Abstract
We have generated a panel of transgenic mice expressing HLA-A*01:03, -A*24:02, -B*08:01, -B*27:05, -B*35:01, -B*44:02, or -C*07:01 as chimeric monochain molecules (i.e., appropriate HLA α1α2 H chain domains fused with a mouse α3 domain and covalently linked to human β2-microglobulin). Whereas surface expression of several transgenes was markedly reduced in recipient mice that coexpressed endogenous H-2 class I molecules, substantial surface expression of all human transgenes was observed in mice lacking H-2 class I molecules. In these HLA monochain transgenic/H-2 class I null mice, we observed a quantitative and qualitative restoration of the peripheral CD8+ T cell repertoire, which exhibited a TCR diversity comparable with C57BL/6 WT mice. Potent epitope-specific, HLA-restricted, IFN-γ–producing CD8+ T cell responses were generated against known reference T cell epitopes after either peptide or DNA immunization. HLA-wise, these new transgenic strains encompass a large proportion of individuals from all major human races and ethnicities. In combination with the previously created HLA-A*02:01 and -B*07:02 transgenic mice, the novel HLA transgenic mice described in this report should be a versatile preclinical animal model that will speed up the identification and optimization of HLA-restricted CD8+ T cell epitopes of potential interest in various autoimmune human diseases and in preclinical evaluation of T cell–based vaccines.
Human leukocyte Ag class I transgenic mice (as reviewed in Ref. 1) are useful preclinical tools for the identification of T cell epitopes of human relevance in infectious, tumor, and autoimmune pathological situations (2–5). As a model of human immune responses, HLA class I transgenic mice enable, if needed, a careful optimization of the immunogenicity of human CD8+ T cell epitopes, and a controlled and reproducible comparative preclinical evaluation of epitope-based vaccine candidates (6, 7). However, the first generation of HLA class I transgenic mice with introduction of unmodified (H chain α3 domain of human origin) HLA class I gene with and without association to the human β2-microglobulin (β2-m) gene did not provide, except possibly for few alleles (8–10), the expected versatile animal models for the study of HLA class I–restricted responses despite significant cell-surface expression of the HLA molecules (11–13). The low affinity of mouse CD8 molecule for most allelic forms of HLA class I molecules and the identification in the α3 domain of MHC class I molecules of the major site of interaction with CD8 molecules (14–16) led to the development of HLA class I transgenic mice expressing chimeric molecules in which the mouse α3 domain was substituted for the human one (17, 18). These mice made much better use of the transgenic chimeric molecules, and the α3 substitution strategy has been subsequently applied by many laboratories (19–22). Alternatively, HLA class I transgenic mice expressing additionally human CD8 α/β accessory molecules were created with comparable improvement of their HLA-restricted responses (23). Because in mice coexpressing H-2 and HLA class I molecules, the H-2–restricted cytolytic responses hamper the development of the HLA-restricted ones (24), further optimization of the transgenic molecule usage has been achieved after destruction by homologous recombination of H-2 class Ia genes (25) and HLA class I transgenesis (26, 27). As an example, expression of α3 chimeric HLA-B*07:02 molecule in an H-2 Kb, Db double-knockout (KO) environment has produced an excellent mouse model for the study of HLA-B*07:02–restricted responses (27). However, these mice still express a full set of H-2 class Ib molecules that contribute to the thymic positive selection of a H-2–restricted CD8+ T cell repertoire (28), and many of these class Ib molecules are endowed with the capacity to present H-2–restricted epitopes and stimulate “mouse” CD8+ T cell responses (29). Therefore, the observed CD8+ cytolytic responses in such mice are not necessarily HLA class I restricted. To further constrain the mouse CD8+ T cells to make better usage of the HLA class I transgenic molecules and generate more functional HLA-restricted CD8+ T cell responses, we created H-2 Db and mouse β2-m double-KO mice expressing an α3 chimeric HLA-A*02:01 H chain covalently linked to human β2-m by a peptidic arm (HHD II mice) (30). The destruction of the H-2 Db gene, in addition to that of the mouse β2-m one, was motivated by the fact that, in the absence of β2-m, a substantial fraction of functionally conformed H-2 Db heavy chains reaches the cell surface (31–34). Despite the fact that immunocompetent β2-m–free H-2 Kb heavy chains are also (although to a lesser extent than H-2 Db heavy chains) expressed on cell surface (35–37), in our experience, the cytolytic responses of these mice have consistently been found restricted by the HLA-A*02:01 monochain, and these mice have since been successfully used by many laboratories (38, 39).
Although the HLA-A*02 Ag is the most frequent in the Caucasoid and Southeast Asian ethnic groups, it is represented in only ~50% of individuals and in the black population only 30% of individuals are HLA-A*02+. Therefore, for a better coverage of all ethnic groups, to further reduce the possibility of generating H-2 class I molecule–restricted CD8+ T cell repertoires in the HLA transgenic mouse model, and to further improve the usage the transgenic HLA monochains, in this study, we developed seven novel HLA class I monochain transgenic mouse strains in an H-2 Kb, Db, and mouse β2-m triple KO (H-2 class I null) context by selecting HLA molecules of cumulative high frequencies in all human populations, with several of these HLA molecules being also associated with autoimmune diseases, in particular, type 1 diabetes.
Phenotypic and functional analyses of the CD8+ T cell repertoires generated in each of the seven new HLA transgenic strains demonstrated that they efficiently use the transgenic HLA monochains at both the thymic and peripheral effector levels.
Materials and Methods
HLA class I monochain constructs
The α1 α2 domain coding sequences were extracted from either cDNAs (HLA-B*08:01, -B*35:01) or genes in a genomic configuration (HLA-A*01:03, -A24*:02, -B*27:05, -B*44:01, -C*07:01) and substituted for the α1 α2 domain coding sequences of a HLA-A*02:01 monochain plasmid construct (30). All monochain constructs have the same overall organization as illustrated in Supplemental Fig. 1: the HLA-A*02:01 promotor and the coding sequence of a chimeric molecule composed of the HLA-A*02:01 leader segment, the human β2-m in its mature form, a 15-aa (GGGGS) × 3 linker peptide, the α1 α2 HLA class I domains, and the H-2 Db α3 to cytoplasmic domains. Except for HLA-C*07:01 with a Cysteine1 > Glycine substitution, the amino acid sequences of the α1 α2 domains of all other monochains were unaltered. The linkage of the coding exons of the α1 α2 domains with H-2 Db exons 4 (α3 domain) to 8 was achieved for the monochains of the HLA-B series by fusion of a natural BssHII restriction site at the 3′ end of exon 3 with an artificial BssHII site introduced by site-directed mutagenesis at the 5′ end of H-2 Db exon 4. For other monochains, either a natural (HLA-A*01:03, -C*07:01) or artificial (HLA-A*24:02) intronic BglII site was linked to a natural BamHI site of H-2 Db intron 3. The capacity of the seven monochains to exhibit significant cell-surface expression was verified transfecting β2-m–deficient mouse EL4 S3− Rob cells.
Mice
C57BL/6 × SJL fertilized eggs were microinjected with the linearized (SalI, NotI) monochain constructs and reimplanted in pseudopregnant female mice. Founder mice were PCR-identified using a common 3′ reverse H-2 Db primer, 5′-GCATATGTACATGAATGTATTCACTTCAT-3′, and either an HLA-A*01:03/-A*24:02 common forward primer, 5′-TGCGC-TCTTGGACCGCGGCGGACATG-3′ (amplicon of 1535 bp, A*01:03 or 1539 bp, A*24:02), an HLA-C*07:01 forward primer, 5′-TGCGCTCCT-GGACCGCCGCGGACACC-3′ (amplicon of 1574 bp), an HLA-B*08:01 forward primer, 5′-TGCGCTCCTGGACCGCGGCGGACACC-3′ (amplicon of 1031 bp), an HLA-B*27:05 forward primer, 5′-CTGAGCTCCTGGAC-CGCCGCGGACAC-3′ (amplicon of 1035 bp), or an HLA-B*35:01/-B*44:02 common forward primer, 5′-TGAGCTCCTGGACCGCGGCGGACACC-3′ (amplicon of 1031 bp B*35:01, 1034 bp B*44:02).
After five backcrosses with H-2 Kb, Db, β2-m triple-KO C57BL/6 mice (40), heterozygous monochain transgenic, homozygous H-2 Kb, Db, β2-m triple-KO mice were intercrossed and monochain transgenic homozygous/ H-2 class I KO mice identified in a progeny test, crossbreeding them with OF1 mice (Charles River, L’Arbresle, France). These homozygous back-crossed mice were used in all experiments. HHD and H-2 Kb, Db, β2-m triple-KO control mice have been previously described (30, 40). Mice were maintained in our animal facilities at the Institut Pasteur. All protocols were reviewed by the Institut Pasteur competent authority for compliance with the French and European Regulations on Animal Welfare and with Public Health Service recommendations.
Flow cytometry analyses
Ficoll-purified splenocytes from experimental and control mice were labeled with either anti-human β2-m (clone Tü 99; BD Pharmingen, San Diego, CA), anti-mouse CD3ε (clone 145-2C11; BD Pharmingen), anti-mouse CD4 (clone RM4-5; BD Pharmingen), anti-mouse CD8α (clone 53-6-7; BD Pharmingen), anti-mouse CD11b (clone M1/70; BD Pharmingen), anti-mouse CD11c (clone HL3; BD Pharmingen), anti-mouse B220 (clone RA3-6B2; BD Pharmingen) allophycocyanin-, PE-, PerCP-, or FITC-conjugated mAbs and analyzed with a BD LSRFortessa apparatus (BD Pharmingen). Data were analyzed using the FlowJo program (Tree Star, Ashland, OR).
