Summary
Mammalian cells generate citrate by decarboxylating pyruvate in the mitochondria to supply the tricarboxylic acid (TCA) cycle. In contrast, hypoxia and other impairments of mitochondrial function induce an alternative pathway that produces citrate by reductively carboxylating α-ketoglutarate (AKG) via NADPH-dependent isocitrate dehydrogenase (IDH). It is unknown how cells generate reducing equivalents necessary to supply reductive carboxylation in the setting of mitochondrial impairment. Here we identified shared metabolic features in cells using reductive carboxylation. Paradoxically, reductive carboxylation was accompanied by concomitant AKG oxidation in the TCA cycle. Inhibiting AKG oxidation decreased reducing equivalent availability and suppressed reductive carboxylation. Interrupting transfer of reducing equivalents from NADH to NADPH by nicotinamide nucleotide transhydrogenase increased NADH abundance and decreased NADPH abundance while suppressing reductive carboxylation. The data demonstrate that reductive carboxylation requires bidirectional AKG metabolism along oxidative and reductive pathways, with the oxidative pathway producing reducing equivalents used to operate IDH in reverse.
INTRODUCTION
Proliferating cells support their growth by converting abundant extracellular nutrients like glucose and glutamine into precursors for macromolecular biosynthesis. A continuous supply of metabolic intermediates from the tricarboxylic acid (TCA) cycle is essential for cell growth, because many of these intermediates feed biosynthetic pathways to produce lipids, proteins and nucleic acids (Deberardinis et al., 2008). This underscores the dual roles of the TCA cycle for cell growth: it generates reducing equivalents for oxidative phosphorylation by the electron transport chain (ETC), while also serving as a hub for precursor production. During rapid growth, the TCA cycle is characterized by large influxes of carbon at positions other than acetyl-CoA, enabling the cycle to remain full even as intermediates are withdrawn for biosynthesis. Cultured cancer cells usually display persistence of TCA cycle activity despite robust aerobic glycolysis, and often require mitochondrial catabolism of glutamine to the TCA cycle intermediate AKG to maintain rapid rates of proliferation (Icard et al., 2012, Hiller and Metallo, 2013).
Some cancer cells contain severe, fixed defects in oxidative metabolism caused by mutations in the TCA cycle or the ETC. These include mutations in fumarate hydratase (FH) in renal cell carcinoma and components of the succinate dehydrogenase (SDH) complex in pheochromocytoma, paraganglioma, and gastrointestinal stromal tumors (Tomlinson et al., 2002, Astuti et al., 2001, Baysal et al., 2000, Killian et al., 2013, Niemann and Muller, 2000). All of these mutations alter oxidative metabolism of glutamine in the TCA cycle. Recently, analysis of cells containing mutations in FH, ETC Complexes I or III, or exposed to the ETC inhibitors metformin and rotenone or the ATP synthase inhibitor oligomycin revealed that turnover of TCA cycle intermediates was maintained in all cases (Mullen et al., 2012). However, the cycle operated in an unusual fashion characterized by conversion of glutamine-derived AKG to isocitrate through a reductive carboxylation reaction catalyzed by NADP+/NADPH-dependent isoforms of isocitrate dehydrogenase (IDH). As a result, a large fraction of the citrate pool carried five glutamine-derived carbons. Citrate could be cleaved to produce acetyl-CoA to supply fatty acid biosynthesis, and oxaloacetate (OAA) to supply pools of other TCA cycle intermediates. Thus, reductive carboxylation enables biosynthesis by enabling cells with impaired mitochondrial metabolism to maintain pools of biosynthetic precursors that would normally be supplied by oxidative metabolism. Reductive carboxylation is also induced by hypoxia and by pseudo-hypoxic states caused by mutations in the von Hippel-Lindau (VHL) tumor suppressor gene (Metallo et al., 2012, Wise et al., 2011).
Interest in reductive carboxylation stems in part from the possibility that inhibiting the pathway might induce selective growth suppression in tumor cells subjected to hypoxia or containing mutations that prevent them from engaging in maximal oxidative metabolism. Hence, several recent studies have sought to understand the mechanisms by which this pathway operates. In vitro studies of IDH1 indicate that a high ratio of NADPH/NADP+ and low citrate concentration activate the reductive carboxylation reaction (Leonardi et al., 2012). This is supported by data demonstrating that reductive carboxylation in VHL-deficient renal carcinoma cells is associated with a low concentration of citrate and a reduced ratio of citrate:AKG, suggesting that mass action can be a driving force to determine IDH directionality (Gameiro et al., 2013b). Moreover, interrupting the supply of mitochondrial NADPH by silencing the nicotinamide nucleotide transhydrogenase (NNT) suppresses reductive carboxylation (Gameiro et al., 2013a). This mitochondrial transmembrane protein catalyzes the transfer of a hydride ion from NADH to NADP+ to generate NAD+ and NADPH. Together, these observations suggest that reductive carboxylation is modulated in part through the mitochondrial redox state and the balance of substrate/products.
Here we used metabolomics and stable isotope tracing to better understand overall metabolic states associated with reductive carboxylation in cells with defective mitochondrial metabolism, and to identify sources of mitochondrial reducing equivalents necessary to induce the reaction. We identified high levels of succinate in some cells using reductive carboxylation, and determined that most of this succinate was formed through persistent oxidative metabolism of AKG. Silencing this oxidative flux by depleting the mitochondrial enzyme AKG dehydrogenase substantially altered the cellular redox state and suppressed reductive carboxylation. The data demonstrate that bidirectional/branched AKG metabolism occurs during reductive carboxylation in cells with mitochondrial defects, with oxidative metabolism producing reducing equivalents to supply reductive metabolism.
RESULTS
Shared metabolomic features among cell lines with cytb or FH mutations
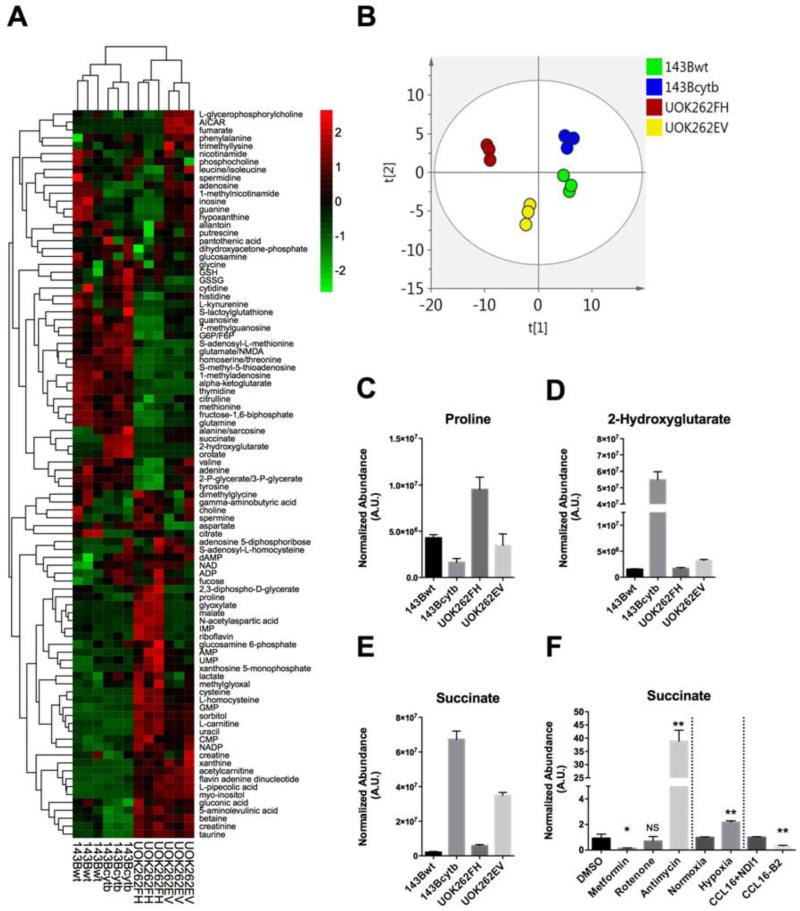
To identify conserved metabolic features associated with reductive carboxylation in cells harboring defective mitochondrial metabolism, we analyzed metabolite abundance in isogenic pairs of cell lines in which one member displayed substantial reductive carboxylation and the other did not. We used a pair of previously described cybrids derived from 143B osteosarcoma cells, in which one cell line contained wild-type mitochondrial DNA (143Bwt) and the other contained a mutation in the cytb gene (143Bcytb), severely reducing complex III function (Rana et al., 2000, Weinberg et al., 2010). The 143Bwt cells primarily use oxidative metabolism to supply the citrate pool while the 143Bcytb cells use reductive carboxylation (Mullen et al., 2012). The other pair, derived from FH-deficient UOK262 renal carcinoma cells, contained either an empty vector control (UOK262EV) or a stably re-expressed wild-type FH allele (UOK262FH). Metabolites were extracted from all four cell lines and analyzed by triple-quadrupole mass spectrometry. We first performed a quantitative analysis to determine the abundance of AKG and citrate in the four cell lines. Both 143Bcytb and UOK262EV cells had less citrate, more AKG, and lower citrate:AKG ratios than their oxidative partners (Fig. S1A-C), consistent with findings from VHL-deficient renal carcinoma cells (Gameiro et al., 2013b).
