Abstract
The TGF-beta pathway is an evolutionarily conserved signal transduction module that mediates diverse biological processes in animals. In Drosophila, both the BMP and Activin branches are required for viability. Studies rooted in classical and molecular genetic approaches continue to uncover new developmental roles for TGF-beta signaling. We present an overview of the secreted ligands, transmembrane receptors and cellular Smad transducer proteins that comprise the core pathway in Drosophila. An assortment of tools have been developed to conduct tissue-specific loss and gain-of-function experiments for these pathway components. We discuss the deployment of these reagents, with an emphasis on appropriate usage and limitations of the available tools. Throughout, we note reagents that are in need of further improvement of development, and signaling features requiring further study. A general theme is that comparison of phenotypes for ligands, receptors, and Smads can be used to map tissue interactions, and to separate canonical and non-canonical signaling activities. Core TGF-beta signaling components are subject to multiple layers of regulation, and are coupled to context-specific inputs and outputs. In addition to fleshing out how TGF-beta signaling serves the fruit fly, we anticipate that future studies will uncover new regulatory nodes and modes and will continue to advance paradigms for how TGF-beta signaling regulates general developmental processes.
Keywords: TGF-beta, Activin, BMP, Drosophila
1. Introduction: An ancient signaling pathway
The core canonical TGF-β pathway is deceptively simple. Extracellular ligands form a complex with Type II and Type I transmembrane receptors, which phosphorylate Smad effector proteins to regulate their transcription factor activity. There are two pathway branches that effectively run in parallel but share several components. Numerous regulatory mechanisms that modulate TGF-β signaling have been discovered, but the strong relationship between ligands, receptors and Smads provides the framework for much research into TGF-β signaling.
TGF-β pathway signaling is a hallmark feature of metazoan cell communication. All metazoan genomes studied to data encode a suite of core TGF-β pathway proteins, including multiple ligands, at least three Type I receptors, at least one Type II receptor, and four or more Smads [1–3]. It can be difficult to categorize ligands and receptors by sequence alone, but the presence of BMP- and TGF-β/Activin-type Receptor-regulated Smads (R-Smads) in all genomes suggests that a double-barreled pathway is common to metazoans. This clouds the nature of the “original” pathway, but stokes the exciting possibility that evolution of animals and the crystallization of an elaborated TGF-β pathway have a shared natural history. The original assembly of the components into a functional signal transduction module and the implied duplications of factors is lost to history, but it seems likely that the ancestral role of TGF-β signaling in animals was early embryonic patterning. For the modern researcher, a relevant perspective is that some features of the TGF-β pathway have been around since the dawn of animals, but millions of years of speciation and regulatory ornamentation have produced impressive variety. Researchers must therefore maintain an open mind when studying TGF-β signaling.
Strongly connected nodes in regulatory networks tend to be highly conserved between species [4]. The factors then become stationary targets in evolutionary terms, allowing new cell- and species-specific connections to form. The deep conservation of the core TGF-β signaling pathway in metazoans thus allows the diverse regulation of the pathway inputs and outputs. An overarching theme is that studies of TGF-β signaling typically leverage general knowledge of the canonical players while attempting to explain specific pathway inputs and outputs for a given biological process. In many cases using model organisms has helped identify not only the major players for modulation of TGF-β signaling activity but also their molecular activities. The best examples of this to date are the study of BMP morphogen gradients and movement of signaling molecules [5–6]. Facilitated diffusion of Dpp in the early embryo is one of the classic cases of morphogen action pattern changing over developmental time. This involves the activity of many ligand and receptor interacting proteins. Similarly, the Dpp gradient in the wing imaginal disc has served as a model system for testing morphogen diffusion, gradient scaling, and other potential transport mechanisms such as transcytosis [7] and cytoneme delivery [8].
In this chapter we describe the TGF-β signaling network in Drosophila and resources available to researchers who wish to study the role of the signaling pathway in their biological process of interest. Of course, many studies will employ both traditional Drosophila genetics, as well as more recent molecular genetic approaches. These include classical mutant analysis, RNAi-based gene silencing, and genome engineering methodologies. Still other experiments will require specialized approaches based on the biological process under study. For example, NMJ studies require electrophysiology measurements and metabolic studies require collection of metabolomic datasets. Here we focus on the tools available to study the activities of TGF-β signaling components and how they are used to explain intracellular signaling.
2. Overview of core TGF-β pathway factors of Drosophila
The term TGF-β is used in several contexts of varying scale. It can be used as a shorthand for denoting the signaling events associated with all TGF-β superfamily ligands, or it many indicate a more narrow context that refers to the Activin/TGF-β branch of the pathway that acts through Smad2/3, or it might refer to a particular vertebrate TGF-β ligand (e.g. TGF-β1). As Drosophila lacks sensu strictu TGF-β ligands, for simplicity here we assign the term TGF-β to the entire superfamily signaling network, and we refer to the two specific branches found in Drosophila as the BMP and Activin pathways.
The Drosophila genome encodes a compact set of components that is overall a good representative for the metazoan TGF-β toolkit [9]. The genes encoding ligands, receptors, and Smads are presented in Table 1. Several deviations are notable because they illustrate areas of evolutionary plasticity. The Scw ligand represents a specialized BMP-type factor found only in higher dipterans that participates in the highly derived syncytial early embryo patterning [10–11]. The Maverick ligand cannot be easily categorized based on sequence alignment [12], illustrating the trend that ligands tend to have greater sequence diversity than other players. Finally, the Baboon Type I receptor has three isoforms that differ only in the ligand binding domain [13–14] thereby potentially supplying signaling specificity for three different ligands; it is not clear how prevalent this strategy is in different animals. A suite of regulatory molecules that are often considered part of the network are also encoded in the fruit fly genome. These include Follistatin [15] and the interacting group of proteins Sog(Chordin), Twisted gastrulation and Tolloid [16,17].
Table 1.
TGF-β factors of Drosophila. The names of the ligands, receptors and Smads encoded in the fruit fly genome are listed, along with their molecular type within the signaling pathway. Representative roles are listed based on prominent developmental defects observed in mutants. Note that many other roles have been described, especially for the BMP branch. See Flybase for a more complete list of references concerning the study of TGF-β pathway mutations.
| Factor Name (Symbol) | Category | Representative Roles |
|---|---|---|
| Decapentaplegic (Dpp) | BMP branch ligand | D/V embryo patterning [18] Inductive events (e.g. gut) [107] Disc patterning [19] Germline stem cell [45] |
| Glass Bottom Boat (Gbb) | BMP branch ligand | NMJ homeostasis [119] Fine-tuning disc patterning [120] Fat body metabolism [121] |
| Screw (Scw) | BMP branch ligand | D/V embryo patterning [122] |
| Activin-β (Actβ) | Activin branch ligand | Neuroblast proliferation [27] Mutants lethal with escapers |
| Dawdle (Daw) | Activin branch ligand | Axon guidance [49] Promotes on-time development |
| Myoglianin (Myo) | Activin branch ligand | Axon pruning in mushroom body [28] Required for pupal development |
| Maverick (Mav) | Activin branch ligand? | Glia signaling at NMJ [23] |
| Thickveins (Tkv) | BMP Type I receptor | Primary receptor for Dpp [123] |
| Saxophone (Sax) | BMP Type I receptor | Gbb receptor, dimerizes with Tkv [17] |
| Baboon [a,b,c] (Babo) | Activin Type I receptor | Axon guidance and pruning [20] Cell proliferation [27] |
| Punt (Put) | Type II receptor | Primary type II receptor for BMPs and Activins (?) [75] |
| Wishful Thinking (Wit) | Type II receptor | NMJ function [76] Neurosecretory cell functional [77] Eggshell patterning [78] |
| Mothers Against Dpp (Mad) | BMP R-Smad | Transcriptional activator and repressor in multiple tissues [124] |
| Smad on X (dSmad2/Smox) | Activin R-Smad | Neuroblast proliferation [20] Prothoracic Gland function [112] Inhibits Baboon activity in wing disc [22] |
| Medea (Med) | Common Smad (Co-Smad) | Transcriptional co-factor for Mad and dSmad2 [99,125] May mediate balance between branches [93] |
| Daughters Against Dpp (Dad) | Inhibitory Smad (I-Smad) | Feedback inhibitor for Mad [46] |
In comparison to mammalian genomes, Drosophila has far fewer ligands, fewer receptors, and fewer Smad proteins [9]. This suggests that many roles of TGF-β signaling in vertebrates may be specific to this phylum and will need to be studied directly in those organisms to determine the details of signal regulation. On the positive side, the compact genome of Drosophila facilitates functional studies with less obstruction from redundancy. The study of TGF-β signaling in Drosophila stems from identification of mutants with defective development.
After several decades of study, BMP signaling is well known for its instructional roles in patterning. Without Dpp signaling, Drosophila development fails early and often. Dpp is famously required for dorsal-ventral patterning of the embryo, and later for the formation, growth and patterning of imaginal discs [18–19]. The Activin signaling branch is also required for viability. Mutants in the shared pathway members accumulate developmental defects and fail to pupate properly [14,20,21]. The requirement for viability underscores the importance of the pathways, but the lethality can obscure pleiotropic signaling roles in development and homeostatic functions such as metabolism and tissue repair. For these investigations, conditional and tissue-specific manipulations are required to reveal the function of TGF-β signaling in different tissues. Below we discuss the tools used to study ligands, receptors and Smad proteins.
3. Ligands
Under normal conditions, ligands initiate the signal cascade by binding to receptors. Four fundamental aspects for studying ligands are: identifying producing cells, determining a ligand’s distribution surrounding its source, identifying the mechanism of ligand spread, and characterizing ligand binding to receptors on receiving cells (See Figure 1A).
