Abstract
A systematic study involving the use and optimization of gas phase modifiers in quantitative differential mobility- mass spectrometry (DMS-MS) analysis is presented using mucleoside-adduct biomarkers of DNA damage as an important reference point for analysis in complex matrices. Commonly used polar protic and polar aprotic modifiers have been screened for use against two deoxyguanosine adducts of DNA: N-(deoxyguanosin-8-yl)-4-aminobiphenyl (dG-C8-4-ABP) and N-(deoxyguanosin-8-y1)-2-amino-l-methyl-6-phenylimidazo[4,5-b]pyridine (dG-C8-PhIP). Particular attention was paid to compensation voltage (CoV) shifts, peak shapes and product ion signal intensities while optimizing the DMS-MS conditions. The optimized parameters were then applied to rapid quantitation of the DNA adducts in calf thymus DNA. After a protein precipitation step, adduct levels corresponding to less than one modification in 106 normal DNA bases were detected using the DMS-MS platform. Based on DMS fundamentals and ab-initio thermochemical results we interpret the complexity of DMS modifier responses in terms of thermal activation and the development of solvent shells. At very high bulk gas temperature, modifier dipole moment may be the most important factor in cluster formation and cluster geometry in mobility differences, but at lower temperatures multi-neutral clusters are important and less predictable. This work provides a useful protocol for targeted DNA adduct quantitation and a basis for future work on DMS modifier effects.
Introduction
Differential Mobility is a rapidly evolving new analytical technology which offers rapid gas phase ion separation/ filtration prior to mass analysis, and the advantages of this platform such as improved signal to noise ratio, separation of closely related compounds, and removal of interferences have been well demonstrated[1–6].
The mobility of charged ion species in an applied electric field forms the basis for ion separation in a DMS cell. Krylov and Nazarov [7] have studied three different models of ionneutral interactions in the applied electric field: (a) rigid sphere scattering, (b) long-range ion–dipole attraction and (c) clustering. They concluded that ion-neutral clustering is the most relevant phenomenon to explain the dependence of ion mobility on field strength that is the source of DMS selectivity. The mobility of ions between the electrodes in a differential mobility cell has been well described in a paper by Schneider et al [4] and more extensively in monographs by Shvartsburg [8], Eiceman and Karpas[9], and in a discussion on the fundamentals of ion mobility by Mason and McDaniel [10]. We present here a brief discussion of these principles that provides a framework for understanding the role of modifiers in DMS separations as they particularly pertain to the analysis of selected model DNA adducts, biomarkers indicative of DNA damage from carginogens.
Ion mobility as used in DMS is a high pressure phenomenon in which ions in an electric field quickly reach a limiting speed, described by a field-dependent ion mobility coefficient, K(E),
| (1) |
where v⃗d(E) is the drift velocity of the ion, E the electric field strength, and α(E) contains the deviation from low-field behavior, and is known as the alpha parameter, or the differential mobility. K(0) is the low-field mobility coefficient, with a dependence on the ion’s chemical identity, but also depending on pressure, temperature, and chemical environment. Low field mobility, K(0), is the property measured in ion mobility spectrometry (IMS) under the same conditions. However, the ion separations used in differential mobility spectrometry (DMS) are determined solely by the differential mobility parameter, α(E), [11, 12] because of the DMS filter condition which determines the relationship between the DMS separation peak voltage, SV , and the DMS compensation voltage, CoV, for a particular DMS waveform shape, f(t) [13]. Ion transmission in a planar DMS geometry requires that the ion remain along the axis of the DMS analytical region after each f(t) waveform cycle of period Tf,
| (2) |
As a result of this filter condition, there is complete equivalence between the observed DMS compensation voltages (CoV) for a range of field amplitudes (SV) and the differential mobility, α(E) [4].
The mobility coefficient, K(E), acquires a dependence on field strength, E = |E⃗|, as a result of interactions of the ion with its chemical environment [4, 7, 14], and also varies with pressure and temperature in the DMS analytical region [15, 16]. Ion mobility loses its dependence on pressure if the field used in ion mobility is expressed in density-normalized Townsend units for the electric field [10]:
| (3) |
where ETd = E/N is the ion mobility field in Townsend units [15](1 Td = 10−17 volt/cm2). Density scaling provides pressure correction for analytical instruments such as the AB SCIEX SelexION system [17] , but is not useful for temperature variation. The ion mobility coefficient, even under cluster-free conditions, still retains a dependence on temperature that can be minimized but not eliminated by temperature scaling. Temperature scaling leads to a reduced mobility, K0 (corrected to 0°C, 1 atm) that still depends weakly on temperature, but that variation is dwarfed in DMS by dynamic cluster size variation. Although it is not important in low pressure IMS, or in traveling wave IMS [18, 19], the importance of clustering is well known in atmospheric pressure IMS, where a cluster-free mass-mobility correlation can only be obtained by the use of high drift tube temperatures, typically 200°C or higher [9, 20–26]. Except in special cases, low pressure IMS, and travelling wave IMS, operate with pure, non-polar drift gases and determine accurate cross-sections for a wide range of molecules, while DMS modifier effects depend on polar molecules with a long range attractive potential [27].
The dependence on chemical composition of the transport gas has been found to be the largest contributor to differential mobility. This is both a hinderance and an advantage. Ions of similar molecular weight and related structure can usually be separated with the assistance of a modifier and peak capacity is greatly increased, but the underlying cause is still obscure. While useful for improved resolution and reduced chemical noise, the modifier effect is the most difficult to predict; however, it is important to recognize that the mobility coefficient is directly related to the effective size of the ion, whether it is a bare ion or a ion-neutral cluster (effective size is referred to as the cross-section for collisions, between the ionic species and molecules in the neutral transport gas). The simplest expression for the mobility coefficient that presents that connection in a way that is valid even at the very high DMS field strengths is from the two-temperature theory in momentum-transfer form (section 6-2.C (eq. 6-2-25), [10])
| (4) |
In this expression, µ is the ion-neutral reduced mass (µ =mM/(m+M), m the ion mass, and M the neutral mass), vrel is the well-known expression for the relative speed of particles in a Maxwell-Boltzmann distribution, while Ω is an average cross-section. Other symbols in equation (4) are N (gas density), q (ion charge magnitude), kB (Boltzmann constant), and αK (a small correction < 0.02). From this it is clear that the ion mobility is inversely related to the loss of momentum by collisions, since the momentum, µvrel (Teff), multiplies a cross-section Ω̄(Teff) giving a density-scaled rate. Both relative speed and cross-section are evaluated at a special temperature, Teff, that varies dynamically with the DMS field and depends jointly on the bulk temperature, pressure, and electric field.
Early on in the application of DMS technology, it was realized that the use of modifiers in the transport gas is an important step for analytical applications [28, 29]. Prior works from our laboratory and several others have demonstrated the advantages of introducing drift gas modifiers to aid in DMS separations [30, 31]. In a recent publication by Schneider et al., the effects of transport gas modifiers have been extensively documented with particular attention to resulting peak capacity [32] and other groups are active as well in areas like Green Chemistry [31, 33]. These results are part of a growing interest in the use of modifiers with the DMS-MS platform as it offers a promising alternative for rapid analytical applications as well as possible field applications such as those proposed for NASA applications [34, 35].
Our group has long been involved in the development of DMS technology for analytical applications [14, 30, 36] and in recent years, we have been particularly interested in the use of DMS-MS for quantitative analysis with reduced sample cleanup and minimal or zero LC time [37]. There are a number of advantages to the use of DMS in rapid quantitative biomarker analysis, many previously discussed, as in Coy et al. [2], but one point is of special interest when DMS-MS quantitative analysis is compared to LC-MS: LC-MS as well as drift-time or travelling-wave IMS require integration of an ion current over an elution or drift time profile, but DMS-MS intensities for quantitation by infusion are obtained continuously by monitoring the CoV for peak ion transmission. Under conditions of flow injection or LC-DMS-MS, a time profile is integrated, but without scanning CoV [38, 39]. The DMS analytical region is a continuous ion filter that transmits with minimum diffusion losses at characteristic (SV,CoV) values. Because the width of the peak in CoV units is inversely related to the low field ion mobility of the ion, integrating over the DMS peak width would contaminate the observed data with an unknown scale factor, one which has a variable environmental dependence. Thus, DMS at the peak CoV value provides a continuous measurement of ion concentration, limited in response time only by the transit time of the ion through the DMS analytical region, and the agility of the electronics of the mass spectrometer.
Prior works from our laboratory have demonstrated that the judicious use of modifiers provided conditions for rapid quantitation of selected analytes in matrices of varying complexities [36, 37]. We have shown that, in addition to the rapid analysis time afforded by use of the DMS, sample preparation steps can be minimized or even eliminated by taking advantage of the unique post-electrospray selective ion filtration capabilities. As with any other analytical platform, proper method development is necessary to achieve optimal sensitivity and efficacy. In the work presented here, we demonstrate the process of stepwise evaluation of modifier effects from a method development perspective in order to establish the optimal conditions for quantitation of selected DNA damage biomarker analytes by differential mobility- mass spectrometry.
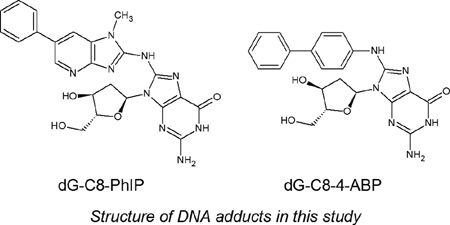
DNA adducts provide direct evidence of genetic exposure and damage in cells and monitoring their levels from a health perspective is important. So-called bulky DNA adducts share common structural features in that, other than the carcinogen, they all contain a deoxyribose moiety along with the nucleobase, typically a guanine. In ths regard, the compounds selected for this study provide excellent models for the purpose of evaluating the DMS conditions that might be appropriate for the DMS – MS analysis of this class of analytes. N-(2-deoxyguanosine-8-yl)-4-ABP (dG-C8-4-ABP), the deoxyguanosine adduct of the bladder carcinogen 4-aminobiphenyl (4-ABP) is a known carcinogen found in cigarette smoke, paints, food colors, hair dyes and fumes from heated oils and fuels[40–43]. The heterocyclic aromatic amine 2-amino-1-methyl-6-phenylimidazo[4,5]pyridine (PhIP) is found in grilled meats [44]. PhiP is a known foodborne carcinogen which has been implicated in mammary gland tumors in rodents [45]. These two adducts have been related to lifestyles of people and we have used them as model analytes to explore and optimize the gas phase interactions in DMS and develop differential mobility- mass spectrometry as a rapid quantitative platform.
Experimental Section
Chemicals and Standards
Caution: 2-Hydroxyamino-1-methyl-6-phenylimidazo[4,5]pyridine and 4-Aminobiphenyl and its derivatives are carcinogenic and should be handled carefully.
