Abstract
Objective: Hepatocellular carcinoma (HCC) represents the third leading cause of cancer-related death worldwide. Increasing evidence suggests that microRNAs, a novel class of non-coding RNAs that function as endogenous suppressors of gene expression, are deregulated in HCC. Although microRNA-222 (miR-222) overexpression has been described in HCC, the role of miR-222 and its target genes in the proliferation of hepatocellular carcinoma cells remain poorly elucidated. Methods: HepG2 cells were transfected with miR-222 mimic, inhibitor or their negative controls. Cell proliferation was assessed by Cell Counting Kit-8 (CCK-8) and EdU incorporation assay. Flow cytometry was performed to assess the effects of miR-222 on HepG2 cell cycle progression. MiR-222 and putative targets genes (p27 and p57) expression levels were determined using qRT-PCR and/or Western blot. Results: MiR-222 overexpression induced an enhancement of HepG2 cell proliferation in vitro, paralleling with an altered cell cycle progression via increased cell population in S phase. P27 expression, other than p57, was negatively regulated by miR-222 overexpression at post-transcriptional level in HepG2 cells. Transfection of either small interfering RNA (siRNA) for p27 or miR-222 mimic increased HepG2 cell proliferation rate, whereas co-transfection of p27 siRNA and miR-222 mimic did not further enhance HepG2 cell proliferation in comparison with the cells transfected with p27 siRNA or miR-222 mimic alone, validating that p27 is a target gene of miR-222 during HepG2 cell proliferation. Conclusion: This study suggests that miR-222 overexpression promotes HepG2 cell proliferation by downregulating p27.
Keywords: Hepatocellular carcinoma, MicroRNA-222, proliferation, P27
Introduction
Hepatocellular carcinoma (HCC) that accounts for a major form of primary liver cancer in adults is the third leading cause of cancer-related death worldwide [1,2]. Therapeutic strategy for HCC must take into account the tumor stage and the clinical status of patients [3,4]. The presence of chronic liver diseases, and notably cirrhosis, makes treatment even more challenging [5,6]. Although liver transplantation is one of best ideal treatments for early stage of HCC, it is unfortunately limited to the lack of donors [7,8]. Thus, the identification of new therapeutic target for HCC is an urgent requirement.
MicroRNAs (miRNAs, miRs), a class of non-coding RNAs of ~22 nucleotides in length, function as endogenous suppressors of gene expression, mainly by binding to 3’-untranslated region (3’-UTR) of target messenger RNA (mRNA) that induces mRNA degradation and/or translational repression [9,10]. To date, more than 1000 miRNAs have been identified in human and each miRNA can control hundreds of genes [11]. Since miRNAs play important roles in a wide range of cell functions like cell division, differentiation, proliferation and apoptosis [12,13], deregulated miRNAs are involved in the pathogenesis of many human diseases, including cancers [14-16]. Although increasing evidence reveals miRNAs involvement in HCC, the biological and molecular functions affected by miRNAs in HCC are far from exploited [17-19].
Deregulated cell cycle and aberrant proliferation of hepatocytes are critically implicated in the initiation and development of HCC, via the deregulation of multiple signaling pathways that is associated with the abnormal activation of growth factors and the aberrantly expressed oncogenes and tumor suppressor genes [20-23]. Recent advances demonstrate that various miRNAs deregulation have been identified in HCC, including down-regulation of miR-1 [24], -125a [25,26], -125b [27,28], -195 [29] and -214 [30], and up-regulation of miR-21 [31], -221 [32,33] and -224 [34], which can lead hepatocytes to proliferate and disrupt the homeostasis between cell growth and apoptosis. MiR-221 and miR-222, encoded in tandem on the X chromosome in human, share a high degree of homology [35]. Among the deregulated miRNAs in HCC, the up-regulation of miR-221/222 was also reported in other types of cancers, comprising breast [36], pancreatic [37], kidney [38], prostate [39] and thyroid cancer [40]. Moreover, overexpression of miR-221 was shown to enhance cancer cell proliferation, via altering the expression of cyclin-dependent kinase inhibitor (CDKI) CDKN1B/p27 and CDKN1C/p57 [32]. However, the role of miR-222 and its target genes in the proliferation of hepatocellular carcinoma cells remain poorly elucidated.
Materials and methods
Cell culture
The human hepatocellular carcinoma HepG2 cells were purchased from the Cell Bank of Chinese Academy of Sciences (Shanghai, China). The cells were cultured in high glucose-Dulbecco’s Modified Eagle Medium (Hyclone, USA) supplemented with 10% fetal calf serum (Hyclone, USA) and 1% penicillin/ streptomycin in a humidified atmosphere of 5% CO2 at 37°C.
Cell transfection
The miR-222 mimic, inhibitor and their negative controls (NC) were purchased from RiboBio (China). HepG2 cells were transfected with miR-222 mimic (50 nM), inhibitor (100 nM) or their negative controls for 48 h using Lipofectamine 2000 (Invitrogen, USA) according to the manufacturer’s instructions.
Cell proliferation assay
HepG2 cells seeded at a density of 3×104 per well into 96-well plates were transfected with miR-222 mimic (50 nM), inhibitor (100 nM) or their negative controls. Forty-eight hours after transfection, a cell proliferation assay was performed using Cell Counting Kit-8 (CCK-8) (Dojindo, Japan) according to the manufacturer’s instructions. The absorbance was read at 450 nm with a microplate reader (Bio-Rad).