Next, to identify other perturbations, we profiled the relative abundance of more than 90 metabolites from glycolysis, the pentose phosphate pathway, one-carbon/nucleotide metabolism, the TCA cycle, amino acid degradation, and other pathways (Tables S1 and S2). Each metabolite was normalized to protein content, and relative abundance was determined between cell lines from each pair. Hierarchical clustering (Fig 1A) and principal component analysis (Fig 1B) revealed far greater metabolomic similarities between the members of each pair than between the two cell lines using reductive carboxylation. Only three metabolites displayed highly significant (p<0.005) differences in abundance between the two members of both pairs, and in all three cases the direction of the difference (i.e. higher or lower) was shared in the two cell lines using reductive carboxylation. Proline, a nonessential amino acid derived from glutamine in an NADPH-dependent biosynthetic pathway, was depleted in 143Bcytb and UOK262EV cells (Fig. 1C). 2-hydroxyglutarate (2HG), the reduced form of AKG, was elevated in 143Bcytb and UOK262EV cells (Fig. 1D), and further analysis revealed that while both the L- and D-enantiomers of this metabolite were increased, L-2HG was quantitatively the predominant enantiomer (Fig. S1D). It is likely that 2HG accumulation was related to the reduced redox ratio associated with cytb and FH mutations. Although the sources of 2HG are still under investigation, promiscuous activity of the TCA cycle enzyme malate dehydrogenase produces L-2HG in an NADH-dependent manner (Rzem et al., 2007). Both enantiomers are oxidized to AKG by dehydrogenases (L-2HG dehydrogenase and D-2HG dehydrogenase). It is therefore likely that elevated 2-HG is a consequence of a reduced NAD+/NADH ratio. Consistent with this model, inborn errors of the ETC result in 2-HG accumulation (Reinecke et al., 2011). Exposure to hypoxia (<1% O2) has also been demonstrated to reduce the cellular NAD+/NADH ratio (Santidrian et al., 2013) and to favor modest 2HG accumulation in cultured cells (Wise et al., 2011), although these levels were below those noted in gliomas expressing 2HG-producing mutant alleles of isocitrate dehydrogenase-1 or -2 (Dang et al., 2009).
Figure 1. Metabolomic features of cells using reductive carboxylation.
(A) Relative abundance of metabolites extracted from triplicate samples of 143Bwt, 143Bcytb, UOK262FH and UOK262EV cells. Peak areas of each metabolite were normalized to protein abundance. The color reflects a log2 scale.
(B) Principal component analysis for the twelve extracts used to generate the heat map in (A).
(C-E) Metabolites demonstrating consistent and significant alterations in abundance in cells using reductive carboxylation. In all cases, p<0.005 for the comparison between cells lines in each pair.
(F) Abundance of succinate in several models of reductive carboxylation induced by impaired ETC activity. High doses of metformin and rotenone inhibit complex I. Antimycin inhibits complex III. Hypoxia functions as an inhibitor of complex IV. These treatments were all applied to 143Bwt cells. CCL16-B2 are Chinese hamster fibroblast cells with impaired Complex I activity as a consequence of NDUFA1 mutation. CCL16-NDI1 cells were generated by stably infecting CCL16- B2 with yeast NADH quinone oxidoreductase (NDI1) which restores oxidative capacity and eliminates reductive carboxylation. Data are the average and S.D. of three independent cultures. *p<0.05; **p<0.005, Student's t-test. NS, not significant.
Finally, the TCA cycle intermediate succinate was markedly elevated in both cell lines (Fig. 1E). We tested additional factors previously reported to stimulate reductive AKG metabolism, including a genetic defect in ETC Complex I, exposure to hypoxia, and chemical inhibitors of the ETC (Mullen et al., 2012, Wise et al., 2011, Metallo et al., 2012). These factors had a variable effect on succinate, with impairments of Complex III or IV strongly inducing succinate accumulation, while impairments of Complex I either had little effect or suppressed succinate (Fig. 1F).
Oxidative glutamine metabolism is the primary route of succinate formation
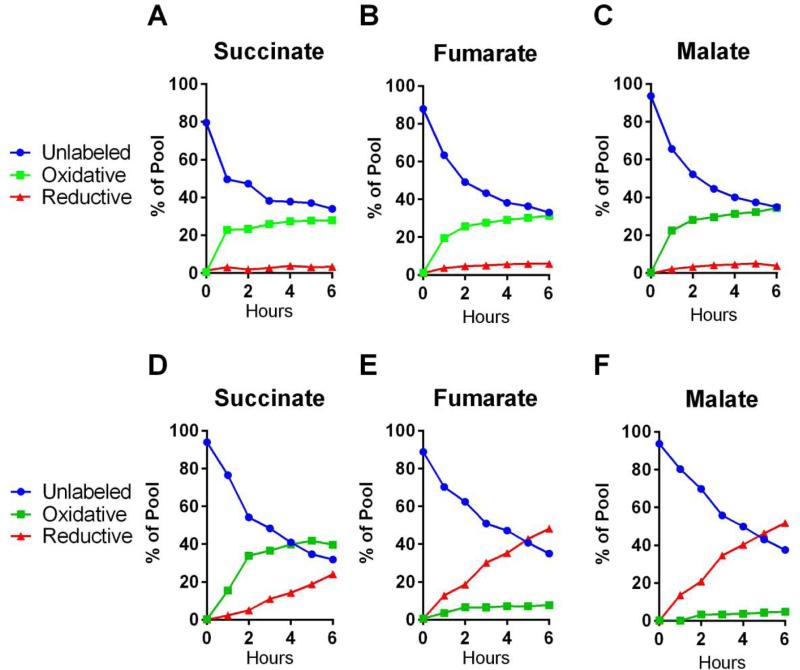
UOK262EV cells lack FH activity and accumulate large amounts of fumarate (Frezza et al., 2011); elevated succinate was therefore not surprising in these cells, because succinate precedes fumarate by one reaction in the TCA cycle. On the other hand, TCA cycle perturbation in 143Bcytb cells results from primary ETC dysfunction, and reductive carboxylation is postulated to be a consequence of accumulated AKG (Anastasiou and Cantley, 2012, Fendt et al., 2013). Accumulation of AKG is not predicted to result in elevated succinate. We previously reported that 143Bcytb cells produce succinate through simultaneous oxidative and reductive glutamine metabolism (Mullen et al., 2012). To determine the relative contributions of these two pathways, we cultured 143Bwt and 143Bcytb with [U-13C]glutamine and monitored time-dependent 13C incorporation in succinate and other TCA cycle intermediates. Oxidative metabolism of glutamine generates succinate, fumarate and malate containing four glutamine-derived 13C nuclei on the first turn of the cycle (m+4), while reductive metabolism results in the incorporation of three 13C nuclei in these intermediates (Fig. S2). As expected, oxidative glutamine metabolism was the predominant source of succinate, fumarate and malate in 143Bwt cells (Fig. 2A-C). In 143Bcytb, fumarate and malate were produced primarily through reductive metabolism (Fig. 2E-F). Conversely, succinate was formed primarily through oxidative glutamine metabolism, with a minor contribution from the reductive carboxylation pathway (Fig. 2D). Notably, this oxidatively-derived succinate was detected prior to that formed through reductive carboxylation. This indicated that 143Bcytb cells retain the ability to oxidize AKG despite the observation that most of the citrate pool bears the labeling pattern of reductive carboxylation. Together, the labeling data in 143Bcytb cells revealed bidirectional metabolism of carbon from glutamine to produce various TCA cycle intermediates.
Figure 2. Oxidative glutamine metabolism is the primary route of succinate formation in cells using reductive carboxylation to generate citrate.
(A-C) 143Bwt cells were cultured in medium containing unlabeled glucose and [U-13C]glutamine, and labeling patterns in succinate, fumarate and malate were followed. Isotopologues with four 13C atoms (m+4) are formed through oxidative metabolism (green), and isotopologues with three 13C atoms (m+3) are formed through reductive metabolism (red). See Fig. S2 for labeling scheme.
(D-F) 143Bcytb cells were cultured in medium containing unlabeled glucose and [U-13C]glutamine, and labeling patterns in succinate, fumarate and malate were followed as in (A-C). Note that succinate formation is primarily oxidative, but that fumarate and malate formation is almost exclusively reductive, indicating bidirectional metabolism of glutamine.
