Abstract
Mature human adenovirus particles contain four minor capsid proteins, in addition to the three major capsid proteins (penton base, hexon and fiber) and several proteins associated with the genomic core of the virion. Of the minor capsid proteins, VI plays several crucial roles in the infection cycle of the virus, including hexon nuclear targeting during assembly, activation of the adenovirus proteinase (AVP) during maturation and endosome escape following cell entry. VI is translated as a precursor (pVI) that is cleaved at both the N- and C-termini by AVP. Whereas the role of the C-terminal fragment of pVI, pVIc, is well established as an important co-factor of AVP, the role of the N-terminal fragment, pVIn, is currently elusive. In fact, the fate of pVIn following proteolytic cleavage is completely unknown. Here, we use a combination of proteomics-based peptide identification, native mass spectrometry and hydrogen-deuterium exchange mass spectrometry to show that pVIn is associated with mature human adenovirus, where it binds at the base of peripentonal hexons in a pH-dependent manner. Our findings suggest a possible role for pVIn in targeting pVI to hexons for proper assembly of the virion and timely release of the membrane lytic mature VI molecule.
Keywords: Virus maturation, mass spectrometry, native MS, hydrogen-deuterium exchange, adenovirus proteinase
Introduction
There are more than 60 known types of human adenovirus (HAdV), approximately one-third of which cause acute infection in humans. Infection is generally self-limiting, except in immunocompromised patients. Replication-defective HAdV is also used in numerous gene therapy and vaccine delivery applications 1, 2. HAdV is one of the largest known non-enveloped dsDNA viruses. It consists of a ~36 kb genome that is encapsidated in a pseudo T=25 icosahedral capsid 3–6. The HAdV virion is approximately 90 nm in diameter, with a total mass estimated at 150 MDa. The capsid of HAdV is composed of 240 trimeric hexons, 12 pentameric penton base molecules that occupy the vertices, and 12 trimeric fiber proteins that extend outward from the penton base and interact with host receptors to facilitate cell attachment. In addition, the mature capsid contains four cement proteins (IIIa, VI, VIII and IX), five proteins associated with the genomic core of the virion (V, VII, µ, IVa2 and terminal protein), as well as a few copies of the adenovirus proteinase (AVP).
As is common in dsDNA viruses, HAdV assembles with the aid of scaffolding proteins and subsequently matures into the infectious virion. AVP plays an essential role in HAdV maturation by processing many of the accessory proteins through cleavage at two consensus motifs, (I,L,M)XGG/X and (I,L,M)XGX/G 7. Upon assembly in the immature virus particle, AVP is inactive and bound to DNA 8. Complete activation of AVP requires both DNA-binding and pVI 8, 9. In immature virus particles, pVI slides along the viral genome through interactions of the c-terminus of pVI with the DNA 10, 11. As pVI encounters AVP, AVP cleaves the 11 C-terminal residues from pVI (pVIc), which then binds and covalently links to AVP via a disulfide bridge yielding maximum AVP activity 8, 10–16. AVP then processes the precursors of IIIa, VI, VII, VIII, µ and terminal protein into their mature forms 7. In addition to cleaving pVIc, AVP also cleaves the 33 amino-terminal residues of pVI to generate mature VI and pVIn (i.e. residues 1–33 of pVI, see Figure 1) 17. In mature virions, VI is likely bound to peripentonal hexons on the capsid interior, though clear density for the molecule has not been definitively identified in current structural models 3, 4. The precursor pVI was shown to bind to hexon trimers with nanomolar affinity in vitro with a stoichiometry of 3:3 15. Whether this stoichiometry also reflects the mode of binding in the virion is currently not known; however the disparity in copy number for hexon (720) and VI (360) monomers suggests this is not the case, at least in the mature virion. In addition to activation of AVP, pVI also plays an important role in nuclear import of the hexon 18. There are two nuclear localization (NLS) and two nuclear export signals (NES) on the full sequence of pVI. One particular NLS is located entirely on the sequence corresponding to pVIc, and this NLS is crucial for nuclear import of the hexon, in an importin α/β dependent pathway.
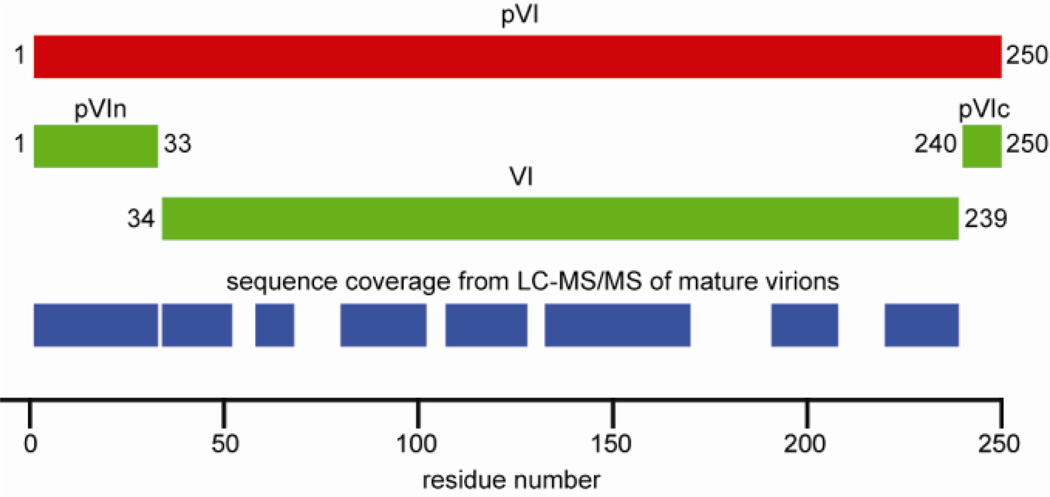
Figure 1.
Schematic of pVI maturation cleavage and the obtained sequence coverage from LC-MS/MS analysis of mature virions. The 250 aa precursor pVI (red) is cleaved by AVP between residues 33/34 and 239/240 to form pVIn, VI and pVIc (green). Approximately 70% of the pVI sequence is covered by peptide identifications from mature virions, including the full pVIn sequence (blue).
Whereas pVI (pVIc in particular) plays an essential role in virus maturation, thereby priming the virion for efficient uncoating 19, VI is essential at a later stage during infection. HAdV enters host cells through receptor-mediated endocytosis. Following fiber-mediated cell attachment, subsequent interactions of the penton base with cell-surface integrins trigger endocytosis of the virion 20–24. Partial disassembly of the virion is initiated at the cell surface with release of the fiber 25, 26. Integrin binding weakens the vertex region of HAdV, which is thought to facilitate subsequent release of the penton base in early endosomes, triggered by the mildly acidic conditions encountered in that environment 21, 27. Release of the penton base opens the virion at its vertices, allowing release of VI into the endosome. VI was shown to have membrane lytic activity and is essential for escape from the endosome into the cytosol 28, 29. In addition to promoting endosome escape, VI was also reported to promote adenovirus gene expression by counteracting the Daxx-mediated host defense that suppresses viral gene expression 30.
