Abstract
Tazarotene induced gene 3 (TIG3) is a tumor suppressor protein. In normal human epidermis, TIG3 is present in the differentiated, suprabasal layers and regulates terminal differentiation. TIG3 level is reduced in hyperproliferative diseases, including psoriasis and skin cancer, suggesting that loss of TIG3 is associated with enhanced cell proliferation. Moreover, transient expression of TIG3 leads to terminal differentiation in normal keratinocytes and apoptosis in skin cancer cells. In both cell types, TIG3 distributes to the cell membrane and to the centrosome. At the cell membrane, TIG3 interacts with and activates type I transglutaminase (TG1) to enhance keratinocyte terminal differentiation. TIG3 at the centrosome acts to inhibit centrosome separation during mitosis and to alter microtubule function. These findings argue that TIG3 is involved in control of keratinocyte differentiation and that loss of TIG3 in transformed cells contributes to the malignant phenotype.
Keywords: TIG3, tumor suppressor, epidermis, keratinocyte, centrosome, tazarotene, H-rev107
The epidermis and terminal differentiation
The epidermis is a multi-layered tissue designed to provide protection from the environment. The major epidermal cell types if the keratinocyte which differentiates to form basal, spinous, granular and cornified layers (Eckert et al., 1997). Formation of these layers involves a process called differentiation. The innermost epidermal layer, the basal layer, contains undifferentiated/proliferative cells that undergo regulated cell division (Pincelli and Marconi, 2010). A fraction of basal cells undergo differentiation to form the spinous, granular and cornified layers. The layer immediately adjacent the basal layer is the spinous layer, which is distinguished by a spinous appearance. The next layer, the granular layer, consists of cells having intracellular membrane-bound granules, which contain proteins and lipids required for formation of the epidermal barrier. Basal layer cells have proliferative potential, but cells in the spinous and granular layers, although viable, do not proliferate. This change in differentiation status is associated with stage-specific changes in protein expression. For example, basal cells express keratins 5 and 14, while suprabasal cells express differentiation markers including type I transglutaminase (TG1), involucrin, loricrin, filaggrin, keratin 1, and keratin 10 (Eckert et al., 1997). Cells in the outermost “cornified layer” undergo terminal differentiation which involves loss of the nuclei and assembly of the cornified envelope. These fully-differentiated cells are called corneocytes which consist of a network of stabilized keratin surrounded by a covalently crosslinked envelop of protein. Many envelope precursor proteins have been identified, including involucrin, cystatin-α, loricrin, elafin, small proline-rich proteins (SPRs), filaggrin, and keratin (Eckert et al., 1997). These precursor proteins are covalently-crosslinked to form the cornified envelope by TG1 which is anchored to the inner surface of the plasma membrane and catalyzes formation of ε-(γ-glutamyl)lysine protein-protein crosslinks (Chakravarty and Rice, 1989). The ultimate function of the epidermis is to provide a protective barrier. This process is tightly controlled, and a current area of interest is identifying proteins that control this process. TIG3 is an important regulator of cell survival that plays an important role in this process (Eckert et al., 2009).
Tazarotene induced gene 3
TIG3, also known as retinoid induced gene 1 (RIG1) and retinoic acid receptor responder 3 (RARRES3), is a tumor suppressor protein composed of 164 amino acids (Ou et al., 2008; Jiang et al., 2005; Tsai et al., 2007; Huang et al., 2000). TIG3 was discovered as expressed in keratinocyte treated with the synthetic retinoid, tazarotene (DiSepio et al., 1998). TIG3 shares sequence similarity with members of the H-rev107 family of tumor suppressors, and is over 50% identical to human and rat H-rev107 (DiSepio et al., 1998). H-rev107 family members also show homology with the lecithin retinol acyltransferase family and NlpC/P60 super family (Anantharaman and Aravind, 2003; Husmann et al., 1998; Hajnal et al., 1994; Jahng et al., 2003). The H-rev107 family of proteins shares a common structure that includes N-terminal hydrophilic and C-terminal hydrophobic domains. The TIG3 N-terminal domain comprises 134-amino acids and the C-terminal region 30-amino acids (Fig. 1). The N-terminus includes several conserved elements, including the N-terminal homology domain, and the NCEHFV and LRYG motifs. The C-terminus contains fewer conserved elements and mutagenesis studies suggest that is serves as a membrane-anchoring domain (Fig. 1) (Deucher et al., 2000).
Fig. 1. Structure of TIG3 and interaction with transglutaminase.