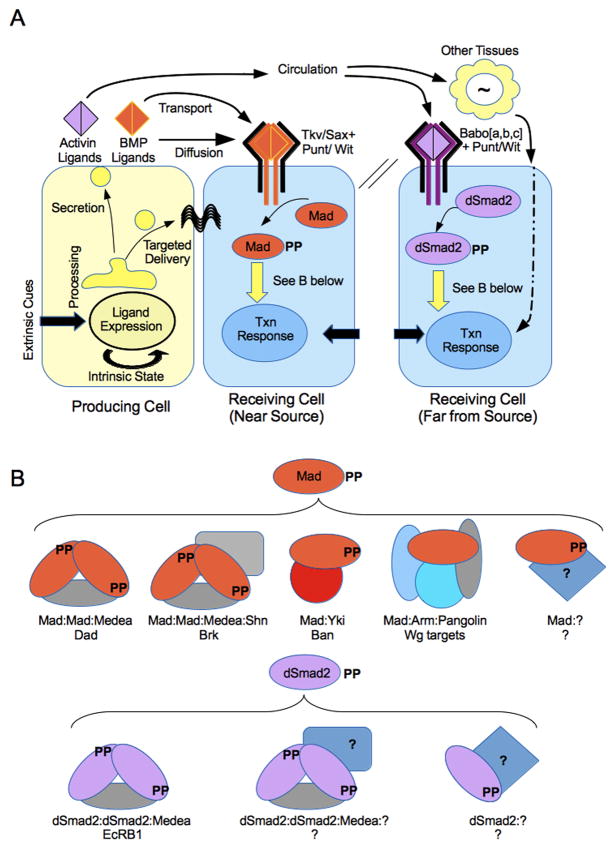
Figure 1. TGF-β ligand delivery and canonical transcriptional response of receiving cells.
(A) TGF-β ligands of the BMP (orange) and Activin (purple) classes are expressed and processed in source cells. Delivery can occur by targeted delivery to neighboring cells or by secretion. Ligand movement outside of the cell can occur by local diffusion, active transport, or widespread circulation. Responding cells assemble a complex of ligand and Type I and Type II receptors that phosphorylate R-Smads. BMP ligands and Type I receptors lead to Mad phosphorylation and sometimes dSmad2 phosphorylation, whereas Activin ligands and TypeI receptor isoforms are only known to phosphorylate dSmad2. Each cell’s response will be a function of many factors, including receptors expressed, co-receptors and surface proteins, transcriptional cofactors, and input from other signaling factors. The flower represents the general concept that TGF-β ligands can act indirectly on a target cell, using other tissues to relay information. (B) Activated Mad and dSmad2 as multi-modal transcription factors. The best studied complexes regulate Activator Elements and Silencer Elements. Mad:Medea without Shn activate targets such as Dad. Mad:Medea:Shn complexes repress SE-containing targets, principally Brk. Mad also complexes with Yorkie(Yki) to regulate bantam, and with Armadillo(Arm) and Pangolin to regulate Wg targets. Unknown Mad complexes are likely to regulate different genes. dSmad2 is less characterized, but a canonical dSmad2:Medea complex likely regulates EcRB1. By analogy to other Smads, there are probably undiscovered cofactors that work with dSmad2 with or without Medea. Not depicted are blended complexes containing Mad and dSmad2 that may form and have different transcription factor activities.
3.1 Ligand expression
Expression of TGF-β ligands in a particular cell may depend on the cell’s intrinsic state (incorporating cell type and history of the cell) and extrinsic cues including exposure to signaling molecules. The expression of dpp illustrates these concepts. Consistent with Dpp’s vital role in conveying positional information during development, the spatio-temporal expression control of dpp has been extensively studied. Even in cases were Dpp is expressed in a defined position, such as the wing imaginal disc, the expression level can be modulated by the growth state of the tissue [22]. Dpp production can also be conditional, as shown in the midgut where dpp expression is induced by tissue damage. Gbb acts as a retrograde signal to neurons to control NMJ size, and its expression in muscles is controlled by Maverick signaling from associated glial cells [23]. Inputs for the expression of other TGF-β ligands remain to be described in terms of their developmental controls and environmental inputs.
In principle, identifying ligand-producing cells and tissues is relatively straightforward using either immunohistochemistry (IHC), in situ hybridization (ISH), or tissue-specific RT-PCR, and this information can provide important clues for where a ligand functions. For example, the Myoglianin ligand is named for its prominent expression in muscle and glial cells. Expression reporters are also useful to identify producing cells and to monitor changes due to genetic perturbation. Useful reagents include Dpp-LacZ [24], Dpp-Gal4 [25], Actβ-Gal4 [26], Dawdle-GAL4 [27] and Myo-Gal4 [28]. It is important to bear in mind that it is not trivial to capture the full range of endogenous expression. ISH and expression reporters may miss signaling tissues if the expression is weak, or expression is induced only under certain conditions. In addition, enhancer traps and regulatory fusions many not contain all the relevant enhancers. The dpp locus is large and contains well-characterized enhancers such as those mediating imaginal disc expression, but new enhancers and expression domains continue to be identified. Examples include feedforward dpp expression domains that pattern the cardiac field and the posterior spiracles [29–30], autocrine dpp expression in the corpus allatum that regulates Juvenile Hormone signaling [31], and visceral muscle expression that supports a role for Dpp in limiting gut stem cell proliferation [32].
To directly influence other cells, the ligand must leave the cell, so secretion is an important step. This topic has received limited attention, but conceptually the secretion rate may be tunable to modulate signal strength, and secretion may be targeted to limited membrane structures. As an alternative to traditional secretion, membrane extensions called cytonemes have been proposed as a mechanism to directly transfer Dpp between cells [8]. A recent study of Dpp trafficking in the germ stem cell niche found a specific secretion pathway in the source cell that spatially restricted receptor activation in the receiving cell [33].
3.2 Ligand detection reagents
Additional experiments are required to infer the range of action of a ligand expressed in a given locale. Protein detection reagents are needed to directly follow the ligand and antibodies against several ligands have been generated. Dpp antibodies were developed years ago [34], but have largely been supplanted by detection of tagged forms, as described below. More recently, an antibody against Gbb was used to show that two forms of Gbb are produced in vivo by differential processing [35] and another antibody was used to monitor exposed Gbb protein at the NMJ [36]. An antibody against Maverick detected puncta near the NMJ to support a signaling role for glial cells [23]. Validated antibodies for the remaining ligands have not been reported. Several antibodies raised against Drosophila TGF-β proteins are available from commercial outlets, but as with any antibody study, rigorous tests in the absence of antigen must be conducted to support specificity of detection in Western or IHC applications.
3.3 Tagged ligands
A useful alternative to native antibodies is the use of tagged expression constructs. In addition to detecting ligands for which no antibody is available, tagged constructs can permit live analysis and allow detection of mutant variants. For mature BMP ligands, bioactive tagged versions are available for Dpp (HA and GFP) [37–39], Gbb (Flag, GFP and Venus) [40–42] and Scw (Flag) [40]. The processing and secretion of TGF-β ligands may impose restrictions on the location of useful tags for some ligands. For Activin ligands, pro-domain tags have been used to monitor ligand processing [43], but the only reported tagged mature protein is a C-terminally tagged Maverick construct, which behaves similarly to the untagged version [23]. Detection reagents for the other mature ligands would be valuable additions.
In cases where signaling targets are known, the range of action for ligands can be inferred from monitoring the expression of the transcriptional targets. In practice, this approach has been almost entirely limited to BMP targets. In the wing disc several positive and negative targets respond to a gradient of BMP signaling [44]. Among these, dad expression is a particularly useful readout of BMP signaling in a variety of tissues, such as gonadal stem cell niches [45] and the gut [32]. dad-LacZ has been used as a reporter [46] and more recently GFP and RFP reporters were generated [47–48]. EcRB1 expression is a readout for Activin signaling in the mushroom body, but whether it is a direct target is not known. Clearly, further study is needed to identify additional direct targets for both the BMP and Activin branches in other tissues and at other times in development.
3.4 BMP ligand distribution
The range of action has been studied extensively for Dpp with constructs that express bioactive tagged Dpp. In both the early embryo and the wing discs, the expression pattern of Dpp is dramatically modified to produce the extracellular ligand distribution that patterns surrounding cells. In the embryo, dpp is expressed in a broad dorsal domain, but the distribution of Dpp protein is refined by a network of factors that concentrate Dpp at the dorsal midline by facilitated diffusion [17]. In the wing disc, a narrow stripe of cells located at the A/P compartment boundary secrete Dpp that spreads within the epithelial cell layer to form a gradient that decays into each compartment with different characteristics [38,39]. These two systems illustrate that ligand distribution is tunable to meet the specific tissue patterning context. These systems have also been fertile ground for mathematical modeling of morphogen action. Experiments and modeling reveal that robust patterning requires many factors. In the embryo, an orchestra of ligand binding proteins, proteases, and surface proteins are required to form a sharp dorsal stripe of Dpp. In the wing disc, a gradient that scales with the growing tissue requires a network of positive and negative feedback loops [6]. Dpp signaling is also critical for germ cell proliferation and renewal in the ovary and testis. Studies in these organs uncovered multiple mechanisms that limit ligand production, distribution and reception by surrounding cells [45].
