Abstract
In our previous study, the upregulation of adipophilin in lung adenocarcinoma were identified compared with normal lung tissues by quantitative proteomics. In this study, our aim was to verify the result from quantitative proteomics, further investigate the relationship between adipophilin expression and clinicopathologic factors of lung cancer patients. The expression levels of adipophilin were examined in 10 pairs of lung adenocarcinoma and normal lung tissues using western blotting and the expression and cellular distribution of adipophilin were determined by IHC in 62 formalin-fixed and paraffin embedded primary lung cancer specimens. Adipophilin expression was significantly higher in lung adenocarcinoma specimens than in normal tissues and lung squamous cell carcinomas (P<0.05). There were no significant difference of adipophilin expression between lung squamous cell carcinomas and normal lung tissues. The expression of adipophilin in lung cancer did not correlate with any clinicopathologic factors such as lymph node metastasis, patients’ age, gender, tumor size, grade, and TNM stage. In Conclusion, Adipophilin was upregulated in lung adenocarcinoma, suggesting that adipophilin play an important role in tumorigenesis of lung adenocarcinoma and may serve as a potential marker for lung adenocarcinoma.
Keywords: Adipophilin, lung cancer, western blotting, immunohistochemistry
Introduction
Lung cancer is currently one of the most common types of cancer and remains the leading cause of cancer‑related mortality worldwide. Lung cancer accounts for 13% of the total cancer cases and 18% of the total cancer deaths [1]. Adenocarcinoma is the most common histologic type of lung cancer, accounting for almost half of all lung cancers [2]. Most lung cancer patients are initially diagnosed at an advanced stage. Although there has been considerable progress in treatment for advanced lung cancer, its prognosis remains poor, with the 5-year survival of about 16% [3]. Lung adenocarcinoma tend to grow and spread faster than lung squamous cell carcinoma and the pathogenesis remains unclear. Therefore, it is urgent to investigate the molecular mechanism of carcinogenesis and development in lung adenocarcinoma.
Proteomics approaches are important and very useful in the large-scale study of proteins, including differential protein expression, post-translational modifications. Recently, there has been a tendency to focus on organellar proteomics to provide information about the protein contents of organelles, substructures, or compartments isolated from cells [4]. The iTRAQ-labeling combined with two-dimensional liquid chromatography tandem mass spectrometry (2D-LC-MS/MS) technology has been widely used in many proteomics study including subcellular proteomics such as membrane proteomics and mitochondrial proteomics [5-8].
In our previous study, we performed iTRAQ labeling combined with 2D LC-MS/MS to identify differential protein expression profiles of cell membrane from lung adenocarcinoma and matched normal lung tissue samples. The result showed Adipophilin (alternative name(s): perilipin-2; adipose differentiation-related protein)was significantly upregulated in lung adenocarcinoma (2.46-fold) compared with normal lung tissues [9]. The MS/MS spectra on adipophilin is listed in Figure 1. Adipophilin was first characterized as a novel protein induced early during the process of differentiation [10,11]. Several studies shown that adipophilin was expressed at high levels in adipose tissue and also in a variety of cells including murine MA-10 Leydig cells, Chinese hamster ovary (CHO) fibroblasts, human HepG2 hepatoma cells and restrictedly expressed in lactating mammary epithelial cells, adrenal cortex cells, Sertoli and Leydig cells of the male reproductive system, and steatosis or fatty change hepatocytes in tissues [12,13]. In recent years, it has been shown that adipophilin is involved in carcinogenesis [14,15].
Figure 1.
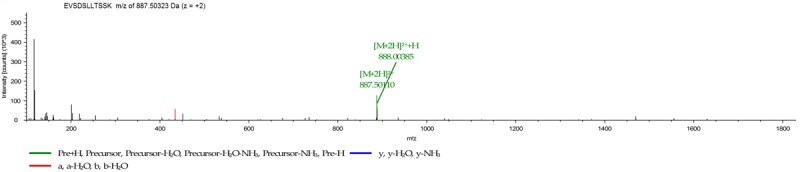
MS/MS spectra of identified peptides for adipophilin. The MS spectra of Sequence: EVSDSLLTSSK with m/z of 887.50323 Da (z = +2).
However, up to now, the clinical impact of adipophilin expression in lung cancer remains yet unknown. In the present study, we verified the upregulation of adipophilin in lung cancer samples and further investigated the relationships between adipophilin expression and clinicopathologic factors of lung cancer patients. The study may provide important information on the role of adipophilin in carcinogenesis and development of lung cancer.