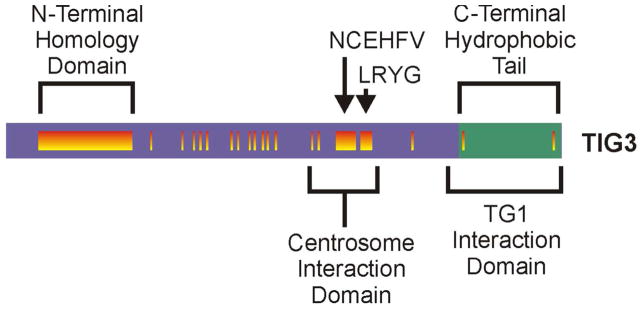
A TIG3 is a 164 amino acid protein that is divided into an N-terminal (purple, amino acids 1–134) and C-terminal (green, amino acids 135–164) domains. The N-terminal hydrophilic domain includes regions (orange vertical bars) that are highly conserved among members of the H-rev107 family of tumor suppressor proteins (Deucher et al., 2000). The type I transglutaminase interaction domain and the centrosome interaction domain are indicated, as are the N-terminal homology domain, and the NCEHFV and LRYG motifs.
TIG3 regulates keratinocyte terminal differentiation
TIG3 is selectively expressed in differentiating cells and during programmed cell death. As such, TIG3 is expressed at vanishingly-low levels in proliferating keratinocyte in monolayer culture, but is expressed at high levels in differentiated human epidermis and keratinocytes grown as epidermal equivalents (Sturniolo et al., 2003; Duvic et al., 2000; Jans et al., 2008; Duvic et al., 1997). This pattern of expression suggests that TIG3 is involved in differentiation and survival processes. Transient expression of TIG3 in keratinocytes, at levels within the physiological range observed in differentiated keratinocytes (Jans et al., 2008) is associated with a marked morphological change. TIG3 positive cells display a morphology that resembles that of a terminal corneocyte (Sturniolo et al., 2003). This morphological change is associated with reduced cell survival (Deucher et al., 2000; Sturniolo et al., 2005; Jans et al., 2008; Sturniolo et al., 2003; Eckert et al., 2009). In fact, expression of TIG3 in cultures of normal keratinocytes results in nearly quantitative cornification (Sturniolo et al., 2005; Sturniolo et al., 2003), suggesting that TIG3 is an inducer of keratinocyte differentiation. Consistent with a potential role in enhancing keratinocyte differentiation, TIG3 levels are reduced in epidermal hyperproliferative disease and cancer (Eckert et al., 1997; DiSepio et al., 1998; Deucher et al., 2000; Duvic et al., 2000; Jans et al., 2008; Duvic et al., 2003).
Several studies have examined the role of TIG3 in controlling intracellular processes and how this correlates with intracellular localization. These studies show that TIG3 localizes at the keratinocyte plasma membrane via a mechanism that requires the TIG3 C-terminal hydrophilic membrane-anchoring domain (Eckert et al., 2009; Sturniolo et al., 2005; Sturniolo et al., 2003). A key finding is that membrane-associated TIG3 interacts with type I transglutaminase (TG1) at this location (Eckert et al., 2009; Sturniolo et al., 2005; Sturniolo et al., 2003). TG1, is anchored to the inner surface of the plasma membrane by a lipid anchor (Chakravarty and Rice, 1989; Phillips et al., 1993), and is responsible for catalyzing formation of the protein-protein crosslinks required for cornified envelope assembly. It has been proposed that newly-synthesized TIG3 inserts in the plasma membrane, translocates in the membrane to locate TG1, and that a TIG3/TG1 interaction drives activation of TG1. Indeed, confocal studies reveal that TIG3 co-localizes with TG1 in keratinocytes (Jans et al., 2008; Sturniolo et al., 2005; Sturniolo et al., 2003), and that TG1 activity is observed at these sites. Moreover, biochemical pull-down experiments reveal that TIG3 co-precipitation with TG1 (Jans et al., 2008; Sturniolo et al., 2005; Sturniolo et al., 2003). This requires the full-length TIG3 protein, as removal of the C-terminal membrane anchoring domain from TIG3 results in a loss of interaction (Sturniolo et al., 2003; Sturniolo et al., 2005). These studies suggest that the inability of the TIG3 C-terminal truncation mutant to interact with TG1 is because the mutant does not localize at the plasma membrane (Deucher et al., 2000; Jans et al., 2008; Sturniolo et al., 2003). Additional studies identify a TIG3 region at the junction between the N-terminal domain and the C-terminal membrane anchoring domain as being required for interaction with TG1 (Jans et al., 2008) (Fig. 1). Interaction of TIG3 with TG1 leads to increased TG1 activity (Sturniolo et al., 2005; Sturniolo et al., 2003) and the increase in TG1 activity is associated with reduced cell survival. The reduction in cell survival can be partially inhibited by monodansylcadaverine, a TG1 substrate competitive inhibitor (Sturniolo et al., 2005). TIG3 is one of only a few proteins that interact with TG1 to increase activity (Eckert et al., 2009; Jans et al., 2008). Like most proteins that interact with TG1, TIG3 also functions as a substrate (Sturniolo et al., 2005). As expected, the TIG3 mutant lacking the membrane-localization domain does not interact with TG1 and is not a substrate (Sturniolo et al., 2005; Sturniolo et al., 2003). These findings support the hypothesis that TIG3 regulates key events during terminal keratinocyte differentiation and that interaction at the cell membrane is a requirement for TIG3 function. We propose that TIG3 is produced, moves to the membrane where it is anchored by the C-terminal hydrophobic domain, and that it then moves in the membrane until it contacts and activates TG1. This ultimately leads to crosslinking of cornified envelop precursors and envelope assembly (Fig. 2). We further propose that removing the C-terminal region (green) results in an inability of TIG3 to drive activation, due to its inability to localize with TG1.