Peripheral CD8+ T cell repertoire analyses
Total RNAs were extracted from microbead-purified (Macs Separation columns; Miltenyi, Gladbach, Germany) peripheral (spleen and mesenteric lymph nodes) CD8+ or CD4+ T cells (>95% purity) from individual mice using the Gene Elute Mammalian Total RNA Miniprep kit (Sigma-Aldrich, St. Louis, MO) and then reverse transcribed. Determination of AV and BV gene usage were performed as described previously (41). In brief, real-time quantitative PCRs were performed individually for each TCR BV and AV segment family with a complete set of TCR BV (42) and TCR AV (Supplemental Table I) forward primers at a final concentration of 400 nM/l each. When necessary (DNA sequence polymorphism of AV family segment members), appropriate forward primers were mixed. PCRs were carried out on an ABI-7300 system (Applied Biosystem, Carlsbad, CA) combining reverse constant TCR BC (5′-GGTA-GCCTTTTGTTTGTTTGCAA-3′) and reverse constant TCR AC (5′-GGC-ACATTGATTTCCTGGCTATTGC-3′) primers (400 nM/l final concentration) and BC (5′-AGCCATCAAAAGCAA-3′) or AC (5′-CTTCTGCCTGTTCA-CCGA-3′) MGB-TaqMan probes at a 200 nM/l final concentration (Life Technology, Carlsbad, CA) as previously described (42). The relative usage in percentage of each BV and AV family was calculated according to the following formula:
and
where Ct(x) is the fluorescent threshold cycle number measured for each individual BV and AV family.
AV9 CDR3 sequencing was performed after RT-PCR amplification and cloning in a pCR2-TOPO vector (Invitrogen, Carlsbad, CA), using for amplification an AV9-specific forward primer (5′-ACCCAGTGGTT-CAAGGAGTGAA-3′) and the earlier mentioned TCR AC reverse primer.
Immunization protocols and analyses of CD8+ T cell responses
For peptide immunization, mice were injected s.c. at the base of the tail with 100 μg of the HLA class I–restricted peptide and 140 μg of the H-2 IAb–restricted HBV core 128–140 helper peptide (TPPAYRPPNAPIL) coemulsified in IFA (Sigma-Aldrich). Seven days later, splenocytes were tested in an IFN-γ ELISPOT assay (U-CyTech Biosciences, Utrecht, The Netherlands).
For DNA immunization, complementary oligonucleotides coding for each selected reference CD8+ T cell epitope (listed in Table III) were introduced between the more 5′ EcoRI and the more 3′ XhoI restriction sites of the pre-S2 segment of the HBV glycoprotein gene inserted in a pCMV-B10 expression vector (43). Anesthetized mice were first injected i.m. in tibialis anterior muscles with 10 μM cardiotoxin (Latoxan, Rosans, France), then 8 and 22 d later similarly injected with 50 μg per leg of purified recombinant plasmid (PureLink columns; Invitrogen). Eight days after the second DNA injection, epitope-specific CD8+ T cell responses were evaluated in an IFN-γ ELISPOT assay.
Table III.
CD8+ T cell IFN-γ responses after peptide immunization
| Peptides | HLA Restriction | Protein Source | IFN-γ Spots/106 Cellsa |
|---|---|---|---|
| VSDGGPNLY | A*01 | Influenza A basic polymerase I 591–599 (46) | 201/134/71/59 |
| EADPTGHSY | A*01 | MAGE-1 161–169 (47) | 287/212/123/27 |
| RYLKDQQLL | A*24 | HIV I gp41 583–591 (48), | 330/162/74/62/56 |
| TYQIYQEPF | A*24 | HIV I polymerase 493–501 (49) | 83/44/45/35/12 |
| DSRLAFHHM | A*24 | HIV nef 186–194 (50) | 0/0/0/0 |
| RAKFKQLL | B*08 | EBV lytic cycle Ag BZLF1 190–197 (51) | 70/63/60/53 |
| WLKIKRDYL | B*08 | Vaccinia virus, Putative A50R (52) | 524/103/97/74/64/0 |
| SRYWAIRTR | B*27 | Influenza A nucleoprotein 383–391 (53) | 567/459/110/0 |
| LRGKWQRRYR | B*27 | EBV EBNA 3C 249–258 (54) | 113/61/54/23 |
| HPNIEEVAL | B*35 | HCV NS3 protein 1359–1367 (55) | 322/282/173/138 |
| TPEGIIPTL | B*35 | Dengue NS3 protein 500–508 (56) | 459/310/253/6 |
| EENLLDFVRF | B*44 | EBV EBNA 3C 335–343 (57) | 54/28/27/20/0 |
| FEDLRVLSF | B*44 | Influenza A nucleoprotein 338–346 (58) | 167/61/30/17/8 |
| AENGWGFYF | B*44 | Brucella abortus, hypothetical glyoxalase 201–209b | 60/41/29/12/9 |
| RRRPVTRPL | C*07 | Candidatus solibacter usitatus Ellin6076b | 619/599/177/0 |
| RRARYWLTY | C*07 | Giardia intestinalisb | 643/640/133/97/92 |
| PLADLSPFA | C*07 | Protein 5T4 (59) | 0/0/0/0 |
| EGDCAPEEK | C*07 | Mage 2,3,6,12 (60) | 0/0/0/0 |
| RNGYRALMDKS | C*07 | MelanA/Mart1 (61) | 0/0/0/0 |
IFN-γ T lymphocyte responses from the seven HLA class I monochain transgenic mice were evaluated in an ELISPOT assay after in vivo priming and in vitro restimulation with peptides corresponding to published CD8+ T cell epitopes.
Four to six mice were tested individually. Data are given for each mouse as the mean of triplicate wells.
Immune Epitope Data Base (http://www.iedb.org), epitopes 1096, 109862, 93763.
Ficoll-purified, serially diluted splenocytes (106 to 105 per well) in 200 μl RPMI 1640 medium supplemented with 10% FCS, 5 × 10−5 M 2-ME, 50 IU/ml penicillin, 50 μg/ml streptomycin were stimulated overnight with appropriate peptides (10 μg/ml) in 96-well plates (Multiscreen HTS; Millipore, Billerica, MA) coated with rat anti-mouse IFN-γ mAb (10 μg/ml, clone R4-6A2; BD Pharmingen). Eighteen hours later, after cell lysis, complementary biotinylated rat anti-mouse IFN-γ mAb (1 μg/ml, clone XMG.1.2; BD Pharmingen) was added and spots revealed after subsequent incubation with extravidin-alkaline phosphatase and 5-bromo-chloro-3-indolyl-phosphate/nitro blue tetrazolium/NBT substrate (Sigma-Aldrich) according to the manufacturer’s recommendations.
Peptide–HLA-I stability
The stability of peptide–HLA-C*07:01 complexes was determined by a scintillation proximity assay (44). In brief, recombinant, biotinylated, and denatured HLA-C*07:01 HC was diluted to a final concentration of 50 nM in renaturing buffer containing 10 μM peptide and [125I]-radiolabeled β2-m in a 384-well streptavidin-coated FlashPlate (Perkin Elmer SMP103). The plate was incubated overnight at 18°C to obtain HLA-I complex folding. Peptide–HLA-I dissociation was initiated by addition of excess of unlabeled β2-m, and the plate was read continually in a Topcount NXT liquid scintillation counter at 37°C for 24 h. Peptide–HLA-I half-life was calculated by fitting dissociation data to a one-phase dissociation model.
Results
Cell-surface expression of the transgenic HLA class I monochains in a H-2 class I null and in a H-2 class I WT context
Cell-surface expression of the seven transgenic monochains was evaluated by flow cytometry analysis of Ficoll-purified splenocytes using in all cases FITC-conjugated TÜ 99 anti-human β2-m mAb. Expression of these seven transgenic monochains was compared with that of HLA-A*02:01 monochains in HHD II mice. C57BL/6 WT mice were used as negative control.
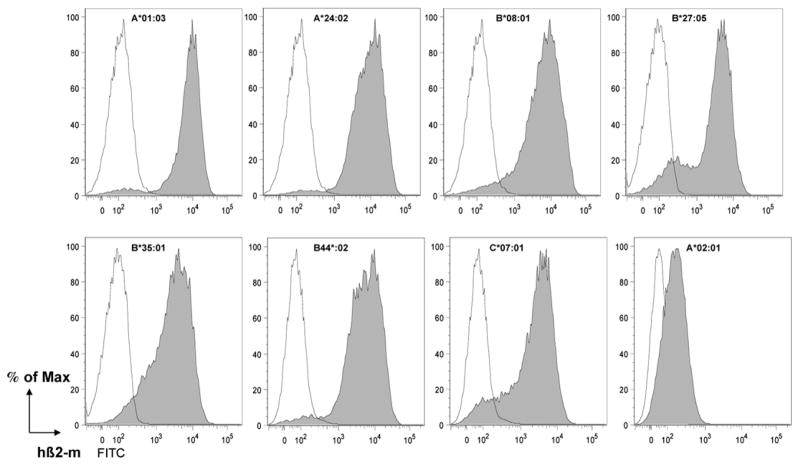
As illustrated in Fig. 1, in a H-2 Kb, Db, mouse β2-m triple-KO context, cell-surface expression was observed in all strains that in all cases was substantially higher (roughly 10-fold) than the expression of the HLA-A*02:01 monochains in HHD II mice. A bimodal profile was consistently observed with HLA-B*27:05 monochain transgenic mice suggestive of a variegation phenomenon (45).
FIGURE 1.
Cell-surface expression of the seven HLA monochains in a H-2 class I null context. Ficoll-purified splenocytes from individual HLA class I monochain transgenic, H-2 class I null (closed gray histograms), and H-2 Kb, Db, mouse β2-m triple-KO (negative control, open histograms) mice (n = 1) were labeled with anti–β2-m mAb Tü 99 and flow cytometrically analyzed. This experiment was performed more than five times.