Calf-thymus DNA, nuclease p1 from Penicillium citrinium, deoxyribonuclease 1 (DNase I) type 2 from bovine pancreas, alkaline phosphatase from Escherichia coli (type IIIs), ethanol, dimethyl sulfoxide (DMSO) were purchased from Sigma–Aldrich Chemical Co. (St. Louis, MO). Snake venom phosphodiesterase was purchased from USB Corporation (Cleveland, OH). N-(2-deoxyguanosine-8-yl)-4-ABP (dG-C8-4-ABP) was purchased from Toronto Research Chemicals (Toronto, ON, Canada, D239600, CAS 84283-08-9, C22H22N6O4). N-(deoxyguanosine-8-yl)-2-amino-1-methyl-6-phenylimidazo[4,5-β]pyridine (dG-C8-PhIP) were purchased from Toronto Research Chemicals (Toronto, ON, Canada, D239630, CAS 142784-25-6, C23H23N9O4). Formic acid solution was purchased from Sigma–Aldrich Chemical Co. (St. Louis, MO). HPLC grade water and methanol were purchased from Fisher Scientific (Fair Lawn, NJ).
DNA quantification, enzymatic digestion and protein precipitation
DNA quantification was done using an Invitrogen Corporation (Carlsbad, CA) Quant-IT™ double strand (ds) DNA BR Assay kit with a Qubit fluorometer. Aliquots containing 2 ug DNA (dissolved in 5mM Tris-Cl/ 10 mM ZnCl2) were removed for digestion and analysis for each sample point.
Calf thymus DNA was hydrolyzed similarly to a method previously described[46]. Samples were incubated at 98 °C for 3–5 min and chilled in the freezer down to room temperature. 0.3 units of nuclease P1 (0.3 units µL−1 solution of 5 mM Tris–Cl, pH 7.4) and 3.1 Kunits of DNase I (1 µg µL−1 solution in 5 mM TRIS/10 mM MgCl2, pH 7.4) were then added per µg of DNA and incubated in a water bath maintained at 37 °C. After 5 hours, 0.003 units of phosphodiesterase (100 ng µL−1 in 5 mM TRIS/10 mM MgCl2, pH 7.4), and 0.002 units of alkaline phosphatase per µg of DNA were added and the mixture was further incubated at 37 °C for 18 h.
Protein precipitation was done by adding five volumes of ice cold ethanol and centrifuging at 10000 rpm for 15 minutes. The samples were dried down and stored at −80°C until analysis.
Instrumentation
The fundamental goals of the proposed approach to the optimization of modifiers in quantitative analysis by DMS-MS was tested using two different planar DMS systems, one attached to a 3-D ion trap (Thermo Finnigan LCQ Classic) (San Jose, CA) and the second interfaced to an AB SCIEX API 3000 triple quadrupole MS [AB SCIEX, Framingham, MA]. The DMS filters varied only in terms of their dimensions, and this comparison provided an assessment of the general effect and applicability of the DMS configuration to such analyses.
DMS-Ion Trap Mass Spectrometer
A planar DMS developed by Sionex Corporation (Bedford, MA) with a filter gap 0.5mm high × 3.0mm wide × 10.0mm long which was positioned at the entrance of the heated capillary of a Thermo-Finnigan, LCQ Classic mass spectrometer was used for the work . Sionex Expert software was used to set the DMS parameters. The SV could be set at zero or scanned in the range from 500 V to 1500 V, and the CoV could be set or scanned from −43 to +15 volts. Although the system in use here is no longer commercially available, the underlying electronics technology has been described, and successor commercial instrumentation (SelexION, AB SCIEX) has become commercially available.
Electrospray was performed using coated 10 µm PicoTip emitters from New Objective (Woburn, MA). The syringe was connected to the emitter tip using a 150 um (ID) capillary tubing and the ESI voltage was applied to the union at the liquid-liquid junction between the capillary and the tip. Samples were introduced at the rate of 300 nl/min using a Harvard Apparatus syringe pump (Holliston, MA). The desolvation gas (ultra high purity nitrogen) was introduced at a flow rate of approximately 100 cc/min into the desolvation region at a temperature of 100°C. The vacuum drag of the mass spectrometer was measured to be 500 cc/min. External air flow of approximately 400 cc/min also merged in with the desolvation gas into the DMS. The bulk gas temperature was estimated to be 45°C. Modifiers were introduced into the desolvation region along with the nitrogen gas. The electrospray emitter voltage was held at 2 kV throughout the analysis.
DMS-Triple Quadrupole Mass Spectrometer
A prototype DMS- API 3000 Triple Quadrupole mass spectrometer (AB SCIEX, Concord, Ontario. Canada) which has an integrated DMS filter online in front of the vacuum orifice was used for this study. The dimensions of the DMS analytical region were 1 mm × 10 mm × 15 mm. A modified version of the software Analyst version 1.5 which included parameters for SV and CoV was used. The SV could be varied from 0 to 5000 volts and the CoV could be varied from −100 to +100 volts. Electrospray was performed using stainless steel 30 µm id emitter from Proxeon (Thermo Fisher,West Palm Beach, FL). Samples were introduced at the rate of 400 nl/min using a Harvard Apparatus syringe pump. Electrospray was held constant at 3500 volts throughout the analysis. Modifiers were introduced into the curtain gas (nitrogen, 1.1 L/min, 600 cc/min vacuum drag 500 cc/min curtain gas outflow at 40° C) using a second Harvard Apparatus syringe pump.
DMS Electric Fields
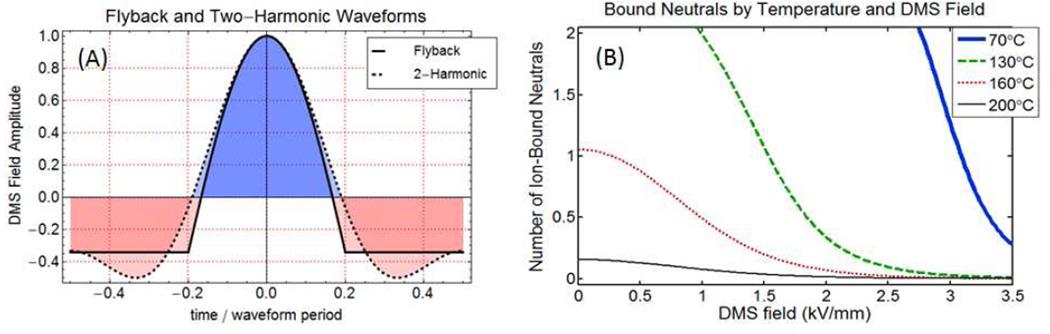
The principal types of waveforms in use for DMS are described in a technical instrumentation paper by Krylov et al. [13], and include a flyback waveform consisting of a truncated sinusoidal pulse connected to a flat baseline, and a two-harmonic design in which phased first and second harmonic voltages are applied to opposing electrodes. The waveform applied on the ion trap MS was of the flyback type at 1.25 MHz where the voltages are applied across a 0.5 mm gap, and are measured in terms of zero to peak separation voltages with CoV =0. The waveform applied in the DMS-API-3000 and on the commercial AB SCIEX SelexION systems is a two-harmonic design applied across a 1.0 mm gap, with frequencies 3 MHz and 6 MHz in 2:1 voltage ratio and the separation voltage measured as peak to peak. Because of the differing gap dimension, and the reporting of flyback SV as zero-to-peak and 2-harmonic SV as peak-to-peak, there is a factor of 3 between the same-field flyback and two-harmonic SV voltage values. For instance, the ion trap system can apply a peak voltage of up to 1500 V (zero-to-peak) over the 0.5 mm gap. The DMS-API 3000 at a setting of 4500 volts SV (peak-to-peak) applies the same peak field to the 1.0 mm DMS region. Flyback and two-harmonic shapes are the most feasible electronics designs for DMS, and have similar DMS efficiencies, with the flyback shape slightly superior, depending on α(E). Flyback and two-harmonic fields at identical peak voltage values are illustrated in Figure 6A, in the discussion section, where clustering mechanisms are considered in more detail.
Figure 6.
(A) Flyback and two-harmonic waveforms. The two-harmonic shape is used by AB SCIEX in the DMS API 3000, and the flyback shape on the ion trap. (B) Using ab-initio thermochemical values, the mean number of bound neutrals is shown as a function of DMS field and transport gas temperature at 1 atm. The maximum neutral count is limited to 3 because shell thermochemistry has not been calculated. These results for R-alphamethylhistamine, which has a polar core similar to dG, with 1.5% isopropanol show that bulk gas temperature is an essential controlling parameter in modifier DMS selectivity and that multiple clustering is common even at relatively high bulk gas temperatures.
RESULTS AND DISCUSSION
Effect of modifiers on CoV shifts
In an effort to understand and characterize the gas phase molecular interactions, varying percentages of modifiers were introduced into the transport gas and the CoV shifts were recorded with increasing separation voltage. Voltages corresponding to the apex of extracted ion chromatograms are reported here as CoV values. The goal of using the modifiers in this work is to shift the analyte of interest selectively to a CoV value free of interferences where DMS can be exploited to perform rapid quantitation in the presence of a complex matrix.
Effect of Modifier on CoV of dG-C8-4-ABP
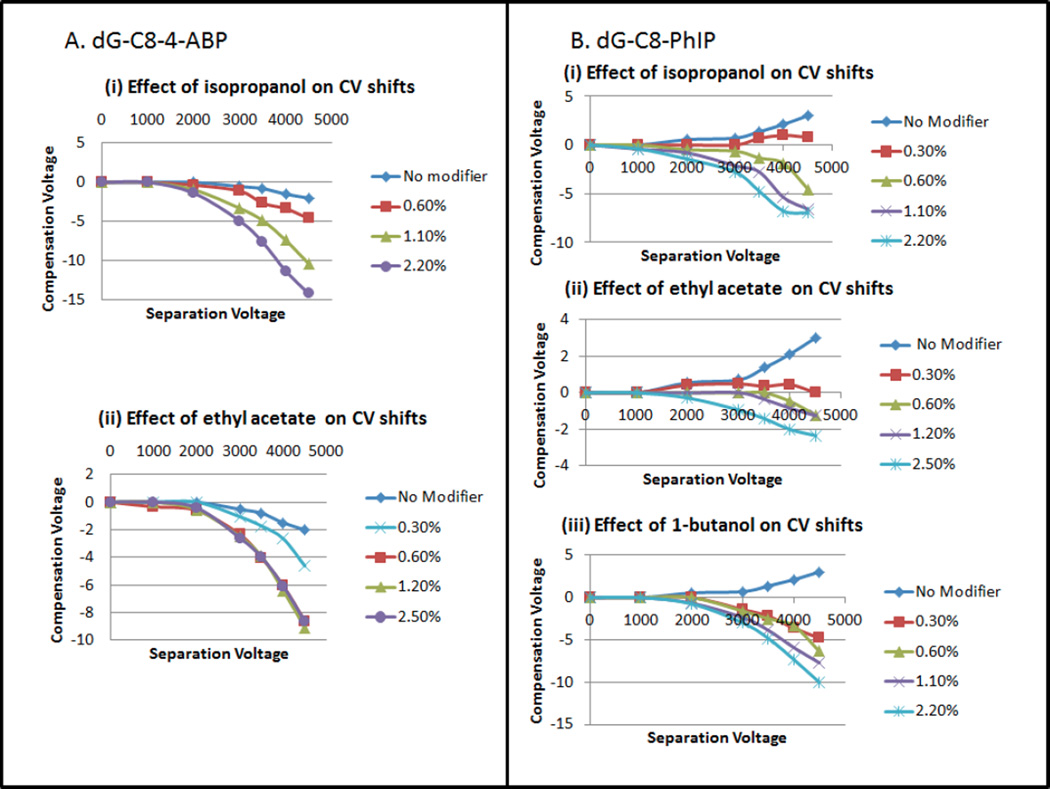
The clustering effects of two gaseous modifiers of differing size and polarity, ethyl acetate (C4H8O2, mw 88.11, µ=4.325 D, Q=−6.619 D·Ǻ) and isopropanol (C3H8O, mw 60.10, µ=1.560 D, Q=3.242 D·Ǻ) [47], on dG-C8-4-ABP at varying concentrations were investigated by monitoring the CoV shift as a function of separation voltage (SV). As shown in Figure 1A, in the absence of a modifier, the analyte ion does not exhibit any shifts in CoV at SV values up to 2000 volts. Even when the SV is increased beyond 2000 volts, the CoV starts shifting only slightly towards the negative voltage reaching a maximum of −2 volts at SV=4500 volts.
