Abstract
Background
Wilson disease (WD) is characterized by hepatic copper accumulation with progressive liver damage to cirrhosis. This study aimed to characterize the toxic milk mouse from The Jackson Laboratory (Bar Harbor, ME, USA) (tx-j) mouse model of WD according to changes over time in hepatic copper concentrations, methionine metabolism, global DNA methylation, and gene expression from gestational day 17 (fetal) to adulthood (28 weeks).
Methods
Included liver histology and relevant biochemical analyses including hepatic copper quantification, S-adenosylmethionine (SAM) and S-adenosylhomocysteine (SAH) liver levels, qPCR for transcript levels of genes relevant to methionine metabolism and liver damage, and DNA dot blot for global DNA methylation.
Results
Hepatic copper was lower in tx-j fetuses but higher in weanling (three weeks) and adult tx-j mice compared to controls. S-adenosylhomocysteinase transcript levels were significantly lower at all time points, except at three weeks, correlating negatively with copper levels and with consequent changes in the SAM:SAH methylation ratio and global DNA methylation.
Conclusion
Compared to controls, methionine metabolism including S-adenosylhomocysteinase gene expression is persistently different in the tx-j mice with consequent alterations in global DNA methylation in more advanced stages of liver disease. The inhibitory effect of copper accumulation on S-adenosylhomocysteinase expression is associated with progressively abnormal methionine metabolism and decreased methylation capacity and DNA global methylation.
Keywords: Wilson disease, copper, DNA, methylation, gene expression
1. Introduction
Wilson disease (WD) is an autosomal recessive disorder characterized by mutations in the ATP7B gene which is responsible for copper (Cu) metabolism and excretion [1], with Cu accumulation in liver [2], and brain [3], that leads to progressive liver damage, as well as neurological and psychiatric manifestations [4,5]. The toxic milk mouse model of WD from The Jackson Laboratory (tx-j) has a G712D missense mutation in the second transmembrane region of ATP7B, which leads to hepatic Cu accumulation similar to the disease described in humans [6]. This Cu accumulation in hepatocytes results in microvesicular lipid droplets in association with damage to mitochondria [5], nuclei [7], and endoplasmic reticulum (ER) [8]. In addition, hepatic Cu accumulation correlates with down-regulation of gene transcripts related to lipid metabolism [9] including cholesterol synthesis [10].
Previous studies with the tx-j mouse model of WD have shown a progressive accumulation of liver Cu over 12 months, along with increases in hepatic apoptotic cells and hepatocyte metallothionein levels [11]. Apoptotic cell damage in the first six months of life was associated with increased levels of Commd1, a Cu binding protein, while decreased levels of proteins associated with inhibition of apoptosis were observed by eight months [4]. Reductions in liver Cu can be achieved in tx-j mice using the chelator tetrathiomolybdate [12].
We have previously demonstrated a close relationship between Cu accumulation and methionine metabolism in tx-j mice [9], as others have shown in other animal models of WD [13].
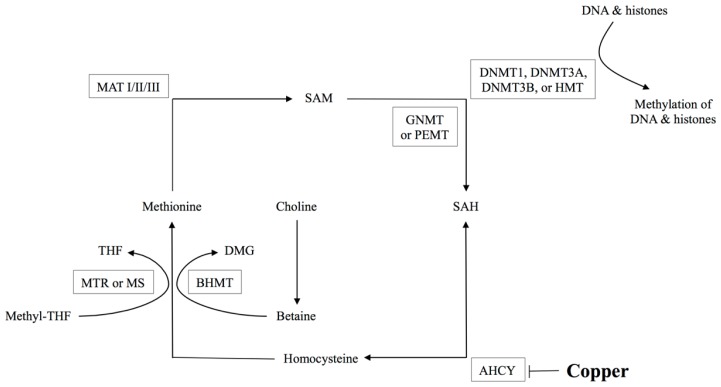
As shown in Figure 1, methionine metabolism is essential for the production of methyl groups used in transmethylation reactions.
Figure 1.
Methionine metabolism. S-adenosylmethionine (SAM) is the principal methyl donor for DNA and histone methylation reactions, whereas S-adenosylhomocysteine (SAH) inhibits all SAM-dependent methylation reactions. S-adenosylhomocysteinase (AHCY) is the bi-directional enzyme that hydrolyzes SAH to generate homocysteine. Methionine synthase (MTR or MS) remethylates homocysteine to form methionine that is in turn converted to SAM via the enzyme methionine adenosyltransferase (isoenzymes MATI/II/III). Homocysteine can also be remethylated to methionine via the betaine homocysteine methyltransferase (BHMT) catalyzed reaction that utilizes betaine. Dimethylglycine (DMG) is a byproduct of the BHMT catalyzed reaction and is also a potent inhibitor of BHMT. Tetrahydrofolate (THF); DNA methyltransferase (DNMT); histone methyltransferase (HMT); glycine N-methyltransferase (GNMT); phosphatidylethanolamine N-methyltransferase (PEMT); DNMTs, GNMT, and PEMT are major consumers of SAM in the liver [14].
Methionine is an essential amino acid that must be ingested to ensure adequate provisions. In addition, methionine is generated from homocysteine via methionine synthase (MS or methionine transferase reductase (MTR) and BHMT-catalyzed reactions and is converted into S-adenosylmethionine (SAM), the main methyl donor for transmethylation reactions. DNA methyltransferases (DNMTs) catalyze DNA methylation reactions with production of S-adenosylhomocysteine (SAH). The maintenance of stable and balanced levels of SAM and SAH is crucial for cell physiology, as SAM is the main substrate for methylation reactions [15], whereas SAH is the major inhibitor for the same reactions. The ratio of SAM:SAH may be considered a relative index of methylation capacity [16], which in normal mouse liver tissue has been previously described to be in the range of 3 to 4 [14]. SAH regenerates homocysteine through S-adenosylhomocysteinase (AHCY), a bidirectional enzyme that favors the generation of SAH if the products homocysteine and adenosine are not removed. AHCY can be inhibited by Cu accumulation that results in elevated levels of SAH [9,17,18]. We recently associated hepatic Cu accumulation with reduced Ahcy gene expression and activity, subsequent elevations of SAH levels, reductions in the SAM: SAH ratio, and reduced DNA methylation. The above were associated with altered expression of genes relevant to liver injury [9]. This study also demonstrated that transcript levels of select genes related to methionine metabolism are down-regulated in tx-j mice and respond to choline supplementation with restoration of gene transcripts to control levels in fetal livers [19]. Others have shown that methionine metabolism and the need for methyl groups change over time, with the requirement for methyl groups being highest during gestational life [20].
In order to study the progression of WD, the present study examined the development of liver histology, methionine metabolism, transcript levels of selected genes central to lipid and methionine metabolism and global DNA methylation levels in tx-j mice from gestational day 17 (fetal) to postnatal week 28 (28 weeks). The study provides new temporal insights into relationships between hepatic Cu accumulation and liver damage, altered methionine metabolism, hepatic global DNA methylation, and regulation of gene expression.
2. Results and Discussion
2.1. Body and Liver Weights, Copper and Iron Status, SAM and SAH Levels, and Liver Histology
As shown in Table 1, body weights of the tx-j mice were significantly lower at three through 28 weeks of age than those of controls. Liver weights in the tx-j mice at three and 12 weeks of age were lower than those of controls. There was a significant increase in the liver/body weight ratio in the tx-j group at 20 and 28 weeks of age that coincided with a worsening of liver histology.
Table 1.