Pyruvate carboxylation contributes to the TCA cycle in cells using reductive carboxylation
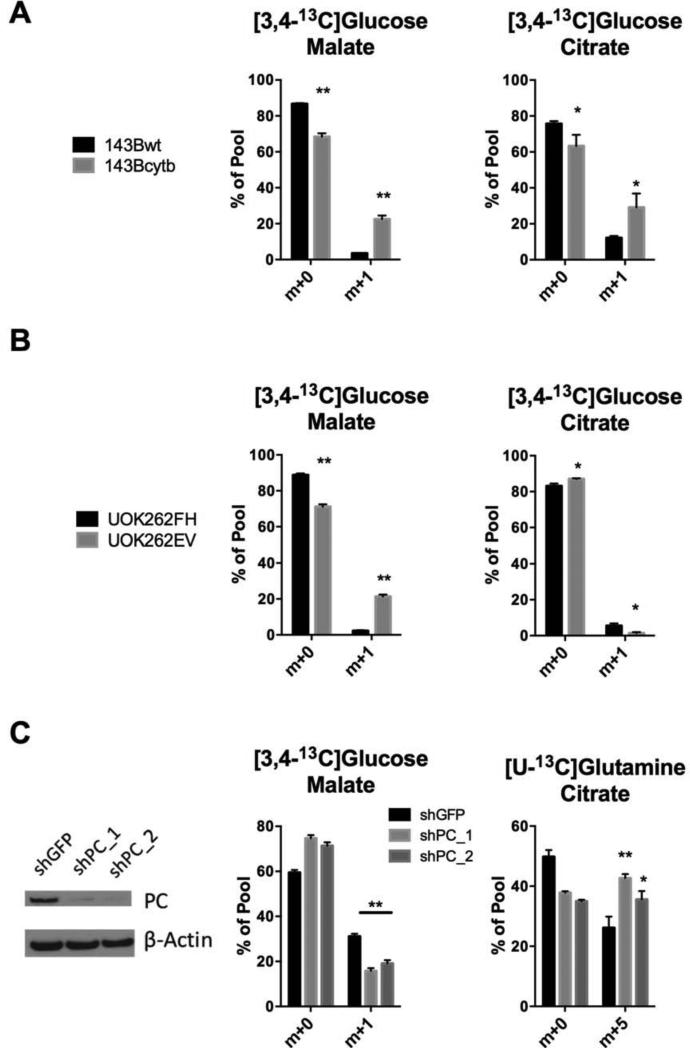
Because of the persistence of oxidative metabolism, we determined the extent to which other routes of metabolism besides reductive carboxylation contributed to the TCA cycle. We previously reported that silencing the glutamine-catabolizing enzyme glutaminase (GLS) depletes pools of fumarate, malate and OAA, eliciting a compensatory increase in pyruvate carboxylase (PC) to supply the TCA cycle (Cheng et al., 2011). In cells with defective oxidative phophorylation, production of OAA by PC may be preferable to glutamine oxidation because it diminishes the need to recycle reduced electron carriers generated by the TCA cycle. Citrate synthase (CS) can then condense PC-derived OAA with acetyl-CoA to form citrate. To examine the contribution of PC to the TCA cycle, cells were cultured with [3,4-13C]glucose. In this labeling scheme, glucose-derived pyruvate is labeled in carbon 1 (Fig. S3). This label is retained in OAA if pyruvate is carboxylated, but removed as CO2 during conversion of pyruvate to acetyl-CoA by pyruvate dehydrogenase (PDH). Thus, labeling of TCA cycle intermediates from [3,4-13C]glucose indicates activity of PC. Both cell lines contained label in citrate and other TCA cycle intermediates from [3,4-13C]glucose, but the fractional contribution was significantly greater in 143Bcytb (Fig. 3A). UOK262EV cells also displayed PC-mediated labeling of TCA cycle intermediates, particularly malate; citrate labeling was barely above baseline in both these cell lines (Fig. 3B). Malate labeling from PC was suppressed in UOK262FH cells because FH-dependent production of fumarate, malate and OAA diluted the PC-dependent labeling. Silencing PC expression in 143Bcytb cells reduced PC-dependent transfer of carbon from glucose to the TCA cycle, and increased the contribution of reductive glutamine metabolism to the citrate pool (Fig. 3C). Together, the data demonstrate that PC-dependent oxidative glucose metabolism contributes to the TCA cycle in these models of glutamine-dependent reductive carboxylation.
Figure 3. Pyruvate carboxylase contributes to citrate formation in cells using reductive carboxylation.
(A-B) 143B and UOK262 cells were cultured with [3,4-13C]glucose and unlabeled glutamine and labeling patterns in malate and citrate were analyzed. Data are the average and S.D. of three independent cultures. *p<0.05; ** p<0.005, Student's t-test.
(C) Left, abundance of pyruvate carboxylase (PC) protein in 143Bcytb cells stably expressing non-targeting (shGFP) or two independent hairpins targeting PC (shPC_1, shPC_2). Cells were cultured with [3,4-13C]glucose and unlabeled glutamine (middle), or with unlabeled glucose and [U- 13C]glutamine, and labeling patterns were analyzed. Data are the average and S.D. of three independent cultures. *p<0.05; ** p<0.005, Student's t-test.
Oxidative metabolism of AKG is required for reductive carboxylation
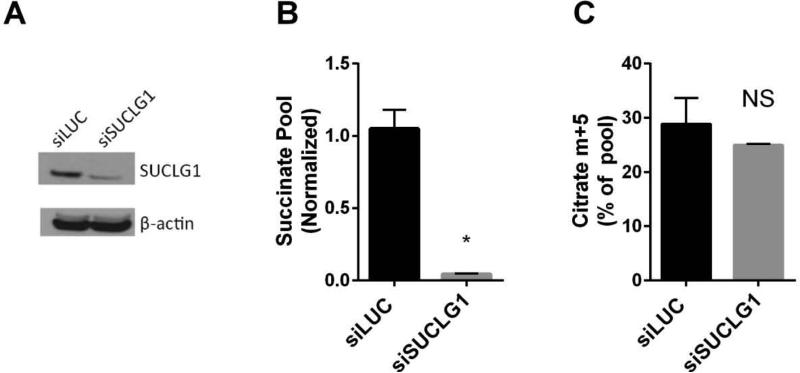
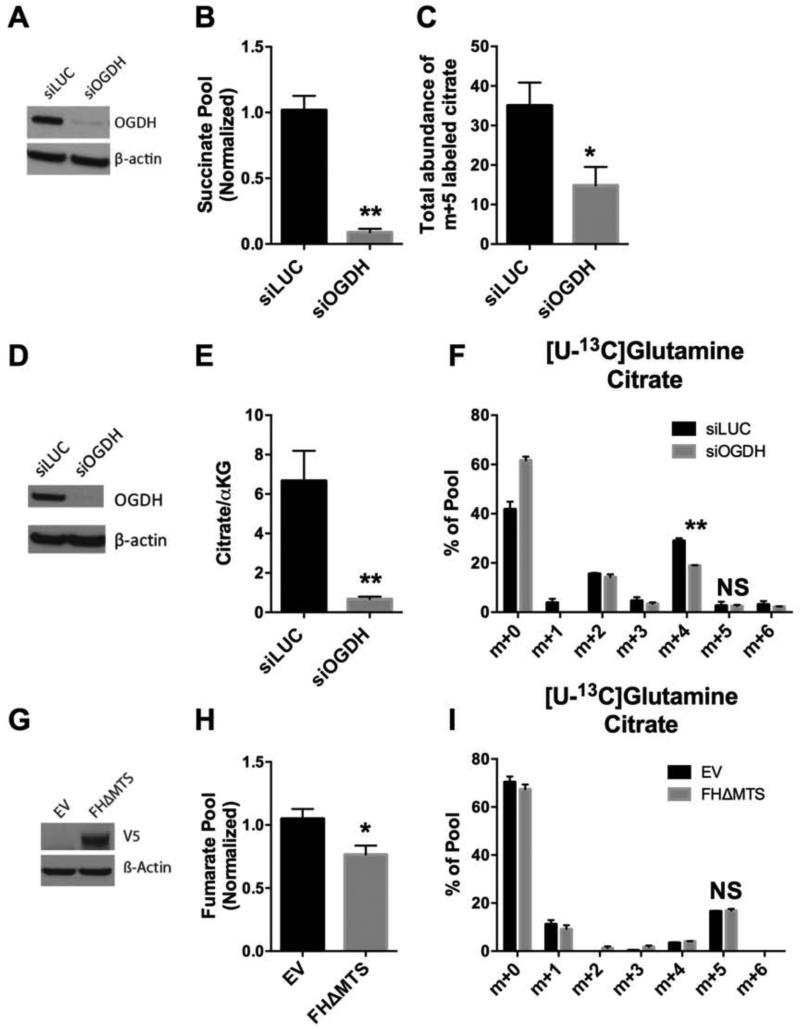
Oxidative synthesis of succinate from AKG requires two reactions: the oxidative decarboxylation of AKG to succinyl-CoA by AKG dehydrogenase, and the conversion of succinyl-CoA to succinate by succinyl-CoA synthetase. In tumors with mutations in the succinate dehydrogenase (SDH) complex, large accumulations of succinate are associated with epigenetic modifications of DNA and histones to promote malignancy (Kaelin and McKnight, 2013, Killian et al., 2013). We therefore tested whether succinate accumulation per se was required to induce reductive carboxylation in 143Bcytb cells. We used RNA interference directed against the gene encoding the alpha subunit (SUCLG1) of succinyl-CoA synthetase, the last step in the pathway of oxidative succinate formation from glutamine (Fig. 4A). Silencing this enzyme greatly reduced succinate levels (Fig. 4B), but had no effect on the labeling pattern of citrate from [U-13C]glutamine (Fig. 4C). Thus, succinate accumulation is not required for reductive carboxylation.
