Abstract
The nuclear transcription factor Stat3 has recently been reported to have a localized mitochondrial regulatory function. Current data suggest that mitochondrial Stat3 (mitoStat3) is necessary for maximal mitochondrial activity and for Ras-mediated transformation independent of Stat3 nuclear activity. We have previously shown that Stat3 plays a pivotal role in epithelial carcinogenesis. Therefore, the aim of the current study was to determine the role of mitoStat3 in epidermal keratinocytes. Herein, we show that normal and neoplastic keratinocytes contain a pool of mitoStat3. EGF and TPA induce Stat3 mitochondrial translocation mediated through phosphorylation of Stat3 at Ser727. In addition, we report that mitoStat3 binds mitochondrial DNA (mtDNA) and associates with the mitochondrial transcription factor TFAM. Furthermore, Stat3 ablation resulted in an increase of mitochondrial encoded gene transcripts. An increase in key nuclear-encoded metabolic genes, PGC-1α and NRF-1, was also observed in Stat3 null keratinocytes, however no changes in nuclear-encoded ETC gene transcripts or mtDNA copy number were observed. Collectively, our findings suggest a heretofore-unreported function for mitoStat3 as a potential mitochondrial transcription factor in keratinocytes. This mitoStat3-mtDNA interaction may represent an alternate signaling pathway that could alter mitochondrial function and biogenesis and play a role in tumorigenesis.
INTRODUCTION
The signal transducers and activators of transcription (STATs) proteins (i.e. STATs 1, 2, 3, 4, 5a, 5b and 6) regulate numerous cellular functions, including survival, proliferation, migration and differentiation in response to a wide-spectrum of stimuli (Levy and Darnell 2002). Classically, activation of STATs occurs after cytokine stimulation of cell surface receptors via Janus associated kinases (JAKs), thereby coined the JAK-STAT pathway. Tyrosine phosphorylation of STATs by JAKs facilitates homodimerization, and in some cases heterodimers, followed by nuclear translocation, where STAT dimers bind to and regulate expression of target genes (Levy and Darnell 2002). Stat3 was originally identified as an IL-6-dependent transcription factor that promotes acute phase gene expression (Zhong et al. 1994). Subsequent studies have shown Stat3 activation by various cytokines and growth factors, including leukemia inhibitory factor, epidermal growth factor, hepatocyte growth factor, and the hormone leptin (Bromberg and Darnell 2000). In addition, there is strong evidence correlating Stat3 activation and cancer. Stat3 is found constitutively activated in cells transformed by the oncogenes v-Src and v-Abl, as well as in various human cancers, including hematologic, pancreatic, breast, head and neck, and prostate cancer (Bowman et al. 2000; Turkson and Jove 2000). While most reports have attributed the oncogenic function of Stat3 to Tyr705 activation, serine phosphorylation at residue 727 is necessary for maximal Stat3 transcriptional activity in vivo and is also linked with certain cancers (Wen et al. 1995; Shen et al. 2004; Yang et al. 2005; Yeh et al. 2006; Qin et al. 2008; Hazan-Halevy et al. 2010).
Previous work from our group has shown that Stat3 plays an important role in epithelial carcinogenesis, including both two-stage chemical and UVB-induced skin carcinogenesis through its ability to regulate a number of nuclear encoded genes (Chan et al. 2004; Chan et al. 2004; Kim et al. 2007; Chan et al. 2008; Sano et al. 2008; Kim et al. 2009; Kim et al. 2009). Apart from its well-documented role as a nuclear transcription factor, a role for Stat3 has recently emerged in the mitochondria (Wegrzyn et al. 2009). Stat3 was found to interact with electron transport chain (ETC) components, complex I and II, (Wegrzyn et al. 2009). In this regard, tissue specific knockout of Stat3 in cardiomyocytes and astrocytes reduces activity of complex I and II of the mitochondrial ETC (Boengler et al. 2010; Sarafian et al. 2010; Szczepanek et al. 2012). Impaired mitochondrial function is rescued by add back of mitochondrial targeted Stat3 (MLS-Stat3) in primary cells (Wegrzyn et al. 2009). Phosphorylation of Stat3 at Ser727 residue appears to be necessary for Stat3 mitochondrial localization and its capacity to modulate mitochondrial respiration (Wegrzyn et al. 2009). However, the ratio of Stat3 to ETC components (I/II) is approximately 105 in cardiomyocytes and a direct protein interaction with electron transport chain components in regulation of mitochondrial respiration has been deemed unlikely (Phillips et al. 2010). In still other studies, Gough et al. reported that mitochondrial Stat3 plays a role in Ras-mediated cellular transformation (Gough et al. 2009). Collectively, the current data suggest that Stat3 has a role in promoting mitochondrial function and cellular metabolism and may play a role in tumorigenesis.