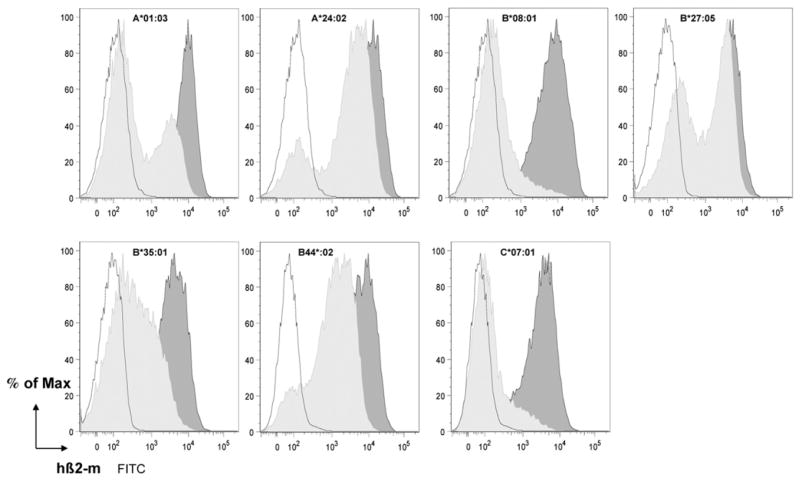
Cell-surface expression was also similarly assessed during the progeny homozygosity test in which the HLA monochain mice were intercrossed with outbred OF1 animals. A reduction of cell-surface expression of the monochains was observed in all cases when coexpressed with H-2 class I molecules in monochain × OF1 F1 hybrid mice. Whereas cell-surface expression of HLA-A*24:02 and -B*27:05 monochains was only slightly altered, that of HLA-B*44:02 was more significantly reduced, whereas expression of HLA-A*01:03, -B*08:01, -B*35:01, and -C*07:01 monochains was almost abrogated (Fig. 2). It is worth noting the bimodal shape of the labeling curves, which is particularly clear for HLA-A*01:03 × OF1 F1 mice. Double-staining experiments of HLA-A*01:03 × OF1 F1 splenocytes (Supplemental Fig. 2) showed that the reduction of the cell-surface expression of the HLA-A*01:03 monochains was less pronounced on macrophages, dendritic cells, and B lymphocytes than on T lymphocytes, in which case surface expression was almost totally lost. These data suggested that these monochains were outcompeted in F1 animals by H-2 class I molecules at some stages of their folding and peptide-loading processes in the endoplasmic reticulum (ER).
FIGURE 2.
Cell-surface expression of the seven monochains in a H-2 class I+ context. Ficoll-purified splenocytes from individual HLA class I monochain transgenic H-2 class I null (dark gray histograms), HLA class I monochain transgenic × OF1 F1 hybrid (light gray histograms), and H-2 Kb, Db, mouse β2-m triple-KO (negative control, open histograms) mice (n = 1) were labeled with anti–β2-m mAb Tü 99 and flow cytometrically analyzed. This experiment was performed thrice.
Significant HLA class I monochain-driven quantitative restoration of the peripheral CD8+ T cell repertoire
Flow cytometry analyses of Ficoll-purified splenocytes from the seven new HLA class I monochain transgenic strains using FITC-conjugated anti-mouse CD8α mAb in conjunction with allophy-cocyanin-conjugated anti-mouse CD3ε mAb were performed to evaluate the size of the CD8+ T cell compartment educated at the thymic level and maintained in the periphery by the HLA class I monochains. H-2 Kb, Db, mouse β2-m C57BL/6 triple-KO and C57BL/6 WT mice were used side by side as negative and positive controls, respectively.
The results are summarized in Table I. Substantial quantitative restoration of CD8+ T cell peripheral compartment was documented in all seven strains with, as expected, some differences between the seven strains. The lowest level (11.7%) of restoration as compared with CD8+ T cell peripheral compartment in C57BL/6 WT mice was observed in HLA-B*08:01 mice; the highest and most significant restoration (71%) was observed in the HLA-B*35:01 mice, even though, as shown in Fig. 1, the levels of cell-surface expression of the HLA-B*08:01 and HLA-B*35:01 monochains were similar between the two strains.
Table I.
Quantitative restoration of the CD8+ T cell compartment
| Alleles | CD8+ T Cells versus Total Splenocytes (%) | Restoration of the CD8+ T Cell Compartment (%) |
|---|---|---|
| HLA-A*01:03 | 4.45 | 44.5 |
| HLA-A*24:02 | 3.4 | 34 |
| HLA-B*08:01 | 1.17 | 11.7 |
| HLA-B*27:05 | 4.77 | 47.7 |
| HLA-B*35:01 | 7.1 | 71 |
| HLA-B*44:02 | 3.75 | 37.5 |
| HLA-C*07:01 | 4.16 | 41.6 |
Ficoll-purified splenocytes of HLA monochain homozygous mice were immunostained with FITC-labeled anti-mouse CD8α mAb using as a reference strain C57BL/6 mice. This experiment was performed three times.
TCR diversity of the HLA class I monochain-selected peripheral CD8+ T cell repertoire
As a first step to evaluate the TCR structural diversity of the peripheral CD8+ T cells selected by the HLA class I monochains, quantitative RT-PCRs were performed to estimate the relative peripheral representation of the 20 families of AV and 19 families of BV variable segments in purified CD8+ T cell population of 4 mice tested individually from each of the 7 monochain transgenic strains. Similar analysis was performed in the CD4+ T cell population from the same individual mice as an internal control. The AV and BV RT-PCR profiles were compared with those derived from C57BL/6 WT mice and are illustrated in Figs. 3 and 4, respectively.
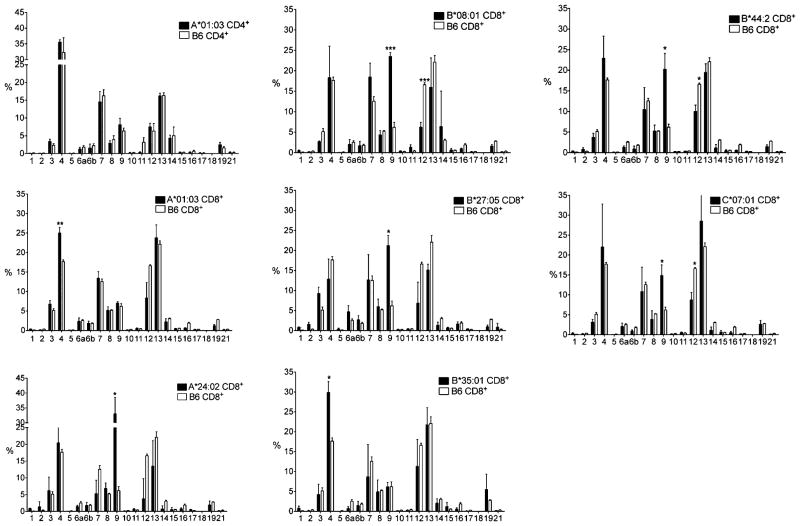
FIGURE 3.
CD8+ and CD4+ T cells TCR AV peripheral repertoire. Relative peripheral representation in percentage of the AV segment families (International Immunogenetics Information System nomenclature, http://www.imgt.org) as evaluated by quantitative RT-PCR in purified CD8+ and CD4+ T cells from HLA-A*01.03, -A*24:02, -B*08:01, -B*27:05, -B*33:01, -B*44:02, and -C*07:01 monochain transgenic and from C57BL/6 (B6) mice. Open and closed bars correspond to C57BL/6 and monochain strains, respectively. CD4+ T cell AV profiles of A*01:03 versus C57BL/6 mice are illustrated in the upper left panel, similar CD4+ AV profiles (data not shown) being obtained with the six additional monochains. The results are means of the values from at least four individually tested mice. Statistical analyses of the data were carried out with the unpaired t test with Welch’s correction, *p < 0.05, **p < 0.005, ***p < 0.0005.
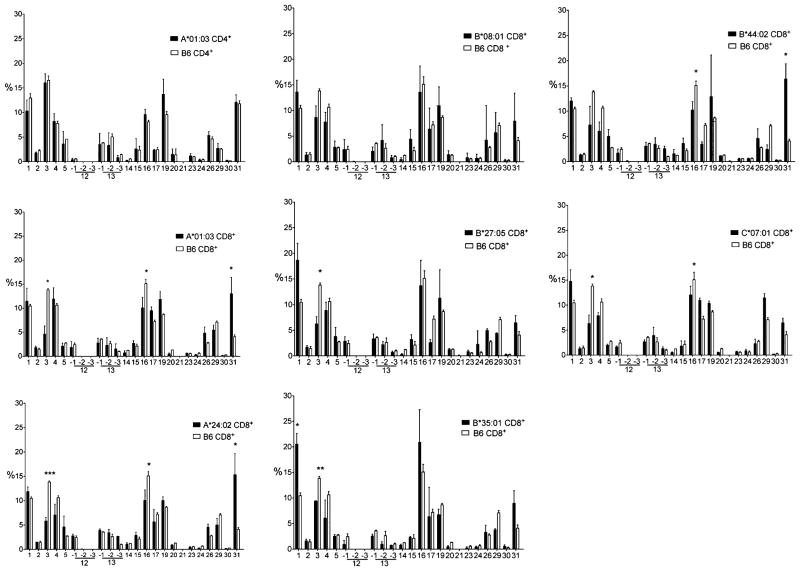
FIGURE 4.
CD8+ and CD4+ T cells TCR BV peripheral repertoire. Relative peripheral representation in percentage of the BV segment families (International Immunogenetics Information System nomenclature, http://www.imgt.org) as evaluated by quantitative RT-PCR in purified CD8+ and CD4+ T cells from HLA-A*01:03, -A*24:02, -B*08:01, -B*27:05, -B*35:01, -B*44:02, and -C*07:01 monochain transgenic and from C57BL/6 (B6) mice. Open and closed bars correspond to C57BL/6 and monochain strains, respectively. CD4+ T cell BV profiles of A*01:03 versus C57BL/6 mice are illustrated in the upper left panel, similar CD4+ BV profiles (not shown) being obtained with the six additional monochains. The results are means of the values from at least four individually tested mice. Statistical analyses of the data were carried out with the unpaired t test with Welch’s correction, *p < 0.05, **p < 0.005, ***p < 0.0005.
It should first be stressed that the AV and BV quantitative RT-PCR profiles of CD4+ T cell populations from the seven HLA monochain transgenics (as shown for one of them; Figs. 3, 4, top left profile) and from C57BL/6 WT mice were strictly superimposable, as expected, because all these mice express the same and single species of class II molecules, H-2 IAb.
The relative peripheral representation of the AVand BV segment families in CD8+ T cells from the seven monochain transgenic strains, as illustrated in Figs. 3 and 4, is globally similar to the AV and BV relative peripheral representation in CD8+ T cells from C57BL/6 WT mice. Note the predominance in all HLA class I monochain transgenic as in C57BL/6 WT mice of the AV4, 7, 8, 9, 12, and 13 family segments (which are among the largest AV segment families). Similarly, the BV1, 4, 13, 15, 16, 17, and 19 family segments are abundantly and similarly represented in CD8+ T cells from monochain transgenics and C57BL/6 mice.