Figure 4. Elevated succinate abundance is dispensable for reductive carboxylation.
(A) Abundance of the alpha subunit of succinate-CoA ligase (encoded by SUCLG1) in 143Bcytb cells transfected with a control siRNA (siLUC) or siRNA directed against SUCLG1 (siSUCLG1).
(B) Relative abundance of succinate in 143Bcytb following transient knockdown of LUC or SUCLG1. Data are the average and S.D. of three independent cultures. *p<0.05, Student's t-test.
(C) Fraction of citrate containing five glutamine-derived 13C nuclei (m+5) following transient knockdown of LUC or SUCLG1 and culture with [U-13C]glutamine and unlabeled glucose. Data are the average and S.D. of three independent cultures. NS, not significant.
We next tested whether the proximal step in the oxidative pathway was required for reductive carboxylation. In 143Bcytb cells, we transiently silenced expression of the E1 component of the AKG dehydrogenase complex encoded by OGDH (Fig. 5A). OGDH silencing greatly diminished succinate abundance, validating effective suppression of the pathway (Fig. 5B). Surprisingly, both the abundance of citrate and the fraction that contained five glutamine-derived carbons from [U-13C]glutamine were suppressed when OGDH was silenced, indicating a suppression of reductive citrate formation (Fig. 5C). Stable silencing of OGDH with either of two shRNAs produced a similar metabolic effect (Fig. S4A-C), as did transient OGDH silencing in UOK262EV cells (Fig. S4E,F). This was unexpected given the association of a low citrate/AKG ratio with induction of reductive carboxylation via mass action. To test whether altering the citrate/AKG ratio by manipulating AKG dehydrogenase activity could stimulate reductive carboxylation in cells with normal ETC function, OGDH was silenced in 143Bwt cells (Fig. 5D). Knockdown of OGDH significantly lowered the citrate/AKG ratio to levels previously reported to induce reductive carboxylation (Fig. 5E) (Fendt et al., 2013, Gameiro et al., 2013a). However, culture with [U- 13C]glutamine revealed a suppression of citrate formed through oxidative metabolism (m+4) and an increase in unlabeled citrate (m+0), but no evidence for an increase in reductive carboxylation (m+5, Fig. 5F). Together the data demonstrate that a reduced citrate/AKG ratio is not sufficient to induce reductive carboxylation in the absence of AKG dehydrogenase function, and imply that additional factors account for the role of AKG dehydrogenase in reductive carboxylation. To determine whether AKG dehydrogenase suppression impaired growth of 143Bcytb cells, OGDH was stably silenced and cells were challenged to form colonies in soft agar. Over the two week assay, OGDH-depleted cells formed colonies as effectively as control cells (Fig. S4D). Thus, the levels of AKG dehydrogenase suppression achievable through stable RNA interference against OGDH was not sufficient to reduce cell growth in this model.
Figure 5. AKG dehydrogenase is required for reductive carboxylation.
(A) Abundance of OGDH protein in 143Bcytb cells transfected with a control siRNA (siLUC) or siRNA directed against OGDH (siOGDH).
(B) Relative abundance of succinate in 143Bcytb cells following transient knockdown of siLUC or siOGDH. Data are average and S.D. of three independent cultures. **p<0.005, Student's t-test.
(C) Abundance of m+5 labeled citrate in 143Bcytb cells after transient knockdown of LUC or OGDH cultured with [U-13C]glutamine and unlabeled glucose. Abundance of labeled citrate was calculated by multiplying the relative citrate pool size by the percent citrate m+5. Data are the average and S.D. of three independent cultures. *p< 0.05, Student's t-test.
(D) Abundance of OGDH protein in 143Bwt cells transiently transfected with LUC or OGDH.
(E) Ratio of citrate to AKG in 143Bwt cells following knockdown of LUC or OGDH. Data are the average and S.D. of four independent cultures. **p<0.005, Student's t-test.
(F) Mass isotopomer distribution of citrate in 143Bwt cells following transient knockdown of LUC or OGDH and cultured with [U-13C]glutamine and unlabeled glucose. Data are the average and S.D. of three independent cultures. **p<0.005, Student's t-test.
(G) Western blot demonstrating expression of fumarate hydratase lacking its mitochondrial targeting sequence and containing a V5 epitope tag (FHΔMTS), in UOK262 cells. EV, empty vector.
(H) Relative fumarate abundance in UOK262 cells containing or lacking FHΔMTS. Data are the average and S.D. of three independent cultures. *p< 0.05, Student's t-test.
(I) Mass isotopomer distribution of citrate in UOK262 cells containing or lacking FHΔMTS. Data are the average and S.D. of three independent cultures. NS, not significant.
As a further examination of the role of mitochondrial dysfunction in reductive carboxylation, we transfected UOK262EV cells with an allele of FH that lacked a mitochondrial targeting sequence and was confined to the cytosol (FHΔMTS). This form of FH was previously shown to reduce fumarate levels and reverse some of the effects of FH deficiency in murine kidney cells (O'Flaherty et al., 2010). Transient expression of FHΔMTS in UOK262EV cells reduced fumarate abundance, but had no effect on the fraction of citrate formed through reductive carboxylation (Fig. 5G-I).
AKG dehydrogenase and NNT maintain a redox state favorable for reductive carboxylation
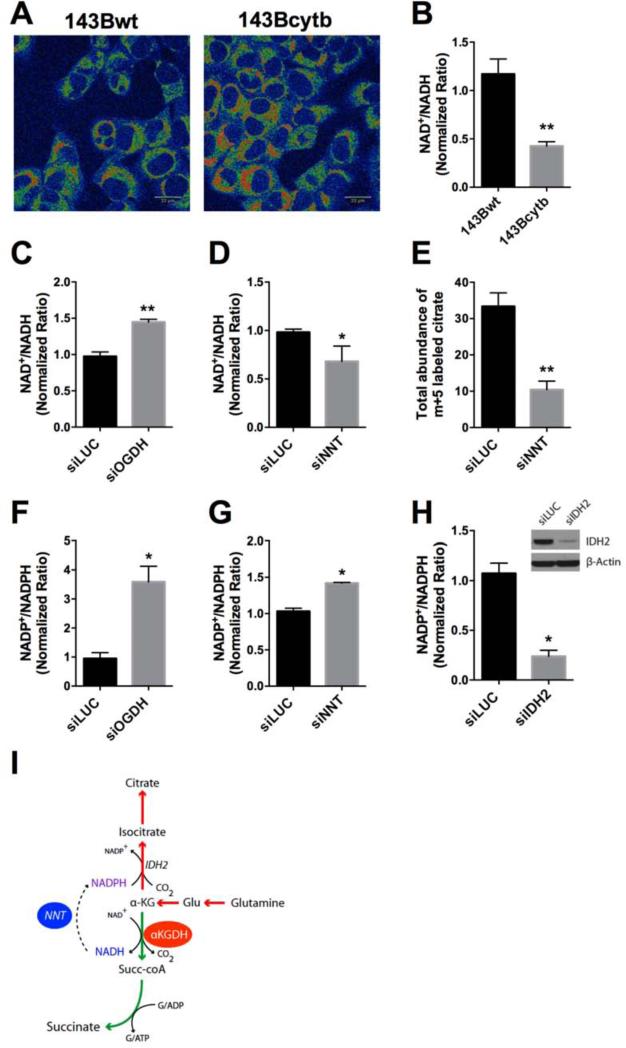
In addition to converting AKG to succinyl-CoA, AKG dehydrogenase also generates reducing equivalents as NADH. We therefore tested whether AKG dehydrogenase supported reductive carboxylation by contributing to a low NAD+/NADH ratio. Two-photon fluorescence microscopy revealed enhanced abundance of NADH in the mitochondria of 143Bcytb cells compared to 143Bwt cells (Fig. 6A). Enzymatic assays performed on whole cell lysates also demonstrated a reduced total NAD+/NADH ratio in 143Bcytb cells (Fig. 6B). Silencing OGDH resulted in an increase in the NAD+/NADH ratio, indicating that AKG dehydrogenase contributes to the low NAD+/NADH ratio in 143Bcytb cells. Importantly, NADPH rather than NADH is the immediate source of reducing equivalents for the reductive carboxylation reaction. Mitochondrial NADPH can be produced from NADH by the enzyme NNT, and this enzyme contributes to reductive glutamine metabolism in some cancer cells (Sazanov and Jackson, 1994, Gameiro et al., 2013a). We therefore tested the impact of silencing NNT on redox balance and reductive carboxylation. Transfection with siRNAs directed against NNT reduced abundance of the transcript by more than 80% and substantially reduced NNT protein levels (Fig. S5A,B). This resulted in a reduction of the NAD+/NADH ratio that was similar in size to the increase noted upon OGDH silencing (Fig. 6D). Culture of these cells with [U-13C]glutamine revealed a significant suppression of reductive citrate formation compared to cells transfected with control siRNA (Fig. 6E). Transient NNT silencing also reduced reductive citrate formation in UOK262EV cells (Fig. S5C,D).
