Abstract
Supramolecular modification of nanoparticle surfaces through threading of cucurbit[7]uril (CB[7]) onto surface ligands is used to regulate protein-nanoparticle interactions.
Nanoparticles (NPs) provide tailorable surface chemistries that allow facile modulation of protein-NP interactions. 1 These interactions are useful tools for modulating enzymatic activity,2 delivering proteins 3 and sensing cancerous cells. 4 Furthermore, regulating protein-NP assemblies offers an effective route to construct novel hybrid materials and devices.5
Supramolecular host-guest chemistry provides an alternative to covalent approaches6 to modulating NP surface functionality.7 The properties of monolayers on NP surface can be efficiently tailored through the complexation with guest molecules, where the physicochemical properties (e.g., hydrophobicity and charge) of the guest molecules are imparted to the NP surface. In reported studies, tailored surface charge, 8 hydrophilicity/phobicity 9 and redox potential 10 of NPs have been achieved through the reversible threading/dethreading of the guest molecules on the NP surface. This supramolecular tailoring approach provides an important “post-synthetic” strategy for surface modification of NPs to regulate molecular recognition and binding strength of the target molecules.8
We report here the use of supramolecular host-guest chemistry to modulate protein-NP interactions through control of the hydrophilicity/phobicity of the NP surface. Gold nanoparticles (AuNPs) functionalized with a diaminohexane motif were modified using cucurbit[7]uril (CB[7])11 to form pseudorotaxane structures (Scheme 1a). Binding of CB[7] to the NP surface modulated the surface properties of NPs, with concomitant regulation of protein-NP interactions. The complexation of the CB[7]-diaminohexane motifs on NP surface was quantified by matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS) and the protein-NP interactions were studied through fluorescence titrations. Increased binding constants (Ks) and binding stoichiometries (n) of the protein-NP complexes were observed with increasing amounts of CB[7] bound on NP surface (Scheme 1b), demonstrating that the CB[7] molecules act as non-covalent regulators for controlling protein-NP interactions in a specific and reversible fashion.
Scheme 1.
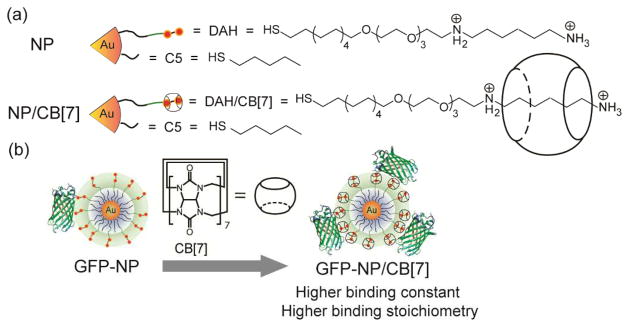
(a) Structure of a mixed monolayer-protected AuNP featuring pentanethiol (C5) and diaminohexane (DAH) terminated thiol ligand. The DAH moiety on the NP surface is a recognition unit for complexation with CB[7]. (b) Schematic of the protein-NP interactions in the absence and presence of the guest molecules CB[7].
Cationic AuNPs featuring diaminohexane (DAH) functional groups were prepared following the reported procedure.12 Briefly, pentanethiol (C5) capped AuNPs (~2 nm core diameter) were synthesized through Brust-Schiffrin two-phase method13 and used for ligand exchange reactions.14 Different amounts of DAH ligand were used in the ligand exchange process to generate two AuNPs with different ligand coverages (NP1 and NP2) (See ESI†). These two NPs allowed us to investigate the role of ligand coverage in the protein-NP interactions. The coverage of DAH ligand on each NP was determined by laser desorption/ionization mass spectrometry (LDI-MS)15 (Fig. S1, ESI†). It was observed that the coverage ratio of DAH ligand on NP1 was 0.73 and that of NP2 was 0.95.
The DAH moiety on the NP surface serves as a recognition unit for complexation with cucurbit[7]uril (CB[7]) (association constant ~1 × 108 M−1).16 Matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS) was used to quantify the number of CB[7] units around a single NP by titrating CB[7] into NP solution (Fig. S2, ESI†). The CB[7] binding amounts per NP were ~ 50 and ~ 100 for NP1 and NP2, respectively.