Cell cycle analysis
HepG2 cells seeded at a density of 6×105 per well into 6-well plates were transfected with miR-222 mimic (50 nM), miR-222 inhibitor (100 nM) or their negative controls. Forty-eight hours after transfection, cells were detached using 0.025% trypsin, washed once with PBS, and fixed in 70% ethanol at 4°C overnight. Cellular DNA content was stained using propidium iodide (PI) (Sigma, USA) and analyzed using MoFlo XDP Cell Sorter (Beckman Coulter). Cell number in each phase of the cell cycle was determined using FlowJo software (Treestar Inc., USA).
EdU incorporation assay
HepG2 cells were seeded at 1.5×105 cells/well in 24-well plates. After transfecting HepG2 cells with miR-222 mimic (50 nM), miR-222 inhibitor (100 nM) or their negative controls for 48 h, the incorporation of 5-ethynyl-2’-deoxyuridine (EdU) into actively proliferating HepG2 cells was evaluated using a Cell-Light™ EdU Cell Proliferation Detection kit (RiboBio, China) following the manufacturer’s instructions. Cellular immunostaining was observed with an epifluorescence microscope (Leica, Germany). Digital images were acquired and analyzed with Image J software.
RNA extraction and quantitative reverse transcription-polymerase chain reaction (qRT-PCR)
Total RNA was isolated using miRNeasy Mini Kit (Qiagen, Germany). For mRNA analysis, cDNA was synthesized using Bio-Rad iScripTM cDNA Synthesis Kit (Bio-Rad). A template equivalent of 400 ng of total RNA was subjected to 40 cycles of quantitative PCR with Takara SYBR Premix Ex TaqTM (Tli RNaseH Plus, Japan) in CFX96TM Real-Time PCR Detection System (Bio-Rad). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal control. Primers sequences (forward and reverse) were designed as follows: p27, CAGGTCTCCAAGACGACATAGA and CGCCTTTTCGATTCATGTACTGC; p57, AGGTAGCGAGGTGGATCTGTC and GGCCTCTGATTCCCGAGGA. For miRNA analysis, the Bulge-LoopTM miRNA qPCR Primer Set (RiboBio, China) was used to detect miR-222 expression by qRT-PCR with Takara SYBR Premix Ex TaqTM in CFX96TM Real-Time PCR Detection System. 5S ribosomal RNA (5S rRNA) was used to normalize target miRNA expression. Relative expression levels for each mRNA and miRNA were determined using the 2-ΔΔCt method.
Western blot analysis
Cells were lysed using RIPA lysis buffer (Beyotime Institute of Biotechnology, China) containing 1% phenylmethanesulfonyl fluoride (PMSF). Equal amounts of 20 to 40 μg of total protein were subjected to electrophoreses on 10% SDS-Page gels, transferred to PVDF membranes and incubated with the appropriate primary antibodies as follows: anti-p27 (Bioworld, 1:1000 dilution), anti-p57 (Bioworld, 1:1000 dilution), and GAPDH (Bioworld, 1:5000 dilution). After incubated with the corresponding HRP-conjugated secondary antibodies, protein bands were visualized using enhanced chemiluminescence (ECL) system (Pierce Biotechnology Inc., Rockford, IL, USA) with the ChemiDoc XRS Plus luminescent image analyzer (Bio-Rad). Densitometric analysis of protein bands was performed using Image Lab software (Bio-Rad). Loading volume of each sample was normalized by GAPDH protein band density.
Target gene validation
P27 and p57, two cell cycle-related genes which function to negatively control cell cycle progression, were chosen as candidate target genes of miR-222 in HepG2 cells. The small interfering RNA (siRNA) for p27 and the negative control siRNA were obtained from RiboBio (China). First, miR-222 mimic (50 nM), miR-222 inhibitor (100 nM) or their negative controls were transfected to HepG2 cells. Forty-eight hours after transfection, qRT-PCR and Western blot were performed to evaluate mRNA and protein expression levels of p27 and p57. Second, as p27 was found endogenously regulated by miR-222 in HepG2 cells, co-transfection of p27 siRNA (75 nM) and miR-222 mimic (50 nM) was performed to determine if miR-222 takes effects through p27 in HepG2 cells.
Statistical analysis
All analyses in the study were evaluated using SPSS software (version 19.0). Data are expressed as mean ± SEM. An independent-samples t-test or one-way ANOVA was conducted to evaluate the one-way layout data. If a significant difference was observed, Bonferroni’s post-hoc test was conducted to identify groups with significant differences. P-value less than 0.05 was considered statistically significant.
Results
MiR-222 overexpression enhances HepG2 cell proliferation
To determine the potential cellular effects of miR-222 on HepG2 cells, HepG2 cells were transfected with miR-222 mimic, inhibitor and their negative controls for 48 h. Using qRT-PCR, we confirmed that miR-222 expression level was significantly increased by miR-222 mimic, while decreased by miR-222 inhibitor, confirming that miR-222 mimic and inhibitor successfully regulated miR-222 expression level in HepG2 cells (Figure 1A).
Figure 1.
