SUMMARY
Astrocytes, the most abundant cells in the central nervous system, promote synapse formation and help refine neural connectivity. Although they are allocated to spatially distinct regional domains during development, it is unknown whether region-restricted astrocytes are functionally heterogeneous. Here we show that postnatal spinal cord astrocytes express several region-specific genes, and that ventral astrocyte-encoded Semaphorin3a (Sema3a) is required for proper motor neuron and sensory neuron circuit organization. Loss of astrocyte-encoded Sema3a led to dysregulated α–motor neuron axon initial segment orientation, markedly abnormal synaptic inputs, and selective death of α–but not of adjacent γ–motor neurons. Additionally, a subset of TrkA+ sensory afferents projected to ectopic ventral positions. These findings demonstrate that stable maintenance of a positional cue by developing astrocytes influences multiple aspects of sensorimotor circuit formation. More generally, they suggest that regional astrocyte heterogeneity may help to coordinate postnatal neural circuit refinement.
INTRODUCTION
Developing neural circuits must form and maintain appropriate regional connections in a rapidly expanding central nervous system (CNS). Although astrocytes (AS) are increasingly recognized as general regulators of synapse formation1, little is known about whether they encode heterogeneous positional signals involved in local neural circuit formation and/or maintenance. Recent studies indicate that AS develop and are regionally allocated in murine brain and spinal cord (SC) according to an embryonic segmental template2–4. AS derived from embryonic radial glia5 migrate in the trajectory of these fibers and proliferate locally6,7, yielding clonally-related populations2,8 that retain spatial restriction into adulthood.
Here we tested whether AS allocated to discrete dorsal-ventral (DV) SC domains might be functionally adapted to support specific neural circuits and neuronal sub-types9. The SC sensorimotor circuit has well-defined organization in the DV axis (Fig. 1a). The ventral horn contains two types of motor neurons (MN), called α-MN and γ-MN, whose axons exit the ventral root to project to extrafusal (α) and intrafusal (γ) muscle fibers10. During development, afferent sensory fibers entering from the dorsal root ganglion (DRG) include Type-1a proprioceptive afferents that synapse directly on ventral α-MN, and TrkA+ sensory axons that synapse in the dorsal gray matter11.
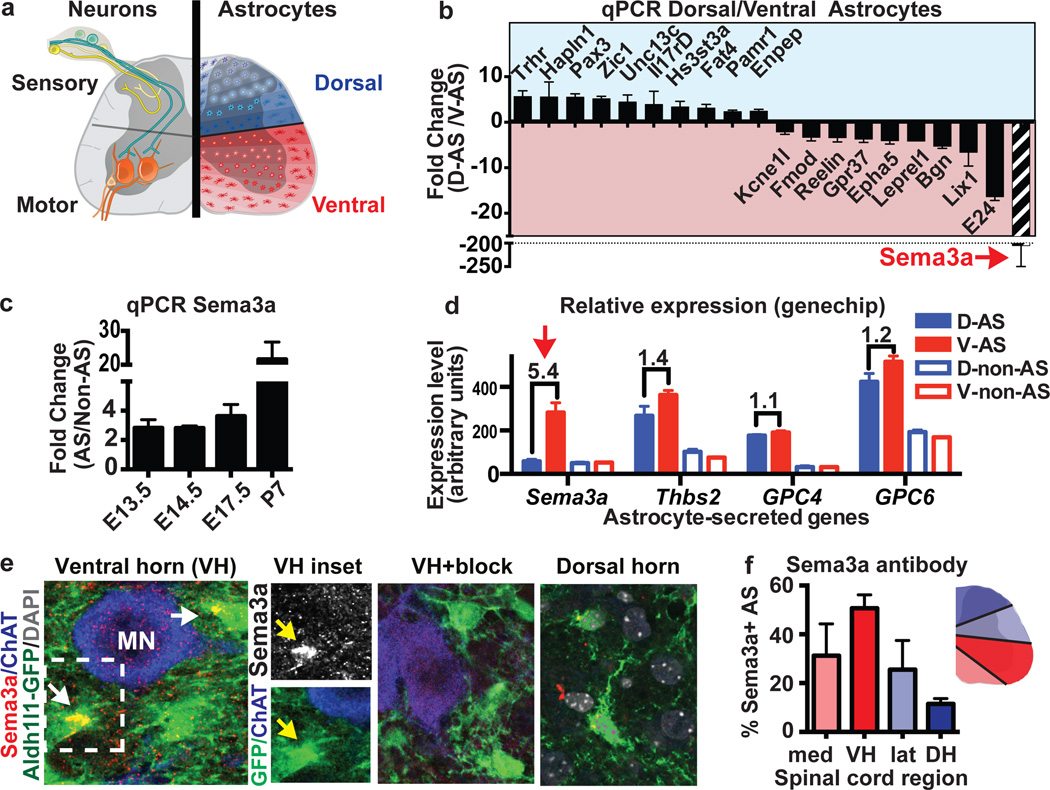
Figure 1. AS express region-specific genes.
a) Concept of dorsal/ventral AS functional specialization. b) qPCR validation of differentially expressed genes in Aldh1L1-GFP+ dorsal or ventral SC AS (2/4 samples had undetectable dorsal Sema3a). c) qPCR of Aldh1L1-GFP AS versus non-AS (purity >95%). d) Regional expression of Sema3a versus thrombospondin 2 (Thbs2), and glypicans-4/6. e) Sema3a proteins in ventral but not dorsal SC Aldh1L1-GFP+ protoplasmic AS (arrows, inset) (antibody blocking peptide “block”), f) Graded DV expression of Sema3a protein as percent of Sema3a+/Aldh1L1-GFP+ AS at E18.5; VH=ventral horn, DH=dorsal horn. (All data mean ±s.e.m.; β-actin as HK gene in qPCRs; n=4 (b) or 3(c–f) biological replicates/bar).
Although programs that control MN diversification and connectivity are well established9,12, comparatively little is known about non-neuronal signals that influence local circuit formation11,13. We report that ventral AS-encoded Semaphorin3a (Sema3a), a secreted molecule that signals through PlexinA/Neuropilin1 receptor (Nrp1) complexes14–16, plays critical roles in orienting MN axon initial segments (AIS), synapse regulation, MN sub-type survival and normal patterning of a subset of TrkA+ sensory neurons. These findings establish a discrete molecular function for region-restricted AS.
RESULTS
AS express DV-restricted positional cues
To identify regionally distinct molecular differences we purified fibrous and protoplasmic AS from microdissected dorsal and ventral postnatal day 7 (P7) SC by flow cytometry using the Aldh1L1-GFP transgene reporter2,7,17,18 (Fig. 1a, Extended data Fig. 1). Gene expression profiling and bioinformatic analysis identified 38 genes that were differentially expressed (Extended data Fig 1), and we validated these results by quantitative PCR. As shown, (Fig. 1b) several genes encoded extracellular matrix molecules19 (Hapln1, Pamr1, Enpep, Bgn) or factors with positional roles in brain development, including Reelin20 and EPH receptor A521. Of these, Sema3a was the most highly expressed ventral AS-specific gene, showing over three-fold higher levels in radial glia and AS (versus non-AS) from E13.5-P7 (Fig. 1c), consistent with Sema3a expression by in situ (Extended data Fig 2a–c.) 15. In contrast to Sema3a, genes for other AS-secreted molecules, including thrompospondin2 (Thbs2), and glypican-4 and −6, were expressed without positional distinction (Fig. 1d.). Sema3B, C, D, E and F were low or undetectable in AS (data not shown).
Extended Data Figure 1. Flow cytometry gating strategy and microarray.
a) Schematic indicating microdissection of Aldh1l1-GFP positive P7 spinal cord and isolation by flow cytometry using scatter gates, doublet exclusion (not shown) and sorting for GFP positive cells with live/dead exclusion by DAPI staining. Percent Aldh1l1-GFP cells not significantly different between dorsal and ventral (not shown.) b) Summary of differentially expressed genes in astrocytes (AS), whole cord, or both using the analysis parameters indicated. c) Heatmap of all 39 genes differentially expressed between dorsal and ventral cord, highlighting astrocyte-enriched genes with known roles in neural circuit development (red) or extracellular matrix (blue.)