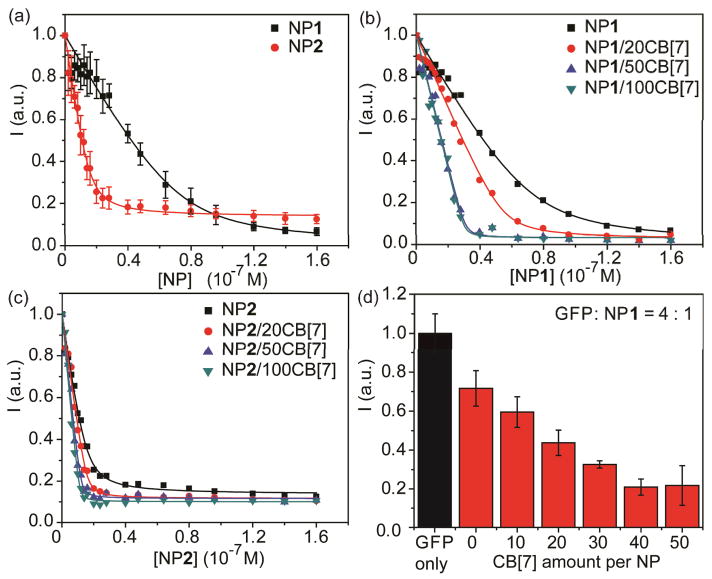
The effect of supramolecularly modified NPs on protein-NP interactions was investigated by studying the binding with a model protein, green fluorescent protein (GFP). In this study, anionic GFP (pI 6.0 at pH 7.4) 17 efficiently bound with positively charged AuNPs, with concomitant fluorescence quenching by AuNPs.4 From the fluorescence titrations, it was clear that the fluorescence intensity of GFP decreased upon the addition of NPs, where NP2 displayed more efficient quenching ability (Fig. 1a). The higher quenching efficacy of NP2 was expected based on the higher number of cationic ligands available to electrostatically bind to GFP surface. These results were consistent with the reported studies demonstrating that the particles with higher cationic ligand coverage can interact more strongly with proteins.18
Fig. 1.
(a) Fluorescence titration of GFP with NP1 and NP2, where the abbreviation “I” represents the fluorescence intensity of GFP. (b) NP1 and (c) NP2 in the presence of different CB[7] amounts on NP surface were titrated with GFP. (d) Fluorescence intensity of the GFP-NP1 complexes in the presence of different CB[7] amounts on NP1 surface at a fixed GFP to NP1 ratio of 4. Each titration experiment was performed in three replicates.
To investigate the effect of CB[7] on the GFP-NP complexations, CB[7]-threaded NPs (NP/CB[7]) were prepared with varying CB[7] to NP ratios. When the NP/CB[7] complexes were titrated with GFP, the particles quenched the fluorescence of GFP in a CB[7] concentration-dependent manner (Fig. 1b and 1c). The binding parameters such as binding constant (Ks) and binding stoichiometry (n) of the GFP-NP conjugates were determined through nonlinear least-squares curve-fitting of the titration data.19 The correlation plots of Ks and n of GFP-NP and the corresponding GFP-NP/CB[7] complexes were shown in Fig. 2. Both the Ks and n values of GFP-NP/CB[7] complexes were higher compared to that of GFP-NP complexes, indicating that the NPs/CB[7] presented an increased protein binding affinity as well as the amount of protein bound. This CB[7]-responsive binding behavior of GFP-NP complexes were further confirmed by the higher fluorescence quenching with increasing CB[7] amounts at a fixed GFP:NP1 ratio (4:1) (Fig. 1d). Taken together, the NP/CB[7] complexes exhibited a greater GFP binding efficiency than the NPs only. In addition, controlling the amount of CB[7] enabled the tuning of protein-NP interactions.
Fig. 2.