Whereas the roles of pVIc and VI are emerging, it is currently unclear what role the 33-residue pVIn peptide has in the replication cycle of HAdV or if it remains part of the mature virion. Here we identify pVIn in mature virions by mass spectrometry (MS) based proteomics. We show with native MS that pVIn is associated with peripentonal hexons, which are released in complex from heat-disrupted virions. It is demonstrated that the interaction between pVIn and hexon is strongly pH-dependent. Finally, using hydrogen-deuterium exchange (HDX) MS, we show that pVIn binds to hexons in a region spanning residues 32–65 near the base of the hexon trimer in the capsid interior. Our findings pinpoint a previously unknown anchoring point for pVI during assembly of the virion within the peripentonal hexons.
Results
pVIn is part of mature HAdV
The virions used for mass spectrometry analyses were derived from double-banded cesium chloride ultracentrifugation that separates immature from mature virions (see “Materials and Methods). We confirmed the purity of mature virions by performing SDS-PAGE to ensure the absence of uncleaved precursor polypeptides that are present in immature particles. In addition we determine the 260/280 ratio of the purified particles that is ~1.3 for fully mature particles containing a complete viral genome.
To identify the parts of pVI that are in the mature virion, we analyzed mature HAdV particles by MS-based proteomics. HAdV particles were proteolytically digested and subsequently analyzed by LC-MS/MS. All 13 proteins of the HAdV virion could be identified from this analysis (data not shown), including pVI. Of the full pVI sequence, ~70% was covered from peptide identifications (see Figure 1). This included most of the mature VI but also the full first 33 residues of the pVI sequence, i.e. pVIn: MEDINFASLAPRHGSRPFMGNWQDIGTSNMSGG (pVIn peptide identifications are listed in Supplementary Table S1). The protease (trypsin) used for digestion of the sample cleaves after lysine and arginine, but the C-terminal pVIn peptide terminates with a glycine residue. The observation that both the C-terminal half of pVIn and the N-terminus of VI were identified from semi-tryptic peptides, containing the AVP consensus terminal motif, suggests that the peptides that cover pVIn originate from the endogenously processed product. No peptides corresponding to pVIc were observed. This might simply mean that pVIc yields unobservable peptides in our analytical procedure and the lack of identification does not exclude that pVIc is still contained in the mature virion.
Endogenous pVIn is released from HAdV during in vitro partial capsid disassembly
In the mature virion, VI is likely located near the capsid vertices, interacting with the interior side of the peripentonal hexons 3, 4. The protein is released in early endosomes following partial capsid disassembly, triggered by the acidic pH of the compartment 28, 29 and perhaps also by association with αv integrins 21, 27. Moderate heating of mature virions results in a similar disassembly of the particle, with nearly complete release of fiber and penton base, and partial release of IIIa, VI, VIII and peripentonal hexons 31–33. We separated the released components from the remaining viral cores using density centrifugation and analyzed the relative abundance of the pVIn peptides generated from heated virions using LC-MS/MS. Judging from peptide spectrum matches with pVII as an internal standard, approximately 80% of pVIn was found to be released from the heated virions (see Supplementary Table S2).
Endogeneous pVIn is bound to released hexon trimers
The fraction of components that is released upon heating of mature HAdV was prepared for analysis by native MS in order to identify the subcomplexes that disassemble from the particle. Native MS is an emerging technique for the detailed mass analysis of non-covalent protein complexes 34, 35. Protein assemblies are transferred to an MS compatible buffer and analyzed by nano-electrospray ionization to yield highly precise and accurate mass information with unmatched resolving power.
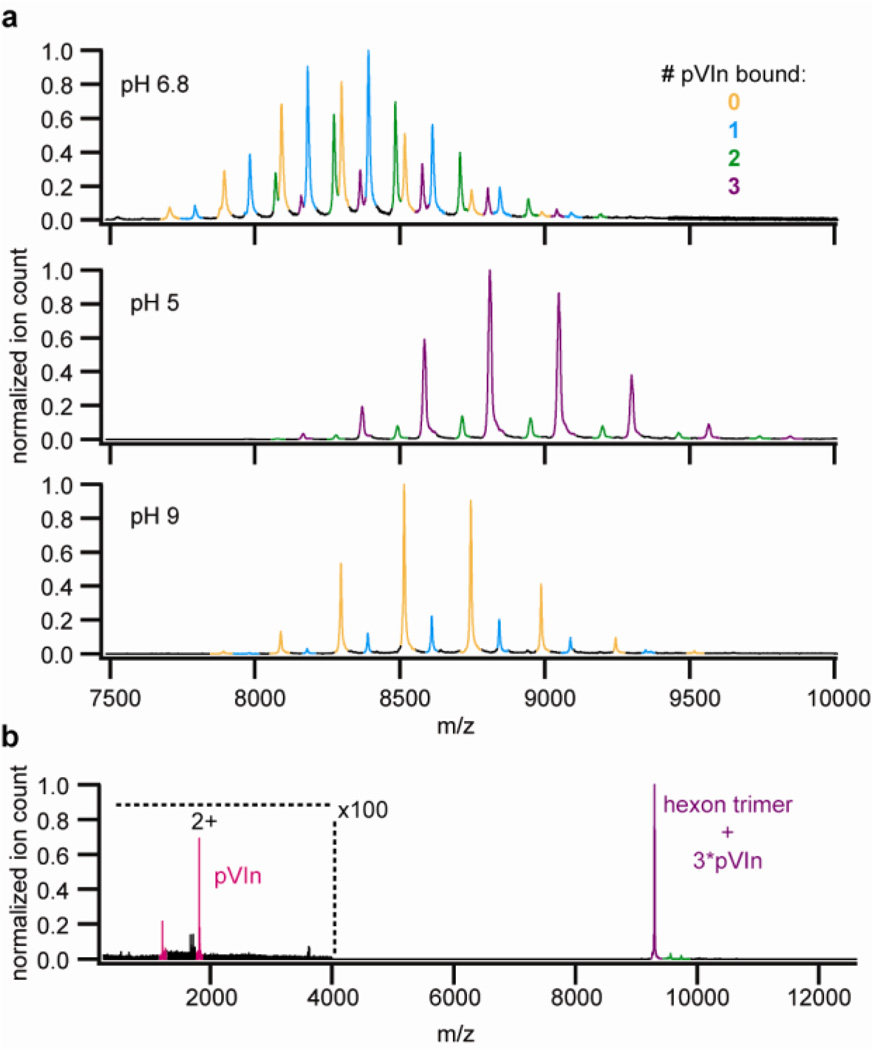
Hexon trimers were the primary molecules detected in these preparations when analyzed by native MS. We did not observe signals for the concomitantly released penton base and fibers, which may be due to losses occurring during the sample preparation for native MS and/or their relative lower abundance in the fraction of released proteins, as all released components (penton base, fiber, hexon, VI, pVIn, IIIa and VIII) could be identified from LC-MS/MS analysis of the same fraction (data not shown). The accurate mass measured for the hexon trimers (323.62 kDa, within 0.025% of the expected mass, see Supplementary table S3) allowed an unambiguous assignment. In addition to free hexon trimers, we observed up to three different hexon trimer complexes with an excess mass that matched within 0.025%, to the binding of 1, 2 or 3 copies of pVIn to the hexon trimer (see Figure 2a and Supplementary Table S3). Tandem MS of these putative hexon-pVIn complexes confirmed that the mass of the bound peptide corresponds closely to that of pVIn (see Figure 2b). At a collision voltage in excess of 100V, we detected two charge states at low m/z that correspond to a mass of 3624.1 Da, compared to a theoretical pVIn mass of 3624.0 Da. When the released fraction was transferred to a pH 5 solution, nearly complete binding was observed, i.e. 3 pVIn molecules/hexon trimer. At pH 9, however, only free hexon trimers were recovered and detected by native MS. The sensitivity of the interaction to pH was utilized to isolate the bound pVIn peptide from recovered hexon trimers. LC-MS/MS analysis of the isolated peptide confirmed that it is indeed pVIn, as the full sequence could be uniquely defined from the observed fragment ions (see Supplementary Figures S1 and S2). Release of pVIn during partial capsid disassembly might suggest a possible role in endosome escape. However, no direct membrane lytic activity of pVIn was detected on SulfoB- loaded liposomes (see Supplementary Figure S3). We cannot exclude the possibility that pVIn might have an indirect role in endosome escape or perhaps in subsequent events in HAdV infection.
