Abstract
Background
Dengue virus (DENV) is responsible for up to approximately 300 million infections and an increasing number of deaths related to severe manifestations each year in affected countries throughout the tropics. It is critical to understand the drivers of this emergence, including the role of vector-virus interactions. When a DENV-infected Aedes aegypti mosquito bites a vertebrate, the virus is deposited along with a complex mixture of salivary proteins. However, the influence of a DENV infection upon the expectorated salivary proteome of its vector has yet to be determined.
Methods
Therefore, we conducted a proteomic analysis using 2-D gel electrophoresis coupled with mass spectrometry based protein identification comparing the naturally expectorated saliva of Aedes aegypti infected with DENV-2 relative to that of uninfected Aedes aegypti.
Results
Several proteins were found to be differentially expressed in the saliva of DENV-2 infected mosquitoes, in particular proteins with anti-hemostatic and pain inhibitory functions were significantly reduced. Hypothetical consequences of these particular protein reductions include increased biting rates and transmission success, and lead to alteration of transmission potential as calculated in our vectorial capacity model.
Conclusions
We present our characterizations of these changes with regards to viral transmission and mosquito blood-feeding success. Further, we conclude that our proteomic analysis of Aedes aegypti saliva altered by DENV infection provides a unique opportunity to identify pro-viral impacts key to virus transmission.
Keywords: Ae. aegypti, Dengue, Arbovirus infection, Transmission, Mosquito saliva, Salivary proteins, Transmission enhancement, Vectorial capacity modeling
Background
Arboviral diseases are major burdens on the health of individuals and economies throughout the tropics and subtropics [1]. Dengue virus (DENV) is critically responsible for this impact, as it results in lost economic and academic productivity due to millions of cases of dengue fever, and it is the leading cause of childhood hospitalizations due to the severe manifestations of dengue hemorrhagic fever and dengue shock syndrome each year [2,3]. Due to the establishment of Aedes aegypti (Ae. aegypti) on the Portuguese island of Madeira, throughout the Black Sea coastal region, south Florida and several cities along the Gulf Coast in Texas and Louisiana; the potential for both DENV and Ae. aegypti to spread north as temperatures rise due to climate change is a serious threat [4-6]. Indeed, recently, autochthonous DENV transmission has been detected in Texas and Florida, as well as in France, Portugal, and Croatia [7-11].
In order to better characterize vector-viral interactions that might explain the expansion of DENV activity, several studies have determined the vector competence of Ae. aegypti with regards to DENV [12-14]. These determinations are very important because they have allowed researchers to parameterize the potential for transmission of various mosquito and viral combinations have, although the mechanisms behind observed differences remain elusive. Consequently, in order to characterize these interactions, some researchers have focused on exploring the effect a DENV infection has upon Ae. aegypti transcription, while others have focused on elucidating the impacts within an immune-response context [15-17]. Still other researchers have explored those interactions by massive computational efforts, such as in silico data analysis utilizing a systems biology approach [18]. While the body of information created in those efforts advance our understanding of the molecular events underpinning infection outcome within Ae. aegypti, characteristics of vector-pathogen interactions that directly impact DENV transmission requires detailed consideration of the impact of infection upon vector saliva.
When a DENV-infected Ae. aegypti bites a vertebrate, the virus is deposited along with a complex mixture of salivary proteins with diverse functions to facilitate blood-feeding. Those proteins are known to be anti-hemostatics, inhibitors of platelet aggregation, and anti-vasoconstricitves; along with allergens and immune-modulatory compounds [19-22]. Of particular importance to the virus-vector-vertebrate interface is the role the infection has on the salivary glands themselves, whose protein expression has been shown to be altered [23,24]. It may be that the DENV infection of Ae. aegypti salivary glands leads to an altered salivary expectorate, which when delivered to the bite site along with virus may enhance transmission success.
Accordingly, we investigated the ability of a DENV infection to change the quality of Ae. aegypti saliva. We characterized the proteins present in uninfected and DENV-infected mosquitoes and compared the relative abundance of matched proteins in each cohort. Herein, we describe these analyses and provide detailed consideration of the possible impacts DENV infection has upon its vector leading to transmission enhancement.
Methods
Virus
Dengue virus serotype-2 strain 1232 (DENV-2), originally isolated from a human patient in Jakarta, Indonesia in 1978 and provided by the World Reference Center of Emerging Viruses and Arboviruses, was previously passaged 6 times through African green monkey kidney (vero) cells before being used for this experiment. Subsequently, it was inoculated on vero cells grown at 37°C and 5% CO2 in Medium-199 with Earle’s salts, Penicillin/Streptomycin/Amphotericin B, and 10% fetal bovine serum [24]. After 5 days, the supernatant was harvested, titrated by plaque assay and qRT-PCR, and used at a concentration of 2.76x106 plaque-forming units per mL [25].
Mosquitoes
Laboratory strain Ae. aegypti (Rockefeller) were maintained under constant environmental conditions (28°C with a 16:8 light:dark photoperiod). The mosquitoes were allowed to feed on bovine blood in Alsever’s anticoagulant via Hemotek feeding device (Discovery Workshops, Lancashire, England), after which the blood-fed females were sorted and allowed to digest the blood meal for 4 days. Six cartons, each containing approximately 80 previously blood-fed females, were then intrathoracically-inoculated using an EntoSphynx Minucie (BioQuip, Rancho Dominguez, CA) dissecting needle dipped in viral stock. The control group of equal size (6 cartons of ~80 mosquitoes each) received an inoculation of media without virus.
