. The current contention in the field holds that ROS are not the sole determinant of aging, but may contribute. At present, the archetypal ROS theory of aging has such scant support, it may follow in the footsteps of phlogiston and phrenology.
Evidence that heat shock proteins contribute to longevity is more compelling. As organisms age, misfolded proteins accumulate but the capacity to induce heat shock proteins decreases, suggesting that the ability to engage heat shock proteins contributes to long-term cellular maintenance [86, 87]. Enhanced thermotolerance correlates with enhanced longevity in insulin/IGF-1 signaling mutants, and expression of hsp-16.2 predicts longevity within isogenic populations of C. elegans [88, 89]. Loss of the C. elegans heat shock regulatory transcription factor HSF-1 abrogates the daf-2 mutant and dietary deprivation-induced lifespan extension phenotypes, while overexpression of hsf-1 extends the lifespan of wild-type animals [27, 90]. Exposure to a mild heat pretreatment induces enhanced thermotolerance and also extends lifespan in C. elegans, as well as Drosophila melanogaster [88, 90]. These findings are supported by transcriptional profiling of daf-2 mutants [47, 48]. Inactivation of HSF-1 targets hsp-16.1, hsp-16.49, hsp-12.6, and sip-1 partially reduces the extension of lifespan conferred by daf-2 mutation or by hsf-1 overexpression. daf-16 is required for thermotolerance and the expression of heat shock response genes, suggesting that HSF-1 and DAF-16 act cooperatively to regulate transcription of stress-responsive targets [90].
Not all chaperones are regulated by HSF-1; some are highly specialized to specific functions or organelles. The mitochondrial unfolded protein response (Mt UPR) and the endoplasmic reticulum unfolded protein response (ER UPR) are examples. In C. elegans, as in nearly all eukaryotes, the accumulation of unfolded proteins in the ER activates an endonuclease, IRE-1, which cleaves an intron from the mRNA of the x-box transcription factor xpb-1. This cleavage results in the expression of compartment-specific chaperones and ER-associated degradation proteins. Lifespan extension in daf-2 mutants is dependent upon both ire-1 and xbp-1, suggesting that genes involved in ER proteostasis may contribute to daf-2 mutant lifespan [91, 92].
In many organisms, susceptibility to infection increases with age, but daf-2 mutants are resistant to diverse pathogens, suggesting a role for immune function in longevity [39, 93, 94]. daf-2 mutants survive five to six fold longer than controls when exposed to the Gram-positive pathogens E. faecalis and S. aureus, and are also resistant to the Gram-negative pathogen P. aeruginosa [39]. This resistance is mediated by daf-16 [39, 95]. Insulin/IGF-1 signaling mutants express probable antimicrobial effectors at elevated levels [47, 48, 93, 96]. Effector classes include peptidoglycan-degrading lysozymes (LYS), lipid-degrading or pore-forming saposin-like proteins (SPP), bacteriocin motif-containing neuropeptide-like proteins (NLP), and c-type lectins (CLEC), which may act in pathogen recognition or directly in defense [93, 97–101]. DAF-16 positively regulates target genes from each of these classes [48].
Chaperones and antioxidants may also contribute to the pathogen resistance of daf-2 mutants. Observations of protein aggregation in the intestine suggest that infection is accompanied by proteotoxicity [102]. Consistent with this finding, hsf-1 is required for resistance to infection, while overexpression of hsf-1 or heat shock pretreatment are protective [103]. The interplay of daf-16 and hsf-1 may also be relevant to these phenotypes [103]. The enhanced pathogen resistance of daf-2 mutants also requires the daf-16-regulated antioxidant enzymes sod-3 and ctl-2, which has been attributed to the use of a reactive oxygen burst as a defensive strategy [95, 104–107]. Alternatively, however, some pathogens utilize ROS to weaken the host organism [108–110]. In addition, ROS defenses may contribute to proteostasis, since proteins can be damaged by ROS and inactivation of superoxide dismutases increases infection-induced aggregation, while treatment with organic antioxidants reduces it [102].
The upregulation of detoxification mechanisms is another aspect of daf-2 mutant cytoprotection. Detoxification is a two-stage process consisting of the addition of a reactive group to the toxin (Phase I) with subsequent conjugation of a water-soluble moiety to the reactive site (Phase II). Phase I relies heavily on Cytochrome P450’s (CYPs), while Phase II utilizes enzymes such as UDP-glucuronosyltransferases (UGTs), glutathione S-transferases (GSTs), sulfotransferases, and acetyltransferases [50]. The C. elegans complement of these enzymes includes 86 CYPs, 72 UGTs, and 48 GSTs [111]. In daf-2 mutants, daf-16 is required for the upregulation of 9 Cytochrome P450s, 3 UGTs, and 1 GST, gst-4 in one study [48]; in a similar study, 21 CYPs, 22 UGTs, and 16 GSTs are up-regulated in daf-2 mutant and/or dauer animals [50]. Similar enzymes are upregulated in GH/IGF-1-deficient mice, as well as mice that are calorically restricted [112]. Detoxification could contribute to longevity through the clearance of toxins generated by endogenous processes, such as metabolism, and lipophilic byproducts [113]. Though detoxification mechanisms may contribute to aging in this way, their endogenous function may be in defense against xenobiotics generated by microbial cohabitants of C. elegans’ natural environment, which includes soil and rotting vegetation. Microbial toxins and virulence factors may also impact mammalian aging in the same way.
The C. elegans gene skn-1 is a homolog of mammalian Cap‘n’Collar transcriptional regulators of phase II detoxification and oxidative stress response. Consistent with the proposed roles of these processes in lifespan regulation, loss of skn-1 decreases the longevity of daf-2 adults, while skn-1 overexpression extends wild-type lifespan. Loss of skn-1 also suppresses the oxidative stress tolerance of daf-2 mutants. Like DAF-16, SKN-1 is sequestered in the nucleus by AKT-1/AKT-2/SGK-1 phosphorylation, but translocates to the nucleus when insulin/IGF-1 signaling is disrupted [114–117]. Either transcription factor can localize to the nucleus and activate target gene transcription without the other, suggesting that they function independently. However, they are at least partially interdependent in lifespan extension, since skn-1 contributes to daf-2 lifespan and daf-16 contributes to lifespan extension when skn-1 is overexpressed [116]. Transcriptional targets of DAF-16 and SKN-1 are also partially overlapping. Genes regulated by SKN-1 include numerous GST, UGT, and CYP class genes as well as the major antioxidants ctl-2, ctl-3, and sod-1 [118, 119].
Decreased insulin/IGF-1 signaling activates autophagy, a means to scavenge resources from non-essential processes when food is limited. Autophagy may also rejuvenate cells by selectively degrading old and damaged proteins [120]. Without bec-1, a highly conserved gene required for autophagosome assembly, dauer formation and adult lifespan extension are disrupted [50]. Unlike most functions required for daf-2 lifespan, autophagy is not downstream of daf-16. Because loss of daf-16 is sufficient to abrogate lifespan extension, autophagy alone is not sufficient to extend lifespan. This result indicates that the role of autophagy may be to channel scavenged resources toward somatic maintenance pathways regulated by daf-16 [121].
daf-16-Independent Cytoprotective Pathways to Lifespan Extension
Several high throughput RNAi screens performed to identify gene inactivations that extend C. elegans longevity revealed that the disruption of core cellular functions such as metabolism and translation extends lifespan [122–124]. Inactivation of numerous genes in both functional groups extends longevity, suggesting that lifespan extension is a response to the general malfunction of these processes. Both extend lifespan in a daf-16-independent manner, suggesting mechanisms distinct from insulin-like signaling.
Since heat shock proteins contribute to the lifespan of daf-2 mutants, the mitochondria-specific chaperones induced by the mitochondrial unfolded protein response (Mt UPR) might contribute to the long life of animals with disrupted mitochondrial function. Two chaperones in particular, hsp-6 and hsp-60, have been identified as C. elegans effectors of this conserved protective response [125–127]. They are upregulated when the accumulation of unfolded proteins in the mitochondria is recognized by the protease ClpP, precipitating the activation of the transcription factor ZC376.7 and the downstream transcription regulatory complex of DVE-1 and UBL-5 [125–131]. Inactivation of the Mt UPR transcriptional coactivator ubl-5 abrogates lifespan extension in a well-studied mitochondrial mutant, isp-1, with minimal effect upon the lifespan of daf-2 or wild-type animals.
Xenobiotic detoxification mechanisms may mediate the disposal of toxic metabolic byproducts and are upregulated by mitochondrial dysfunction. The transcriptional profiles of long-lived isp-1, clk-1, and cyc-1 mitochondrial mutants reveal upregulation of cytochrome P450, UGT, and GST class genes in all three. This is most apparent in cyc-1, in which 22 CYP, 23 UGT, and 9 GST genes are transcriptionally activated [132]. Additionally, mitochondrial mutants are resistant to diverse toxins, suggesting that the up-regulation of detoxification pathways in a mitochondrial mutant activates a general detoxification of toxins [133]. This, too, may contribute to lifespan extension following disruption of mitochondrial function.