Figure 2.
Native MS analysis of hexons from heat-disrupted virions show that pVIn is bound to peripentonal hexons in a pH-dependent manner. A) Hexon-pVIn complexes recovered from heat-disrupted virions were analyzed at the indicated pH. B) Tandem MS of 3:3 hexon:pVIn complex. The measured masses of the different detected hexon-pVIn assemblies are provided in supplementary table S2.
The hexon-pVIn interaction is strongest at endosomal pH
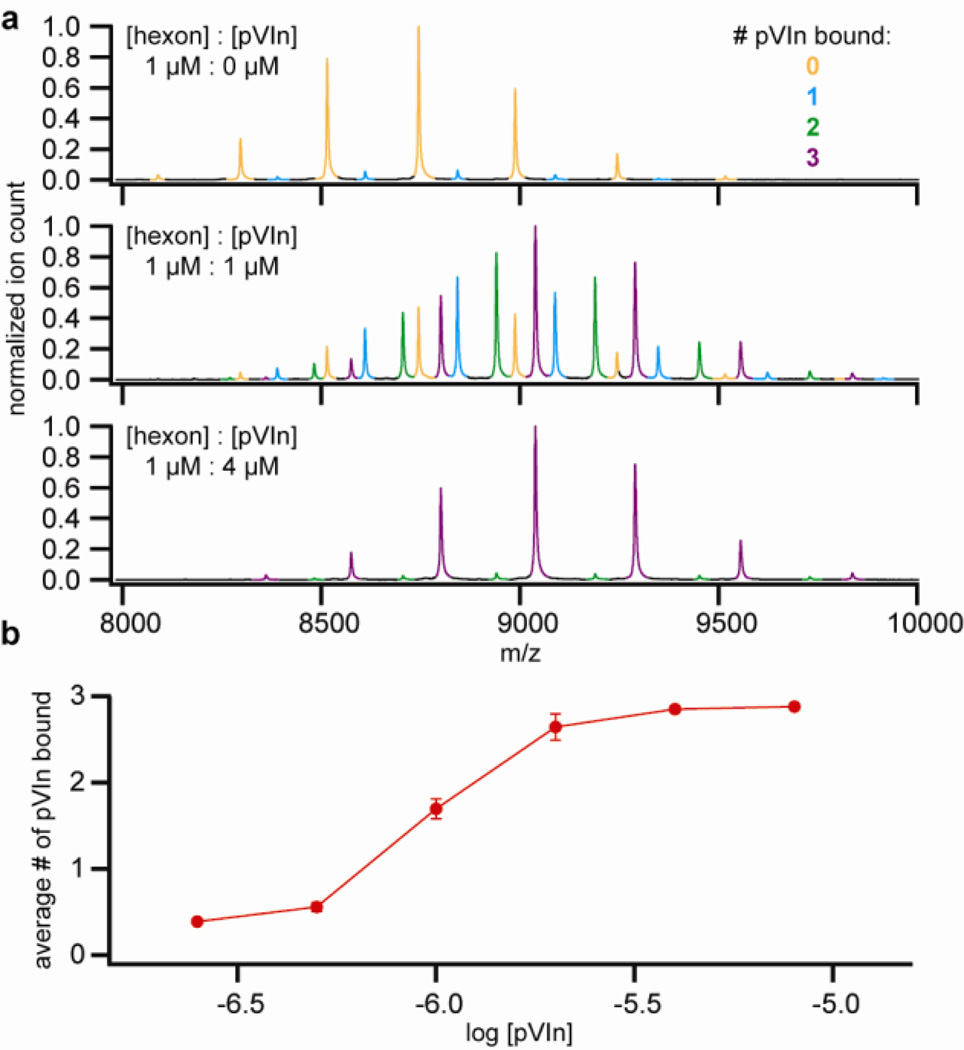
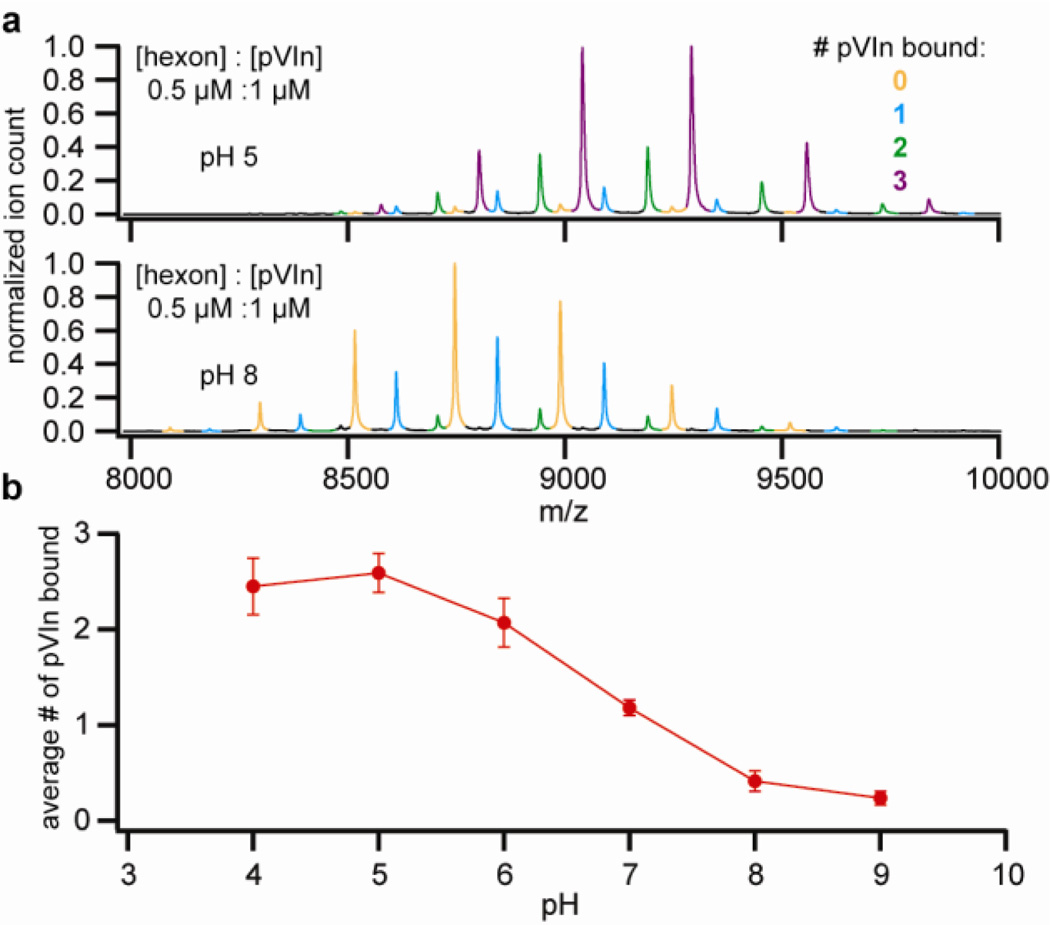
The observation that more hexon-pVIn complex could be recovered at acidic pH than at neutral or high pH strongly suggests that the affinity of the interaction is pH dependent. To test this more systematically, we reconstituted the hexon-pVIn complex from purified hexons and synthetic pVIn (see Figure 3). In a titration experiment, where the relative intensities of the differently bound forms are used as a measure of binding, we observed increased binding of pVIn with increasing concentration. Up to, but no more than, three copies of pVIn were bound to hexon trimers, even when pVIn was added in high excess, indicating that the interaction is highly specific. We next tested the fraction of bound pVIn in a 1:2 mixture of hexon:pVIn in the pH-range of 4–9 (see Figure 4). We observed half-maximum binding at pH 6.5–7 and near-complete dissociation towards pH 8. These findings indicate that the interaction is strongest at endosomal pH but relatively weak at cytosolic pH. There are several basic residues in the sequence of pVIn, including one histidine at position 13, which may contribute to the observed effect of pH on the hexon-pVIn interaction.
Figure 3.
Reconstitution of hexon-pVIn using purified hexon and synthetic pVIn. A) Native MS spectra of hexon-pVIn mixtures. Note that some residual endogenous pVIn is already bound to the purified hexon. B) Average number of pVIn bound to hexon trimer, as a function of pVIn concentration. Points represent average ± standard deviation from duplicate experiments.
Figure 4.
Strength of the hexon-pVIn interaction is pH-dependent. A) Native MS of 1:2 hexon:pVIn mixtures at the indicated pH. B) Average number of pVIn (1 µM) bound to hexon trimer (0.5 µM) as a function of pH. Points represent average ± standard deviation from triplicate experiments.