Developmental changes (fetal–28 weeks) in body and liver weights, hepatic copper and iron, and SAM and SAH concentrations. Values are expressed as mean ± SD.
| Analysis | Fetal | 3 weeks | 12 weeks | 20 weeks | 28 weeks | |||||
|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|
||||||
| Control (n = 8) | Tx-j (n = 5) | Control (n = 11) | Tx-j (n = 10) | Control (n = 8) | Tx-j (n = 7) | Control (n = 9) | Tx-j (n = 6) | Control (n = 7) | Tx-j (n = 7) | |
| Body weight (g) | ND | ND | 9.29 ± 1.03 | 7.91 ± 0.84 * | 32.3 ± 2.42 | 27.1 ± 1.45 * | 34.8 ± 2.14 | 28.3 ± 1.10 * | 38.7 ± 4.31 | 32.2 ± 2.58 * |
| Liver weight (g) | ND | ND | 0.40 ± 0.060 | 0.35 ± 0.045 * | 1.65 ± 0.12 | 1.40 ± 0.068 * | 1.74 ± 0.14 | 1.72 ± 0.089 | 1.84 ± 0.27 | 1.68 ± 0.27 |
| Liver/body ratio | ND | ND | 0.043 ± 0.0035 | 0.044 ± 0.0024 | 0.051 ± 0.0017 | 0.052 ± 0.0024 | 0.050 ± 0.0026 | 0.061 ± 0.0030 * | 0.047 ± 0.0021 | 0.052 ± 0.0045 * |
| Copper (μg/g) ** | 7.83 ± 0.72 | 1.49 ± 0.48 * | 12.17 ± 1.72 | 30.65 ± 7.73 * | 5.17 ± 0.99 | 223.16 ± 52.8 * | 7.44 ± 1.84 | 351 ± 40.5 * | 3.86 ± 0.19 | 190 ± 23.4 *,• |
| Iron (μg/g) ** | 53.34 ± 7.73 | 48.94 ± 6.08 | 23.54 ± 3.39 | 26.16 ± 5.68 | 68.76 ± 11.54 | 79.52 ± 35.84 | 84.60 ± 13.27 | 91.66 ± 3.42 | 97.58 ± 14.92 | 124.07 ± 7.89 *,• |
| SAM (nmol/g) | 105.19 ± 24.83 | 82.13 ± 14.52 | 86.97 ± 17.82 | 122.55 ± 50.37 * | 84.9 ± 11.9 | 76.6 ± 26.3 | 82.7 ± 17.2 | 99.9 ± 10.6 | 74.4 ± 13.2 | 77.2 ± 7.91 |
| SAH (nmol/g) | 4.04 ± 1.21 | 4.67 ± 2.88 | 25.94 ± 8.86 | 36.55 ± 13.35 * | 42.5 ± 10.4 | 57.2 ± 15.7 * | 56.6 ± 26.3 | 67.7 ± 13.4 | 28.2 ± 16.8 | 34.5 ± 11.4 |
| SAM/SAH ratio | 27.59 ± 10.16 | 23.17 ± 12.77 | 3.89 ± 1.84 | 3.39 ± 0.89 | 2.14 ± 0.69 | 1.41 ± 0.59 * | 1.82 ± 1.13 | 1.51 ± 0.26 | 3.76 ± 1.33 | 2.12 ± 0.52 |
Values with * are significantly different (p < 0.05) between tx-j vs. control at each time point; ** Copper and iron concentrations at the three weeks time points are from livers of 8 controls and 5 tx-j mice; Values with • are significantly different (p < 0.05) within the same genotype between 20 and 28 weeks. ND = not determined. Fetal and three weeks data were previously published [19].
Liver Cu concentrations in fetal tx-j mice were significantly lower than in controls but by three weeks postnatal and thereafter, values were 2–50 times higher than in controls. Liver Cu concentration peaked at 20 weeks of age with significant age and genotype interaction (both p < 0.0001) indicating that liver Cu concentrations differed significantly over time between the tx-j and control mice. Liver iron concentrations in tx-j mice were similar to control levels at most time points, although values in the tx-j mice were 1.2 times higher than in controls at 28 weeks.
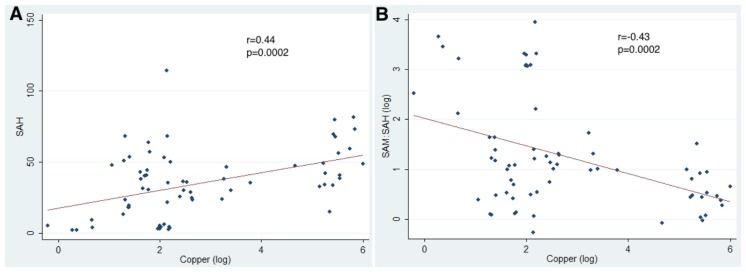
Postnatal hepatic SAM levels were higher in tx-j mice than in controls at three weeks with a significant interaction of age with genotype (p = 0.01) when examined over all time points. Liver SAH levels were similar in both groups at all time points except at three and 12 weeks with an overall significant interaction between age and genotype (p < 0.05) over time. The ratio of SAM to SAH (SAM:SAH ratio) was significantly lower at 12 weeks in the tx-j mice compared to ratios in the controls. When data from all time points were pooled, hepatic Cu concentrations were positively correlated with SAH levels (r = 0.44; p = 0.0002) and negatively correlated with SAM:SAH methylation ratio (r = −0.43; p = 0.0002) (Figure 2). These findings are compatible with the known inhibitory effect of Cu on Ahcy expression and enzymatic activity [9] with a predictive decrease in methylation capacity.
Figure 2.
(A) Correlation between hepatic Cu concentration and SAH levels; (B) Correlation between hepatic Cu concentration and SAM:SAH levels (using data from all time points).
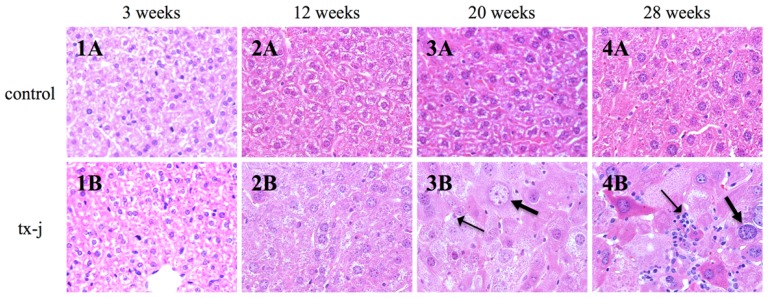
Although there were no changes at 12 weeks, we observed an increase in lymphocyte and PMN infiltration and in hepatocyte size in the tx-j group at 20 weeks. At 28 weeks, all the tx-j mice had increased hepatocyte sizes with giant nuclei, necrosis, increased lymphocytes, and PMNs. There were no histological changes in the control mice at all time points (Figure 3).
Figure 3.
Histology images of tx-j and control livers from three to 28 weeks of age. All hematoxylin and eosin stained, 436×. Whereas liver histology was normal in tx-j mice at both three and 12 weeks of age (1B and 2B), there was an increase in inflammatory infiltrates at 20 and 28 weeks (3B and 4B, thin arrows), in association with giant nuclei and markedly increased cell size of the hepatocytes (3B and 4B, thick arrows). Note that in tx-j mice hepatocyte cell diameters are 2.5 times and nuclear diameters are about two times larger than control mice [19]. Control mice had normal liver histology at all time points (1A, 2A, 3A and 4A).