Saliva collection
After a 10-day extrinsic incubation period, mosquitoes were allowed to probe and feed on 1 mL of 1× phosphate-buffered saline (PBS) and 10 mM adenosine triphosphate (ATP) solution at 37°C in a Hemotek device for 1 hour; a variation on the technique developed by Ribeiro, Rossignol, and Spielman [19]. This was repeated after 72 hours to obtain a larger volume of PBS/ATP/saliva for downstream processing, approximately 12 mL. After the collection of saliva, the inoculated cohort was taken down and their legs were removed and placed in 900 μL of BA-1 diluent for viral RNA extraction and detection via qRT-PCR [26]. Disseminated infections were confirmed in >95% (n = 398/415) of the mosquitoes which supplied the ‘infected’ saliva solutions. From that dilute mixture, 200 μL fractions of the PBS/ATP/saliva solution were added to 800 μL of acetone chilled to −80°C and allowed to incubate at −20°C overnight. The tubes were then centrifuged at 4°C at 10,000 rpm for 10 minutes to pellet the precipitated protein. The supernatant was discarded and the resulting pellet was allowed to air-dry for 5 minutes at room temperature. Once translucent, the pellet was then reconstituted in 100 μL of 2-D rehydration buffer (Bio-Rad) consisting of: 8 M urea, 2% CHAPS, 50 mM dithiothreitol, 0.2% Bio-Lyte 3/10 ampholytes, and 0.001% bromophenol blue. The 100 μL of reconstituted protein solution from the first tube was then used to reconstitute the second pellet, further concentrating the protein solution. This serial reconstitution was performed no more than 5 times to minimize loss of recovery from over-saturation. The concentrated protein solution still contained too much salt for 1st dimension focusing; therefore the samples were processed through a ReadyPrep 2-D cleanup kit (Bio-Rad) and resuspended again in 2-D rehydration buffer. Protein concentrations were determined with a NanoDrop spectrophotometer (Thermo Scientific).
2-D gel electrophoresis
30 μg of protein diluted in previously mentioned 2-D rehydration buffer to a final volume of 200 μL was loaded per sample per 11 cm, pH 3–10 nonlinear IPG strip (Bio-Rad) overlaid with approximately 2 mL of molecular biology-grade mineral oil (Bio-Rad) to prevent evaporation. First and second dimension electrophoresis conditions, gel staining, and imaging methods were performed as described previously [24].
Image analysis
Two gels from each experimental condition (for a total of 4 gels) were analyzed to obtain both gel-to-gel differences between biological replicates and between experimental conditions, “infected saliva” and “non-infected saliva.” Pooling and sample size determination was performed according to acceptable practices in 2D SDS-PAGE and 2D DIGE proteomic analyses [24,27-31]. The normalized spot density values for both gels from each condition were used to determine the experimentally induced fold changes. Gels were normalized as previously described [24]. It is important to note that although ‘speckling’ did occur during staining and imaging, a known artifact associated with SYPRO® Ruby staining, the ‘speckle filter’ function of PDQuest was used to eliminate the small pixel intensity values from being included in the normalization calculations [32]. This allowed determination of the changes in protein spot abundance due to our experimental treatment. The relative expression levels of the individual spots were evaluated by calculating a fold-change value per spot as the infected spot intensity divided by the uninfected control spot intensity to determine the change relative to infection with DENV-2. A representative gel image has been provided with the spots excised for mass spectrometry analysis marked and numbered for reference (Figure 1).
Figure 1.
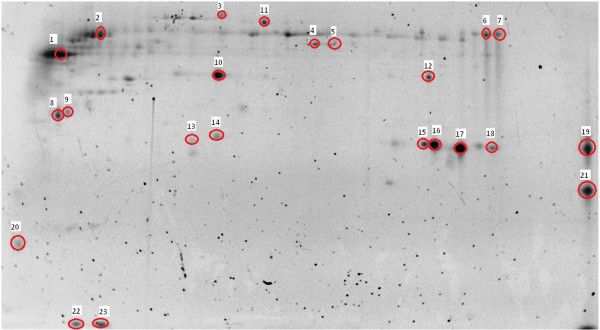
Analyzed protein spots. Representative Ae. aegypti saliva 2-D gel image (12.5% Tris–HCl in TGS buffer using a pH 3–10 non-linear IPG strip) with the spots that were cut circled and numbered to match the IDs in Table 1.
Mass spectrometry
After image analysis, a representative gel from each group containing all of the spots of interest was sent to the Nevada Proteomics Center at the University of Nevada, Reno for robotic spot excision, trypsin digestion and liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis utilizing a Thermo LTQ Orbitrap XL mass spectrometer with ETD coupled to a Michrom Paradigm MDLC and Michrom CaptiveSpray (Thermo Scientific). All MS/MS samples were analyzed using Sequest (Thermo Fisher Scientific, San Jose, CA, USA; version v.27, rev. 11). Sequest was set up to search an Ae. aegypti database (35667 entries) downloaded from Vectorbase.org and the non-redundant database ‘nr’ from NCBI for confirmation, assuming the digestion enzyme trypsin [33]. Sequest was searched with a fragment ion mass tolerance of 1.00 Da and a parent ion tolerance of 0.0068 Da to 0.041 Da, depending on the spot analyzed. Iodoacetamide derivative of cysteine was specified in Sequest as a fixed modification. Oxidation of methionine was specified in Sequest as a variable modification.
Scaffold (version Scaffold_3.4.3, Proteome Software Inc., Portland, OR) was used to validate MS/MS based peptide and protein identifications. Peptide identifications were accepted if they could be established at greater than 95.0% probability as specified by the Peptide Prophet algorithm [34]. Protein identifications were accepted if they could be established at greater than 95.0% probability and contained at least 2 identified peptides. Protein probabilities were assigned by the Protein Prophet algorithm [35]. Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony.
Transmission modeling
To demonstrate a potential consequence of altered salivary quality upon transmission measures, two parameters were derived to account for the hypothetical difference in the biting rate of infected versus uninfected mosquitoes and enhancement of transmission success. Using the framework of vectorial capacity- a quantity that is used to estimate the number of secondary infectious bites resulting from a single, primary infectious bite- measures the force of infection of mosquito to human transmission [12,36]. The parameter values and vectorial equations are given in Additional file 1: SI1. To isolate the potential effects of the altered feeding environment (rendered by the reduction of pain-inhibitory and anti-hemostatic proteins in Ae. aegypti saliva), the daily biting rate of uninfected mosquitoes was set at a = .63[37], while the biting rate of infected mosquitoes was investigated over the range a INF = [.63-1.63]. Similarly, the altered probability of transmission success rendered by additional probing (and presumably deposition of virus into the skin) was investigated by adding the parameter t, which was varied from .7 to 1 [38]. The addition of this parameter is based on the knowledge that not all infectious bites result in a productive infection, and our hypothesis that additional feeding (through increased salivation or separate bites) increases the probability of such. The change in vectorial capacity was calculated by taking the difference between traditional vectorial capacity calculation (where a = aINF) and this new modified vectorial capacity (where a < aINF). The magnitude of the potential enhancement of transmission success was investigated over the range of t and the alteration of vectorial capacity expressed as the difference from tmin = .5 [38].