Materials and methods
Tissue specimens
For western blotting, a total of 10 fresh primary lung adenocarcinoma and matched adjacent normal lung tissues undergoing surgical resection were collected from the second affiliated hospital, Xi’an Jiaotong University, China and stored at -80°C until use. None of the patients received chemotherapy, radiotherapy prior to surgery. The formalin-fixed and paraffin embedded 62 primary lung cancer specimens included 41 lung adenocarcinoma and 21 lung squamous cell carcinoma and 24 cases of normal lung tissues were used for immunohistochemical analysis. These paraffin embedded samples were obtain from the second affiliated hospital, Xi’an Jiaotong University and shaanxi cancer hospital, China. All lung cancer samples were confirmed by histopathology. The normal lung tissues were collected at least 5 cm far away from the cancer. Informed consent was obtained from each patient and the study was approved by the local ethnics committee. Patients were staged according to the TNM staging system of the American Joint Committee on Cancer (AJCC) and the International Union for Cancer Control (UICC). The tumors were histologically subtyped and graded according to the World Health Organization guidelines.
Western blotting
We detected the expression of adipophilin in 10 pairs of fresh lung adenocarcinoma and normal lung tissues by western blotting. In brief, 60 μg of protein were separated by SDS-PAGE, and then electroblotted onto membranes. The membranes were blocked with 5% nonfat dry milk in TBST buffer for 2 h at room temperature, and then incubated with the anti-adipophilin antibody (1:400) overnight at 4°C, followed by incubation with the horseradish peroxidase-conjugated secondary antibody (1:4000) after three washes with TBST. The signals were visualized using the enhanced chemiluminescence method. The expression of β-actin was used as loading control.
Immunohistochemistry
Immunohistochemistry (IHC) was performed using standard SP methods. In Brief, 4 um thick paraffin embedded sections were dewaxed, rehydrated with graded alcohol, and antigen was retrieved with a microwave. The intrinsic peroxidase activity was blocked using 3% hydrogen peroxide solution at room temperature for 10 min. After incubation with normal goat serum, the sections were incubated with anti-adipophilin antibody (1:200) overnight at 4°C. IHC was carried out using the SP9001 Rabbit kit (Zhongshan Jinqiao biotech company, Beijing, China). The immunoreaction was visualized using 3, 3’-diaminobenzidine (DAB) staining. Finally, then sections were counterstained with hematoxylin, dehydrated. For negative controls, the primary antibodies were replaced with PBS. All sections were examined microscopically and immunostaining were evaluated as described previously [16]. Intensity was graded as follows: neg, negative stain; 1+, focally weakly positive stain; 2+, focally intensely or diffusely weakly positive stain; 3+, diffusely intensely positive stain.
Statistical analysis
Statistical analysis was performed using SPSS (Version 16.0; Chicago, IL, USA). The associations between the adipophilin expression status and clinicopathological factors were analyzed using Mann–Whitney U test or Kruskal-Wallis test. The significant difference of adipophilin protein expression levels between lung cancer and normal tissue were analyzed using Mann–Whitney U test. In all tests, two-sided P-values <0.05 were considered statistically significant.
Results
Validation of adipophilin expression by western blotting
To confirm the differential expression of adipophilin, we performed western blotting analysis to detect the differential expression in 10 pairs of lung adenocarcinoma and normal lung tissues. Consistent with the previous findings, adipophilin is significantly increased in tumor tissues compared with adjacent normal tissues (P<0.01, Figure 2).
Figure 2.
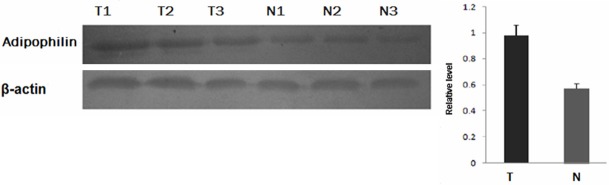
Representative results of the Western blotting of the expressions of adipophilin in tumor tissues and the matched normal tissues. β-actin was used as a loading control. T = tumor tissues; N = normal tissues.