Figure 1.
(A): Effect on compensation voltage of varying concentrations of modifiers (i) Isopropanol and (ii) Ethyl acetate on dG-C8-4-ABP CoV shifts. (B) Effect of varying concentrations of modifiers (i) Isopropanol, (ii) Ethyl acetate, (iii)1-butanol on dG-C8-PhIP CoV shifts
With ethyl acetate as a modifier (Figure 1A(ii)), dG-C8-4-ABP shifts rapidly to negative CoV values, as expected for charge-dipole interactions[7], reaching −5V at only 0.30% ethyl acetate. Interestingly, at higher ethyl acetate concentrations of 0.6%, 1.20% and 2.50%, identical CoV responses are observed, as indicated by the overlapping CoV curves. This strongly suggests that steric and thermodynamic effects limit the maximum number of coordinated ethyl acetate molecules to a value which is reached between 0.3% and 0.6% by volume, under the AB-SCIEX 3000 triple-quad conditions. It can also be noted that the CoV shifts for all SV values were intermediate between those of no modifier and higher modifier concentrations.
With isopropanol as a modifier, in comparison to ethyl acetate, the CoV value of the adduct shifted to even greater negative values with isopropanol than for ethyl acetate, and the effect was even more substantial with increasing concentrations (Figure 1A(i)). For example, going from no modifier to progressively higher modifier concentrations of 0.60% , 1.10% and 2.20%, the respective CoV values at SV=4,500V increased to −4.6, −10.4 and −14.2 V. For isopropanol the greatest CoV shift observed at 2.20% was −14.2 volts compared to ethyl acetate’s maximum shift of −9 volts. The observation of a greater shift toward negative CoV values with isopropanol than with ethyl acetate, even though isopropanol is of lower molecular weight and of smaller geometric cross-section, can be interpreted in different ways. On one hand, it might indicate that the limiting coordination number with isopropanol is greater than with ethyl acetate, or, alternatively, thermochemical effects related to the free energy changes on cluster formation, and the related role of hydrogen bonding in cluster formation in the two systems may be important. This is discussed further below.
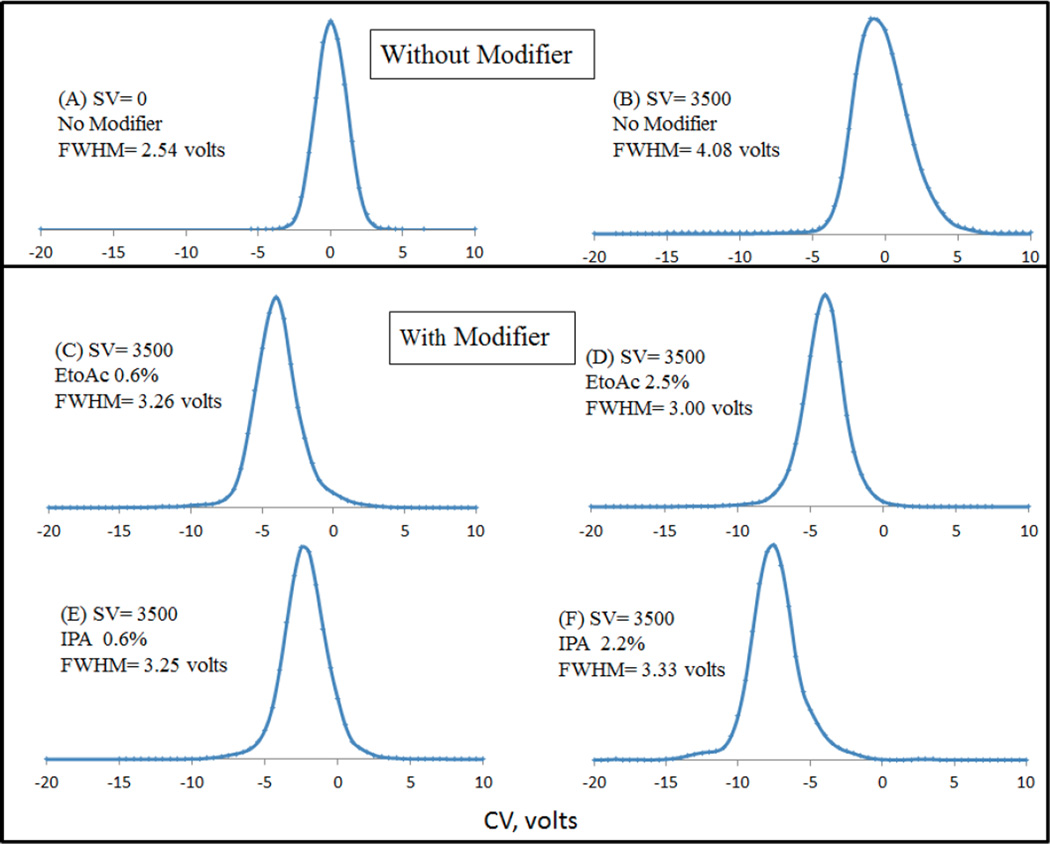
It has been reported that the modifier effect is sensitive to small variations in ion structure, so that even closely related structures may be separated (citrate/isocitrate, [2]; ephedrine / pseudoephedrine [3], and others [48]). Prior works in the field of DMS have established the importance of modifiers to not only enhance CoV shifts, i.e. selectivity but, using the analogy to chromatography, to improve peak capacity in the DMS analysis [4], as well as to suppress chemical noise. We thus next examined the peak shapes of the analyte of interest dG-C8-4-ABP at different DMS conditions. Selected extracted ion chromatograms from the CoV scan are shown in Figure 2. For dG-C8-4-ABP, the improvements in peak shape by the introduction of modifiers can be understood by comparing the FWHM (full width at half maximum) at a constant SV of 3500 volts in Figure 2 (no modifier: Figure 2B, Ethyl acetate modifier at 0.6%: Figure 2C and Isopropanol modifier at 0.6%: Figure 2E). The FWHM without modifier is ∼4 volts compared to ∼3 volts in presence of ethyl acetate and ∼3 volts in presence of isopropanol, which highlights the role of DMS and gas phase clustering in the presence of modifiers on peak shape improvements during DMS analysis. It should also be pointed out that that the FWHM increased initially by 1.5 volts when the SV was set to 3500 volts (Figure 2B) as compared to zero SV (Figure 2A) which was subsequently improved by introducing modifiers. Much of this effect may be due to DMS suppression of chemical noise due to ions of higher molecular weight, which are detected with a wider peak width due to lower mobility.
Figure 2.
Comparison of peak shapes of dG-C8-4-ABP with and without modifier. Improvements in peak shape by the introduction of modifiers can be realized by comparing the FWHM (full width at half maximum) at a constant SV of 3500 volts
It is interesting to point out the effect of increasing modifier percentages on peak shape. Specifically, when ethyl acetate modifier is introduced at 2.5% (Figure 2D) which is roughly 4x the amount of the same modifier introduced at 0.6% (Figure 2C), not only do the CoV shifts overlap, but the peak shapes look very similar which supports our notion of “saturation effect” as discussed above. However, a 4-fold increase in the isopropanol concentration (Figure 2E and Figure 2F) also shows that the peak shape can begin to suffer with larger CoV shifts. It is reasonable to surmise that formation of larger clusters with increasing isopropanol concentration may have caused the transmission of analyte ion dG-C8-4-ABP over a slightly larger CoV range as expected from the higher cluster molecular weight (DMS peak width ∼ (1/K(E)). The broadening effect is seen to be greater near the baseline than at half-height, as would be expected for some fraction of larger clusters with broader peaks, that are in rapid equilibrium with single adducts. It is therefore deemed necessary to optimize the modifier percentage being introduced to avoid excessive peak broadening during DMS-MS analysis.
Effect of Modifier on CoV of dG-C8-PhIP
The principal DNA adduct of PhIP, a diet related carcinogen, is also formed by covalent bonding at the C-8 position of guanine. In that sense this adduct provides an interesting example for comparison with dG-C8-4-ABP since the two compounds share the 2’-deoxyguanosine structure (C10H13N5O4, mw = 267.2413) and differ since the PhIP moiety is larger, richer in heteroatoms, and more polar than ABP. In evaluating the modifier effects on CoV, ethyl acetate and isopropanol were tested as before, and 1-butanol (C4H10O, mw= 74.1216, µ=1.584 D (1.660 D expt.) [47] ) was also screened for use in DMS-MS analysis of the adduct and the results are presented in Figure 1B.
In the absence of a modifier, the dG-C8-PhIP CoV values started shifting in the positive direction reaching +3 volts at SV value of 4500 volts (Figure 1B). As discussed by Krylov, a positive CoV shift is associated with short-range collisions, while long-range, charge–dipole interactions result in negative CoV shifts. Upon the introduction of modifiers, the CoV values reversed direction to the negative side and this trend increased significantly as a function of modifier concentrations and SV. Ethyl acetate was the least effective in shifting the CoV to a negative direction while 1-butanol exhibited larger shifts than isopropanol, reaching a maximum peak value of −10 volts at 2.20%. It is conceivable that the stronger action of 1-butanol compared to isopropanol might be attributable to the larger geometric size of 1-butanol, to the end position of the -OH which increases the cluster profile, and to lower steric inhibition for the end-attached –OH by the alpha methyl group, especially since the dipole moments of the two alcohols are nearly identical and thus have the same long-range attractive interaction with an ion.