Results
Seventy-four spots were matched across the 4 gels created from the cleaned-up, naturally-expectorated saliva. Of those 74 spots, 23 spots were chosen to be analyzed by mass spectrometry due to their being differentially expressed or present in very high quantities, which would be informative for land-marking purposes. The proteins identified within those 23 excised and analyzed spots were: a DEAD-box ATP-dependent RNA helicase, beta chain spectrin, the hypothetical secreted protein AAEL000748, adenosine deaminase, a putative adenosine deaminase, apyrase, a putative apyrase, an inosine-uridine preferring nucleoside hydrolase, a putative purine hydrolase, a salivary anti-FXa serpin, the hypothetical protein AAEL000732, a putative serine protease inhibitor (Serpin-4), an angiopoietin-like protein variant, a putative 34kD family secreted salivary protein, D7 (3 spots), a putative D7, a low density lipoprotein receptor, a putative 30kD secreted protein (‘short-form aegyptin’), a venom allergen/antigen 5, and a putative C-type lectin (2 spots). Their respective fold-changes in the infected saliva, along with accession numbers, are located in Table 1. The locations of these proteins in the gel can be found in Figure 1.
Table 1.
Identified proteins with accession numbers and fold change information
|
Ae. aegypti salivary proteins identified by mass spectrometry with fold change due to DENV infection | |||
|---|---|---|---|
| GenBank ID | Protein name | Spot | Spot fold change |
| gi|108875535| |
DEAD-Box ATP-Dependent RNA Helicase‡ |
1 |
−1.5 |
| gi|108877982| |
Beta Chain Spectrin‡ |
2 |
+1.0 |
| gi|157109431| |
Hypothetical Secreted Protein AAEL000748‡ |
3 |
−4.0 |
| gi|108878609| |
Adenosine Deaminase |
4 |
−2.8* |
| gi|18568326| |
Putative Adenosine Deaminase |
5 |
−5.2* |
| gi|1094353| |
Apyrase‡ |
6 |
−1.1 |
| gi|108877845| |
Putative Apyrase‡ |
7 |
+1.1 |
| gi|108877687| |
Inosine-Uridine Preferring Nucleoside Hydrolase |
8 |
−7.9* |
| gi|18568280| |
Putative Purine Hydrolase |
9 |
−3.8* |
| gi|94468358| |
Salivary Anti-FXa Serpin |
10 |
−4.8* |
| gi|157109433| |
Hypothetical Protein AAEL000732 |
11 |
−7.7* |
| gi|157131306| |
Putative Serine Protease Inhibitor (Serpin-4) |
12 |
−19.4* |
| gi|94468352| |
Angiopoietin-Like Protein Variant‡ |
13 |
−1.8 |
| gi|94468642| |
Putative 34kD Family Secreted Salivary Protein‡ |
14 |
−1.3 |
| gi|222447044| |
D7‡ |
15 |
−4.5 |
| gi|222447044| |
D7‡ |
16 |
−1.3 |
| gi|222447044| |
D7‡ |
17 |
−1.8 |
| gi|108877064| |
Low Density Lipoprotein Receptor |
18 |
−11.3* |
| gi|157113327| |
Putative D7‡ |
19 |
−3.5 |
| gi|18568322| |
Putative 30kD Secreted Protein; ‘Short-Form Aegyptin’ |
20 |
−14.1* |
| gi|157110207| |
Antigen-5/Venom Allergen‡ |
21 |
−2.5 |
| gi|18568318| |
Putative C-Type Lectin‡ |
22 |
N/A1 |
| gi|18568318| | Putative C-Type Lectin‡ | 23 | N/A1 |
*denotes significance at the p ≤ 0.05 level via Student’s t-test of the mean density of each spot between experimental conditions (n = 2 per condition).
1quantity at these spots were unable to be calculated due to the location of the bromophenol blue dye front in two of the four gels.
‡denotes proteins from spots chosen for mass spectrometry identification for the purposes of landmarking.
In addition, all mass spectrometry related data has been included in the Additional file 2– Mass_Spec_Supplement.To account for differences in the salivary proteins of infected mosquitoes involved in anti-hemostatic and pain responses at the bite site, modifications to the vectorial capacity equation were made. The biting rate, which is usually assumed to be the same for all mosquitoes, was estimated separately based on infection status. Accounting for the increase in biting rate of infected mosquitoes relative to uninfected mosquitoes resulted in a linear increase in vectorial capacity. The potential enhancement of transmission success had a similarly linear effect, though since this value is a probability, it is necessarily bounded by 1, and thus its impact is also bounded. Taken together, these changes in proteins could result in as many as two additional infectious bites previously unaccounted for in traditional parameterizations of vectorial capacity (Figure 2).
Figure 2.
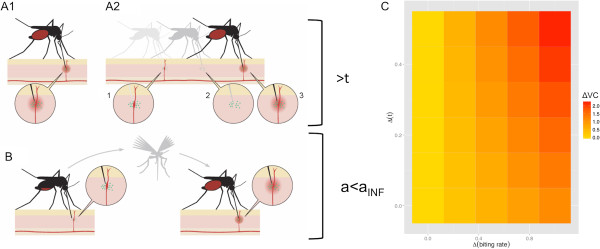
The manifold potential effects of altered salivary proteins on mosquito feeding and DENV transmission. In order to achieve a successful blood feeding, an infected mosquito might: A1) Increase its salivary, and consequentially viral, inoculum in order to restore a normal level of anti-hemostatic and pain-reducing salivary proteins relative to an uninfected mosquito; A2) given reduced anti-hemostatic and pain-reducing salivary proteins, attempt refeeding if (1) the pain perception at the bite site alerts the host or leads to a clot-induced disruption of feeding (2) causing the mosquito to seek another bite site, increasing overall viral inoculum (3) represented by increasing t (the probability of transmission success) in our vectorial capacity (VC) equation; B) Alternatively, if this interrupted female moved on to another host in order to acquire a sufficient blood-meal after a clot-induced or host-triggered interruption, then a subsequent transmission event could occur, even though DENV transmission had previously occurred during the failed previous feeding attempt, represented by aINF in our VC equation. C) The impacts of these potential transmission enhancements due to changes in transmission success probability and daily biting rate could yield an increase in the vectorial capacity of the mosquito (ΔVC) relative to a baseline calculation of VC, and is represented by increasingly darker colors. The x-axis is the difference in probability of transmission success relative to baseline (tmin = .5) and the y-axis is the difference in in biting rate between uninfected mosquitoes and infected mosquitoes (a-aINF) = Δ(daily biting rate). Thus the coordinates (0,0) refer to t = .7 (Δ transmission success of .2) and a = aINF (no enhancement to biting rate).