pVIn binds in the cavity of the hexon trimer
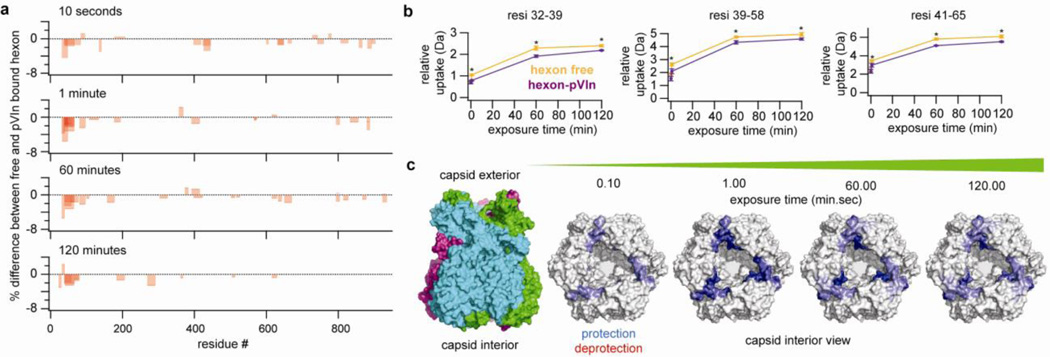
In an attempt to localize the binding site of pVIn on the hexon trimer, we performed an HDX-MS experiment to monitor the changes in deuterium uptake in the hexon trimer upon binding of pVIn. After incubation of free hexon or hexon-pVIn in D20, the reaction was quenched, followed by online pepsin digestion and LC-MS/MS analysis. Peptides were identified from hexon in MilliQ, yielding 236 peptides of sufficient quality for quantifying deuterium uptake, covering 95% of the full hexon sequence (see Supplementary Figure S4). Exposure was carried out for 10 seconds and 1, 60 and 120 minutes in triplicate and changes in deuterium uptake were filtered to p<0.05 using unpaired two-tailed Student’s t-tests. Out of the over 200 peptides analyzed, we observed three peptides that showed a consistent, statistically significant change between the free hexon and hexon-pVIn (See Figure 5). For comparison, the deuterium uptake of the peptides that are affected by pVIn binding is shown alongside representative unaffected peptides in Supplementary Figure S5. The three peptides that exhibit protection upon pVIn binding localize to the same region of the hexon sequence and collectively span amino acid residues 32–65 of the hexon. These residues are in the extended N-terminal domain that interacts with the neighboring hexon monomer of the trimer, and line the rim of the hexon cavity on the capsid interior (Figure 5). These results provide evidence that residues 32–65 span the pVIn-binding region of the hexon.
Figure 5.
The pVIn binding site of hexon is located within the region spanning residues 32–65 at the base of the hexon cavity. A) Overview of peptides that exhibit significantly changed deuterium uptake between free and pVIn-bound hexon. Bars are transparent, such that darker regions represent overlapping peptides. B) Deuterium uptake as a function of time of peptides displaying a consistent and statistically relevant differential uptake between free hexon and hexon-pVIn assemblies. Points represent average ± standard deviation from triplicate experiments. Asterisks indicate that p<0.05 in unpaired two-tailed Student’s t-test between two states at that time point. C) Changes in deuterium uptake mapped to the crystal structure of the hexon (PDB code: 1p30).