Also especially interesting was the limited effect exhibited by ethyl acetate with a maximum peak value of −2.2 volts at 2.50%. This may be interpreted in two radically different ways: (1) difference in hydrogen-bonding donor-acceptor characteristics may cause ethyl acetate (acceptor) to cluster less strongly than the alcohols (donor), or (2) the much greater dipole moment of ethyl acetate and the greater polarity of the PhIP part of dG-C8-PhIP may cause the clustering with dG-C8-PhIP to be so strong that the complex remains fully saturated throughout the DMS waveform, quenching the differential mobility effect. Ab-initio calculations of Gibbs free energies of cluster formation do show significantly stronger binding with ethyl acetate than with isopropanol as expected from the charge-dipole interaction (manuscript in preparation). The stronger effect of 1-butanol compared to isopropanol may be due to the larger geometric size of 1-butanol so that the cross-section increases more on binding 1-butanol. Steric inhibition of the methyl groups located alpha to the hydroxylic carbon might also play a role in determining the free energy change on complex formation since isopropanol is disadvantaged by the mid-chain position of the hydroxyl group.
Use of modifiers in separation
The ability of gaseous modifiers to control the compensation voltage of an analyte provides the means for enhancing separations during DMS analysis. Even more importantly, in a targeted analysis, control of the CoV shift can provide a means for isolating the analyte of interest into a transmission zone that is free of matrix interferences, thereby improving signal to noise ratio and sensitivity. In the studies of the biomarkers of DNA damage, the matrix is a DNA enzymatic digest comprised of normal deoxynucleosides and enzyme-related breakdown products. Ensuring the separation of the adduct from normal (unmodified) nucleosides is of particular importance since they have the common deoxynucleoside functional group.
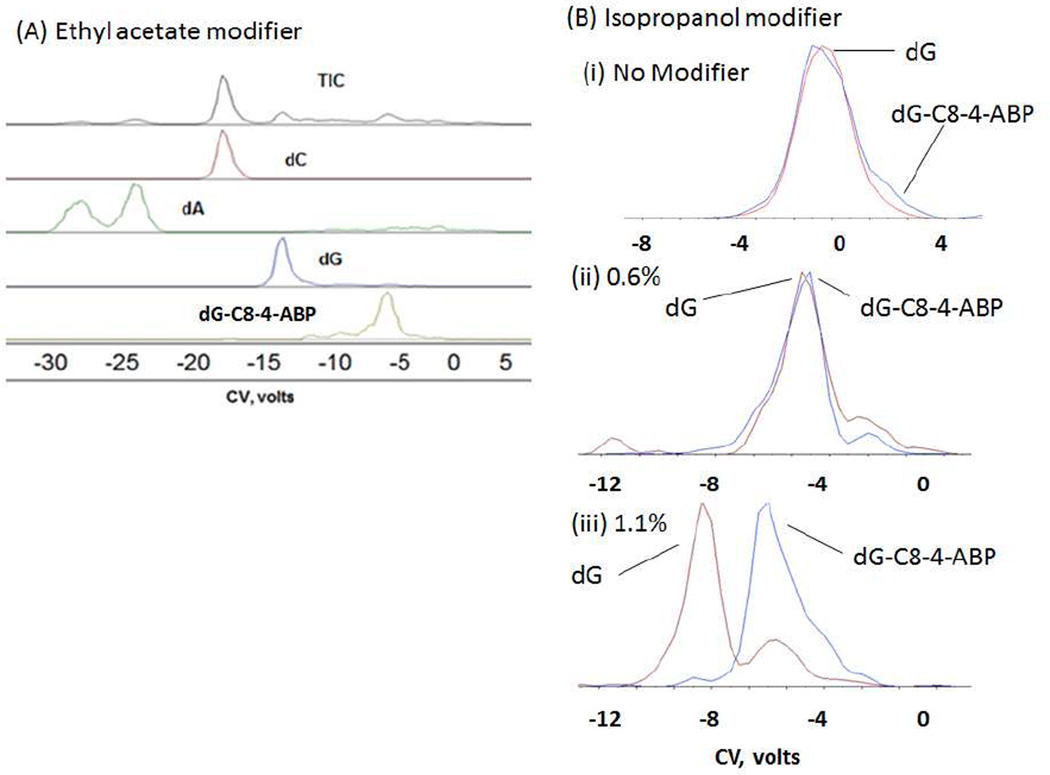
As an initial test, a mixture of 1 ng of each deoxyguanosine (dG), deoxycytosine (dC), deoxyadenosine (dA) and 100 pg of the DNA adduct dG-C8-4-ABP was prepared to investigate the role of modifiers on separation. Two modifiers: ethyl acetate and isopropanol were evaluated for separation of the dG-C8-4-ABP adduct from the excess unmodified DNA bases. CoV scans at a fixed SV value were performed and the analytes of interest were extracted from the TIC. It should be noted that effective separations were not observed in the absence of a modifier.
From Figure 3A, we can see that the four component nucleoside mixture is separated well by the DMS using ethyl acetate as the modifier. As expected, dG-C8-4-ABP being the bulkiest molecule in the mixture, moves the least towards the negative compensation voltage compared to other unmodified nucleosides because its mobility (∼1/cross-section) will be less modified by cluster formation. dA appears as a double peak whose characteristics have not been investigated further. Ethyl acetate and isopropanol have been successfully applied as effective modifiers for other applications with DMS, so we decided to screen isopropanol for separation purposes as well with dG-C8-4-ABP. Introduction of isopropanol as the modifier, provided the necessary conditions for separation of the normal nucleosides dA and dC but failed to resolve dG from dG-C8-4-ABP as the latter two were transmitted at the same CoV value. It was therefore decided to ascertain if an increase in modifier concentration could be used in order to achieve desired separations. As can be seen from (Figure 3B(i) and (ii)), dG and dG-C8-4-ABP could not be separated without modifier or when the modifier isopropanol was introduced at a lower rate of 0.6% into the curtain gas. However, when the modifier was introduced at the rate of 1.1%, separation was achieved between dG and dG-C8-4-ABP. The small dG signal at the dG-C8-4-ABP CoV is interpreted as arising from fragmentation of dG-C8-4-ABP to dG after the DMS and in the mass spectrometer transition to vacuum (Figure 3B(iii)). Using the example of dG-C8-4-ABP and unmodified dG, we have demonstrated here that the percentage of modifier being introduced can be used as an another tool in achieving or enhancing separations using a DMS based platform.
Figure 3.
(A) Separation of a mixture of dC, dA, dG and dG-C8-4-ABP in the presence of ethyl acetate modifier at a fixed concentration done on the DMS- ion trap. (B) Separation of dG and dG-C8-4-ABP (i) without modifier, and using IPA modifier at two different modifier concentration done on DMS-triple quadrupole (ii) 0.60% and (iii) 1.10%
Effect of separation voltages and modifiers on signal intensities in MRM mode
In the experiments discussed above, the focus was on the CoV shifts of dG-C8-4-ABP and dG-C8-PhIP. These studies established that the use of organic modifiers can have a dramatic effect on analytical performance. Given our goal of using the DMS-MS platform to perform rapid quantitation of these biomarkers, we examined next the influence of modifiers on signal intensity in order to integrate all of these variables into a comprehensive analytical protocol. The rapid scanning features of the DMS-MS platform for performing rapid quantitation can be best appreciated by using it in the CoV stepping mode and the signal intensities monitored in the MRM mode. Under those conditions, once the CoV apex for any given analyte has been identified, this voltage value can be set for subsequent quantitative analysis. Setting the CoV value allows the user to filter out matrix contaminants and selectively transmit analyte(s) of interest continuously into the mass spectrometer without physically adding time for the mass spectral acquisition. With the DMS-on, signal intensities in the MRM mode were thus monitored for a fixed concentration of each compound to optimize the best DMS conditions for selected analyte ion transmission.
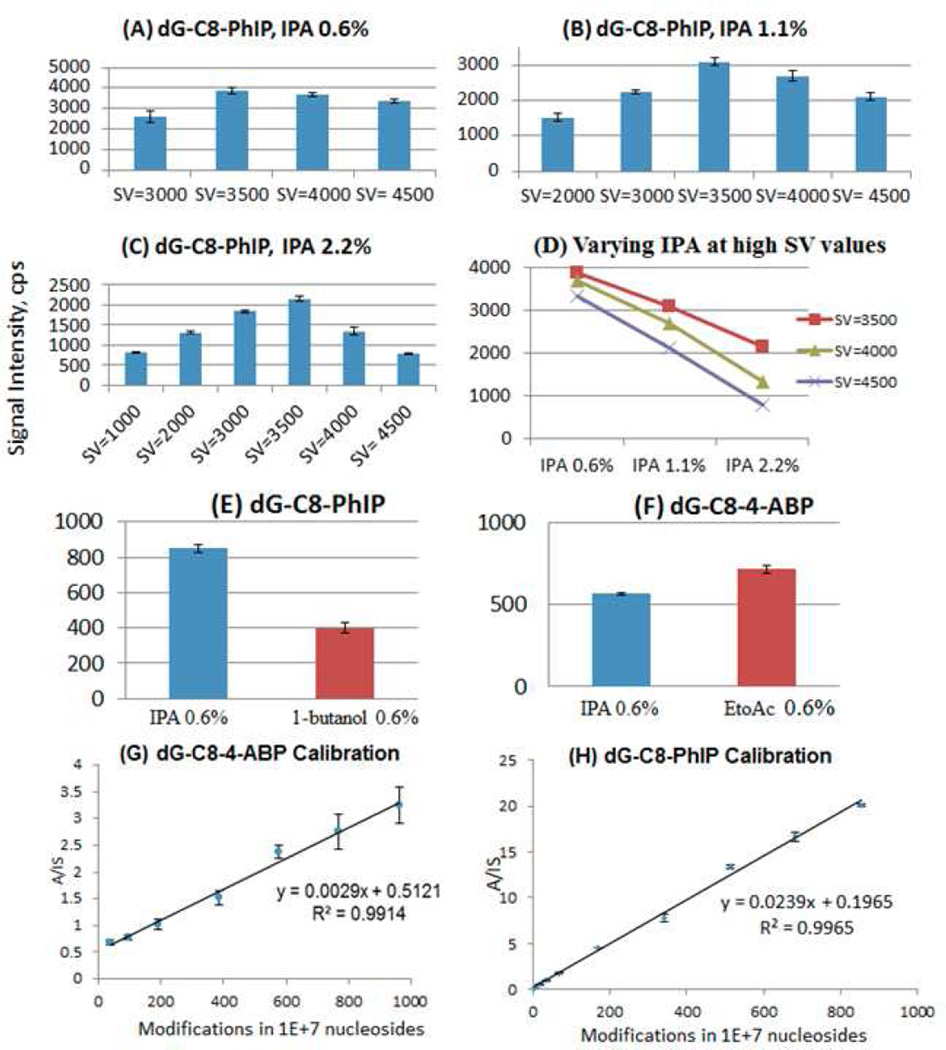
As mentioned above, in light of the variations in CoV shifts as a function of modifier concentration, it was deemed useful to monitor the effect of modifier choice on the product ion signal intensity. Accordingly, samples containing 0.100 picomoles of dG-C8-PhIP were analyzed and, initially, CoV scans were performed at different values of separation voltages and modifier percentages in order to identify the peak apex values. Once these values had been identified, product ion counts were monitored in the MRM mode at the respective operating values of separation voltage, compensation voltage and modifier percentage (only combinations of separation voltages and modifier percentages that exhibited shift away from the zero compensation voltage were investigated). It is shown that, with increasing separation voltages at all three isopropanol modifier percentages (0.6%, 1.1% and 2.2%) examined, the general trend on the intensity of the product ion is the same (Figure 4 A,B,C,D). Specifically, the product ion intensity value increases initially, peaks off at SV=3500 volts and then starts decreasing with application of higher separation voltage. The initial increase with the application of the DMS field is generally described as a DMS focusing effect, but may also involve enhanced desolvation of the electrospray plume due to RF heating by the DMS field. The transmission of the analyte ion through the DMS electrodes is optimal at the separation voltage value of 3500 volts.