Discussion
The majority of the protein identifications determined by mass spectrometry were for known or putative salivary proteins, a total of 16 out of 19 unique IDs [39,40]. The remaining three proteins with previously undetected salivary roles (the DEAD-box ATP-dependent RNA helicase, beta chain spectrin, and the low density lipoprotein receptor) could be present in this salivary sample due to normal cell death in the salivary gland, as has been seen previously [41]. Alternatively, these proteins may be being utilized in non-traditional roles to facilitate blood-feeding as kratagonists, proteins that scavenge host hemostatic or inflammatory system components [42,43]. For instance, the ATP-binding motif present in the DEAD-box ATP-dependent RNA helicase may be fortifying the function of other ATP-degrading salivary proteins like apyrase [39,44].
Likewise, the low density lipoprotein receptor (LDL-r) family has been shown to include members who are involved in a diverse array of cellular functions, including the LDL-r related protein (LRP)-type receptors which regulate proteolytic processes involved in fibrinolysis and coagulation [45]. It is interesting to note that in the saliva of DENV-2 infected Ae. aegypti, this protein’s expression was reduced 11.3 fold, which was statistically significant between treatment groups (p ≤ 0.05). In fact, the Ae. aegypti low density lipoprotein receptor (gi|108877064|) is more structurally similar to the mammalian LRP receptor than the archetypal mammalian LDL-r, according to the Conserved Domain Archetecture Retrieval Tool (cDART) through the National Center for Biotechnology Information (NCBI) [46]. This would suggest that the Ae. aegypti LDL-r may share similar binding properties of other LRP-type receptors and therefore could disrupt mammalian LRP-mediated proteolytic regulation in a kratagonistic fashion. Additionally, a secreted LRP receptor, referred to as sLRP-1 for ‘shed LRP-1’, has recently been found in human plasma, cerebrospinal fluid, and the brain [47]. This soluble receptor is produced from the full-length LRP-1 protein that is cleaved extracellularly in response to inflammation [48]. Further research has determined that injected sLRP-1 decreases inflammation and neuropathic pain in a mouse model [49]. Whether the Ae. aegypti salivary form of this protein deposited at the bite site is behaving the same way remains to be seen.
Another group of proteins whose expression was significantly decreased in DENV-2 infected saliva were the adenosine deaminases, the archetypal protein and a putative version, 2.8 fold and 5.2 fold, respectively (p ≤ 0.05). An amino acid sequence comparison between the two proteins reveals that the putative version shares 97% identity with adenosine deaminase and is slightly larger by 0.692 kDa, in agreement with the almost imperceptible migration difference in the 2-D gel. Adenosine deaminase has been shown to convert adenosine at the bite site into inosine and ammonia, which acts to prevent peripheral pain signaling [50]. Ribiero et al. noted that the reduction in adenosine at the bite site by adenosine deaminase was perplexing due to the fact that adenosine is a vasodialator and platlet aggregation inhibitor, functions that facilitate blood-feeding; therefore they concluded that the secondary characteristic of pain initiation must trump those functions for a diurnal-feeding mosquito such as Ae. aegypti.
Mosquito salivary nucleosidases, such as the inosine-uridine preferring nucleoside hydrolase (reduced 7.9 fold, p ≤ 0.05) and the purine hydrolase (reduced 3.8 fold, p ≤ 0.05) found during our mass spectrometry-based identification effort, convert nucleosides into D-ribose and their respective purine or pyrimidine base with the addition of water and occasionally the assistance of a divalent cation, such as calcium [51]. While the inosine-uridine preferring nucleoside hydrolase will hydrolyze both purines and pyrimidines, the purine hydrolase is specific for inosine, adenosine, and guanosine [52]. With the assistance of adenosine deaminase, any adenosine at the bite site would likely be converted into inosine and then subject to either of the two hydrolases above, leading to the complete removal of any mast cell degranulation-inducing products [53]. Given the synergy of these hydrolases and adenosine deaminase, it is interesting that all three were reduced in the DENV-2 infected saliva.
Two serine protease inhibitors (serpins) were identified via mass spectrometry and were reduced in the DENV-2 infected saliva. This finding mirrors that of Bonizzoni et al., who found a reduction in transcripts for serine proteases in DENV-infected Aedes aegypti midguts [17]. Both serpins are very similar, sharing 98% identity when compared at the amino acid level, therefore they are likely exhibit similar physiochemical properties. The protein identified as a salivary anti-coagulation factor Xa (FXa) serpin was reduced 4.8 fold (p ≤ 0.05) while the protein identified as serpin-4 was reduced 19.4 fold (p ≤ 0.05). The anti-coagulant salivary anti-FXa serpin was first identified by Stark and James as a ‘specific, reversible, noncompetitive, proteinaceous inhibitor of FXa’ [54,55]. This particular serpin is very similar to an Ae. albopictus-derived anti-FXa serpin termed ‘alboserpin’ [56]. Due to the detailed biochemical work done by Calvo et al. on alboserpin demonstrating binding affinities to heparin and phosolipid vesicles, interactions with phosphotidlycholine and phosphotydlethanolamine, and inhibiting FXa; the Ae. aegypti produced serpins may also share these properties.