2.2. Transcript Levels of Selected Genes Related to Methionine and Lipid Metabolism
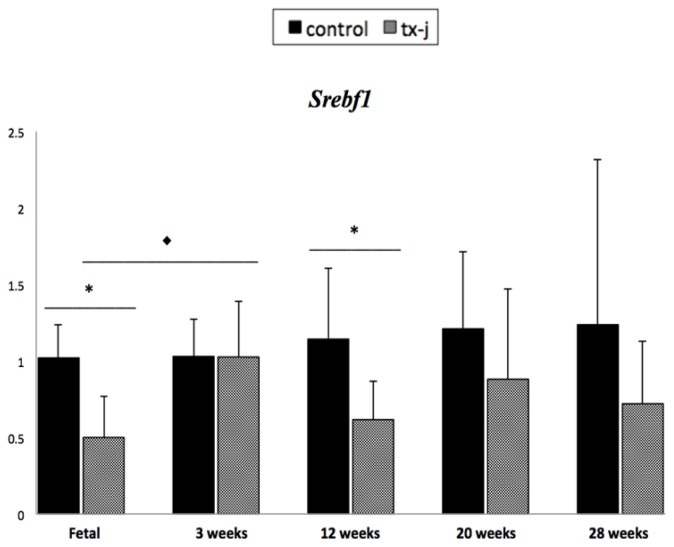
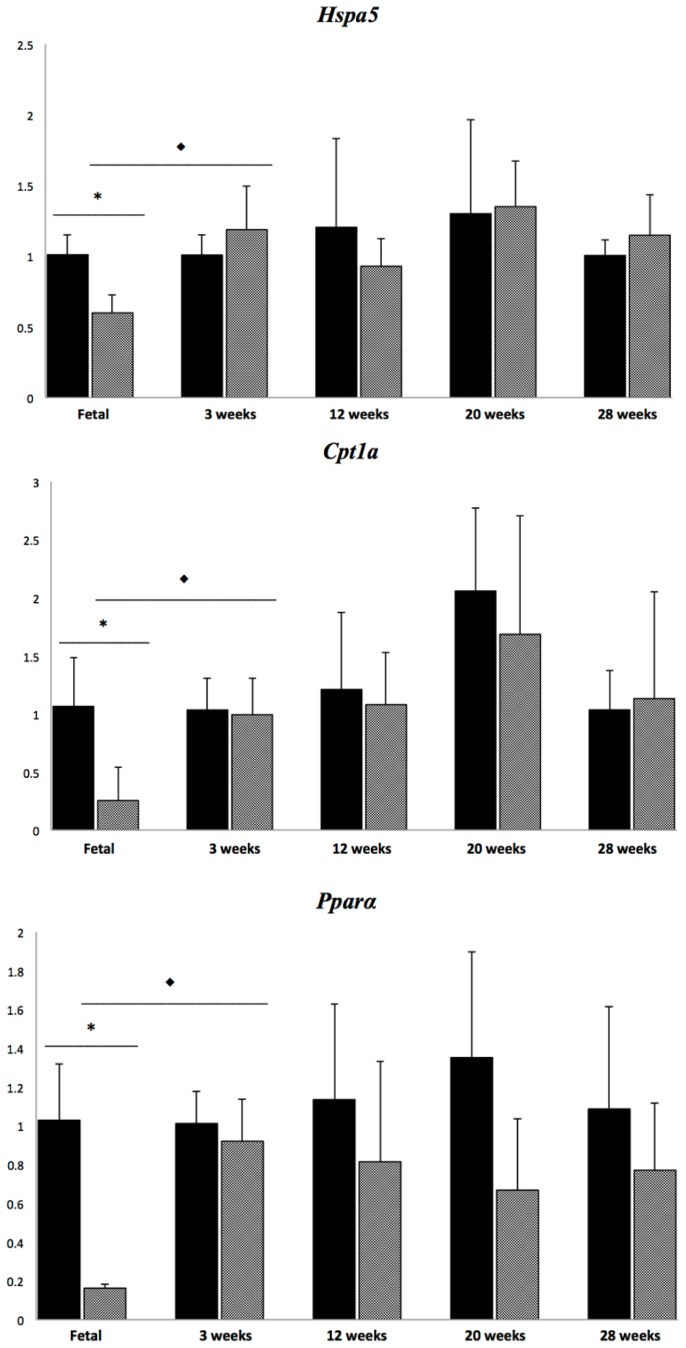
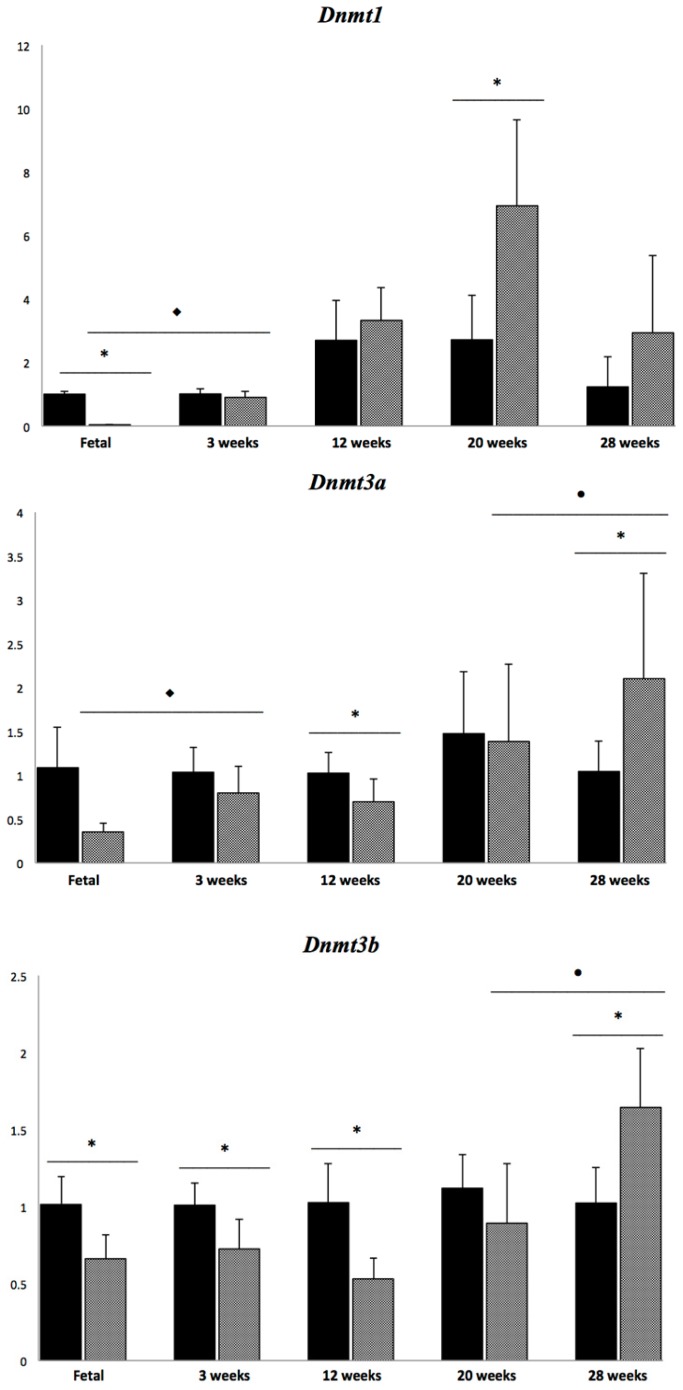
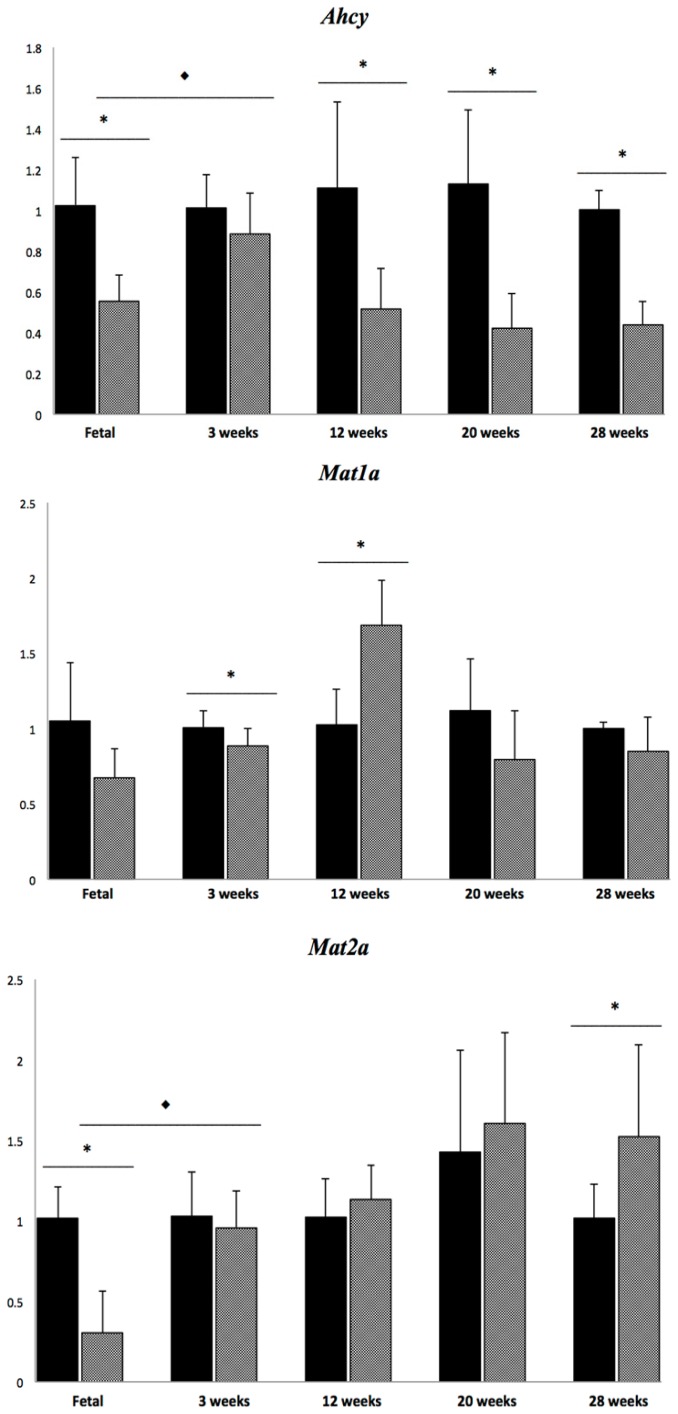
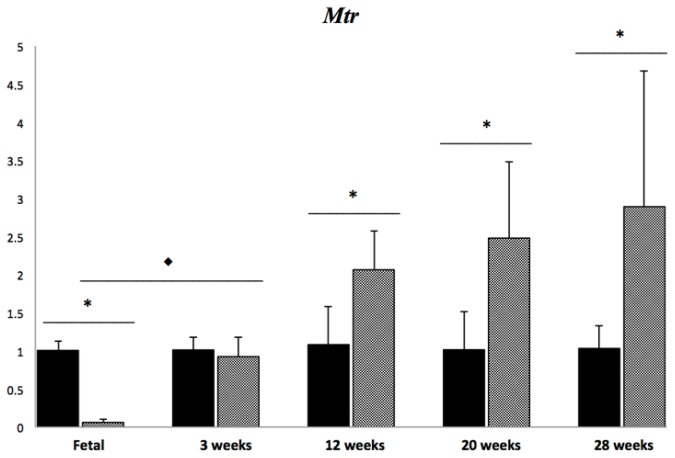
Transcript levels of 11 different genes were quantified including genes central to lipogenesis, fatty acid oxidation, methionine metabolism, and DNA methylation (Figure 4).
Figure 4.
Transcript levels of genes related to lipid and methionine metabolism. x-axis: time points; y-axis: mean ± SD of relative expression of target genes normalized to Gapdh. Values with * are significantly different (p < 0.05) between groups within each time point. Values with ♦ are significantly different (p < 0.05) within the same genotype between fetal and three weeks. Values with ● are significantly different (p < 0.05) within the same genotype between 20 and 28 weeks. Genes include Srebf1 (sterol regulatory element-binding protein) for lipogenesis, Hspa5 (heat shock protein 5) for ER stress, Cpt1A (carnitine palmitoyl transferase 1A) and Pparα (peroxisome proliferator-activated receptor α) for fatty acid oxidation, Dnmt1 (DNA methyltransferase 1), Dnmt3a (DNA methyltransferase 3a), Dnmt3b (DNA methyltransferase 3b), Ahcy (S-adenosylhomocysteinase), Mat1a (methionine adenosyltransferase 1a), Mat2a (methionine adenosyltransferase 2a), and Mtr (methionine transferase reductase or methionine synthase) for methionine metabolism and DNA methylation. Fetal and three weeks data were previously published [19].
As previously reported [19], most of the fetal liver genes that were studied were down-regulated in the tx-j in comparison to control mice, with the exceptions of Dnmt3a and Mat1a. Subsequently, when comparing differences between gene transcript levels in control and tx-j mice at different time points, Ahcy was persistently down-regulated in tx-j mice except at three weeks, while Srebf1 and Dnmt3a were down-regulated in tx-j mice at 12 weeks. Dnmt1 was up-regulated at 20 weeks and Dnmt3b was down-regulated at three and 12 weeks. Both Dnmt3a and Dnmt3b were