3.5 Range of action for Activin ligands
For the Activin branch, little is known about ligand distribution. A recent study of Maverick signaling at the larval NMJ observed the delivery of Maverick-GFP from glial cells to muscles [23]. For Activin-β and Myoglianin, direct experimentation is still required to determine the range of action. The Dawdle ligand is notable because it has been modeled as a circulating factor based on genetic tests wherein ectopic Dawdle expression restores viability to otherwise lethal dawdle mutant animals [49,50]. Indeed, Dawdle was detected in a recent survey of larval hemolymph proteins [51]. Other TGF-β ligands were not detected, but this negative result is inconclusive because only a subset of known circulating proteins were identified. Production of soluble ligands from S2 cells has been used for cell culture signaling assays. All three BMP ligands and the Dawdle ligand can be readily obtained from conditioned media [16,17,13]. Other Activin ligands can be processed and secreted in cell culture, but do not activate downstream signaling [43]. It is not clear if this is due to altered ligand production, or the lack of cofactors that might be required for the ligand to activate receptors.
Although localized action may seem the default expectation considering the prominent roles of BMP ligands in patterning events, the Dawdle example above, as well as multiple examples in vertebrates of the presence of ligands in blood, indicate that various ranges of activity must be considered and addressed experimentally. The factors that influence the diffusion properties of a ligand remain to be explored, but they could include binding to cell surfaces, or binding to circulating proteins or particles. These properties may be influenced by the amino acid sequence of the exposed surfaces of the ligand, or by post-translational modifications. Like most secreted proteins, TGF-β ligands are (or are predicted to be) glycosylated. The modifications are not required for Dpp activity [52], but it is conceivable that the modifications influence stability or diffusibility. The range of action of other prominent signaling molecules such as Hedgehog and Wnt are modulated by lipidation; it is possible that similar modifications alter the properties of TGF-β superfamily ligands. If modifications or processing events occur in a cell-type specific fashion, a ligand can have different signal properties based on its cell of origin. An example of this concept is the processing of Gbb, which is secreted as a large form retaining much of the prodomain or a smaller fully processed ligands. The two forms exhibit different signal ranges and the amount of each form produced varies by tissue [35]. Dpp processing also generates several mature ligand forms, and the proteins display different signaling ranges in vivo [53].
In addition to the limiting cases of paracrine signaling to neighboring cells or general delivery of ligands through the circulatory system, other fluid compartments can result from cell biology and tissue arrangements. For example, in most imaginal discs a lumen is formed between the apical surfaces of epithelial cells of the disc proper and peripodial membrane. This presents a situation where signaling within and between cell layers may be differentially regulated. Another division of spaces that may allow a ligand to be both diffusible and compartmentalized is the blood-brain barrier formed by glial cells. Myoglianin is expressed in glia, and a recent paper suggested that mushroom body neurons compete for a pool of Myo on the brain side of the blood-brain-barrier [54].
In cases where TGF-β signaling is suspected to mediate an endocrine function or multi-tissue signal, two broad possibilities need to be considered. One or more TGF-β ligands may indeed be the circulating factor that acts directly on target tissue, or intermediary factors may act as relays of the TGF-β signal. These questions need to be addressed on a case by case basis through tissue specific gain- and loss-of-function studies for both ligands and downstream components described in later sections. Other modes of action-at-a-distance regulation may come from known interacting factors such as the circulating Tolloid-related protease [43] or from unexpected sources such as the fatbody-derived cv-d factor that promotes crossvein formation in the wing [55].
3.6 Heteromeric ligands
In the vertebrate world, many types of heteromeric ligand species have been identified. These include heterodimers between various BMPs as well as Activin family members [56–59]. Since the dimeric ligands are covalently linked through a disulfide bridge, heterodimers are assumed to form during the secretion process. Thus their formation likely requires expression of both monomers in a common cell. In Drosophila, heterodimers between Dpp and the two other BMPs, Scw and Gbb, have been identified and proposed to provide specific functions distinct from the homodimers. In the Drosophila Activin branch, no clear example of heterodimerization has been identified within the subfamily nor have cases of cross-family (BMP/Activin) heterodimerization been observed. Nevertheless, the possibility of their formation suggests caution when interpreting new phenotypes associated with ligand mutations.
3.7 Loss-of-function approaches for ligands
The classical approach to study gene function is through analysis of loss-of-function mutant phenotypes. Table 2 shows that mutants available for all TGF-β ligand genes except maverick. In addition to pleiotropy concerns, interpreting ligand mutant phenotypes is complicated by their expected non cell-autonomous roles. Dominant-negative approaches have also been used to study ligand function, but like other dominant-negative methods they work by titration, and exactly what is being titrated is often not clear. For example, TGF-β ligands are dimers that are generated from a proprotein by proteolytic processing. Dominant negative forms have been made by expressing a noncleavable proprotein that can dimerize with a wildtype precursor but since only one subunit of the dimer is processed, mature ligand is not released [26]. The problem with this strategy is that TGF-β ligands can heterodimerize before the proteolytic maturation step. Thus, a noncleavable proprotein of one ligand might dimerize with different ligand resulting in titration of multiple ligands into inactive complexes. A more targeted strategy is to use RNAi-mediated ligand transcript degradation in defined tissues or defined times. A future approach that combines the cleanness of genetics with spatial control might be to engineer endogenous ligand expression constructs that can be rendered inert by tissue-specific flip-out recombination. Regardless of the method, the results must be interpreted in the context of the range of action of the ligand and the position of target cells.
Table 2.
An overview of the reported tools useful for the study of TGF-β signaling in vivo. Traditional mutants are available for all members except for Mav. RNAi is RNA interference, DN is dominant-negative, OE is over-expression, and CA is constitutively active form. See the text for details and see Flybase for updated lists of reagents.
| Factor | Mutations (see Flybase) | Loss of Function | Gain of Function | Expression Reporters, Other Detection Tools |
|---|---|---|---|---|
| Dpp | Many | RNAi | OE | Reporters, Tagged |
| Gbb | Many | RNAi | OE | Antibodies, Tagged |
| Scw | Many | OE | Tagged | |
| Act-β | One | RNAi, DN | OE | Reporter |
| Daw | Several | RNAi | OE | Reporter |
| Myo | One | RNAi | OE | Reporter |
| Mav | RNAi | OE | Antibody, Mav-GFP | |
| Tkv | Many | RNAi, DN | OE, CA | Antibodies, Tagged |
| Sax | Many | RNAi, DN | OE, CA | Antibodies, Tagged |
| Babo | Many | RNAi, DN | OE, CA | |
| Punt | Many | RNAi, DN | OE | Antibody |
| Wit | Several | RNAi, DN | OE | Antibody, Tagged |
| Mad | Many | RNAi | OE, CA(?) | Tagged, P-Antibodies |
| dSmad2 | Several | RNAi | OE, CA(?) | Antibodies, Tagged |
| Medea | Many | RNAi | OE | Antibodies |
| Dad | Several | OE | Reporters |
3.8 Gain-of-function approaches for ligands
While loss-of-function experiments establish the necessity of a factor for a particular biological process, gain-of-function experiments can sometimes be used to demonstrate sufficiency, and in the case of ligands, can provide a tool for analysis of ligand spread from a source cells. The classic example is the use of flip-out technology to activate a clone of Dpp-expressing cells in the wing disc and examining the temporal activation of downstream target genes. This methodology helped demonstrate the morphogen properties of Dpp [60]. Similar flip-out activating expression constructs for other ligands have not been reported, but these could prove useful, and additional temporal control can also be achieved through use of Geneswitch methodologies [61]. In a related approach, Gbb constructs engineered to produce different isoforms were expressed in portions of the wing disc. The observed downstream response of distant cells differed, indicating the isoforms differ in their range of action [35].
4. Receptors
The formation of active signaling complexes requires the physical interaction of ligands and receptors. In addition to the extracellular ligand transport and diffusion mechanisms considered above, clues are emerging that formation of the receptor complex is tightly controlled. Here we summarize the known activities of the receptors, and develop the argument that more research is needed to understand the mechanisms that limit and facilitate formation of active signaling complexes.
4.1 Two receptor types
TGF-β receptors are classified into Type I and Type II receptors. Both are single pass transmembrane proteins with a cytoplasmic Ser/Thr kinase domain. Based on molecular genetic and structural studies, the active receptor complex is a tetramer comprising two Type I and two Type II molecules. Type II receptors are constitutively active kinases that, when complexed with a Type I through ligand binding, phosphorylate the Type I’s GS box to activate the Type I kinase [62]. Type I receptors play the critical role of transmitting information from the outside of the cell to the inside; they accomplish this by binding specific extracellular ligands and phosphorylating specific Smad intracellular transducers (See Figure 1A).
4.2 BMP Type I receptors
The two type I receptors of the BMP branch have been extensively studied. Mutation of thickveins or saxophone individually leads to lethality [63,64]. However, ubiquitous production of Thickveins can rescue saxophone mutants [64]. This and other studies led to the general view that Thickveins is the dominant receptor for developmental signaling, and Saxophone modulates signaling levels. Thickveins is broadly but not uniformly expressed. The best studied case for the importance of thickveins expression control is in the wing disc, where patterned expression is controlled by a regulatory network linked to Dpp signaling. Expression has been monitored by antibody staining for Tkv and using a LacZ reporter [65].
An important finding from studies of the two BMP Type I receptors is that heteromeric ligand dimers and heteromeric receptor complexes are often important for patterns and strengths of signaling in both the embryo and the wing disc [17,66]. In the embryo in particular the heterodimer of Dpp/Scw provides a much more potent signal than either homodimer and this requires both Sax and Tkv [17]. The molecular mechanism responsible for generating the synergistic signal remains unexplained. In all published reports, Thickveins and/or Saxophone signaling leads only to Mad activation, illustrating the role of Type I receptors as a specificity determinant for BMP versus Activin signaling.