Inhibition of translation is another decrement of essential function that extends lifespan in RNAi screens. Inactivation by RNAi of the translation initiation factor eIF-5A, encoded by iff-1, extends lifespan in C. elegans [122]. Late life inactivation by RNAi of at least ten other components of the translation machinery extends mean lifespan by as much as 55% [124]. Inhibition by RNAi of cap-binding complex proteins extends mean lifespan by up to 36% [134]. Consistent with the view that translational competence is tightly monitored and coupled to longevity regulation, translation components emerged as a major axis of regulation of yeast replicative aging [135].
Like other forms of longevity extension, inhibition of translation by RNAi increases resistance to starvation, ultraviolet radiation, or heat, though reports are partially conflicted [134, 136, 137]. Most sources confirm that lifespan extension by inhibition of translation is daf-16-independent [122–124, 134, 136–138]. Instead, inhibition of translation extends lifespan through the detoxification and antioxidant response regulatory transcription factor skn-1 [75, 139]. Inhibition by RNAi of initiation factor eIF1 (eif-1), S6 Kinase (rsks-1), and 3 other components of translation machinery induces SKN-1 transcriptional targets (gcs-1 and gst-4) and confers oxidative stress tolerance [140]. In a study including inactivation of translation initiation factors ifg-1 and eif-1, oxidative stress tolerance was dependent upon skn-1, but not daf-16, whereas lifespan could only be fully extended in the presence of both [140].
Translational profiling and microarray comparisons to quantify the translation of mRNAs in ifg-1(RNAi) animals reveal increased translation of 51 stress-responsive genes, including daf-16 and skn-1 [141]. Moreover, the open reading frames, 3′ UTRs, and 5′ UTRs of genes selectively translated during stress are over two-fold longer than those of genes downregulated under the same conditions. Essential and stress response genes are overrepresented amongst the long mRNAs, perhaps defining a basic mechanism for transcript triage during stress [141].
Studies isolating individual tissues may elaborate the cell-non-autonomous and potentially neuroendocrine nature of stress signals in long-lived animals, such as mitochondrial and insulin/IGF-1 signaling mutants. In animals with lifespan-extending tissue-specific neuronal inactivation of the mitochondrial component cco-1, the mitochondrial stress response is induced in the intestine, suggesting a cell-non-autonomous signal. Longevity and cytoprotective signals may be uncoupled by tissue-specific lifespan-extending treatments, such as intestinal inactivation of cco-1, but further support is required to establish this possibility [62, 142, 143].
Cytoprotection Links Diverse Treatments that Extend Longevity
Genetic studies have identified over 50 mutations that extend lifespan and each is resistant to one or more stressors, such as oxidative damage, heat stress or irradiation [62, 142]. The degree of stress tolerance is directly correlated with the degree of longevity extension [62]. Gene inactivations found to extend longevity in large-scale RNAi screens have further confirmed this association.
The association of stress tolerance and lifespan is so intimate that screens to identify mutants or gene inactivations that are stress-resistant efficiently identify perturbations that extend longevity. A genetic screen for enhanced thermotolerance in C. elegans isolated 63 lines, of which 49 (80%) were long-lived by at least 15% [144]. In a screen for enhanced ROS tolerance, six mutants were isolated and three increased lifespan by at least 15% [65]. A similar RNAi screen identified 608 gene inactivations by RNAi that increase ROS tolerance, of which 84 increased mean lifespan by at least 10% [64]. This surrogate primary enrichment for longevity mutants has also been successful in yeast and flies [145, 146].
Conditions that extend lifespan could be grouped into two classes: those that activate stress response pathways through direct genetic perturbation, and those that activate stress response pathways indirectly because they are stress stimuli (Fig. 1). Mutation of daf-2 is paradigmatic of the first class, while disruption of metabolism and translation may fall into the second. Treatments in both classes activate stress response mechanisms that protect the extant components of the cell, including protein chaperones, antioxidant enzymes, putative antimicrobial effectors, and detoxification mechanisms.
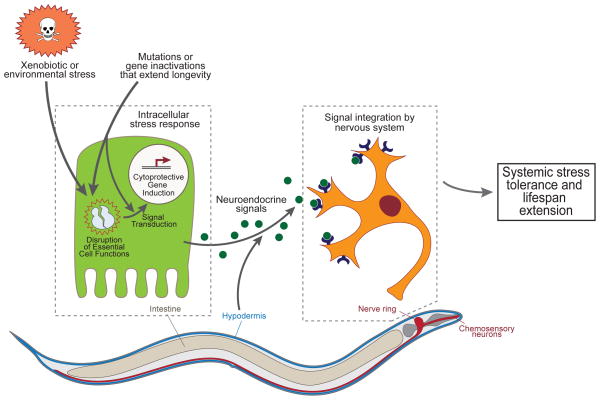
Figure 1. Cytoprotective Signaling Results in Lifespan Extension.
Mutations and gene inactivations that extend lifespan may be classified into those that activate cytoprotective pathways through the disruption of essential cell functions, such as metabolism or translation, and those that do so through direct roles in stress signaling pathways. Cytoprotective pathways evolved to combat environmental stressors, such as pathogens and xenobiotics. Exposed tissues, such as the intestine, hypodermis, or some neurons, are the expected point of first contact with these threats. Non-autonomous signals, such as neuroendocrine signals, are predicted to orchestrate the systemic aspect of stress tolerance, inducing cytoprotective pathways that mitigate the accumulation of cellular damage and mediate lifespan extension.
The induction of improved stress tolerance and increased lifespan by treatments that are ostensibly deleterious, such as disruption of mitochondrial function or translation, is counterintuitive. The paradox is consistent with hormesis, the protective activation of cytoprotective pathways in response to stress stimuli. While not yet fully understood molecularly, hormesis is a compensatory response that may involve maintenance, repair, or both. The ability to buffer damage is ultimately enhanced [147, 148]. Because of hormesis, pretreatment with a low, sub-toxic exposure to stress enhances tolerance of a subsequent lethal dose exposure [62, 149]. Induced stress tolerance has been observed in response to heat, oxidative stress, radiation, and other treatments [149–152]. Pretreatment with one of these stressors induces subsequent tolerance of others [150, 153]. Whether this cross-functionality results from the activation of multiple cytoprotective mechanisms or an unappreciated breadth of protection conferred by individual pathways remains largely untested.
The C. elegans response to heat shock and mitochondrial dysfunction illustrates the role of hormesis in lifespan extension. Treatment of C. elegans with a brief but intense heat shock (1 hour at 35°C) results in a mean longevity increase over 15%, while sustained exposure is lethal. Subsequent treatments administered every third day increasingly extend lifespan, to a maximum of nearly 40% after five treatments [154]. This pattern is reiterated in the response to inactivation of the mitochondrial ATPase atp-3. A 60% reduction in atp-3 mRNA reduces the lifespan of a WT N2 worm from ~14 to ~8 days, but with a 20–40% reduction in atp-3 mRNA lifespan is extended by up to 70% [155]. In both the heat shock and atp-3 studies, cytoprotective mechanisms buffer damage at low doses, but are overwhelmed at high doses. The importance of dosage, when considered against the variable efficacy of RNAi, may explain why functionally related inactivations do not necessarily induce comparable longevity phenotypes.
Exposure to xenobiotics, like environmental stress and genetic perturbation of essential cell functions, can also induce a hormetic response. Antimycin, a lethal mitochondrial inhibitor, can extend longevity by 23% when applied in the appropriate measure [156]. Paraquat, a potent ROS generator, extends longevity by 40–50% when C. elegans are treated continuously with a low dose [157]. The same has been observed using the ROS-generating agent juglone, inducing a lifespan extension of nearly 30% [151]. Efforts to catalogue hormetic effects across diverse model organisms and treatments identify thousands of examples from bacteria to mammals [147, 148].
Hormesis may be adaptive in the wild. If the first signs of stress often forewarn of worse to come, a hormetic response would prove prescient. Although cytoprotective functions are energetically expensive, they insure against the potentially greater expenses of repair, replacement, and death. Cytoprotective pathways would not be expected to be constitutively active, as their requirements would drain resources from growth and reproduction. In fact, reproduction tends to be suspended under stressful conditions, perhaps by endocrine signals that orchestrate energy allocation and defer reproduction under non-optimal conditions. Ecologically, an organism would benefit most from limiting the base-level activity of cytoprotective pathways to a minimum optimized to meet the likely duration of its existence, while retaining inducible capacity for emergency defense. Total cytoprotective capacity may determine the upper limits of organismal lifespan.