Detection of the expression of adipophilin in lung cancer and normal lung tissues by IHC
The expression and cellular distribution of adipophilin were determined by IHC in 62 lung cancer samples including 41 lung adenocarcinoma specimens and 21 lung squamous cell carcinomas and 24 normal lung tissues. In 12 lung adenocarcinoma tissues, adipophilin were diffusely intensely or weakly stain in the cytoplasm or the vicinity of the cell membrane in most tumor cells (Figure 3A and 3B). However, adipophilin were only focally or weakly positive stain in most of lung squamous cell carcinomas and normal lung specimens (Figure 3C and 3D). Compared with normal specimens, the expression level of adipophilin in adenocarcinoma was significantly increased (P=0.001). The upregulation of adipophilin were also observed in the lung cancer (lung adenocarcinoma + squamous cell carcinoma) compared with normal lung specimens (P=0.007) (Table 1). While there were no significant difference of adipophilin expression between lung squamous cell carcinoma and normal lung specimens. Furthermore, we analyzed the association between clinicopathological characteristics and adipophilin expression in lung cancer. The correlation between the clinicopathological characteristics and adipophilin expression is listed in Table 2. The results indicated the upregulation of adipophilin in lung adenocarcinoma compared with squamous cell carcinoma (P=0.026). The expression of adipophilin did not correlated with other clinicopathologic factors such as lymph node metastasis, patients’ age, gender, tumor size, grade, and TNM stage.
Figure 3.
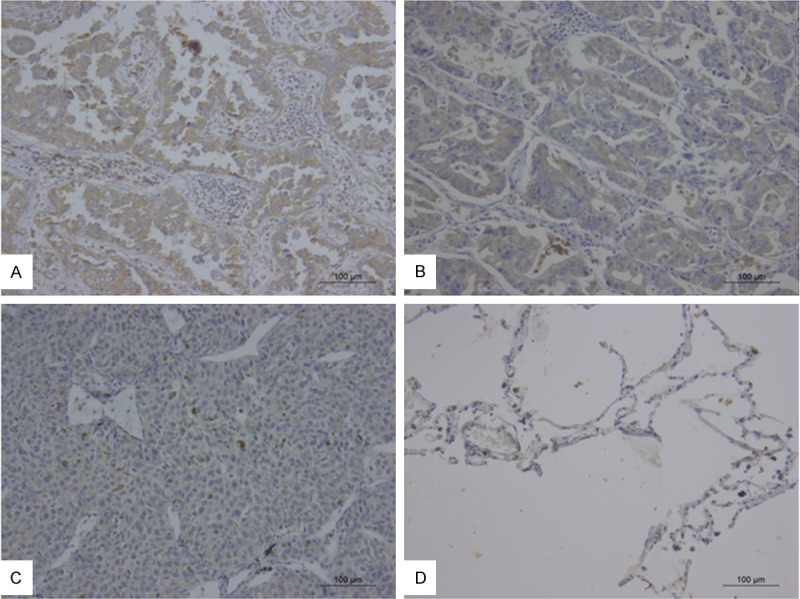
Immunohistochemical staining of adipophilin in lung adenocarcinoma (A, B), lung squamous carcinomas (C), and normal lung tissue (D) (IHC ×200).
Table 1.
Adipophilin expression in lung cancer and normal lung tissues
| Groups | N | Expressional levels | P* | |||
|---|---|---|---|---|---|---|
|
| ||||||
| neg | 1+ | 2+ | 3+ | |||
| LAC | 41 | 6 | 23 | 7 | 5 | 0.001a |
| LSCC | 21 | 7 | 12 | 2 | 0 | 0.335b |
| Normal lung tissues | 24 | 11 | 12 | 1 | 0 | 0.007c |
LAC versus normal lung tissue;
LSCC versus normal lung tissue;
Lung cancer versus normal lung tissue;
P<0.05 by Mann–Whitney U test;
LAC, lung adenocarcinoma; LSCC, lung squamous cell carcinoma; Lung cancer, LAC+LSCC.
Table 2.
Correlation between clinicopathological characteristics and adipophilin expression in lung cancer
| Parameters | N | Expressional levels | P* | |||
|---|---|---|---|---|---|---|
|
| ||||||
| neg | 1+ | 2+ | 3+ | |||
| Age | ||||||
| <60 | 32 | 7 | 18 | 5 | 2 | 0.802 |
| ≥60 | 30 | 6 | 17 | 4 | 3 | |
| Gender | ||||||
| Male | 41 | 8 | 23 | 8 | 2 | 0.722 |
| Female | 21 | 5 | 12 | 1 | 3 | |
| Grade | ||||||
| G1+G2 | 34 | 9 | 19 | 4 | 2 | 0.171 |
| G3 | 28 | 4 | 16 | 5 | 3 | |
| Histology | ||||||
| LAC | 41 | 6 | 23 | 7 | 5 | 0.026 |
| LSCC | 21 | 7 | 12 | 2 | 0 | |
| Lymphatic invasion | ||||||
| N0 | 33 | 7 | 17 | 7 | 2 | 0.666 |
| N+ | 29 | 6 | 18 | 2 | 3 | |
| Tumor size | ||||||
| ≤3 | 22 | 3 | 13 | 5 | 1 | 0.654 |
| 3<T≤7 | 31 | 8 | 17 | 2 | 4 | |
| >7 | 9 | 2 | 5 | 2 | 0 | |
| TNM stage | ||||||
| I+II | 35 | 7 | 19 | 7 | 2 | 0.675 |
| III+IV | 27 | 6 | 16 | 2 | 3 | |
P<0.05 by Mann–Whitney U test.