Figure 6. AKG dehydrogenase and NNT contribute to NAD+/NADH ratio.
(A) Two-photon fluorescence imaging of NADH in 143Bwt and 143Bcytb. Red indicates areas of increased NADH abundance. Scale bar 22μm.
(B) NAD+/NADH ratio in 143Bwt and 143Bcytb cells. Data are the average and S.D. of 3-4 independent cultures. **p<0.005, Student's t-test.
(C) NAD+/NADH ratio in 143Bcytb following transient transfection with siRNAs against Luciferase (siLUC) or OGDH (siOGDH). Data are the average and S.D. of 3-4 independent experiments. **p<0.005, Student's t-test.
(D) NAD+/NADH ratio in 143Bcytb following transfection with siLUC) or an siRNA pool directed against niconatinomide nucleotide transhydrogenase (siNNT). Data are the average and S.D. of 3-4 biological replicates. *p< 0.05, Student's t-test.
(E) Abundance of m+5 labeled citrate in 143Bcytb cells transfected with siLUC or siNNTand cultured with [U-13C]glutamine. Abundance of labeled citrate was calculated by multiplying the relative abundance of total citrate by the fractional contribution of the m+5 isotopomer. Data are the average and S.D. of three independent cultures. **p<0.005, Student's t-test.
(F) NADP+/NADPH ratio in 143Bcytb cells following transient transfection with siLUC or siOGDH. Data are the average and S.D. of three independent cultures. *p< 0.05, Student's t-test.
(G) NADP+/NADPH ratio in 143Bcytb cells following transient transfection with siLUC or siNNT. Data are the average and S.D. of three independent cultures. *p< 0.05, Student's t-test.
(H) NADP+/NADPH ratio in 143Bcytb cells following transient transfection with siLUC or siIDH2. Data are the average and S.D. of three independent cultures. Inset is western blot depicting bundance of IDH2 protein in both conditions. *p< 0.05, Student's t-test.
(I) Model for induction of reductive carboxylation in 143Bcytb cells. Flux through AKG pdehydrogenase generates succinate and NADH, which is dissipated by NNT to generate NADPH during reductive carboxylation.
Finally, we tested whether these enzymes also controlled the NADP+/NADPH ratio in 143Bcytb cells. Silencing either OGDH or NNT increased the NADP+/NADPH ratio (Fig. 6F,G), whereas silencing IDH2 reduced it (Fig. 6H). Together, these data are consistent with a model in which persistent metabolism of AKG by AKG dehydrogenase produces NADH that supports reductive carboxylation by serving as substrate for NNT-dependent NADPH formation, and that IDH2 is a major consumer of NADPH during reductive carboxylation (Fig. 6I).
DISCUSSION
Reductive carboxylation of AKG initiates a non-conventional form of metabolism that produces TCA cycle intermediates when oxidative metabolism is impaired by mutations, drugs or hypoxia. Because NADPH-dependent isoforms of IDH are reversible, supplying supra-physiological pools of substrates on either side of the reaction drives function of the enzyme as a reductive carboxylase or an oxidative decarboxylase. Thus, in some circumstances reductive carboxylation may operate in response to a mass effect imposed by drastic changes in the abundance of AKG and isocitrate/citrate. However, reductive carboxylation cannot occur without a source of reducing equivalents to produce NADPH. The current work demonstrates that AKG dehydrogenase, an NADH-generating enzyme complex, is required to maintain a low NAD+/NADH ratio for reductive carboxylation of AKG. Thus, reductive carboxylation not only coexists with oxidative metabolism of AKG, but depends on it. Furthermore, silencing NNT, a consumer of NADH, also perturbs the redox ratio and suppresses reductive formation of citrate. These observations suggest that the segment of the oxidative TCA cycle culminating in succinate is necessary to transmit reducing equivalents to NNT for the reductive pathway (Fig 6I).
It is clear from the data that the cells studied here demonstrate persistent oxidative metabolism despite their mutations in oxidative enzymes and their citrate pools largely bearing the labeling hallmarks of reductive metabolism. First, isotope labeling studies with [U-13C]glutamine revealed flow of carbon from glutamine to succinate in a pattern identical to the conventional TCA cycle. Second, silencing SUCLG1 reduced the abundance of succinate, which would not be expected in the absence of oxidative succinate formation. Third, silencing OGDH suppressed the formation of succinate and increased the NAD+/NADH ratio. A recent study used computational flux modeling to provide evidence that oxidative TCA cycle metabolism occurs in the context of either hypoxia or cytB mutation, and that oxidation of acetyl-CoA produces a fraction of cellular succinate and may account for the net direction of IDH (Fan et al., 2013). Here we used specific molecular approaches to demonstrate that much of the succinate pool is derived from oxidation of glutamine rather than acetyl-CoA, and that interrupting this pathway elicits changes in both the cellular redox state and in the pathways available to produce citrate. Specifically, AKG dehydrogenase, which is required to produce succinate through oxidative metabolism, is required for maximal activity of the reductive pathway. Thus, the persistent oxidative metabolism of glutamine is required for both redox homeostasis and for maintenance of TCA cycle metabolite pools in cells harboring mitochondrial defects.
An induction of pyruvate carboxylation was also observed in cells lacking cytb or FH. PC contributes to the TCA cycle, although other than requiring acetyl-CoA as an activator, it does not require concomitant oxidative metabolism to function. Pyruvate carboxylation produces OAA without the need for glutamine metabolism or reductive carboxylation, and thus its induction in the cells observed here may signify a role as a complementary source of TCA cycle metabolites in addition to these other pathways. In most glutamine-dependent cancer cells studied to date, pyruvate carboxylase is dispensable (Cheng et al., 2011), although it seems to be active in several types of tumors in vivo (Marin-Valencia et al., 2012, Fan et al., 2009). Although PC-dependent contribution of carbon to the TCA cycle was induced by mutation of cytb or FH, it accounted for only a minority of TCA cycle metabolites, and silencing PC increased the fractional contribution from glutamine. In other cases of severe mitochondrial dysfunction, PC or alternative pathways to citrate formation may play a more prominent role than they do in the cell lines studied here. For example, although human FH-deficient renal carcinoma cells show evidence of reductive carboxylation, mouse embryonic fibroblasts homozygous null for FH do not appear to use this pathway (Adam et al., 2013). These cells have been demonstrated to covalently modify a number of metabolic enzymes through the non-enzymatic addition of fumarate to cysteine residues, creating an S-(2-succino) cysteine group in a process termed succination (Ternette et al., 2013). The mitochondrial isoform of aconitase is inhibited by succination in FH-deficient MEFs, and this inhibition may prevent isocitrate produced through mitochondrial reductive carboxylation from being converted to citrate.
Succinate accumulation was observed in cells with cytb or FH mutations. However, this accumulation was dispensable for reductive carboxylation, because silencing SUCLG1 expression had no bearing on the pathway as long as AKG dehydrogenase was active. Furthermore, succinate accumulation was not a universal finding of cells using reductive carboxylation. Rather, high succinate levels were observed in cells with distal defects in the ETC (complex III: antimycin, cytb mutation; complex IV: hypoxia) but not defects in complex I (rotenone, metformin, NDUFA1 mutation). These differences reflect the known suppression of SDH activity when downstream components of the ETC are impaired, and the various mechanisms by which succinate may be formed through either oxidative or reductive metabolism. Succinate has long been known as an evolutionarily conserved anaerobic end product of amino acid metabolism during prolonged hypoxia, including in diving mammals (Hochachka and Storey, 1975, Hochachka et al., 1975). The terminal step in this pathway is the conversion of fumarate to succinate using the NADH-dependent “fumarate reductase” system, essentially a reversal of succinate dehydrogenase/ETC complex II (Weinberg et al., 2000, Tomitsuka et al., 2010). However, this process requires reducing equivalents to be passed from NADH to complex I, then to Coenzyme Q, and eventually to complex II to drive the reduction of fumarate to succinate. Hence, producing succinate through reductive glutamine metabolism would require functional complex I. Interestingly, the fumarate reductase system has generally been considered as a mechanism to maintain a proton gradient under conditions of defective ETC activity. Our data suggest that the system is part of a more extensive reorganization of the TCA cycle that also enables reductive citrate formation.