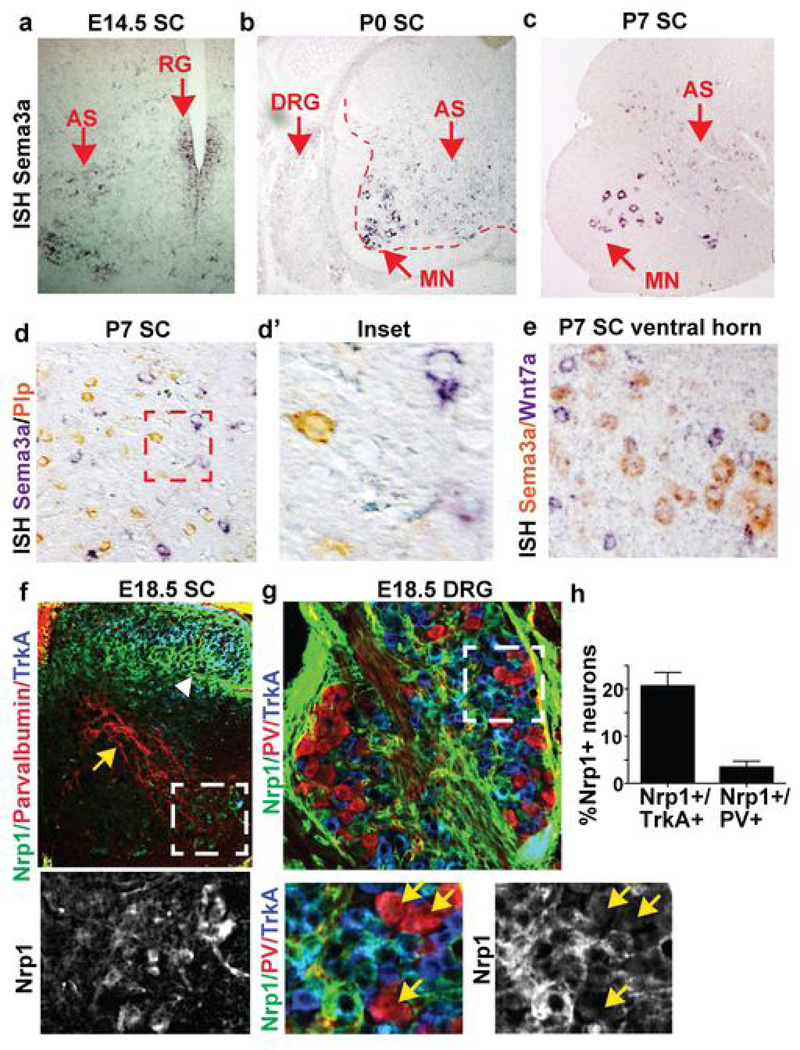
Extended Data Figure 2. Coordinate expression of Sema3a and Nrp1 in astrocytes and neurons.
a-c)Sema3a mRNA is expressed in radial glia (RG) and in protoplasmic cells that are NeuN negative throughout the embryonic and early postnatal period. Sema3a not detected in DRG or in SC white matter (b). d) Sema3a is segregated from Plp-positive oligodendrocytes. e) MN Sema3a expression is detected in α−but not γ-MN in cervical SC. f-g) High levels of Nrp1 expression in TrkA+ fibers and cell bodies (white arrowhead) and in MN, but not in PV positive fibers and cell bodies (yellow arrows;) h) Quantification of percent Nrp1+ neurons/condition.
AS Sema3a protein expression showed graded expression, with lowest numbers of Sema3a+ cells in the dorsal horn and highest numbers in the ventral horn. In ventral AS, Sema3a proteins appeared oriented towards MN soma (Fig. 1e,f). Although Sema3a mRNA transcripts were highly expressed in α-MN, we did not detect corresponding Sema3a protein levels (Fig. 1e, Extended data Fig. 2e). MN express the obligate semaphorin receptor Nrp115,22,23 (Extended data Fig. 2f). In addition, dorsal root ganglion (DRG) TrkA+ sensory neurons, but not parvalbumin (PV)+ proprioceptive afferents, express high levels of Nrp1 (Extended data Fig. 2f–h). These findings suggested potential neuronal subtype-specific functions for AS-encoded Sema3a.
AS Sema3a restricts α–MN AIS orientation
We first investigated ventral astrocytic Sema3a function during early postnatal MN development (Fig. 1e). To conditionally target Sema3a in AS, we crossed Sema3afl/fl mice24 to hGFAPcre25, chosen because: (1) cre activity commences in late stage glial progenitors, (2) it targets protoplasmic AS, and (3) cre activity segregates from SC MN26 and interneurons (Extended data Fig. 3a). hGFAPcre:Sema3afl/fl animals survived postnatally in near-normal numbers.
Extended Data Figure 3. Fate map of conditional astrocyte deletion lines used in this study.
a)hGFAPcre fate map labels fibrous and a subset of protoplasmic astrocytes but not MN or interneurons in P10 SC. b) Aldh1L1cre fate maps to astrocytes but not to neurons in P10 SC, including α-MN (purple), γ-MN (blue), and interneurons (red.)
Previous work indicates Sema3a has roles in supporting dendrite versus axon identity27 and hippocampal neuron axon repulsion and dendrite growth in vitro28. To assess a potential role for AS-encoded Sema3a in orienting MN axons in vivo we used ankyrin G29 to define the direction of the AIS relative to the ventral root. By P7, large α-MN and smaller γ-MN normally exist in a 2:1 ratio in most MN pools10. To determine AIS orientation of both α-MN and γ-MN following loss of AS-encoded Sema3a, we used AnkG to identify the AIS and NeuN staining to distinguish α–MN (Chat+/NeuN+) from γ–MN (Chat+/NeuN-)10. This analysis was performed at both lumbar and cervical levels and the results were plotted on a positional grid (Fig. 2a–c, Extended data Fig. 4). In control animals, we found that α–MN AIS were oriented an average of 39±33° (SD) degrees from the ventral root. In contrast, we observed marked disorganization of α–MN AIS orientation in hGFAPcre:Sema3afl/fl animals, with increases in both mean angle and variability (62±46°(SD); p<0.001) (Fig. 2d; Extended data Tab. 1) at all topographic positions (Fig. 2c, Extended data Fig 4.) Notably, we did not detect any differences in AIS orientation in γ–MN (control: 52±31° vs. mutant 49±29°, p=ns; Fig. 2e). Furthermore, these prominent differences at P7 were no longer detectable by P30 (Fig. 2f).
Figure 2. Axon-repulsive effects of AS-encoded Sema3a maintain α-MN AIS orientation.
a) P7 lumbar SC ChAT/NeuN+ α-MNs with AnkG+ AIS. Ventral root (*) and orientation vector towards it for selected α-MN (white arrows). b) Insets of yellow boxed areas in (a). Yellow arrow denotes AnkG+ AIS junction, the angle between these vectors determines AIS orientation. c) Overlay of all lumbar AIS orientation angles demonstrates that misoriented MN are topographically distributed. d) Mean angle and variability is significantly increased in P7 α-MN in absence of AS-encoded Sema3a (0°=towards ventral root). e) No difference in AIS orientation in γ-MN. f) Scatter plot of data generated as in panel (c) shows no misorientation of α- or γ-MN by P30. g) AIS angles in Aldh1L1cre:Sema3afl/fl are normal at E14.5 and significantly misoriented by P0. h) Deletion of Sema3a from MN with Hb9cre does not affect MN AIS orientation at E17.5. i) AS-MN coculture protocol. j,k) Increased MN axon overlap with Ade-cre deletion of AS Sema3afl/fl (white arrowhead: proximal axon). l) Summary. Statistics and error bars: mean±S.D. Watson’s U2 test, except k: mean±s.e.m. student’s t-test. d,e,g) >100 neurons, n=3–4/genotype, except at E14.5 n=2/genotype; f) >40 neurons n=4/genotype; h) >30 neurons n=2/genotype; k >20 neurons/condition; 3 independent experiments. *p<0.05, **p<0.01, ***p<0.001.