Discussion
We performed iTRAQ-based quantitative proteomic analysis to identify the differentially expressed membrane proteins between lung adenocarcinoma and matched lung normal tissues. Adipophilin was significantly upregulated for 2.46 fold in lung adenocarcinoma tissues compared with normal tissues. In this study, we confirmed adipophilin expression level in the two types tissues, then further evaluated the association between adipophilin expression and clinicopathological factors in lung cancer patients.
Adipophilin is a member of PAT (PERILIPIN, ADRP, and TIP47) family. It was first identified in 1246 Cells during adipocyte differentiation and found as a membrane-associated protein by investigating localization [10,11]. Adipophilin was expressed not only at high levels in adipose tissue but also at lower level in many different types of cells and various tissues such as lung, liver, testes, etc [12,13].
Several studies suggested that adipophilin might be involved in uptake of long chain fatty acids, the formation or stabilization of lipid droplets in adipocytes and serve as a saturable transport component for long chain fatty acids [17,18]. Adipophilin also play an important role in this novel mechanism for the transfer of lipids from lipid droplet to the type 2 lung epithelial cells for the production of surfactant phospholipids, but such a role has not been substantiated [19,20]. The study by Torday et al [21] shows that adipophilin derived from lipofibroblast coordinates alveolar type II epithelial’ synthesis of surfactant phospholipid and surfactant protein-B. Recently, few studies showed adipophilin significantly expressed in clear-cell renal carcinoma and colorectal cancer cells [14-16].
In this study, we performed western blotting analysis to verify the result from quantitative proteomics. Consistent with our previous findings, the result showed that adipophilin in lung adenocarcinoma was significantly higher than in normal lung tissues. Furthermore, we evaluated the association between adipophilin and clinicopathological characteristics using immunohistochemistry. The results showed that the expression of adipophilin was increased in lung adenocarcinomas compared with normal lung tissues and lung squamous cell carcinomas. While we did not found significant difference of adipophilin expression between lung squamous cell carcinomas and normal lung tissues. In addition, our results indicated the expression of adipophilin in lung cancer did not correlate with other clinicopathologic factors such as lymph node metastasis, age, gender, tumor size, grade, and TNM stage. Few studies reported adipophilin was upregulated in clear-cell renal carcinoma and colorectal cancer cells and associated with the differentiated grade [14,16], but we did not found the association with grade. Our data indicated that adipophilin might play an important role in carcinogenesis of lung adenocarcinoma. To our knowledge, this is the first report to investigate adipophilin expression in lung cancer and indicate the correlation with pathological type of lung cancer. It has been demonstrated that adipophilin are inducible by hypoxia-inducible factor 1 (HIF-1), a heterodimer of HIF-1α and ARNT (Ah receptor nuclear translocator; HIF-1β) [22]. In addition, HIF-1α plays an important role in lung cancer progression and metastasis through activation of many target genes which are involved in important aspects of cancer biology [23]. Although the exact mechanism of the interactions between adipophilin and HIF in cancer has not been investigated, we speculated adipophilin may play a potential role in tumorigenesis through adaptation to hypoxia which is essential for tumor progression. Several studies showed dietary fat including dairy products, saturated fats, and lipids increased the risk for lung cancer [24-26] and adipophilin might be involved in uptake of long chain fatty acids. Taken together, we hypothesize adipophilin may play a possible role in carcinogenesis by involving in fatty acid uptake and transport in lung.
However, there are some limitations in the present study. Firstly, only few IIIB and IV stages specimens were used because surgery was mostly involved in patients with early stage lung cancer, which may results in selection bias, secondly our results may be still preliminary results because of relatively small sample sizes and experimental investigations involving a larger group of patients is needed to further evaluate.