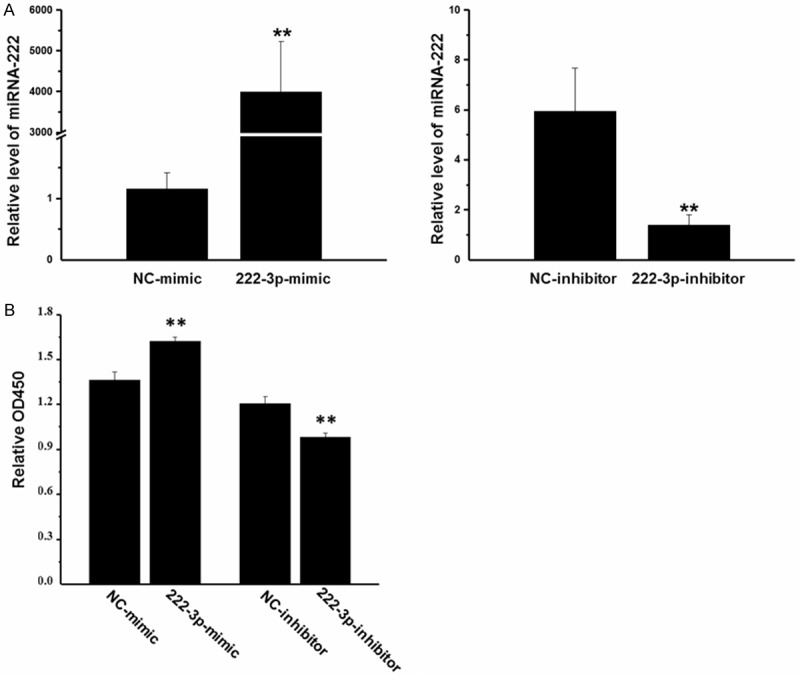
MiR-222 overexpression promotes HepG2 cell proliferation. A: The expression level of miR-222 was significantly increased by miR-222 mimic, while decreased by miR-222 inhibitor in HepG2 cells (n = 4). B: MiR-222 mimic increased HepG2 cell proliferation, while miR-222 inhibitor decreased their proliferation (n = 6). **P < 0.01.
As determined by CCK-8 cell proliferation assay, miR-222 mimic significantly increased HepG2 cell proliferation, while miR-222 inhibitor decreased their proliferation (Figure 1B). A quantitative analysis of EdU-positive cells further showed that miR-222 overexpressing cells were 39% EdU-positive compared with 26% in negative control cells (Figure 2A), while miR-222 inhibiting cells were 16% EdU-positive compared with 27% in negative control cells (Figure 2B). These results further confirmed that overexpression of miR-222 promoted HepG2 cell proliferation while inhibition of miR-222 suppressed HepG2 cell proliferation.
Figure 2.
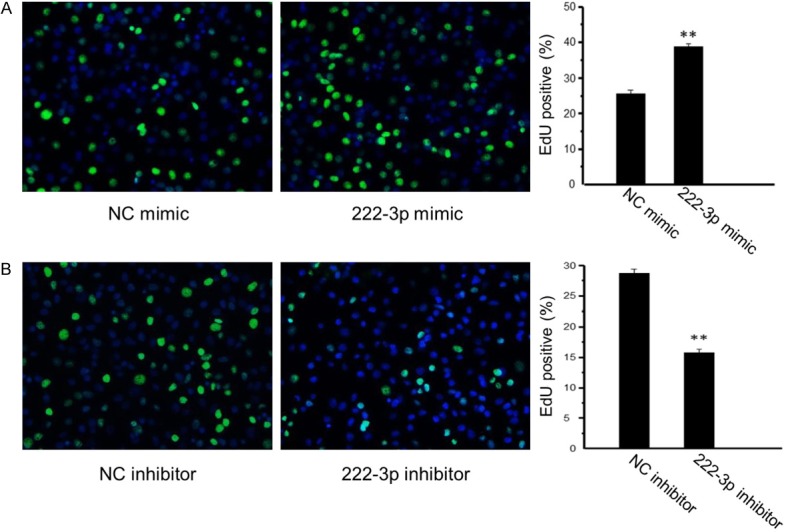
Quantitative analysis of EdU-positive cells confirms that miR-222 overexpression promotes HepG2 cell proliferation. A: MiR-222 overexpressing cells were 39% EdU-positive compared with 26% in negative control cells (n = 4). B: MiR-222 inhibiting cells were 16% EdU-positive compared with 27% in negative control cells (n = 4). **P < 0.01.
MiR-222 regulates HepG2 cell cycle progression
As enhanced cell proliferation is associated with altered cell cycle, flow cytometry was performed to assess the effects of miR-222 on progression of HepG2 cell cycle. The percentage of HepG2 cells in S phase was higher but that in G2 phase was lower by miR-222 mimic (Figure 3A), which paralleled with a decreased percentage of cells in S phase and an increased percentage of cells in G2 phase by miR-222 inhibitor (Figure 3B). These results suggested that miR-222 can regulate HepG2 cell cycle progression via an increase in the population of HepG2 cells in S phase.
Figure 3.
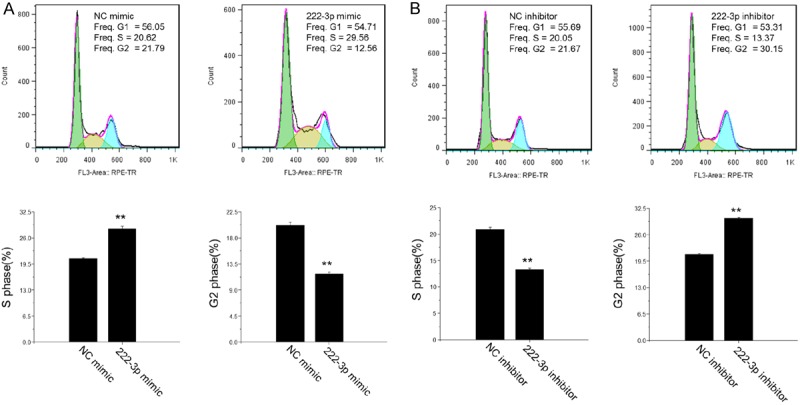
MiR-222 regulates HepG2 cell cycle progression. A: MiR-222 mimic increased the percentage of HepG2 cells in S phase and decreased that in G2 phase (n = 3). B: MiR-222 inhibitor decreased the percentage of HepG2 cells in S phase and increased that in G2 phase (n = 3). **P < 0.01.