Figure 4.
Effect of separation voltage and modifier isopropanol (IPA) concentration on the MS/MS signal intensity of the DNA adduct dG-C8-PhIP (A) at 0.6% (B) 1.1% (C) 2.2% (D) Trends of AB SCIEX DMS API 3000 signal intensities at separation voltages of 3500, 4000, 4500 volts with increasing modifier percentages. Effect of modifiers on the product ion intensity at a fixed separation voltage of 3500 volts: (E) Modifiers Isopropanol (IPA) and 1-butanol on the DNA adduct dG-C8-PhIP & (F) Modifiers Isopropanol (IPA) and Ethyl acetate (EtoAc) on the DNA adduct dG-C8-4-ABP. Calibration curves: (G) dG-C8-4-ABP with ethyl acetate (0.6%), SV(3500V) and (H)dG-C8-PhIP with IPA(0.6%), SV 3500V. Selection of optimum conditions of SV and modifier allow accurate calibration of low adduct concentrations as indicated by calibration curves.
Further increase in separation voltage results in a reduction in analyte ion count which is evident at each modifier concentration, but becomes more pronounced as modifier concentration increases. This has previously been illustrated through the use of a dispersion plot [36], where the drop in signal intensity is shown in 2-D DMS SV-CoV scanning mode. Figure 4(D) shows the comparison of the product ion intensity at separation voltages of 3500, 4000 and 4500 volts with varying isopropanol modifier percentages of 0.6%, 1.1% and 2.2%. Signal intensity of the product ion of dG-C8-PhIP falls both with increased modifier concentration and with SV above 3500 V. This effect can be understood in terms of two contributions: (1) higher modifier contributions result in more frequent ion-modifier collisions, leading to higher probability of loss of charge from the ion or ion cluster, and (2) higher SV results in higher velocity and more energetic and more destructive collisions between the ion and the transport gas, again leading to a higher rate of charge loss, or even to reactive collisions[49].
Referring back to Figure 1B, it may be noted that in the case of dG-C8-PhIP, the shifts have been significantly influenced by both IPA and 1-butanol with the latter giving larger CoV shifts. It was thus important in the context of quantitative analysis to also compare the effect of these two modifiers on the signal intensities. Samples of dG-C8-PhIP (20 femtomoles), using isopropanol and 1-butanol at 0.6% were screened at the optimal separation voltage value of 3500 volts (Figures 4E and 4F). Signal intensities were 830 for isopropanol and 400 for 1-butanol (2.1 : 1). For dG-C8-PhIP, use of isopropanol as the modifier gave much better signal intensity than 1-butanol by more than a factor of 2. For dG-C8-4-ABP, a sample containing 58 femtomoles of dG-C8-4-ABP was analyzed in the MRM mode at a separation voltage value of 3500 volts at 0.6% concentration of modifiers isopropanol and ethyl acetate in two separate runs. Signal intensities were 570 for IPA and 910 for ethyl acetate (1 : 1.60) Thus, for dG-C8-4-ABP, ethyl acetate as the modifier produced 60% higher signal intensity in the MRM mode when compared to isopropanol. The same experiment was repeated at different concentrations of both the analytes (dG-C8-4-ABP and dG-C8-PhIP) and the same trend was observed for all concentrations.
Since generation of ion signal is directly dependent on the abundance of the respective [M+H]+ ions, the simplest assumption is to attribute these differences in signal intensity to a competition for the proton between the analyte ion or ion-neutral clusters and the modifiers. The proton affinity of ethyl acetate is 835.7 kJ/mol, isopropanol is 793.0 kJ/mol and 1-butanol is 789.2 kJ/mol respectively [50]. Had the difference in modifier proton affinity been the dominant factor, then ethyl acetate with a greater proton affinity value than isopropanol would have produced lower signal intensity for dG-C8-4-ABP. For dG-C8-PhIP, although the proton affinity value of 1-butanol is almost the same as that of isopropanol, the difference in signal intensity between the two modifiers is large, with isopropanol giving higher signal intensity. Methyl groups on the isopropanol introduce some steric hindrance that reduce the contribution of H-bonding to the clustering process, whereas the hydroxyl group is more exposed in 1-butanol which may have facilitated clustering. In an alternative model, the fact that isopropanol and 1-butanol have very similar dipole moments, while ethyl acetate dipole moment is much larger could lead to alternative conclusions. None of these approximations can lead to a full and accurate prediction of modifier behavior; only accurate thermochemical free energy values based on all these effects can be expected to be of help. Thus, screening of potential modifiers against the analyte(s) of interest is necessary and can provide users a better understanding of the modifier selection process and optimization of the DMS parameters for analysis. Signal intensities in detail must be examined experimentally, although the general trends with modifier concentration and with SV amplitude are understood.
Quantitation of DNA adducts in calf-thymus DNA
The gas phase modifier selection and optimization studies were performed in order to facilitate rapid quantitation of DNA adducts: dG-C8-4-ABP and dG-C8-PhIP, with the results shown in Figure panels 4(G) and 4(H). Ethyl acetate at 0.6% was established as the modifier of choice for dG-C8-4-ABP analysis and isopropanol at 0.6% was established as the modifier of choice for dG-C8-PhIP analysis. Separation voltage value at 3500 volts was used for analysis of both the adducts. For dG-C8-4-ABP quantitation, the transition from the precursor ion [M+H]+ of dG-C8-4-ABP (m/z 435) to the product ion (m/z 319) [M+H-116]+ was monitored for the analyte and the transition from the precursor ion [M+H]+ of dG-C8-4-ABP-d9 (m/z 444) to the product ion (m/z 328) [M+H-116]+ was monitored for the internal standard respectively. For dG-C8-PhIP quantitation, the transition from the precursor ion [M+H]+ of dG-C8-PhIP (m/z 490) to the product ion (m/z 374) [M+H-116]+ for the analyte and transition from the precursor ion [M+H]+ of dG-C8-4-ABP-d9 (m/z 493) to the product ion (m/z 377) [M+H-116]+ for the internal standard were monitored. 2 micrograms of calf thymus DNA was used as the matrix for each sample point and calibration curves were prepared in the DMS-on mode. The utility of DMS to remove matrix ion interferences to produce a linear calibration curve has previously been demonstrated [37]. DNA modifications of less than 1 in 106 nucleosides were detected with both of the calibration curves showing excellent linearity (R2>0.99) through 1000 modifications in 107 nucleosides after a simple protein precipitation step. It is interesting to note that, whereas the calibration curve for dG-C8-PhIP had essentially a zero intercept, the one for dG-C8-4-ABP crossed the y-axis at a higher value, most likely due to background interferences at the ion masses monitored.
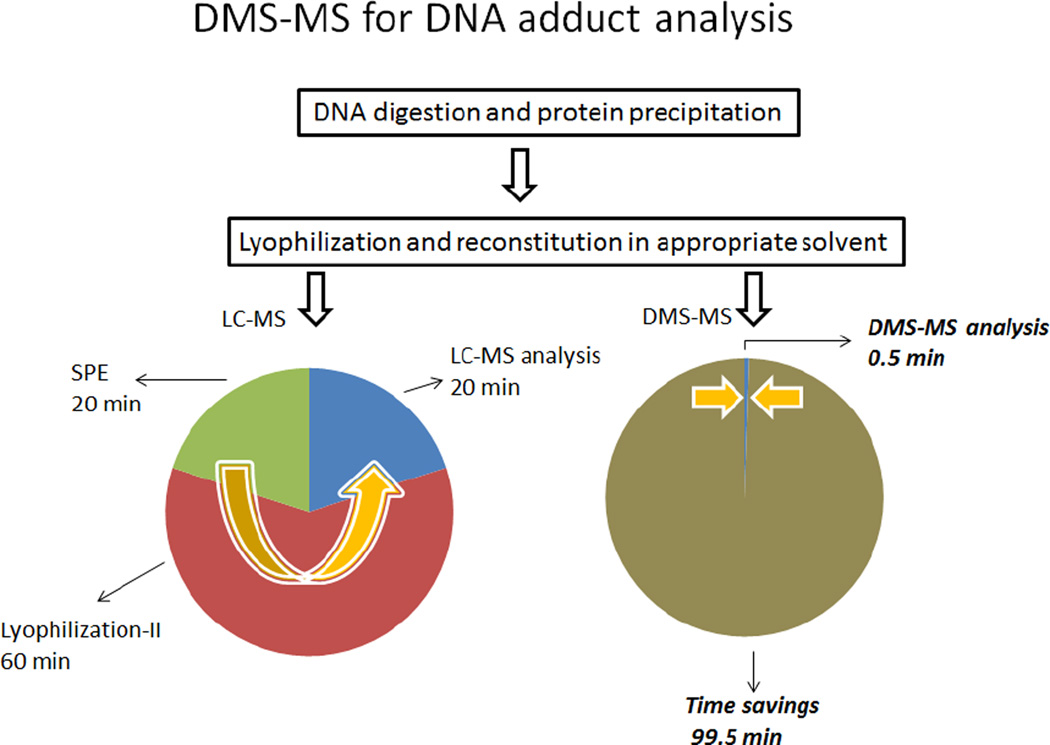
In reviewing the calibration curves in Figure 4, it is important to further consider the broader significance of the results and the implications not only to the field of DNA adduct analysis but also to other related analytical applications that may be conducted by DMS-MS/MS. To begin with, these calibration curves were generated in approximately three hours and consisted of 7–9 points each analyzed in triplicate. In addition, they included several blanks in order to account for any carry-over contamination. This is only a fraction of the time normally consumed using our traditional LC-MS/MS approach in which additional valuable time is also expended waiting for column equilibration before each chromatographic run. Moreover, the value of DMS-MS/MS should also be considered in the context of the broader picture of an analysis of biological specimens which typically involves several sample preparation steps, before injection into the LC-MS. The procedure for the DNA adducts of interest here is outlined in Figure 5 and also shows the approximate time period associated with each one of the steps. As indicated, following a simple protein precipitation lyophilized samples can be reconstituted and directly analyzed by DMS/MS bypassing other preparative steps. In the DMS-on mode, the analyte of interest can be selectively introduced into the mass spectrometer by selecting the appropriate modifier and setting the CV to a unique voltage for that particular compound. Once the parameters for DMS/MS analysis have been set, data were acquired in just 30 s and, since DMS is a continuous method, subsequent samples can be introduced without the need for reequilibration. In summary, the example presented here for the analysis of DNA adducts from a biological matrix demonstrates the high throughput analytical capabilities of DMS and the time savings that can be realized in a broad range of bioanalytical applications.