Discussion
Using a variety of primarily MS-based methods, we show here that the N-terminus of pVI, pVIn, is part of mature HAdV particles following cleavage by AVP. We demonstrate that pVIn interacts with residues 32–65 of the hexon, located near the rim of the cavity of the hexon trimer on the capsid interior. Full-length pVI binds with residues 48–74 and 233–239 (in addition to residues from pVIn)17 to hexon trimers with high affinity 15. Our experiments indicate that the affinity of the hexon pVIn interaction is highest at endosomal pH but decreases significantly at higher pH. Given the pH-dependence of the interaction, it is perhaps surprising that pVIn stays bound to hexon, despite the relatively high pH of the extracellular environment, cytosol and nucleus. However, considering the fact that pVIn is enclosed in the small inner volume of the capsid, local pH and concentration is hard to estimate. However, we expect that the local concentrations of both pVIn and hexon are effectively high, especially considering the volume that is excluded by the densely packed genome and additional core components of the virion. Partial disassembly of HAdV in early endosomes would increase the volume in which pVIn is free to diffuse, thereby effectively decreasing the local concentrations and possibly allowing release of free pVIn into the endosome. However, we found that at pH 5, endogenous pVIn appears to still be fully retained on hexon trimers. If pVIn is retained on hexon trimers in the endosome, the transition to higher pH (~ 7.4) following endosome escape likely results in dissociation of the hexon-pVIn complex. It is currently unclear whether pVIn has any effect on other steps in the replication cycle of HAdV.
The exact copy number of pVI and for that matter pVIn is currently not well defined. The near-complete occupancy of endogenous pVIn-binding sites on hexons released from heated virions would suggest that the copy number is on the order of 180, assuming that the hexons that are recovered in our experiment are all peripentonal, and that no additional pVIn is present in heated particles (approximately 80% is released according to our LC-MS/MS analysis). Our native MS data do show that hexon trimers with sub-stoichiometric pVIn are also stable configurations, and it is thus possible that the actual copy number is lower. This estimate of ~180 copies of pVIn in mature virions also suggests that a subset of pVIn molecules are lost prior to full maturation, as the VI copy number is estimated to be 340–360. On the other hand, it is possible that additional copies of pVIn dissociate from heated particles without being bound to hexon, and would therefore not be detected in our native MS experiment, in which case the actual copy number of pVIn might be higher.
It is interesting to note that the purified hexons in our experiments had a very small residual amount of endogenous pVIn bound (see Figure 2a, approximately 3% of maximum occupancy). These hexons, isolated from virus-infected cells, are produced in excess of what is actually assembled into complete virions, hence suggesting that (partial) processing of pVI can possibly take place outside of the assembled virion, or that these represent “debris” from broken/disassembled virions.
The binding site of pVIn that we observed in our HDX-MS experiments had not been previously identified as a pVI binding site on hexons. Two cryo-electron microscopy reconstructions of the maturation defective ts1 mutant of HAdV did show additional density in the hexon cavity, which was tentatively assigned to pVI, but no additional density in the vicinity of residues 32–65 of the hexon monomer has been reported 36, 37. This small 33 residue portion of pVI might simply not have been visible at the moderate resolution of these cryo-EM reconstructions, but rearrangement of pVIn upon cleavage is also a possible scenario.
In conclusion, we have shown that pVIn is a component of mature HAdV virions. The fragment resides in the cavity of hexon trimers and is released in an in vitro model of partial capsid disassembly. The interaction of pVIn with hexon points to a role for this peptide in AdV assembly. This is supported by the high sequence conservation of both the pVIn fragment and hexon (residues 32–65) from both human and non-human AdVs. We speculate that cleavage by AVP between residues 33/34 of pVI is necessary for efficient release of the membrane lytic VI molecule in the endosome during entry. This might contribute to the uncoating defect of the maturation defective ts1 mutant. In this regard, mutagenesis of the AVP cleavage site for pVIn in a WT HAdV background should clarify the role of AVP cleavage of pVI in efficient endosome escape. In the context of mature HAdV particles, pVIn might simply be a remnant of hexon-pVI assembly, that is cleaved from pVI for efficient release of the mature product VI.
Materials and Methods
Purification of Ad5F35 and hexon
Replication-defective Ad5F35 and Ad5 hexon, were grown and isolated as previously described with some modification 38, 39. Briefly, a Nunc cell factory of 293β5 cells was infected with 300–500 Ad5F35 particles/cell 28. Between 48–60 hours post-infection, cells were harvested and resuspended in 8 ml of 10 mM Bis-Tris pH 6.5 and flash-frozen in liquid nitrogen. Cells were thawed at 37°C and 1 ml of Freon-113 (Sigma) was added. Complete lysis was achieved with two additional rounds of freeze/thaw. Clarified lysates were separated on 15–40% cesium chloride density gradients and the hexon and virus fractions were collected. Virus was dialyzed into DX10/10 buffer (40 mM Tris pH 8.1, 500 mM NaCl, 10% glycerol, 10% ethylene glycol, 2% sucrose and 1% mannitol). Hexon was dialyzed into 10 mM Bis-Tris pH 7.0 and further purified by ion exchange on a ResourceQ (GE Healthcare) column in 10 mM Bis-Tris pH 7.0 with a multi-step NaCl gradient to 1 M. Hexon fractions were pooled and concentrated in an Amicon centrifugal filter unit (Millipore) to 7–12 mg/ml. Aliquots of purified virus and hexon were frozen in liquid nitrogen and stored at −80°C until use.
In vitro partial capsid disassembly
Large-scale partial disassembly of HAdV capsids suitable for native mass spectrometry was adapted from previously published protocols 31, 40. Ad5F35 (5 mg) was diluted in 7.5 mM HEPES pH 7.4, 50 mM NaCl to a final volume of 3 ml. To ensure complete disassembly, the virus was divided into six 500 µl aliquots and heated to 55°C for 15 min. The aliquots were pooled together and loaded onto four 30–80% discontinuous Histodenz (Sigma) gradients dissolved in 20 mM Tris pH 7.4 and centrifuged at 198,000xg (ave) for 3 hrs. The supernatant fraction, containing the subset of Ad5F35 proteins released from the virions, was collected with a pipette from the top of the gradient in two 1 ml fraction, which were subsequently pooled together. Samples were stored frozen until analysis.
Membrane lytic activity of pVIn
Synthetic pVIn peptide was dissolved at high concentration (>20 mg/ml) in MilliQ water. Recombinant protein VI114 (residues 34–114) was purified as described 41 and used as a positive control for membrane lytic activity. The L40Q mutant of VI114 has decreased membrane lytic activity compared to the wild-type protein and is used to show an intermediate phenotype 40. Bovine serum albumin (BSA, Sigma) was used as a negative control. Serial dilutions of protein were done in 50 mM Tris pH 8.0, 150 mM NaCl. The membrane lytic activity of pVIn and VI114 was measured in a liposome lysis assay as previously described 41.