In summary, we demonstrated that branched AKG metabolism is required to sustain levels of reductive carboxylation observed in cells with mitochondrial defects. The organization of this branched pathway suggests that it serves as a relay system to maintain the redox requirements for reductive carboxylation, with the oxidative arm producing reducing equivalents at the level of AKG dehydrogenase and NNT linking this activity to the production of NADPH to be used in the reductive carboxylation reaction. Hence, impairment of the oxidative arm prevents maximal engagement of reductive carboxylation. As both NNT and AKG dehydrogenase are mitochondrial enzymes, the work emphasizes the flexibility of metabolic systems in the mitochondria to fulfill requirements for redox balance and precursor production even when the canonical oxidative function of the mitochondria is impaired.
EXPERIMENTAL PROCEDURES
Cell lines, culture and reagents
143Bwt, 143Bcytb, CCL16-B2 and CCL16-NDI1 cells were cultured with DMEM supplemented with penicillin/streptomycin, 10% fetal bovine serum (FBS), L-glutamine (6 mM), uridine (100 μg/mL) and sodium-pyruvate (1 mM). To generate vector controlled UOK262 cells (UOK262EV), PCDNA3.1 empty vector plasmid was transfected with Fugene (Promega) to UOK262 cells. To isolate stable integration of the plasmid, cells were continuously grown in G418 (300 μg/mL; Invitrogen). Both cell lines were then cultured with 143B media supplemented with G418.
Metabolomics
Subconfluent culture dishes were incubated for two hours in DMEM which contained 15 mM glucose and 2 mM glutamine supplemented with 10% dialyzed FBS. Following this, cells were washed twice with ice cold saline, then overlaid with 500 μl of cold methanol/water (50/50, v/v). Cells were transferred to an eppendorf tube and subjected to three freeze-thaw cycles. After rigorous vortexing, the debris was pelleted by centrifugation at 16,000 × g and 4°C for 15 min. Pellets were used for protein quantitation (BCA Protein Assay, Thermo). The supernatant was transferred to a new tube and evaporated to dryness using a SpeedVac concentrator (Thermo Savant, Holbrook, NY). Metabolites were reconstituted in 100 μL of 0.03% formic acid in analytical-grade water, vortex-mixed and centrifuged to remove debris. Thereafter, the supernatant was transferred to a HPLC vial for the metabolomics study.
Targeted metabolite profiling was performed using a liquid chromatography-mass spectrometry/mass spectrometry (LC/MS/MS) approach. Separation was achieved on a Phenomenex Synergi Polar-RP HPLC column (150 × 2 mm, 4 μm, 80 Å) using a Nexera Ultra High Performance Liquid Chromatograph (UHPLC) system (Shimadzu Corporation, Kyoto, Japan). The mobile phases employed were 0.03% formic acid in water (A) and 0.03% formic acid in acetonitrile (B). The gradient program was as follows: 0-3 min, 100% A; 3-15 min, 100% - 0% A; 15-21 min, 0% A; 21-21.1 min, 0% - 100% A; 21.1-30 min, 100% A. The column was maintained at 35°C and the samples kept in the autosampler at 4°C. The flow rate was 0.5 mL/min, and injection volume 10 μL. The mass spectrometer was an AB QTRAP 5500 (Applied Biosystems SCIEX, Foster City, CA) with electrospray ionization (ESI) source in multiple reaction monitoring (MRM) mode. Sample analysis was performed in positive/negative switching mode. Declustering potential (DP) and collision energy (CE) were optimized for each metabolite by direct infusion of reference standards using a syringe pump prior to sample analysis. The MRM MS/MS detector conditions were set as follows: curtain gas 30 psi; ion spray voltages 5000 V (positive) and -1500 V (negative); temperature 650°C; ion source gas 1 50 psi; ion source gas 2 50 psi; interface heater on; entrance potential 10 V. In total, 173 water soluble endogenous metabolites were targeted, with 92 metabolites confidently detected above the baseline set by cell-free samples. The MRM transitions (m/z), DPs (V) and CEs (V) of the detected metabolites are listed in Table S1. Dwell time for each transition was set at 3 msec. Cell samples were analyzed in a randomized order, and MRM data was acquired using Analyst 1.6.1 software (Applied Biosystems SCIEX, Foster City, CA).
Chromatogram review and peak area integration were performed using MultiQuant software version 2.1 (Applied Biosystems SCIEX, Foster City, CA). Although the numbers of cells were very similar and each sample was processed identically and randomly, the peak area for each detected metabolite was normalized against the protein content of that sample to correct any variations introduced from sample handling through instrument analysis. The normalized area values were used as variables for the multivariate and univariate statistical data analysis. The chromatographically co-eluted metabolites with shared MRM transitions were shown in a grouped format, i.e., alanine/sarcosine. All multivariate analyses and modeling on the normalized data were carried out using SIMCA-P (version 13.0.1, Umetrics, Umeå, Sweden). The pre-processed datasets were mean-centered and unit-variance scaled, and then evaluated by principal component analysis (PCA) to visualize the clustering trend, as well as to detect and exclude outlier datasets. The hierarchical clustering of metabolites in different cell lines was analyzed using the R Statistical Package. Univariate statistical differences of the metabolites between two groups were analyzed using Student's t-test.
Quantitation of citrate, alpha-ketoglutarate and succinate
For the quantitation of citrate, alpha-ketoglutarate and succinate, a 50 μL aliquot of an internal standard (IS) mixture containing [13C6]-citrate (1 μM), [13C4]-alpha-ketoglutarate (10 μM) and [13C2]-succinate (1 μM) prepared in analytical-grade water was added to 50 μL of the scraped cells. After adding 500 μL of pre-cooled methanol/water (50/50, v/v), the analyte/IS mix was extracted and prepared using the same procedures as described above. To construct a calibration curve for absolute quantitation, working solutions containing all the three analytes were prepared in 1 × PBS at concentrations of 0.2, 0.5, 1, 2, 5, 10, 20, 50 and 100 μM, and then spiked with 50 μL of the same IS mixture. Thereafter, the standard samples were processed as described under “Metabolomics.” For LC/MS/MS analysis, only the MRM transitions of the three analytes and their ISes were monitored, which were 191 > 111 for citrate, 197 > 116 for [13C6]-citrate, 145 > 57.1 for alpha-ketoglutarate, 149 > 60 for [13C4]-alpha-ketoglutarate, 117 > 73.2 for succinate, and 119 > 74.2 for [13C2]-succinate, respectively. Dwell time for each transition was set at 30 msec.
Chromatogram review, peak area integration and concentration calculation of each metabolites was performed using MultiQuant software version 2.1 (Applied Biosystems SCIEX, Foster City, CA). The concentration (micromole per microliter) of each detected metabolite was normalized against the protein amount of that sample to achieve the final units of micromoles per milligram of protein. The normalized concentrations were compared between different cell lines by using Student's t-test. Differences with p < 0.05 were considered as statistically significant.
Quantitation of (L)-2HG and (D)-2HG
. Measurement of D-2HG and L-2HG was performed as described previously (Rakheja et al., 2011b, Rakheja et al., 2011a). Briefly, the metabolite extracts were derivatization with (+)-Di-O-acetyl-L-tartaric anhydride and injected for chromatographic separation on an Agilent Hypersil ODS 4.0×250 mm, 5 μm column (Santa Clara, CA), followed by detection and measurement using API 3000 triple–quadrupole mass spectrometer equipped with an electrospray ionization source (Applied Biosystems, Foster City, CA). MRM transitions were monitored at 363.2 > 147.2 for both L-2HG and D-2HG.
Isotope labeling experiments
Isotope labeling experiments were essentially preformed as described previously (Mullen et al., 2012). Briefly, DMEM with 10% dialyzed FBS was supplemented with isotopically labeled glucose or glutamine at a concentration of 15 mM and 2 mM, respectively. Unless specified, cells were incubated for exactly two hours. After this, dishes were washed twice with cold saline and metabolites were extracted in 50% methanol followed by three freeze-thaw cycles. Extracts were centrifuged and supernatant was dried. Metabolites were derivitized with Trisil (Thermo) and analyzed on an Agilent 6970 gas chromatograph networked to an Agilent 5973 mass selective detector. A detailed list of all masses monitored were described previously (Cheng et al., 2011, Mullen et al., 2012).
RNA interference
Transient gene silencing experiments were performed using commercial siRNA pools as described (Mullen et al., 2012). Briefly, siRNA oligos targeting SUCLG1, OGDH or NNT (siGenome, Thermo) were transfected into 143B cells with DharmaFECT transfection reagent (Thermo); oligos targeting luciferase were used as a negative control (siGenome, Thermo). All experiments took place 72 hours later and were carried out as described above. For stable gene silencing, lentiviral-mediated shRNAs targeting PC or OGDH from the Mission shRNA pLKO.1-puro library (Sigma) were used to infect 143Bcytb cells according to supplied protocol. 143B cells were infected with an individual shRNA hairpin and stable integrants were selected with Puromycin (Invitrogen).
To monitor protein abundance cells were lysed in RIPA buffer and protein separated on NuPAGE® Novex® 4-12% Bis-Tris gel (Invitrogen). Protein was transferred to Immobilon transfer membranes (Millipore). Protein was detected using commercially available antibodies against SUCLG1 (Cell Signaling), OGDH (Sigma) or PC (Santa Cruz Biotechnology). NNT knockdown was quantified using qPCR. Briefly, RNA was extracted in Trizol (Invitrogen) and isolated according to manufacturer's protocol. cDNA was generated using iScript synthesis kit (Biorad) and transcript abundance was measured on a Thermo qPCR instrument.