up-regulated in tx-j mice at 28 weeks. Mat1a was down-regulated at three weeks but was up-regulated at 12 weeks, and Mat2a was up-regulated in tx-j mice at 28 weeks. Mtr was up-regulated in tx-j mice at 12, 20, and 28 weeks. When comparing transcript levels in tx-j mice at different ages, transcript levels of almost all genes (except for Dnmt3b and Mat1a) were up-regulated at three weeks compared to fetal livers, whereas Dnmt3a and Dnmt3b levels were up-regulated at 28 weeks compared to 20 weeks. Consistent with our proposed interaction of Cu with methionine metabolism while considering all time points, Ahcy transcript levels were negatively correlated with hepatic Cu concentration (r = −0.58; p < 0.0001), and hepatic Cu was positively correlated with the expressions of Dnmt1 and Dnmt3a (r = 0.68, p < 0.0001 and r = 0.28, p = 0.01, respectively).
2.3. Hepatic Global DNA Methylation
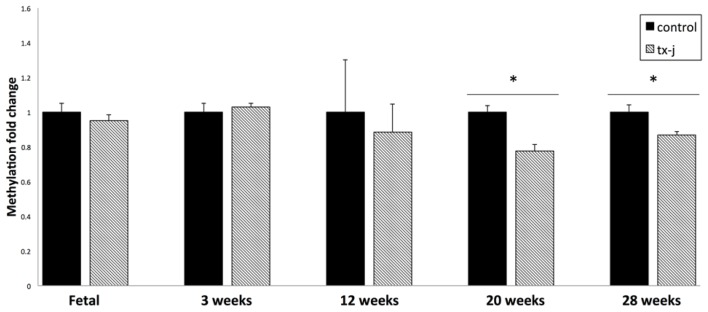
DNA dot blots for global DNA methylation showed that there were no group differences in DNA methylation in fetal, three weeks, or 12 weeks old livers between tx-j and control mice. Starting at 20 weeks of age, tx-j mice DNA showed global hypomethylation compared to control mice with a significant interaction with age (p < 0.001), and this pattern persisted at 28 weeks (Figure 5). Pooling all time points, global DNA methylation levels were negatively correlated with Dnmt3a and Dnmt3b transcript levels (r = −0.26, p = 0.26; r = −0.27, p = 0.01) and were positively correlated with Mat1a transcript levels (r = 0.46, p < 0.001).
Figure 5.
Hepatic global DNA methylation. Values with * are significantly different (p < 0.05) between tx-j vs. control within each time point. Values are expressed as fold change ± SD. Fetal and three weeks data were previously published [19].
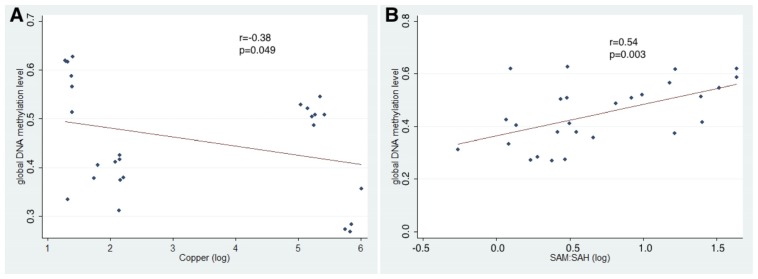
In addition, when using data only from 20 and 28 weeks old tx-j and control mice, global DNA methylation was negatively correlated with Cu concentrations (r = −0.38; p = 0.049) and SAH levels (r = −0.69; p ≤ 0.0001) and positively correlated with SAM:SAH ratio (r = 0.54; p = 0.003) (Figure 6).
Figure 6.