The short form of aegyptin, also referred to as SAAG-4 by Boppana et al., was identified previously as being down-regulated in the salivary gland extract of Ae. aegypti infected with DENV-2 [24,57]. In agreement with the previous observation, this same aegyptin was found to be reduced in DENV-2 infected saliva by 14.1 fold (p ≤ 0.05). Beyond aegyptin’s role as an allergen, Calvo and others have thoroughly analyzed the physiological and biochemical capacities of the archetypal aegyptin molecule and found that it binds to collagen, inhibiting its interaction with platelet glycoprotein IV, integrin α2β1, and vonWillenbrand factor leading to an overall inhibition of coagulation [20,58-62]. It is important to note that this is not the only anti-coagulant protein reduced in the saliva of DENV-infected Ae. aegypti.
With three known salivary proteins related to the adenosine degrading complex of Ae. aegypti reduced in the DENV-2 infected saliva, along with a novel low density lipoprotein receptor that may be involved in the reduction of pain, it would appear that there is a trend towards the greater likelihood of both mast cell degranulation and pain perception compared to the same volume of uninfected mosquito saliva. Considering that the prevention of detection during feeding is a priority for Ae. aegypti, due to the fact that it possesses redundant machinery to remove adenosine, any reduction in expression of salivary proteins involved in that process could alert the host to the feeding attempt and lead to feeding interruptions causing failure to reach repletion. When combined with a reduction in salivary proteins involved in the prevention of clotting, such as the two serpins and the short form of aegyptin, yet another feeding pressure would be placed upon the infected mosquito compared to an uninfected one. If the saliva of an infected mosquito contains less anti-hemostatics on a per unit volume basis, as suggested from our data, there would be a greater chance of clot formation occurring during feeding which could also lead to an interruption in feeding with a failure to reach repletion. These two mechanisms which would hinder an infected mosquito during feeding could be the physiological basis for the interruptions or delays in feeding by DENV-infected mosquitoes reported by Platt et al. and Maciel-de-Freitas et al. [63,64].
These changes in salivary composition led us to revise the method of calculating vectorial capacity- a measure of transmission potential- of Ae. aegypti, a significant modifier of which is the biting rate of a vector on the pathogen-relevant vertebrate population (a) [65,66]. Typical formulations of the equation assume that the biting rate of mosquitoes is unchanged, regardless of infection status. However, the reduction in specific proteins identified in this research, combined with the changes in feeding behavior of DENV-infected mosquitoes seen by others, would indicate that this assumption may be violated in nature and would have consequences for transmission [63,64,67]. Thus, a separate parameter of aINF to account for this alteration in vector biting rate due to infection status is appropriate. Indeed, the transmission differences due to the value of this new parameter aINF are not inconsequential. Further, the reduction of these particular proteins may lead to 1) difficultly in immediate blood-meal acquisition and thus, additional probing by a foraging mosquito and 2) vertebrate host interruption of feeding by the mosquito. The combination of these two things could enhance the transmission success of DENV by 1) additional deposition of virus due to increased probing or 2) additional transmission events to a new vertebrate host due to probing interruption. The magnitude of this transmission success probability will require further experimental investigation, but given the estimated effects of this measure, this would merit investigation (Figure 2).
Finally, the remaining protein significantly reduced in the DENV-2 infected saliva 7.7 fold (p ≤ 0.05) was the hypothetical protein known through VectorBase.org as AAEL000732 [33]. Using cDART, no known conserved domains were identified and VectorBase.org has no information on suspected function. It would appear that the role of this protein in the saliva of Ae. aegypti remains to be determined.
Conclusions
In summary, our findings indicate that DENV-2 infection alters the expression of various salivary proteins in Ae. aegypti, in particular proteins involved in anti-hemostatic and pain-reducing capacities. These changes may confer a fitness advantage upon the virus by enhancing viral establishment in the vertebrate or by increasing the number of transmission events. While this work is an important beginning, much remains to be characterized. In particular, the exact roles these salivary components have at the bite site within the context of viral deposition remains to be detailed. Restoration of reduced proteins and the resulting viral dynamics and host responses are currently being investigated and will likely be of use for vaccine development, treatment options, and a better understanding of the role of these critical vector components in arboviral transmission.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
DMC designed the experiment, infected the mosquitoes, collected and purified saliva, performed 2-D gel electrophoresis, image analysis, data analysis, and contributed to the manuscript. RCC created the viral stocks, infected the mosquitoes, contributed to the manuscript, and performed statistical analysis and mathematic modeling. MKM assisted with data analysis and contributed to the manuscript. AMJ collected and purified saliva, extracted RNA and performed qRT-PCR for infection verification. BLL assisted with data analysis and contributed to the manuscript. CNM designed the experiment, assisted with data analysis, and contributed to the manuscript. All authors have read and approved the final version of the manuscript.
Supplementary Material
SI1. Description of parameters and equations used to model impacts of altered mosquito salivary expectorate on the vectorial capacity of Ae. aegypti mosquitoes.
Mass_Spec_Supplement. Excel formatted spreadsheet with additional information obtained from mass spectrometry analysis.
Contributor Information
Daniel M Chisenhall, Email: dchisenh@lsu.edu.
Rebecca C Christofferson, Email: rcarri1@lsu.edu.
Michael K McCracken, Email: mmccra4@lsu.edu.
Ann-Marie F Johnson, Email: ajohnson2211@gmail.com.
Berlin Londono-Renteria, Email: berlin@lsu.edu.
Christopher N Mores, Email: cmores@lsu.edu.
Acknowledgements
This work was supported by the National Institutes of Health P20GM103458 and U01GM097661.