Proteomics of mature Ad5F35
Both the intact and heated HAdV particles were prepared for LC-MS/MS analysis by resuspending them in 50mM ammonium bicarbonate, 5% (w/v) sodium deoxycholate (SDC, Sigma Aldrich) and heating at 90° for 5 minutes. Proteins (~200µg) for each enzymatic digestion, were first reduced for 30 minutes at 56° and then alkylated for 1h in the dark. Proteins were first digested with Lys-C (Roche) for 4h at 37°. After diluting the samples to a final SDC concentration of 0.5% (w/v), enzymatic digestion was performed overnight at 37° adding Trypsin (Promega) in a substrate/enzyme ratio of 50:1 (w/w). Digestion was quenched by acidification with Formic acid (FA) to a final concentration of 10% and peptides were desalted by solid phase extraction (Seppack Vac C18 cartridges, Waters). Peptides were eluted in 80% acetonitrile, dried in a speedvac and then resuspended in 10% formic acid solution. Each adenovirus sample was analyzed by LC-MS/MS in triplicate. An EASY-nLC 1000 (Thermo Fischer Scientific) was equipped with a 20 mm Aqua C18 (Phenomenex) trapping column (packed in-house, 100 µm i.d., 5 µm particle size) and a 400 mm Zorbax SB-C18 (Agilent) analytical column (packed in-house, either 50 µm i.d. or 75 µm i.d., 1.8 µm particle size). Trapping and washing was performed at 10 µl/min for 4 min with solvent A (0.1 M acetic acid in water). Subsequently, peptides were transferred to the analytical column at about 150 nl/min and eluted with a gradient of 3–40% (v/v) solvent B (0.1 M acetic acid in 80% ACN) in 45 min. The eluent was sprayed via distal coated emitter tips butt-connected to the analytical column and the ion spray voltage was set to 1.7 kV. The mass spectrometers were operated in data-dependent mode, automatically switching between MS and MS/MS. The full-scan MS spectra (from m/z 350 to 1500) were acquired in the Orbitrap analyzer with a resolution of 60,000 FHMW at 400 m/z after accumulation to target value of 1e6 in the linear ion trap (maximum injection time was 250 ms). After the survey scans, the ten most intense precursor ions at a threshold above 5000 were selected for MS/MS with an isolation width of 1.5 Da. Peptide fragmentation was carried out by using a decision tree performing higher collision dissociation (HCD) or electron transfer dissociation (ETD) depending on their charge state and m/z. HCD fragment ions readout was performed in the orbitrap analyzer with a resolution of 15,000 FHMW, activation time was of 0.1 ms and normalized collision energy (CE) of 32. ETD fragment ions readout was performed in the linear ion trap analyzer with an activation time of 50ms.
MS raw data from the shotgun LC-MS/MS analyses were processed by Proteome Discoverer (version 1.3, Thermo Electron). Peptide identification was performed with Mascot 2.3 (Matrix Science) against a concatenated forward-decoy UniPROT database including the HAdV protein sequences and supplemented with all the frequently observed contaminants in MS. The following parameters were used: 6 ppm precursor mass tolerance, 0.6 Da (for ETD) and 0.05 Da (for HCD) fragment ion tolerance. In order to evaluate the viral protease activity we allowed the identification of peptides containing one non-specific cleavage and a maximum of 2 missed cleavages. Carbamidomethyl cysteine was allowed as fixed modification, while oxidized methionine and protein N-terminal acetylation were set as variable modifications. Finally, results were filtered using the following criteria: (i) mass deviations of ±5 p.p.m., (ii) Mascot Ion Score of at least 20, (iii) a minimum of 6 amino-acid residues per peptide and (iv) position rank 1 in Mascot search. As a result, we obtained peptide FDRs below 1% for all the three peptide mixtures analyzed. Relative quantification was performed by spectral counts, which relies on the number of peptide spectrum matches (PSMs) specific for a certain protein (or protein domain). As an internal control for data normalization, we used the two DNA interacting proteins pVII and pTP as housekeeping proteins.
Native MS of hexon-pVIn
The supernatant fraction from heat-disrupted virion density centrifugation or purified hexon trimer were buffer exchanged to 150 mM ammonium acetate at the indicated pH using Vivaspin 500K 10 kDa MWCO centrifugal filter units. Reconstituted hexon-pVIn from synthetic pVIn was prepared by mixing hexon in ammonium acetate with pVIn dissolved in MilliQ water. Samples were loaded into gold-coated boro-silicate capillaries, prepared in-house, for nano-electrospray ionization. Mass spectrometry was performed on a QToF II instrument (Waters), modified for optimal transmission of high-mass ions (MS Vision) 42, 43. The instrument was operated at 10 mbar in the source region, and 1.5*10−2 mbar in the collision cell using xenon as collision gas. Capillary voltage was set at 1400–1500 V, sample cone voltage at 160 V. Hexon-pVIn complexes were analyzed at a collision voltage of 100 V, and tandem MS of hexon-pVIn was performed at a collision voltage of 120 V. Spectra were calibrated with cesium iodide.
Isolation and identification of endogenous pVIn from hexon-pVIn complexes
We utilized the pH-dependence of the hexon-pVIn interaction to isolate pVIn from the complex. Hexon-pVIn was trapped on C18 ZipTip resin. The trapped complex was dissociated with a washing step, using 150 mM ammonium acetate pH 9, thereby trapping free pVIn on the C18 resin. After one additional washing step with 0.1% formic acid in water, pVIn was eluted from the resin with 80% acetonitrile/0.1% formic acid in water, while hexon was retained on the ZipTip. The sample was dried in a speedvac and redissolved in 10% formic acid/5% DMSO in water. The sample was analyzed by reversed phase LC coupled on-line to an LTQ or LTQ-Orbitrap Velos II for MS/MS analysis. The nano-LC consists of an Agilent 1200 series LC system equipped with a 20 mm ReproSil- Pur C18-AQ (Dr. Maisch GmbH) trapping column (packed in-house, i.d., 100 µm; resin, 5 µm) and a 400 mm ReproSil-Pur C18-AQ (Dr. Maisch GmbH) analytical column (packed in-house, i.d., 50 µm; resin, 3 µm). The flow was passively split to 100 nl/min. We used a standard 45 minute gradient from 7–30% acetonitrile in aqueous 0.1% formic acid. For MS/MS analysis on the LTQ-Orbitrap XL, all precursors were sequenced with both CID and ETD fragmentation and MS analysis in the ion trap. The data were searched with Mascot against a custom database containing all HAdV5 protein sequences, except for the fiber, which was replaced with the HAdV35 fiber sequence.