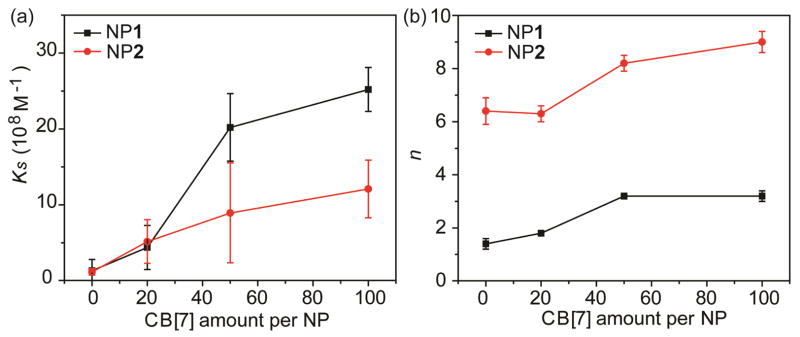
Correlation between (a) Ks and (b) n values of the GFP-NP complexes in the presence of different CB[7] amounts on NP surface
A significant difference in the GFP-NP interactions was reflected in the larger change in the slope of the titration curve for NP1 than that of NP2 at the same amount of CB[7]. At a certain amount of CB[7] the change in Ks for NP1 was much greater than that of NP2, such as the change of Ks for NP1 was ~ 20-fold compared to that for NP2 (~ 8-fold) at the CB[7] to NP ratio of 50. These results demonstrated that a greater impact of CB[7] on regulating the GFP-NP complexations was observed for NP1, indicating that the NPs with lower cationic ligand coverage possessed a broader modulation window. Therefore, the cationic ligand coverage on NPs not only influenced the GFP-NP interactions but also determined the impact of CB[7] on regulating protein-NP binding.
The CB[7] moiety is a good synthetic receptor for amino acids (e.g., tryptophan and phenylalanine), peptides and proteins.20 Thus, it can potentially affect GFP-NP interactions by binding to GFP. To test the effect of CB[7] on the present GFP-NP/CB[7] binding, we used trimethylamine-functionalized NP (NPTMA) as a negative control, wherein CB[7] did not bind to trimethylamine terminal group (Fig. S3a and S3b, ESI†). A minor change in the titration curve of the GFP-NPTMA complexes was observed in the presence of free CB[7] in solution (Fig. S3c, ESI†). From these control studies it can be inferred that CB[7] molecules did not have significant impact on the GFP-NP interactions through binding to surface functionality of GFP.
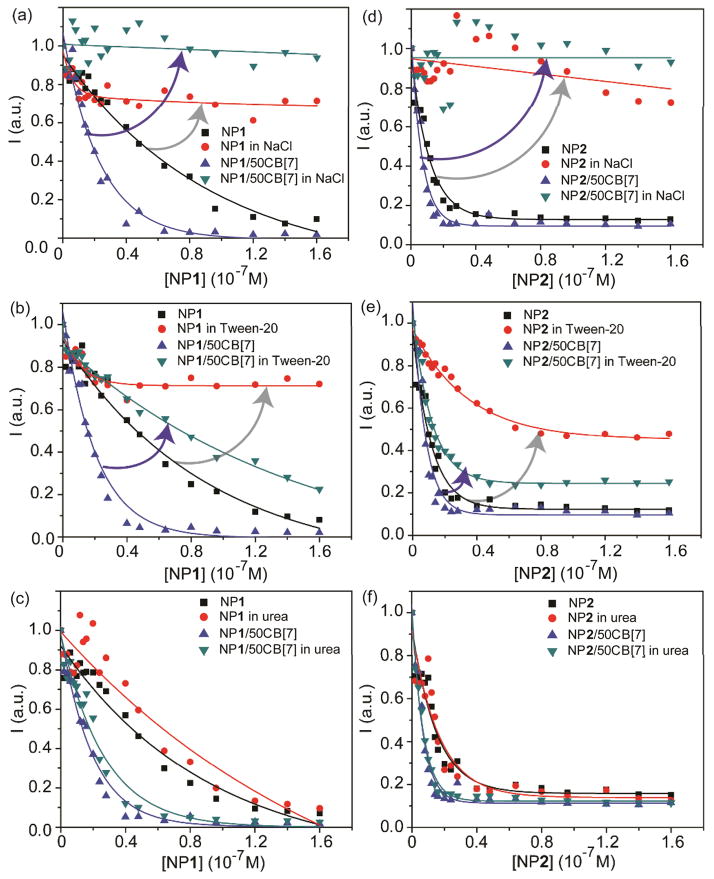
Reversibility of host-guest binding using CB[7] chemistry provides an important tool to modulate surface properties of the supramolecular complexes. We utilized the competitive disruption of the NP/CB[7] complexes by 1-adamantylamine (ADA) to tune the surface properties of the NPs. ADA was used as a competitive guest molecule to dethread CB[7] from NP surface to form more favorable ADA-CB[7] complexes (association constant ~1.7 × 1012 M−1).12,16 After adding ADA to the solution of GFP-NP1/50CB[7] complexes, the fluorescence titration curve of GFP-NP1/50CB[7] was very similar to that of GFP-NP1 (Fig. 3a). A similar result was observed for GFP-NP2/50CB[7] complexes (Fig. 3b), demonstrating the utility of this mode of protein binding regulation.