P27 is a target gene of miR-222 in HepG2 cells
P27 and p57, two well-known targets of miR-222 in multiple types of cells, are members of the Cip/Kip family of cyclin-dependent kinase inhibitors and function to negatively control cell cycle progression. Consequently, we assessed the effect of miR-222 on endogenous expressions of p27 and p57 in HepG2 cells by qRT-PCR and Western blot. P27 expression was endogenously regulated by miR-222 mimic or inhibitor at protein level (Figure 4B) but not at mRNA level (Figure 4A). However, miR-222 mimic or inhibitor exerted no effects on p57 expression neither at mRNA level nor at protein level (Figure 4A and 4B). These data indicated that p27 expression was post-transcriptionally regulated by miR-222 in HepG2 cells.
Figure 4.
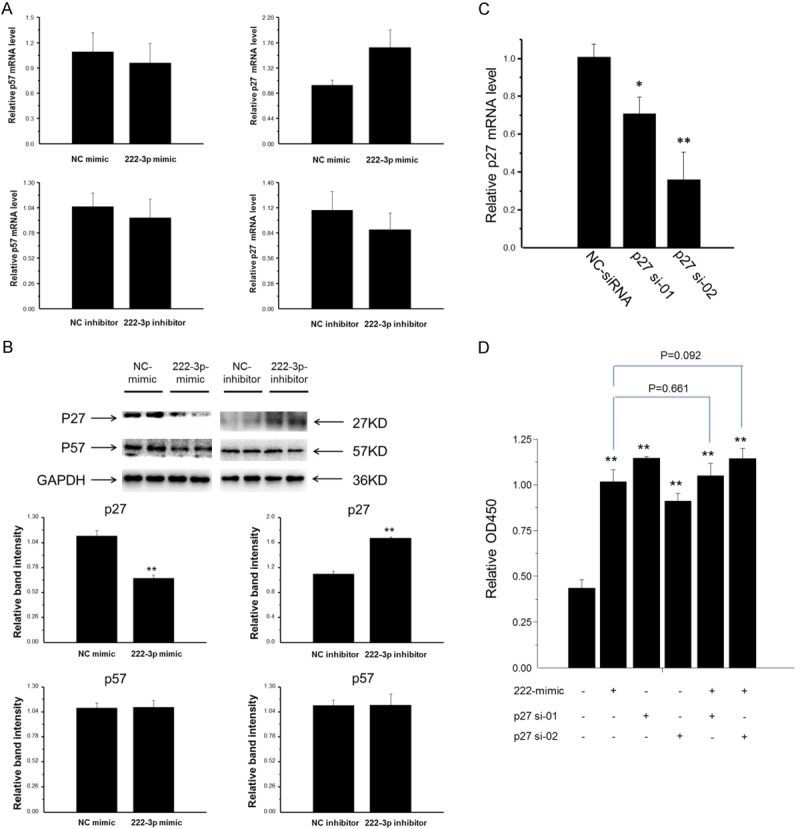
P27 is a target gene of miR-222 in HepG2 cells. A: MiR-222 mimic or inhibitor did not regulate p27 or p57 expression at mRNA level (n = 4). B: P27 protein expression, other than p57, was negatively regulated by miR-222 in HepG2 cells (n = 4). C: Transfection of p27 siRNA led to efficient knockdown of p27 in HepG2 cells (n = 4). D: The cells transfected with either p27 siRNA or miR-222 mimic had higher proliferation rate than negative control cells, while co-transfection of p27 siRNA and miR-222 mimic did not further enhance cell proliferation compared with the cells transfected with p27 siRNA or miR-222 mimic alone (n = 6). **P < 0.01.
The p27 siRNAs, p27 si-01 and p27 si-02, were then tested in HepG2 cells. As expected, the transfection of either p27 si-01 or p27 si-02 led to efficient knockdown of p27 as detected by qRT-PCR (Figure 4C). To further determine the potential role of p27 in HepG2 cell proliferation induced by miR-222, we used co-transfection of p27 siRNA (75 nM) and miR-222 mimic (50 nM) to HepG2 cells for 48 h, and assessed cell proliferation by CCK-8 assay. The cells transfected with either p27 siRNA (p27 si-01 or p27 si-02) or miR-222 mimic had higher proliferation rate than negative control cells, while co-transfection of p27 siRNA and miR-222 mimic did not further enhance cell proliferation compared with the cells transfected with p27 siRNA or miR-222 mimic alone (Figure 4D), indicating that miR-222 promoted HepG2 cell proliferation, at least in part, via p27 targeting.
Discussion
MiR-221 and miR-222, two highly homologous microRNAs, have been described in various types of human cancers [35], whose overexpression is responsible for tumor initiation and progression via regulating cancer cell differentiation, proliferation, survival and metastasis [37-41]. Moreover, increasing evidence indicates that miR-221 overexpression contributes to liver tumorigenesis [32,33,42]. However, the possible roles of miR-222 and its associated target genes in HCC are largely unexplored. The present study shows that miR-222 overexpression promotes HepG2 cell proliferation, at least in part, by downregulating its target gene, p27.