Extended Data Figure 4. Motor neuron AIS orientation defects in cervical spinal cord.
a) Representative images of cervical spinal cord confocal sections stained to distinguish α and γ MN and identify their proximal axon segment (asterisk=ventral root). b) Inset shows high-magnification view of representative MN with identifiable AIS and a schematic of their location with respect to the ventral root. c) Overlay of all cervical a-MN angles measured to generate data summarized in Fig. 2c, with positional information preserved, demonstrates that misoriented AIS can be seen at all dorsoventral positions.
Extended Data Table 1.
| Control (cre-) | hGFAPcre: Sema3afl/fl | p-value | ||
|---|---|---|---|---|
| MN axon orient ation, P7 (° from ventral root) | All MN | 42±33°(SD) | 58±42°(SD) | ** |
| Cervical MN | 42±34°(SD) | 62±49°(SD) | ** | |
| Lumbar MN | 42±32°(SD) | 55±42°(SD) | * | |
| α-MN (NeuN+) | 39±33°(SD) | 62±46°(SD) | *** | |
| γ-MN (NeuN−) | 52±31°(SD) | 49±29°(SD) | ns | |
| MN soma size, P30 (µM2) | All MN | 664±14 | 483±10 | **** |
| Cervical MN | 599±24 | 497±16 | *** | |
| Lumbar MN | 687±17 | 469±14 | **** | |
| #vGlut1/MN | All MN | 4.8±0.26 | 2.9±0.20 | **** |
| Cervical MN | 4.3±0.29 | 2.9±0.19 | *** | |
| Lumbar MN | 5.1±0.33 | 3.3±0.33 | *** | |
| α-MN (>500 µM) | 5.6±0.25 | 4.1±0.24 | *** | |
| γ-MN (<500 µM) | 2.27±0.28 | 1.95±0.19 | ns | |
| #VGAT/MN | All MN | 21±0.74 | 27±0.96 | **** |
| Cervical MN | 19.88±1.1 | 24.5±1.5 | * | |
| Lumbar MN | 21.8±1.0 | 27.8±1.4 | ** | |
| α-MN (>500 µM) | 25.7±1.1 | 37.5±1.3 | **** | |
| γ-MN (<500 µM) | 11.4±0.82 | 22.7±1.0 | *** | |
| #vGlut2/MN | All MN | 18.29±0.93 | 16.11±0.88 | ns |
| Cervical MN | 15.0±0.82 | 13.0±0.68 | ns | |
| Lumbar MN | 23.5±1.4 | 21.7±1.4 | ns | |
| α-MN (>500 µM) | 22.98±1.0 | 21.21±1.2 | ns | |
| γ-MN (<500 µM) | 12.18±0.81 | 11.7±0.57 | ns |
To determine whether this AIS orientation defect reflected aberrant initial positioning or a failure to maintain MN orientation during later growth, we targeted radial glia and AS using Aldh1L1cre, which targets gliogenic radial glia at E12.5–13.57 . Aldh1L1cre fate maps most AS, some oligodendrocytes (that are Sema3a negative, Extended data Fig. 2d), but excludes SC MN and interneurons (Extended data Fig. 3b). Aldh1L1cre: Sema3afl/fl mice were perinatal lethal (28% expected survival at P1-P5, n=112), but with normal numbers of AnkG+ MN, suggesting that axon specification is not affected in vivo (AnkG+ MN per confocal section: 42±5% vs. 46±4%, p=ns). At E14.5 (before the major period of AS expansion7), we found no evidence of abnormal MN cell body positioning (Extended data Fig. 5) or MN AIS orientation in Aldh1L1cre:Sema3afl/fl animals (control: 29±21° SD vs. mutant: 32±30°, p=ns; Fig. 2g). By P0, AIS orientation defects were evident and similar in magnitude to those observed in P7 hGFAPcre:Sema3afl/fl (37±25°(SD) vs. 66±38°(SD); p<0.001; Fig 2g.) As a control, we deleted Sema3a in MN using Hb9cre30,31. This did not lead to defects in MN AIS orientation (31±25° control, 30±33° Hb9cre:Sema3afl/fl, p=ns ; Fig. 2h). These findings demonstrate that early developmental events -- including initial positioning of MN soma and exit of MN axons from the ventral root -- occur normally despite loss of AS-encoded Sema3a. However, with loss of AS-encoded Sema3a, many α–MN fail to properly maintain AIS orientation towards the ventral root at early postnatal stages.
Extended Data Figure 5. No evidence of abnormal MN cell body positioning with loss of astrocyte-encoded Sema3a.
a) Representative FoxP1/Islet1/2 colabeling at three rostrocaudal levels in control and mutant animals shows no differences between control and mutant. b) Similar stainings using Scip (a PMC and LMC marker.) c-d) No obvious differences in dorsoventral or mediolateral boundaries of ChaT+ MN at comparable cervical or lumbar levels at P0 (using Aldh1L1-cre to delete Sema3a) and P7 (with hGFAPcre), both time periods where misorientation of AIS is clearly evident.
To test whether AS-encoded Sema3a acts directly on MN without other cellular intermediates we co-cultured ventral SC AS from Sema3afl/fl animals at sub-confluent density with embryonic rat MN32(Fig. 2i), and added adenoviral cre (Ade-cre) to some wells to delete Sema3a. We found that the length of proximal segment overlap in MN adjacent to AS was significantly increased after Ade-cre mediated deletion of Sema3a from ventral AS (50±9 µm vs. 111±16 µm, p<0.001; Fig. 2j,k), suggesting a failure to properly repel the axon. Thus, AS-encoded Sema3a directly repels MN axons both in vivo and in vitro.
AS Sema3a is needed for α–MN survival
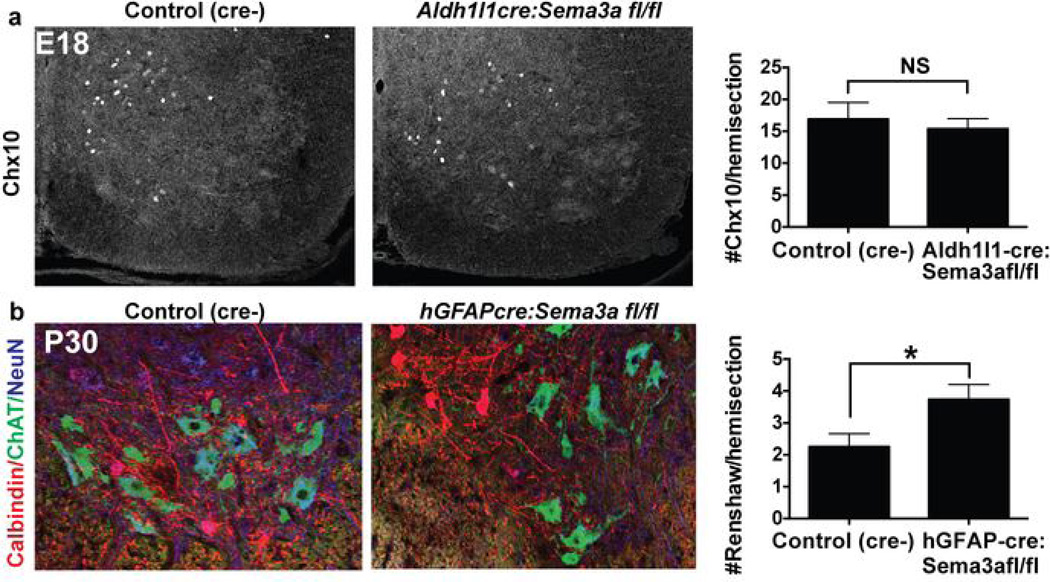
Given these findings, we investigated whether ventral AS-encoded Sema3a might also have later roles in maintenance of the sensorimotor circuit. At P7, numbers of both α- and γ-MN were normal in hGFAPcre:Sema3afl/fl mice (Fig. 3a–b10 and data not shown33). In contrast, by P30 we found a significant (p < 0.05) two-fold reduction in numbers of surviving Err3-negative10 α–MN in hGFAPcre:Sema3afl/fl mice (Fig. 3c–d). This was supported by a dose-dependent reduction in the average size of MN soma reflecting selective loss of a large MN population (Fig. 3e–f; Extended data Tab. 1). In contrast, γ–MN persisted in normal numbers. Additionally, two ventral interneuron populations (Chx10+ interneurons and calbindin+ Renshaw cells) did not show Sema3a–dependent depletion. In fact, the number of Renshaw cells was significantly increased at P30 (Extended data Fig. 6.)