In conclusions, we confirmed the result from quantitative proteomics that adipophilin were upregulated in lung adenocarcinoma by western blotting and immunohistochemistry. Our results indicate that the upregulation of adipophilin was potentially involved in the carcinogenesis of lung adenocarcinoma and adipophilin may serve as a potential marker for distinguishing between lung adenocarcinoma and lung squamous cell carcinoma. However, a study of larger populations is required to further confirm our results.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (No. 81350032).
Disclosure of conflict of interest
None.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Travis WD, Brambilla E, Noguchi M, Nicholson AG, Geisinger K, Yatabe Y, Powell CA, Beer D, Riely G, Garg K, Austin JH, Rusch VW, Hirsch FR, Jett J, Yang PC, Gould M. American Thoracic Society: International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society: international multidisciplinary classification of lung adenocarcinoma: executive summary. Proc Am Thorac Soc. 2011;8:381–385. doi: 10.1513/pats.201107-042ST. [DOI] [PubMed] [Google Scholar]
- 3.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 4.Drissi R, Dubois ML, Boisvert FM. Proteomics methods for subcellular proteome analysis. FEBS J. 2013;280:5626–5634. doi: 10.1111/febs.12502. [DOI] [PubMed] [Google Scholar]
- 5.Boylan KL, Andersen JD, Anderson LB, Higgins L, Skubitz A. Quantitative proteomic analysis by iTRAQ(R) for the identification of candidate biomarkers in ovarian cancer serum. Proteome Sci. 2010;8:31. doi: 10.1186/1477-5956-8-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen JS, Chen KT, Fan CW, Han CL, Chen YJ, Yu JS, Chang YS, Chien CW, Wu CP, Hung RP, Chan EC. Comparison of membrane fraction proteomic profiles of normal and cancerous human colorectal tissues with gel-assisted digestion and iTRAQ labeling mass spectrometry. FEBS J. 2010;277:3028–3038. doi: 10.1111/j.1742-4658.2010.07712.x. [DOI] [PubMed] [Google Scholar]
- 7.Zhang Z, Zhang L, Hua Y, Jia X, Li J, Hu S, Peng X, Yang P, Sun M, Ma F, Cai Z. Comparative proteomic analysis of plasma membrane proteins between human osteosarcoma and normal osteoblastic cell lines. BMC Cancer. 2010;10:206. doi: 10.1186/1471-2407-10-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Song G, Hu C, Zhu H, Li X, Zhao L, Zhou R, Zhang X, Zhang F, Wu L, Li Y. Comparative proteomics study on liver mitochondria of primary biliary cirrhosis mouse model. BMC Gastroenterol. 2013;13:64. doi: 10.1186/1471-230X-13-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang XD, Li W, Hou YL, Niu ZQ, Zhong YJ, Zhang YP, Yang SY. Comparative membrane proteomic analysis between lung adenocarcinoma and normal tissue by iTRAQ labeling mass spectrometry. Am J Transl Res. 2014:6. [Epub ahead of print] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang HP, Harris SE, Serrero G. Molecular cloning of a differentiation-related mRNA in the adipogenic cell line 1246. Cell Growth Differ. 1992;3:21–30. [PubMed] [Google Scholar]
- 11.Jiang HP, Serrero G. Isolation and characterization of a full-length cDNA coding for an adipose differentiation-related protein. Proc Natl Acad Sci U S A. 1992;89:7856–7860. doi: 10.1073/pnas.89.17.7856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brasaemle DL, Barber T, Wolins NE, Serrero G, Blanchette-Mackie EJ, Londos C. Adipose differentiation-related protein is an ubiquitously expressed lipid storage droplet-associated protein. J Lipid Res. 1997;38:2249–2263. [PubMed] [Google Scholar]
- 13.Heid HW, Moll R, Schwetlick I, Rackwitz HR, Keenan TW. Adipophilin is a specific marker of lipid accumulation in diverse cell types and diseases. Cell Tissue Res. 1998;294:309–321. doi: 10.1007/s004410051181. [DOI] [PubMed] [Google Scholar]