Colony formation in soft agar
Cells (10×103 cells/well) were suspended in growth medium containing 0.3% agarose and plated in 6-well dishes onto a base layer composed of growth medium containing 0.6% agarose. The growth medium was replenished every four days, and after 14 days colonies greater than 100 microns in size were counted. Each condition was performed in triplicate wells, and the entire experiment was repeated three times.
NAD+/NADH and NADP+/NADPH measurements
The NAD+/NADH ratio was measured using a commercially available kit (Biovisison, K337-100). Extraction and measurements were performed according to manufacturer's protocol. Briefly, cells were incubated two hours with DMEM supplemented with 10% dialyzed FBS and unlabeled glucose and glutamine at a concentration of 15 mM and 2 mM, respectively. After this cells were washed twice with saline and fresh saline was added so that cells could be scraped off the dishes and pelleted. Next, NAD+ and NADH were extracted in the supplied Extraction Buffer. Samples were subjected to two freeze-thaws and centrifuged. Aliquots of each sample were heated at 60°C for 30 minutes to decompose NAD+. Following this, samples were loaded to 96 well plates and for absorbance measurment at OD450. The NADP+/NADPH ratio was measured using a commercially available kit (Abcam, ab65349). Cells were washed with cold saline, then fresh saline was added so that the cells could be scraped from the dishes and pelleted. Next, NADP+ and NADPH were extracted in the supplied Extraction Buffer. Samples were subjected to two freeze-thaw cycles and centrifuged. Aliquots of each sample were heated at 60°C for 30 minutes to decompose NADP+. Samples were then transferred to 96 well plates for absorbance measurement at OD450.
Two-Photon Fluorescence of NADH
NADH was imaged using 2-photon fluorescence microcopy. Cells were seeded to 35 mm glass-bottom petri dishes. Before imaging, media was replaced with DMEM lacking phenol red. Cells were imaged on a Zeiss LSM 510 META with Chameleon XR NIR laser. NADH fluorescence was imaged after excitation at 775 nm. To better demonstrate the changes in NADH intensity a rainbow color table was applied to the images using Image J (ImageJ, National Institutes of Health, http://rsb.info.nih.gov/ij/).
Statistical Methods
Unless otherwise indicated, data were analyzed in either Microsoft Excel or GraphPad Prism. Statistical significance was established using Student's t-test.
Supplementary Material
Highlights.
Cells with mitochondrial defects use bidirectional metabolism of the TCA cycle.
Glutamine supplies the succinate pool through oxidative and reductive metabolism.
Oxidative TCA cycle metabolism is required for reductive citrate formation.
Oxidative metabolism produces reducing equivalents for reductive carboxylation.
ACKNOWLEDGEMENTS
We thank Hien Nguyen for measurements of AKG, the Live Cell Imaging Core at UT Southwestern for microscopy, and Robert Harris for experimental assistance. Patrick Pollard provided the FHΔMTS construct, and Othon Iliopoulos provided advice on western blots for NNT. The 143B cybrids were supplied by I.F.M. de Coo and Carlos Moraes. R.J.D. was supported by grants from the N.I.H. (R01 CA157996) and the Welch Foundation (I-1733). A.R.M. was supported by an N.I.H. Training Grant (5T32GM083831).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
COMPETING INTERESTS. The corresponding author is a member of the Scientific Advisory Boards of Agios Pharmaceuticals, Inc, and Peloton Therapeutics, Inc.
REFERENCES
- ADAM J, YANG M, BAUERSCHMIDT C, KITAGAWA M, O'FLAHERTY L, MAHESWARAN P, OZKAN G, SAHGAL N, BABAN D, KATO K, SAITO K, IINO K, IGARASHI K, STRATFORD M, PUGH C, TENNANT DA, LUDWIG C, DAVIES B, RATCLIFFE PJ, EL-BAHRAWY M, ASHRAFIAN H, SOGA T, POLLARD PJ. A role for cytosolic fumarate hydratase in urea cycle metabolism and renal neoplasia. Cell Rep. 2013;3:1440–8. doi: 10.1016/j.celrep.2013.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ANASTASIOU D, CANTLEY LC. Breathless cancer cells get fat on glutamine. Cell Res. 2012;22:443–6. doi: 10.1038/cr.2012.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ASTUTI D, LATIF F, DALLOL A, DAHIA PL, DOUGLAS F, GEORGE E, SKOLDBERG F, HUSEBYE ES, ENG C, MAHER ER. Gene mutations in the succinate dehydrogenase subunit SDHB cause susceptibility to familial pheochromocytoma and to familial paraganglioma. Am J Hum Genet. 2001;69:49–54. doi: 10.1086/321282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BAYSAL BE, FERRELL RE, WILLETT-BROZICK JE, LAWRENCE EC, MYSSIOREK D, BOSCH A, VAN DER MEY A, TASCHNER PE, RUBINSTEIN WS, MYERS EN, RICHARD CW, 3RD, CORNELISSE CJ, DEVILEE P, DEVLIN B. Mutations in SDHD, a mitochondrial complex II gene, in hereditary paraganglioma. Science. 2000;287:848–51. doi: 10.1126/science.287.5454.848. [DOI] [PubMed] [Google Scholar]
- CHENG T, SUDDERTH J, YANG C, MULLEN AR, JIN ES, MATES JM, DEBERARDINIS RJ. Pyruvate carboxylase is required for glutamine-independent growth of tumor cells. Proc Natl Acad Sci U S A. 2011;108:8674–9. doi: 10.1073/pnas.1016627108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DANG L, WHITE DW, GROSS S, BENNETT BD, BITTINGER MA, DRIGGERS EM, FANTIN VR, JANG HG, JIN S, KEENAN MC, MARKS KM, PRINS RM, WARD PS, YEN KE, LIAU LM, RABINOWITZ JD, CANTLEY LC, THOMPSON CB, VANDER HEIDEN MG, SU SM. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–44. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DEBERARDINIS RJ, SAYED N, DITSWORTH D, THOMPSON CB. Brick by brick: metabolism and tumor cell growth. Curr Opin Genet Dev. 2008;18:54–61. doi: 10.1016/j.gde.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAN J, KAMPHORST JJ, RABINOWITZ JD, SHLOMI T. Fatty acid labeling from glutamine in hypoxia can be explained by isotope exchange without net reductive IDH flux. J Biol Chem. 2013 doi: 10.1074/jbc.M113.502740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAN TW, LANE AN, HIGASHI RM, FARAG MA, GAO H, BOUSAMRA M, MILLER DM. Altered regulation of metabolic pathways in human lung cancer discerned by (13)C stable isotope-resolved metabolomics (SIRM). Mol Cancer. 2009;8:41. doi: 10.1186/1476-4598-8-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FENDT SM, BELL EL, KEIBLER MA, OLENCHOCK BA, MAYERS JR, WASYLENKO TM, VOKES NI, GUARENTE L, VANDER HEIDEN MG, STEPHANOPOULOS G. Reductive glutamine metabolism is a function of the alpha-ketoglutarate to citrate ratio in cells. Nat Commun. 2013;4:2236. doi: 10.1038/ncomms3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FREZZA C, ZHENG L, FOLGER O, RAJAGOPALAN KN, MACKENZIE ED, JERBY L, MICARONI M, CHANETON B, ADAM J, HEDLEY A, KALNA G, TOMLINSON IP, POLLARD PJ, WATSON DG, DEBERARDINIS RJ, SHLOMI T, RUPPIN E, GOTTLIEB E. Haem oxygenase is synthetically lethal with the tumour suppressor fumarate hydratase. Nature. 2011;477:225–8. doi: 10.1038/nature10363. [DOI] [PubMed] [Google Scholar]
- GAMEIRO PA, LAVIOLETTE LA, KELLEHER JK, ILIOPOULOS O, STEPHANOPOULOS G. Cofactor balance by nicotinamide nucleotide transhydrogenase (NNT) coordinates reductive carboxylation and glucose catabolism in the tricarboxylic acid (TCA) cycle. J Biol Chem. 2013a;288:12967–77. doi: 10.1074/jbc.M112.396796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GAMEIRO PA, YANG J, METELO AM, PEREZ-CARRO R, BAKER R, WANG Z, ARREOLA A, RATHMELL WK, OLUMI A, LOPEZ-LARRUBIA P, STEPHANOPOULOS G, ILIOPOULOS O. In vivo HIF-mediated reductive carboxylation is regulated by citrate levels and sensitizes VHL-deficient cells to glutamine deprivation. Cell Metab. 2013b;17:372–85. doi: 10.1016/j.cmet.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HILLER K, METALLO CM. Profiling metabolic networks to study cancer metabolism. Curr Opin Biotechnol. 2013;24:60–8. doi: 10.1016/j.copbio.2012.11.001. [DOI] [PubMed] [Google Scholar]