Figure 3. AS-encoded Sema3a is required for postnatal α-MN survival.
a,b) Normal numbers of α-MN (Chat+/NeuN+) and γ-MN (Chat+/NeuN-) at P7 in hGFAPcre:Sema3afl/fl cervical and lumbar SC. c, d) Fewer large α-MN and preserved numbers of γ-MN (Err3-bright, arrows.) in hGFAPcre:Sema3afl/fl animals at P28-P33. e, f) Dose-dependent decrease in MN soma area at P28–33 with loss of AS-Sema3a; histograms show relative depletion of large MN (arrow). g) Representative Peripherin+/Map2+ embryonic rat MN cultured in factor-free media or with recombinant Sema3a (red arrow: soma). h) MN survival with recombinant Sema3a with/without Nrp1-blocking antibody. i) Summary. Statistics: mean ±s.e.m. (e,f) one-way mixed effects ANOVA with Tukey’s multiple comparison. (b,c,h) student’s t-test. Data in b,c from 4/genotype from 4 sections/animal; h average of 4 independent experiments. *p<0.05, **p<0.01, ***p<0.001.
Extended Data Figure 6. Quantification of ventral interneuron populations after loss of astrocyte-encoded Sema3a.
a) Chx 10 staining at E18 and quantification. b) Calbindin staining of Renshaw interneurons at P30 and quantification demonstrates a significant increase at this age. Statistics: mean±s.e.m., student’s t-test, data in a from n=2 /group, 4 sxns/animal, data in b from 4/group 4 sxns/animal.
To further assess direct effects of Sema3a on MN survival, we cultured MN in factor free media for 24 hrs (to allow for initial polarization28) then added recombinant Sema3a proteins. We found that exogenous Sema3a promoted MN survival in a dose-dependent manner that was abrogated by preincubating with an Nrp1-blocking antibody (Fig. 3g,h.) Interestingly, the MN that survived in the absence of Sema3a typically had bipolar morphology (No add: 86±7% bipolar, vs. Sema3a: 20±11% , p<0.01). Together, these findings indicate that AS-encoded Sema3a can directly promote MN survival (Fig. 3i) in a manner that is tightly linked to its tropic effects. Notably, abnormal MN AIS orientation was no longer evident in adult hGFAP-cre:Sema3afl/fl mice (Fig. 2f), suggesting misoriented α–MN are lost by adulthood.
AS Sema3a regulates MN function
To test whether MN in hGFAPcre:Sema3afl/fl mutants integrated normally into local synaptic circuits (Fig. 4a), we first counted excitatory vGlut1+ presynaptic puncta on MN soma, which reflect type 1a proprioceptive sensory afferents11. We observed a significant decrease in the number of vGlut1 puncta/MN (Fig. 4b; 4.82±0.26 vs. 2.91±0.2; p<0.0001) in adult mutant animals. These differences remained highly significant even when only large (>500 µm) putative α–MN were counted (5.59±0.25 vs. 4.13±0.23; p<0.001). Numbers of excitatory vGlut2 puncta, which are less dependent on DRG afferent input34–36 were not significantly different in hGFAP-cre:Sema3afl/fl animals versus controls (18.29±0.93 vs. 16.11±0.88 p=ns; Extended data Tab. 1). In contrast, we found significant increases in numbers of vGAT inhibitory presynaptic puncta with loss of AS- encoded Sema3a (Fig. 4b; 21±0.74 vs. 27±0.96; p<0.0001).
Figure 4. AS-encoded Sema3a regulates MN synaptogenesis and function.
a) Schematic of MN synaptic puncta. b) Decreased sensorimotor excitatory puncta (vGlut1+), increased inhibitory puncta (VGAT+), and preserved vGlut2+ inputs in hGFAPcre:Sema3afl/fl animals. c) Electrophysiology schematic. d,e) Representative action potentials and mean rheobase value demonstrate hyper excitable hGFAPcre:Sema3afl/flMN. f,g) Decreased sEPSC frequency and (h,i) increased sIPSC frequency in hGFAPcre:Sema3afl/fl MN. Statistics: mean ±s.e.m. Data in b from cervical and lumbar levels of >4/genotype and >200 MN/ea; data in d-h =5–6/genotype from lumbar slices. (b) one-way mixed effects ANOVA with Tukey’s comparison; vglut2: student’s t-test. (d–h) student’s t-test. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
To test whether these synaptic changes correlated with changes in MN function, we performed electrophysiological recordings of lumbar α–MN at P13–14 from hGFAPcre;Sema3afl/fl:ChAT-GFP reporter mice and cre-negative controls. We performed whole-cell patch clamp recording37 using ChAT-GFP expression to identify intact MN able to generate action potentials (Fig. 4c, d.) As shown (Fig. 4f–i), loss of AS-encoded Sema3a conferred a large shift in the balance of excitation/inhibition with significantly decreased MN sEPSC frequency (control: 3.19 ±0.35 Hz vs. mutant: 1.21±0.38 Hz; p<0.01) and increased sIPSC frequency (control: 0.14±0.04 Hz vs. mutant: 0.84±0.15 Hz; p<0.01). Amplitudes of sEPSC and sIPSC were not changed (Extended data Fig. 7). Interestingly, hGFAPcre:Sema3afl/fl MN were hyperexcitable as reflected by a significant decrease in the rheobase value (control: 90.0±23.0 pA vs. mutant:37.5±7.2 pA, p<0.05; Fig. 4d–e) with unchanged input resistances (Extended data Fig. 7), suggesting compensatory changes in MN function. Together, these findings demonstrate that loss of AS-encoded Sema3a leads to changes in MN excitatory and inhibitory synaptic inputs and has global effects on MN firing properties.
Extended Data Figure 7. Additional data and controls for MN electrophysiology.
(a) 2µM strychnine and 20µM bicuculline block postsynaptic currents (at-55mV) in a ChAT-GFP+ lumbar MN. (b) 20µM 6,7-dinitroquinoxaline-2,3-dione (DNQX) and 50µM (2R)-amino-5-phosphonovaleric acid (AP5) block postsynaptic currents (at −75mV) in a ChAT-GFP+ lumbar MN. (c) No difference in input resistance, sIPSC amplitude, and sEPSC amplitude between control (cre-) and hGFAPcre:Sema3afl/fl (fl/fl) MN. n=5/eA; mean±s.e.m. student’s t-test.
AS regulate sensory axon targeting
Initial studies of Sema3a demonstrated selective chemorepellent activity for sensory axons expressing TrkA or CGRP14,22,38 which label overlapping sets of DRG neurons39. However, the cellular source of Sema3a in SC has never been defined. Sensory axon guidance takes place between E13.5-E18.5 in the mouse SC40. As shown (Fig. 1a), TrkA+ sensory axons normally synapse in the dorsal horn41, whereas PV+ proprioceptive 1a afferents synapse with ventral αMN11.