(A) Correlation between hepatic Cu concentration and global DNA methylation; (B) Correlation between SAM:SAH ratio and global DNA methylation (using data from 20 and 28 week old mice).
The tx-j mice represent a valid model of WD and offer the opportunity to study the relations between Cu accumulation and methionine metabolism and their consequences on DNA methylation and gene expression. The rationale underlying our study was to describe the temporal relations between changes in methionine metabolism, DNA methylation, and gene expression, given that epigenetic marks are very dynamic, subject to influences that start during the early development and continue throughout life as a consequence of continuous environmental changes. The major findings of our study include the following: first that hepatic Cu was significantly lower in fetal livers of tx-j than in controls but postnatal values increased significantly over time with a plateau starting at 20 weeks of age; in contrast there were only minor differences in liver Fe concentrations. The finding that liver Cu concentrations were lower in fetal tx-j than controls may be attributed to the fact that adult tx-j dams are characterized by low circulating ceruloplasmin levels, with a putative Cu transport protein [19]. The observation that liver Cu concentrations reached a plateau around five months of age has been described before in other murine models of hepatic Cu accumulation [21] and may be attributed to adaptation mechanisms to hepatic Cu accumulation [4]. Hepatic Fe accumulation, as shown at 28 weeks, has been described previously in the LEC rat model of WD [22] and it can be speculated that this Fe accumulation plays a role in liver damage progression. Kato et al. [22] demonstrated that Fe-deficient diet could even prevent the development of fulminant hepatitis in LEC rats. While the mechanisms of hepatic Fe accumulation have yet to be elucidated, it is reasonable to speculate that since ceruloplasmin can contribute to Fe transport through hepatocyte basolateral membranes, low levels of this protein in WD may contribute to hepatic Fe accumulation [23]. Second, whereas almost all measured gene transcript levels were down-regulated in the tx-j fetal livers relative to control values, low levels of Ahcy persisted throughout all other time points. As a consequence of Ahcy down-regulation, the SAM:SAH methylation ratio was lower at 12 weeks of age although not consistently at all time points. Similarly, both SAM and SAH did not change consistently over time as expected since SAM was increased only at three weeks and SAH was increased only at three and 12 weeks which may be related to increased Mtr and Mat transcript and activity levels at this time points. A previous study reported transcript levels of enzymes related to methionine metabolism and mean SAM levels of 112.8 ± 12.4 nmol/g, SAH levels of 25.5 ± 3.9 nmol/g and their ratio of 4.4 ± 0.8 in mouse liver tissue [14]. Even though it is difficult to compare different mouse strains and animal ages, our SAM and SAH data in fetal livers are comparable with a previous report indicating very low SAH levels in fetal livers [24]. However, our present postnatal data are similar to the previous report in tx-j and control mice only at three weeks of age, whereas at later time points our SAM:SAH ratio is lower (range 1.8–2) due to progressive reduction of SAM levels relative to progressive increase of SAH levels. In an unrelated study, untreated C57BL/6J mice had similar SAM:SAH ratio of 2 ± 0.6 at five months of age [25]. The effects of Cu levels on methionine metabolism are also emphasized by the positive correlation between Cu and SAH levels, and negative correlation between Cu with Ahcy transcript levels and the SAM:SAH ratio. Two previous studies in toxic milk mice showed that Ahcy has Cu binding properties and its hepatic transcript levels can be reduced up to 42% in association with Cu accumulation [17,26]. Interestingly, Bethin et al. showed that Cu deficiency was also associated with 45% reduction of Ahcy hepatic levels [26], a finding that is similar to our results in fetal tx-j livers and that indicates that Cu metabolism must be strictly regulated to ensure stable Ahcy levels. Others showed that Ahcy deficiency in a 26-year-old man was associated with myopathy and developmental delay. Electron microscopy of the liver biopsy demonstrated extensive cytoplasmic lipid droplets [27]. Mutation of Ahcy in zebrafish was associated with SAH accumulation, increased levels of TNFα, and hepatic steatosis [28]. Third, although gene transcript levels were all down-regulated in fetal livers of tx-j mice, their improvement to control values after cross-fostering with control dams in all but Dnmt3b, Ahcy, and Mat1a at three weeks [19] suggests that maternal milk components may regulate hepatic gene expression to control levels. Of note, Ahcy transcript levels were reduced as well in fetal livers, similarly to all the other studied genes, despite the fact there was no Cu accumulation. We previously hypothesized that fetal hepatocytes in tx-j mice present dysregulation of cell cycle that has consequences on gene transcript levels, including Ahcy, and is corrected by methyl groups provision [19]. Delgado et al. [13] previously conducted a study on nine-week-old LEC rat models of WD, and showed down-regulation of Ahcy transcript levels. Interestingly, their finding of down-regulation of Mtr is opposite to present data and they failed to demonstrate any difference in SAM or SAH levels compared to wild type rats. These data suggest that various other factors may affect methionine metabolites levels.
Although the gene transcript levels were quite variable over time, global DNA hypomethylation was observed at 20 weeks of age and DNA methylation levels correlated negatively with Dnmt3a and Dnmt3b and positively with Mat1a transcript levels over all time points. The positive correlation between SAM:SAH and global DNA methylation supports the concept that Cu concentrations, methionine metabolism, and global DNA methylation can be tightly interrelated with gene expression regulation. We confirmed that Ahcy transcript levels were the most affected with consequences on SAM:SAH ratio and global DNA hypomethylation at 20 and 28 weeks. In addition, noteworthy, hepatic global DNA hypomethylation observed at the later time points was associated with more advanced inflammatory infiltrate. The finding of an association of decreased global DNA methylation at later time points with inflammatory infiltrates supports our previous results that chronic inflammation in WD is associated with an increased demand for methyl groups and consequent global DNA hypomethylation [9]. Previous reports suggested that progressive liver disease with early indication of fibrosis can be associated with changes in Dnmt levels and global DNA methylation in a mouse model of fibrosis induced by carbon tetrachloride [29]. Another study on human liver biopsies from patients with various degrees of severity of liver disease from chronic hepatitis to cirrhosis and hepatocellular carcinoma showed a progressive increase transcript levels of Dnmts in association with more advanced liver disease [30], an observation that is similar to our current study. In addition, previous reports described increased Dnmts transcript levels correlating with reduced global DNA methylation likely as a result of a compensatory mechanism [31,32]. The design of our present study that is based on timed changes in methionine metabolism and DNA methylation does not allow us to determine if DNA methylation is the cause of worsening liver pathology or if it is epiphenomenon or a consequence of it. However, it is possible that both hypotheses may be true. As shown by our data, the changes in methionine metabolism including down-regulation of Ahcy expression in the fetal liver preceded Cu accumulation and inflammation and amplified liver damage, which in turn could increase a requirement for methyl groups. Global DNA hypomethylation has consequences on the regulation of gene expression as shown in a recent study of progressive liver fibrosis [29]. In addition, improvement of global DNA methylation after folate supplementation was associated with decreased inflammation in gastric mucosa infected by Helicobacter Pylori [33]. To summarize, our results indicate that the tx-j mouse model of WD is characterized by changes in methionine metabolism that are evident from the gestational phase of development onwards. Ahcy down-regulation is persistent over time and is negatively correlated with increasing hepatic Cu concentration, a finding that is consistent with the known inhibitory effects of Cu on its expression [9,26]. As a consequence of Ahcy down-regulation, there is a reduction in liver SAM:SAH levels which correlated with global DNA methylation observed at the later time points.
3. Experimental Section
3.1. Animals and Care
The study was conducted using C3HeB/FeJ-Atp7btx-J/J (tx-j) mice and C3HeB/FeJ (control) mice. All mice were bred in-house on the UC Davis campus (Davis, CA, USA). All animals had access to Purina LabDiet 5001 stock (13 μg Cu, 270 μg Fe, 70 μg Zn per g diet, 28% Kcal protein, 12% Kcal fat, and 60% Kcal carbohydrate) and deionized water ad libitum. Animals were group-housed in polycarbonate cages and maintained according to guidelines set forth by the American Association for Accreditation of Laboratory Animal Care IACUC (Institutional Animal Care and Use Committee, UC Davis, Davis, CA, USA). The animal room was maintained at 20–23 °C and 45%–65% relative humidity with a 14 h light/10 h dark light cycle.