MiR-222 has been previously described to enhance cellular invasiveness and motility of hepatocellular carcinoma cells [43]. Meanwhile, increased expression of miR-222 correlates with advanced stage of HCC tumors and shorter disease-free survival of patients [43].Consistently, miR-222 overexpression is associated with enhanced degree of tumor differentiation [44]. In addition, miR-222 has been shown to promote the proliferation of many somatic cells, such as endothelial cells [45], smooth muscle cells [46], Schwann cells [47], glioma cells [48], and cancer cells [49]. These data imply the importance of miR-222 on cell motility, migration, metastasis and proliferation, which prompted us to further investigate its potential roles in liver tumorigenesis by focusing on hepatocellular carcinoma cell proliferation. Our present study shows that miR-222 overexpression induced an enhancement of HepG2 cell proliferation in vitro, paralleling with an altered cell cycle progression, in which the cells in S phase were increased while in G2 phase were decreased. Abnormal cell cycle progression appears to be an essential early event in HCC initiation and accounts for a critical step in HCC development [50]. However, the mechanisms underlying the roles of miR-222 in controlling HepG2 cell proliferation and cell cycle progression remain to be further investigated.
P27 and p57, two key cyclin-dependent kinase inhibitors that can be negatively regulated by miR-221/222 in cancers [32,39,49,51-54], have been chosen as putative target genes of miR-222 in hepatocellular carcinoma cells. Down-regulation of p27 and p57 is associated with advanced tumor stage, lower survival and high proliferation activity in HCC [55-57]. Our present study shows that p27 protein expression, other than its mRNA level, was negatively regulated by miR-222 overexpression in HepG2 cells. However, p57 expression was not modified by miR-222 mimic or inhibitor transfection. These data indicate that p27 expression can be suppressed by miR-222 overexpression in HepG2 cells at post-transcriptional level. We then further investigated the functional role of p27 knockdown on HepG2 cell proliferation by using co-transfection of p27 siRNA and miR-222 mimic to HepG2 cells. The results showed that transfection of either p27 siRNA or miR-222 mimic increased HepG2 cell proliferation rate, whereas co-transfection of p27 siRNA and miR-222 mimic did not further enhance cell proliferation in comparison with the cells transfected with p27 siRNA or miR-222 mimic alone, indicating that the p27 knockdown is, at least in part, involved in the effect of miR-222 overexpression on HepG2 cell proliferation. Taken together, the present results suggest that p27 is a target gene of miR-222 during HepG2 cell proliferation. Further study will be required to identify other putative target genes of miR-222 as well as associated molecular pathways altered in HepG2 cell proliferation.
In conclusion, the present study shows that miR-222 overexpression promotes HepG2 cell proliferation by targeting p27. Out work may provide new evidence for understanding the potential roles of miR-222 in the pathogenesis of HCC and developing novel therapeutic strategy for HCC.
Acknowledgements
This work was supported by the grants from National Natural Science Foundation of China (81070343 and 81370559 to C. Yang; 81200169 to J. Xiao), funds from Shanghai Innovation Program (12431901002 to C. Yang), Innovation Program of Shanghai Municipal Education Commission (13YZ014 to J. Xiao), Foundation for University Young Teachers by Shanghai Municipal Education Commission (year 2012, to J. Xiao), Innovation Foundation of Shanghai University (sdcx2012038, to J. Xiao) and partially by Leading Academic Discipline Project of Shanghai Municipal Education Commission “Molecular Physiology” and Shanghai Municipal Science and Technology Committee (13DZ2272100).
Disclosure of conflict of interest
None.
References
- 1.He L, Zhou X, Qu C, Hu L, Tang Y, Zhang Q, Liang M, Hong J. Musashi2 predicts poor prognosis and invasion in hepatocellular carcinoma by driving epithelial-mesenchymal transition. J Cell Mol Med. 2014;18:49–58. doi: 10.1111/jcmm.12158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bosch FX, Ribes J, Díaz M, Cléries R. Primary liver cancer: worldwide incidence and trends. Gastroenterology. 2004;127:S5–S16. doi: 10.1053/j.gastro.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 3.Lin S, Hoffmann K, Schemmer P. Treatment of Hepatocellular Carcinoma: A Systematic Review. Liver Cancer. 2012;1:144–58. doi: 10.1159/000343828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Salgia R, Singal AG. Hepatocellular carcinoma and other liver lesions. Med Clin North Am. 2014;98:103–18. doi: 10.1016/j.mcna.2013.09.003. [DOI] [PubMed] [Google Scholar]