However, a closer analysis of the RT-PCR profiles brought to light quantitative differences in the peripheral representation of some AV and BV family segments between the transgenic strains and the C57BL/6 WT mice, as well as within the seven monochain transgenic strains themselves. Relative to C57BL/6 WT mice, the AV9 segment family is markedly more abundant in HLA-A*24:02, -B*08:01, -B*27:05, -B*44:02, and -C*07:01 monochain strains but remains at the same level as in C57BL/6 WT mice in HLA-A*01:03 and -B*35:01 monochain mice. In these two latter strains, the AV4 family segment is moderately overrepresented. In contrast, in all the strains tested in this study, the peripheral representation of the AV12 segment family was lower than in C57BL/6 WT mice. Concerning the BV segments, the BV31 segments tend to be overrepresented in all but HLA-B*27:05 of the monochain strains, whereas underrepresentation of the BV3 segment family was observed in all strains.
Whereas these data are clearly in support of a substantial diversity of the peripheral CD8+ TCR repertoires educated by the HLA class I monochains, to more definitively document this diversity, we performed DNA sequence analysis of the CDR3 of randomly selected AV9+ TCRα-chains from each of the seven monochain mice. CDR3 diversity (number of distinct in-frame sequences/total number of in-frame sequences analyzed) was compared with that of AV9+ TCRα-chains from C57BL/6 WT mice. As documented in Table II, in all monochain strains, a similar level of CDR3 diversity was observed as in C57BL/6 WT mice. Thus, the overrepresentation of the AV9 family observed in several monochain strains appears to be of polyclonal nature.
IFN-γ–producing T cell HLA monochain responses to reference epitopes in HLA monochain transgenic mice
We next assessed the functionality of HLA-restricted CD8+ T cells from each strain: 1) by selecting HLA-restricted CD8+ T cell reference epitopes previously identified by several independent laboratories, or 2) by selecting CD8+ T cell epitopes of predicted (BIMAS, SYFPEITHI prediction programs) high affinity for their restricting HLA class I molecule. For HLA-C*07:01–restricted epitopes, additional in-house identified epitopes of high affinity and stabilizing capacity were also tested. Mice were immunized either with the CD8+ reference T cell peptide epitope coemulsified in IFA with the T13L helper peptide or with a DNA plasmid vector encoding a recombinant HBV glycoprotein in which the pre-S2 coding segment has been replaced by the nucleotide sequence corresponding to the CD8+ reference T cell epitopes. In both cases, Ficoll-Hypaque–purified immune splenocytes were tested for HLA-restricted, epitope-specific, IFN-γ–producing CD8+ T cells using an IFN-γ ELISPOT assay.
As summarized in Tables III and IV, IFN-γ responses were obtained in each of the seven monochain transgenic mice following peptide as well as DNA immunization against most of the reference epitopes with the noticeable exceptions of one (HIV nef 186–194) of the three HLA-A*24:02–restricted epitopes and of the three epitopes selected from the literature (59–61) and supposedly restricted by the HLA-C*07:01 molecule. In contrast, in the assay involving HLA-C*07:01 epitopes that had been selected with the NetMHCpan prediction program (62), and shown to stabilize HLA-C*07:01 molecules (Fig. 5), IFN-γ responses were induced following either peptide or DNA immunization.
FIGURE 5.
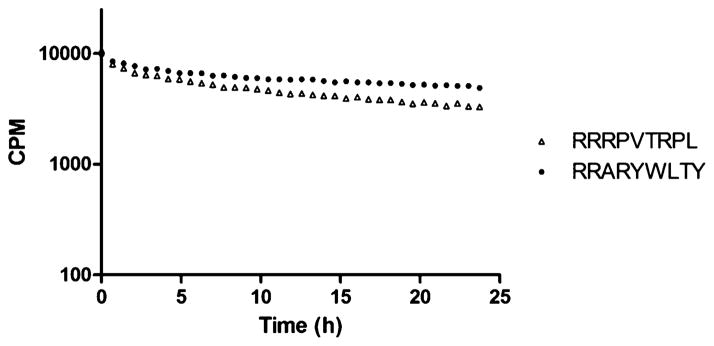
Peptide–HLA-C*07:01 dissociation. Dissociation of predicted HLA-C*07:01 binding peptides RRRPVTRPL (T1/2 16.1 h) and RRARYWLTY (T1/2 29.4 h) from HLA-C*07:01 measured by scintillation proximity assay.
As expected, depending on the epitopes, differences in magnitude of the responses were observed regardless of the immunization strategy. They likely reflect differences in the intrinsic immunogenicity of the epitopes that were not experimentally assessed (except for those presented by HLA-C*07:01) for their stabilizing capacity for their presenting HLA molecule.
Discussion
HLA transgenic mice represent a suitable versatile small-animal model system to quickly evaluate T cell responses specific to human epitopes and complex pathogens. In this report, we generated and characterized seven new HLA class I monochain transgenic, H-2 Kb, Db mouse β2-m triple KO mice that express a significant amount of transgenic molecules at the cell surface resulting in substantial restoration of their CD8+ T cell peripheral compartment, which appears endowed with a well-diversified TCR repertoire. Accordingly, these HLA transgenic mice developed efficient epitope-specific IFN-γ CD8+ T cell responses when immunized with reference human T cell epitopes. The lack of response to the HIV nef 186–194 peptide likely reflects the fact that this peptide is deprived of the HLA-A*24:02 binding motifs (P2, Y/F, P9, F/W/I/L) and, as such, is predicted as a nonbinder peptide by all epitope prediction programs. A similar argument certainly applies to the three published HLA-C*07:01 epitopes that all lack the positively charged residues in P1 and P2 considered as anchoring residues for HLA-C*07:01, but this argument remains somewhat hypothetical because the HLA-C*07:01 peptide-binding motifs have only been partially identified (63).
Compared with previously described HHD II mice (30), in the seven new strains, the additional destruction of the H-2 Kb gene practically abolishes the possibility that the transgenic molecules would not restrict the induced CD8+ T cell responses. When the HHD II mouse strain was created, H-2 Kb/Db/mouse β2-m triple-KO mice were not available. Expression of the HHD II transgene in a H-2 Db (rather than Kb)/mouse β2-m double-KO context was selected because a number of observations suggested that the cell-surface expression of β2-m–free H-2 Db molecules should be more problematic than that of β2-m–free H-2 Kb molecules. Contrary to β2-m–free H-2 Kb molecules (35, 36), β2-m–free H-2 Db molecules can be detected on cell surface with conformation-dependent mAb (31, 32, 34). It was further shown that cell-surface–expressed β2-m–free H-2 Kb molecules are essentially associated with long (>17 aa) atypical peptides lacking H-2 Kb canonical anchor residues, whereas a majority of those associated with β2-m–free H-2 Db molecules have the same size and anchor residues as those presented by β2-m–associated H-2 Db molecules (35, 37). However, it has been reported that virus-specific CD8+ CTLs restricted by β2-m–associated H-2 Kb molecules could also lyse virus-infected cells expressing β2-m–free H-2 Kb molecules (37) inferring presentation by some β2-m–free H-2 Kb molecules of the “correct” viral peptides. Thus, expression of HLA monochains in a H-2 Kb/Db/mouse β2-m triple-KO context minimizes or eliminates any possible contribution of the H-2 Kb molecules in the induced CD8+ T cell responses.
Within several founders of each monochain strain, we observed that, for a given allele, up to a certain point, the size of the peripheral CD8+ T cell compartment increases and correlates positively with the level of cell-surface expression of the monochains (data not shown). However, when distinct HLA class I monochain molecules are considered, the size of the peripheral CD8+ T cell compartment they select does not necessarily correlate with their respective levels of cell-surface expression. Thus, the highest restoration of the peripheral CD8+ T cell compartment was observed with the HLA-B*35:01 strain, whereas the lowest restoration was observed with the HLA-B*08:01 strain even though both expressed similar levels of monochains on their cell surfaces. This may suggest that the interaction of the mouse CD8 accessory molecules with the chimeric HLA-B8.01 monochain molecules is of lower affinity than with the other HLA chimeric monochains, such as HLA-B*35:01. Only human CD8/HLA-A*02:01 and mouse CD8/H-2 Kb cocrystals have been structurally resolved (64, 65). Whereas the mouse CD8 molecule only interacts with the H-2 Kb H chain α3 domain, additional contacts between the human CD8 molecule and the HLA-A*02:01 H chain α2 domain have been documented and proved important for a fully efficient CD8 interaction (66). Similarly, we may assume that a fully efficient CD8 interaction with HLA-B*08:01 molecules is dependent on additional interactions with the HLA-B*08:01 α2 domain, additional interactions that the mouse CD8 molecules might be unable to develop. However, despite their limited number, the HLA-B*08:01–educated CD8+ T cells are efficiently stimulated by Ags.
The RT-PCR and CDR3 sequencing results obtained with the seven new strains clearly support the notion that they have all educated a diversified repertoire of HLA class I–restricted TCRs. It is notable that phylogenetically distant human and mouse MHC class I molecules preferentially select, with few exceptions, the same AV and BV families of variable segments. This certainly reflects the fact that genetic and epigenetic factors (family size, nucleotidic variations in the recombination signal sequence that flanks each V gene or differences in the structure of the chromatin associated with each V gene) are key determinants of V gene rearrangement frequency (67). However, noticeable increases (AV9, BV31) or decreases (AV12, BV3) in peripheral representations of certain segment families (compared with C57BL/6 mice) were also observed that differ in magnitude from one HLA class I monochain to another. Thus, in addition to the earlier-mentioned genetic and epigenetic factors, the AV and BV selection is also likely influenced by the MHC class I molecules themselves. Structural analyses have shed light on the relative contributions of the CDR1, CDR2, and CDR3 of the TCRα- and β-chains in contacting peptide/MHC class I molecules (68). Whereas CDR3 interacts essentially with the presented peptides, CDR1 and CDR2 interact mainly with the MHC class I molecule α helices. Therefore, we hypothesize that these increased or decreased representations of certain families of AV or BV segments in the CD8+ T cell compartment of a monochain mouse reflect the quality of the interaction of their CDR1 and CDR2 segments with the α helices of the unique species of HLA class I molecules expressed. This is reminiscent of certain TCR AV biases serologically documented in naive CD8+ T cell populations in both the mouse (69) and rat (70) species. In rat, the AV8.2 bias of the CD8+ T cell population has been linked to the RT1f class I molecule. This bias is also observed at the CD8+ T cell effector level when RT1f− lymphocytes are stimulated by RT1f+ cells (70). Similarly, preliminary results suggest that HLA class I monochains that overselect certain AV segments at the thymic level also preferentially mobilize T cells expressing the same AV segments when they are acting as stimulating molecules in MLC.