4.3 Activin Type I receptor isoforms
An aspect of receptor biology that is central for Activin branch signaling in Drosophila is that the baboon gene encodes three receptor isoforms with different extracellular domains. The emerging scheme is that different ligands bind and stimulate different Baboon isoforms. Dawdle was reported to signal exclusively through the Baboon-c isoform in S2 cells, and the relationship was also observed in wing discs [13]. Myoglianin signaling has not been reported in cell culture, but genetic evidence strongly implicates Baboon-a as the relevant isoform for Myoglianin [27]. The Baboon isoform preference for Activin-β requires further study, as does the identity of the Type I receptor for Maverick. Whether the simple biochemical and genetic assignments for ligand-receptor isoform interactions holds up, it is still necessary to recognize the complexities that different isoforms present for phenotypic analysis. The isoforms are not amenable to traditional genetic studies because the exons that define the protein variants are very near each other and no alternative exon-specific mutations have been described. However, miRNAs targeting the transcripts of individual isoforms have been reported and these reagents were used to infer a functional relationship between Myoglianin and Baboon-a in mushroom body axon remodeling [28]. These reagents should prove useful for mapping isoform functions to other cell types and tissues, which in turn will implicate which ligands signal to that cell type or tissue. The complete expression patterns of the three receptors have not been reported, but varying ratios of isoform mRNA expression were observed in different larval tissues [13] so we can intuitively expect that differential isoform expression is a key part of the code linking ligands and the tissues competent to respond to them. At present it is unknown what controls the differential splicing patterns of the baboon transcripts, and this remains an attractive avenue for exploration. Another interesting property of Baboon is that it can directly phosphorylate Mad in addition to the expected dSmad2 substrate. This cross-talk activity is readily observed in cell culture [50], and also occurs in vivo but primarily when dSmad2 is absent [21, 67]. Recognizing that this cross-pathway signaling can occur in vivo is important to consider for proper interpretation of some Activin signaling related phenotypes.
4.4 Gain-of-function approaches for Type I receptors
Gain-of-function experiments with receptors are conducted by over-expressing the proteins in a defined set of cells or in clonal patches. Over-expression of Type I receptors does not necessarily produce ectopic signaling in vivo, indicating that other factors, such as ligands and Type II receptors, are limiting. For instance, forced production of Thickveins in the entire wing disc does not lead to excess signaling, but does perturb patterning [65]. Clonal misexpression will exaggerate differences between neighbors, and can reveal a tissue’s response to irregular levels of BMP signaling. An extension of this approach used temporal control of exogenous receptors (Geneswitch) to explore the kinetic response of cells with increased BMP signaling [61].
A stronger way to activate signaling is to express constitutively active receptors. Mutations near the GS box of mammalian Type I receptors were found to cause hyperactivity, overriding the normal requirement for ligand binding [68]. Similar mutations were made for fly receptors and are available for Tkv [69,70], Sax [49], and Babo [14]. These reagents should be used with caution for several reasons. In the obvious sense, the phospho-Smad response will be extreme and may be ectopic depending on the tissue of exogenous expression. Secondly, activated receptors can titrate away other factors, thereby causing an unanticipated loss-of-function situation. For example, in the wing disc, expression of constitutively active Baboon (Babo-CA) causes a loss of phospho-Mad, apparently by titrating away Type II receptors needed for Thickveins and Saxophone signaling to Mad [21,67]. Finally, the output of CA receptors cannot be assumed to result only from signaling through canonical Smad proteins. In vertebrates, many non-Smad signaling pathways have been described and at least one example has been uncovered in Drosophila [71,72]. Despite these caveats, expression of CA receptors can be useful in tissue mapping studies, epistasis tests, and in studying the response of cells or tissues to signaling. For example, the similar phenotype of wing disc cells expressing activated Tkv and those lacking brinker was used to support the critical role of brinker as a BMP target [73].
One additional point concerning the use of activated receptors that is not generally recognized is that even constitutively activated Type I receptors that bypass ligand control still require a Type II receptor [74]. This indicates a structural or activity requirement for Type II receptors beyond altering the GS box of Type I receptors. One likely possibility is that the CA mutation simply facilitates interaction with a Type II in the absence of ligand, which then leads to phosphorylation of Serine residues in the GS box and it is the phosphorylations that lead to activation of the Type I, as normally happens when ligand is bound. That receptors can associate in the absence of ligand is well documented in cell culture, where simple over-expression of Type I or Type II receptors stimulates downstream signaling, presumably because the higher concentrations of receptors leads to productive complex formation despite the low natural affinity. Interestingly, in vivo, widespread over-expression of Type I receptors is remarkably well tolerated. Over-expression of Punt, however, is toxic, resembling expression of constitutively active Type I receptors (unpublished observations). This suggests that the availability of Punt is limiting for active complex formation.
4.5 Type II receptors
Punt and Wit are the two Type II receptors encoded in the Drosophila genome (see Table 1). Punt is broadly expressed [75], and Wit expression is spatially restricted, mainly seen in the nervous system, imaginal discs and some follicle cells [76]. Punt is considered the primary type II receptor responsible for mediating TGF-β signals because of its broad expression, severe loss of function phenotype, and its ability to form complexes with both BMP and Activin ligands [17,43]. In contrast, Wit seems to function in a more limited manner with phenotypes identified only in motoneurons, subsets of neuroendocrine cells, and in follicle cells that help pattern the eggshell [76–78].
The question arises, however, as to whether the two receptors can act interchangeably and whether the apparent specificity is simply a matter of the more restricted expression of Wit compared to Punt. In the case of mushroom body axon remodeling, it seems that Punt and Wit are redundant since loss of either alone is inconsequential, whereas loss of both produces a phenotype similar to loss of the ligand or the type I receptor [20]. In contrast, elimination of Punt in the wing disc leads to loss of epithelial cells [69], but loss of Wit is inconsequential even though both Punt and Wit are expressed in this tissue [76]. The opposite is true in motor neurons. In this case Wit appears to be required as forced expression of Punt does not rescue Wit mutants [76]. These disparities could reflect differences in biochemical properties of the receptors (e.g. ligand affinities) or restraints imposed by the unique cellular architecture of epithelial cells or neurons. For example, in cell culture, Punt but not Wit can support Dawdle signaling through Baboon-c, suggesting that biochemical properties impart specificity in some cases [13]. In summary, the combinations of ligands and receptors that can form active signaling complexes has not been fully explored, but the available examples indicate there is a mixture of promiscuity and specificity of Type II receptors. Finally, a Type II receptor may mediate non-canonical signaling by phosphorylating effectors other than Type I receptors [79], or by influencing the availability or location of binding partners [80].
4.6 Receptor loss-of-function approaches
Mutations have been identified for each Drosophila TGF-β receptor (Table 2). Whole animal mutants readily reveal developmental processes dependent on signaling, and directed experiments can uncover additional roles. Clones of mutant cells have been used to catalog the requirement of BMP signaling in the wing disc. Loss of BMP signaling leads to change in target gene expression, apoptosis, and removal of cells from the epithelium. RNAi-based clones are useful to study polyploid or non-mitotic cells, and can be extended to conduct epistasis tests by RNAi. Dominant-negative receptor constructs have also been used to partially block signaling. These truncated proteins are active in vivo [66] and exhibit some specificity for BMP and Activin branch signaling in cell culture signaling assays [50] but it is unclear how they act mechanistically, for example by titrating ligand or the partner receptor. Therefore, assuming the proper controls are conducted, RNAi approaches offer more specificity and flexibility because they don’t rely on the heavy over-expression (multiple transgene copies) needed for dominant-negative protein effects.
Pharmacological inhibitors of TGF-β receptors have been developed, including SB-431542 and SB-505124 for Activin/TGF-β Type I receptors and Dorsomorphin, LDN-193189, and K02288 for BMP Type I receptors [81–82]. This work is primarily motivated by the goal of using specific inhibitors to treat human diseases involving excessive receptor activity, but the reagents can also be used to dissect signaling mechanisms. The inhibitors bind to the conserved ATP binding pocket [81], and Dorsomorphin has been used to disrupt BMP signaling in vertebrates and sea urchins [83]. These chemicals are thus expected to inhibit Drosophila receptors, but their usage has not been adopted, perhaps owing to the reliance on genetic manipulations. Additionally, all of the inhibitors disrupt the activity of at least several other kinases [81–82], which limits their direct usefulness in discovering new phenotypes caused by loss of receptor activity.
4.7 Regulation of active complex formation at the receptor level
In contrast to the attention given to ligand transport mechanisms, receptor localization and activation is an under-appreciated topic. Receptor endocytosis rates are considered in models of signaling, but there is little direct experimental data for fruit fly TGF-β receptors. One study used receptors tagged with photo-activatable GFP and found that degradation rates of Tkv and Babo change during wing disc development [48]. Mammalian TGF-β receptors have restricted membrane localization in epithelial cells [84], and this would have obvious implications for limiting signaling if Drosophila receptors also exhibit restricted subcellular targeting [85]. Another study took a direct approach to monitor the location of ligand-receptor interaction in several cell types. The binding properties of Tkv and FKBP12 were used to build a reporter that fluoresces only when Tkv is active [33]. In the germline stem cell niche, Tkv activation was restricted to specialized membrane regions. Additional reagents such as antibodies that recognize activated receptors (e.g. phosphorylated form of Type I) could prove useful for following signaling complex internalization and discerning if the receptors are inactivated by dephosphorylation. Mammalian receptors are regulated by post-translational modification and cleavage [86,87], but the conservation of these mechanisms has not been described in the fruit fly counterparts.