Figure 5.
Comparison of LC-MS and DMS-MS speed of analysis for DNA adducts. For SPE-LC-MS, a second lyophilization step is required.
Thermochemical kinetics of modifier-assisted DMS
Ion-neutral clustering has been the subject of considerable study, although mechanisms are complex and not yet fully elucidated. A useful early review is that of Castleman and Keesee, but there are a number of more recent studies, including those from the Leone group[51], and others. A quantitative analysis of comparative modifier effects appears possible based on thermochemical free energy values from computational chemistry, optimized conformations considering energetics and steric hindrance, combined with computations of mobility [22, 52, 53] from predicted structures, and further investigations are in progress. The schema and thermochemistry for ion-cluster / modifier equilibria is summarized in Appendix A.
In order to understand the observed variation in compensation voltage with DMS field strength and modifier concentration, it is necessary to consider three effects of the DMS field on the ion and on ion-modifier clusters: the heating of the ion, the change in the frequency of collision between the ion and the bulk gas, and the change in collision energy. The rapidly varying DMS field does not affect the transport gas because the ions are very dilute in the total gas mixture. However, the DMS field does change the internal and translational energy of the ion / ionic cluster, as well as the collision energy and collision rate for collisions between the ion or ion cluster and the transport gas composition. The heating effect on the ion can be written in terms of an ion effective temperature, Teff, which is different from the bulk gas temperature, T , as follows [7, 10, 16].
| (5) |
In this expression, P is the ambient pressure, N0 the gas number density at 0°C, 1 atm, M the mass of the transport gas neutral molecule. We have first used the reduced mobility relationship NK = N0K0 (exact for pressure variation, approximate for temperature (Ch. 5-1.B, Mason and McDaniel [10])), then the ideal gas law, and we have followed Krylov [16] in introducing ζ,, an inelasticity factor necessary for polyatomic ions. Without an independent measurement of K0, DMS data can determine only the product. For an understanding of the modifier effects in DMS, the most important aspect of Teff is the DMS heating term (Teff – T) which is quadratic in both bulk temperature, T, and applied electric field, E; these effects jointly allow ions in DMS to be strongly heated and become unclustered.
The kinetic limit to the ability of clustering number to track the dynamic effective temperature can be estimated from the collision rate in the two-temperature theory, and comparing it to the amount of time spent at each DMS voltage level (Figure 6(A)). There are different ways to estimate the collision rate, but the different approaches produce similar results. For instance, equation (4) can be rearranged to estimate the A–C collision rate,ξA–C, as
| (6) |
(XC is the neutral mole fraction). A form that differs only in numerical coefficient is given by eq (37) in Viehland and Goeringer[54]. For collisions between 1% isopropanol in a non-polar transport gas and molecular ions in the range from methyl histamine to deoxyguanosine cations, assuming K0=1.5 cm2/V.sec, and T=200°C, the momentum transfer collision rate is generally between 50 and 100 per microsecond. From Figure 6(A), the 2-H waveform spends about 50% of the period near the low field value and 10% near the highest field. For the AB SCIEX Selexion (3 MHz frequency) this gives 0.167 microseconds (<17 collisions) at low field, and 0.033 microseconds (∼ 3 collisions) at high field.
The ability of the internal energy of the ion and its clustered state to remain in dynamic equilibrium with the DMS effective temperature, as determined by the varying electric field, bulk pressure and temperature, is based on the following considerations:
Cluster formation rate. Ion mobility is inversely related to the ion-neutral collisional cross-section. The ion mobility cross-section was first derived by Langevin[55] and more fully developed for ion-polar molecule interactions by Su and coworkers using trajectory calculations and dipole orientation [56–59], and by Turulski and coworkers using transition state theory and other methods [27]. The long-range interaction between an ion and a polar molecule is through the ion - dipole interaction and is strongly attractive (r−2 , [60, 61]). Cluster formation, when the free energy of association is negative, can occur in a very small number of collisions, or even in a single collision, as long as the cluster contains more than a few atoms so that the rovibrational state density allows the collision energy to be retained as internal energy (see especially Lias and Ausloos [62]). Thus, cluster formation between an ion and a polar modifier can proceed at a nearly gas-kinetic rate. Redistribution and stabilization of the internal degrees of freedom within the cluster [63–65] is enhanced by hydrogen bonding between the polar modifier and the ion after they have been attracted by the long range forces.
Thermal equilibration rate. The cluster ion acquires kinetic energy through field acceleration between collisions but loses this energy in collisions, largely with the predominant non-polar bulk gas (N2 in our case). While modifier collisions can lead to cluster formation, the more frequent bulk-gas collisions lead to modifier loss and thermal equilibration at the effective temperature, affecting both the internal state and the size of the cluster ion. This effective temperature is determined with reasonable accuracy by momentum transfer theory, as modified for polyatomic ions by Krylov [7]. The theory developed by Goeringer and Viehland[66, 67] for an analysis of ion swarms in an ion trap can be applied to follow the oscillatory motion of the dynamic cluster ion. Thus the collision rate that determines equilibration of the cluster at the effective temperature is approximately 100 times faster than the rate that controls cluster formation (for 1% modifier). Thermal equilibration of an existing cluster ion at the effective temperature corresponding to the electric field, bulk temperature and pressure occurs rapidly because it is controlled by bulk gas collisions. Because collisions with the polar modifier occur much more infrequently, the dynamic change in cluster number from the DMS low-field period to the DMS high field can be limited by kinetic effects.
Saturation behavior. After the first modifier is in place, modifier shell development is controlled and limited by geometric compatibility, charge and dipole shielding, polarity localization, and hydrogen bonding. It is possible for a second strongly dipolar modifier to add to an existing cluster canceling the dipole moment of the first, but this behavior is dependent on the lock and key relationships between ion and modifier. We have observed such saturation behavior with ethyl acetate, but have not yet performed any related calculations on the decrease in cluster free energy with size. Multiple clustering makes performance less predictable, and appears to be a primary cause of the need for compound-specific method development.
As an example of the cooperative effect of gas temperature and DMS separation field in modulating the cluster number, we extract one result from an extensive set of ab-initio calculations for the change in free energy, enthalpy, and entropy on cluster formation. The full set results will be reported in detail, but are beyond the scope of the current publication. Abinitio structural optimizations and thermochemical calculations were performed for several small organic molecules with and without the modifiers isopropanol or ethyl acetate. The calculations were done with a series of new and intensive G4 “compound” methods which have had good success in predicting complexation reactions[68]. For all molecules studied, ΔG0 values show that isopropanol is less strongly bound than ethyl acetate, much as might be expected from the difference in dipole moment values (IPA (1.560 D), EtOAc (4.325 D)). For R-alphamethylhistamine (C6H11N3, mw 125.17164, PubChem 156615), thermochemical values for clustering of the protonated methyl-histamine molecule with IPA were determined to be (ΔG0 = − 7.45 kcal/mol, ΔH0 = −16.62 kcal/mol, ΔS0= −30.76 cal/mol/K). This compound has some structural similarity to deoxyguanosine in that it has nitrogen heteroatoms both in a 5-membered ring and in an amino group. In order to examine access to the unclustered ion configuration, and in recognition of the kinetic limit discussed above, we have applied a limit of NC=3 for the maximum IPA count. Assuming K0=1.5 cm2/V.sec, 1.5% isopropanol, ζ=0.7, the cluster number dependence both on applied electric field and on bulk gas temperature for (methyl histamine·H+·isopropanol) can be calculated, with the very interesting result of Figure 6(B). The requirement of high combined field and/or bulk gas temperature for ion declustering is not unexpected because of the earlier experience with atmospheric pressure drift-time IMS [22, 25, 26, 69],but it indicates that both bulk gas temperature and DMS field are critical control parameters.
We can also use these results to interpret intensity data. In Figure 4(D), we saw that the DMS intensity fell off at higher DMS field values. An additional mechanism for signal loss in the higher SV range is collision-induced chemistry, either dissociation (CID) or charge detachment. CID in DMS has been reported in DMS [49] and charge detachment in systems like SF6− is well known[70]. At lower DMS field, the heating is quite a bit less so that less declustering will occur, preserving the ion intensity regardless of the modifier concentration. At very high fields very high energy collisions are effective at declustering but can also be destructive, more so for larger clusters because the proton is more likely to go with the cluster of modifiers. Thus, both DMS field and modifier concentration affect the signal intensity in similar ways, as our data shows.
Conclusions
In the past few years, efforts have been made to understand and characterize the gas phase cluster formation between the analyte ion and the modifiers. However, an explicit method of characterizing the interactions and predicting the CoV shifts is not available to date. Screening modifiers against the analyte of interest at different conditions has been the most feasible way to perform method development using differential mobility spectrometry and contribute to the understanding on the use of modifiers.
In this report, we propose theoretical considerations for the phenomena associated with compensation voltage shifts in DMS that explain step-wise clustering of modifiers including “saturation effect” and variations in signal intensities. These results show that high transport gas temperatures increase the DMS effect and that some combinations of temperature and DMS field lead to sharp changes in cluster number with DMS SV field (Figure 6). Conversely, with moderate DMS temperatures, DMS is modulating an already clustered ion, making the effect less predictable and requiring a method development phase. This process is illustrated by the approach we have taken for the selective quantitation of modified nucleosides, dG-C8-4-ABP and dG-C8-PhIP, used as model analytes, from a complex mixture of normal nucleosides using a differential mobility- mass spectrometry-based platform. The potential to eliminate or minimize sample preparation steps while bringing the analysis time down to less than a minute per sample point makes this method highly attractive for further bio-analytical applications. Optimizing the DMS operational parameters including modifier concentration gives us the maximum selectivity and sensitivity for selected applications.
Modifier DMS at lower temperatures is generally modulating the size of an already clustered ion species for which thermochemical information is not available and few calculations have been done. We believe that further progress can be made in understanding and predicting DMS modifier performance by approaching future experimental work from the high temperature limit with modifiers of differing dipole moments and structure, and by performing calculations on the development of solvent shell clusters.
Acknowledgements
The authors consider it an honor to be included in this special issue of JASMS honoring Prof Yinzheng Wang in his receipt of the Biemann Medal. We gratefully acknowledge support from NIH: RO1 CA 069390-16 (Paul Vouros, PI) and R01 AI101798 (Albert J. Fornace, Jr., PI).
Appendix A. Ion-modifier clustering kinetics
The kinetic analysis of ion-neutral clustering can be based on the following scheme describing the chemical equilibria among clustering polar modifier, C, protonated ion, MH+, and ion-neutral clusters of different sizes, (Cn).MH+ in the gas phase.
| (A.1) |
Using an abbreviated notation for concentrations relative to the standard state of 273.15 K, 1 atm, we write concentrations and the total ion concentration as follows.
| (A.2) |
The Gibbs free energy of cluster formation is expected to decrease with increasing cluster size. Because cluster shell thermodynamic properties for ions of significant size are not available, we use a constant value, ΔG0,(f) = ΔG0, independent of cluster number, j. Instead, we limit the maximum number of clustered modifiers to a specified number, NC. This is a modifier shell-size limit corresponding to a number of modifier molecules geometrically and electrically compatible with the conditions. There is also a kinetic limit in DMS to the maximum change in cluster number between low and high field cases, determined by the ratio of the ion-neutral collision rate to the waveform frequency, as discussed later. Assuming a smoothly declining free energy with cluster size, and taking a discrete maximum shell size differ practically in that only the shell model can lead to the dramatic solvent saturation effect we have observed in dG-C8-4-ABP with ethyl acetate, but not with isopropanol.