Fig. 3.
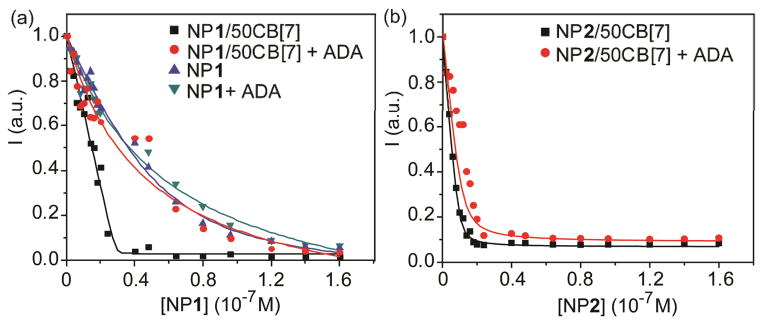
Fluorescence titration curve of (a) GFP-NP1, GFP-NP1/50CB[7] and (b) GFP-NP2/50CB[7] complexes and the responses after the addition of ADA.
We utilized GFP fluorescence to probe the major non-covalent interactions involved in protein-NP and protein-NP/CB[7] complexation, The effect of different chemical additives on the interactions in the GFP-NP and GFP-NP/CB[7] complexes was explored: NaCl (electrostatic attractions), Tween-20 (hydrophobic interactions) and urea (hydrogen bonding). In the presence of these chemical reagents, the fluorescence recovery of GFP was observed when the complexes were treated with NaCl and Tween-20, while the negligible change was shown in the presence of urea (Fig. 4). These results indicated that both electrostatic and hydrophobic interactions contributed in GFP-NP and GFP-NP/CB[7] complexation. It should be noted that NP/CB[7] still presented a stronger GFP affinity compared to the NP in the presence of Tween-20 that disrupted the hydrophobic interactions (Fig. 4b and 4e), suggesting stronger electrostatic interactions involved in GFP-NP/CB[7] complexes than that in GFP-NP complexes.
Fig. 4.
Different chemical agents were used to interfere with the major interactions in the complexes: (a–c) GFP-NP1 and GFP-NP1/50CB[7] complexes; (d–f) GFP-NP2 and GFP-NP2/50CB[7] complexes.
NPs with hydrophobic and charged surface tend to adsorb more proteins due to higher protein binding affinity or the presence of more protein binding sites. 21 CB[n] possesses a hydrophilic exterior and a hydrophobic cavity.11 However, the “equatorial” region of the outer surface of CB[n] is somewhat positively charged as observed in its electrostatic surface potentials22 and is hence able to interact with negatively charged molecules.23 According to our findings, NP/CB[7] presented stronger electrostatic interactions with GFP, implying that the NPs possessed higher cationic surface charge after CB[7] complexation. Therefore, the threading of CB[7] on NP surface may enhance the surface charge of NPs and subsequently increase the protein binding affinity and the amounts of protein binding.
Building on our GFP studies, we next investigated the effect of these supramolecularly tailored NPs on regulating enzyme activity. β-galactosidase (β-gal), a negatively charged protein (pI 4.6 at pH 7.4), can be inhibited by cationic AuNPs through electrostatic complex formation.24 The inhibition of β-gal was studied with NP1, NP1/50CB[7], NP2 and NP2/100CB[7]. The activity of β-gal was monitored by the hydrolysis of the chromogenic substrate chlorophenol red β-D-galactopyranoside (CPRG), where the active β-gal converted yellow substrate into red product to provide a colorimetric readout. As shown in Fig. 5a, the extent of inhibition increased with increasing of NP concentration. It was observed that NP1/50CB[7] and NP2/100CB[7] produced a greater enzyme inhibition than the uncomplexed NP1 and NP2, respectively. As observed with GFP, NP1 exhibited a larger change in the slope of the inhibition curve than that of NP2. NP1 threaded with different amount of CB[7] was further used to inhibit the activity of β-gal at a fixed β-gal:NP ratio (1:15) (Fig. 5b). The β-gal activity was inhibited by NP1 in a CB[7] concentration-dependent manner, indicating the amount of CB[7] threaded on NP1 enabling the fine-tuning of β-gal activity.
Fig. 5.