- 14.Matsubara J, Honda K, Ono M, Sekine S, Tanaka Y, Kobayashi M, Jung G, Sakuma T, Nakamori S, Sata N, Nagai H, Ioka T, Okusaka T, Kosuge T, Tsuchida A, Shimahara M, Yasunami Y, Chiba T, Yamada T. Identification of adipophilin as a potential plasma biomarker for colorectal cancer using label-free quantitativemass spectrometry and protein microarray. Cancer Epidemiol Biomarkers Prev. 2011;20:2195–2203. doi: 10.1158/1055-9965.EPI-11-0400. [DOI] [PubMed] [Google Scholar]
- 15.Yao M, Huang Y, Shioi K, Hattori K, Murakami T, Nakaigawa N, Kishida T, Nagashima Y, Kubota Y. Expression of adipose differentiation-related protein: a predictor of cancer-specific survival in clear cell renal carcinoma. Clin Cancer Res. 2007;13:152–160. doi: 10.1158/1078-0432.CCR-06-1877. [DOI] [PubMed] [Google Scholar]
- 16.Yao M, Tabuchi H, Nagashima Y, Baba M, Nakaigawa N, Ishiguro H, Hamada K, Inayama Y, Kishida T, Hattori K, Yamada-Okabe H, Kubota Y. Gene expression analysis of renal carcinoma: adipose differentiation-related protein as a potential diagnostic and prognostic biomarker for clear-cell renal carcinoma. J Pathol. 2005;205:377–387. doi: 10.1002/path.1693. [DOI] [PubMed] [Google Scholar]
- 17.Steiner S, Wahl D, Mangold BL, Robison R, Raymackers J, Meheus L, Anderson NL, Cordier A. Induction of the adipose differentiation-related protein in liver of etomoxir-treated rats. Biochem Biophys Res Commun. 1996;218:777–782. doi: 10.1006/bbrc.1996.0138. [DOI] [PubMed] [Google Scholar]
- 18.Gao J, Serrero G. Adipose differentiation related protein (ADRP) expressed in transfected COS-7 cells selectively stimulates longchain fatty acid uptake. J Biol Chem. 1999;274:16825–16830. doi: 10.1074/jbc.274.24.16825. [DOI] [PubMed] [Google Scholar]
- 19.Schultz CJ, Torres E, Londos C, Torday JS. Role of adipocyte differentiation-related protein in surfactant phospholipid synthesis by type II cells. Am J Physiol Lung Cell Mol Physiol. 2002;283:L288–296. doi: 10.1152/ajplung.00204.2001. [DOI] [PubMed] [Google Scholar]
- 20.Magra AL, Mertz PS, Torday JS, Londos C. Role of adipose differentiation-related protein in lung surfactant production: a reassessment. J Lipid Res. 2006;47:2367–2373. doi: 10.1194/jlr.M600157-JLR200. [DOI] [PubMed] [Google Scholar]
- 21.Torday J, Rehan V. Neutral lipid trafficking regulates alveolar type II cell surfactant phospholipid and surfactant protein expression. Exp Lung Res. 2011;37:376–386. doi: 10.3109/01902148.2011.580903. [DOI] [PubMed] [Google Scholar]
- 22.Saarikoski ST, Rivera SP, Hankinson O. Mitogen-inducible gene 6 (MIG-6), adipophilin and tuftelin are inducible by hypoxia. FEBS Lett. 2002;530:186–190. doi: 10.1016/s0014-5793(02)03475-0. [DOI] [PubMed] [Google Scholar]
- 23.Ren W, Mi D, Yang K, Cao N, Tian J, Li Z, Ma B. The expression of hypoxia-inducible factor-1α and its clinical significance in lung cancer: a systematic review and metaanalysis. Swiss Med Wkly. 2013;143:w13855. doi: 10.4414/smw.2013.13855. [DOI] [PubMed] [Google Scholar]
- 24.Alavanja MC, Brownson RC, Benichou J. Estimating the effect of dietary fat on the risk of lung cancer in nonsmoking women. Lung Cancer. 1996;14:S63–S74. doi: 10.1016/s0169-5002(96)90211-1. [DOI] [PubMed] [Google Scholar]
- 25.De Stefani E, Fontham ET, Chen V, Correa P, Deneo-Pellegrini H, Ronco A, Mendilaharsu M. Fatty foods and the risk of lung cancer: a case-control study from Uruguay. Int J Cancer. 1997;71:760–766. doi: 10.1002/(sici)1097-0215(19970529)71:5<760::aid-ijc12>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 26.Goodman MT, Kolonel LN, Yoshizawa CN, Hankin JH. The effect of dietary cholesterol and fat on the risk of lung cancer in Hawaii. Am J Epidemiol. 1988;128:1241–1255. doi: 10.1093/oxfordjournals.aje.a115078. [DOI] [PubMed] [Google Scholar]