HDX-MS of hexon-pVIn complexes
Free hexon or hexon-pVIn (at a 1:4 molar ratio) was prepared at a concentration of 10 µM in 150 mM ammonium acetate pH 5. At the start of the exchange reaction, the sample was diluted six-fold into D2O to a final volume of 30 µL. Exposure was carried out for 10 seconds, 1, 60 and 120 minutes in triplicate at 25°C. The reaction was quenched by 1:1 mixing with ice-cold 4 M urea, 200 mM TCEP and adjusted with HCl to give a final pH upon mixing of 2.5. Immediately after quenching, the sample was injected into a Waters HDX/ nanoAcquity system for digestion on an online pepsin column (25 °C, at a flow-rate of 50 µL.min−1) followed by separation on a 10 minute RP-UPLC gradient at 0 °C and MS on a Waters Xevo QToF G2. For peptide identification, hexon was prepared under identical conditions in H2O and analyzed using MSe data acquisition. Data for peptide identification was processed with ProteinLynx Global Server 2.5 software. Deuterium uptake was calculated compared to the control samples in H2O using Waters DynamX 1.0.0 software. Back-exchange was estimated at approximately 30% in our workflow. No corrections for back-exchange were applied since the analysis focuses on relative changes in deuterium uptake, rather than on absolute levels. Observed changes in deuterium uptake were filtered to p<0.05 in an upaired two-tailed Student’s t-test between free and bound hexon.
Supplementary Material
Highlights.
The fate of cleavage products of pVI in human adenovirus is investigated.
The cleaved N-terminus of pVI, pVIn, is retained in mature virus particles.
Native MS shows up to 3 copies of pVIn binding to hexons in a pH-dependent manner.
pVIn binds to the base of peripentonal hexons on the capsid interior.
pVIn is retained in mature adenovirus and binds specifically to hexon.
Acknowledgements
We thank Tina-Marie Mullen for technical assistance. These studies were supported in part by NIH grants HL054352 to G.R.N. and 5T32 AI007354 for C.L.M. J.S., M.B, and A.J.R.H. are supported by the Netherlands Proteomics Centre, and by the European Community‘s Seventh Framework Programme (FP7/2007–2013) by the PRIME-XS project grant agreement number 262067.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kay MA. State-of-the-art gene-based therapies: The road ahead. Nature Reviews Genetics. 2011;12:316–328. doi: 10.1038/nrg2971. [DOI] [PubMed] [Google Scholar]
- 2.Small JC, Ertl HCJ. Viruses - From pathogens to vaccine carriers. Current Opinion in Virology. 2011;1:241–245. doi: 10.1016/j.coviro.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reddy VS, Natchiar SK, Stewart PL, Nemerow GR. Crystal structure of human adenovirus at 3.5 Å resolution. Science. 2010;329:1071–1075. doi: 10.1126/science.1187292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu H, Jin L, Koh SBS, Atanasov I, Schein S, Wu L, Zhou ZH. Atomic structure of human adenovirus by Cryo-EM reveals interactions among protein networks. Science. 2010;329:1038–1043. doi: 10.1126/science.1187433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nemerow GR, Stewart PL, Reddy VS. Structure of human adenovirus. Current Opinion in Virology. 2012;2:115–121. doi: 10.1016/j.coviro.2011.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martín CS. Latest insights on adenovirus structure and assembly. Viruses. 2012;4:847–877. doi: 10.3390/v4050847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diouri M, Keyvani-Amineh H, Geoghegan KF, Weber JM. Cleavage efficiency by adenovirus protease is site-dependent. J. Biol. Chem. 1996;271:32511–32514. doi: 10.1074/jbc.271.51.32511. [DOI] [PubMed] [Google Scholar]
- 8.Mangel WF, McGrath WJ, Toledo DL, Anderson CW. Viral DNA and a viral peptide can act as cofactors of adenovirus virion proteinase activity. Nature. 1993;361:274–275. doi: 10.1038/361274a0. [DOI] [PubMed] [Google Scholar]
- 9.Webster A, Hay RT, Kemp G. The adenovirus protease is activated by a virus-coded disulphide-linked peptide. Cell. 1993;72:97–104. doi: 10.1016/0092-8674(93)90053-s. [DOI] [PubMed] [Google Scholar]
- 10.Blainey PC, Graziano V, Pérez-Berná AJ, McGrath WJ, Flint SJ, Martín CS, Xie XS, Mangel WF. Regulation of a viral proteinase by a peptide and DNA in one-dimensional space IV: Viral proteinase slides along dna to locate and process its substrates. J. Biol. Chem. 2013;288:2092–2102. doi: 10.1074/jbc.M112.407460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Graziano V, Luo G, Blainey PC, Pérez-Berná AJ, McGrath WJ, Flint SJ, Martín CS, Xie XS, Mangel WF. Regulation of a viral proteinase by a peptide and DNA in one-dimensional space: II. adenovirus proteinase is activated in an unusual one-dimensional biochemical reaction. J. Biol. Chem. 2013;288:2068–2080. doi: 10.1074/jbc.M112.407312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baniecki ML, McGrath WJ, McWhirter SM, Li C, Toledo DL, Pellicena P, Barnard DL, Thorn KS, Mangel WF. Interaction of the human adenovirus proteinase with its 11- amino acid cofactor pVIc. Biochemistry (NY) 2001;40:12349–12356. doi: 10.1021/bi0109008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mangel WF, Toledo DL, Brown MT, Martin JH, McGrath WJ. Characterization of three components of human adenovirus proteinase activity in vitro. J. Biol. Chem. 1996;271:536–543. doi: 10.1074/jbc.271.1.536. [DOI] [PubMed] [Google Scholar]
- 14.McGrath JW, Baniecki ML, Li C, McWhirter SM, Brown MT, Toledo DL, Mangel WF. Human adenovirus proteinase: DNA binding and stimulation of proteinase activity by DNA. Biochemistry (N.Y.) 2001;40:13237–13245. doi: 10.1021/bi0111653. [DOI] [PubMed] [Google Scholar]
- 15.Graziano V, McGrath WJ, Suomalainen M, Greber UF, Freimuth P, Blainey PC, Luo G, Xie XS, Mangel WF. Regulation of a viral proteinase by a peptide and DNA in one-dimensional space: I. binding to dna and to hexon of the precursor to protein VI, pVI, of human adenovirus. J. Biol. Chem. 2013;288:2059–2067. doi: 10.1074/jbc.M112.377150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baniecki ML, McGrath WJ, Mangel WF. Regulation of a viral proteinase by a peptide and DNA in one-dimensional space: III. atomic resolution structure of the nascent form of the adenovirus proteinase. J. Biol. Chem. 2013;288:2081–2091. doi: 10.1074/jbc.M112.407429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matthews DA, Russell WC. Adenovirus protein-protein interactions: Molecular parameters governing the binding of protein VI to hexon and the activation of the adenovirus 23K protease. J. Gen. Virol. 1995;76:1959–1969. doi: 10.1099/0022-1317-76-8-1959. [DOI] [PubMed] [Google Scholar]