Ag processing and peptide loading on MHC class I molecules is a multistep process in which functional differences have been documented between human and mouse species as detailed later. This might substantially impact negatively on epitope presentation by HLA class I molecules when expressed in mouse instead of human cells. Most of the peptides bound by MHC class I molecules are produced in the cytosol by proteasomes. The human and mouse proteasomes produce largely overlapping, although not absolutely identical sets of peptides (71). Peptides are then transported in the ER by the TAP pumps. The mouse TAP pump does not import in the ER as efficiently as the human one peptides with a positively charged C terminus. Such peptides are efficiently presented by some allelic forms of HLA class I molecules such as HLA-A*03 and HLA-B*27 (72, 73). However, this transport deficiency appears only relative because by sequencing peptides eluted from an HLA-A*03:01–transfected mouse mastocytoma cell, the capacity of HLA-A*03 molecules to bind peptides with a positively charged C terminus was correctly identified (72). Many of the peptides translocated in the ER are too long to be accommodated by class I molecules unless they are shortened by ER aminopeptidases. In humans, there are two ER aminopeptidases (ERAAP1 and 2) that cooperate to ensure efficient N-terminal peptide trimming. Human ERAAP1 does not remove basic amino acids as efficiently as ERAAP2 (74). Whether the absence of ERAAP2 in the mouse species (75) impacts on peptide trimming remains to be determined. Peptides are finally loaded on MHC class I molecules, a process facilitated by Tapasin that bridges peptide-empty class I molecules with the luminal face of the TAP peptide pumps (76). Some degree of species incompatibility has been documented at this stage between mouse Tapasin and some HLA class I molecules resulting in limited (i.e., HLA-B*27:05) (77, 78) or extensive (i.e., HLA-B*44:02) (79) antigenic peptide presentation defect that, at least in the latter case, can be overcome by overexpressing the mouse Tapasin. The potential impact of these cumulative functional differences between human and mouse Ag processing/peptide loading machineries should always be kept in mind. However, it has been shown by analyzing the peptide pools eluted from HLA-B*27:05 molecules expressed in mouse or human cells that they largely overlap (≈90% conservation once interspecies proteome differences are excluded). Accordingly, there are only very few documented examples of complete lack of presentation by HLA class I–transfected mouse cells of epitopes that are efficiently processed and presented by human cells (80, 81).
A special attention should be paid to the profound reduction of cell-surface expression of HLA class I transgenic monochains when coexpressed with mouse H-2 class I molecules as observed following crosses with OF1 mice. Such observation is reminiscent of the poor cell-surface expression of HLA-B*44:02 molecules in transfected mouse L cells (79). We accordingly found that cell-surface expression of HLA-B*44:02 monochain transgenic molecules was reduced in HLA-B*44:02 × OF1 hybrid mice, albeit moderately, whereas expression of HLA class I HLA-A*01:03, -B*08:01, -B*35:01, and -C*07:01 transgenic monochains in similar OF1 hybrids was almost completely abrogated. By contrast, expression of HLA-A*24:02 and HLA-B*27:05 transgenic monochains was practically unaltered. Such allele-related differential effects lead us to suspect that OF1 H-2 class I molecules out-compete HLA class I transgenic monochains at the Tapasin level, preventing their recruitment in the peptide loading complex. In the absence of such recruitment, Tapasin-“dependent” molecules (e.g., HLA-B*08, -A*01) (76) are not peptide loaded and remain in the ER, whereas Tapasin-“independent” molecules (i.e., HLA-B*27:05) still bind peptides of sufficient (however globally suboptimal) (77) affinity for their exit to the cell surface. From a practical point of view, in particular, when working with HLA class I transgenic mice expressing monochain molecules (the peptide loading of which is likely hampered by the covalent association with β2-m), the complete absence of endogenous H-2 class I competitor molecules is of crucial importance for the mouse strain functionality regarding certain allo/isoforms of HLA class I molecules.
The set of HLA class I monochain transgenic mice described in this report, combined with the two strains (HLA-A*02:01 and HLA-B*07:02 transgenic, H-2 KO) previously created (27, 30), is representative of a high percentage of individuals in the three major ethnic groups. We therefore expect that, together, these HLA transgenic mice will have broad immunology and vaccine preclinical application to a wide range of infectious diseases and cancers that would cover many areas of the world regardless of race and ethnicity. The selection of the HLA-A*01:03 allele frequent only in eastern Africa (http://www.allelefrequencies.net) (82) was to some extent rather unfortunate. However, the only difference at position 97 with the HLA-A*01:01 allele, where a methionine replaces an isoleucine, should not have a profound impact on the set of presented peptides because this residue is in the central part of the floor of the peptide binding groove, away from the D and F pockets that accommodate the major anchor residues P3 and PΩ of HLA-A*01 bound peptides. In fact, the two HLA-A*01–restricted reference antigenic peptides we have tested and that were efficiently presented by the HLA-A*01:03 molecule were identified in Caucasoid patients that almost certainly expressed the HLA-A*01:01 allele. There are still, however, some relatively frequent alleles that would be worth considering, especially in the HLA-A series such as A*03, A*11, A*23, A*30, and A*68. However, our attempt to apply the monochain strategy to the HLA-A*03:01 allele failed because this molecule in a monochain configuration does not reach the cell surface in a significant amount. Therefore, monochain viability should be assessed for each new construct by transfecting β2-m–deficient cells prior transgenesis. We expect the set of HLA class I monochain transgenic mice described in this report to cover a majority of HLA haplotypes in any given population. Although the high degree of HLA polymorphism can be a hindrance to epitope-based vaccines, this can be dealt with by the inclusion of multiple supertype-restricted epitopes that are recognized in the context of diverse related HLA alleles: a combination of nine HLA supertypes has been recently defined to provide an almost perfect coverage (>99%) of the entire repertoire of HLA molecules (83).
As clearly illustrated in this report, in particular for HLA-A*24:02 and -C*07:01 epitopes, monochain HLA-class I transgenic H-2 class I null mice are useful tools to validate or invalidate human CD8 + T cell epitopes of potential clinical interest. The diversified CD8+ TCR repertoire that is educated by each of these transgenic monochains should allow valuable preclinical studies aiming at the development of candidate epitope-based vaccines for infectious diseases and cancer. For example, similar monochain HLA transgenic mice have been used to identify under well-controlled experimental conditions the most immunogenic epitopes from pathogens, and to assess the multiplicity of the elicited responses by grouping these epitopes in a single construct (84). Such mice have also been used to test vaccine strategies aiming at improving epitope immunogenicity in the respect of antigenicity, thereby transforming cryptic tumor-associated epitopes in efficient immunogens (7). These mice are also of interest in autoimmune diseases. Compared with HLA class II alleles, only a few class I alleles (e.g., HLA-B*27:05 and ankylosing spondylitis) have been directly linked to autoimmune disorders. However, HLA class I–restricted T cells are often key autoimmune effectors such as in type 1 diabetes (85). In that case, it will be of interest to identify the epitopes targeted by HLA-A*01–, -C*07:01–, -B*08–restricted CTLs in pancreatic β cell autoantigens. These three HLA class I genes are in strong linkage disequilibrium with HLA-DRB1*03, a HLA class II gene associated with many autoimmune diseases including type 1 diabetes (86). In all these pathological situations, the number of candidate Ags and epitopes are numerous, and the access we have to human samples is limited. Preclinical studies conducted with monochain HLA-class I transgenic, H-2 class I null mice, should allow to speed up and focus on the most promising candidates when performing preclinical vaccine studies with human material.
Table II.
CD8+ T cell AV9+ TCR CDR3 diversity
| Mouse Strains | AV9 Segment Usage | CDR3 Diversity |
|---|---|---|
| C57BL/6 | 42/45a | |
| 9D-4b | 28 | |
| 9-4 | 14 | |
| A*01:03 | 28/28 | |
| 9D-4 | 14 | |
| 9N-3 | 12 | |
| 9N-4 | 1 | |
| 9D-1 | 1 | |
| A*24:02 | 19/20 | |
| 9N-2 | 12 | |
| 9D-1 | 5 | |
| 9N-3 | 2 | |
| B*08:01 | 27/32 | |
| 9N-3 | 17 | |
| 9D-4 | 8 | |
| 9N-4 | 3 | |
| 9D-1 | 3 | |
| 9N-2 | 1 | |
| B*27:05 | 36/37 | |
| 9N-3 | 21 | |
| 9D-4 | 7 | |
| 9N-4 | 6 | |
| 9D-1 | 2 | |
| 9D-2 | 1 | |
| B*35:01 | 20/20 | |
| 9N-3 | 12 | |
| 9N-2 | 7 | |
| 9-4 | 1 | |
| B*44:02 | 19/19 | |
| 9D-4 | 11 | |
| 9N-3 | 4 | |
| 9-4 | 2 | |
| 9N-2 | 1 | |
| 9D-1 | 1 | |
| C*07:01 | 25/32 | |
| 9N-3 | 18 | |
| 9D-4 | 10 | |
| 9D-1 | 4 | |
| 9-4 | 2 |
After purification of CD8+ T cells, the CDR3 subregions of AV9+ TCR α-chains from each of the HLA monochain transgenic strains were RT-PCR amplified, cloned, and sequenced; AV9+ TCR α-chains of C57BL/6 WT mice were similarly analyzed.
Number of different CDR3 sequences versus the total number of in-frame sequences analyzed.
Usage of the individual variable segments of the AV9 family.
Table IV.