In cases where the ligand and the receptors are in proximity, there still may be barriers to binding and active complex formation. Accessory factors have been identified that support ligand-receptor binding, such as the so-called mammalian Type III TGF-β receptor [5]. Several types of “co-receptors” for BMP signaling have been suggested including Dally and Dally-like, two Drosophila glipican homologs, and CV-2, a BMP binding protein that can associate with Punt in some circumstances to facilitate Dpp signaling [6]. The requirement for additional cofactors in Drosophila can also be inferred from contrasting cell culture and in vivo data. For example, Myoglianin acts as a traditional positive signal for canonical signaling in the mushroom body, but it does not signal in S2 cells, despite being able to form a complex with receptors [88]. The recently described plum gene product, a member of the IgG superfamily, may provide the illusive cofactor activity for Myoglianin signaling but at present no direct biochemical data supports this possibility [89]. In summary, the absence or presence of positive co-factors and inhibitors may constitute a permission system for ligands and receptors to interact and form a signaling-competent complex.
Additional tools that detect activated and bulk receptor populations are needed to fully explore the regulation of active signal complexes. These may include receptors tagged with pairs of fluorescent molecules suitable for FRET, or additional tools to detect activated receptors as described above. In motorneuron axons, observation of tagged receptor movement compared to Mad transport led to the model that receptors are transported along the axon to phosphorylate Mad near the cell body rather than at the NMJ [90]. Future studies will shed light on activation and inactivation of receptors in vesicle compartments.
5. Smad proteins
The above sections are concerned with conveying information to a target cell by forming an active receptor complex. The receiving cell must then interpret and act on the cue. In this section we consider the Smad proteins, which act as transcription factors to execute the canonical program (See Figure 1B).
Drosophila has a minimal complement of Smads, with one protein representative each of the BMP R-Smad, Activin R-Smad, shared co-Smad, and I-Smad types (See Table 1). In canonical signaling, R-Smads receive instructions from membrane-associated receptors and act as transcription factors in the nucleus. R-Smad proteins are divided into three domains: The DNA-binding MH1 domain, a linker domain, and the MH2 domain that couples oligomerization and receptor regulation. The co-Smad Medea forms complexes with R-Smads (likely trimers containing two R-Smads and one co-Smad if it follows the vertebrate model) and is subject to regulation by post-translational modification that limits signaling [91,92]. Medea is expected to be used for canonical signaling by both Mad and dSmad2. This means medea loss-of-function experiments can be used to implicate TGF-β signaling, but alone can not distinguish between the branches. The distribution of Medea between Mad and dSmad2 binding may regulate the relative activities of the signaling branches [93]. Finally, the Dad I-Smad is a feedback inhibitor of BMP signaling, but its role in Activin signaling is not well characterized [46,94].
5.1 Detection of activated Smads
R-Smads are activated by phosphorylation, so the direct detection of R-Smad post-translational modifications is an important readout of their activity state. Western blotting for phosphorylated Smads is feasible with commercial antibodies (see list for IHC below). This approach can be used in cell culture signaling assays [43,17], and to monitor the signal status of tissues [95]. In cases where patterns and quantification at the single cell level are relevant, IHC must be used. P-Mad was first detected with the PS1 antibody raised against vertebrate P-Smad1 [96], and several other polyclonal antibodies have been reported [65]. There are now several commercial antibodies that specifically detect C-terminally phosphorylated Mad by IHC (Cell Signaling rabbit monoclonal 9516, Epitomics rabbit monoclonal 1880-1, R&D Systems rabbit polyclonal AB3226). Each antibody detects nuclear P-Mad in a variety of tissues, but substantial variation in the ability to detect P-Mad at the NMJ was reported, with polyclonal antibodies providing better detection than the monoclonal antibodies [90]. Note that antibodies raised against C-terminally phosphorylated mammalian Smad1 or Smad3 can specifically bind to P-Mad, likely because they share the terminal amino acid sequence SSVS. For P-dSmad2, detection by IHC has been problematic, with only one report of specific detection with a PS2 antibody [67]. Two antibodies against bulk dSmad2 are available, and these may be useful for monitoring dSmad2 levels or subcellular localization (rabbit polyclonal from [95] and sheep polyclonal R&D Systems Product AF7948). Another approach called BifC couples complex formation with fluorescence. BifC reagents showed that Mad and Medea interact in a spatial pattern that resembles the phospho-Mad profile [48].
5.2 Mutated Smads
Mutated forms of Smads have been used to stimulate, inhibit, or otherwise interrogate the pathway. Mutation of C-terminal serine residues renders an R-Smad unable to respond to TGF-β receptor kinases. Conversely, phosphomimetic mutations are predicted to simulate activation [50]. Overexpression of phosphomimetic Mad or dSmad2 perturbs patterning consistent with activation, but it is not known if they regulate transcriptional targets in the same fashion as C-terminally phosphorylated proteins. Other mutations that have been tested in cell culture and in vivo include those that block receptor binding [22] and linker mutations that prevent additional modifications [97,98].
5.3 Mad as a multi-modal transcription factor
There are two well-studied modes of Mad transcription factor activity. Mad binding sites have been identified for Silencing Elements and Activating Elements. Repression through the SE depends on Mad, Medea, and Schnurri, and these proteins form a complex [99,100]. Prominent genes regulated by SEs in the wing disc are brk and pent. Mad and Medea without Schnurri bind to AEs to regulate a different set of target genes, notably dad [101]. Other targets of BMP signaling include sal, omb, vg and ltl [102–105]. Another set of targets have distinct D/V patterns in the early embryo, including zen, race, and ush [91,106], and induction of labial expression by Dpp is required for gut development [107]. Even in the wing disc, where much of the patterning and growth-control aspects of Dpp signaling can be explained by expression of target genes under the control of SEs and AEs [44], Mad was found to participate in other complexes (Figure 1B). For example, Mad and Yorkie physically interact and regulate transcription, independent of Medea, through an uncharacterized binding sequence [108]. Mad also forms a complex with Armadillo and Pangolin [98].
Application of ChIP methodologies should shed light on the diversity of Mad binding sites in the genome and identify more target genes in multiple cell types. ChIP of Mad in the early embryo has been reported, but the number of high-confidence binding sites was lower than for a panel of other transcription factors that included the Mad cofactors Medea and Schnurri [109]. The antibody used for the study may not be optimal, and other Mad antibodies may be more suitable for ChIP [110]. Alternatively, epitope-tagging can be employed to validate direct association of Mad with DNA sites [108]. Association of a factor with a DNA region does not guarantee that the binding is relevant for transcriptional regulation, so functional analyses of regulatory regions will be required to validate binding sites and understand how Mad binding controls transcription at different genes. The number of cells required for ChIP-Seq experiments continues do drop, so future studies will likely focus on factor occupancy in spatially defined subsets of cells [111].
5.4 dSmad2 as an undocumented transcription factor
By analogy to the well-studied homologs Smad2 and Smad3, it is reasonable to expect that dSmad2 can act as transcription factor. Tagged dSmad2 was detected in the nuclei of R7 photoreceptors [26] and endogenous nuclear Phospho-dSmad2 was detected in multiple larval tissues, including the wing disc, where P-dSmad2 staining depends on Baboon signaling [67]. EcRB1 expression in the mushroom body depends on a canonical signaling module of Myo, Baboon-a and dSmad2. Several putative candidate genes have been identified in the Prothoracic Gland, including torso and InR [112]. However, direct binding has not been reported for these targets. In fact, very little is known about dSmad2 as a transcription factor. Another unknown property of dSmad2 is what target DNA sequences it binds. There is notable amino acid variation in the MH1 DNA-binding domain of dSmad2, with little conservation in DNA binding residues identified in a human Smad3:DNA crystal structure [113]. Once again, ChIP for dSmad2 will provide an unbiased avenue to study association of dSmad2 with target genes. ChIP-Seq for dSmad2 has not been reported, so it not known if existing antibodies are suitable for this technique. It is likely that dSmad2 binding requires cofactors and that consensus sequences may not resemble consensus sequences for other Smad proteins. By analogy to studies of vertebrate Smads [114], complexes containing Mad and dSmad2 could also regulate transcription.
5.5 Smad linker biology
The role of the DNA-binding MH1 domain and the phospho-regulated MH2 domain are conceptually clear. Despite the maudlin moniker, the linker regions of Smad proteins are also important. The known functions of linker sequences fall under the broad categories of down-regulating Smad function and coordination with other signaling pathways.
The linker region of Mad has been studied extensively and shares conserved features with other Smad1 family proteins. One set of linker modifications leads to degradation of Mad, which serves to dampen active signaling. Another series of modifications revealed a combinatorial code involving Wnt and MAPK, two other major signaling pathways. Mutation of Mad at MAPK/Nemo target sites [97] or GSK-3 target sites [98] activates Mad such that it produces stronger phenotypes upon overexpression compared to wildtype Mad. Several antibodies were developed to monitor these linker specific phospho-forms of Mad [98]. These interactions have been best studied in wing development, but signal integration will be a common theme as TGF-β signaling is considered in the context of regulatory networks in other tissues. Some of these interactions, like the physical association of Mad with Wg pathway components, might be evolutionarily conserved and directly relevant to patterning in other animals.
The linker of Smad2/3 proteins have also been studied and implicated in degradation. However, the dSmad2 linker region is poorly conserved with vertebrates, and is quite divergent even among insects. The stability of dSmad2 was shown to decrease upon phosphorylation [21], so an intriguing possibility is that linker region also regulates protein stability despite the absence of sequence conservation. More generally, we can expect additional Smad post-translational modifications to be discovered by comparison to vertebrates (such as the methylation of Smad6 [115]) and by direct molecular studies of fly Smads.
6. Non-canonical signaling
TGF-β receptors and Smads also conduct non-canonical functions. For the purpose of this chapter, we define non-canonical signaling to mean information flow outside of the conventional ligand-receptor-Smad transduction sequence. This could include receptors acting on non-Smad proteins, cross-talk between the branches, and other activities of Smad proteins.