Dams were anesthetized at gestational day 17 (GD17) via CO2 anesthesia followed by cervical dislocation, and fetal livers were pooled and flash-frozen in liquid nitrogen (n = 8 pools of control fetal livers; n = 5 pools of tx-j fetal livers). All postnatal tx-j pups were cross-fostered to a lactating control dam between post-partum day 0 and 6 due to an insufficient amount of Cu to sustain neonatal development and growth in the milk of a tx-j mouse. Mice at 3 (control n = 11; tx-j n = 10), 12 (control n = 8; tx-j n = 7), 20 (control n = 9; tx-j n = 6), and 28 (control n = 7; tx-j n = 7) weeks of age were euthanized via isoflurane anesthetic followed by exsanguination and cervical dislocation. Sections of postnatal livers were placed in formalin or stored in the −80 °C freezer until analysis. Fetal livers and 3 weeks livers have been previously published [19]. The protocol was approved by UC Davis IACUC (protocol#16172, approved on 21 October 2010).
3.2. Hepatic SAM and SAH
Liver levels of SAM and SAH were measured through high-performance liquid chromatography. Liver tissue was homogenized in cold 0.5 N perchloric acid at a ratio of 50 mg tissue: 400 μL perchloric acid and subsequently centrifuged at 14,000 rpm for 10 min. The supernatant was then filtered through a 0.2 μm syringe filter, aliquoted, and stored in the −80 °C freezer until HPLC analysis could be performed to quantify SAM and SAH [34]. HPLC analysis was done within 4 weeks of tissue collection to ensure sample stability. This method has been confirmed independently in another laboratory [35].
3.3. Hepatic Copper and Iron
Approximately 100 mg of liver tissue was digested with concentrated nitric acid and then wet-ashed for analysis using flame atomic absorption spectroscopy [36].
3.4. Liver Histology
Liver tissue from both control and tx-j mice were prepared by staining sections with hematoxylin and eosin. Images were blind-evaluated for mitosis, nuclei, lymphocytes, PMNs, hepatocyte size, and fibrosis.
3.5. Transcript Levels of Selected Genes by qPCR
RNeasy Mini Kit (QIAGEN, Valencia, CA, USA) was used to isolate total RNA from liver tissue. Purity and concentration of extracted RNA was determined by NanoDrop spectrophotometry (Cole-Parmer, IL, USA) and RNA integrity determined by gel electrophoresis. Samples were stored at −80 °C until analysis. cDNA was synthesized using the SuperScript III First-Strand cDNA synthesis kit (Invitrogen, Carlsbad, CA, USA). SYBR green was used to detect transcript levels of 11 selected genes (Table 2); all samples were run in triplicate. Primers for cDNA sequences were designed using AB Tm calculator (http://www6.appliedbiosystems.com/support/techtools/calc/index.cfm), NCBI Primer-BLAST (http://www.ncbi.nlm.nih.gov/tools/primer-blast/), and Premier Biosoft International Beacon Designer (http://www.premierbiosoft.com/qOligo/Oligo.jsp?PID=1). Efficiency of all primers was >95% and specificity checked via melt curve and gel electrophoresis. All primers were used at a concentration of 300 nM except for Mtr primers which were used at a concentration of 900 nM. qPCR was done on the AB ViiA 7 Real-Time PCR System (Applied Biosystem, Foster City, CA, USA).
Table 2.
qPCR primer sequences of selected genes.
| Gene | Sequence 5′ to 3′ | Accession | Position (bp) | Exon-exon overlap | % Primer efficiency |
|---|---|---|---|---|---|
| Srebf1-F | CTGGCTTGGTGATGCTATGTTG | NM_011480.3 | 3901–3922 | No | 104.9 |
| Srebf1-R | GACCATCAAGGCCCCTCAA | 3978–3960 | No | ||
|
| |||||
| Hspa5-F | GTGGAGATCATAGCCAACG | NM 022310.3 | 383–401 | No | 102.9 |
| Hspa5-R | CACATACGACGGCGTGATGC | 433–414 | No | ||
|
| |||||
| Cpt1a-F | GGAGGAGACAGACACCATCCA | NM 013495.2 | 1044–1064 | Yes | 104.1 |
| Cpt1a-R | CGTCATGGTAGAGCCAGACCTT | 1138–1117 | No | ||
|
| |||||
| Pparα-F | CGATGCTGTCCTCCTTGATGA | NM_011144.6 | 1401–1421 | No | 98.5 |
| Pparα-R | GAAGTCAAACTTGGGTTCCATGAT | 1528–1505 | No | ||
|
| |||||
| Dnmt1-F | CCAGCTGCCAAACGGAGA | NM_001199431.1 | 1060–1077 | Yes | 102 |
| Dnmt1-R | CCTCGGGAGTCTCTGGAGCTA | 1123–1103 | No | ||
|
| |||||
| Dnmt3a-F | CACTGGAGTAGGCGCTGAGAC | NM_007872.4 | 7635–7655 | No | 101.6 |
| Dnmt3a-R | CAGCAAAGGGCCTTCCATAG | 7699–7680 | No | ||
|
| |||||
| Dnmt3b-F | CCGTTCGACTTGGTGATTGG | NM_001003961.4 | 2352–2371 | No | 101.3 |
| Dnmt3b-R | GGGCAGGATTGACGTTAGAGAG | 2412–2391 | No | ||
|
| |||||
| Ahcy-F | ATCCTTGGCCGGCACTTT | NM_016661.3 | 949–966 | Yes | 97.1 |
| Ahcy-R | TTCTTTAGCCAGTAGCGGTCCA | 1100–1079 | No | ||
|
| |||||
| Mat1a-F | TCTGTCCCATACTCACCTCTTCAG | NM_133653 | 1683–1706 | No | 98.4 |
| Mat1a-R | TGCCCTGAGGGTAGAAGGC | 1771–1753 | No | ||
|
| |||||
| Mat2a-F | CAGGAGACCAGGGTTTGATGTT | NM_145569 | 517–538 | Yes | 95.8 |
| Mat2a-R | GCGTAACCAAGGCAATGTACC | 653–633 | No | ||
|
| |||||
| Mtr-F | CTGCAGATGTGGCCAGAAAAG | NM_001081128 | 580–600 | Yes | 98.9 |
| Mtr-R | CAGCCACAAACCTCTTGACTCC | 648–627 | No | ||
Reactions were run at 50 °C for 2 min and 95 °C for 10 min, then 40 cycles at 95 °C for 15 s and 60 °C for 1 min. All Cq expression values were normalized to Gapdh and relative expression was calculated using the equation 2−ΔΔCq, where ΔΔCq = ΔCq (sample) − Cq (calibrator).
3.6. Global DNA Methylation via Dot Blot Analyses
DNA was isolated from liver samples using the QIAGEN DNeasy Blood & Tissue Kit (Valencia, CA, USA). An established method was used to measure relative methylation against 5-methylcytosine [32]. 50 ng of genomic DNA was alkaline denatured and spotted on a nitrocellulose membrane. After UV cross-linking, membranes were blocked in LiCor Odyssey Blocking Buffer (LiCor Biosciences, Lincoln, NE, USA), and incubated with anti-5-methylcytosine (Eurogentec, Fremont, CA, USA) in Blocking Buffer + 0.1% Tween-20 overnight at 4 °C. Membranes were washed in 1× PBS + 0.1% Tween-20, followed by incubation with LiCor 700-IR secondary antibody (LiCor Biosciences, Lincoln, NE, USA). Blots were washed again and imaged using the Licor Odyssey Imager (LiCor Biosciences, Lincoln, NE, USA). Blots were rinsed with 2× SSC buffer then equilibrated in PerfectHyb Plus Hybridization Buffer (Sigma, St. Louis, MO, USA) at 42 °C. Blots were hybridized with heat-denatured biotin-labeled gDNA overnight at 42 °C, washed with a high stringency buffer, then blocked again with Blocking Buffer. Blots were further incubated with LiCor Streptavidin 800-IR secondary antibody (LiCor Biosciences, Lincoln, NE, USA) in Blocking Buffer + 0.1% Tween-20 for 1 h at room temperature, then imaged on the LiCor Odyssey Imager (LiCor Biosciences, Lincoln, NE, USA). LiCor Odyssey software (LiCor Biosciences, Lincoln, NE, USA) was used to analyze the integrated intensities. Methylation signal was normalized to the total DNA signal [9] and results expressed as fold change relative to the control group for each time point.