Surveillance of Cellular Functions Ties Longevity to Detoxification and Innate Immunity
Detoxification and pathogen response may play a central role in the evolution of longevity-regulatory cytoprotective networks. Natural toxins, or xenobiotics, are produced by microbes to inhibit competition for local resources or weaken eukaryotic hosts. Such toxins often evolve to target highly conserved essential cell processes, thereby achieving efficacy against a broad range of competitors or hosts; accordingly, many xenobiotics target conserved components of translation or metabolism. Exposure to natural toxins throughout evolutionary history may have been a driving force behind the development of cytoprotective capabilities. For instance, the natural habitat of C. elegans, within soil and rotting vegetation, is rich with microbes, many of which secrete xenobiotics or transfer diverse virulence factors to eukaryotic hosts. In this hostile milieu, pathogenesis and toxin exposure may be primary causes of mortality. Detoxification is the most direct response to xenobiotics, followed by innate immune defenses against the producing microbes. Transcriptional profiling has revealed that these mechanisms are induced in many tested modes of longevity extension. Further, with striking parity, genetic disruption of the very cell processes targeted by xenobiotics results in potent longevity extension across multiple species [124, 135]. These observations suggest an intimate association between xenobiotic response and lifespan extension.
In support of the link between xenobiotic response and longevity, C. elegans respond similarly to xenobiotics and genetic inactivation of xenobiotic-targeted longevity-modulatory functions. In particular, both induce a behavioral escape response termed ‘aversion’, in which normally sedate animals vacate a bacterial lawn by moving rapidly in long paths. This stress-responsive dispersal is induced by gene inactivations that disrupt metabolism and translation, as well as by xenobiotics that target those processes [158]. Such a response is best suited to the avoidance of a localized stimulus, suggesting that the genetic disruption of essential cell functions is interpreted as a xenobiotic or pathogenic attack, consistent with the transcriptional induction of xenobiotic and pathogen response effectors.
The transcriptional response associated with aversive behavior further supports the possibility that the disruption of essential cell functions activates a pathogen and xenobiotic defense apparatus. Upregulated genes include effectors of detoxification, such as cyp-35B, protein chaperones, and the potent induction of the pathogen-responsive genes clec-60, irg-1, and F35H12.5. The response occurs even when the trigger is dsRNA produced by a non-pathogenic E. coli strain but targeting an essential cell process (e.g., translation, mitochondrial function, vacuolar ATPase activity, or basic metabolic pathways), despite the absence of a pathogen or toxin. Under these conditions, the induction of a transcriptional response normally induced by bacterial pathogens implies that the innate immune apparatus can be triggered by a single signal of dysfunction from the disruption of a core cellular process [158]. Similarly, treatment with the Pseudomonas aeruginosa ribotoxin ToxA induces a transcriptional response congruent with that induced by exposure to the pathogen itself [159]. Thus C. elegans, and most likely all eukaryotes, has evolved a system that surveils essential functions, such as translation, and interprets decrements in those functions as a xenobiotic attack indicative of an adverse microbial presence [158]. We refer to this system of cellular surveillance-activated detoxification and defense as cSADD [158] (Fig. 2).
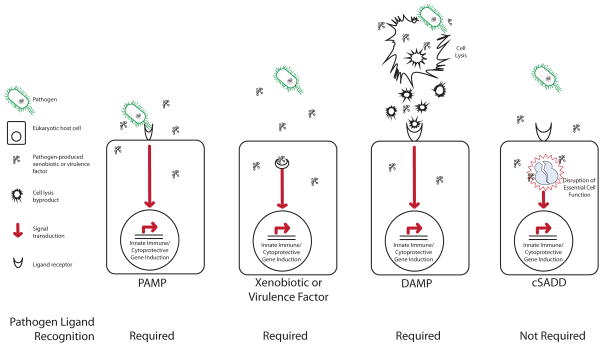
Figure 2. Surveillance of Core Cellular Functions is a Distinct Mechanism of Xenobiotic and Pathogen Detection and Response.
Prevalent models of pathogen and xenobiotic response require the detection of pathogen- or damage-associated ligands by cell surface receptors. Pathogen ligands are referred to as pathogen-associated molecular patterns (PAMPs) and include compounds such as lipid A on the outer membranes of Gram-negative bacteria or conserved proteins such as flagellins. Xenobiotics produced by pathogens may be detected as well. Damage-associated molecular patterns (DAMPs) are the secondary products of pathogen or xenobiotic damage, such as the detritus resulting from the breakdown of compromised cells, an example of which may be DNA released into the extracellular milieu by a pathogen-lysed cell. In each of these models, the receptor-mediated detection of the PAMP or DAMP initiates a signaling cascade that results in the induction of defense responses, such as innate immune gene induction. In contrast, cellular surveillance activated detoxification and defenses (cSADD) are initiated by decrements in the activity of core cellular functions, such as the mitochondria or ribosome, which are explicitly surveilled. Because many diverse pathogens and xenobiotics disrupt a small number of critical cell functions, monitoring core cellular processes provides an efficient mechanism of comprehensively detecting and responding to highly diverse stimuli, escaping the limitations of PAMP or DAMP ligand detection. These models are not mutually exclusive.
Taken together, the transcriptional and behavioral response to essential cell process dysfunction indicates that the status of essential cell functions is directly or indirectly surveilled upstream of a multifaceted cSADD response to xenobiotics/pathogens that integrates detoxification, immunity, proteostatic mechanisms, and other defenses. These functions are consistent with a role in pathogen defense: detoxification to neutralize and purge xenobiotics, antimicrobial defenses to rebuff the producing microbes, and chaperones and antioxidants for maintenance and repair of targeted or secondarily damaged cell structures [124, 158, 160]. These are the same cytoprotective functions that are so tightly linked with longevity extension, and indeed the most potent models of longevity extension induce just such multifaceted cytoprotective suites. Cumulatively, these results indicate that the cytoprotective defenses induced by exposure to xenobiotics may be the same as those that extend lifespan following genetic disruption of translation and metabolism. This model parallels insulin/IGF-1 signaling: mechanisms evolved to serve in emergency cellular defense can, under certain conditions, mediate longevity extension.
The possibility that essential cell processes are explicitly surveilled (cSADD) diverges from the most established view of biohazard detection; namely, that toxins and pathogens are detected by the recognition of ligands, or pathogen-associated molecular patterns (PAMPs), such as lipid A on the outer membranes of Gram-negative bacteria (Fig. 2). A problem with the PAMP hypothesis is that even benign bacteria express these signatures of bacteria, so this model does not simply distinguish between pathogen and harmless or commensal bacteria. An alternative to PAMPs is the recognition of damage-associated molecular patterns (DAMPs), such as the release of DNA into the extracellular milieu by compromised cells. The DAMP concept is consistent with the proposal that hosts monitor internal structures for toxin-indicative signs of distress [161]. The usual way that the DAMP hypothesis is configured is that as the damage to host cells becomes profound enough, their corpses signal an infection. This may be too late to trigger effective immunity and one can envision a selective advantage to earlier and earlier signals of such an infection. At its limit, the sort of cellular surveillance for toxins and virulence factors of the cSADD hypothesis would constitute an effective, evolved DAMP system. Activation of a suite of pathogen defenses by a single signal, such as translation dysfunction, contrasts with the multiplicity of stimuli presented by natural pathogens, which transfer a highly evolved suite of virulence factors and chemical toxins to hosts, and removes many layers of evolved measure/counter measure complexity in the pathogen/host interaction.
In mammals, the best-studied effectors of toxin detection are the nuclear hormone receptors of the CAR and PXR class and aryl hydrocarbon receptors, thought to use promiscuous ligand binding domains to detect particular classes of xenobiotic chemicals. However, the number of receptors is too limited to explain the detection of an almost infinite number of possible chemical toxins (e.g., humans have only 48 NHR genes). It is more likely that these ligand gated transcription factors respond to internally produced alarm ligands, for example signaling molecules that are themselves modified by cytochrome p450s, in analogy to cytochrome p450 modification of mammalian sex hormones. The detection problem of highly variable toxins may be solved if the inhibition of core cellular components are monitored and detected, rather than the toxins themselves. Direct surveillance of xenobiotic targets could provide a means of responding to diverse and unfamiliar xenobiotics or pathogens, perhaps anticipating pathogen invasion, without the limitations of ligand recognition [158, 159]. The greater the load of toxins targeting a given function, the greater the selective advantage of surveilling it. Dysfunction of core cellular processes (such as those mediated by the ribosome, cytoskeleton, or mitochondrion) must then trigger a signal to activate the downstream response apparatus.
The regulatory cascade of xenobiotic response may involve mechanisms for the detection of dysfunction, the transduction and dissemination of resulting signals that an attack is in progress, and the activation of downstream effectors, potentially coordinating systemic defenses through endocrine cues (Fig. 1). To date, studies of xenobiotic response have focused on the identification of the effectors of detoxification (CYP, UGT, and GST class genes and others) more so than the mechanisms by which these effectors are regulated. As a result, the upstream signaling and regulatory factors are poorly understood. One exception, the detoxification transcription factor skn-1, contributes significantly to lifespan extension and extends lifespan when overexpressed [116]. In addition, a Jun kinase-related MAPK pathway is required for pathogen and stress resistance as well as the aversive behavior induced by essential gene inactivation. These pathways may be responsible for transducing damage signals within attacked cells or across cells and tissues [158].