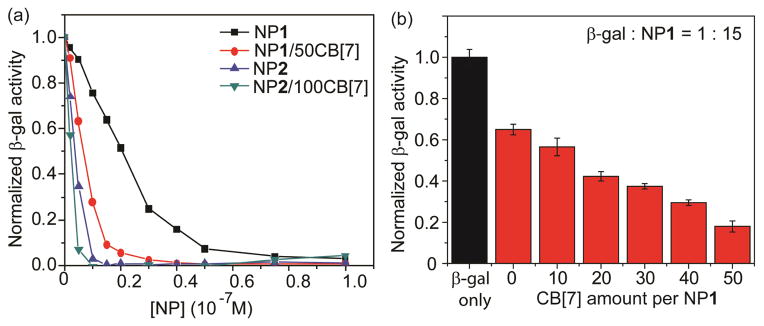
(a) Inhibition of β-gal activity upon addition of different NPs. (b) Inhibition studies of β-gal-NP1 complexes in the presence of different CB[7] amounts on NP1 surface at a fixed NP1 to β-gal ratio of 15. Each titration experiment was performed in three replicates.
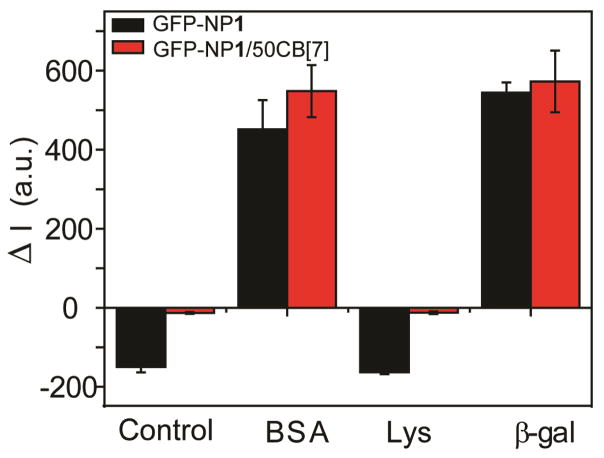
We also investigated the selective protein binding ability of NPs in the absence and presence of CB[7]. Three proteins (i.e. bovine serum albumin (BSA), lysozyme (Lys) and β-gal were added into the GFP-NP1 and GFP-NP1/50CB[7] complex solutions, with different fluorescence responses were generated due to the competitive binding of proteins on NP surface (Fig. 6). The increased fluorescence indicated that GFP was released from NP surface by BSA and β-gal, while the addition of Lys did not show obvious fluorescence change. These results demonstrated that different proteins presented different binding affinities to NP1 and NP1/CB[7]. It also indicated the use of CB[7] can potentially make NPs selectively bind to a target protein, which is a promising property of NPs in the design of biosensors.25
Fig. 6.
Fluorescence response (ΔI) of GFP-NP1 and GFP-NP1/50CB[7] conjugates after the incubation with different proteins at a 100 nM concentration. These GFP-NP conjugates were prepared in a fixed GFP concentration (100 nM) and GFP to NP ratio (1:1). Each titration experiment was performed in three replicates. Protein information: BSA: 66.3 kDa, pI 4.8; Lys: 14.4 kDa, pI 11.0; β-gal: 540 kDa, pI 4.6.
In this study we used NPs with 2 nm core size to demonstrate the tunable surface properties of NPs by adding different amounts of CB[7]. This general strategy can be applied to NPs with different sizes to modulate the NP surface properties as well as the protein-NP interactions. However, the curvature of the NP determines the organization of the self-assembled monolayers (SAMs)26 and protein adsorption27 on NP surface. The addition of CB[7] to the NPs with different curvature may affect the SAMs and regulate the protein-NP interactions. The role of NP curvature along with CB[7] molecules on the SAMs and protein adsorption could be further investigated using computational stimulation28 and analytical tools.29
In summary, we have demonstrated that protein-NP interactions can be efficiently tailored by threading different amounts of CB[7] on NP surface. We showed that supramolecular host-guest chemistry provided a simple and straightforward method to modulate the surface properties of NPs and concomitantly regulate the protein-NP interactions. Taken together, this supramolecular tailoring approach can open new possibilities to effectively control protein stabilization for biotechnology and delivery applications.
Supplementary Material
Acknowledgments
This research was supported by the NIH (EB014277) and the NSF (CHE-1307021). We thank Professor Lyle Isaacs (University of Maryland) for providing cucurbit[7]uril.