- HOCHACHKA PW, OWEN TG, ALLEN JF, WHITTOW GC. Multiple end products of anaerobiosis in diving vertebrates. Comp Biochem Physiol B. 1975;50:17–22. doi: 10.1016/0305-0491(75)90292-8. [DOI] [PubMed] [Google Scholar]
- HOCHACHKA PW, STOREY KB. Metabolic consequences of diving in animals and man. Science. 1975;187:613–21. doi: 10.1126/science.163485. [DOI] [PubMed] [Google Scholar]
- ICARD P, POULAIN L, LINCET H. Understanding the central role of citrate in the metabolism of cancer cells. Biochim Biophys Acta. 2012;1825:111–6. doi: 10.1016/j.bbcan.2011.10.007. [DOI] [PubMed] [Google Scholar]
- KAELIN WG, JR., MCKNIGHT SL. Influence of metabolism on epigenetics and disease. Cell. 2013;153:56–69. doi: 10.1016/j.cell.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KILLIAN JK, KIM SY, MIETTINEN M, SMITH C, MERINO M, TSOKOS M, QUEZADO M, SMITH WI, JR., JAHROMI MS, XEKOUKI P, SZAREK E, WALKER RL, LASOTA J, RAFFELD M, KLOTZLE B, WANG Z, JONES L, ZHU Y, WANG Y, WATERFALL JJ, O'SULLIVAN MJ, BIBIKOVA M, PACAK K, STRATAKIS C, JANEWAY KA, SCHIFFMAN JD, FAN JB, HELMAN L, MELTZER PS. Succinate dehydrogenase mutation underlies global epigenomic divergence in gastrointestinal stromal tumor. Cancer Discov. 2013;3:648–57. doi: 10.1158/2159-8290.CD-13-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEONARDI R, SUBRAMANIAN C, JACKOWSKI S, ROCK CO. Cancer-associated isocitrate dehydrogenase mutations inactivate NADPH-dependent reductive carboxylation. J Biol Chem. 2012;287:14615–20. doi: 10.1074/jbc.C112.353946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MARIN-VALENCIA I, YANG C, MASHIMO T, CHO S, BAEK H, YANG XL, RAJAGOPALAN KN, MADDIE M, VEMIREDDY V, ZHAO Z, CAI L, GOOD L, TU BP, HATANPAA KJ, MICKEY BE, MATES JM, PASCUAL JM, MAHER EA, MALLOY CR, DEBERARDINIS RJ, BACHOO RM. Analysis of tumor metabolism reveals mitochondrial glucose oxidation in genetically diverse human glioblastomas in the mouse brain in vivo. Cell Metab. 2012;15:827–37. doi: 10.1016/j.cmet.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- METALLO CM, GAMEIRO PA, BELL EL, MATTAINI KR, YANG J, HILLER K, JEWELL CM, JOHNSON ZR, IRVINE DJ, GUARENTE L, KELLEHER JK, VANDER HEIDEN MG, ILIOPOULOS O, STEPHANOPOULOS G. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature. 2012;481:380–4. doi: 10.1038/nature10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MULLEN AR, WHEATON WW, JIN ES, CHEN PH, SULLIVAN LB, CHENG T, YANG Y, LINEHAN WM, CHANDEL NS, DEBERARDINIS RJ. Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature. 2012;481:385–8. doi: 10.1038/nature10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NIEMANN S, MULLER U. Mutations in SDHC cause autosomal dominant paraganglioma, type 3. Nat Genet. 2000;26:268–70. doi: 10.1038/81551. [DOI] [PubMed] [Google Scholar]
- O'FLAHERTY L, ADAM J, HEATHER LC, ZHDANOV AV, CHUNG YL, MIRANDA MX, CROFT J, OLPIN S, CLARKE K, PUGH CW, GRIFFITHS J, PAPKOVSKY D, ASHRAFIAN H, RATCLIFFE PJ, POLLARD PJ. Dysregulation of hypoxia pathways in fumarate hydratase-deficient cells is independent of defective mitochondrial metabolism. Hum Mol Genet. 2010;19:3844–51. doi: 10.1093/hmg/ddq305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RAKHEJA D, BORIACK RL, MITUI M, KHOKHAR S, HOLT SA, KAPUR P. Papillary thyroid carcinoma shows elevated levels of 2-hydroxyglutarate. Tumour Biol. 2011a;32:325–33. doi: 10.1007/s13277-010-0125-6. [DOI] [PubMed] [Google Scholar]
- RAKHEJA D, MITUI M, BORIACK RL, DEBERARDINIS RJ. Isocitrate dehydrogenase 1/2 mutational analyses and 2-hydroxyglutarate measurements in Wilms tumors. Pediatr Blood Cancer. 2011b;56:379–83. doi: 10.1002/pbc.22697. [DOI] [PubMed] [Google Scholar]
- RANA M, DE COO I, DIAZ F, SMEETS H, MORAES CT. An out-of-frame cytochrome b gene deletion from a patient with parkinsonism is associated with impaired complex III assembly and an increase in free radical production. Ann Neurol. 2000;48:774–81. [PubMed] [Google Scholar]
- RZEM R, VINCENT MF, VAN SCHAFTINGEN E, VEIGA-DA-CUNHA M. L-2-hydroxyglutaric aciduria, a defect of metabolite repair. J Inherit Metab Dis. 2007;30:681–9. doi: 10.1007/s10545-007-0487-0. [DOI] [PubMed] [Google Scholar]
- SANTIDRIAN AF, MATSUNO-YAGI A, RITLAND M, SEO BB, LEBOEUF SE, GAY LJ, YAGI T, FELDING-HABERMANN B. Mitochondrial complex I activity and NAD+/NADH balance regulate breast cancer progression. J Clin Invest. 2013;123:1068–81. doi: 10.1172/JCI64264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAZANOV LA, JACKSON JB. Proton-translocating transhydrogenase and NAD- and NADP-linked isocitrate dehydrogenases operate in a substrate cycle which contributes to fine regulation of the tricarboxylic acid cycle activity in mitochondria. FEBS Lett. 1994;344:109–16. doi: 10.1016/0014-5793(94)00370-x. [DOI] [PubMed] [Google Scholar]
- TERNETTE N, YANG M, LAROYIA M, KITAGAWA M, O'FLAHERTY L, WOLHULTER K, IGARASHI K, SAITO K, KATO K, FISCHER R, BERQUAND A, KESSLER BM, LAPPIN T, FRIZZELL N, SOGA T, ADAM J, POLLARD PJ. Inhibition of mitochondrial aconitase by succination in fumarate hydratase deficiency. Cell Rep. 2013;3:689–700. doi: 10.1016/j.celrep.2013.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TOMITSUKA E, KITA K, ESUMI H. The NADH-fumarate reductase system, a novel mitochondrial energy metabolism, is a new target for anticancer therapy in tumor microenvironments. Ann N Y Acad Sci. 2010;1201:44–9. doi: 10.1111/j.1749-6632.2010.05620.x. [DOI] [PubMed] [Google Scholar]
- TOMLINSON IP, ALAM NA, ROWAN AJ, BARCLAY E, JAEGER EE, KELSELL D, LEIGH I, GORMAN P, LAMLUM H, RAHMAN S, ROYLANCE RR, OLPIN S, BEVAN S, BARKER K, HEARLE N, HOULSTON RS, KIURU M, LEHTONEN R, KARHU A, VILKKI S, LAIHO P, EKLUND C, VIERIMAA O, AITTOMAKI K, HIETALA M, SISTONEN P, PAETAU A, SALOVAARA R, HERVA R, LAUNONEN V, AALTONEN LA. Germline mutations in FH predispose to dominantly inherited uterine fibroids, skin leiomyomata and papillary renal cell cancer. Nat Genet. 2002;30:406–10. doi: 10.1038/ng849. [DOI] [PubMed] [Google Scholar]
- WEINBERG F, HAMANAKA R, WHEATON WW, WEINBERG S, JOSEPH J, LOPEZ M, KALYANARAMAN B, MUTLU GM, BUDINGER GR, CHANDEL NS. Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity. Proc Natl Acad Sci U S A. 2010;107:8788–93. doi: 10.1073/pnas.1003428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WEINBERG JM, VENKATACHALAM MA, ROESER NF, NISSIM I. Mitochondrial dysfunction during hypoxia/reoxygenation and its correction by anaerobic metabolism of citric acid cycle intermediates. Proc Natl Acad Sci U S A. 2000;97:2826–31. doi: 10.1073/pnas.97.6.2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WISE DR, WARD PS, SHAY JE, CROSS JR, GRUBER JJ, SACHDEVA UM, PLATT JM, DEMATTEO RG, SIMON MC, THOMPSON CB. Hypoxia promotes isocitrate dehydrogenase-dependent carboxylation of alpha-ketoglutarate to citrate to support cell growth and viability. Proc Natl Acad Sci U S A. 2011;108:19611–6. doi: 10.1073/pnas.1117773108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.