Fig. 2. TIG3 interaction with type I transglutaminase.
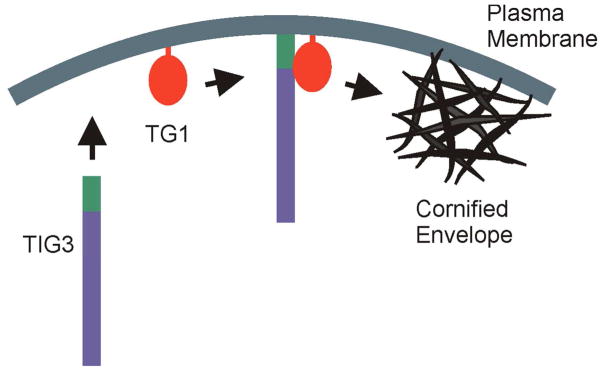
TIG3 is produced in differentiated keratinocytes and is anchored to the plasma membrane via the C-terminal membrane-anchoring domain (green). Transglutaminase type I (TG1) is tethered to the plasma membrane via a lipid anchor (Chakravarty and Rice, 1989). TIG3 interaction with TG1 produces a complex that activates TG1 enzymatic activity which leads to cornified envelope assembly. It is very likely that the TIG3/TG1 complex also includes other proteins.
TIG3 at the centrosome
In addition to localizing at the plasma membrane, TIG3 is also observed at a perinuclear location. Keratinocytes were stained with anti-TIG3 and antibodies that detect organelle-specific proteins including pericentrin (centrosome), γ-tubulin (centrosome), GM130 (cis-Golgi apparatus), mannose-6-phosphate receptor (M6PR, trans-Golgi and late endosome), rab11 (recycling endosome), EEA1 (early endosome), clathrin heavy chain (CHC, intracellular transport vesicle), lamp1 (lysosome), calnexin (endoplasmic reticulum) and Mitotracker Red (mitochondria). TIG3 localizes with pericentrin and γ-tubulin in ninety percent of cells (Scharadin et al., 2011). In addition, some co-localization is observed with the Golgi (GM130 and M6PR) markers which is consistent with the observation that TIG3 localizes at the Golgi in a cervical cancer cells (Scharadin et al., 2012; Tsai et al., 2007). TIG3 localization in the vicinity of the centrosome suggests that TIG3 may impact centrosome-related functions.
TIG3 impact on microtubule distribution and elongation
The centrosome (microtubule organizing center) is a 1–2 μm diameter organelle which functions as a site of microtubule assembly (Tsai et al., 2007; Lim et al., 2009; Doxsey et al., 2005). Microtubules form from α- and β-tubulin heterodimers which nucleate at the centrosome (Doxsey et al., 2005). This microtubule network has two main functions in the cell. In interphase cells, the microtubules serve as a highway to guide and regulate intracellular cargo movement. The cargo includes proteins, vesicles, and organelles. Cargo is bound to the microtubules by molecular motors, called kinesins and dineins, which control anterograde (away from centrosome) and retrograde (toward centrosome) movement. The second role of the centrosome is to assemble the mitotic spindles that guide chromosome movement during mitosis. Both of these functions are absolutely essential for cell division and survival (Doxsey et al., 2005).