CD8+ T cell IFN-γ responses after DNA immunization
| Peptides | HLA Restriction | IFN-γ Spots/106 Cellsa |
|---|---|---|
| VSDGGPNLY | A*01 | 265/169/105 |
| EADPTGHSY | A*01 | 118/91/86/38 |
| RYLKDQQLL | A*24 | 130/90/75/27 |
| TYQYQEPF | A*24 | 208/89/0/0 |
| DSRLAFHHM | A*24 | 0/0/0/0 |
| RAKFKQLL | B*08 | 52/44/43/36 |
| QAKWRLQTL | B*08 | 269/269/44/34 |
| WLKIKRDYL | B*08 | 327/46/39/21/21/11 |
| SRYWAIRTR | B*27 | 917/890/411/241 |
| LRGKWQRRYR | B*27 | 221/213/189/13 |
| HPNIEEVAL | B*35 | 50/30/18/16 |
| TPEGIIPTL | B*35 | 113/79/48/0 |
| EENLLDFVRF | B*44 | 134/87/0/0 |
| FEDLRVLSF | B*44 | 66/44/0/0 |
| AENGWGFYF | B*44 | 91/63/46/0 |
| RRRPVTRPL | C*07 | 33/33/24/1 |
| RRARYWLTY | C*07 | 273/246/42/9 |
| PLADLSPFA | C*07 | 0/0/0/0 |
| EGDCAPEEK | C*07 | 0/0/0/0 |
| RNGYRALMDKS | C*07 | 0/0/0/0 |
IFN-γ T lymphocyte responses of the seven HLA class I monochain transgenic mice to published CD8+ epitopes were evaluated in an ELISPOT assay after in vivo DNA priming with a plasmid vector coding for a chimeric hepatitis B glycoprotein in which the epitope DNA sequence was inserted.
Mice were tested individually. Data are given for each mouse as the mean of triplicates.
Acknowledgments
This work was supported by the Institut Pasteur (Paris), INSERM, the Agence Nationale de la Recherche (ANR 2010 BIOT 008 01), the Ministère de l’Enseignement Supérieur et de la Recherche (H.K.-M.), and the Fondation pour la Recherche Médicale (R.B.).
We thank G. Chauveau-Le Friec, A. Zago, and A. Carbon (Centre d’Ingénierie Génétique Murine, Institut Pasteur) for technical support in microinjection experiments and animal husbandry. We thank the members of INSERM Unit 1016 for scientific support.
Abbreviations used in this article
- ER
endoplasmic reticulum
- KO
knockout
- β2-m
β2-microglobulin
Footnotes
Disclosures
The authors have no financial conflicts of interest.
The online version of this article contains supplemental material.
References
- 1.Pascolo S. HLA class I transgenic mice: development, utilisation and improvement. Expert Opin Biol Ther. 2005;5:919–938. doi: 10.1517/14712598.5.7.919. [DOI] [PubMed] [Google Scholar]
- 2.Cardinaud S, Moris A, Février M, Rohrlich PS, Weiss L, Langlade-Demoyen P, Lemonnier FA, Schwartz O, Habel A. Identification of cryptic MHC I-restricted epitopes encoded by HIV-1 alternative reading frames. J Exp Med. 2004;199:1053–1063. doi: 10.1084/jem.20031869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Szpak Y, Vieville JC, Tabary T, Naud MC, Chopin M, Edelson C, Cohen JHM, Dausset J, de Kozak Y, Pla M. Spontaneous retinopathy in HLA-A29 transgenic mice. Proc Natl Acad Sci USA. 2001;98:2572–2576. doi: 10.1073/pnas.051595998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Toma A, Laïka T, Haddouk S, Luce S, Briand JP, Camoin L, Connan F, Lambert M, Caillat-Zucman S, Carel JC, et al. Recognition of human proinsulin leader sequence by class I-restricted T-cells in HLA-A*0201 transgenic mice and in human type 1 diabetes. Diabetes. 2009;58:394–402. doi: 10.2337/db08-0599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Visseren MJW, van der Burg SH, van der Voort EIH, Brandt RMP, Schrier PI, van der Bruggen P, Boon T, Melief CJM, Kast WM. Identification of HLA-A*0201-restricted CTL epitopes encoded by the tumor-specific MAGE-2 gene product. Int J Cancer. 1997;73:125–130. doi: 10.1002/(sici)1097-0215(19970926)73:1<125::aid-ijc19>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 6.Ishioka GY, Fikes J, Hermanson G, Livingston B, Crimi C, Qin M, del Guercio MF, Oseroff C, Dahlberg C, Alexander J, et al. Utilization of MHC class I transgenic mice for development of minigene DNA vaccines encoding multiple HLA-restricted CTL epitopes. J Immunol. 1999;162:3915–3925. [PubMed] [Google Scholar]
- 7.Tourdot S, Scardino A, Saloustrou E, Gross DA, Pascolo S, Cordopatis P, Lemonnier FA, Kosmatopoulos K. A general strategy to enhance immunogenicity of low-affinity HLA-A2. 1-associated peptides: implication in the identification of cryptic tumor epitopes. Eur J Immunol. 2000;30:3411–3421. doi: 10.1002/1521-4141(2000012)30:12<3411::AID-IMMU3411>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 8.Dill O, Kievits F, Koch S, Ivanyi P, Hämmerling GJ. Immunological function of HLA-C antigens in HLA-Cw3 transgenic mice. Proc Natl Acad Sci USA. 1988;85:5664–5668. doi: 10.1073/pnas.85.15.5664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kievits F, Ivanyi P, Krimpenfort P, Berns A, Ploegh HL. HLA-restricted recognition of viral antigens in HLA transgenic mice. Nature. 1987;329:447–449. doi: 10.1038/329447a0. [DOI] [PubMed] [Google Scholar]
- 10.Kievits F, Boerenkamp WJ, Lokhorst W, Ivanyi P. Specificity and frequency of primary anti-HLA cytotoxic T lymphocytes in normal and HLA-B27.2-, HLA-B27.5-, and HLA-Cw3-transgenic mice. A transgenic model for MHC xenoantigen recognition. J Immunol. 1990;144:4513–4519. [PubMed] [Google Scholar]
- 11.Barra C, Pérarnau B, Gerlinger P, Lemeur M, Gillet A, Gibier P, Lemonnier FA. Analysis of the HLA-Cw3-specific cytotoxic T lymphocyte response of HLA-B7 X human beta 2m double transgenic mice. J Immunol. 1989;143:3117–3124. [PubMed] [Google Scholar]
- 12.Epstein H, Hardy R, May JS, Johnson MH, Holmes N. Expression and function of HLA-A2.1 in transgenic mice. Eur J Immunol. 1989;19:1575–1583. doi: 10.1002/eji.1830190909. [DOI] [PubMed] [Google Scholar]
- 13.Le AX, Bernhard EJ, Holterman MJ, Strub S, Parham P, Lacy E, Engelhard VH. Cytotoxic T cell responses in HLA-A2.1 transgenic mice. Recognition of HLA alloantigens and utilization of HLA-A2.1 as a restriction element. J Immunol. 1989;142:1366–1371. [PubMed] [Google Scholar]
- 14.Connolly JM, Potter TA, Wormstall EM, Hansen TH. The Lyt-2 molecule recognizes residues in the class I alpha 3 domain in allogeneic cytotoxic T cell responses. J Exp Med. 1988;168:325–341. doi: 10.1084/jem.168.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Engelhard VH, Lacy E, Ridge JP. Influenza A-specific, HLA-A2.1-restricted cytotoxic T lymphocytes from HLA-A2.1 transgenic mice recognize fragments of the M1 protein. J Immunol. 1991;146:1226–1232. [PubMed] [Google Scholar]
- 16.Irwin MJ, Heath WR, Sherman LA. Species-restricted interactions between CD8 and the alpha 3 domain of class I influence the magnitude of the xenogeneic response. J Exp Med. 1989;170:1091–1101. doi: 10.1084/jem.170.4.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kalinke U, Arnold B, Hämmerling GJ. Strong xenogeneic HLA response in transgenic mice after introducing an alpha 3 domain into HLA B27. Nature. 1990;348:642–644. doi: 10.1038/348642a0. [DOI] [PubMed] [Google Scholar]
- 18.Vitiello A, Marchesini D, Furze J, Sherman LA, Chesnut RW. Analysis of the HLA-restricted influenza-specific cytotoxic T lymphocyte response in transgenic mice carrying a chimeric human-mouse class I major histocompatibility complex. J Exp Med. 1991;173:1007–1015. doi: 10.1084/jem.173.4.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alexander J, Oseroff C, Sidney J, Wentworth P, Keogh E, Hermanson G, Chisari FV, Kubo RT, Grey HM, Sette A. Derivation of HLA-A11/Kb transgenic mice: functional CTL repertoire and recognition of human A11-restricted CTL epitopes. J Immunol. 1997;159:4753–4761. [PubMed] [Google Scholar]
- 20.Borenstein SH, Graham J, Zhang XL, Chamberlain JW. CD8+ T cells are necessary for recognition of allelic, but not locus-mismatched or xeno-, HLA class I transplantation antigens. J Immunol. 2000;165:2341–2353. doi: 10.4049/jimmunol.165.5.2341. [DOI] [PubMed] [Google Scholar]
- 21.Gotoh M, Takasu H, Harada K, Yamaoka T. Development of HLA-A2402/K(b) transgenic mice. Int J Cancer. 2002;100:565–570. doi: 10.1002/ijc.10509. [DOI] [PubMed] [Google Scholar]
- 22.Bharadwaj M, Sherritt M, Khanna R, Moss DJ. Contrasting Epstein-Barr virus-specific cytotoxic T cell responses to HLA A2-restricted epitopes in humans and HLA transgenic mice: implications for vaccine design. Vaccine. 2001;19:3769–3777. doi: 10.1016/s0264-410x(01)00085-8. [DOI] [PubMed] [Google Scholar]
- 23.LaFace DM, Vestberg M, Yang Y, Srivastava R, DiSanto J, Flomenberg N, Brown S, Sherman LA, Peterson PA. Human CD8 transgene regulation of HLA recognition by murine T cells. J Exp Med. 1995;182:1315–1325. doi: 10.1084/jem.182.5.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barra C, Gournier H, Garcia Z, Marche PN, Jouvin-Marche E, Briand P, Fillipi P, Lemonnier FA. Abrogation of H-2-restricted CTL responses and efficient recognition of HLA-A3 molecules in DBA/2 HLA/A24 responder mice. J Immunol. 1993;150:3681–3689. [PubMed] [Google Scholar]