Non-canonical activities are well studied in mammalian cells [71] but less so in Drosophila. There are two reports of parallel non-canonical signaling between vertebrate and fly TGF-β pathways. BMPRII and Drosophila Wit both bind LIMK through a peptide region C-terminal to the kinase domain. A functional Wit-LIMK interaction was invoked in the control of NMJ growth [116], although the C-terminus of Wit is not required for viability [76]. In mushroom body axon extension, Rho GTPases and LIMK were implicated in the transmission of Smad-independent receptor signals [72].
Other signaling components shown to mediate non-canonical signaling are conserved in Drosophila. These include RTK signaling through ERK, TAK1 activation of JNK/p38, and PI3K-AKT-TOR pathways. However, this does not mean that the interactions are common in both systems. For example, TGF-β can induce EMT in mammalian cells by influencing the activity of factors controlling cytoskeleton and cellular junctions. TGF-β signaling is not known to regulate EMT in Drosophila, so we can predict that the specific molecular interactions are not conserved. This principle of the limits of conservation applies to other proteins that have been shown to impact vertebrate signaling. For example, SARA is important for recruiting vertebrate Smads to receptors [117], but even though there is a homolog in Drosophila, it does not play the same critical role in signaling [118].
One way to identify canonical versus non-canonical activities is to compare the phenotypes of receptors and Smads. For the BMP branch, loss-of-function of Punt or Thickveins generally gives the same phenotype as Mad loss-of-function. For Activin signaling, the available genetic evidence indicates that canonical and non-canonical signaling are both prevalent. For dSmad2 in particular, the null allele [21] displays a different spectrum of phenotypes than the originally described point mutant allele [20]. In cases where loss of baboon or dSmad2 produce similar results, we conclude that canonical signaling is involved. These include neuroblast proliferation [27], mushroom body remodeling via EcRB1 [20], photoreceptor tiling [26], PG function [112], and some aspects of proliferation in the wing disc [67]. For other phenotypes, divergent phenotypes for loss of Baboon or dSmad2 reveal that non-canonical configurations are operative. These include axonal extension in the mushroom body, the namesake baboon enlarged anal pad phenotype, and regulation of BMP-brinker signaling in wing disc. Genetic interactions suggest that Smad-independent receptor functions controlling axon projection likely involve the actin cytoskeleton [72]. In the wing disc, epistasis tests uncovered an anti-signaling configuration wherein dSmad2 inhibits Baboon. A null allele for dSmad2 phenocopied hyperactivation of BMP signaling, but simultaneous loss of baboon nullified this phenotype [22].
7. Perspectives on future studies
There is still much to learn about TGF-β signaling in Drosophila biology. Some of the knowledge gaps are obvious, and we have pointed out several areas of study in this overview. These include developing protein detection reagents to follow the less studied ligands, deciphering the control of babo isoform expression, and discovering new Smad targets by ChIP. Other areas that deserve consideration are direct structural biology studies of Drosophila ligands, investigating the formation and activity of ligand heterodimers and receptor heteromers, and identifying non-canonical targets of Baboon. Other discoveries will undoubtedly follow serendipity. These may begin by observing a curious behavior in a mutant, or with the identification of a TGF-β pathway component in a functional or expression screen. The existing tools can provide important information about which factors are required in which tissues, and point to canonical transcriptional output or more exotic configurations.
Interrogation of the roles of TGF-β signaling in more biological processes will continue the iterative process of discovering how a conserved pathway is deployed in diverse contexts. For any biological process involving TGF-β signaling, knowledge of the pathway components can be leveraged to study the process, and resulting context-specific findings of how the components behave will flesh out the range of action of the pathway. In some cases the results will identify a canonical relationship where a ligand expressed in one cell acts on another cell to activate a transcriptional program. In other cases, we can expect to uncover surprising activities of the molecules and functional interactions with other signaling pathways.
Acknowledgments
We are grateful to Lindsay Moss-Taylor for constructive comments on the manuscript. The authors were supported by the National Institutes of Health (grant R01 GM95746). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Herpin A, Lelong C, Favrel P. Dev Comp Immunol. 2004;28:461–485. doi: 10.1016/j.dci.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 2.Srivastava M, Begovic E, Chapman J, Putnam NH, Hellsten U, Kawashima T, Kuo A, Mitros T, Salamov A, Carpenter ML, Signorovitch AY, Moreno MA, Kamm K, Grimwood J, Schmutz J, Shapiro H, Grigoriev IV, Buss LW, Schierwater B, Dellaporta SL, Rokhsar DS. Nature. 2008;454:955–960. doi: 10.1038/nature07191. [DOI] [PubMed] [Google Scholar]
- 3.Pang K, Ryan JF, Baxevanis AD, Martindale MQ. PLoS One. 2011;6:e24152. doi: 10.1371/journal.pone.0024152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fraser HB, Hirsh AE, Steinmetz LM, Scharfe C, Feldman MW. Science. 2002;296:750–752. doi: 10.1126/science.1068696. [DOI] [PubMed] [Google Scholar]
- 5.Ramel MC, Hill CS. FEBS Lett. 2012;586:1929–1941. doi: 10.1016/j.febslet.2012.02.035. [DOI] [PubMed] [Google Scholar]
- 6.Wharton KA, Serpe M. Curr Opin Genet Dev. 2013;23:374–384. doi: 10.1016/j.gde.2013.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kruse K, Pantazis P, Bollenbach T, Julicher F, Gonzalez-Gaitan M. Development. 2004;131:4843–4856. doi: 10.1242/dev.01335. [DOI] [PubMed] [Google Scholar]
- 8.Roy S, Huang H, Liu S, Kornberg TB. Science. 2014 doi: 10.1126/science.1244624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huminiecki L, Goldovsky L, Freilich S, Moustakas A, Ouzounis C, Heldin CH. BMC Evol Biol. 2009;9:28. doi: 10.1186/1471-2148-9-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van der Zee M, da Fonseca RN, Roth S. Dev Genes Evol. 2008;218:203–213. doi: 10.1007/s00427-007-0179-7. [DOI] [PubMed] [Google Scholar]
- 11.Fritsch C, Lanfear R, Ray RP. Dev Genes Evol. 2010;220:235–250. doi: 10.1007/s00427-010-0341-5. [DOI] [PubMed] [Google Scholar]
- 12.Nguyen M, Parker L, Arora K. Mech Dev. 2000;95:201–206. doi: 10.1016/s0925-4773(00)00338-5. [DOI] [PubMed] [Google Scholar]
- 13.Jensen PA, Zheng X, Lee T, O’Connor MB. Mech Dev. 2009;126:950–957. doi: 10.1016/j.mod.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brummel T, Abdollah S, Haerry TE, Shimell MJ, Merriam J, Raftery L, Wrana JL, O’Connor MB. Genes Dev. 1999;13:98–111. doi: 10.1101/gad.13.1.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bickel D, Shah R, Gesualdi SC, Haerry TE. Mech Dev. 2008;125:117–129. doi: 10.1016/j.mod.2007.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ross JJ, Shimmi O, Vilmos P, Petryk A, Kim H, Gaudenz K, Hermanson S, Ekker SC, O’Connor MB, Marsh JL. Nature. 2001;410:479–483. doi: 10.1038/35068578. [DOI] [PubMed] [Google Scholar]
- 17.Shimmi O, Umulis D, Othmer H, O’Connor MB. Cell. 2005;120:873–886. doi: 10.1016/j.cell.2005.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Irish VF, Gelbart WM. Genes Dev. 1987;1:868–879. doi: 10.1101/gad.1.8.868. [DOI] [PubMed] [Google Scholar]
- 19.Spencer FA, Hoffmann FM, Gelbart WM. Cell. 1982;28:451–461. doi: 10.1016/0092-8674(82)90199-4. [DOI] [PubMed] [Google Scholar]
- 20.Zheng X, Wang J, Haerry TE, Wu AY, Martin J, O’Connor MB, Lee CH, Lee T. Cell. 2003;112:303–315. doi: 10.1016/s0092-8674(03)00072-2. [DOI] [PubMed] [Google Scholar]