We characterized sensory axon guidance in Aldh1L1cre: Sema3afl/fl animals to determine potential AS-encoded functions (Fig. 5). Notably, while Aldh1L1cre activity fate maps to ∼5% of DRG cells, Sema3a expression was undetectable in the DRG15 (Extended data Fig. 2b), and we observed normal numbers of both PV+ and TrkA+ cells in the DRG of Aldh1L1cre:Sema3afl/fl mice (Extended data Fig. 8). DiI labeling of DRGs of Aldh1L1cre:Sema3afl/fl mutants at E18.5-P0 showed abnormal ventrally positioned axons in 7/7 mutants examined versus 0/7 controls (Fig. 5a).
Figure 5. AS-encoded Sema3a regulates DV positioning of sensory axons.
a) DiI labeling of upper thoracic SC demonstrates ectopic ventral fibers in Aldh1L1cre:Sema3afl/fl mice inset (box, arrows) (n=7/group).b) No ectopic proprioceptive 1a afferents (white arrow) in Aldh1L1cre:Sema3afl/flmice; n=4/group. c) Multiple ectopic ventral TrkA+ projections in Aldh1L1cre:Sema3afl/fl mice (box, insets). d) Overlay dot-plot of TrkA+ terminations in cre-negative controls (black circles) and Aldh1L1cre:Sema3afl/fl (red stars) n≥3/genotype. e) Quantification of DV termination index44 (DTI) shows a significant increase in ventral terminations in Aldh1L1cre:Sema3afl/fl mice. f) Medial vs. lateral terminations unchanged. g) Summary. h) Culture schematic. i) Representative TrkA+ DRG neuron grown on SC AS. j) Tracings of wild-type DRG neurons grown on dorsal/ventral SC AS from Sema3afl/fl mice, under WT (no virus) or Sema3a−/− (+Ade-cre) conditions. k) Reduced total neurite length of neurons grown on ventral (red) vs dorsal (blue) AS significantly rescued on Sema3a-/- ventral AS. l,m) Sholl analysis shows significantly less branching on ventral AS (l), rescued withSema3a deletion (m). Statistics: Culture data from 4–6 independent experiments with >10 neurons/condition per experiment. Mean+/-s.e.m; * p<0.05, ** p<0.01, ***p<0.001 student’s t-test).
Extended Data Figure 8. Normal dorsal root ganglia in Aldh1L1 cre:Sema3afl/fl mice.
a) No difference in the number of subtype specific neurons per DRG in control or Aldh1l1cre:Sema3afl/fl mice (n=3 from 4-5 sections per animal; mean±s.e.m.; student’s t-test).
We then investigated the sub-classes of DRG sensory afferents affected in Sema3a mutants. We observed normal dorsoventral positioning of PV+ type 1a proprioceptive afferents albeit with subtle fasciculation defects (Fig. 5b). In contrast, TrkA+ afferents showed numerous abnormal ventral terminations in Aldh1L1cre:Sema3afl/fl mutants (Fig. 5c–f). Thus, AS-encoded Sema3a regulates DV patterning of sensory axon projections in a subtype specific manner in vivo (Fig. 5g), consistent with previous results38 and with the neuronal pattern of Nrp1 expression (Extended data Fig. 2).
To test whether distinctions between regional SC AS exist independent of environmental positional cues, we co-cultured dorsal or ventral AS with dissociated DRG sensory neurons (Fig. 5h) using AS from Sema3afl/fl animals and deleted Sema3a by adding adenoviral cre recombinase (Ade-cre). Interestingly, AS cultures retained their distinct regional expression characteristics for many of the genes prospectively identified in vivo (see Fig. 1b, Extended data Fig. 9), including higher ventral Sema3a levels.
Extended Data Figure 9. Differential expression of regionally heterogeneous astrocyte genes is partly preserved in vitro.
qPCR quantification demonstrates that many regionally heterogenous microarray genes prospectively identified in vivo remain differentially expressed in vitro after 17 days in culture, including ventral Sema3a. Mean±s.e.m., n=3 independent experiments.
We observed that both dorsal and ventral AS cultures preferentially supported the survival of TrkA positive sensory neurons (>85% of neurons were TrkA+; none were PV+ Fig. 5i), with no significant survival differences between D/V cultures. However, ventral SC AS cultures significantly (p <0.01) inhibited neurite outgrowth and complexity relative to dorsal AS (Fig. 5j–m). Ade-cre deletion of Sema3afl/fl normalized differences in neuronal length and complexity conferred by ventral AS (Fig. 5j–m). Together, these findings show that AS encode subtype-specific sensory axon guidance signals.
DISCUSSION
We propose that region-restricted AS comprise a stable “scaffold” that maintains positional information throughout embryonic and postnatal development. This positional code is necessary for proper circuit formation, refinement, and neuronal survival in a subtype-specific manner. Loss-of-Sema3a–function from AS led to a sequence of α–MN specific phenotypes, comprising defective postnatal maintenance of AIS orientation, markedly abnormal inhibitory and excitatory currents in MN and abnormal synapse investment, and finally, α–MN loss. Concomitantly, in more dorsal regions, AS-encoded Sema3a acts to repel TrkA sensory afferent fibers in a subtype-specific manner. It is possible that these phenotypes represent a pathological progression, or alternatively, that ventral AS-encoded Sema3a has multiple coordinated functions that determine structural and functional sensorimotor circuit integrity.
Our in vitro studies further suggest that AS positional identity is at least partly cell intrinsic, as cognate Sema3a–dependent regional AS properties were retained in co-cultures independent of local environmental cues. As such, a testable prediction is that embryonic CNS patterning mechanisms might establish a template for generation of heterogeneous properties of AS 42 . Furthermore, while maintenance of MN axonal orientation represents one tropic effect of AS-encoded Sema3a, further investigation is needed to assess other potential roles of this or other AS-encoded regional cues, such as promoting dendrite growth27,28, maintenance of neuronal soma position, local synaptic strength and/or sensorimotor specificity 13.
Specialized local functions of AS in neural circuit formation may also have significance in human disease. For example, loss of ventral SC MN in amyotrophic lateral sclerosis (ALS) has been associated with mutant superoxide dismutase (SOD) protein in ventral AS in animal models of the disease43. Our findings suggest the possibility that the unique identity of ventral horn AS might lead to deficient local support for MN and disease progression in ALS. More generally, given that AS are regionally patterned throughout the CNS2, the concept of regional AS function and dysfunction has implications for a variety of neurodevelopmental and psychiatric disorders.
Methods summary
Animals were maintained in the University of California San Francisco animal facility. All protocols were IRB approved and in accordance with the Institutional Animal Care and Use Committee guidelines. Circular data and statistics analyzed using Oriana4 software (Kovach Computing Services, UK). Astrocyte monolayers from microdissected P0 mouse spinal cord were cultured for 12–14 days before replating at subconfluent density for MN cocultures and high density for DRG cocultures. Experiments with recombinant Sema3a (Peprotech) were on matrigel in serum-free, growth-factor free media. Nrp1 blockade with 10 µg/ml antibody (R&D AF566) added 30 minutes before Sema3a.
FULL METHODS
Mice
All mouse strains were maintained in the University of California San Francisco specific pathogen–free animal facility, and all animal protocols were approved by and in accordance with the guidelines established by the Institutional Animal Care and Use Committee and Laboratory Animal Resource Center. Mouse strains not otherwise referenced include CAG-GFP (MGI:3849685). For embryonic tissues, plug date was considered embryonic day 0.5. Embryo age was confirmed by morphology and crown-rump length measurements at harvesting.