Footnotes
Electronic Supplementary Information (ESI) available: preparation of nanoparticles and proteins, mass spectrometry instrumentations and conditions, calculations of ligand coverage ratio and CB[7] binding amounts on NPs, and titration experiments of GFP-NPTMA complexes. See DOI: 10.1039/c000000x/
Notes and references
- 1.(a) Hung A, Mwenifumbo S, Mager M, Kuna JJ, Stellacci F, Yarovsky I, Stevens MM. J Am Chem Soc. 2011;133:1438. doi: 10.1021/ja108285u. [DOI] [PubMed] [Google Scholar]; (b) Gonzalez-Campo A, Brasch M, Uhlenheuer DA, Gomez-Casado A, Yang L, Brunsveld L, Huskens J, Jonkheijm P. Langmuir. 2012;28:16364. doi: 10.1021/la303987c. [DOI] [PubMed] [Google Scholar]; (c) Vigderman L, Khanal BP, Zubarev ER. Adv Mater. 2012;24:4811. doi: 10.1002/adma.201201690. [DOI] [PubMed] [Google Scholar]; (d) Rana S, Yeh YC, Rotello VM. Curr Opin Chem Biol. 2010;14:828. doi: 10.1016/j.cbpa.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Lynch I, Dawson KA. Nano Today. 2008;3:40. [Google Scholar]; (f) Tellechea E, Wilson KJ, Bravo E, Hamad-Schifferli K. Langmuir. 2012;28:5190. doi: 10.1021/la2050866. [DOI] [PubMed] [Google Scholar]; (g) Jamison JA, Bryant EL, Kadali SB, Wong MS, Colvin VL, Matthews KS, Calabretta MK. J Nanopart Res. 2011;13:625. [Google Scholar]; (h) White KA, Rosi NL. Nanomedicine. 2008;7:543. doi: 10.2217/17435889.3.4.543. [DOI] [PubMed] [Google Scholar]; (i) Gentilini C, Pasquato L. J Mater Chem. 2010;20:1403. [Google Scholar]
- 2.(a) Wu Z, Zhang B, Yan B. Int J Mol Sci. 2009;10:4198. doi: 10.3390/ijms10104198. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) You CC, De M, Han G, Rotello VM. J Am Chem Soc. 2005;127:12873. doi: 10.1021/ja0512881. [DOI] [PubMed] [Google Scholar]; (b) Karim Z, Adnan R, Ansari MS. PLoS One. 2012;7:e41422. doi: 10.1371/journal.pone.0041422. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Fischer NO, Verma A, Goodman CM, Simard JM, Rotello VM. J Am Chem Soc. 2003;125:13387. doi: 10.1021/ja0352505. [DOI] [PubMed] [Google Scholar]; (d) Bayir A, Jordan BJ, Verma A, Pollier MA, Cooke G, Rotello VM. Chem Commun. 2006;13:4033. doi: 10.1039/b608928c. [DOI] [PubMed] [Google Scholar]
- 3.(a) Chen YP, Chen CT, Hung Y, Chou CM, Liu TP, Liang MR, Chen CT, Mou CY. J Am Chem Soc. 2013;135:1516. doi: 10.1021/ja3105208. [DOI] [PubMed] [Google Scholar]; (b) Sun X, Zhao Y, Lin VSY, Slowing II, Trewyn BG. J Am Chem Soc. 2011;133:18554. doi: 10.1021/ja2080168. [DOI] [PubMed] [Google Scholar]; (c) Ghosh P, Yang X, Arvizo R, Zhu ZJ, Agasti SS, Mo Z, Rotello VM. J Am Chem Soc. 2010;132:2642. doi: 10.1021/ja907887z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.(a) Rana S, Singla AK, Bajaj A, Elci SG, Miranda OR, Mout R, Yan B, Jirik FR, Rotello VM. ACS Nano. 2013;6:8233. doi: 10.1021/nn302917e. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Bajaj A, Rana S, Miranda OR, Yawe JC, Jerry DJ, Bunz UHF, Rotello VM. Chem Sci. 2010;1:134. [Google Scholar]