References
- Gubler DJ. The global emergence/resurgence of arboviral diseases as public health problems. Arch Med Res. 2002;33:330–342. doi: 10.1016/S0188-4409(02)00378-8. [DOI] [PubMed] [Google Scholar]
- Jarman RG, Holmes EC, Rodpradit P, Klungthong C, Gibbons RV, Nisalak A, Rothman AL, Libraty DH, Ennis FA, Mammen MP Jr, Endy TP. Microevolution of dengue viruses circulating among primary school children in Kamphaeng Phet, Thailand. J Virol. 2008;82:5494–5500. doi: 10.1128/JVI.02728-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wettstein ZS, Fleming M, Chang AY, Copenhaver DJ, Wateska AR, Bartsch SM, Lee BY, Kulkarni RP. Total economic cost and burden of dengue in Nicaragua: 1996–2010. Am J Trop Med Hyg. 2012;87:616–622. doi: 10.4269/ajtmh.2012.12-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anez G, Rios M. Dengue in the United States of America: a worsening scenario? Biomed Res Int. 2013;2013:678645. doi: 10.1155/2013/678645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffner F, Bellini R, Petric D, Scholte EJ, Zeller H, Rakotoarivony LM. Development of guidelines for the surveillance of invasive mosquitoes in Europe. Parasit Vectors. 2013;6:209. doi: 10.1186/1756-3305-6-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seixas G, Salgueiro P, Silva AC, Campos M, Spenassatto C, Reyes-Lugo M, Novo MT, Ribolla PE, da Silva Pinto JP, Sousa CA. Aedes aegypti on Madeira Island (Portugal): genetic variation of a recently introduced dengue vector. Mem Inst Oswaldo Cruz. 2013;108(Suppl 1):3–10. doi: 10.1590/0074-0276130386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locally acquired dengue--Key West, Florida, 2009–2010. MMWR Morb Mortal Wkly Rep. 2010;59:577–581. [PubMed] [Google Scholar]
- Dengue. [ http://www.floridahealth.gov/diseases-and-conditions/dengue/index.html]
- Murray KO, Rodriguez LF, Herrington E, Kharat V, Vasilakis N, Walker C, Turner C, Khuwaja S, Arafat R, Weaver SC, Martinez D, Kilborn C, Bueno R, Reyna M. Identification of dengue fever cases in Houston, Texas, with evidence of autochthonous transmission between 2003 and 2005. Vector borne and zoonotic diseases. 2013;13:835–845. doi: 10.1089/vbz.2013.1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasello D, Schlagenhauf P. Chikungunya and dengue autochthonous cases in Europe, 2007–2012. Travel Med Infect Dis. 2013;11:274–284. doi: 10.1016/j.tmaid.2013.07.006. [DOI] [PubMed] [Google Scholar]
- Marchand E, Prat C, Jeannin C, Lafont E, Bergmann T, Flusin O, Rizzi J, Roux N, Busso V, Deniau J, Noel H, Vaillant V, Leparc-Goffart I, Six C, Paty MC. Autochthonous case of dengue in France, October 2013. Euro Surveill. 2013;18:20661. doi: 10.2807/1560-7917.es2013.18.50.20661. [DOI] [PubMed] [Google Scholar]
- Christofferson RC, Mores CN. Estimating the magnitude and direction of altered arbovirus transmission due to viral phenotype. PLoS One. 2011;6:e16298. doi: 10.1371/journal.pone.0016298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards SL, Anderson SL, Alto BW. Vector competence of Aedes aegypti and Aedes albopictus (Diptera: Culicidae) for dengue virus in the Florida Keys. J Med Entomol. 2012;49:942–946. doi: 10.1603/ME11293. [DOI] [PubMed] [Google Scholar]
- Bennett KE, Olson KE, Munoz Mde L, Fernandez-Salas I, Farfan-Ale JA, Higgs S, Black WC, Beaty BJ. Variation in vector competence for dengue 2 virus among 24 collections of Aedes aegypti from Mexico and the United States. Am J Trop Med Hyg. 2002;67:85–92. doi: 10.4269/ajtmh.2002.67.85. [DOI] [PubMed] [Google Scholar]
- Sanchez-Vargas I, Scott JC, Poole-Smith BK, Franz AW, Barbosa-Solomieu V, Wilusz J, Olson KE, Blair CD. Dengue virus type 2 infections of Aedes aegypti are modulated by the mosquito’s RNA interference pathway. PLoS Pathog. 2009;5:e1000299. doi: 10.1371/journal.ppat.1000299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim S, Ramirez JL, Dimopoulos G. Dengue virus infection of the Aedes aegypti salivary gland and chemosensory apparatus induces genes that modulate infection and blood-feeding behavior. PLoS Pathog. 2012;8:e1002631. doi: 10.1371/journal.ppat.1002631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonizzoni M, Dunn WA, Campbell CL, Olson KE, Marinotti O, James AA. Complex modulation of the Aedes aegypti transcriptome in response to dengue virus infection. PLoS One. 2012;7:e50512. doi: 10.1371/journal.pone.0050512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Xu Y, Bian G, Pike AD, Xie Y, Xi Z. Response of the mosquito protein interaction network to dengue infection. BMC Genomics. 2010;11:380. doi: 10.1186/1471-2164-11-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro JM, Rossignol PA, Spielman A. Role of mosquito saliva in blood vessel location. J Exp Biol. 1984;108:1–7. doi: 10.1242/jeb.108.1.1. [DOI] [PubMed] [Google Scholar]
- Peng Z, Estelle F, Simons R. Mosquito allergy and mosquito salivary allergens. Protein Pept Lett. 2007;14:975–981. doi: 10.2174/092986607782541088. [DOI] [PubMed] [Google Scholar]
- Peng Z, Yang M, Simons FE. Immunologic mechanisms in mosquito allergy: correlation of skin reactions with specific IgE and IgG antibodies and lymphocyte proliferation response to mosquito antigens. Ann Allergy Asthma Immunol. 1996;77:238–244. doi: 10.1016/S1081-1206(10)63262-0. [DOI] [PubMed] [Google Scholar]
- Surasombatpattana P, Patramool S, Luplertlop N, Yssel H, Misse D. Aedes aegypti saliva enhances dengue virus infection of human keratinocytes by suppressing innate immune responses. J Invest Dermatol. 2012;132:2103–2105. doi: 10.1038/jid.2012.76. [DOI] [PubMed] [Google Scholar]