The presence of TIG3 in the vicinity of the centrosome suggests that TIG3 may alter centrosome function. Recent reports support this hypothesis. Keratinocytes which transiently overexpress TIG3 display mark changes in cell division and status of the microtubule network. In control cells the microtubule network radiates from the centrosome throughout the cell. In contrast, in TIG3 expressing cells, the microtubules redistribute to the cell periphery where they form a thick band adjacent the cell membrane that is linked to the centrosome by thin microtubule “spokes” (Scharadin et al., 2012). This rearrangement is accompanied by microtubule stability-inducing post-translational modification of α-tubulin (Scharadin et al., 2011) including increased acetylation of lysine 40 and detyrosination of the C-terminal tyrosine of α-tubulin (Ikegami and Setou, 2010; Quinones et al., 2011; Hammond et al., 2008). The latter modification results in formation of Δ2-α-tubulin, a highly stable form of α-tubulin. These modifications markedly increase microtubule stability (Gundersen et al., 1987). However, in spite of the presence of these stabilizing modifications in TG3-positive cells, the peripheral microtubule band remains susceptible to disruption by nocodazole (Scharadin et al., 2012) and reforms upon nocodazole removal (Scharadin et al., 2012). Thus, the impact of TIG3 on microtubule dynamics requires further study. The changes in microtubule organization are consistent with an impact of TIG3 on centrosome function. This is supported by studies of microtubule elongation. Microtubule anterograde growth can be measured in real-time in live cells using the plus-end binding protein, EB1 (Dixit et al., 2009; Piehl et al., 2004; Jaworski et al., 2008). Fluorescently-labeled EB1 studies in control cells reveal abundant anterograde microtubule growth and show that this growth is markedly attenuated in TIG3 expressing cells. In contrast, although initiation of microtubule formation is maintained in TIG3-positive cells, subsequent elongation of these microtubules is severely attenuated (Scharadin et al., 2012). This suggests that TIG3 may suppress centrosome-dependent intracellular cargo distribution.
TIG3 impact on centrosome separation
TIG3 is also impacts the centrosome during mitosis. During normal cell division the centrosome is duplicated along with the chromosomes during S phase of the cell cycle. Subsequently, in early prophase, the daughter centrosomes separate and initiate formation of radial arrays of microtubules, known as asters. The asters then move to opposite poles of the cell during metaphase and serve to form the mitotic spindle which interacts with the chromosomes throughout mitosis. A critical and necessary step is separation of the daughter centrosomes, and their migration to the spindle poles. The impact of TIG3 on centrosome function during mitosis was studied by monitoring centrosome separation (Doxsey et al., 2005). In control keratinocytes, sixteen percent of cells display separated daughter centrosomes, which corresponds to the percentage of dividing cells. In comparison, only one percent of TIG3-positive cells display centrosome separation (Scharadin et al., 2012). This difference is not due to TIG3-associated suppression of new centrosome formation, as centrosome synthesis is unchanged in control and TIG3-positive cells (Scharadin et al., 2012). This inhibition of centrosome separation impacts cell division. Keratinocytes which express TIG3 display a significant reduction in the uptake of the S-phase cell cycle marker, bromodeoxyuridine. Approximately fourteen percent of TIG3-negative keratinocytes, versus one percent of TIG3-expressing keratinocytes, are in S-phase (Scharadin et al., 2012). These findings support the hypothesis that TIG3 distribution to the centrosome inhibits centrosome separation and secondarily inhibits cell division.
TIG3 intracellular localization and control of cell function
A key question is how the cell regulates TIG3 distribution between the plasma membrane and the centrosome. The mechanism is not well understood. Recent studies used a series of TIG3 mutants fused to enhanced green fluorescent protein (EGFP). These studies identified amino acids 102 to 125 as sufficient to mobilize EGFP to the centrosome (Scharadin et al., 2013). The presence of a centrosome localization sequence at amino acids 102–125 is particularly interesting, as this region encodes amino acids that are highly conserved when TIG3 is compared to other members of the H-rev107 tumor suppressor family (Deucher et al., 2000), suggesting that centrosome-associated action may be a property of other family members. Moreover, TIG3 peptides, derived from within this region (amino acids 111–123), are able to reduce cell survival in some cell types (Simmons et al., 2006).
The microtubule rearrangement displayed in cultured TIG3-expressing keratinocytes may have an important in vivo correlate. In the basal cells of the mouse epidermis, the microtubule network emanates from the centrosome. In suprabasal differentiated cells, in contrast, the microtubules preferentially distribute to cell-cell junctions and form a microtubule band at the cell periphery (Scharadin et al., 2011; Scharadin et al., 2012; Lechler and Fuchs, 2007). It has been proposed that this microtubule redistribution is driven by ninein, a microtubule anchoring protein. Ninein moves to the cell periphery in differentiated cells (Lechler and Fuchs, 2007; Dammermann and Merdes, 2002; Delgehyr et al., 2005; Mogensen et al., 2000). Preliminary studies indicate that TIG3 expression in human keratinocytes does not alter ninein distribution in human keratinocytes (Scharadin and Eckert, unpublished). Though further studies are necessary to elucidate the mechanism whereby TIG3 alters microtubule distribution, it is intriguing that TIG3 causes microtubules to distribute to the cell periphery in a pattern similar to that observed in suprabasal keratinocytes in epidermis (Scharadin et al., 2013). Thus, elevated TIG3 in suprabasal epidermis may contribute to the microtubule redistribution observed in differentiated cells in vivo.