- 5.Barone C, Koeberle D, Metselaar H, Parisi G, Sansonno D, Spinzi G. Multidisciplinary approach for HCC patients: hepatology for the oncologists. Ann Oncol Off J Eur Soc Med Oncol ESMO. 2013;24(Suppl 2):ii15–23. doi: 10.1093/annonc/mdt053. [DOI] [PubMed] [Google Scholar]
- 6.Vivarelli M, Montalti R, Risaliti A. Multimodal treatment of hepatocellular carcinoma on cirrhosis: An update. World J Gastroenterol. 2013;19:7316–26. doi: 10.3748/wjg.v19.i42.7316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Belghiti J, Fuks D. Liver Resection and Transplantation in Hepatocellular Carcinoma. Liver Cancer. 2012;1:71–82. doi: 10.1159/000342403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reddy MS, Varghese J, Venkataraman J, Rela M. Matching donor to recipient in liver transplantation: Relevance in clinical practice. World J Hepatol. 2013;5:603–11. doi: 10.4254/wjh.v5.i11.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van Rooij E. The art of microRNA research. Circ Res. 2011;108:219–34. doi: 10.1161/CIRCRESAHA.110.227496. [DOI] [PubMed] [Google Scholar]
- 10.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–73. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 11.Liang M. MicroRNA: a new entrance to the broad paradigm of systems molecular medicine. Physiol Genomics. 2009;38:113–5. doi: 10.1152/physiolgenomics.00080.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 13.Xiao J, Liang D, Zhang H, Liu Y, Zhang D, Liu Y, Pan L, Chen X, Doevendans PA, Sun Y, Liang X, Sluijter JP, Chen YH. MicroRNA-204 is required for differentiation of human-derived cardiomyocyte progenitor cells. J Mol Cell Cardiol. 2012;53:751–9. doi: 10.1016/j.yjmcc.2012.08.024. [DOI] [PubMed] [Google Scholar]
- 14.Garofalo M, Condorelli G, Croce CM. MicroRNAs in diseases and drug response. Curr Opin Pharmacol. 2008;8:661–7. doi: 10.1016/j.coph.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 15.Xu J, Zhao J, Evan G, Xiao C, Cheng Y, Xiao J. Circulating microRNAs: novel biomarkers for cardiovascular diseases. J Mol Med (Berl) 2012;90:865–75. doi: 10.1007/s00109-011-0840-5. [DOI] [PubMed] [Google Scholar]
- 16.Li J, Xu J, Cheng Y, Wang F, Song Y, Xiao J. Circulating microRNAs as mirrors of acute coronary syndromes: MiRacle or quagMire? J Cell Mol Med. 2013;17:1363–70. doi: 10.1111/jcmm.12148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khare S, Zhang Q, Ibdah JA. Epigenetics of hepatocellular carcinoma: role of microRNA. World J Gastroenterol. 2013;19:5439–45. doi: 10.3748/wjg.v19.i33.5439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hou W, Bonkovsky HL. Non-coding RNAs in hepatitis C-induced hepatocellular carcinoma: Dysregulation and implications for early detection, diagnosis and therapy. World J Gastroenterol. 2013;19:7836–45. doi: 10.3748/wjg.v19.i44.7836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giordano S, Columbano A. MicroRNAs: new tools for diagnosis, prognosis, and therapy in hepatocellular carcinoma? Hepatology. 2013;57:840–7. doi: 10.1002/hep.26095. [DOI] [PubMed] [Google Scholar]
- 20.Berasain C, Avila MA. The EGFR signalling system in the liver: from hepatoprotection to hepatocarcinogenesis. J Gastroenterol. 2014;49:9–23. doi: 10.1007/s00535-013-0907-x. [DOI] [PubMed] [Google Scholar]
- 21.Zheng YW, Nie YZ, Taniguchi H. Cellular reprogramming and hepatocellular carcinoma development. World J Gastroenterol. 2013;19:8850–60. doi: 10.3748/wjg.v19.i47.8850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Uematsu S, Higashi T, Nouso K, Kariyama K, Nakamura SI, Suzuki M, Nakatsukasa H, Kobayashi Y, Hanafusa T, Tsuji T, Shiratori Y. Altered expression of vascular endothelial growth factor, fibroblast growth factor-2 and endostatin in patients with hepatocellular carcinoma. J Gastroenterol Hepatol. 2005;20:583–8. doi: 10.1111/j.1440-1746.2005.03726.x. [DOI] [PubMed] [Google Scholar]
- 23.Fischer ANM, Fuchs E, Mikula M, Huber H, Beug H, Mikulits W. PDGF essentially links TGF-beta signaling to nuclear beta-catenin accumulation in hepatocellular carcinoma progression. Oncogene. 2007;26:3395–405. doi: 10.1038/sj.onc.1210121. [DOI] [PubMed] [Google Scholar]
- 24.Li D, Yang P, Li H, Cheng P, Zhang L, Wei D, Su X, Peng J, Gao H, Tan Y, Zhao Z, Li Y, Qi Z, Rui Y, Zhang T. MicroRNA-1 inhibits proliferation of hepatocarcinoma cells by targeting endothelin-1. Life Sci. 2012;91:440–7. doi: 10.1016/j.lfs.2012.08.015. [DOI] [PubMed] [Google Scholar]