- 21.Peterson AJ, Jensen PA, Shimell M, Stefancsik R, Wijayatonge R, Herder R, Raftery LA, O’Connor MB. PLoS One. 2012;7:e36548. doi: 10.1371/journal.pone.0036548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peterson AJ, O’Connor MB. Development. 2013;140:649–659. doi: 10.1242/dev.085605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fuentes-Medel Y, Ashley J, Barria R, Maloney R, Freeman M, Budnik V. Curr Biol. 2012;22:1831–1838. doi: 10.1016/j.cub.2012.07.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Minami M, Kinoshita N, Kamoshida Y, Tanimoto H, Tabata T. Nature. 1999;398:242–246. doi: 10.1038/18451. [DOI] [PubMed] [Google Scholar]
- 25.Morimura S, Maves L, Chen Y, Hoffmann FM. Dev Biol. 1996;177:136–151. doi: 10.1006/dbio.1996.0151. [DOI] [PubMed] [Google Scholar]
- 26.Ting CY, Herman T, Yonekura S, Gao S, Wang J, Serpe M, O’Connor MB, Zipursky SL, Lee CH. Neuron. 2007;56:793–806. doi: 10.1016/j.neuron.2007.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu CC, Boone JQ, Jensen PA, Hanna S, Podemski L, Locke J, Doe CQ, O’Connor MB. Development. 2008;135:513–521. doi: 10.1242/dev.010876. [DOI] [PubMed] [Google Scholar]
- 28.Awasaki T, Huang Y, O’Connor MB, Lee T. Nat Neurosci. 2011;14:821–823. doi: 10.1038/nn.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson AN, Burnett LA, Sellin J, Paululat A, Newfeld SJ. Genetics. 2007;176:1609–1624. doi: 10.1534/genetics.107.073569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takaesu NT, Bulanin DS, Johnson AN, Orenic TV, Newfeld SJ. Dev Biol. 2008;313:829–843. doi: 10.1016/j.ydbio.2007.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang J, Tian L, Peng C, Abdou M, Wen D, Wang Y, Li S, Wang J. Development. 2011;138:2283–2291. doi: 10.1242/dev.057687. [DOI] [PubMed] [Google Scholar]
- 32.Guo Z, Driver I, Ohlstein B. J Cell Biol. 2013;201:945–961. doi: 10.1083/jcb.201302049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Michel M, Raabe I, Kupinski AP, Perez-Palencia R, Bokel C. Nat Commun. 2011;2:415. doi: 10.1038/ncomms1426. [DOI] [PubMed] [Google Scholar]
- 34.Panganiban GE, Rashka KE, Neitzel MD, Hoffmann FM. Mol Cell Biol. 1990;10:2669–2677. doi: 10.1128/mcb.10.6.2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Akiyama T, Marques G, Wharton KA. Sci Signal. 2012;5:ra28. doi: 10.1126/scisignal.2002549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dani N, Nahm M, Lee S, Broadie K. PLoS Genet. 2012;8:e1003031. doi: 10.1371/journal.pgen.1003031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Akiyama T, Kamimura K, Firkus C, Takeo S, Shimmi O, Nakato H. Dev Biol. 2008;313:408–419. doi: 10.1016/j.ydbio.2007.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Entchev EV, Schwabedissen A, Gonzalez-Gaitan M. Cell. 2000;103:981–991. doi: 10.1016/s0092-8674(00)00200-2. [DOI] [PubMed] [Google Scholar]
- 39.Teleman AA, Cohen SM. Cell. 2000;103:971–980. doi: 10.1016/s0092-8674(00)00199-9. [DOI] [PubMed] [Google Scholar]
- 40.Fritsch C, Sawala A, Harris R, Maartens A, Sutcliffe C, Ashe HL, Ray RP. J Biol Chem. 2012;287:5942–5953. doi: 10.1074/jbc.M111.316745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nahm M, Kim S, Paik SK, Lee M, Lee S, Lee ZH, Kim J, Lee D, Bae YC. J Neurosci. 2010;30:8138–8150. doi: 10.1523/JNEUROSCI.0256-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Matsuda S, Shimmi O. Dev Biol. 2012;366:153–162. doi: 10.1016/j.ydbio.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 43.Serpe M, O’Connor MB. Development. 2006;133:4969–4979. doi: 10.1242/dev.02711. [DOI] [PubMed] [Google Scholar]
- 44.Affolter M, Basler K. Nat Rev Genet. 2007;8:663–674. doi: 10.1038/nrg2166. [DOI] [PubMed] [Google Scholar]
- 45.Chen S, Wang S, Xie T. Curr Opin Genet Dev. 2011;21:684–689. doi: 10.1016/j.gde.2011.07.008. [DOI] [PubMed] [Google Scholar]
- 46.Tsuneizumi K, Nakayama T, Kamoshida Y, Kornberg TB, Christian JL, Tabata T. Nature. 1997;389:627–631. doi: 10.1038/39362. [DOI] [PubMed] [Google Scholar]
- 47.Hamaratoglu F, de Lachapelle AM, Pyrowolakis G, Bergmann S, Affolter M. PLoS Biol. 2011;9:e1001182. doi: 10.1371/journal.pbio.1001182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wartlick O, Mumcu P, Kicheva A, Bittig T, Seum C, Julicher F, Gonzalez-Gaitan M. Science. 2011;331:1154–1159. doi: 10.1126/science.1200037. [DOI] [PubMed] [Google Scholar]
- 49.Parker L, Ellis JE, Nguyen MQ, Arora K. Development. 2006;133:4981–4991. doi: 10.1242/dev.02673. [DOI] [PubMed] [Google Scholar]
- 50.Gesualdi SC, Haerry TE. Fly (Austin) 2007;1:212–221. doi: 10.4161/fly.5116. [DOI] [PubMed] [Google Scholar]
- 51.Handke B, Poernbacher I, Goetze S, Ahrens CH, Omasits U, Marty F, Simigdala N, Meyer I, Wollscheid B, Brunner E, Hafen E, Lehner CF. PLoS One. 2013;8:e67208. doi: 10.1371/journal.pone.0067208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Groppe J, Rumpel K, Economides AN, Stahl N, Sebald W, Affolter M. J Biol Chem. 1998;273:29052–29065. doi: 10.1074/jbc.273.44.29052. [DOI] [PubMed] [Google Scholar]
- 53.Kunnapuu J, Bjorkgren I, Shimmi O. Proc Natl Acad Sci U S A. 2009;106:8501–8506. doi: 10.1073/pnas.0809885106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu XM, Gutman I, Mosca TJ, Iram T, Ozkan E, Garcia KC, Luo L, Schuldiner O. Neuron. 2013;78:456–468. doi: 10.1016/j.neuron.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen J, Honeyager SM, Schleede J, Avanesov A, Laughon A, Blair SS. Development. 2012;139:2170–2176. doi: 10.1242/dev.073817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Israel DI, Nove J, Kerns KM, Kaufman RJ, Rosen V, Cox KA, Wozney JM. Growth Factors. 1996;13:291–300. doi: 10.3109/08977199609003229. [DOI] [PubMed] [Google Scholar]
- 57.Chang H, Brown CW, Matzuk MM. Endocr Rev. 2002;23:787–823. doi: 10.1210/er.2002-0003. [DOI] [PubMed] [Google Scholar]
- 58.Little SC, Mullins MC. Nat Cell Biol. 2009;11:637–643. doi: 10.1038/ncb1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Butler SJ, Dodd J. Neuron. 2003;38:389–401. doi: 10.1016/s0896-6273(03)00254-x. [DOI] [PubMed] [Google Scholar]
- 60.Burke R, Basler K. Development. 1996;122:2261–2269. doi: 10.1242/dev.122.7.2261. [DOI] [PubMed] [Google Scholar]
- 61.Rogulja D, Irvine KD. Cell. 2005;123:449–461. doi: 10.1016/j.cell.2005.08.030. [DOI] [PubMed] [Google Scholar]
- 62.Wrana JL, Attisano L, Wieser R, Ventura F, Massague J. Nature. 1994;370:341–347. doi: 10.1038/370341a0. [DOI] [PubMed] [Google Scholar]
- 63.Xie T, Finelli AL, Padgett RW. Science. 1994;263:1756–1759. doi: 10.1126/science.8134837. [DOI] [PubMed] [Google Scholar]
- 64.Brummel TJ, Twombly V, Marques G, Wrana JL, Newfeld SJ, Attisano L, Massague J, O’Connor MB, Gelbart WM. Cell. 1994;78:251–261. doi: 10.1016/0092-8674(94)90295-x. [DOI] [PubMed] [Google Scholar]
- 65.Crickmore MA, Mann RS. Science. 2006;313:63–68. doi: 10.1126/science.1128650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Haerry TE, Khalsa O, O’Connor MB, Wharton KA. Development. 1998;125:3977–3987. doi: 10.1242/dev.125.20.3977. [DOI] [PubMed] [Google Scholar]
- 67.Hevia CF, de Celis JF. Dev Biol. 2013;377:138–153. doi: 10.1016/j.ydbio.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 68.Wieser R, Wrana JL, Massague J. Embo j. 1995;14:2199–2208. doi: 10.1002/j.1460-2075.1995.tb07214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nellen D, Burke R, Struhl G, Basler K. Cell. 1996;85:357–368. doi: 10.1016/s0092-8674(00)81114-9. [DOI] [PubMed] [Google Scholar]
- 70.Hoodless PA, Haerry T, Abdollah S, Stapleton M, O’Connor MB, Attisano L, Wrana JL. Cell. 1996;85:489–500. doi: 10.1016/s0092-8674(00)81250-7. [DOI] [PubMed] [Google Scholar]
- 71.Zhang YE. Cell Res. 2009;19:128–139. doi: 10.1038/cr.2008.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ng J. Development. 2008;135:4025–4035. doi: 10.1242/dev.028209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schwank G, Restrepo S, Basler K. Development. 2008;135:4003–4013. doi: 10.1242/dev.025635. [DOI] [PubMed] [Google Scholar]
- 74.Le VQ, Wharton KA. Dev Dyn. 2012;241:200–214. doi: 10.1002/dvdy.22779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Letsou A, Arora K, Wrana JL, Simin K, Twombly V, Jamal J, Staehling-Hampton K, Hoffmann FM, Gelbart WM, Massague J, et al. Cell. 1995;80:899–908. doi: 10.1016/0092-8674(95)90293-7. [DOI] [PubMed] [Google Scholar]