Astrocyte isolation by flow cytometry
Postnatal day 7 spinal cords were microdissected using an “open book” preparation to separate dorsal and ventral halves. DRGs and meninges removed, then dissociated with papain 20 U/ml (Worthington) for 80 minutes at 33°C as previously described 17. Aldh1l1-positive and -negative cells were sorted as previously described18on a BD Facs Aria II and gated on forward/side scatter, live/dead by DAPI exclusion, and GFP, using GFP-negative and DAPI-negative controls to set gates for each experiment. In some cases GFP-positive populations were re-sorted using the same gates to >95% purity.
RNA isolation
RNA was isolated using TRIZOL reagent (Invitrogen) with glycogen added as carrier, DNAse digested to remove genomic DNA contamination, and further purified using the RNAeasy Kit (Qiagen). For microarray analysis, RNA samples were amplified using the Nugen Pico WT Ovation Kit and hybridized to Affymetrix Mouse Gene 1.0 ST arrays.
Bioinformatics
Microarray data were preprocessed in R using the Bioconductor suite of software packages. The "oligo" package was used to background correct, normalize, and summarize 1,102,500 probes on mouse gene 1.0 ST arrays via the Robust Multi-array Analysis (RMA) algorithm. After non-specific filtering was applied to remove low-intensity (<100 FU in at least 75% of the arrays), low-variance, and un-annotated probesets, 7,799 probesets remained. The Limma package was used to compare and assess differential expression between different groups of samples using the “treat” algorithm. Dorsal astrocytes, ventral astrocytes, dorsal non-astrocytes, and ventral non-astrocytes were directly compared for the “Dorsal vs Ventral” analysis. To generate an “astrocyte vs non-astrocyte” dataset, Dorsal and ventral astrocyte probesets were assigned equal weight and compared against Dorsal and ventral non-astrocyte probesets. With Fold-change>1.2, FDR=0.15, we identified 5158 genes differentially expressed between astrocytes and non-astrocytes, and 38 genes (39 transcripts) differentially expressed between dorsal and ventral astrocytes.
qPCR analysis
cDNA was generated from purified RNA using Superscript III (Invitrogen) and random decamers. Primers were designed for amplicons of 75–150 bp using Primer 3 and are available upon request. qPCR was done on a Roche lightcycler 480 using Sybr Green Master Mix (Roche.) Melt curves were analyzed for each experiment to ensure primer specificity. In most cases both β-actin and GAPDH were used as housekeeping genes for normalization with similar results, Aldh1L1 used as HK gene in vitro.
Immunohistochemistry/In Situ Hybridization
Most images collected using a Leica SP5 confocal microscope. Antibodies used included mouse NeuN (Millipore), rabbit Sema3a (EMD biosciences), goat ChAT (Millipore), rabbit TrkA (gift of L. Reichardt, UCSF), chick GFP (Aveslabs), mouse Parvalbumin (Sigma), chick Neurofilament (Encor), Rabbit Ankyrin G (Santa Cruz), mouse Err3 (PPMX), chick peripherin (Millipore), rabbit Map2 (Millipore) mouse Isl 1/2 (DSHB), sheep Chx10 (abcam) and rabbit calbindin (swant). Rabbit FoxP1 and guinea pig Scip provided by Dr. Jeremy Dasen. In most cases staining was done overnight at 4°C in 5% serum/0.4% Triton, following heat-mediated antigen retrieval for 2 minutes at 95°C in 0.1 M citrate buffer pH 6.0. For GFP labeling Ag retrieval was 10 minutes at 70°C. For TrkA staining, no retrieval was used, and slides were stained overnight at room temperature in 5% serum/1% triton. In situ hybridization performed using standard protocols. Sema3a probe against the full length rat Sema3a transcript provided by Dr. Alex Kolodkin, Wnt7a probe from A. McMahon. Plp probe from Ian Griffiths (Edinburg.)
Polarity analysis in vivo
Measurements of motor neuron orientation relative to the ventral root were calculated by measuring the angle between the following: 1) the vector from the MN nucleus to the axon hillock, marked by AnkG staining of the proximal axon segment. 2) The vector from the MN nucleus and the exit point of the ventral root from the gray matter. Angle measurements ranged from 0° (axon pointing towards the ventral root) to 180° (axon pointing directly away from ventral root.) Circular data was analyzed using Oriana4 software (Kovach Computing Services, UK), and statistical analyses performed using a Watson’s U2 test.
Synapse and soma diameter counts
Counts of synaptic puncta on MN soma performed at cervical (C4–5) and lumbar (L3–4) levels. Counts in main figure represent pooled unbiased data from all levels with no size cutoff, however all data was analyzed in histogram format and sorted by MN soma area to determine whether results were likely to be biased by altered ratios of MN subtypes. For some subgroup analyses (Extended data Figure 1), putative α-MN and γ-MN were identified by size based on histogram analyses of soma area.
Astrocyte cell culture
Dorsal and ventral spinal cords from P0-P1 mice were isolated and dissociated as above. Cells were plated at a density of >1×106 per 25 cm2 flask in DMEM-hi glucose with 10% FCS/ 10 µM hydrocortisone, 5 µg/ml N-acetylcysteine, 2 µg /ml insulin, and 20 ng/ml EGF. Six days after plating cells, flasks were shaken to remove oligodendrocyte contamination. At 8 days, AraC was added to kill rapidly proliferating cells. For DRG cocultures, 10–12 days after initial plating cells were replated into assay containers which consisted of 8 well glass chamber slides (BD) coated with poly-D-lysine and recombinant human fibronectin (Biomedical Technologies) to promote astrocyte adhesion, plating 30,000 cells/well. In most cases, cultures were established using Sema3afl/fl mice, and adenoviral cre-recombinase (Vector Biolabs) was added to some wells 2–4 hrs after replating. For MN cocultures, astrocytes were replated at 2,000 cells/well onto a reduced GF Matrigel substrate (BD) diluted 1:25 in DMEM. Astrocyte monolayers were then cultured for 2–3 days prior to adding neurons.
DRG isolation and coculture
DRGs from E13.5–14.5 mouse embryos were isolated and dissociated for 45 minutes in 0.25% trypsin/EDTA (Invitrogen). 500 cells per well were plated onto astrocyte monolayers in minimal neural growth media containing DMEM:F12, 10% FCS, N2 and B27 supplements (Invitrogen,) and cocultured for 48 hours before fixation in 4% PFA and immunolabeling.
Motor neuron isolation and coculture
Spinal cord neurons were isolated from embryonic rat spinal cords based on previous protocols32. Briefly, spinal cords were dissected from E15 rat embryos, dissociated in 0.25% trypsin (Gibco) for 15 minutes and triturated to form a single cell suspension in L-15 plus 10% FBS media (Gibco). The suspension was immunopanned in a series of negative selection plates against rat neural antigen-2 (Ran2) and galactocerebroside (GC), and then motor neurons were positively selected for on a final p75NTR panning plate. Adherent cells were released from the plate with a brief application of 0.05% trypsin (Gibco) and resuspended in growth media (DMEM, B27, N2, Pen-Strep (Gibco) prior to culturing. At replating (onto matrigel-plated subconfluent astrocytes for cocultures, or matrigel coated wells for recombinant Sema3a experiments), fresh media was added consisting of DMEM hi-glucose supplemented with N2 and B27 supplements 5 µg/ml N-acetylcysteine, 5 µg /ml insulin, and 5 µM forskolin.
Recombinant Sema3a addition and Nrp1 blockade in vitro
MN were plated at 1000 cells per well as above. 12 hours after plating, they were assessed by light microscopy, at which point most could be seen to have budded polar processes. 24 hours after plating, recombinant human Sema3a was added (Peprotech) at indicated concentrations. For Nrp1 blockade, a Nrp1- blocking antibody validated for this purpose (R&D systems AF566) was added at a concentration of 10 µg/ml and incubated with MN for 30 minutes prior to Sema3a addition. Goat IgG control added separately to control for nonspecific effects (not shown.) Cells were cultured an additional 36 hours before fixation and immunolabeling.