TIG3 in hyperproliferative disease
As mentioned above, TIG3 is observed at reduced levels in skin cancer and epidermal hyperproliferative disease (DiSepio et al., 1998). This suggests that absence of TIG3 may be a permissive event in skin carcinogenesis. Based on this information, restoration of TIG3 expression in hyperproliferative diseases would be predicted to reduce cell proliferation and survival. Endogenous TIG3 levels can be increased by treating cells with tazarotene or other retinoids (Jiang et al., 2005; Huang et al., 2000; DiSepio et al., 1998; Higuchi et al., 2003). Retinoid-dependent increased TIG3 expression is also observed in retinoid-treated psoriatic lesions (Duvic et al., 2000; Duvic et al., 2003; Talpur et al., 2009). This is consistent with observations in cultured keratinocytes. Transient expression of TIG3 in normal keratinocytes leads to a reduction in cell proliferation (Deucher et al., 2000; Sturniolo et al., 2005; Jans et al., 2008; Sturniolo et al., 2003; Eckert et al., 2009). Further, transient expression of TIG3 suppresses growth in cell lines created from several types of cancers, including lung, head and neck, gastric, cervical, and prostate cancer (Huang et al., 2000; DiSepio et al., 1998; Deucher et al., 2000; Higuchi et al., 2003; Huang et al., 2002; Kawakami et al., 2006). Consistent with a role in suppressing proliferation/increasing cell death, enhanced TIG3 expression is observed in tau neuropathies where it is associated with increased neuronal cell death (Wilhelmus et al., 2011). TIG3 suppression of cell survival was also studied using epidermis-derived squamous cell carcinoma cells. These tumor cells proliferate rapidly and restoring TIG3 expression slows cell proliferation (Scharadin et al., 2011). Expression of TIG3 in cancer cells activates caspase 3 and 9, and PARP-cleavage leading to apoptosis (Scharadin et al., 2011). In addition, TIG3 reduces the level of G1 and G1/S phase cell cycle regulatory proteins including cyclin D, cyclin E, cdk4 and cdk6. This leads to a cell cycle block at the G1/S phase transition (Scharadin et al., 2011). The G1/S block is associated with increased expression of p21Cip1 mRNA and protein (Scharadin et al., 2011). Elevated TIG3 expression in other keratinocyte cell lines, including the immortalized HaCaT keratinocyte cell line and the transformed A431 squamous cell carcinoma cell line, yields similar results (Scharadin and Eckert, unpublished). The observation that TIG3 causes apoptosis in cancer cells is in contrast to the findings in normal human keratinocytes where it causes differentiation (Scharadin et al., 2011; Eckert et al., 2009; Jans et al., 2008; Sturniolo et al., 2005; Sturniolo et al., 2003). As mentioned above, TIG3 enhances normal keratinocyte terminal differentiation, in part through binding to and activation of type I transglutaminase (TG1), but does not activate apoptosis (Jans et al., 2008; Sturniolo et al., 2003). Interestingly, TIG3 is distributed to the cell membrane and the centrosome in normal keratinocytes and skin cancer cell lines (Scharadin et al., 2011). Thus, although TIG3 displays a similar pattern of localization in cancer and normal cells, the net outcome (differentiation or apoptosis) is different in the two cell types. Understanding the mechanistic basis for this difference will require additional study.
Summary
TIG3 is a regulator of keratinocyte proliferation and terminal differentiation that is expressed in the suprabasal differentiated epidermis. We propose that onset of TIG3 expression in the suprabasal epidermis is an integral event in the process that reduces cell proliferation and enhances terminal differentiation. In this role, recent studies suggest that TIG3 operates via interaction at two major subcellular locations. The first location is the plasma membrane where it interacts with type I transglutaminase to increase activity leading to increased cornified envelope assembly (Sturniolo et al., 2005; Duvic et al., 2000; Jans et al., 2008; Sturniolo et al., 2003) (Fig. 3C). The second location is the centrosome where TIG3 acts to alter microtubule function and inhibit centrosome separation. TIG3 presence promotes a redistribution of the microtubules to form a peripheral band near the plasma membrane (Fig. 3B). The α-tubulin subunit in these microtubules is altered by covalent modifications that reduce microtubule turnover and increase stability (Scharadin et al., 2011; Scharadin et al., 2012). TIG3 also suppresses daughter centrosome separation which is associated with reduced cell proliferation (Scharadin et al., 2011; Scharadin et al., 2012) (Fig. 3A). Finally, consistent with enhanced cell survival, TIG3 levels are reduced in hyperproliferative skin disorders, including psoriasis and skin cancer (Duvic et al., 2000; Duvic et al., 2003; Talpur et al., 2009; Duvic et al., 1997). We propose that TIG3 has a dual role to drive growth cessation and differentiation in maturing epidermis. It acts at the centrosome to inhibit centrosome separation and promote microtubule rearrangement to impede cell division, and it also acts at the plasma membrane to activate transglutaminase to stimulate cornified envelop formation.