- 76.Marqués G, Bao H, Haerry TE, Shimell MJ, Duchek P, Zhang B, O’Connor MB. Neuron. 2002;33:529–543. doi: 10.1016/s0896-6273(02)00595-0. [DOI] [PubMed] [Google Scholar]
- 77.Marqués G, Haerry TE, Crotty ML, Xue M, Zhang B, O’Connor MB. Development. 2003;130:5457–5470. doi: 10.1242/dev.00772. [DOI] [PubMed] [Google Scholar]
- 78.Marmion RA, Jevtic M, Springhorn A, Pyrowolakis G, Yakoby N. Dev Biol. 2013;375:45–53. doi: 10.1016/j.ydbio.2012.12.011. [DOI] [PubMed] [Google Scholar]
- 79.Ozdamar B, Bose R, Barrios-Rodiles M, Wang HR, Zhang Y, Wrana JL. Science. 2005;307:1603–1609. doi: 10.1126/science.1105718. [DOI] [PubMed] [Google Scholar]
- 80.Lee-Hoeflich ST, Causing CG, Podkowa M, Zhao X, Wrana JL, Attisano L. Embo j. 2004;23:4792–4801. doi: 10.1038/sj.emboj.7600418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sanvitale CE, Kerr G, Chaikuad A, Ramel MC, Mohedas AH, Reichert S, Wang Y, Triffitt JT, Cuny GD, Yu PB, Hill CS, Bullock AN. PLoS One. 2013;8:e62721. doi: 10.1371/journal.pone.0062721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Vogt J, Traynor R, Sapkota GP. Cell Signal. 2011;23:1831–1842. doi: 10.1016/j.cellsig.2011.06.019. [DOI] [PubMed] [Google Scholar]
- 83.Luo YJ, Su YH. PLoS Biol. 2012;10:e1001402. doi: 10.1371/journal.pbio.1001402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Murphy SJ, Shapira KE, Henis YI, Leof EB. Mol Biol Cell. 2007;18:3788–3799. doi: 10.1091/mbc.E06-10-0930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gibson MC, Lehman DA, Schubiger G. Dev Cell. 2002;3:451–460. doi: 10.1016/s1534-5807(02)00264-2. [DOI] [PubMed] [Google Scholar]
- 86.Mu Y, Sundar R, Thakur N, Ekman M, Gudey SK, Yakymovych M, Hermansson A, Dimitriou H, Bengoechea-Alonso MT, Ericsson J, Heldin CH, Landstrom M. Nat Commun. 2011;2:330. doi: 10.1038/ncomms1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liu C, Xu P, Lamouille S, Xu J, Derynck R. Mol Cell. 2009;35:26–36. doi: 10.1016/j.molcel.2009.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lee-Hoeflich ST, Zhao X, Mehra A, Attisano L. FEBS Lett. 2005;579:4615–4621. doi: 10.1016/j.febslet.2005.06.088. [DOI] [PubMed] [Google Scholar]
- 89.Yu XM, Gutman I, Mosca TJ, Iram T, Ozkan E, Garcia KC, Luo L, Schuldiner O. Neuron. 2013;78:456–468. doi: 10.1016/j.neuron.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Smith RB, Machamer JB, Kim NC, Hays TS, Marques G. J Cell Sci. 2012;125:3752–3764. doi: 10.1242/jcs.094292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Miles WO, Jaffray E, Campbell SG, Takeda S, Bayston LJ, Basu SP, Li M, Raftery LA, Ashe MP, Hay RT, Ashe HL. Genes Dev. 2008;22:2578–2590. doi: 10.1101/gad.494808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Stinchfield MJ, Takaesu NT, Quijano JC, Castillo AM, Tiusanen N, Shimmi O, Enzo E, Dupont S, Piccolo S, Newfeld SJ. Development. 2012;139:2721–2729. doi: 10.1242/dev.077206. [DOI] [PubMed] [Google Scholar]
- 93.Takaesu NT, Hyman-Walsh C, Ye Y, Wisotzkey RG, Stinchfield MJ, O’Connor B, Wotton MD, Newfeld SJ. Genetics. 2006;174:1299–1313. doi: 10.1534/genetics.106.064956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kamiya Y, Miyazono K, Miyazawa K. FEBS Lett. 2008;582:2496–2500. doi: 10.1016/j.febslet.2008.05.052. [DOI] [PubMed] [Google Scholar]
- 95.Bai H, Kang P, Hernandez AM, Tatar M. PLoS Genet. 2013;9:e1003941. doi: 10.1371/journal.pgen.1003941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Persson U, Izumi H, Souchelnytskyi S, Itoh S, Grimsby S, Engstrom U, Heldin CH, Funa K, ten Dijke P. FEBS Lett. 1998;434:83–87. doi: 10.1016/s0014-5793(98)00954-5. [DOI] [PubMed] [Google Scholar]
- 97.Zeng YA, Rahnama M, Wang S, Sosu-Sedzorme W, Verheyen EM. Development. 2007;134:2061–2071. doi: 10.1242/dev.02853. [DOI] [PubMed] [Google Scholar]
- 98.Eivers E, Demagny H, Choi RH, De Robertis EM. Sci Signal. 2011;4:ra68. doi: 10.1126/scisignal.2002034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Müller B, Hartmann B, Pyrowolakis G, Affolter M, Basler K. Cell. 2003;113:221–233. doi: 10.1016/s0092-8674(03)00241-1. [DOI] [PubMed] [Google Scholar]
- 100.Pyrowolakis G, Hartmann B, Müller B, Basler K, Affolter M. Dev Cell. 2004;7:229–240. doi: 10.1016/j.devcel.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 101.Weiss A, Charbonnier E, Ellertsdóttir E, Tsirigos A, Wolf C, Schuh R, Pyrowolakis G, Affolter M. Nat Struct Mol Biol. 2010;17:69–76. doi: 10.1038/nsmb.1715. [DOI] [PubMed] [Google Scholar]
- 102.de Celis JF, Barrio R, Kafatos FC. Nature. 1996;381:421–424. doi: 10.1038/381421a0. [DOI] [PubMed] [Google Scholar]
- 103.Grimm S, Pflugfelder GO. Science. 1996;271:1601–1604. doi: 10.1126/science.271.5255.1601. [DOI] [PubMed] [Google Scholar]
- 104.Kim J, Sebring A, Esch JJ, Kraus ME, Vorwerk K, Magee J, Carroll SB. Nature. 1996;382:133–138. doi: 10.1038/382133a0. [DOI] [PubMed] [Google Scholar]
- 105.Szuperak M, Salah S, Meyer EJ, Nagarajan U, Ikmi A, Gibson MC. Development. 2011;138:715–724. doi: 10.1242/dev.059477. [DOI] [PubMed] [Google Scholar]
- 106.Rushlow C, Colosimo PF, Lin MC, Xu M, Kirov N. Genes Dev. 2001;15:340–351. doi: 10.1101/gad.861401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Panganiban GE, Reuter R, Scott MP, Hoffmann FM. Development. 1990;110:1041–1050. doi: 10.1242/dev.110.4.1041. [DOI] [PubMed] [Google Scholar]
- 108.Oh H, Irvine KD. Dev Cell. 2011;20:109–122. doi: 10.1016/j.devcel.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.MacArthur S, Li XY, Li J, Brown JB, Chu HC, Zeng L, Grondona BP, Hechmer A, Simirenko L, Keranen SV, Knowles DW, Stapleton M, Bickel P, Biggin MD, Eisen MB. Genome Biol. 2009;10:R80. doi: 10.1186/gb-2009-10-7-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chen X, Xu L. Mol Cell Biol. 2010;30:4022–4034. doi: 10.1128/MCB.00124-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Slattery M, Ma L, Negre N, White KP, Mann RS. PLoS One. 2011;6:e14686. doi: 10.1371/journal.pone.0014686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gibbens YY, Warren JT, Gilbert LI, O’Connor MB. Development. 2011;138:2693–2703. doi: 10.1242/dev.063412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Chai J, Wu JW, Yan N, Massague J, Pavletich NP, Shi Y. J Biol Chem. 2003;278:20327–20331. doi: 10.1074/jbc.C300134200. [DOI] [PubMed] [Google Scholar]
- 114.Gronroos E, Kingston IJ, Ramachandran A, Randall RA, Vizan P, Hill CS. Mol Cell Biol. 2012;32:2904–2916. doi: 10.1128/MCB.00231-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Xu J, Wang AH, Oses-Prieto J, Makhijani K, Katsuno Y, Pei M, Yan L, Zheng YG, Burlingame A, Bruckner K, Derynck R. Mol Cell. 2013;51:5–19. doi: 10.1016/j.molcel.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Eaton BA, Davis GW. Neuron. 2005;47:695–708. doi: 10.1016/j.neuron.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 117.Tsukazaki T, Chiang TA, Davison AF, Attisano L, Wrana JL. Cell. 1998;95:779–791. doi: 10.1016/s0092-8674(00)81701-8. [DOI] [PubMed] [Google Scholar]
- 118.Bokel C, Schwabedissen A, Entchev E, Renaud O, Gonzalez-Gaitan M. Science. 2006;314:1135–1139. doi: 10.1126/science.1132524. [DOI] [PubMed] [Google Scholar]
- 119.McCabe BD, Marques G, Haghighi AP, Fetter RD, Crotty ML, Haerry TE, Goodman CS, O’Connor MB. Neuron. 2003;39:241–254. doi: 10.1016/s0896-6273(03)00426-4. [DOI] [PubMed] [Google Scholar]
- 120.Wharton KA, Cook JM, Torres-Schumann S, de Castro K, Borod E, Phillips DA. Genetics. 1999;152:629–640. doi: 10.1093/genetics/152.2.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ballard SL, Jarolimova J, Wharton KA. Dev Biol. 2010;337:375–385. doi: 10.1016/j.ydbio.2009.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Arora K, Levine MS, O’Connor MB. Genes Dev. 1994;8:2588–2601. doi: 10.1101/gad.8.21.2588. [DOI] [PubMed] [Google Scholar]
- 123.Affolter M, Nellen D, Nussbaumer U, Basler K. Development. 1994;120:3105–3117. doi: 10.1242/dev.120.11.3105. [DOI] [PubMed] [Google Scholar]
- 124.Newfeld SJ, Chartoff EH, Graff JM, Melton DA, Gelbart WM. Development. 1996;122:2099–2108. doi: 10.1242/dev.122.7.2099. [DOI] [PubMed] [Google Scholar]
- 125.Zheng X, Zugates CT, Lu Z, Shi L, Bai JM, Lee T. Embo j. 2006;25:615–627. doi: 10.1038/sj.emboj.7600962. [DOI] [PMC free article] [PubMed] [Google Scholar]