- Wasinpiyamongkol L, Patramool S, Thongrungkiat S, Maneekan P, Sangmukdanan S, Misse D, Luplertlop N. Protein expression in the salivary glands of dengue-infected Aedes aegypti mosquitoes and blood-feeding success. Southeast Asian J Trop Med Public Health. 2012;43:1346–1357. [PubMed] [Google Scholar]
- Chisenhall DM, Londono BL, Christofferson RC, McCracken MK, Mores CN. Effect of dengue-2 virus infection on protein expression in the salivary glands of Aedes aegypti mosquitoes. Am J Trop Med Hyg. 2014;90:431–437. doi: 10.4269/ajtmh.13-0412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christofferson RC, McCracken MK, Johnson AM, Chisenhall DM, Mores CN. Development of a transmission model for dengue virus. Virol J. 2013;10:127. doi: 10.1186/1743-422X-10-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisenhall DM, Mores CN. Diversification of West Nile virus in a subtropical region. Virol J. 2009;6:106. doi: 10.1186/1743-422X-6-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J, Person MD, Zhu J, Abbruzzese JL, Li D. Protein expression profiles in pancreatic adenocarcinoma compared with normal pancreatic tissue and tissue affected by pancreatitis as detected by two-dimensional gel electrophoresis and mass spectrometry. Cancer Res. 2004;64:9018–9026. doi: 10.1158/0008-5472.CAN-04-3262. [DOI] [PubMed] [Google Scholar]
- Hamada A, Sharma R, du Plessis SS, Willard B, Yadav SP, Sabanegh E, Agarwal A. Two-dimensional differential in-gel electrophoresis-based proteomics of male gametes in relation to oxidative stress. Fertil Steril. 2013;99:1216–1226. doi: 10.1016/j.fertnstert.2012.11.046. e1212. [DOI] [PubMed] [Google Scholar]
- Kultima K, Scholz B, Alm H, Skold K, Svensson M, Crossman AR, Bezard E, Andren PE, Lonnstedt I. Normalization and expression changes in predefined sets of proteins using 2D gel electrophoresis: a proteomic study of L-DOPA induced dyskinesia in an animal model of Parkinson’s disease using DIGE. BMC Bioinformatics. 2006;7:475. doi: 10.1186/1471-2105-7-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey MJ, Shield-Artin KL, Oliva K, Ayhan M, Reisman S, Rice GE. Stage-specific analysis of plasma protein profiles in ovarian cancer: Difference in-gel electrophoresis analysis of pooled clinical samples. J Carcinog. 2013;12:10. doi: 10.4103/1477-3163.114216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Zhao Q, Das Singla L, Min J, He S, Cong H, Li Y, Su C. Differential proteomic profiles from distinct toxoplasma gondii strains revealed by 2D-difference gel electrophoresis. Exp Parasitol. 2013;133:376–382. doi: 10.1016/j.exppara.2013.01.009. [DOI] [PubMed] [Google Scholar]
- Chevalier F, Rofidal V, Vanova P, Bergoin A, Rossignol M. Proteomic capacity of recent fluorescent dyes for protein staining. Phytochemistry. 2004;65:1499–1506. doi: 10.1016/j.phytochem.2004.04.019. [DOI] [PubMed] [Google Scholar]
- Megy K, Emrich SJ, Lawson D, Campbell D, Dialynas E, Hughes DS, Koscielny G, Louis C, Maccallum RM, Redmond SN, Sheehan A, Topalis P, Wilson D. VectorBase: improvements to a bioinformatics resource for invertebrate vector genomics. Nucleic Acids Res. 2012;40:D729–734. doi: 10.1093/nar/gkr1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- Reisen WK. Estimation of vectorial capacity: introduction. Bull Soc Vector Ecol. 1989;14:39–40. [Google Scholar]
- Scott TW, Amerasinghe PH, Morrison AC, Lorenz LH, Clark GG, Strickman D, Kittayapong P, Edman JD. Longitudinal studies of Aedes aegypti (Diptera: Culicidae) in Thailand and Puerto Rico: blood feeding frequency. J Med Entomol. 2000;37:89–101. doi: 10.1603/0022-2585-37.1.89. [DOI] [PubMed] [Google Scholar]
- Siler JF, Hall MW, Hitchens AP. Dengue: its history, epidemiology, mechanism of transmission, etiology, clinical manifestations, immunity, and prevention. Manila: Bureau of Printing; 1926. [Google Scholar]
- Ribeiro JM. Role of saliva in blood-feeding by arthropods. Annu Rev Entomol. 1987;32:463–478. doi: 10.1146/annurev.en.32.010187.002335. [DOI] [PubMed] [Google Scholar]
- Ribeiro JM, Arca B, Lombardo F, Calvo E, Phan VM, Chandra PK, Wikel SK. An annotated catalogue of salivary gland transcripts in the adult female mosquito, Aedes aegypti. BMC Genomics. 2007;8:6. doi: 10.1186/1471-2164-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly EM, Moon DC, Bowers DF. Apoptosis in mosquito salivary glands: Sindbis virus-associated and tissue homeostasis. J Gen Virol. 2012;93:2419–2424. doi: 10.1099/vir.0.042846-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro JMC, Arcà B. In: Advances in Insect Physiology. Volume, Volume Volume 37. Stephen JS, Jeacuterocircme C, editor. Oxford: Academic Press; 2009. Chapter 2 From Sialomes to the Sialoverse: An Insight into Salivary Potion of Blood-Feeding Insects; pp. 59–118. [Google Scholar]
- Mans BJ. Evolution of vertebrate hemostatic and inflammatory control mechanisms in blood-feeding arthropods. J Innate Immun. 2011;3:41–51. doi: 10.1159/000321599. [DOI] [PubMed] [Google Scholar]
- Smartt CT, Kim AP, Grossman GL, James AA. The Apyrase gene of the vector mosquito, Aedes aegypti, is expressed specifically in the adult female salivary glands. Exp Parasitol. 1995;81:239–248. doi: 10.1006/expr.1995.1114. [DOI] [PubMed] [Google Scholar]
- Nykjaer A, Willnow TE. The low-density lipoprotein receptor gene family: a cellular Swiss army knife? Trends Cell Biol. 2002;12:273–280. doi: 10.1016/S0962-8924(02)02282-1. [DOI] [PubMed] [Google Scholar]