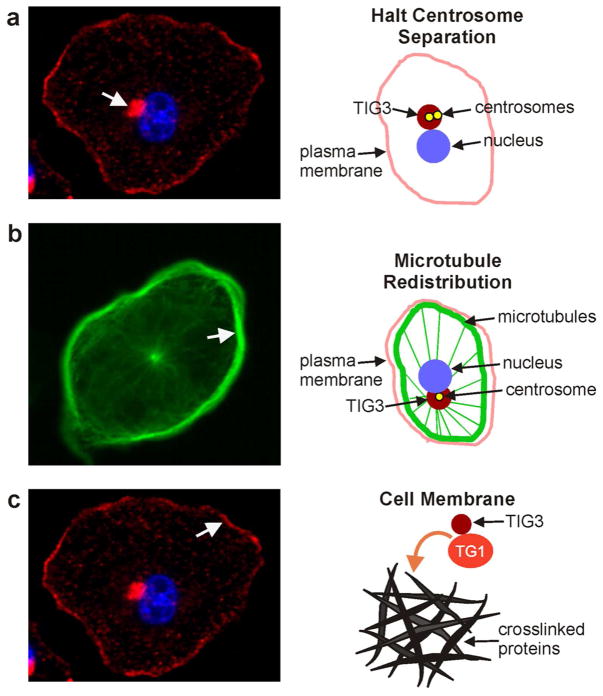
Fig. 3. Role of TIG3 in keratinocytes.
A TIG3 halts centrosome separation. Cultured normal foreskin keratinocytes were infected with TIG3-encoding adenovirus and after 24 h stained with anti-TIG3. The arrow indicates TIG3 distribution at the centrosome. The schematic indicates the distribution of the centrosomes, the nucleus, the plasma membrane and TIG3. The daughter centrosomes are produced during G2 of interphase of the cell cycle, but in the presence of TIG3 they never separate in prophase and move to the poles. B Impact of TIG3 on microtubule distribution. TIG3 causes a redistribution of the microtubules from a uniform array that extends throughout the cell to the distribution shown in this image stained with anti-β-tubulin (arrow indicates marginal microtubule distribution). Note the presence of the thin microtubule spokes emanating from the centrosome. The schematic shows the location of various cellular structures and the localization of TIG3. C TIG3 activates transglutaminase type I. The image shows that some of TIG3 distributes to the plasma membrane (arrow). The schematic shows that it interacts with the plasma membrane where it interacts with and activates TG1 which leads to crosslinking of envelope precursor proteins to form the cornified envelope.
Acknowledgments
This work was supported by National Institute of Health grants (CA184027, AR049713) awarded to Richard L. Eckert.
Abbreviations
- TG1
type I transglutaminase
- TIG3
tazarotene induced gene 3
Footnotes
Conflict of Interest
The authors have no conflict of interest.
References
- 1.Anantharaman V, Aravind L. Evolutionary history, structural features and biochemical diversity of the NlpC/P60 superfamily of enzymes. Genome Biol. 2003;4:R11. doi: 10.1186/gb-2003-4-2-r11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chakravarty R, Rice RH. Acylation of keratinocyte transglutaminase by palmitic and myristic acids in the membrane Anchorage region. J Biol Chem. 1989;264:625–629. [PubMed] [Google Scholar]
- 3.Dammermann A, Merdes A. Assembly of centrosomal proteins and microtubule organization depends on PCM-1. J Cell Biol. 2002;159:255–266. doi: 10.1083/jcb.200204023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Delgehyr N, Sillibourne J, Bornens M. Microtubule nucleation and anchoring at the centrosome are independent processes linked by ninein function. J Cell Sci. 2005;118:1565–1575. doi: 10.1242/jcs.02302. [DOI] [PubMed] [Google Scholar]
- 5.Deucher A, Nagpal S, Chandraratna RA, et al. The carboxy-terminal hydrophobic domain of TIG3, a class II tumor suppressor protein, is required for appropriate cellular localization and optimal biological activity. Int J Oncol. 2000;17:1195–1203. doi: 10.3892/ijo.17.6.1195. [DOI] [PubMed] [Google Scholar]
- 6.DiSepio D, Ghosn C, Eckert RL, et al. Identification and characterization of a retinoid-induced class II tumor suppressor/growth regulatory gene. Proc Natl Acad Sci U S A. 1998;95:14811–14815. doi: 10.1073/pnas.95.25.14811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dixit R, Barnett B, Lazarus JE, et al. Microtubule plus-end tracking by CLIP-170 requires EB1. Proc Natl Acad Sci U S A. 2009;106:492–497. doi: 10.1073/pnas.0807614106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doxsey S, Zimmerman W, Mikule K. Centrosome control of the cell cycle. Trends Cell Biol. 2005;15:303–311. doi: 10.1016/j.tcb.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 9.Duvic M, Helekar B, Schulz C, et al. Expression of a retinoid-inducible tumor suppressor, Tazarotene-inducible gene-3, is decreased in psoriasis and skin cancer. Clin Cancer Res. 2000;6:3249–3259. [PubMed] [Google Scholar]
- 10.Duvic M, Nagpal S, Asano AT, et al. Molecular mechanisms of tazarotene action in psoriasis. J Am Acad Dermatol. 1997;37:S18–S24. [PubMed] [Google Scholar]