Culture image analysis
For DRG cocultures, neurons were identified with NF-H (Encor), in some cases, TrkA and parvalbumin immunostaining was used to identify sensory neuron subtypes. MN were identified with peripherin and traced with an overlay of peripherin and Map2 (to label distal dendrites). Spatially distinct neurons were photographed and analyzed using NeuronJ plugin/Image J to trace total neurite length for each neuron. Sholl analyses were performed using the Sholl analysis plugin after thresholding each neuron using Image J (parameters: start 25 uM, step size 10 uM). >10 neurons per condition per experiment per condition were analyzed. For axon overlap in vitro, only MN directly abutting an astrocyte were included in the analysis, and overlap of proximal axon (defined as the axon segment proximal to the first branch point) was quantified using ImageJ. For neurite length, statistical analyses were performed on means of each experiment, in Sholl analyses, and axon overlap, t-tests were performed on pooled data.
Whole-Cell Patch Clamp Recordings
Acute fresh lumbar (L3–4) spinal cord slices were prepared from hGFAP-cre:Sema3afl/fl:ChAT-GFP mice and cre negative controls from postnatal day 12–14 using previously described protocols and solutions37. Briefly, transverse slices (350-µm thick) were cut with a vibratome (Leica Microsystems) in a chamber filled with ice cold sucrose cutting solution followed by a brief (60 second) incubation in polythethylene glycol (Mn = 1,900–2,200). The slices were then incubated in cutting solution at 35°C for 30 min followed by 30 min in artificial cerebrospinal fluid then equilibrated to room temperature. Whole-cell recordings were made using patchclamp amplifiers (Multiclamp 700B) under infrared–differential interference contrast microscop. Data acquisition and analysis were performed using digitizers (DigiData 1440A) and analysis software pClamp 10 (Molecular Devices). Signals were filtered at 6 kHz and sampled at 20 kHz. Spontaneous EPSCs (sEPSCs) were recorded from motor neurons at −75 mV in voltage-clamp mode where the chloride reversal potential was 0mV. Spontaneous IPSCs (sIPSCs) were recorded at −55mV where chloride currents are positive deflections in voltage clamp. Glass pipettes with a resistance of 2.5–4MΩ were filled with a K-methanesulfonate internal solution. To ensure currents measured were sEPSCs and sIPSCs, control recordings were performed using standard pharmacology (Extended data Figure 7). Series resistance (15–25 MΩ) was monitored throughout the whole-cell recording and data were discarded if the change in series resistance was >20% during the course of the experiment.
DiI labeling of sensory afferents
Spinal cords from E18.5-P0 mice were fixed for at least 48 hours in 4% paraformaldehyde, then dissected from spinal column with DRGs intact. The ventral root was severed and a crystal of DiI (Molecular probes) was applied to the DRG with a needle. Cords were incubated 12 days at 37°C, then 300 µm sections were cut by vibratome, mounted in PBS, and imaged by collecting confocal z-stacks at 2 µM spacing.
Sensory neuron termination analysis
Confocal sections of cervical spinal cords were overlaid with a uniform grid using Adobe Illustrator and terminations per image were labeled. When more than one break in a process was visible, only the distal-most termination was labeled. Data from all sections and animals were overlaid on a normalized grid and the number of terminations per quadrant calculated.
Statistical analysis
Consultation obtained from the UCSF department of Biostatistics. Coefficient of variance (standard error as a percentage of the mean) was used to calculate minimum sample sizes. In most cases, sample sizes were well in excess. Student’s t-test’s were two-tailed and based on Gaussian distributions. In all cases, replicates refer to biological rather than technical replicates. Blinding during analysis was used whenever possible for all in vivo studies. Most statistics analyzed using Graphpad Prism software (La Jolla, CA.) Circular data in Figure 2 was analyzed using Oriana4 software (Kovach Computing Services, UK).
Acknowledgements
We thank Drs. J. Flanagan, T. Jessell, J. de Nooj, N. Balaskas, M. Hancock, Shinya Ohata, and R. Krencik for comments on the manuscript and technical suggestions. We are grateful to K. Sabeur, M. Wong and the UCSF Flow Cytometry and Genomics core facilities for expert technical help, Drs. A. Kolodkin (Johns Hopkins) for Sema3a probe construct, L. Reichardt (UCSF) for the TrkA antibody, J. Dasen (NYU) for FoxP1 and Scip antibodies, and N. Heintz and J. Dougherty (Rockefeller University) for Aldh1L1-cre mice. A.V.M. is supported by an NIMH Training Grant (5T32MH089920-04) and an APA/Pfizer MD/PhD Psychiatric Research Fellowship. K.W.K is supported by the California Institute for Regenerative Medicine (TG2-01153). S.A.R. is supported by a Ruth L. Kirschstein NRSA FNS081905A. This work was supported by grants from the NINDS (to D.H.R. R01 NS059893, and J.R.C.) and NIMH (to E.M.U.) D.H.R. is a HHMI Investigator.
Footnotes
Author Contributions:
A.V.M. performed most experiments and data analysis. K.W.K performed electrophysiology under supervision of E.M.U. H.H.T. contributed to data analysis and experimental design. S.A.R. performed MN purification under supervision of J.R.C. S.C performed mouse genotyping. L.M. and S.E.B. performed bioinformatics data processing and analysis. A.V.M and D.H.R. designed the experiments and wrote the manuscript.
Author information:
Microarray data has been deposited to GEO under accession number GSE55054. The authors declare no competing financial interests. Correspondence and requests for materials should be addressed to D.H.R. at rowitchd@peds.ucsf.edu.
REFERENCES
- 1.Clarke LE, Barres BA. Emerging roles of astrocytes in neural circuit development. Nat Rev Neurosci. 2013;14:311–321. doi: 10.1038/nrn3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tsai H-H, et al. Regional astrocyte allocation regulates CNS synaptogenesis and repair. Science. 2012;337:358–362. doi: 10.1126/science.1222381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hochstim C, Deneen B, Lukaszewicz A, Zhou Q, Anderson DJ. Identification of Positionally Distinct Astrocyte Subtypes whose Identities Are Specified by a Homeodomain Code. Cell. 2008;133:510–522. doi: 10.1016/j.cell.2008.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Muroyama Y, Fujiwara Y, Orkin SH, Rowitch DH. Specification of astrocytes by bHLH protein SCL in a restricted region of the neural tube. Nature. 2005;438:360–363. doi: 10.1038/nature04139. [DOI] [PubMed] [Google Scholar]
- 5.Rowitch DH, Kriegstein AR. Developmental genetics of vertebrate glial-cell specification. Nature. 2010;468:214–222. doi: 10.1038/nature09611. [DOI] [PubMed] [Google Scholar]
- 6.Ge W-P, Miyawaki A, Gage FH, Jan Y-N, Jan LY. Local generation of glia is a major astrocyte source in postnatal cortex. Nature. 2012;484:376–380. doi: 10.1038/nature10959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tien A-C, et al. Regulated temporal-spatial astrocyte precursor cell proliferation involves BRAF signalling in mammalian spinal cord. Development. 2012;139:2477–2487. doi: 10.1242/dev.077214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Magavi S, Friedmann D, Banks G, Stolfi A, Lois C. Coincident generation of pyramidal neurons and protoplasmic astrocytes in neocortical columns. J Neurosci. 2012;32:4762–4772. doi: 10.1523/JNEUROSCI.3560-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jessell TM. Neuronal specification in the spinal cord: inductive signals and transcriptional codes. Nat Rev Genet. 2000;1:20–29. doi: 10.1038/35049541. [DOI] [PubMed] [Google Scholar]