3.7. Statistical Analysis
Statistical analysis was performed using a two-way ANOVA with an interaction term. Where the overall ANOVA was significant, we identified genotypes (tx-j vs. control) and cross-sectional time points (fetal and 3, 12, 20, or 28 weeks of age) that differed significantly using Tukey’s multiple comparison procedure and maintained the family-wise error rate at 0.05. Pearson correlation coefficient and its p-value for significance of correlation were calculated to assess the magnitude and direction of an association between two given variables. For data that were highly skewed, we applied a natural log transformation to achieve normality prior to statistical analysis and significance testing was done on a log-transformed scale. All reported p-values are based on two-sided tests. A p-value <0.05 was considered significant. All statistical analyses were performed using SAS, Version 9.4 (SAS Institute, Cary, NC, USA).
4. Conclusions
In the present study, we observed that transcript levels of genes related to methionine metabolism are aberrant in the liver of the tx-j mouse model of WD from late gestation to adult life in parallel with increasing levels of hepatic Cu and abnormal histopathology. The accumulation of hepatic Cu correlated with decreasing expression of Ahcy, with consequent increases in SAH levels and reduction in SAM:SAH methylation ratios at several time points. In more advanced phases of liver disease, tx-j mice presented global DNA hypomethylation which in turn correlated with the SAM:SAH ratio. The interaction between Cu accumulation and methionine metabolism is a crucial mechanism of disease onset and progression in WD indicating that there is a close connection between genetic and epigenetic mechanisms that ultimately determinates the phenotypic expression of this condition.
Acknowledgments
This research was supported by grant number K08DK084111 from the National Institute of Diabetes and Digestive and Kidney Diseases, by Department and Division funds, and by the National Center for Research Resources (NIH) through grant #UL1 RR024146 (to V.M.), by a Veterans Health Administration Biomedical Laboratory Research and Development National Merit Review grant award BX001155 (to K.K.K.) and by NIH/NIAAA P50-011999 Morphology core (to S.W.F.). V.M. is a full member of the University of California San Francisco Liver Center (Liver Center grant number P30 DK026743). The content is the sole responsibility of the authors and does not necessarily represent the official views of the National Institute of Health.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Author Contributions
Anh Le: conducted part of the experiments and assays, analyzed and interpreted the data, performed the literature search and drafted the manuscript.
Noreene M. Shibata: conducted the animal and laboratory experiments, analyzed the data, contributed to the preparation of the manuscript, and approved the final version of the manuscript.
Samuel W. French: performed histological analysis, contributed to the preparation of the manuscript, and approved the final version of the manuscript.
Kyoungmi Kim: performed the statistical analysis, contributed to the data analysis and manuscript preparation, and approved the final version of the manuscript.
Kusum K. Kharbanda: performed part of the assays, contributed to the manuscript preparation, and approved the final version of the manuscript.
Mohammad S. Islam: performed part of the assays, contributed to the manuscript preparation, and approved the final version of the manuscript.
Janine M. LaSalle: contributed to the data interpretation, manuscript preparation, and approved the final version of the manuscript.
Charles H. Halsted: contributed to the data interpretation, contributed to the manuscript preparation, and approved the final version of the manuscript.
Carl L. Keen: contributed to the experiments and laboratory assays, contributed to the manuscript preparation, and approved the final version of the manuscript.
Valentina Medici: developed the original idea of the study, interpreted the data, performed part of the literature search, drafted the manuscript, and approved the final version of the manuscript.
References
- 1.Geng J., Wang J., Yao R.E., Liu X.Q., Fu Q.H. Identification of one novel and nine recurrent mutations of the ATP7B gene in 11 children with Wilson disease. World J. Pediatr. 2013;9:158–162. doi: 10.1007/s12519-012-0388-7. [DOI] [PubMed] [Google Scholar]
- 2.Faa G., Nurchi V., Demelia L., Ambu R., Parodo G., Congiu T., Sciot R., van Eyken P., Silvagni R., Crisponi G. Uneven hepatic copper distribution in Wilson’s disease. J. Hepatol. 1995;22:303–308. doi: 10.1016/0168-8278(95)80283-5. [DOI] [PubMed] [Google Scholar]
- 3.Faa G., Lisci M., Caria M.P., Ambu R., Sciot R., Nurchi V.M., Silvagni R., Diaz A., Crisponi G. Brain copper, iron, magnesium, zinc, calcium, sulfur and phosphorus storage in Wilson’s disease. J. Trace Elem. Med. Biol. 2001;15:155–160. doi: 10.1016/S0946-672X(01)80060-2. [DOI] [PubMed] [Google Scholar]
- 4.Roberts E.A., Lau C.H., da Silveira T.R., Yang S. Developmental expression of Commd1 in the liver of the Jackson toxic milk mouse. Biochem. Biophys. Res. Commun. 2007;363:921–925. doi: 10.1016/j.bbrc.2007.09.059. [DOI] [PubMed] [Google Scholar]
- 5.Roberts E.A., Robinson B.H., Yang S. Mitochondrial structure and function in the untreated Jackson toxic milk (tx-j) mouse, a model for Wilson disease. Mol. Genet. Metab. 2008;93:54–65. doi: 10.1016/j.ymgme.2007.08.127. [DOI] [PubMed] [Google Scholar]
- 6.Coronado V., Nanji M., Cox D.W. The Jackson toxic milk mouse as a model for copper loading. Mamm. Genome. 2001;12:793–795. doi: 10.1007/s00335-001-3021-y. [DOI] [PubMed] [Google Scholar]
- 7.Howell J.M., Mercer J.F. The pathology and trace element status of the toxic milk mutant mouse. J. Comp. Pathol. 1994;110:37–47. doi: 10.1016/s0021-9975(08)80268-x. [DOI] [PubMed] [Google Scholar]
- 8.Biempica L., Rauch H., Quintana N., Sternlieb I. Morphologic and chemical studies on a murine mutation (toxic milk mice) resulting in hepatic copper toxicosis. Lab. Investig. 1988;59:500–508. [PubMed] [Google Scholar]
- 9.Medici V., Shibata N.M., Kharbanda K.K., LaSalle J.M., Woods R., Liu S., Engelberg J.A., Devaraj S., Torok N.J., Jiang J.X., et al. Wilson’s disease: Changes in methionine metabolism and inflammation affect global DNA methylation in early liver disease. Hepatology. 2013;57:555–565. doi: 10.1002/hep.26047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huster D., Purnat T.D., Burkhead J.L., Ralle M., Fiehn O., Stuckert F., Olson N.E., Teupser D., Lutsenko S. High copper selectively alters lipid metabolism and cell cycle machinery in the mouse model of Wilson disease. J. Biol. Chem. 2007;282:8343–8355. doi: 10.1074/jbc.M607496200. [DOI] [PubMed] [Google Scholar]
- 11.Deng D.X., Ono S., Koropatnick J., Cherian M.G. Metallothionein and apoptosis in the toxic milk mutant mouse. Lab. Investig. 1998;78:175–183. [PubMed] [Google Scholar]