- 25.Bi Q, Tang S, Xia L, Du R, Fan R, Gao L, Jin J, Liang S, Chen Z, Xu G, Nie Y, Wu K, Liu J, Shi Y, Ding J, Fan D. Ectopic expression of MiR-125a inhibits the proliferation and metastasis of hepatocellular carcinoma by targeting MMP11 and VEGF. PLoS One. 2012;7:e40169. doi: 10.1371/journal.pone.0040169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim JK, Noh JH, Jung KH, Eun JW, Bae HJ, Kim MG, Chang YG, Shen Q, Park WS, Lee JY, Borlak J, Nam SW. Sirtuin7 oncogenic potential in human hepatocellular carcinoma and its regulation by the tumor suppressors MiR-125a-5p and MiR-125b. Hepatology. 2013;57:1055–67. doi: 10.1002/hep.26101. [DOI] [PubMed] [Google Scholar]
- 27.Fan DNY, Tsang FHC, Tam AHK, Au SLK, Wong CCL, Wei L, Lee JM, He X, Ng IO, Wong CM. Histone lysine methyltransferase, suppressor of variegation 3-9 homolog 1, promotes hepatocellular carcinoma progression and is negatively regulated by microRNA-125b. Hepatology. 2013;57:637–47. doi: 10.1002/hep.26083. [DOI] [PubMed] [Google Scholar]
- 28.Liang L, Wong CM, Ying Q, Fan DNY, Huang S, Ding J, Yao J, Yan M, Li J, Yao M, Ng IO, He X. MicroRNA-125b suppressesed human liver cancer cell proliferation and metastasis by directly targeting oncogene LIN28B2. Hepatology. 2010;52:1731–40. doi: 10.1002/hep.23904. [DOI] [PubMed] [Google Scholar]
- 29.Xu T, Zhu Y, Xiong Y, Ge YY, Yun JP, Zhuang SM. MicroRNA-195 suppresses tumorigenicity and regulates G1/S transition of human hepatocellular carcinoma cells. Hepatology. 2009;50:113–21. doi: 10.1002/hep.22919. [DOI] [PubMed] [Google Scholar]
- 30.Wang X, Chen J, Li F, Lin Y, Zhang X, Lv Z, Jiang J. MiR-214 inhibits cell growth in hepatocellular carcinoma through suppression of β-catenin. Biochem Biophys Res Commun. 2012;428:525–31. doi: 10.1016/j.bbrc.2012.10.039. [DOI] [PubMed] [Google Scholar]
- 31.Xu G, Zhang Y, Wei J, Jia W, Ge Z, Zhang Z, Liu X. MicroRNA-21 promotes hepatocellular carcinoma HepG2 cell proliferation through repression of mitogen-activated protein kinase-kinase 3. BMC Cancer. 2013;13:469. doi: 10.1186/1471-2407-13-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fornari F, Gramantieri L, Ferracin M, Veronese A, Sabbioni S, Calin GA, Grazi GL, Giovannini C, Croce CM, Bolondi L, Negrini M. MiR-221 controls CDKN1C/p57 and CDKN1B/p27 expression in human hepatocellular carcinoma. Oncogene. 2008;27:5651–61. doi: 10.1038/onc.2008.178. [DOI] [PubMed] [Google Scholar]
- 33.Pineau P, Volinia S, McJunkin K, Marchio A, Battiston C, Terris B, Mazzaferro V, Lowe SW, Croce CM, Dejean A. miR-221 overexpression contributes to liver tumorigenesis. Proc Natl Acad Sci U S A. 2010;107:264–9. doi: 10.1073/pnas.0907904107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y, Takahashi S, Tasaka A, Yoshima T, Ochi H, Chayama K. Involvement of microRNA-224 in cell proliferation, migration, invasion, and anti-apoptosis in hepatocellular carcinoma. J Gastroenterol Hepatol. 2013;28:565–75. doi: 10.1111/j.1440-1746.2012.07271.x. [DOI] [PubMed] [Google Scholar]
- 35.Garofalo M, Quintavalle C, Romano G, Croce CM, Condorelli G. miR221/222 in Cancer: Their Role in Tumor Progression and Response to Therapy. Curr Mol Med. 2012;12:27–33. doi: 10.2174/156652412798376170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen WX, Hu Q, Qiu MT, Zhong SL, Xu JJ, Tang JH, Zhao JH. miR-221/222: promising biomarkers for breast cancer. Tumour Biol. 2013;34:1361–70. doi: 10.1007/s13277-013-0750-y. [DOI] [PubMed] [Google Scholar]
- 37.Zhang Y, Li M, Wang H, Fisher WE, Lin PH, Yao Q, Chen C. Profiling of 95 microRNAs in pancreatic cancer cell lines and surgical specimens by real-time PCR analysis. World J Surg. 2009;33:698–709. doi: 10.1007/s00268-008-9833-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gottardo F, Liu CG, Ferracin M, Calin GA, Fassan M, Bassi P, Sevignani C, Byrne D, Negrini M, Pagano F, Gomella LG, Croce CM, Baffa R. Micro-RNA profiling in kidney and bladder cancers. Urol Oncol. 2007;25:387–92. doi: 10.1016/j.urolonc.2007.01.019. [DOI] [PubMed] [Google Scholar]
- 39.Galardi S, Mercatelli N, Giorda E, Massalini S, Frajese GV, Ciafrè SA, Farace MG. miR-221 and miR-222 expression affects the proliferation potential of human prostate carcinoma cell lines by targeting p27Kip1. J Biol Chem. 2007;282:23716–24. doi: 10.1074/jbc.M701805200. [DOI] [PubMed] [Google Scholar]
- 40.Pallante P, Visone R, Ferracin M, Ferraro A, Berlingieri MT, Troncone G, Chiappetta G, Liu CG, Santoro M, Negrini M, Croce CM, Fusco A. MicroRNA deregulation in human thyroid papillary carcinomas. Endocr Relat Cancer. 2006;13:497–508. doi: 10.1677/erc.1.01209. [DOI] [PubMed] [Google Scholar]