- 5.(a) Katz E, Willner I. Angew Chem Int Ed. 2004;43:6042. doi: 10.1002/anie.200400651. [DOI] [PubMed] [Google Scholar]; (b) Rosi NL, Mirkin CA. Chem Rev. 2005;105:1547. doi: 10.1021/cr030067f. [DOI] [PubMed] [Google Scholar]; (c) Hu MH, Qian LP, Brinas RP, Lymar ES, Hainfeld JF. Angew Chem Int Ed. 2007;46:5111. doi: 10.1002/anie.200701180. [DOI] [PubMed] [Google Scholar]; (d) Cheung-Lau JC, Liu D, Pulsipher KW, Liu W, Dmochowski IJ. J Inorg Biochem. 2014;130:59. doi: 10.1016/j.jinorgbio.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 6.(a) Plunkett KN, Mohraz A, Haasch RT, Lewis JA, Moore JS. J Am Chem Soc. 2005;127:14574. doi: 10.1021/ja054666a. [DOI] [PubMed] [Google Scholar]; (b) Fischer NO, Paulini R, Drechsler U, Rotello VM. Chem Commun. 2004;11:2866. doi: 10.1039/b408972c. [DOI] [PubMed] [Google Scholar]; (c) Patil US, Qu H, Caruntu D, O’Connor CJ, Sharma A, Cai Y, Tarr MA. Bioconjugate Chem. 2013;24:1562. doi: 10.1021/bc400165r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cooke G, Garety JF, Hewage SG, Rabani G, Rotello VM, Woisel P. Chem Commun. 2006;13:4119. doi: 10.1039/b608543a. [DOI] [PubMed] [Google Scholar]
- 8.Szarpak A, Burgess C, De Cola L, Huskens J. Chem Eur J. 2013;19:14925. doi: 10.1002/chem.201302153. [DOI] [PubMed] [Google Scholar]
- 9.(a) Mondal A, Jana NR. Chem Commun. 2012;48:7316. doi: 10.1039/c2cc33410k. [DOI] [PubMed] [Google Scholar]; (b) Dorokhin D, Tomczak N, Han M, Reinhoudt DN, Velders AH, Vancso GJ. ACS Nano. 2009;3:661. doi: 10.1021/nn8006515. [DOI] [PubMed] [Google Scholar]
- 10.Klajn R, Fang L, Coskun A, Olson MA, Wesson PJ, Stoddart JF, Grzybowski BA. J Am Chem Soc. 2009;131:4233. doi: 10.1021/ja9001585. [DOI] [PubMed] [Google Scholar]
- 11.(a) Kaifer AE, Li W, Yi S. Isr J Chem. 2011;51:496. [Google Scholar]; (b) Lee JW, Samal S, Selvapalam N, Kim HJ, Kim K. Acc Chem Res. 2003;36:621. doi: 10.1021/ar020254k. [DOI] [PubMed] [Google Scholar]; (c) Das D, Scherman OA. Isr J Chem. 2011;51:537. [Google Scholar]; (d) Masson E, Ling X, Joseph R, Kyeremeh-Mensah L, Lu X. RSC Adv. 2012;2:1213. [Google Scholar]; (e) Lee JW, Samal S, Selvapalam N, Kim HJ, Kim K. Acc Chem Res. 2003;36:621. doi: 10.1021/ar020254k. [DOI] [PubMed] [Google Scholar]
- 12.Kim C, Agasti SS, Zhu Z, Isaacs L, Rotello VM. Nat Chem. 2010;2:962. doi: 10.1038/nchem.858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brust M, Walker M, Bethell D, Schiffrin DJ, Whyman R. J Chem Soc, Chem Commun. 1994;801 [Google Scholar]
- 14.Templeton AC, Wuelfing WP, Murray RW. Acc Chem Res. 2000;33:27. doi: 10.1021/ar9602664. [DOI] [PubMed] [Google Scholar]
- 15.(a) Yan B, Zhu ZJ, Miranda OR, Chompoosor A, Rotello VM, Vachet RW. Anal Bioanal Chem. 2010;396:1025. doi: 10.1007/s00216-009-3250-6. [DOI] [PubMed] [Google Scholar]; (b) Yan B, Jeong Y, Mercante L, Tonga G, Kim C, Zhu ZJ, Vachet RW, Rotello VM. Nanoscale. 5:5063. doi: 10.1039/c3nr01384g. [DOI] [PubMed] [Google Scholar]
- 16.Liu S, Ruspic C, Mukhopadhyay P, Chakrabarti S, Zavalij PY, Isaacs L. J Am Chem Soc. 2005;127:15959. doi: 10.1021/ja055013x. [DOI] [PubMed] [Google Scholar]