- 12.Czachor J.D., Cherian M.G., Koropatnick J. Reduction of copper and metallothionein in toxic milk mice by tetrathiomolybdate, but not deferiprone. J. Inorg. Biochem. 2002;88:213–222. doi: 10.1016/s0162-0134(01)00383-x. [DOI] [PubMed] [Google Scholar]
- 13.Delgado M., Perez-Miguelsanz J., Garrido F., Rodriguez-Tarduchy G., Perez-Sala D., Pajares M.A. Early effects of copper accumulation on methionine metabolism. Cell. Mol. Life Sci. 2008;65:2080–2090. doi: 10.1007/s00018-008-8201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen N.C., Yang F., Capecci L.M., Gu Z., Schafer A.I., Durante W., Yang X.F., Wang H. Regulation of homocysteine metabolism and methylation in human and mouse tissues. FASEB J. 2010;24:2804–2817. doi: 10.1096/fj.09-143651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu S.C., Mato J.M. S-adenosylmethionine in liver health, injury, and cancer. Physiol. Rev. 2012;92:1515–1542. doi: 10.1152/physrev.00047.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clarke S., Banfield K. S-adenosylmethionine-dependent methyltransferases. In: Carmel R., Jacobsen D.W., editors. Homocysteine in Health and Disease. Cambridge University Press; Cambridge, UK: 2001. pp. 63–78. [Google Scholar]
- 17.Bethin K.E., Cimato T.R., Ettinger M.J. Copper binding to mouse liver S-adenosylhomocysteine hydrolase and the effects of copper on its levels. J. Biol. Chem. 1995;270:20703–20711. doi: 10.1074/jbc.270.35.20703. [DOI] [PubMed] [Google Scholar]
- 18.Li M., Li Y., Chen J., Wei W., Pan X., Liu J., Liu Q., Leu W., Zhang L., Yang X., et al. Copper ions inhibit S-adenosylhomocysteine hydrolase by causing dissociation of NAD+ cofactor. Biochemistry. 2007;46:11451–11458. doi: 10.1021/bi700395d. [DOI] [PubMed] [Google Scholar]
- 19.Medici V., Shibata N.M., Kharbanda K.K., Islam M.S., Keen C.L., Kim K., Tillman B., French S.W., Halsted C.H., Lasalle J.M. Maternal choline modifies fetal liver copper, gene expression, DNA methylation, and neonatal growth in the tx-j mouse model of Wilson disease. Epigenetics. 2013;9:286–296. doi: 10.4161/epi.27110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zeisel S.H. Nutrition in pregnancy: The argument for including a source of choline. Int. J. Women’s Health. 2013;5:193–199. doi: 10.2147/IJWH.S36610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buiakova O.I., Xu J., Lutsenko S., Zeitlin S., Das K., Das S., Ross B.M., Mekios C., Scheinberg I.H., Gilliam T.C. Null mutation of the murine ATP7B (Wilson disease) gene results in intracellular copper accumulation and late-onset hepatic nodular transformation. Hum. Mol. Genet. 1999;8:1665–1671. doi: 10.1093/hmg/8.9.1665. [DOI] [PubMed] [Google Scholar]
- 22.Kato J., Kobune M., Kohgo Y., Sugawara N., Hisai H., Nakamura T., Sakamaki S., Sawada N., Niitsu Y. Hepatic iron deprivation prevents spontaneous development of fulminant hepatitis and liver cancer in Long-Evans Cinnamon rats. J. Clin. Investig. 1996;98:923–929. doi: 10.1172/JCI118875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pfeiffenberger J., Gotthardt D.N., Herrmann T., Seeßle J., Merle U., Schirmacher P., Stremmel W., Weiss K.H. Iron metabolism and the role of HFE gene polymorphisms in Wilson disease. Liver Int. 2012;32:165–170. doi: 10.1111/j.1478-3231.2011.02661.x. [DOI] [PubMed] [Google Scholar]
- 24.Tsang V., Fry R.C., Niculescu M.D., Rager J.E., Saunders J., Paul D.S., Zeisel S.H., Waalkes M.P., Styblo M., Drobna Z. The epigenetic effects of a high prenatal folate intake in male mouse fetuses exposed in utero to arsenic. Toxicol. Appl. Pharmacol. 2012;264:439–450. doi: 10.1016/j.taap.2012.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Medici V., Schroeder D., Woods R., LaSalle J.M., Geng Y., Shibata N.M., Peerson J., Hodzic E., Dayal S., Tsukamoto H., et al. Methylation and Gene Expression Responses to Ethanol Feeding and Betaine Supplementation in the Cystathionine Beta Synthase-Deficient Mouse. Alcohol. Clin. Exp. Res. 2014 doi: 10.1111/acer.12405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bethin K.E., Petrovic N., Ettinger M.J. Identification of a major hepatic copper binding protein as S-adenosylhomocysteine hydrolase. J. Biol. Chem. 1995;270:20698–20702. doi: 10.1074/jbc.270.35.20698. [DOI] [PubMed] [Google Scholar]
- 27.Buist N.R., Glenn B., Vugrek O., Wagner C., Stabler S., Allen R.H., Pogribny I., Schulze A., Zeisel S.H., Baric I., et al. S-adenosylhomocysteine hydrolase deficiency in a 26-year-old man. J. Inherit. Metab. Dis. 2006;29:538–545. doi: 10.1007/s10545-006-0240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matthews R.P., Lorent K., Manoral-Mobias R., Huang Y., Gong W., Murray I.V., Blair I.A., Pack M. TNFalpha-dependent hepatic steatosis and liver degeneration caused by mutation of zebrafish S-adenosylhomocysteine hydrolase. Development. 2009;136:865–875. doi: 10.1242/dev.027565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Komatsu Y., Waku T., Iwasaki N., Ono W., Yamaguchi C., Yanagisawa J. Global analysis of DNA methylation in early-stage liver fibrosis. BMC Med. Genomics. 2012;5:12. doi: 10.1186/1755-8794-5-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oh B., Kim H., Park H., Shim Y., Choi J., Park C., Park Y.N. DNA methyltransferase expression and DNA methylation in human hepatocellular carcinoma and their clinicopathological correlation. Int. J. Mol. Med. 2007;20:65–73. [PubMed] [Google Scholar]
- 31.Wolstenholme J.T., Taylor J.A., Shetty S.R., Edwards M., Connelly J.J., Rissman E.F. Gestational exposure to low dose bisphenol A alters social behavior in juvenile mice. PLoS One. 2011;6:e25448. doi: 10.1371/journal.pone.0025448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Woods R., Vallero R.O., Golub M.S., Suarez J.K., Ta T.A., Yasui D.H., Chi L.H., Kostyniak P.J., Pessah I.N., Berman R.F., et al. Long-lived epigenetic interactions between perinatal PBDE exposure and Mecp2308 mutation. Hum. Mol. Genet. 2012;21:2399–2411. doi: 10.1093/hmg/dds046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gonda T.A., Kim Y.I., Salas M.C., Gamble M.V., Shibata W., Muthupalani S., Sohn K.J., Abrams J.A., Fox J.G., Wang T.C., et al. Folic acid increases global DNA methylation and reduces inflammation to prevent Helicobacter-associated gastric cancer in mice. Gastroenterology. 2012;142:824–833.e7. doi: 10.1053/j.gastro.2011.12.058. [DOI] [PubMed] [Google Scholar]
- 34.Kharbanda K.K., Rogers D.D., 2nd, Mailliard M.E., Siford G.L., Barak A.J., Beckenhauer H.C., Sorrell M.F., Tuma D.J. Role of elevated S-adenosylhomocysteine in rat hepatocyte apoptosis: Protection by betaine. Biochem. Pharmacol. 2005;70:1883–1890. doi: 10.1016/j.bcp.2005.09.021. [DOI] [PubMed] [Google Scholar]
- 35.Bottiglieri T. Isocratic high performance liquid chromatographic analysis of S-adenosylmethionine and S-adenosylhomocysteine in animal tissues: The effect of exposure to nitrous oxide. Biomed. Chromatogr. 1990;4:239–241. doi: 10.1002/bmc.1130040606. [DOI] [PubMed] [Google Scholar]
- 36.Clegg M.S., Keen C.L., Lonnerdal B., Hurley L.S. Influence of ashing techniques on the concentration of trace elements in animal tissues. I: Wet ashing. Biol. Trace Elem. Res. 1981;3:107–115. doi: 10.1007/BF02990451. [DOI] [PubMed] [Google Scholar]