Another set of new regulatory candidates has emerged from a systematic screen for upstream components of cytoprotective gene induction following exposure to various toxins. The screen explored four cytoprotective effectors upregulated in long-lived animals (hsp-4, hsp-6, sod-3 and gst-4) by probing their induction in response to xenobiotic and genetic stimuli. Identified regulatory genes include the import in alpha nuclear factor ima-3 and the osmotic stress response kinase wnk-1, each required for the induction of organelle-specific chaperones, the mevalonate synthesis gene phi-50, required for oxidative stress and detoxification responses, and other regulatory genes functioning in processes such as phosphorylation, deacetylation, and protein degradation. Inactivation of these cytoprotective regulatory genes compromises survival following toxin exposure and disrupts lifespan extension in each of three tested models: the long-lived insulin/IGF-1 receptor mutant daf-2, the feeding defective mutant eat-2, and the mitochondrial mutant isp-1 [160]. These findings support the causality of cytoprotective functions in longevity extension and contribute to a growing body of knowledge suggesting that such functions underpin longevity phenotypes across diverse paradigms of lifespan extension. It follows that further exploration of xenobiotic response is likely to unveil new detoxification and lifespan-regulatory pathways.
Human Variation in Xenobiotic Surveillance
Xenobiotic surveillance has implications in medicine, since suites of drug detoxification genes are induced by medicinal drugs as well [162, 163]. The phase I and phase II detoxification effectors that modify and export small molecule toxins or drugs are numerous and highly variable [164]. Variation in detoxification genes and their regulatory cascades may be the result of the history of pathogen and toxin exposures in the lineage of each animal and plant species, further intensified in humans by the dramatic dietary and behavioral shifts that accompanied the advent of agriculture. Detoxification mechanisms may also continuously evolve to evade anti-surveillance measures of microbes, developed in the billion-year arms race against pathogens, perhaps including factors that neutralize surveillance components, block neuroendocrine signals, or disrupt the induction of defenses. These pressures must vary across diets and habitats. Within this variation, detoxification mechanisms must be selected for sensitivity, since a system responding to insignificant stimuli would be inefficient while a system responding only to extreme stimuli would be ineffective [164].
Given the challenge of responding to many xenobiotic and pathogenic stimuli with a high-variation detection and response apparatus, and the additional variability of genetic backgrounds, it is likely that some variants in some backgrounds are nonfunctional, hyper-responsive, or maladaptively responsive. Loss of appetite, nausea, and headaches may be common physiological phenotypes of the human detoxification response. Toxic responses to therapeutic drugs, such as nausea, may be exemplary of a detoxification response. Variation of such a system of xenobiotic surveillance may contribute to variation in the efficacy or tolerance of particular drugs or to normally benign chemicals or foods. For patients manifesting diseases of essential cell process dysfunction, such as mitochondrial diseases, variation in the sensitivity of the surveillance system may explain distinct clinical courses taken by individual patients. Even the experience of malaise associated with illness may be a function of the detoxification program, perhaps downstream of surveillance mechanisms. The molecular components of these surveillance and detoxification pathways may constitute new drug targets to suppress the nausea and loss of appetite induced by many therapeutic drugs, or the toxic side effects that derail many drugs in development.
The evolution of detoxification may also explain observed gender differences in drug response, such as the elevated frequency of adverse drug reactions observed in females as compared to males [165]. Females, particularly in placental mammals such as humans, may have evolved a more fulminant or sensitive response to toxins than males as a necessity of protecting gestating offspring from the developmental consequences of toxication. A low female set point for the induction of antimicrobial responses could also explain the gender bias in autoimmune disorders: more fulminant immune responses would increase the propensity for autoimmune reactions.
Hormesis, Disruption of Essential Cell Functions, and Human Longevity
Induction of cytoprotective pathways extends longevity in model organisms and may do so in humans. The inhibition of translation by treatment with rapamycin or mutation of the nutrient sensor TOR extends lifespan in yeast, worms, flies, and mice [166–168]. Human trials of rapamycin have been proposed [169, 170]. Similarly, drugs that inhibit metabolism and extend lifespan in model systems, such as antimycin, might also extend mammalian lifespan. A more practicable application to human longevity, however, will be the identification of therapeutics that induce cytoprotective pathways directly, rather than through the inhibition of essential cell functions. Such compounds could engage constitutive utilization of cytoprotective capacity normally induced only by stress stimuli, imparting to cells the robustness of maintenance and repair essential to lifespan extension.
Concluding Remarks
Recent advances in the study of longevity have elaborated the relationships between cytoprotective mechanisms and longevity phenotypes. The co-regulation of diverse cytoprotective mechanisms is driven by a shared role in balancing cellular defense, damage, and repair. While there are many paths to lifespan extension, research increasingly highlights thematic convergence on cytoprotection, and even on particular cytoprotective effectors and regulatory genes such as those described above. Ongoing research will continue to push beyond the piecemeal study of individual stress response functions or models of lifespan extension to address the unifying role and network characteristics of cytoprotection. Maximal lifespan extension may demand the co-activity of multiple cytoprotective functions. Appreciation of the conceptual parity between lifespan extension and hormesis will allow each of these fields to benefit from the advances of the other.
In C. elegans, the potent induction of cytoprotection that results from severe dysfunction of core cellular processes results in growth arrest and other pleiotropies, including behavioral aversion, while low to moderate dysfunction results in lifespan extension. The cytoprotective mechanisms that confer longevity extension when induced by moderate disruption of surveilled processes remain poorly understood. Analysis of the phenotypes associated with damaging levels of dysfunction may provide an avenue for probing these cytoprotective pathways. For instance, new longevity regulators may emerge from screens to identify animals that fail to arrest in the presence of a toxic dosage of xenobiotic, screens to identify animals that fail to disperse in the presence of a toxin or gene inactivation that disrupts an essential function, or studies of toxin dosages that extend lifespan. As the molecular pathways for cellular surveillance emerge from genetic analyses in C. elegans, we can expect new longevity pathways to emerge as well.
Highlights.
Maximal lifespan extension requires the co-activity of multiple cytoprotective mechanisms.
Longevity-modulatory cytoprotective mechanisms participate in complex regulatory networks.
Toxin/pathogen response is triggered by the disruption of surveilled core cellular processes.
Induction of this cytoprotective toxin/pathogen response program may contribute to lifespan extension.
Acknowledgments
We thank Sean Curran, Justine Melo, Yan Qi, Ying Liu, and Amaranath Govindan for advice and contributions to the evolving model of cytoprotection in aging. We thank Fred Ausubel and members of the Ausubel lab, Deb McEwan, Natasha Kirienko, and Read Pukkila-Worley for insights into bacterial virulence. We thank Rusty Howson for the figure.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lyman CP, et al. Hibernation and longevity in the Turkish hamster Mesocricetus brandti. Science. 1981;212:668–670. doi: 10.1126/science.7221552. [DOI] [PubMed] [Google Scholar]
- 2.Tatar M, et al. Negligible Senescence during Reproductive Dormancy in Drosophila melanogaster. Am Nat. 2001;158:248–258. doi: 10.1086/321320. [DOI] [PubMed] [Google Scholar]
- 3.Houthoofd K, et al. Ageing is reversed, and metabolism is reset to young levels in recovering dauer larvae of C. elegans. Exp Gerontol. 2002;37:1015–1021. doi: 10.1016/s0531-5565(02)00063-3. [DOI] [PubMed] [Google Scholar]
- 4.Wilkinson G, South J. Life history, ecology and longevity in bats. Aging Cell. 2002;1:124–155. doi: 10.1046/j.1474-9728.2002.00020.x. [DOI] [PubMed] [Google Scholar]