The results for the populations of each ion-neutral cluster size, Aj, and the average cluster number, n̄c, in terms of the product KC = K1C (the equilibrium constant times the neutral concentration relative to the standard state, giving the density ratio of larger cluster to smaller) are shown here.
| (A.3) |
As a first approximation, the stepwise equilibrium constants are taken to be independent of cluster number, Kj = K1 for 1 ≤ j ≤ NC , where NC is the maximum number of clustered neutrals. The concentrations are relative to 1 atm, at T0=273.15 K, so that 1% modifier at 1 atm corresponds to C ≅ 0.01 N0, or at a different temperature or pressure to , where N is the total number density, with either N0 under standard conditions (T0=273.15 K, 1 atm), or N at a different pressure and temperature.
Contributor Information
Amol Kafle, Department of Chemistry and Chemical Biology and Barnett Institute, Northeastern University, Boston, MA..
Stephen L. Coy, Department of Chemistry and Chemical Biology and Barnett Institute, Northeastern University, Boston, MA.
Bryan M. Wong, Department of Chemistry and Department of Materials Science & Engineering, Drexel University, Philadelphia, PA 19104.
Albert J. Fornace, Jr., Georgetown University, Department of Biochemistry & Molecular & Cell Biology, Washington, DC 20057 USA, and King Abdulaziz Univ, Center of Excellence in Genomic Medical Research, Jeddah 21413, Saudi Arabia.
James J. Glick, Department of Chemistry and Chemical Biology and Barnett Institute, Northeastern University, Boston, MA.
Paul Vouros, Department of Chemistry and Chemical Biology and Barnett Institute, Northeastern University, Boston, MA..
References
- 1.Schneider BB, Covey TR, Coy SL, Krylov EV, Nazarov EG. Control of chemical effects in the separation process of a differential mobility mass spectrometer system. European Journal of Mass Spectrometry. 2010;16(1):57–71. doi: 10.1255/ejms.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coy SL, Krylov EV, Schneider BB, Covey TR, Brenner DJ, Tyburski JB, Patterson AD, Krausz KW, Fornace AJ, Nazarov EG. Detection of Radiation-Exposure Biomarkers by Differential Mobility Prefiltered Mass Spectrometry (DMS-MS) Int J Mass Spectrom. 2010;291(3):108–117. doi: 10.1016/j.ijms.2010.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schneider B, Covey T, Coy S, Krylov E, Nazarov E. Planar differential mobility spectrometer as a pre-filter for atmospheric pressure ionization mass spectrometry. International Journal of Mass Spectrometry. 2010;298(1–3):45–54. doi: 10.1016/j.ijms.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schneider BB, Covey TR, Coy SL, Krylov EV, Nazarov EG. Chemical Effects in the Separation Process of a Differential Mobility/Mass Spectrometer System. Analytical Chemistry. 2010;82(5):1867–1880. doi: 10.1021/ac902571u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hall A, Coy S, Kafle A, Glick J, Nazarov E, Vouros P. Extending the Dynamic Range of the Ion Trap by Differential Mobility Filtration. J. Am. Soc. Mass Spectrom. 2013:1428–1436. doi: 10.1007/s13361-013-0655-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hall AB, Coy SL, Nazarov EG, Vouros P. Rapid Separation and Characterization of Cocaine and Cocaine Cutting Agents by Differential Mobility Spectrometry-Mass Spectrometry. Journal of Forensic Sciences. 2012;57(3):750–756. doi: 10.1111/j.1556-4029.2011.02033.x. [DOI] [PubMed] [Google Scholar]
- 7.Krylov EV, Nazarov EG. Electric field dependence of the ion mobility. International Journal of Mass Spectrometry. 2009;285(3):149–156. [Google Scholar]
- 8.Shvartsburg AA. Differential Ion Mobility Spectrometry: Nonlinear Ion Transport and Fundamentals of FAIMS. Boca Raton, FL: CRC Press, Taylor & Francis Group; 2008. p. 322. [Google Scholar]
- 9.Eiceman GA, Karpas Z. Ion mobility spectrometry / G.A. Eiceman, Z. Karpas. Boca Raton, FL: Taylor & Francis/CRC Press; 2005. p. 350. [Google Scholar]
- 10.Mason EA, McDaniel EW. Transport Properties of Ions in Gases. New York: Wiley-Interscience; 1988. [Google Scholar]
- 11.Buryakov IA, Krylov EV, Makas AL, Nazarov EG, Pervukhin VV, Rasulev UK. Drift Spectrometer For The Control Of Amine Traces In The Atmosphere. Journal of Analytical Chemistry. 1993;48(1):114–121. [Google Scholar]
- 12.Buryakov IA, Krylov EV, Nazarov EG, Rasulev UK. A new method of separation of multiatomic ions by mobility at atmospheric pressure using a high-frequency amplitude-asymmetric strong electric field. International Journal of Mass Spectrometry and Ion Processes. 1993;128(3):143–148. [Google Scholar]
- 13.Krylov E, Coy S, Vandermey J, Schneider B, Covey T, Nazarov E. Selection and generation of waveforms for differential mobility spectrometry. Review of Scientific Instruments. 2010;81(2):024101. doi: 10.1063/1.3284507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levin DS, Vouros P, Miller RA, Nazarov EG, Morris JC. Characterization of gas-phase molecular interactions on differential mobility ion behavior utilizing an electrospray ionizationdifferential mobility-mass spectrometer system. Analytical Chemistry. 2006;78(1):96–106. doi: 10.1021/ac051217k. [DOI] [PubMed] [Google Scholar]
- 15.Nazarov EG, Coy SL, Krylov EV, Miller RA, Eiceman GA. Pressure effects in differential mobility spectrometry. Analytical Chemistry. 2006;78(22):7697–7706. doi: 10.1021/ac061092z. [DOI] [PubMed] [Google Scholar]
- 16.Krylov EV, Coy SL, Nazarov EG. Temperature effects in differential mobility spectrometry. International Journal of Mass Spectrometry. 2009;279(2–3):119–125. [Google Scholar]
- 17.ABSCIEX. [cited 2013 Aug, 2013];selexION technology. 2013 Available from: http://www.absciex.com/Documents/Downloads/Literature/SelexION-Technology-New-Solution-Selectivity-Challenges-TN%20P-2960211-01.pdf.
- 18.Campuzano I, Bush MF, Robinson CV, Beaumont C, Richardson K, Kim H, Kim HI. Structural Characterization of Drug-like Compounds by Ion Mobility Mass Spectrometry: Comparison of Theoretical and Experimentally Derived Nitrogen Collision Cross Sections. Analytical Chemistry. 2012;84(2):1026–1033. doi: 10.1021/ac202625t. [DOI] [PubMed] [Google Scholar]
- 19.Salbo R, Bush M, Naver H, Campuzano I, Robinson C, Pettersson I, Jorgensen T, Haselmann K. Traveling-wave ion mobility mass spectrometry of protein complexes: accurate calibrated collision cross-sections of human insulin oligomers. Rapid Communications in Mass Spectrometry. 2012;26(10):1181–1193. doi: 10.1002/rcm.6211. [DOI] [PubMed] [Google Scholar]
- 20.Kim H, Johnson P, Beegle L, Beauchamp J, Kanik I. Electrospray ionization ion mobility spectrometry of carboxylate anions: Ion mobilities and a mass-mobility correlation. Journal of Physical Chemistry A. 2005:7888–7895. doi: 10.1021/jp051274h. [DOI] [PubMed] [Google Scholar]
- 21.Johnson P, Kim H, Beegle L, Kanik I. Electrospray ionization ion mobility spectrometry of amino acids: Ion mobilities and a mass-mobility correlation. Journal of Physical Chemistry A. 2004:5785–5792. doi: 10.1021/jp051274h. [DOI] [PubMed] [Google Scholar]
- 22.Kim H, Kim H, Johnson P, Beegle L, Beauchamp J, Goddard W, Kanik I. Experimental and theoretical investigation into the correlation between mass and ion mobility for choline and other ammonium cations in N-2. Analytical Chemistry. 2008:1928–1936. doi: 10.1021/ac701888e. [DOI] [PubMed] [Google Scholar]
- 23.Berant Z, Karpas Z. Mass-Mobility Correlation Of Ions In View Of New Mobility Data. Journal of the American Chemical Society. 1989;111(11):3819–3824. [Google Scholar]
- 24.Berant Z, Karpas Z, Shahal O. Effects Of Temperature And Clustering On Mobility Of Ions In CO2. Journal of Physical Chemistry. 1989;93(21):7529–7532. [Google Scholar]
- 25.Berant Z, Shahal O, Karpas Z. Correlation Between Measured And Calculated Mobilities Of Ions - Sensitivity Analysis Of The Fitting Procedure. Journal of Physical Chemistry. 1991;95(19):7534–7538. [Google Scholar]
- 26.Karpas Z, Berant Z, Shahal O. Effect Of Temperature On The Mobility Of Ions. Journal of the American Chemical Society. 1989;111(16):6015–6018. [Google Scholar]
- 27.Turulski J, Forys M. ION-POLAR MOLECULE COLLISION FREQUENCY - THERMODYNAMIC TREATMENT. Journal of Physical Chemistry. 1979;83(22):2815–2817. [Google Scholar]
- 28.Eiceman GA, Krylov EV, Krylova NS, Nazarov EG, Miller RA. Separation of Ions from Explosives in Differential Mobility Spectrometry by Vapor-Modified Drift Gas. Analytical Chemistry. 2004;76(17):4937–4944. doi: 10.1021/ac035502k. [DOI] [PubMed] [Google Scholar]
- 29.Levin DS, Vouros P, Miller RA, Nazarov EG, Morris JC. Characterization of Gas-Phase Molecular Interactions on Differential Mobility Ion Behavior Utilizing an Electrospray Ionization Differential Mobility-Mass Spectrometer System. Anal. Chem. 2006;78:96–106. doi: 10.1021/ac051217k. [DOI] [PubMed] [Google Scholar]
- 30.Levin DS, Vouros P, Miller RA, Nazarov EG. Using a nanoelectrospray-differential mobility spectrometer-mass spectrometer system for the analysis of oligosaccharides with solvent selected control over ESI aggregate ion formation. Journal of the American Society for Mass Spectrometry. 2007;18(3):502–511. doi: 10.1016/j.jasms.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rorrer III LC, Yost RA. Solvent vapor effects on planar high-field asymmetric waveform ion mobility spectrometry. International Journal of Mass Spectrometry. 2011;300(2):173–181. [Google Scholar]