- 41.Chen G. miR-222 is upregulated in epithelial ovarian cancer and promotes cell proliferation by downregulating P27kip1. Oncol Lett. 2013;6:507–512. doi: 10.3892/ol.2013.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gramantieri L, Fornari F, Ferracin M, Veronese A, Sabbioni S, Calin GA, Grazi GL, Croce CM, Bolondi L, Negrini M. MicroRNA-221 Targets Bmf in Hepatocellular Carcinoma and Correlates with Tumor Multifocality. Clin Cancer Res. 2009;15:5073–81. doi: 10.1158/1078-0432.CCR-09-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wong QWL, Ching AKK, Chan AWH, Choy KW, To KF, Lai PBS, Wong N. MiR-222 overexpression confers cell migratory advantages in hepatocellular carcinoma through enhancing AKT signaling. Clin Cancer Res. 2010;16:867–75. doi: 10.1158/1078-0432.CCR-09-1840. [DOI] [PubMed] [Google Scholar]
- 44.Murakami Y, Yasuda T, Saigo K, Urashima T, Toyoda H, Okanoue T, Shimotohno K. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene. 2006;25:2537–45. doi: 10.1038/sj.onc.1209283. [DOI] [PubMed] [Google Scholar]
- 45.Liu X, Cheng Y, Yang J, Xu L, Zhang C. Cell-specific effects of miR-221/222 in vessels: molecular mechanism and therapeutic application. J Mol Cell Cardiol. 2012;52:245–55. doi: 10.1016/j.yjmcc.2011.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu X, Cheng Y, Zhang S, Lin Y, Yang J, Zhang C. A necessary role of miR-221 and miR-222 in vascular smooth muscle cell proliferation and neointimal hyperplasia. Circ Res. 2009;104:476–87. doi: 10.1161/CIRCRESAHA.108.185363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu B, Zhou S, Wang Y, Qian T, Ding G, Ding F, Gu X. miR-221 and miR-222 promote Schwann cell proliferation and migration by targeting LASS2 after sciatic nerve injury. J Cell Sci. 2012;125:2675–83. doi: 10.1242/jcs.098996. [DOI] [PubMed] [Google Scholar]
- 48.Zhang J, Han L, Ge Y, Zhou X, Zhang A, Zhang C, Zhong Y, You Y, Pu P, Kang C. miR-221/222 promote malignant progression of glioma through activation of the Akt pathway. Int J Oncol. 2010;36:913–20. doi: 10.3892/ijo_00000570. [DOI] [PubMed] [Google Scholar]
- 49.Sun C, Li N, Zhou B, Yang Z, Ding D, Weng D, Meng L, Wang S, Zhou J, Ma D, Chen G. miR-222 is upregulated in epithelial ovarian cancer and promotes cell proliferation by downregulating P27(kip1. ) Oncol Lett. 2013;6:507–12. doi: 10.3892/ol.2013.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gramantieri L, Fornari F, Callegari E, Sabbioni S, Lanza G, Croce CM, Bolondi L, Negrini M. MicroRNA involvement in hepatocellular carcinoma. J Cell Mol Med. 2008;12:2189–204. doi: 10.1111/j.1582-4934.2008.00533.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Le Sage C, Nagel R, Egan DA, Schrier M, Mesman E, Mangiola A, Anile C, Maira G, Mercatelli N, Ciafrè SA, Farace MG, Agami R. Regulation of the p27(Kip1) tumor suppressor by miR-221 and miR-222 promotes cancer cell proliferation. EMBO J. 2007;26:3699–708. doi: 10.1038/sj.emboj.7601790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Visone R, Russo L, Pallante P, De Martino I, Ferraro A, Leone V, Borbone E, Petrocca F, Alder H, Croce CM, Fusco A. MicroRNAs (miR)-221 and miR-222, both overexpressed in human thyroid papillary carcinomas, regulate p27Kip1 protein levels and cell cycle. Endocr Relat Cancer. 2007;14:791–8. doi: 10.1677/ERC-07-0129. [DOI] [PubMed] [Google Scholar]
- 53.Miller TE, Ghoshal K, Ramaswamy B, Roy S, Datta J, Shapiro CL, Jacob S, Majumder S. MicroRNA-221/222 confers tamoxifen resistance in breast cancer by targeting p27Kip1. J Biol Chem. 2008;283:29897–903. doi: 10.1074/jbc.M804612200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fu X, Wang Q, Chen J, Huang X, Chen X, Cao L, Tan H, Li W, Zhang L, Bi J, Su Q, Chen L. Clinical significance of miR-221 and its inverse correlation with p27Kip1 in hepatocellular carcinoma. Mol Biol Rep. 2011;38:3029–35. doi: 10.1007/s11033-010-9969-5. [DOI] [PubMed] [Google Scholar]
- 55.Ito Y, Takeda T, Sakon M, Tsujimoto M, Monden M, Matsuura N. Expression of p57/Kip2 protein in hepatocellular carcinoma. Oncology. 2001;61:221–5. doi: 10.1159/000055378. [DOI] [PubMed] [Google Scholar]
- 56.Tannapfel A, Grund D, Katalinic A, Uhlmann D, Köckerling F, Haugwitz U, Wasner M, Hauss J, Engeland K, Wittekind C. Decreased expression of p27 protein is associated with advanced tumor stage in hepatocellular carcinoma. Int J Cancer. 2000;89:350–5. doi: 10.1002/1097-0215(20000720)89:4<350::aid-ijc6>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 57.Nakai S, Masaki T, Shiratori Y, Ohgi T, Morishita A, Kurokohchi K, Watanabe S, Kuriyama S. Expression of p57(KIP2) in hepatocellular carcinoma: relationship between tumor differentiation and patient survival. Int J Oncol. 2002;20:769–75. [PubMed] [Google Scholar]