- 5.Cassada RC, Russell RL. The dauerlarva, a post-embryonic developmental variant of the nematode Caenorhabditis elegans. Dev Biol. 1975;46:326–342. doi: 10.1016/0012-1606(75)90109-8. [DOI] [PubMed] [Google Scholar]
- 6.Bitterman KJ, et al. Longevity regulation in Saccharomyces cerevisiae: linking metabolism, genome stability, and heterochromatin. Microbiol Mol Biol Rev. 2003;67:376–399. doi: 10.1128/MMBR.67.3.376-399.2003. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fabrizio P, Longo VD. The chronological life span of Saccharomyces cerevisiae. Aging Cell. 2003;2:73–81. doi: 10.1046/j.1474-9728.2003.00033.x. [DOI] [PubMed] [Google Scholar]
- 8.Longo VD, Fabrizio P. Regulation of longevity and stress resistance: a molecular strategy conserved from yeast to humans? Cell Mol Life Sci. 2002;59:903–908. doi: 10.1007/s00018-002-8477-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Werner-Washburne M, et al. Stationary phase in the yeast Saccharomyces cerevisiae. Microbiol Rev. 1993;57:383–401. doi: 10.1128/mr.57.2.383-401.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stuart JA, Brown MF. Energy, quiescence and the cellular basis of animal life spans. Comp Biochem Physiol A Mol Integr Physiol. 2006;143:12–23. doi: 10.1016/j.cbpa.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 11.Bieber C, Ruf T. Summer dormancy in edible dormice (Glis glis) without energetic constraints. Naturwissenschaften. 2009;96:165–171. doi: 10.1007/s00114-008-0471-z. [DOI] [PubMed] [Google Scholar]
- 12.Storey KB. Out cold: biochemical regulation of mammalian hibernation - a mini-review. Gerontology. 2010;56:220–230. doi: 10.1159/000228829. [DOI] [PubMed] [Google Scholar]
- 13.Yarwood EA, Hansen EL. Dauer Larvae of Caenorhabditis briggsae in Axenic Culture. J Nematol. 1969;1:184–189. [PMC free article] [PubMed] [Google Scholar]
- 14.Klass M, Hirsh D. Non-ageing developmental variant of Caenorhabditis elegans. Nature. 1976;260:523–525. doi: 10.1038/260523a0. [DOI] [PubMed] [Google Scholar]
- 15.Klass MR. A method for the isolation of longevity mutants in the nematode Caenorhabditis elegans and initial results. Mech Ageing Dev. 1983;22:279–286. doi: 10.1016/0047-6374(83)90082-9. [DOI] [PubMed] [Google Scholar]
- 16.Friedman DB, Johnson TE. A mutation in the age-1 gene in Caenorhabditis elegans lengthens life and reduces hermaphrodite fertility. Genetics. 1988;118:75–86. doi: 10.1093/genetics/118.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Riddle DL, et al. Interacting genes in nematode dauer larva formation. Nature. 1981;290:668–671. doi: 10.1038/290668a0. [DOI] [PubMed] [Google Scholar]
- 18.Swanson MM, Riddle DL. Critical periods in the development of the Caenorhabditis elegans dauer larva. Dev Biol. 1981;84:27–40. doi: 10.1016/0012-1606(81)90367-5. [DOI] [PubMed] [Google Scholar]
- 19.Albert PS, et al. Sensory control of dauer larva formation in Caenorhabditis elegans. J Comp Neurol. 1981;198:435–451. doi: 10.1002/cne.901980305. [DOI] [PubMed] [Google Scholar]
- 20.Thomas JH, et al. Evidence for parallel processing of sensory information controlling dauer formation in Caenorhabditis elegans. Genetics. 1993;134:1105–1117. doi: 10.1093/genetics/134.4.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gottlieb S, Ruvkun G. daf-2, daf-16 and daf-23: genetically interacting genes controlling Dauer formation in Caenorhabditis elegans. Genetics. 1994;137:107–120. doi: 10.1093/genetics/137.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kenyon C. The first long-lived mutants: discovery of the insulin/IGF-1 pathway for ageing. Philos Trans R Soc Lond B Biol Sci. 2011;366:9–16. doi: 10.1098/rstb.2010.0276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morris J, et al. A phosphatidylinositol-3-OH kinase family member regulating longevity and diapause in Caenorhabditis elegans. Nature. 1996;382:536–545. doi: 10.1038/382536a0. [DOI] [PubMed] [Google Scholar]
- 24.Johnson TE, et al. Comparing mutants, selective breeding, and transgenics in the dissection of aging processes of Caenorhabditis elegans. Genetica. 1993;91:65–77. doi: 10.1007/BF01435988. [DOI] [PubMed] [Google Scholar]
- 25.Malone EA, et al. Genetic analysis of the roles of daf-28 and age-1 in regulating Caenorhabditis elegans dauer formation. Genetics. 1996;143:1193–1205. doi: 10.1093/genetics/143.3.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kimura KD, et al. daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science. 1997;277:942–946. doi: 10.1126/science.277.5328.942. [DOI] [PubMed] [Google Scholar]
- 27.Greer EL, Brunet A. Different dietary restriction regimens extend lifespan by both independent and overlapping genetic pathways in C. elegans. Aging Cell. 2009;8:113–127. doi: 10.1111/j.1474-9726.2009.00459.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roush W. Worm longevity gene cloned. Science. 1997;277:897–898. doi: 10.1126/science.277.5328.897. [DOI] [PubMed] [Google Scholar]
- 29.Morris JZ, et al. A phosphatidylinositol-3-OH kinase family member regulating longevity and diapause in Caenorhabditis elegans. Nature. 1996;382:536–539. doi: 10.1038/382536a0. [DOI] [PubMed] [Google Scholar]
- 30.Ogg S, et al. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature. 1997;389:994–999. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- 31.Paradis S, Ruvkun G. Caenorhabditis elegans Akt/PKB transduces insulin receptor-like signals from AGE-1 PI3 kinase to the DAF-16 transcription factor. Genes Dev. 1998;12:2488–2498. doi: 10.1101/gad.12.16.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin K, et al. daf-16: An HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science. 1997;278:1319–1322. doi: 10.1126/science.278.5341.1319. [DOI] [PubMed] [Google Scholar]
- 33.O’Brien RM, et al. Hepatic nuclear factor 3- and hormone-regulated expression of the phosphoenol pyruvate carboxykinase and insulin-like growth factor-binding protein 1 genes. Mol Cell Biol. 1995;15:1747–1758. doi: 10.1128/mcb.15.3.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O’Brien RM, et al. Insulin-regulated gene expression. Biochem Soc Trans. 2001;29:552–558. doi: 10.1042/bst0290552. [DOI] [PubMed] [Google Scholar]
- 35.Finch CE, Ruvkun G. The genetics of aging. Annu Rev Genomics Hum Genet. 2001;2:435–462. doi: 10.1146/annurev.genom.2.1.435. [DOI] [PubMed] [Google Scholar]
- 36.Onuma H, et al. Correlation between FOXO1a (FKHR) and FOXO3a (FKHRL1) binding and the inhibition of basal glucose-6-phosphatase catalytic subunit gene transcription by insulin. Mol Endocrinol. 2006;20:2831–2847. doi: 10.1210/me.2006-0085. [DOI] [PubMed] [Google Scholar]
- 37.Kenyon C, et al. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- 38.Barsyte D, et al. Longevity and heavy metal resistance in daf-2 and age-1 long-lived mutants of Caenorhabditis elegans. Faseb J. 2001;15:627–634. doi: 10.1096/fj.99-0966com. [DOI] [PubMed] [Google Scholar]
- 39.Garsin DA, et al. Long-lived C. elegans daf-2 mutants are resistant to bacterial pathogens. Science. 2003;300:1921. doi: 10.1126/science.1080147. [DOI] [PubMed] [Google Scholar]
- 40.Lamitina ST, Strange K. Transcriptional targets of DAF-16 insulin signaling pathway protect C. elegans from extreme hypertonic stress. Am J Physiol Cell Physiol. 2005;288:C467–474. doi: 10.1152/ajpcell.00451.2004. [DOI] [PubMed] [Google Scholar]
- 41.Lin K, et al. Regulation of the Caenorhabditis elegans longevity protein DAF-16 by insulin/IGF-1 and germline signaling. Nat Genet. 2001;28:139–145. doi: 10.1038/88850. [DOI] [PubMed] [Google Scholar]
- 42.Mendenhall AR, et al. Glyceraldehyde-3-phosphate dehydrogenase mediates anoxia response and survival in Caenorhabditis elegans. Genetics. 2006;174:1173–1187. doi: 10.1534/genetics.106.061390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Murakami S, Johnson TE. A genetic pathway conferring life extension and resistance to UV stress in Caenorhabditis elegans. Genetics. 1996;143:1207–1218. doi: 10.1093/genetics/143.3.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Honda Y, Honda S. The daf-2 gene network for longevity regulates oxidative stress resistance and Mn-superoxide dismutase gene expression in Caenorhabditis elegans. Faseb J. 1999;13:1385–1478. [PubMed] [Google Scholar]
- 45.Cherkasova V, et al. Diverse Caenorhabditis elegans genes that are upregulated in dauer larvae also show elevated transcript levels in long-lived, aged, or starved adults. J Mol Biol. 2000;300:433–481. doi: 10.1006/jmbi.2000.3880. [DOI] [PubMed] [Google Scholar]
- 46.Yu H, Larsen P. DAF-16-dependent and independent expression targets of DAF-2 insulin receptor-like pathway in Caenorhabditis elegans include FKBPs. J Mol Biol. 2001;314:1017–1045. doi: 10.1006/jmbi.2000.5210. [DOI] [PubMed] [Google Scholar]
- 47.Halaschek-Wiener J, et al. Analysis of long-lived C. elegans daf-2 mutants using serial analysis of gene expression. Genome Res. 2005;15:603–615. doi: 10.1101/gr.3274805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Murphy C, et al. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature. 2003;424:277–360. doi: 10.1038/nature01789. [DOI] [PubMed] [Google Scholar]