- 17.Shaner NC, Steinbach PA, Tsien RY. Nat Methods. 2005;2:905. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 18.Patil S, Sandberg A, Heckert E, Self W, Seal S. Biomaterials. 2007;28:4600. doi: 10.1016/j.biomaterials.2007.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Phillips RL, Miranda OR, Mortenson DE, Subramani C, Rotello VM, Bunz UHF. Soft Matter. 2009;5:607. [Google Scholar]
- 20.(a) Logsdon LA, Schardon CL, Ramalingam V, Kwee SK, Urbach AR. J Am Chem Soc. 2011;133:17087. doi: 10.1021/ja207825y. [DOI] [PubMed] [Google Scholar]; (b) Urbach AR, Ramalingam V. Isr J Chem. 2011;51:664. [Google Scholar]
- 21.(a) Walkey CD, Chan WCW. Chem Soc Rev. 2012;41:2780. doi: 10.1039/c1cs15233e. [DOI] [PubMed] [Google Scholar]; (b) Lindman S, Lynch I, Thulin E, Nilsson H, Dawson KA, Linse S. Nano Lett. 2007;7:914. doi: 10.1021/nl062743+. [DOI] [PubMed] [Google Scholar]; (c) Cedervall T, Lynch I, Lindman S, Berggard T, Thulin E, Nilsson H, Dawson KA, Linse S. Proc Natl Acad Sci U S A. 2007;104:2050. doi: 10.1073/pnas.0608582104. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Gessner A, Lieske A, Paulke BR, Müller RH. Eur J Pharm Biopharm. 2002;54:165. doi: 10.1016/s0939-6411(02)00081-4. [DOI] [PubMed] [Google Scholar]; (e) Gessner A, Waicz R, Lieske A, Paulke BR, Mäder K, Müller RH. Int J Pharm. 2000;196:245. doi: 10.1016/s0378-5173(99)00432-9. [DOI] [PubMed] [Google Scholar]
- 22.Jeon WS, Moon K, Park SH, Chun H, Ko YH, Lee JY, Lee ES, Samal S, Selvapalam N, Rekharsky MV, Sindelar V, Sobransingh D, Inoue Y, Kaifer AE, Kim K. J Am Chem Soc. 2005;127:12984. doi: 10.1021/ja052912c. [DOI] [PubMed] [Google Scholar]
- 23.Choi TS, Ko JY, Heo SW, Ko YH, Kim K, Kim HI. J Am Soc Mass Spectrom. 2012;23:1786. doi: 10.1007/s13361-012-0443-6. [DOI] [PubMed] [Google Scholar]
- 24.Miranda OR, Li X, Garcia-Gonzalez L, Zhu Z-J, Yan B, Bunz UHF, Rotello VM. J Am Chem Soc. 2011;133:9650. doi: 10.1021/ja2021729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.(a) De M, Rana S, Akpinar H, Miranda OR, Arvizo RR, Bunz UHF, Rotello VM. Nat Chem. 2009;1:461. doi: 10.1038/nchem.334. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) You CC, Miranda OR, Gider B, Ghosh PS, Kim IK-B, Erdogan B, Krovi SA, Bunz UHF, Rotello VM. Nat Nanotech. 2007;2:318. doi: 10.1038/nnano.2007.99. [DOI] [PubMed] [Google Scholar]
- 26.Hill HD, Millstone JE, Banholzer MJ, Mirkin CA. ACS Nano. 2009;24:418. doi: 10.1021/nn800726e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lundqvist M, Sethson I, Jonsson B-H. Langmuir. 2004;20:10639. doi: 10.1021/la0484725. [DOI] [PubMed] [Google Scholar]
- 28.(a) Ghorai PK, Glotzer SC. J Phys Chem C. 2010;114:19182. [Google Scholar]; (b) Lehn RCV, Alexander-Katz A. Soft Matter. 2011;7:11392. [Google Scholar]
- 29.(a) Ong QK, Reguera J, Silva PJ, Moglianetti M, Harkness K, Longobardi M, Mali KS, Renner C, Feyter SD, Stellacci F. ACS Nano. 2013;7:8529. doi: 10.1021/nn402414b. [DOI] [PubMed] [Google Scholar]; (b) Harkness KM, Balinski A, McLean JA, Cliffel DE. Angew Chem Int Ed. 2011;50:10554. doi: 10.1002/anie.201102882. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Liu X, Yu M, Kim H, Mameli M, Stellacci F. Nat Commun. 2012;3:1182. doi: 10.1038/ncomms2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.