- 25.Pérarnau B, Saron M-F, Reina San Martin B, Bervas N, Ong H, Soloski MJ, Smith AG, Ure JM, Gairin J-E, Lemonnier FA. Single H2Kb, H2Db and double H2KbDb knockout mice: peripheral CD8+ T cell repertoire and anti-lymphocytic choriomeningitis virus cytolytic responses. Eur J Immunol. 1999;29:1243–1252. doi: 10.1002/(SICI)1521-4141(199904)29:04<1243::AID-IMMU1243>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 26.Cheuk E, D’Souza C, Hu N, Liu Y, Lang H, Chamberlain JW. Human MHC class I transgenic mice deficient for H2 class I expression facilitate identification and characterization of new HLA class I-restricted viral T cell epitopes. J Immunol. 2002;169:5571–5580. doi: 10.4049/jimmunol.169.10.5571. [DOI] [PubMed] [Google Scholar]
- 27.Rohrlich PS, Cardinaud S, Firat H, Lamari M, Briand P, Escriou N, Lemonnier FA. HLA-B*0702 transgenic, H-2KbDb double-knockout mice: phenotypical and functional characterization in response to influenza virus. Int Immunol. 2003;15:765–772. doi: 10.1093/intimm/dxg073. [DOI] [PubMed] [Google Scholar]
- 28.Laouini D, Casrouge A, Dalle S, Lemonnier F, Kourilsky P, Kanellopoulos J. V β T cell repertoire of CD8+ splenocytes selected on nonpolymorphic MHC class I molecules. J Immunol. 2000;165:6381–6386. doi: 10.4049/jimmunol.165.11.6381. [DOI] [PubMed] [Google Scholar]
- 29.Bouwer HG, Seaman MS, Forman J, Hinrichs DJ. MHC class Ib-restricted cells contribute to antilisterial immunity: evidence for Qa-1b as a key restricting element for Listeria-specific CTLs. J Immunol. 1997;159:2795–2801. [PubMed] [Google Scholar]
- 30.Pascolo S, Bervas N, Ure JM, Smith AG, Lemonnier FA, Pérarnau B. HLA-A2.1-restricted education and cytolytic activity of CD8 (+) T lymphocytes from beta2 microglobulin (beta2m) HLA-A2.1 monochain transgenic H-2Db beta2m double knockout mice. J Exp Med. 1997;185:2043–2051. doi: 10.1084/jem.185.12.2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Allen H, Fraser J, Flyer D, Calvin S, Flavell R. β 2-microglobulin is not required for cell surface expression of the murine class I histocompatibility antigen H-2Db or of a truncated H-2Db. Proc Natl Acad Sci USA. 1986;83:7447–7451. doi: 10.1073/pnas.83.19.7447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bix M, Raulet D. Functionally conformed free class I heavy chains exist on the surface of beta 2 microglobulin negative cells. J Exp Med. 1992;176:829–834. doi: 10.1084/jem.176.3.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Glas R, Franksson L, Ohlén C, Höglund P, Koller B, Ljunggren HG, Kärre K. Major histocompatibility complex class I-specific and -restricted killing of beta 2-microglobulin-deficient cells by CD8+ cytotoxic T lymphocytes. Proc Natl Acad Sci USA. 1992;89:11381–11385. doi: 10.1073/pnas.89.23.11381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zijlstra M, Bix M, Simister NE, Loring JM, Raulet DH, Jaenisch R. Beta 2-microglobulin deficient mice lack CD4-8+ cytolytic T cells. Nature. 1990;344:742–746. doi: 10.1038/344742a0. [DOI] [PubMed] [Google Scholar]
- 35.Joyce S, Kuzushima K, Kepecs G, Angeletti RH, Nathenson SG. Characterization of an incompletely assembled major histocompatibility class I molecule (H-2Kb) associated with unusually long peptides: implications for antigen processing and presentation. Proc Natl Acad Sci USA. 1994;91:4145–4149. doi: 10.1073/pnas.91.10.4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Machold RP, Andrée S, Van Kaer L, Ljunggren HG, Ploegh HL. Peptide influences the folding and intracellular transport of free major histocompatibility complex class I heavy chains. J Exp Med. 1995;181:1111–1122. doi: 10.1084/jem.181.3.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schell TD, Mylin LM, Tevethia SS, Joyce S. The assembly of functional beta(2)-microglobulin-free MHC class I molecules that interact with peptides and CD8(+) T lymphocytes. Int Immunol. 2002;14:775–782. doi: 10.1093/intimm/dxf041. [DOI] [PubMed] [Google Scholar]
- 38.Gallez-Hawkins G, Villacres MC, Li X, Sanborn MC, Lomeli NA, Zaia JA. Use of transgenic HLA A*0201/Kb and HHD II mice to evaluate frequency of cytomegalovirus IE1-derived peptide usage in eliciting human CD8 cytokine response. J Virol. 2003;77:4457–4462. doi: 10.1128/JVI.77.7.4457-4462.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramage JM, Metheringham R, Moss R, Spendlove I, Rees R, Durrant LG. Comparison of the immune response to a self antigen after DNA immunisation of HLA*A201/H-2Kb and HHD transgenic mice. Vaccine. 2004;22:1728–1731. doi: 10.1016/j.vaccine.2004.01.034. [DOI] [PubMed] [Google Scholar]
- 40.Grigoriadou K, Ménard C, Pérarnau B, Lemonnier FA. MHC class Ia molecules alone control NK-mediated bone marrow graft rejection. Eur J Immunol. 1999;29:3683–3690. doi: 10.1002/(SICI)1521-4141(199911)29:11<3683::AID-IMMU3683>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 41.Lim A, Baron V, Ferradini L, Bonneville M, Kourilsky P, Pannetier C. Combination of MHC-peptide multimer-based T cell sorting with the Immunoscope permits sensitive ex vivo quantitation and follow-up of human CD8+ T cell immune responses. J Immunol Methods. 2002;261:177–194. doi: 10.1016/s0022-1759(02)00004-2. [DOI] [PubMed] [Google Scholar]
- 42.Bouvier I, Jusforgues-Saklani H, Lim A, Lemaitre F, Lemercier B, Auriau C, Nicola MA, Leroy S, Law HK, Bandeira A, et al. Immunization route dictates cross-priming efficiency and impacts the optimal timing of adjuvant delivery. Front Immunol. 2011;2:71. doi: 10.3389/fimmu.2011.00071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Le Borgne S, Mancini M, Le Grand R, Schleef M, Dormont D, Tiollais P, Rivière Y, Michel ML. In vivo induction of specific cytotoxic T lymphocytes in mice and rhesus macaques immunized with DNA vector encoding an HIV epitope fused with hepatitis B surface antigen. Virology. 1998;240:304–315. doi: 10.1006/viro.1997.8942. [DOI] [PubMed] [Google Scholar]
- 44.Harndahl M, Rasmussen M, Roder G, Buus S. Real-time, high-throughput measurements of peptide-MHC-I dissociation using a scintillation proximity assay. J Immunol Methods. 2011;374:5–12. doi: 10.1016/j.jim.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mintz B, Bradl M. Mosaic expression of a tyrosinase fusion gene in albino mice yields a heritable striped coat color pattern in transgenic homozygotes. Proc Natl Acad Sci USA. 1991;88:9643–9647. doi: 10.1073/pnas.88.21.9643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.DiBrino M, Tsuchida T, Turner RV, Parker KC, Coligan JE, Biddison WE. HLA-A1 and HLA-A3 T cell epitopes derived from influenza virus proteins predicted from peptide binding motifs. J Immunol. 1993;151:5930–5935. [PubMed] [Google Scholar]
- 47.Traversari C, van der Bruggen P, Luescher IF, Lurquin C, Chomez P, Van Pel A, De Plaen E, Amar-Costesec A, Boon T. A nonapeptide encoded by human gene MAGE-1 is recognized on HLA-A1 by cytolytic T lymphocytes directed against tumor antigen MZ2-E. J Exp Med. 1992;176:1453–1457. doi: 10.1084/jem.176.5.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dai LC, West K, Littaua R, Takahashi K, Ennis FA. Mutation of human immunodeficiency virus type 1 at amino acid 585 on gp41 results in loss of killing by CD8+ A24-restricted cytotoxic T lymphocytes. J Virol. 1992;66:3151–3154. doi: 10.1128/jvi.66.5.3151-3154.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ikeda-Moore Y, Tomiyama H, Miwa K, Oka S, Iwamoto A, Kaneko Y, Takiguchi M. Identification and characterization of multiple HLA-A24-restricted HIV-1 CTL epitopes: strong epitopes are derived from V regions of HIV-1. J Immunol. 1997;159:6242–6252. [PubMed] [Google Scholar]
- 50.Autran B, Letvin NL. HIV epitopes recognized by cytotoxic T-lymphocytes. AIDS. 1991;5(Suppl 2):S145–S150. doi: 10.1097/00002030-199101001-00020. [DOI] [PubMed] [Google Scholar]
- 51.Bogedain C, Wolf H, Modrow S, Stuber G, Jilg W. Specific cytotoxic T lymphocytes recognize the immediate-early transactivator Zta of Epstein-Barr virus. J Virol. 1995;69:4872–4879. doi: 10.1128/jvi.69.8.4872-4879.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jing L, Chong TM, McClurkan CL, Huang J, Story BT, Koelle DM. Diversity in the acute CD8 T cell response to vaccinia virus in humans. J Immunol. 2005;175:7550–7559. doi: 10.4049/jimmunol.175.11.7550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tussey LG, Rowland-Jones S, Zheng TS, Androlewicz MJ, Cresswell P, Frelinger JA, McMichael AJ. Different MHC class I alleles compete for presentation of overlapping viral epitopes. Immunity. 1995;3:65–77. doi: 10.1016/1074-7613(95)90159-0. [DOI] [PubMed] [Google Scholar]
- 54.Brooks JM, Murray RJ, Thomas WA, Kurilla MG, Rickinson AB. Different HLA-B27 subtypes present the same immunodominant Epstein-Barr virus peptide. J Exp Med. 1993;178:879–887. doi: 10.1084/jem.178.3.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ibe M, Sakaguchi T, Tanaka K, Saito S, Yokota S, Tanaka T, Shimotohno K, Chujoh Y, Shiratori Y, Omata M, et al. Identification and characterization of a cytotoxic T cell epitope of hepatitis C virus presented by HLA-B*3501 in acute hepatitis. J Gen Virol. 1998;79:1735–1744. doi: 10.1099/0022-1317-79-7-1735. [DOI] [PubMed] [Google Scholar]