- 49.McElwee J, et al. Transcriptional outputs of the Caenorhabditis elegans forkhead protein DAF-16. Aging Cell. 2003;2:111–121. doi: 10.1046/j.1474-9728.2003.00043.x. [DOI] [PubMed] [Google Scholar]
- 50.McElwee JJ, et al. Shared transcriptional signature in Caenorhabditis elegans Dauer larvae and long-lived daf-2 mutants implicates detoxification system in longevity assurance. J Biol Chem. 2004;279:44533–44543. doi: 10.1074/jbc.M406207200. [DOI] [PubMed] [Google Scholar]
- 51.Harman D. Aging: a theory based on free radical and radiation chemistry. J Gerontol. 1956;11:298–300. doi: 10.1093/geronj/11.3.298. [DOI] [PubMed] [Google Scholar]
- 52.Bokov A, et al. The role of oxidative damage and stress in aging. Mech Ageing Dev. 2004;125:811–826. doi: 10.1016/j.mad.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 53.Golden TR, et al. Oxidative stress and aging: beyond correlation. Aging Cell. 2002;1:117–123. doi: 10.1046/j.1474-9728.2002.00015.x. [DOI] [PubMed] [Google Scholar]
- 54.Perez-Campo R, et al. The rate of free radical production as a determinant of the rate of aging: evidence from the comparative approach. J Comp Physiol B. 1998;168:149–158. doi: 10.1007/s003600050131. [DOI] [PubMed] [Google Scholar]
- 55.Robert KA, et al. Testing the ‘free radical theory of aging’ hypothesis: physiological differences in long-lived and short-lived colubrid snakes. Aging Cell. 2007;6:395–404. doi: 10.1111/j.1474-9726.2007.00287.x. [DOI] [PubMed] [Google Scholar]
- 56.Salmon AB, et al. The long lifespan of two bat species is correlated with resistance to protein oxidation and enhanced protein homeostasis. Faseb J. 2009;23:2317–2326. doi: 10.1096/fj.08-122523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ungvari Z, et al. Extreme longevity is associated with increased resistance to oxidative stress in Arctica islandica, the longest-living non-colonial animal. J Gerontol A Biol Sci Med Sci. 2011;66:741–750. doi: 10.1093/gerona/glr044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brown JC, et al. Examining the mechanisms responsible for lower ROS release rates in liver mitochondria from the long-lived house sparrow (Passer domesticus) and big brown bat (Eptesicus fuscus) compared to the short-lived mouse (Mus musculus) Mech Ageing Dev. 2009;130:467–476. doi: 10.1016/j.mad.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 59.Brunet-Rossinni AK. Reduced free-radical production and extreme longevity in the little brown bat (Myotis lucifugus) versus two non-flying mammals. Mech Ageing Dev. 2004;125:11–20. doi: 10.1016/j.mad.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 60.Ku HH, et al. Relationship between mitochondrial superoxide and hydrogen peroxide production and longevity of mammalian species. Free Radic Biol Med. 1993;15:621–627. doi: 10.1016/0891-5849(93)90165-q. [DOI] [PubMed] [Google Scholar]
- 61.Delaney JR, et al. Stress profiling of longevity mutants identifies Afg3 as a mitochondrial determinant of cytoplasmic mRNA translation and aging. Aging Cell. 2013;12:156–166. doi: 10.1111/acel.12032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Johnson TE, et al. Relationship between increased longevity and stress resistance as assessed through gerontogene mutations in Caenorhabditis elegans. Exp Gerontol. 2001;36:1609–1617. doi: 10.1016/s0531-5565(01)00144-9. [DOI] [PubMed] [Google Scholar]
- 63.Lee SS, et al. A systematic RNAi screen identifies a critical role for mitochondria in C. elegans longevity. Nat Genet. 2003;33:40–48. doi: 10.1038/ng1056. [DOI] [PubMed] [Google Scholar]
- 64.Kim Y, Sun H. Functional genomic approach to identify novel genes involved in the regulation of oxidative stress resistance and animal lifespan. Aging Cell. 2007;6:489–992. doi: 10.1111/j.1474-9726.2007.00302.x. [DOI] [PubMed] [Google Scholar]
- 65.de Castro E, et al. Isolation of long-lived mutants in Caenorhabditis elegans using selection for resistance to juglone. Free Radic Biol Med. 2004;37:139–184. doi: 10.1016/j.freeradbiomed.2004.04.021. [DOI] [PubMed] [Google Scholar]
- 66.Perez VI, et al. Is the oxidative stress theory of aging dead? Biochim Biophys Acta. 2009;1790:1005–1014. doi: 10.1016/j.bbagen.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Knight JA. The biochemistry of aging. Adv Clin Chem. 2000;35:1–62. doi: 10.1016/s0065-2423(01)35014-x. [DOI] [PubMed] [Google Scholar]
- 68.Larsen P. Aging and resistance to oxidative damage in Caenorhabditis elegans. Proc Natl Acad Sci USA. 1993;90:8905–8914. doi: 10.1073/pnas.90.19.8905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Loew O. A New Enzyme of General Occurrence in Organismis. Science. 1900;11:701–702. doi: 10.1126/science.11.279.701. [DOI] [PubMed] [Google Scholar]
- 70.McCord J, Fridovich I. Superoxide dismutase. An enzymic function for erythrocuprein (hemocuprein) J Biol Chem. 1969;244:6049–6104. [PubMed] [Google Scholar]
- 71.Phillips JP, et al. Null mutation of copper/zinc superoxide dismutase in Drosophila confers hypersensitivity to paraquat and reduced longevity. Proc Natl Acad Sci USA. 1989;86:2761–2765. doi: 10.1073/pnas.86.8.2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tawe WN, et al. Identification of stress-responsive genes in Caenorhabditis elegans using RT-PCR differential display. Nucleic Acids Res. 1998;26:1621–1627. doi: 10.1093/nar/26.7.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tolmasoff JM, et al. Superoxide dismutase: correlation with life-span and specific metabolic rate in primate species. Proc Natl Acad Sci USA. 1980;77:2777–2781. doi: 10.1073/pnas.77.5.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vanfleteren JR. Oxidative stress and ageing in Caenorhabditis elegans. Biochem J. 1993;292 (Pt 2):605–608. doi: 10.1042/bj2920605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.An JH, Blackwell TK. SKN-1 links C. elegans mesendodermal specification to a conserved oxidative stress response. Genes Dev. 2003;17:1882–1893. doi: 10.1101/gad.1107803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Doonan R, et al. Against the oxidative damage theory of aging: superoxide dismutases protect against oxidative stress but have little or no effect on life span in Caenorhabditis elegans. Genes Dev. 2008;22:3236–3241. doi: 10.1101/gad.504808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Van Raamsdonk JM, Hekimi S. Deletion of the mitochondrial superoxide dismutase sod-2 extends lifespan in Caenorhabditis elegans. PLoS Genet. 2009;5:e1000361. doi: 10.1371/journal.pgen.1000361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schriner SE, Linford NJ. Extension of mouse lifespan by overexpression of catalase. Age (Dordr) 2006;28:209–218. doi: 10.1007/s11357-006-9010-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schriner SE, et al. Extension of murine life span by overexpression of catalase targeted to mitochondria. Science. 2005;308:1909–1911. doi: 10.1126/science.1106653. [DOI] [PubMed] [Google Scholar]
- 80.Keaney M, et al. Superoxide dismutase mimetics elevate superoxide dismutase activity in vivo but do not retard aging in the nematode Caenorhabditis elegans. Free Radic Biol Med. 2004;37:239–250. doi: 10.1016/j.freeradbiomed.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 81.Melov S, et al. Extension of life-span with superoxide dismutase/catalase mimetics. Science. 2000;289:1567–1569. doi: 10.1126/science.289.5484.1567. [DOI] [PubMed] [Google Scholar]
- 82.Pun PB, et al. Ageing in nematodes: do antioxidants extend lifespan in Caenorhabditis elegans? Biogerontology. 2010;11:17–30. doi: 10.1007/s10522-009-9223-5. [DOI] [PubMed] [Google Scholar]
- 83.Shibamura A, et al. A method for oral administration of hydrophilic substances to Caenorhabditis elegans: Effects of oral supplementation with antioxidants on the nematode lifespan. Mech Ageing Dev. 2009;130:652–655. doi: 10.1016/j.mad.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 84.Snell TW, et al. Antioxidants can extend lifespan of Brachionus manjavacas (Rotifera), but only in a few combinations. Biogerontology. 2012;13:261–275. doi: 10.1007/s10522-012-9371-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wilson M, et al. Blueberry polyphenols increase lifespan and thermotolerance in Caenorhabditis elegans. Aging Cell. 2006;5:59–127. doi: 10.1111/j.1474-9726.2006.00192.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fargnoli J, et al. Decreased expression of heat shock protein 70 mRNA and protein after heat treatment in cells of aged rats. Proc Natl Acad Sci USA. 1990;87:846–850. doi: 10.1073/pnas.87.2.846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Udelsman R, et al. Vascular heat shock protein expression in response to stress. Endocrine and autonomic regulation of this age-dependent response. J Clin Invest. 1993;91:465–473. doi: 10.1172/JCI116224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lithgow GJ, et al. Thermotolerance and extended life-span conferred by single-gene mutations and induced by thermal stress. Proc Natl Acad Sci USA. 1995;92:7540–7544. doi: 10.1073/pnas.92.16.7540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rea S, et al. A stress-sensitive reporter predicts longevity in isogenic populations of Caenorhabditis elegans. Nat Genet. 2005;37:894–902. doi: 10.1038/ng1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hsu AL, et al. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science. 2003;300:1142–1147. doi: 10.1126/science.1083701. [DOI] [PubMed] [Google Scholar]