- 11.Duvic M, Ni X, Talpur R, et al. Tazarotene-induced gene 3 is suppressed in basal cell carcinomas and reversed in vivo by tazarotene application. J Invest Dermatol. 2003;121:902–909. doi: 10.1046/j.1523-1747.2003.12488.x. [DOI] [PubMed] [Google Scholar]
- 12.Eckert RL, Crish JF, Robinson NA. The epidermal keratinocyte as a model for the study of gene regulation and cell differentiation. Physiol Rev. 1997;77:397–424. doi: 10.1152/physrev.1997.77.2.397. [DOI] [PubMed] [Google Scholar]
- 13.Eckert RL, Sturniolo MT, Jans R, et al. TIG3: a regulator of type I transglutaminase activity in epidermis. Amino Acids. 2009;36:739–746. doi: 10.1007/s00726-008-0123-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gundersen GG, Khawaja S, Bulinski JC. Postpolymerization detyrosination of alpha-tubulin: a mechanism for subcellular differentiation of microtubules. J Cell Biol. 1987;105:251–264. doi: 10.1083/jcb.105.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hajnal A, Klemenz R, Schafer R. Subtraction cloning of H-rev107, a gene specifically expressed in H-ras resistant fibroblasts. Oncogene. 1994;9:479–490. [PubMed] [Google Scholar]
- 16.Hammond JW, Cai D, Verhey KJ. Tubulin modifications and their cellular functions. Curr Opin Cell Biol. 2008;20:71–76. doi: 10.1016/j.ceb.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Higuchi E, Chandraratna RA, Hong WK, et al. Induction of TIG3, a putative class II tumor suppressor gene, by retinoic acid in head and neck and lung carcinoma cells and its association with suppression of the transformed phenotype. Oncogene. 2003;22:4627–4635. doi: 10.1038/sj.onc.1206235. [DOI] [PubMed] [Google Scholar]
- 18.Huang SL, Shyu RY, Yeh MY, et al. Cloning and characterization of a novel retinoid-inducible gene 1(RIG1) deriving from human gastric cancer cells. Mol Cell Endocrinol. 2000;159:15–24. doi: 10.1016/s0303-7207(99)00207-5. [DOI] [PubMed] [Google Scholar]
- 19.Huang SL, Shyu RY, Yeh MY, et al. The retinoid-inducible gene I: effect on apoptosis and mitogen-activated kinase signal pathways. Anticancer Res. 2002;22:799–804. [PubMed] [Google Scholar]
- 20.Husmann K, Sers C, Fietze E, et al. Transcriptional and translational downregulation of H-REV107, a class II tumour suppressor gene located on human chromosome 11q11–12. Oncogene. 1998;17:1305–1312. doi: 10.1038/sj.onc.1202060. [DOI] [PubMed] [Google Scholar]
- 21.Ikegami K, Setou M. Unique post-translational modifications in specialized microtubule architecture. Cell Struct Funct. 2010;35:15–22. doi: 10.1247/csf.09027. [DOI] [PubMed] [Google Scholar]
- 22.Jahng WJ, Xue L, Rando RR. Lecithin retinol acyltransferase is a founder member of a novel family of enzymes. Biochemistry. 2003;42:12805–12812. doi: 10.1021/bi035370p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jans R, Sturniolo MT, Eckert RL. Localization of the TIG3 transglutaminase interaction domain and demonstration that the amino-terminal region is required for TIG3 function as a keratinocyte differentiation regulator. J Invest Dermatol. 2008;128:517–529. doi: 10.1038/sj.jid.5701035. [DOI] [PubMed] [Google Scholar]
- 24.Jaworski J, Hoogenraad CC, Akhmanova A. Microtubule plus-end tracking proteins in differentiated mammalian cells. Int J Biochem Cell Biol. 2008;40:619–637. doi: 10.1016/j.biocel.2007.10.015. [DOI] [PubMed] [Google Scholar]
- 25.Jiang SY, Wu MS, Chen LM, et al. Identification and characterization of the retinoic acid response elements in the human RIG1 gene promoter. Biochem Biophys Res Commun. 2005;331:630–639. doi: 10.1016/j.bbrc.2005.03.214. [DOI] [PubMed] [Google Scholar]
- 26.Kawakami S, Suzuki S, Yamashita F, et al. Induction of apoptosis in A549 human lung cancer cells by all-trans retinoic acid incorporated in DOTAP/cholesterol liposomes. J Control Release. 2006;110:514–521. doi: 10.1016/j.jconrel.2005.10.030. [DOI] [PubMed] [Google Scholar]