- 32.Schneider B, Nazarov E, Covey T. Peak capacity in differential mobility spectrometry: effects of transport gas and gas modifiers. International Journal for Ion Mobility Spectrometry. 2012;15(3):141–150. [Google Scholar]
- 33.Tsai CW, Yost RA, Garrett TJ. High-field asymmetric waveform ion mobility spectrometry with solvent vapor addition: a potential greener bioanalytical technique. Bioanalysis. 2012;4(11):1363–1375. doi: 10.4155/bio.12.110. [DOI] [PubMed] [Google Scholar]
- 34.Coy SL, Krylov EV, Nazarov EG, Fornace AJJ, Kidd RD. Differential mobility spectrometry with nanospray ion source as a compact detector for small organics and inorganics. Int. J. Ion Mobility Spectrometry. 2013;16:217–227. doi: 10.1007/s12127-013-0135-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnson P, Beegle L, Kim H, Eiceman G, Kanik I. Ion mobility spectrometry in space exploration. International Journal of Mass Spectrometry. 2007:1–15. [Google Scholar]
- 36.Levin DS, Miller RA, Nazarov EG, Vouros P. Rapid Separation and Quantitative Analysis of Peptides Using a New Nanoelectrospray- Differential Mobility Spectrometer-Mass Spectrometer System. Analytical Chemistry. 2006;78(15):5443–5452. doi: 10.1021/ac060003f. [DOI] [PubMed] [Google Scholar]
- 37.Kafle A, Klaene J, Hall AB, Glick J, Coy SL, Vouros P. A differential mobility spectrometry/mass spectrometry platform for the rapid detection and quantitation of DNA adduct dG-ABP. Rapid Communications in Mass Spectrometry. 2013;27(13):1473–1480. doi: 10.1002/rcm.6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Varesio E, Le Blanc JCY, Hopfgartner G. Real-time 2D separation by LC × differential ion mobility hyphenated to mass spectrometry. Analytical and Bioanalytical Chemistry. 2012;402(8):2555–2564. doi: 10.1007/s00216-011-5444-y. [DOI] [PubMed] [Google Scholar]
- 39.Jasak J, Le Blanc Y, Speer K, Billian P, Schoening RM. Analysis of Triazole-Based Metabolites in Plant Materials Using Differential Mobility Spectrometry to Improve LC/MS/MS Selectivity. Journal of Aoac International. 2012;95(6):1768–1776. doi: 10.5740/jaoacint.12-073. [DOI] [PubMed] [Google Scholar]
- 40.Garrigos MC, Reche F, Pernias K, Sanchez A, Jimenez A. Determination of some aromatic amines in finger-paints for children's use by supercritical fluid extraction combined with gas chromatography. J Chromatogr A. 1998;819(1–2):259–66. doi: 10.1016/s0021-9673(98)00432-4. [DOI] [PubMed] [Google Scholar]
- 41.Oh SW, Kang MN, Cho CW, Lee MW. Detection of carcinogenic amines from dyestuffs or dyed substrates. Dyes and Pigments. 1997;33(2):119–135. [Google Scholar]
- 42.Tokiwa H, Nakagawa R, Horikawa K. Mutagenic/carcinogenic agents in indoor pollutants; the dinitropyrenes generated by kerosene heaters and fuel gas and liquefied petroleum gas burners. Mutat Res. 1985;157(1):39–47. doi: 10.1016/0165-1218(85)90047-3. [DOI] [PubMed] [Google Scholar]
- 43.Turesky RJ, Freeman JP, Holland RD, Nestorick DM, Miller DW, Ratnasinghe DL, Kadlubar FF. Identification of aminobiphenyl derivatives in commercial hair dyes. Chem Res Toxicol. 2003;16(9):1162–73. doi: 10.1021/tx030029r. [DOI] [PubMed] [Google Scholar]
- 44.Sinha R, Rothman N, Salmon CP, Knize MG, Brown ED, Swanson CA, Rhodes D, Rossi S, Felton JS, Levander OA. Heterocyclic amine content in beef cooked by different methods to varying degrees of doneness and gravy made from meat drippings. Food Chem Toxicol. 1998;36(4):279–87. doi: 10.1016/s0278-6915(97)00162-2. [DOI] [PubMed] [Google Scholar]
- 45.Sugimura T, Wakabayashi K, Nakagama H, Nagao M. Heterocyclic amines: Mutagens/carcinogens produced during cooking of meat and fish. Cancer Sci. 2004;95(4):290–9. doi: 10.1111/j.1349-7006.2004.tb03205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gangl ET, Turesky RJ, Vouros P. Determination of in vitro- and in vivo-formed DNA adducts of 2-amino-3-methylimidazo[4,5-f]quinoline by capillary liquid chromatography/microelectrospray mass spectrometry. Chem Res Toxicol. 1999;12(10):1019–27. doi: 10.1021/tx990060m. [DOI] [PubMed] [Google Scholar]
- 47.Fathi F, Tabrizchi M, Farrokhpour H. Two-dimensional optogalvanic spectroscopy. Journal of Quantitative Spectroscopy & Radiative Transfer. 2012;113(3):226–231. [Google Scholar]
- 48.Campbell J, Le Blanc J, Schneider B. Probing Electrospray Ionization Dynamics Using Differential Mobility Spectrometry: The Curious Case of 4-Aminobenzoic Acid. Analytical Chemistry. 2012;84(18):7857–7864. doi: 10.1021/ac301529w. [DOI] [PubMed] [Google Scholar]
- 49.Kendler S, Lambertus GR, Dunietz BD, Coy SL, Nazarov EG, Miller RA, Sacks RD. Fragmentation pathways and mechanisms of aromatic compounds in atmospheric pressure studied by GC-DMS and DMS-MS. International Journal of Mass Spectrometry. 2007;263(2–):137–147. [Google Scholar]
- 50.NIST Chemistry Webbook. National Institute of Standards and Technology (NIST) 2012 http://webbook.nist.gov/chemistry/.
- 51.Ikonomou MG, Blades AT, Kebarle P. Investigations of the electrospray interface for liquid chromatography/mass spectrometry. Analytical Chemistry. 1990;62(9):957–967. [Google Scholar]
- 52.Campuzano I, Bush M, Robinson C, Beaumont C, Richardson K, Kim H, Kim H. Structural Characterization of Drug-like Compounds by Ion Mobility Mass Spectrometry: Comparison of Theoretical and Experimentally Derived Nitrogen Collision Cross Sections. Analytical Chemistry. 2012;84(2):1026–1033. doi: 10.1021/ac202625t. [DOI] [PubMed] [Google Scholar]
- 53.Kim H, Johnson P, Beegle L, Beauchamp J, Kanik I. Electrospray ionization ion mobility spectrometry of carboxylate anions: Ion mobilities and a mass-mobility correlation. Journal of Physical Chemistry a. 2005;109(35):7888–7895. doi: 10.1021/jp051274h. [DOI] [PubMed] [Google Scholar]
- 54.Viehland L, Goeringer D. Moment theory of ion motion in traps and similar devices: I. General theories. Journal of Physics B-Atomic Molecular and Optical Physics. 2005;38(22):3987–4009. [Google Scholar]
- 55.Langevin P. A fundamental formula of kinetic theory. Annales De Chimie Et De Physique. 1905;5:245–288. [Google Scholar]
- 56.Su T. KINETIC-ENERGY DEPENDENCE OF ION-POLAR MOLECULE COLLISION RATE CONSTANTS BY TRAJECTORY CALCULATIONS. Journal of Chemical Physics. 1985;82(4):2164–2166. [Google Scholar]
- 57.Su T, Chesnavich WJ. PARAMETRIZATION OF THE ION-POLAR MOLECULE COLLISION RATE-CONSTANT BY TRAJECTORY CALCULATIONS. Journal of Chemical Physics. 1982;76(10):5183–5185. [Google Scholar]
- 58.Su T, Viggiano AA, Paulson JF. THE EFFECT OF THE DIPOLE-INDUCED DIPOLE POTENTIAL ON ION POLAR MOLECULE COLLISION RATE CONSTANTS. Journal of Chemical Physics. 1992;96(7):5550–5551. [Google Scholar]
- 59.Su T, Su ECF, Bowers MT. ION-POLAR MOLECULE COLLISIONS - CONSERVATION OF ANGULAR-MOMENTUM IN AVERAGE DIPOLE ORIENTATION THEORY - AADO THEORY. Journal of Chemical Physics. 1978;69(5):2243–2250. [Google Scholar]
- 60.Barker RA, Ridge DP. ION-POLAR NEUTRAL MOMENTUM-TRANSFER COLLISION FREQUENCIES - THEORETICAL APPROACH. Journal of Chemical Physics. 1976;64(11):4411–4416. [Google Scholar]
- 61.Chesnavich WJ, Su T, Bowers MT. COLLISIONS IN A NON-CENTRAL FIELD - VARIATIONAL AND TRAJECTORY INVESTIGATION OF ION-DIPOLE CAPTURE. Journal of Chemical Physics. 1980;72(4):2641–2655. [Google Scholar]
- 62.Lias SG, Ausloos P. COMMENTS ON ENTROPY CHANGE IN ION-MOLECULE EQUILIBRIA. Journal of the American Chemical Society. 1977;99(14):4831–4833. [Google Scholar]
- 63.Rice SA, Wolynes PG. An overview of the dynamics of intramolecular transfer of vibrational energy. Advances in Chemical Physics. 1981:117–200. [Google Scholar]
- 64.Gruebele M, Bigwood R. Molecular vibrational energy flow: beyond the Golden Rule. International Reviews in Physical Chemistry. 1998;17(2):91–145. [Google Scholar]
- 65.Gruebele M, Wolynes PG. Vibrational energy flow and chemical reactions. Accounts of Chemical Research. 2004;37(4):261–267. doi: 10.1021/ar030230t. [DOI] [PubMed] [Google Scholar]
- 66.Goeringer D, Viehland L. Moment theory of ion motion in traps and similar devices: III. Two-temperature treatment of quadrupole ion traps. Journal of Physics B-Atomic Molecular and Optical Physics. 2005;38(22):4027–4044. [Google Scholar]
- 67.Viehland L, Danailov D, Goeringer D. Moment theory of ion-neutral reactions in traps and similar devices. Journal of Physical Chemistry a. 2007;111(15):2820–2829. doi: 10.1021/jp066096m. [DOI] [PubMed] [Google Scholar]
- 68.Wong B, Lacina D, Nielsen I, Graetz J, Allendorf M. Thermochemistry of Alane Complexes for Hydrogen Storage: A Theoretical and Experimental Investigation. Journal of Physical Chemistry C. 2011;115(15):7778–7786. doi: 10.1021/jp112258s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Beegle L, Kanik I, Matz L, Hill H. Effects of drift-gas polarizability on glycine peptides in ion mobility spectrometry. International Journal of Mass Spectrometry. 2002:257–268. [Google Scholar]
- 70.Lifshitz C. Energy Entropy Trade-Offs In The Unimolecular Decompositions Of SF6. Journal of Physical Chemistry. 1983;87(18):3474–3479. [Google Scholar]