- 91.Calfon M, et al. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- 92.Henis-Korenblit S, et al. Insulin/IGF-1 signaling mutants reprogram ER stress response regulators to promote longevity. Proc Natl Acad Sci USA. 2010;107:9730–9735. doi: 10.1073/pnas.1002575107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kurz CL, Tan MW. Regulation of aging and innate immunity in C. elegans. Aging Cell. 2004;3:185–193. doi: 10.1111/j.1474-9728.2004.00108.x. [DOI] [PubMed] [Google Scholar]
- 94.Laws TR, et al. Age influences resistance of Caenorhabditis elegans to killing by pathogenic bacteria. FEMS Microbiol Lett. 2004;234:281–287. doi: 10.1016/j.femsle.2004.03.034. [DOI] [PubMed] [Google Scholar]
- 95.Chavez V, et al. Oxidative stress enzymes are required for DAF-16-mediated immunity due to generation of reactive oxygen species by Caenorhabditis elegans. Genetics. 2007;176:1567–1577. doi: 10.1534/genetics.107.072587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Alper S, et al. Specificity and complexity of the Caenorhabditis elegans innate immune response. Mol Cell Biol. 2007;27:5544–5553. doi: 10.1128/MCB.02070-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Couillault C, et al. TLR-independent control of innate immunity in Caenorhabditis elegans by the TIR domain adaptor protein TIR-1, an ortholog of human SARM. Nat Immunol. 2004;5:488–494. doi: 10.1038/ni1060. [DOI] [PubMed] [Google Scholar]
- 98.Ewbank JJ, Zugasti O. C. elegans: model host and tool for antimicrobial drug discovery. Dis Model Mech. 2011;4:300–304. doi: 10.1242/dmm.006684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Leipe DD, et al. Did DNA replication evolve twice independently? Nucleic Acids Res. 1999;27:3389–3401. doi: 10.1093/nar/27.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mallo G, et al. Inducible antibacterial defense system in C. elegans. Curr Biol. 2002;12:1209–1223. doi: 10.1016/s0960-9822(02)00928-4. [DOI] [PubMed] [Google Scholar]
- 101.O’Rourke D, et al. Genomic clusters, putative pathogen recognition molecules, and antimicrobial genes are induced by infection of C. elegans with M. nematophilum. Genome Res. 2006;16:1005–1016. doi: 10.1101/gr.50823006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mohri-Shiomi A, Garsin D. Insulin signaling and the heat shock response modulate protein homeostasis in the Caenorhabditis elegans intestine during infection. J Biol Chem. 2008;283:194–395. doi: 10.1074/jbc.M707956200. [DOI] [PubMed] [Google Scholar]
- 103.Singh V, Aballay A. Heat-shock transcription factor (HSF)-1 pathway required for Caenorhabditis elegans immunity. Proc Natl Acad Sci USA. 2006;103:13092–13099. doi: 10.1073/pnas.0604050103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Beaman L, Beaman BL. The role of oxygen and its derivatives in microbial pathogenesis and host defense. Annu Rev Microbiol. 1984;38:27–48. doi: 10.1146/annurev.mi.38.100184.000331. [DOI] [PubMed] [Google Scholar]
- 105.Ha EM, et al. A direct role for dual oxidase in Drosophila gut immunity. Science. 2005;310:847–850. doi: 10.1126/science.1117311. [DOI] [PubMed] [Google Scholar]
- 106.Ha EM, et al. An antioxidant system required for host protection against gut infection in Drosophila. Dev Cell. 2005;8:125–132. doi: 10.1016/j.devcel.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 107.Rada B, Leto TL. Oxidative innate immune defenses by Nox/Duox family NADPH oxidases. Contrib Microbiol. 2008;15:164–187. doi: 10.1159/000136357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bolm M, et al. Hydrogen peroxide-mediated killing of Caenorhabditis elegans: a common feature of different streptococcal species. Infect Immun. 2004;72:1192–1194. doi: 10.1128/IAI.72.2.1192-1194.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Jansen WT, et al. Hydrogen peroxide-mediated killing of Caenorhabditis elegans by Streptococcus pyogenes. Infect Immun. 2002;70:5202–5207. doi: 10.1128/IAI.70.9.5202-5207.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Moy TI, et al. Cytotoxicity of hydrogen peroxide produced by Enterococcus faecium. Infect Immun. 2004;72:4512–4520. doi: 10.1128/IAI.72.8.4512-4520.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lindblom TH, Dodd AK. Xenobiotic detoxification in the nematode Caenorhabditis elegans. J Exp Zool A Comp Exp Biol. 2006;305:720–730. doi: 10.1002/jez.a.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Steinbaugh MJ, et al. Activation of genes involved in xenobiotic metabolism is a shared signature of mouse models with extended lifespan. American journal of physiology Endocrinology and metabolism. 2012;303:E488–495. doi: 10.1152/ajpendo.00110.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gems D, McElwee JJ. Broad spectrum detoxification: the major longevity assurance process regulated by insulin/IGF-1 signaling? Mech Ageing Dev. 2005;126:381–387. doi: 10.1016/j.mad.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 114.Sykiotis GP, Bohmann D. Stress-activated cap‘n’collar transcription factors in aging and human disease. Sci Signal. 2010;3:re3. doi: 10.1126/scisignal.3112re3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sykiotis GP, et al. The role of the antioxidant and longevity-promoting Nrf2 pathway in metabolic regulation. Curr Opin Clin Nutr Metab Care. 2011;14:41–48. doi: 10.1097/MCO.0b013e32834136f2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Tullet JM, et al. Direct inhibition of the longevity-promoting factor SKN-1 by insulin-like signaling in C. elegans. Cell. 2008;132:1025–1038. doi: 10.1016/j.cell.2008.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Walker AK, et al. A conserved transcription motif suggesting functional parallels between Caenorhabditis elegans SKN-1 and Cap’n’Collar-related basic leucine zipper proteins. J Biol Chem. 2000;275:22166–22171. doi: 10.1074/jbc.M001746200. [DOI] [PubMed] [Google Scholar]
- 118.Oliveira RP, et al. Condition-adapted stress and longevity gene regulation by Caenorhabditis elegans SKN-1/Nrf. Aging Cell. 2009;8:524–541. doi: 10.1111/j.1474-9726.2009.00501.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Park SK, et al. Oxidative stress and longevity in Caenorhabditis elegans as mediated by SKN-1. Aging Cell. 2009;8:258–269. doi: 10.1111/j.1474-9726.2009.00473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Toth ML, et al. Longevity pathways converge on autophagy genes to regulate life span in Caenorhabditis elegans. Autophagy. 2008;4:330–338. doi: 10.4161/auto.5618. [DOI] [PubMed] [Google Scholar]
- 121.Hansen M, et al. A role for autophagy in the extension of lifespan by dietary restriction in C. elegans. PLoS Genet. 2008;4:e24. doi: 10.1371/journal.pgen.0040024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hamilton B, et al. A systematic RNAi screen for longevity genes in C. elegans. Genes Dev. 2005;19:1544–1599. doi: 10.1101/gad.1308205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hansen M, et al. New genes tied to endocrine, metabolic, and dietary regulation of lifespan from a Caenorhabditis elegans genomic RNAi screen. PLoS Genet. 2005;1:119–147. doi: 10.1371/journal.pgen.0010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Curran S, Ruvkun G. Lifespan regulation by evolutionarily conserved genes essential for viability. PLoS Genet. 2007;3:e56. doi: 10.1371/journal.pgen.0030056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Jazwinski SM. The retrograde response links metabolism with stress responses, chromatin-dependent gene activation, and genome stability in yeast aging. Gene. 2005;354:22–27. doi: 10.1016/j.gene.2005.03.040. [DOI] [PubMed] [Google Scholar]
- 126.Parikh VS, et al. The mitochondrial genotype can influence nuclear gene expression in yeast. Science. 1987;235:576–580. doi: 10.1126/science.3027892. [DOI] [PubMed] [Google Scholar]
- 127.Yoneda T, et al. Compartment-specific perturbation of protein handling activates genes encoding mitochondrial chaperones. J Cell Sci. 2004;117:4055–4066. doi: 10.1242/jcs.01275. [DOI] [PubMed] [Google Scholar]
- 128.Benedetti C, et al. Ubiquitin-like protein 5 positively regulates chaperone gene expression in the mitochondrial unfolded protein response. Genetics. 2006;174:229–239. doi: 10.1534/genetics.106.061580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Haynes CM, et al. ClpP mediates activation of a mitochondrial unfolded protein response in C. elegans. Dev Cell. 2007;13:467–480. doi: 10.1016/j.devcel.2007.07.016. [DOI] [PubMed] [Google Scholar]