- 27.Lechler T, Fuchs E. Desmoplakin: an unexpected regulator of microtubule organization in the epidermis. J Cell Biol. 2007;176:147–154. doi: 10.1083/jcb.200609109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lim HH, Zhang T, Surana U. Regulation of centrosome separation in yeast and vertebrates: common threads. Trends Cell Biol. 2009;19:325–333. doi: 10.1016/j.tcb.2009.03.008. [DOI] [PubMed] [Google Scholar]
- 29.Mogensen MM, Malik A, Piel M, et al. Microtubule minus-end anchorage at centrosomal and non-centrosomal sites: the role of ninein. J Cell Sci. 2000;113 ( Pt 17):3013–3023. doi: 10.1242/jcs.113.17.3013. [DOI] [PubMed] [Google Scholar]
- 30.Ou CC, Hsu SC, Hsieh YH, et al. Downregulation of HER2 by RIG1 involves the PI3K/Akt pathway in ovarian cancer cells. Carcinogenesis. 2008;29:299–306. doi: 10.1093/carcin/bgm263. [DOI] [PubMed] [Google Scholar]
- 31.Phillips MA, Qin Q, Mehrpouyan M, et al. Keratinocyte transglutaminase membrane anchorage: analysis of site-directed mutants. Biochemistry. 1993;32:11057–11063. doi: 10.1021/bi00092a015. [DOI] [PubMed] [Google Scholar]
- 32.Piehl M, Tulu US, Wadsworth P, et al. Centrosome maturation: measurement of microtubule nucleation throughout the cell cycle by using GFP-tagged EB1. Proc Natl Acad Sci U S A. 2004;101:1584–1588. doi: 10.1073/pnas.0308205100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pincelli C, Marconi A. Keratinocyte stem cells: friends and foes. J Cell Physiol. 2010;225:310–315. doi: 10.1002/jcp.22275. [DOI] [PubMed] [Google Scholar]
- 34.Quinones GB, Danowski BA, Devaraj A, et al. The posttranslational modification of tubulin undergoes a switch from detyrosination to acetylation as epithelial cells become polarized. Mol Biol Cell. 2011;22:1045–1057. doi: 10.1091/mbc.E10-06-0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scharadin TM, Adhikary G, et al. Pericentrosomal localiation of the TIG3 tumor suppressor requires an N-terminal hydrophilic region motif. J Invest Dermatol. 2013 doi: 10.1038/jid.2013.533. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scharadin TM, Jiang H, Jans R, et al. TIG3 Tumor Suppressor-Dependent Organelle Redistribution and Apoptosis in Skin Cancer Cells. PLoS One. 2011;6:e23230. doi: 10.1371/journal.pone.0023230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scharadin TM, Jiang H, Martin S, et al. TIG3 interaction at the centrosome alters microtubule distribution and centrosome function. J Cell Sci. 2012;125:2604–2614. doi: 10.1242/jcs.096495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simmons DP, Peach ML, Friedman JR, et al. Evidence that sequence homologous region in LRAT-like proteins possesses anti-proliferative activity and DNA binding properties: translational implications and mechanism of action. Carcinogenesis. 2006;27:693–707. doi: 10.1093/carcin/bgi235. [DOI] [PubMed] [Google Scholar]
- 39.Sturniolo MT, Chandraratna RA, Eckert RL. A novel transglutaminase activator forms a complex with type 1 transglutaminase. Oncogene. 2005;24:2963–2972. doi: 10.1038/sj.onc.1208392. [DOI] [PubMed] [Google Scholar]
- 40.Sturniolo MT, Dashti SR, Deucher A, et al. A novel tumor suppressor protein promotes keratinocyte terminal differentiation via activation of type I transglutaminase. J Biol Chem. 2003;278:48066–48073. doi: 10.1074/jbc.M307215200. [DOI] [PubMed] [Google Scholar]
- 41.Talpur R, Cox K, Duvic M. Efficacy and safety of topical tazarotene: a review. Expert Opin Drug Metab Toxicol. 2009;5:195–210. doi: 10.1517/17425250902721250. [DOI] [PubMed] [Google Scholar]
- 42.Tsai FM, Shyu RY, Jiang SY. RIG1 suppresses Ras activation and induces cellular apoptosis at the Golgi apparatus. Cell Signal. 2007;19:989–999. doi: 10.1016/j.cellsig.2006.11.005. [DOI] [PubMed] [Google Scholar]
- 43.Wilhelmus MM, de JM, Rozemuller AJ, et al. Transglutaminase 1 and its regulator tazarotene-induced gene 3 localize to neuronal tau inclusions in tauopathies. J Pathol. 2011 doi: 10.1002/path.2984. [DOI] [PMC free article] [PubMed] [Google Scholar]