- 18.Wodrich H, Guan T, Cingolani G, Von Seggern D, Nemerow G, Gerace L. Switch from capsid protein import to adenovirus assembly by cleavage of nuclear transport signals. EMBO J. 2003;22:6245–6255. doi: 10.1093/emboj/cdg614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perez-Berna AJ, Ortega-Esteban A, Menendez-Conejero R, Winkler DC, Menendez M, Steven AC, Flint SJ, de Pablo PJ, San Martin C. The Role of Capsid Maturation on Adenovirus Priming for Sequential Uncoating. J. Biol. Chem. 2012;287:31582–31595. doi: 10.1074/jbc.M112.389957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mathias P, Galleno M, Nemerow GR. Interactions of soluble recombinant integrin αvβ5 with human adenoviruses. J. Virol. 1998;72:8669–8675. doi: 10.1128/jvi.72.11.8669-8675.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lindert S, Silvestry M, Mullen T, Nemerow GR, Stewart PL. Cryo-electron microscopy structure of an adenovirus-integrin complex indicates conformational changes in both penton base and integrin. J. Virol. 2009;83:11491–11501. doi: 10.1128/JVI.01214-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stewart PL, Nemerow GR. Cell integrins: commonly used receptors for diverse viral pathogens. Trends Microbiol. 2007;15:500–507. doi: 10.1016/j.tim.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 23.Wickham TJ, Mathias P, Cheresh DA, Nemerow GR. Integrins α(v)β3 and α(v)β5 promote adenovirus internalization but not virus attachment. Cell. 1993;73:309–319. doi: 10.1016/0092-8674(93)90231-e. [DOI] [PubMed] [Google Scholar]
- 24.Chiu CY, Mathias P, Nemerow GR, Stewart PL. Structure of adenovirus complexed with its internalization receptor, α(v)β5 integrin. J. Virol. 1999;73:6759–6768. doi: 10.1128/jvi.73.8.6759-6768.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakano MY, Boucke K, Suomalainen M, Stidwill RP, Greber UF. The first step of adenovirus type 2 disassembly occurs at the cell surface, independently of endocytosis and escape to the cytosol. J. Virol. 2000;74:7085–7095. doi: 10.1128/jvi.74.15.7085-7095.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burckhardt CJ, Suomalainen M, Schoenenberger P, Boucke K, Hemmi S, Greber UF. Drifting motions of the adenovirus receptor CAR and immobile integrins initiate virus uncoating and membrane lytic protein exposure. Cell Host and Microbe. 2011;10:105–117. doi: 10.1016/j.chom.2011.07.006. [DOI] [PubMed] [Google Scholar]
- 27.Snijder J, Reddy VS, May ER, Roos WH, Nemerow GR, Wuite GJL. Integrin and defensin modulate the mechanical properties of adenovirus. J. Virol. 2013;87:2756–2766. doi: 10.1128/JVI.02516-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith JG, Silvestry M, Lindert S, Lu W, Nemerow GR, Stewart PL. Insight into the mechanisms of adenovirus capsid disassembly from studies of defensin neutralization. PLoS Pathogens. 2010;6:e1000959. doi: 10.1371/journal.ppat.1000959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wiethoff CM, Wodrich H, Gerace L, Nemerow GR. Adenovirus protein VI mediates membrane disruption following capsid disassembly. J. Virol. 2005;79:1992–2000. doi: 10.1128/JVI.79.4.1992-2000.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schreiner S, Martinez R, Groitl P, Rayne F, Vaillant R, Wimmer P, Bossis G, Sternsdorf T, Marcinowski L, Ruzsics Z, Dobner T, Wodrich H. Transcriptional activation of the adenoviral genome is mediated by capsid protein VI. PLoS Pathogens. 2012;8:e1002549. doi: 10.1371/journal.ppat.1002549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith JG, Nemerow GR. Mechanism of Adenovirus Neutralization by Human α- Defensins. Cell Host and Microbe. 2008;3:11–19. doi: 10.1016/j.chom.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 32.Rexroad J, Wiethoff CM, Green AP, Kierstead TD, Scott MO, Middaugh CR. Structural stability of adenovirus type 5. J. Pharm. Sci. 2003;92:665–678. doi: 10.1002/jps.10340. [DOI] [PubMed] [Google Scholar]
- 33.Russell WC, Valentine RC, Pereira HG. The effect of heat on the anatomy of the adenovirus. J. Gen. Virol. 1967;1:509–522. doi: 10.1099/0022-1317-1-4-509. [DOI] [PubMed] [Google Scholar]
- 34.Van Den Heuvel RHH, Heck AJR. Native protein mass spectrometry: From intact oligomers to functional machineries. Curr. Opin. Chem. Biol. 2004;8:519–526. doi: 10.1016/j.cbpa.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 35.Sharon M. Structural MS pulls its weight. Science. 2013;340:1059–1060. doi: 10.1126/science.1236303. [DOI] [PubMed] [Google Scholar]
- 36.Pérez-Berná AJ, Marabini R, Scheres SHW, Menéndez-Conejero R, Dmitriev IP, Curiel DT, Mangel WF, Flint SJ, Martín CS. Structure and Uncoating of Immature Adenovirus. J. Mol. Biol. 2009;392:547–557. doi: 10.1016/j.jmb.2009.06.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Silvestry M, Lindert S, Smith JG, Maier O, Wiethoff CM, Nemerow GR, Stewart PL. Cryo-electron microscopy structure of adenovirus type 2 temperature-sensitive mutant 1 reveals insight into the cell entry defect. J. Virol. 2009;83:7375–7383. doi: 10.1128/JVI.00331-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reddy VS, Natchiar SK, Gritton L, Mullen T, Stewart PL, Nemerow GR. Crystallization and preliminary X-ray diffraction analysis of human adenovirus. Virology. 2010;402:209–214. doi: 10.1016/j.virol.2010.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rux JJ, Burnett RM. Large-scale purification and crystallization of adenovirus hexon. Methods Mol. Med. 2007;131:231–250. doi: 10.1007/978-1-59745-277-9_17. [DOI] [PubMed] [Google Scholar]
- 40.Moyer CL, Wiethoff CM, Maier O, Smith JG, Nemerow GR. Functional genetic and biophysical analyses of membrane disruption by human adenovirus. J. Virol. 2011;85:2631–2641. doi: 10.1128/JVI.02321-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moyer CL, Nemerow GR. Disulfide-bond formation by a single cysteine mutation in adenovirus protein VI impairs capsid release and membrane lysis. Virology. 2012;428:41–47. doi: 10.1016/j.virol.2012.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Van Den Heuvel RHH, Van Duijn E, Mazon H, Synowsky SA, Lorenzen K, Versluis C, Brouns SJJ, Langridge D, Van Der Oost J, Hoyes J, Heck AJR. Improving the performance of a quadrupole time-of-flight instrument for macromolecular mass spectrometry. Anal. Chem. 2006;78:7473–7483. doi: 10.1021/ac061039a. [DOI] [PubMed] [Google Scholar]
- 43.Lorenzen K, Versluis C, van Duijn E, van den Heuvel RHH, Heck AJR. Optimizing macromolecular tandem mass spectrometry of large non-covalent complexes using heavy collision gases. International Journal of Mass Spectrometry. 2007;268:198–206. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.