- 130.Haynes CM, Ron D. The mitochondrial UPR - protecting organelle protein homeostasis. J Cell Sci. 2010;123:3849–3855. doi: 10.1242/jcs.075119. [DOI] [PubMed] [Google Scholar]
- 131.Haynes CM, et al. The matrix peptide exporter HAF-1 signals a mitochondrial UPR by activating the transcription factor ZC376.7 in C. elegans. Mol Cell. 2010;37:529–540. doi: 10.1016/j.molcel.2010.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Cristina D, et al. A regulated response to impaired respiration slows behavioral rates and increases lifespan in Caenorhabditis elegans. PLoS Genet. 2009;5:e1000450. doi: 10.1371/journal.pgen.1000450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zubovych IO, et al. Mitochondrial dysfunction confers resistance to multiple drugs in Caenorhabditis elegans. Mol Biol Cell. 2010;21:956–969. doi: 10.1091/mbc.E09-08-0673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Pan KZ, et al. Inhibition of mRNA translation extends lifespan in Caenorhabditis elegans. Aging Cell. 2007;6:111–119. doi: 10.1111/j.1474-9726.2006.00266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Smith ED, et al. Quantitative evidence for conserved longevity pathways between divergent eukaryotic species. Genome Res. 2008;18:564–570. doi: 10.1101/gr.074724.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hansen M, et al. Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans. Aging Cell. 2007;6:95–110. doi: 10.1111/j.1474-9726.2006.00267.x. [DOI] [PubMed] [Google Scholar]
- 137.Li X, et al. Specific SKN-1/Nrf stress responses to perturbations in translation elongation and proteasome activity. PLoS Genet. 2011;7:e1002119. doi: 10.1371/journal.pgen.1002119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ching TT, et al. drr-2 encodes an eIF4H that acts downstream of TOR in diet-restriction-induced longevity of C. elegans. Aging Cell. 2010;9:545–557. doi: 10.1111/j.1474-9726.2010.00580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kahn NW, et al. Proteasomal dysfunction activates the transcription factor SKN-1 and produces a selective oxidative-stress response in Caenorhabditis elegans. Biochem J. 2008;409:205–213. doi: 10.1042/BJ20070521. [DOI] [PubMed] [Google Scholar]
- 140.Wang J, et al. RNAi screening implicates a SKN-1-dependent transcriptional response in stress resistance and longevity deriving from translation inhibition. PLoS Genet. 2010;6:e1001048. doi: 10.1371/journal.pgen.1001048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Rogers AN, et al. Life span extension via eIF4G inhibition is mediated by posttranscriptional remodeling of stress response gene expression in C. elegans. Cell Metab. 2011;14:55–66. doi: 10.1016/j.cmet.2011.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Johnson TE, et al. Longevity genes in the nematode Caenorhabditis elegans also mediate increased resistance to stress and prevent disease. J Inherit Metab Dis. 2002;25:197–206. doi: 10.1023/a:1015677828407. [DOI] [PubMed] [Google Scholar]
- 143.Durieux J, et al. The cell-non-autonomous nature of electron transport chain-mediated longevity. Cell. 2011;144:79–91. doi: 10.1016/j.cell.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Munoz M, Riddle D. Positive selection of Caenorhabditis elegans mutants with increased stress resistance and longevity. Genetics. 2003;163:171–251. doi: 10.1093/genetics/163.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Fabrizio P, et al. Regulation of longevity and stress resistance by Sch9 in yeast. Science. 2001;292:288–378. doi: 10.1126/science.1059497. [DOI] [PubMed] [Google Scholar]
- 146.Vettraino J, et al. Direct selection for paraquat resistance in Drosophila results in a different extended longevity phenotype. J Gerontol A Biol Sci Med Sci. 2001;56:B415–425. doi: 10.1093/gerona/56.10.b415. [DOI] [PubMed] [Google Scholar]
- 147.Calabrese EJ, Blain RB. The hormesis database: the occurrence of hormetic dose responses in the toxicological literature. Regul Toxicol Pharmacol. 2011;61:73–81. doi: 10.1016/j.yrtph.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 148.Kaiser J. Hormesis. Sipping from a poisoned chalice Science. 2003;302:376–385. doi: 10.1126/science.302.5644.376. [DOI] [PubMed] [Google Scholar]
- 149.Cypser JR, et al. Hormesis and aging in Caenorhabditis elegans. Exp Gerontol. 2006;41:935–939. doi: 10.1016/j.exger.2006.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Cypser JR, Johnson TE. Multiple stressors in Caenorhabditis elegans induce stress hormesis and extended longevity. J Gerontol A Biol Sci Med Sci. 2002;57:B109–114. doi: 10.1093/gerona/57.3.b109. [DOI] [PubMed] [Google Scholar]
- 151.Heidler T, et al. Caenorhabditis elegans lifespan extension caused by treatment with an orally active ROS-generator is dependent on DAF-16 and SIR-2.1. Biogerontology. 2010;11:183–195. doi: 10.1007/s10522-009-9239-x. [DOI] [PubMed] [Google Scholar]
- 152.Przybysz AJ, et al. Increased age reduces DAF-16 and SKN-1 signaling and the hormetic response of Caenorhabditis elegans to the xenobiotic juglone. Mech Ageing Dev. 2009;130:357–369. doi: 10.1016/j.mad.2009.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Yanase S, et al. Oxidative stress pretreatment increases the X-radiation resistance of the nematode Caenorhabditis elegans. Mutat Res. 1999;426:31–39. doi: 10.1016/s0027-5107(99)00079-2. [DOI] [PubMed] [Google Scholar]
- 154.Wu D, et al. Multiple mild heat-shocks decrease the Gompertz component of mortality in Caenorhabditis elegans. Exp Gerontol. 2009;44:607–612. doi: 10.1016/j.exger.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Rea S, et al. Relationship between mitochondrial electron transport chain dysfunction, development, and life extension in Caenorhabditis elegans. PLoS Biol. 2007:5. doi: 10.1371/journal.pbio.0050259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Dillin A, et al. Rates of behavior and aging specified by mitochondrial function during development. Science. 2002;298:2398–2401. doi: 10.1126/science.1077780. [DOI] [PubMed] [Google Scholar]
- 157.Lee SJ, et al. Inhibition of respiration extends C. elegans life span via reactive oxygen species that increase HIF-1 activity. Curr Biol. 2010;20:2131–2137. doi: 10.1016/j.cub.2010.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Melo JA, Ruvkun G. Inactivation of Conserved C. elegans Genes Engages Pathogen- and Xenobiotic-Associated Defenses. Cell. 2012;149:452–466. doi: 10.1016/j.cell.2012.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.McEwan DL, et al. Host Translational Inhibition by Pseudomonas aeruginosa Exotoxin A Triggers an Immune Response in Caenorhabditis elegans. Cell Host Microbe. 2012;11:364–374. doi: 10.1016/j.chom.2012.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Shore D, et al. Induction of cytoprotective pathways is central to the extension of lifespan conferred by multiple longevity pathways. PLoS Genet. 2012;8:e1002792. doi: 10.1371/journal.pgen.1002792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Vance RE, et al. Patterns of pathogenesis: discrimination of pathogenic and nonpathogenic microbes by the innate immune system. Cell Host Microbe. 2009;6:10–21. doi: 10.1016/j.chom.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Kakizaki S, et al. New insights on the xenobiotic-sensing nuclear receptors in liver diseases--CAR and PXR. Curr Drug Metab. 2008;9:614–621. doi: 10.2174/138920008785821666. [DOI] [PubMed] [Google Scholar]
- 163.Li X, et al. Molecular mechanisms of metabolic resistance to synthetic and natural xenobiotics. Annu Rev Entomol. 2007;52:231–253. doi: 10.1146/annurev.ento.51.110104.151104. [DOI] [PubMed] [Google Scholar]
- 164.Thomas JH. Rapid birth-death evolution specific to xenobiotic cytochrome P450 genes in vertebrates. PLoS Genet. 2007;3:e67. doi: 10.1371/journal.pgen.0030067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Zopf Y, et al. Gender-based differences in drug prescription: relation to adverse drug reactions. Pharmacology. 2009;84:333–339. doi: 10.1159/000248311. [DOI] [PubMed] [Google Scholar]
- 166.Bjedov I, Partridge L. A longer and healthier life with TOR down-regulation: genetics and drugs. Biochem Soc Trans. 2011;39:460–925. doi: 10.1042/BST0390460. [DOI] [PubMed] [Google Scholar]
- 167.Harrison D, et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460:392–397. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.McCormick M, et al. TOR and ageing: a complex pathway for a complex process. Philos Trans R Soc Lond B Biol Sci. 2011;366:17–44. doi: 10.1098/rstb.2010.0198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Blagosklonny M. Increasing healthy lifespan by suppressing aging in our lifetime: preliminary proposal. Cell cycle. 2010;9:4788–4882. doi: 10.4161/cc.9.24.14360. [DOI] [PubMed] [Google Scholar]
- 170.Sharp Z, Richardson A. Aging and cancer: can mTOR inhibitors kill two birds with one drug? Targeted oncology. 2011;6:41–92. doi: 10.1007/s11523-011-0168-7. [DOI] [PubMed] [Google Scholar]