- 56.Livingston PG, Kurane I, Dai LC, Okamoto Y, Lai CJ, Men R, Karaki S, Takiguchi M, Ennis FA. Dengue virus-specific, HLA-B35-restricted, human CD8+ cytotoxic T lymphocyte (CTL) clones. Recognition of NS3 amino acids 500 to 508 by CTL clones of two different serotype specificities. J Immunol. 1995;154:1287–1295. [PubMed] [Google Scholar]
- 57.Burrows SR, I, Misko S, Sculley TB, Schmidt C, Moss DJ. An Epstein-Barr virus-specific cytotoxic T-cell epitope present on A- and B-type transformants. J Virol. 1990;64:3974–3976. doi: 10.1128/jvi.64.8.3974-3976.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.DiBrino M, Parker KC, Margulies DH, Shiloach J, Turner RV, Biddison WE, Coligan JE. Identification of the peptide binding motif for HLA-B44, one of the most common HLA-B alleles in the Caucasian population. Biochemistry. 1995;34:10130–10138. doi: 10.1021/bi00032a005. [DOI] [PubMed] [Google Scholar]
- 59.Redchenko I, Harrop R, Ryan MG, Hawkins RE, Carroll MW. Identification of a major histocompatibility complex class I-restricted T-cell epitope in the tumour-associated antigen, 5T4. Immunology. 2006;118:50–57. doi: 10.1111/j.1365-2567.2006.02338.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Breckpot K, Heirman C, De Greef C, van der Bruggen P, Thielemans K. Identification of new antigenic peptide presented by HLA-Cw7 and encoded by several MAGE genes using dendritic cells transduced with lentiviruses. J Immunol. 2004;172:2232–2237. doi: 10.4049/jimmunol.172.4.2232. [DOI] [PubMed] [Google Scholar]
- 61.Larrieu P, Renaud V, Godet Y, Jotereau F, Fonteneau J-F. A HLA-Cw*0701 restricted Melan-A/MART1 epitope presented by melanoma tumor cells to CD8+ tumor infiltrating lymphocytes. Cancer Immunol Immunother. 2008;57:745–752. doi: 10.1007/s00262-007-0436-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hoof I, Peters B, Sidney J, Pedersen LE, Sette A, Lund O, Buus S, Nielsen M. NetMHCpan, a method for MHC class I binding prediction beyond humans. Immunogenetics. 2009;61:1–13. doi: 10.1007/s00251-008-0341-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dionne SO, Lake DF, Grimes WJ, Smith MH. Identification of HLA-Cw6.02 and HLA-Cw7.01 allele-specific binding motifs by screening synthetic peptide libraries. Immunogenetics. 2004;56:391–398. doi: 10.1007/s00251-004-0710-1. [DOI] [PubMed] [Google Scholar]
- 64.Gao GF, Tormo J, Gerth UC, Wyer JR, McMichael AJ, Stuart DI, Bell JI, Jones EY, Jakobsen BK. Crystal structure of the complex between human CD8α(α) and HLA-A2. Nature. 1997;387:630–634. doi: 10.1038/42523. [DOI] [PubMed] [Google Scholar]
- 65.Kern PS, Teng MK, Smolyar A, Liu JH, Liu J, Hussey RE, Spoerl R, Chang HC, Reinherz EL, Wang JH. Structural basis of CD8 coreceptor function revealed by crystallographic analysis of a murine CD8alphaalpha ectodomain fragment in complex with H-2Kb. Immunity. 1998;9:519–530. doi: 10.1016/s1074-7613(00)80635-4. [DOI] [PubMed] [Google Scholar]
- 66.Sun J, Leahy DJ, Kavathas PB. Interaction between CD8 and major histocompatibility complex (MHC) class I mediated by multiple contact surfaces that include the alpha 2 and alpha 3 domains of MHC class I. J Exp Med. 1995;182:1275–1280. doi: 10.1084/jem.182.5.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Feeney AJ. Genetic and epigenetic control of V gene rearrangement frequency. Adv Exp Med Biol. 2009;650:73–81. doi: 10.1007/978-1-4419-0296-2_6. [DOI] [PubMed] [Google Scholar]
- 68.Rudolph MG, I, Wilson A. The specificity of TCR/pMHC interaction. Curr Opin Immunol. 2002;14:52–65. doi: 10.1016/s0952-7915(01)00298-9. [DOI] [PubMed] [Google Scholar]
- 69.Sim BC, Zerva L, Greene MI, Gascoigne NR. Control of MHC restriction by TCR Valpha CDR1 and CDR2. Science. 1996;273:963–966. doi: 10.1126/science.273.5277.963. [DOI] [PubMed] [Google Scholar]
- 70.Torres-Nagel N, Mehling B, LeRolle AF, Joly E, Hünig T. Genetic control of peripheral TCRAV usage by representation in the preselection repertoire and MHC allele-specific overselection. Int Immunol. 2001;13:63–73. doi: 10.1093/intimm/13.1.63. [DOI] [PubMed] [Google Scholar]
- 71.Sesma L, Alvarez I, Marcilla M, Paradela A, López de Castro JA. Species-specific differences in proteasomal processing and tapasin-mediated loading influence peptide presentation by HLA-B27 in murine cells. J Biol Chem. 2003;278:46461–46472. doi: 10.1074/jbc.M308816200. [DOI] [PubMed] [Google Scholar]
- 72.Maier R, Falk K, Rötzschke O, Maier B, Gnau V, Stevanović S, Jung G, Rammensee HG, Meyerhans A. Peptide motifs of HLA-A3, -A24, and -B7 molecules as determined by pool sequencing. Immunogenetics. 1994;40:306–308. doi: 10.1007/BF00189978. [DOI] [PubMed] [Google Scholar]
- 73.Jardetzky TS, Lane WS, Robinson RA, Madden DR, Wiley DC. Identification of self peptides bound to purified HLA-B27. Nature. 1991;353:326–329. doi: 10.1038/353326a0. [DOI] [PubMed] [Google Scholar]
- 74.Saveanu L, Carroll O, Lindo V, Del Val M, Lopez D, Lepelletier Y, Greer F, Schomburg L, Fruci D, Niedermann G, van Endert PM. Concerted peptide trimming by human ERAP1 and ERAP2 aminopeptidase complexes in the endoplasmic reticulum. Nat Immunol. 2005;6:689–697. doi: 10.1038/ni1208. [DOI] [PubMed] [Google Scholar]
- 75.Serwold T, Gonzalez F, Kim J, Jacob R, Shastri N. ERAAP customizes peptides for MHC class I molecules in the endoplasmic reticulum. Nature. 2002;419:480–483. doi: 10.1038/nature01074. [DOI] [PubMed] [Google Scholar]
- 76.Ortmann B, Copeman J, Lehner PJ, Sadasivan B, Herberg JA, Grandea AG, Riddell SR, Tampé R, Spies T, Trowsdale J, Cresswell P. A critical role for tapasin in the assembly and function of multimeric MHC class I-TAP complexes. Science. 1997;277:1306–1309. doi: 10.1126/science.277.5330.1306. [DOI] [PubMed] [Google Scholar]
- 77.Sesma L, Galocha B, Vázquez M, Purcell AW, Marcilla M, McCluskey J, López de Castro JA. Qualitative and quantitative differences in peptides bound to HLA-B27 in the presence of mouse versus human tapasin define a role for tapasin as a size-dependent peptide editor. J Immunol. 2005;174:7833–7844. doi: 10.4049/jimmunol.174.12.7833. [DOI] [PubMed] [Google Scholar]
- 78.Williams AP, Peh CA, Purcell AW, McCluskey J, Elliott T. Optimization of the MHC class I peptide cargo is dependent on tapasin. Immunity. 2002;16:509–520. doi: 10.1016/s1074-7613(02)00304-7. [DOI] [PubMed] [Google Scholar]
- 79.Peh CA, Laham N, Burrows SR, Zhu Y, McCluskey J. Distinct functions of tapasin revealed by polymorphism in MHC class I peptide loading. J Immunol. 2000;164:292–299. doi: 10.4049/jimmunol.164.1.292. [DOI] [PubMed] [Google Scholar]
- 80.Braud VM, McMichael AJ, Cerundolo V. Differential processing of influenza nucleoprotein in human and mouse cells. Eur J Immunol. 1998;28:625–635. doi: 10.1002/(SICI)1521-4141(199802)28:02<625::AID-IMMU625>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 81.Street MD, Doan T, Herd KA, Tindle RW. Limitations of HLA-transgenic mice in presentation of HLA-restricted cytotoxic T-cell epitopes from endogenously processed human papillomavirus type 16 E7 protein. Immunology. 2002;106:526–536. doi: 10.1046/j.1365-2567.2002.01442.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Poland GA, Sohni Y, Domanico M, Kroning CM, DeGoey SR, Jimale M, Jacobson RM, Moore SB. High frequency of HLA-A*0103 allele in a Somali population. Hum Immunol. 2001;62:197–200. doi: 10.1016/s0198-8859(00)00238-x. [DOI] [PubMed] [Google Scholar]
- 83.Sidney J, Peters B, Frahm N, Brander C, Sette A. HLA class I supertypes: a revised and updated classification. BMC Immunol. 2008;9:1–15. doi: 10.1186/1471-2172-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Firat H, Tourdot S, Ureta-Vidal A, Scardino A, Suhrbier A, Buseyne F, Rivìere Y, Danos O, Michel ML, Kosmatopoulos K, Lemonnier FA. Design of a polyepitope construct for the induction of HLA-A0201-restricted HIV 1-specific CTL responses using HLA-A*0201 transgenic, H-2 class I KO mice. Eur J Immunol. 2001;31:3064–3074. doi: 10.1002/1521-4141(2001010)31:10<3064::aid-immu3064>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 85.Wong FS, Karttunen J, Dumont C, Wen L, Visintin I, Pilip IM, Shastri N, Pamer EG, Janeway CA., Jr Identification of an MHC class I-restricted autoantigen in type 1 diabetes by screening an organ-specific cDNA library. Nat Med. 1999;5:1026–1031. doi: 10.1038/12465. [DOI] [PubMed] [Google Scholar]
- 86.Price P, Witt C, Allcock R, Sayer D, Garlepp M, Kok CC, French M, Mallal S, Christiansen F. The genetic basis for the association of the 8.1 ancestral haplotype (A1, B8, DR3) with multiple immunopathological diseases. Immunol Rev. 1999;167:257–274. doi: 10.1111/j.1600-065x.1999.tb01398.x. [DOI] [PubMed] [Google Scholar]