- 10.Friese A, et al. Gamma and alpha motor neurons distinguished by expression of transcription factor Err3. Proc Natl Acad Sci USA. 2009;106:13588–13593. doi: 10.1073/pnas.0906809106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arber S. Motor Circuits in Action: Specification, Connectivity, and Function. Neuron. 2012;74:975–989. doi: 10.1016/j.neuron.2012.05.011. [DOI] [PubMed] [Google Scholar]
- 12.Dasen JS, Jessell TM. Hox networks and the origins of motor neuron diversity. Curr Top Dev Biol. 2009;88:169–200. doi: 10.1016/S0070-2153(09)88006-X. [DOI] [PubMed] [Google Scholar]
- 13.Sürmeli G, Akay T, Ippolito GC, Tucker PW, Jessell TM. Patterns of spinal sensory-motor connectivity prescribed by a dorsoventral positional template. Cell. 2011;147:653–665. doi: 10.1016/j.cell.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Messersmith EK, et al. Semaphorin III can function as a selective chemorepellent to pattern sensory projections in the spinal cord. Neuron. 1995;14:949–959. doi: 10.1016/0896-6273(95)90333-x. [DOI] [PubMed] [Google Scholar]
- 15.Kolodkin AL, et al. Neuropilin is a semaphorin III receptor. Cell. 1997;90:753–762. doi: 10.1016/s0092-8674(00)80535-8. [DOI] [PubMed] [Google Scholar]
- 16.Pasterkamp RJ, Giger RJ. Semaphorin function in neural plasticity and disease. Curr Opin Neurobiol. 2009;19:263–274. doi: 10.1016/j.conb.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cahoy JD, et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci. 2008;28:264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Molofsky AV, et al. Expression profiling of Aldh1l1-precursors in the developing spinal cord reveals glial lineage-specific genes and direct Sox9-Nfe2l1 interactions. Glia. 2013;61:1518–1532. doi: 10.1002/glia.22538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barros CS, Franco SJ, Müller U. Extracellular matrix: functions in the nervous system. Cold Spring Harb Perspect Biol. 2011;3:a005108. doi: 10.1101/cshperspect.a005108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tissir F, Goffinet AM. Reelin and brain development. Nat Rev Neurosci. 2003;4:496–505. doi: 10.1038/nrn1113. [DOI] [PubMed] [Google Scholar]
- 21.Dufour A, et al. Area specificity and topography of thalamocortical projections are controlled by ephrin/Eph genes. Neuron. 2003;39:453–465. doi: 10.1016/s0896-6273(03)00440-9. [DOI] [PubMed] [Google Scholar]
- 22.Gu C, et al. Neuropilin-1 conveys semaphorin and VEGF signaling during neural and cardiovascular development. Dev Cell. 2003;5:45–57. doi: 10.1016/s1534-5807(03)00169-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cohen S, et al. A semaphorin code defines subpopulations of spinal motor neurons during mouse development. Eur J Neurosci. 2005;21:1767–1776. doi: 10.1111/j.1460-9568.2005.04021.x. [DOI] [PubMed] [Google Scholar]
- 24.Taniguchi M, et al. Disruption of semaphorin III/D gene causes severe abnormality in peripheral nerve projection. Neuron. 1997;19:519–530. doi: 10.1016/s0896-6273(00)80368-2. [DOI] [PubMed] [Google Scholar]
- 25.Zhuo L, et al. hGFAP-cre transgenic mice for manipulation of glial and neuronal function in vivo. Genesis. 2001;31:85–94. doi: 10.1002/gene.10008. [DOI] [PubMed] [Google Scholar]
- 26.McCall MA, et al. Targeted deletion in astrocyte intermediate filament (Gfap) alters neuronal physiology. Proc Natl Acad Sci USA. 1996;93:6361–6366. doi: 10.1073/pnas.93.13.6361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nishiyama M, et al. Semaphorin 3A induces CaV2.3 channel-dependent conversion of axons to dendrites. Cell Biol. 2011;13:676–685. doi: 10.1038/ncb2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shelly M, et al. Semaphorin3A regulates neuronal polarization by suppressing axon formation and promoting dendrite growth. Neuron. 2011;71:433–446. doi: 10.1016/j.neuron.2011.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Duflocq A, Chareyre F, Giovannini M, Couraud F, Davenne M. Characterization of the axon initial segment (AIS) of motor neurons and identification of a para-AIS and a juxtapara-AIS, organized by protein 4.1B. BMC Biol. 2011;9:66. doi: 10.1186/1741-7007-9-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arber S, et al. Requirement for the homeobox gene Hb9 in the consolidation of motor neuron identity. Neuron. 1999;23:659–674. doi: 10.1016/s0896-6273(01)80026-x. [DOI] [PubMed] [Google Scholar]
- 31.Huber AB, et al. Distinct roles for secreted semaphorin signaling in spinal motor axon guidance. Neuron. 2005;48:949–964. doi: 10.1016/j.neuron.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 32.Camu W, Henderson CE. Purification of embryonic rat motoneurons by panning on a monoclonal antibody to the low-affinity NGF receptor. J Neurosci Methods. 1992;44:59–70. doi: 10.1016/0165-0270(92)90114-s. [DOI] [PubMed] [Google Scholar]
- 33.Ashrafi S, et al. Wnt7A Identifies Embryonic -Motor Neurons and Reveals Early Postnatal Dependence of -Motor Neurons on a Muscle Spindle-Derived Signal. J Neurosci. 2012;32:8725–8731. doi: 10.1523/JNEUROSCI.1160-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brumovsky P, Watanabe M, Hökfelt T. Expression of the vesicular glutamate transporters-1 and −2 in adult mouse dorsal root ganglia and spinal cord and their regulation by nerve injury. Neuroscience. 2007;147:469–490. doi: 10.1016/j.neuroscience.2007.02.068. [DOI] [PubMed] [Google Scholar]
- 35.Brumovsky PR. VGLUTs in Peripheral Neurons and the Spinal Cord: Time for a Review. ISRN Neurology. 2013;2013:1–28. doi: 10.1155/2013/829753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alvarez FJ, Villalba RM, Zerda R, Schneider SP. Vesicular glutamate transporters in the spinal cord, with special reference to sensory primary afferent synapses. J Comp Neurol. 2004;472:257–280. doi: 10.1002/cne.20012. [DOI] [PubMed] [Google Scholar]
- 37.Mitra P, Brownstone RM. An in vitro spinal cord slice preparation for recording from lumbar motoneurons of the adult mouse. J Neurophysiol. 2012;107:728–741. doi: 10.1152/jn.00558.2011. [DOI] [PubMed] [Google Scholar]
- 38.Behar O, Golden JA, Mashimo H, Schoen FJ, Fishman MC. Semaphorin III is needed for normal patterning and growth of nerves, bones and heart. Nature. 1996;383:525–528. doi: 10.1038/383525a0. [DOI] [PubMed] [Google Scholar]
- 39.Averill S, McMahon SB, Clary DO, Reichardt LF, Priestley JV. Immunocytochemical localization of trkA receptors in chemically identified subgroups of adult rat sensory neurons. Eur J Neurosci. 1995;7:1484–1494. doi: 10.1111/j.1460-9568.1995.tb01143.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ozaki S, Snider WD. Initial trajectories of sensory axons toward laminar targets in the developing mouse spinal cord. J Comp Neurol. 1997;380:215–229. [PubMed] [Google Scholar]
- 41.Liu Y, Ma Q. Generation of somatic sensory neuron diversity and implications on sensory coding. Curr Opin Neurobiol. 2011;21:52–60. doi: 10.1016/j.conb.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Freeman MR, Rowitch DH. Evolving concepts of gliogenesis: a look way back and ahead to the next 25 years. Neuron. 2013;80:613–623. doi: 10.1016/j.neuron.2013.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nagai M, et al. Astrocytes expressing ALS-linked mutated SOD1 release factors selectively toxic to motor neurons. Nat Neurosci. 2007;10:615–622. doi: 10.1038/nn1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen AI, de Nooij JC, Jessell TM. Graded activity of transcription factor Runx3 specifies the laminar termination pattern of sensory axons in the developing spinal cord. Neuron. 2006;49:395–408. doi: 10.1016/j.neuron.2005.12.028. [DOI] [PubMed] [Google Scholar]