- Geer LY, Domrachev M, Lipman DJ, Bryant SH. CDART: protein homology by domain architecture. Genome Res. 2002;12:1619–1623. doi: 10.1101/gr.278202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorovoy M, Gaultier A, Campana WM, Firestein GS, Gonias SL. Inflammatory mediators promote production of shed LRP1/CD91, which regulates cell signaling and cytokine expression by macrophages. J Leukoc Biol. 2010;88:769–778. doi: 10.1189/jlb.0410220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn KA, Grimsley PG, Dai YP, Tapner M, Chesterman CN, Owensby DA. Soluble low density lipoprotein receptor-related protein (LRP) circulates in human plasma. J Biol Chem. 1997;272:23946–23951. doi: 10.1074/jbc.272.38.23946. [DOI] [PubMed] [Google Scholar]
- Gaultier A, Arandjelovic S, Li X, Janes J, Dragojlovic N, Zhou GP, Dolkas J, Myers RR, Gonias SL, Campana WM. A shed form of LDL receptor-related protein-1 regulates peripheral nerve injury and neuropathic pain in rodents. J Clin Invest. 2008;118:161–172. doi: 10.1172/JCI32371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro JM, Charlab R, Valenzuela JG. The salivary adenosine deaminase activity of the mosquitoes Culex quinquefasciatus and Aedes aegypti. J Exp Biol. 2001;204:2001–2010. doi: 10.1242/jeb.204.11.2001. [DOI] [PubMed] [Google Scholar]
- Versees W, Steyaert J. Catalysis by nucleoside hydrolases. Curr Opin Struct Biol. 2003;13:731–738. doi: 10.1016/j.sbi.2003.10.002. [DOI] [PubMed] [Google Scholar]
- Parkin DW. Purine-specific nucleoside N-ribohydrolase from Trypanosoma brucei brucei. Purification, specificity, and kinetic mechanism. J Biol Chem. 1996;271:21713–21719. [PubMed] [Google Scholar]
- Ribeiro JM, Valenzuela JG. The salivary purine nucleosidase of the mosquito, Aedes aegypti. Insect Biochem Mol Biol. 2003;33:13–22. doi: 10.1016/S0965-1748(02)00078-4. [DOI] [PubMed] [Google Scholar]
- Stark KR, James AA. A factor Xa-directed anticoagulant from the salivary glands of the yellow fever mosquito Aedes aegypti. Exp Parasitol. 1995;81:321–331. doi: 10.1006/expr.1995.1123. [DOI] [PubMed] [Google Scholar]
- Stark KR, James AA. Isolation and characterization of the gene encoding a novel factor Xa-directed anticoagulant from the yellow fever mosquito, Aedes aegypti. J Biol Chem. 1998;273:20802–20809. doi: 10.1074/jbc.273.33.20802. [DOI] [PubMed] [Google Scholar]
- Calvo E, Mizurini DM, Sa-Nunes A, Ribeiro JM, Andersen JF, Mans BJ, Monteiro RQ, Kotsyfakis M, Francischetti IM. Alboserpin, a factor Xa inhibitor from the mosquito vector of yellow fever, binds heparin and membrane phospholipids and exhibits antithrombotic activity. J Biol Chem. 2011;286:27998–28010. doi: 10.1074/jbc.M111.247924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boppana VD, Thangamani S, Adler AJ, Wikel SK. SAAG-4 is a novel mosquito salivary protein that programmes host CD4 T cells to express IL-4. Parasite Immunol. 2009;31:287–295. doi: 10.1111/j.1365-3024.2009.01096.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons FE, Peng Z. Mosquito allergy: recombinant mosquito salivary antigens for new diagnostic tests. Int Arch Allergy Immunol. 2001;124:403–405. doi: 10.1159/000053771. [DOI] [PubMed] [Google Scholar]
- Peng Z, Simons FE. Mosquito allergy: immune mechanisms and recombinant salivary allergens. Int Arch Allergy Immunol. 2004;133:198–209. doi: 10.1159/000076787. [DOI] [PubMed] [Google Scholar]
- Calvo E, Tokumasu F, Marinotti O, Villeval JL, Ribeiro JM, Francischetti IM. Aegyptin, a novel mosquito salivary gland protein, specifically binds to collagen and prevents its interaction with platelet glycoprotein VI, integrin alpha2beta1, and von Willebrand factor. J Biol Chem. 2007;282:26928–26938. doi: 10.1074/jbc.M705669200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo E, Tokumasu F, Mizurini DM, McPhie P, Narum DL, Ribeiro JM, Monteiro RQ, Francischetti IM. Aegyptin displays high-affinity for the von Willebrand factor binding site (RGQOGVMGF) in collagen and inhibits carotid thrombus formation in vivo. FEBS J. 2010;277:413–427. doi: 10.1111/j.1742-4658.2009.07494.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizurini DM, Francischetti IM, Monteiro RQ. Aegyptin inhibits collagen-induced coagulation activation in vitro and thromboembolism in vivo. Biochem Biophys Res Commun. 2013;436:235–239. doi: 10.1016/j.bbrc.2013.05.082. [DOI] [PubMed] [Google Scholar]
- Platt KB, Linthicum KJ, Myint KS, Innis BL, Lerdthusnee K, Vaughn DW. Impact of dengue virus infection on feeding behavior of Aedes aegypti. Am J Trop Med Hyg. 1997;57:119–125. doi: 10.4269/ajtmh.1997.57.119. [DOI] [PubMed] [Google Scholar]
- Maciel-de-Freitas R, Sylvestre G, Gandini M, Koella JC. The influence of dengue virus serotype-2 infection on Aedes aegypti (Diptera: Culicidae) motivation and avidity to blood feed. PLoS One. 2013;8:e65252. doi: 10.1371/journal.pone.0065252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett-Jones C. Prognosis for interruption of malaria transmission through assessment of the mosquito’s vectorial capacity. Nature. 1964;204:1173–1175. doi: 10.1038/2041173a0. [DOI] [PubMed] [Google Scholar]
- Dye C. Vectorial capacity: must we measure all its components? Parasitol Today. 1986;2:203–209. doi: 10.1016/0169-4758(86)90082-7. [DOI] [PubMed] [Google Scholar]
- Luz PM, Lima-Camara TN, Bruno RV, Castro MG, Sorgine MH, Lourenco-de-Oliveira R, Peixoto AA. Potential impact of a presumed increase in the biting activity of dengue-virus-infected Aedes aegypti (Diptera: Culicidae) females on virus transmission dynamics. Mem Inst Oswaldo Cruz. 2011;106:755–758. doi: 10.1590/S0074-02762011000600017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SI1. Description of parameters and equations used to model impacts of altered mosquito salivary expectorate on the vectorial capacity of Ae. aegypti mosquitoes.
Mass_Spec_Supplement. Excel formatted spreadsheet with additional information obtained from mass spectrometry analysis.