Given the role of Stat3 in epithelial carcinogenesis and the emerging role of mitochondrial localized Stat3 (hereafter referred to as mitoStat3), we asked whether mitoStat3 plays a role in epithelial carcinogenesis. In order to address this question, the current study was undertaken to explore the function of mitoStat3 in keratinocytes.
EXPERIMENTAL RESULTS
Stat3 in keratinocyte mitochondria
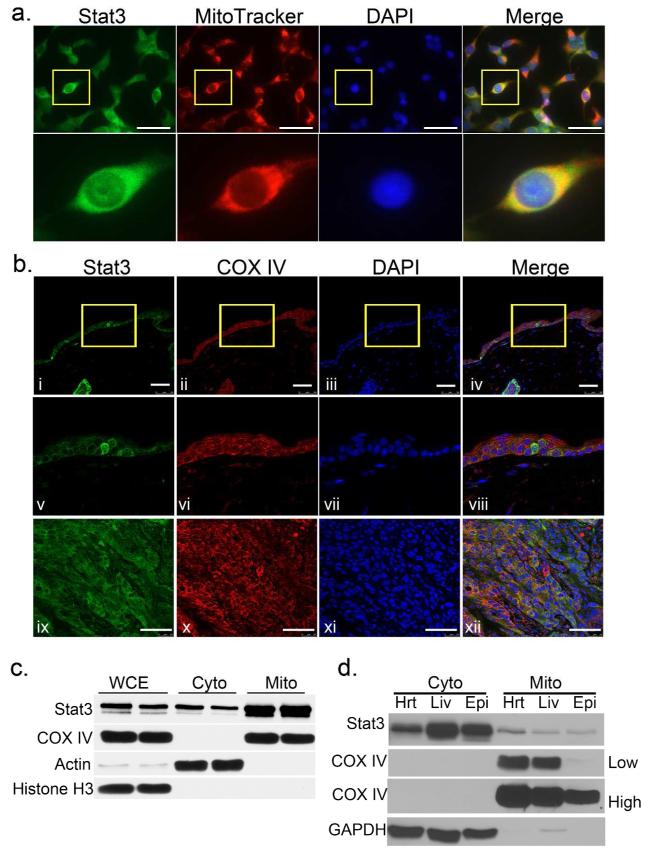
To investigate if keratinocytes contain mitoStat3, we initially utilized MitoTracker red and stained for Stat3 in methanol fixed mouse 3PC keratinocytes grown in culture. As shown in Figure 1A, Stat3 colocalized with MitoTracker red in 3PC cells as shown by cells with an orange to yellow hue. To determine if Stat3 localizes to mitochondria in vivo, we conducted double immunocytochemical staining for the mitochondrial marker cytochrome c oxidase subunit IV (COX IV) and Stat3 in paraffin embedded sections of untreated mouse skin. As detected by confocal microscopy, Stat3 and COX IV were colocalized in epidermal keratinocytes (Figure 1B i-viii). Moreover, Stat3 colocalized with COX IV in the epithelial cells of SCCs derived from a two-stage (DMBA/TPA) chemical carcinogenesis protocol (Figure 1B ix-xii). In order to test these initial observations in a more rigorous fashion, western blot analyses were performed on lysates from mitochondrial-enriched fractions isolated via a differential centrifugation method. Consistent with our immunocytochemical experiments, western blot analyses showed retention of Stat3 in mitochondrial fractions isolated from adult mouse primary keratinocyte cultures (Figure 1C) and in vivo from mouse epidermal scrapings (Figure 1D). In addition, mitochondrial fractions from liver and heart tissues showed retention of Stat3, consistent with previously published reports (Gough et al. 2009; Wegrzyn et al. 2009; Boengler et al. 2010). It is important to note that despite having a relatively low mitochondrial load (COX IV), relative to heart and liver, epidermal keratinocytes contained a comparable amount of mitoStat3 in vivo (Figure 1D).
Figure 1. Stat3 in keratinocyte mitochondria.
(A) Mitochondrial localization of Stat3. Mouse 3PC keratinocyte cells were incubated with MitoTracker for 15 minutes, then permeabilized and stained for Stat3 (green). Scale bars = 50 μm
(B) Colocalization of Stat3 and mitochondrial marker COX IV in vivo. Mouse skin (i-viii) and SCC (ix-xii) sections were co-immunostained with mouse anti-Stat3 (green) and rabbit anti-COX IV (red). Scale bars = 50 μm.
(C) Detection of Stat3 in mitochondrial enriched fractions. Western blot analysis of mitochondrial fractions from primary culture adult mouse keratinocytes and (D) Analysis of mitoStat3 in vivo. Mitochondrial fractions from mouse heart (Hrt), liver (Liv) and skin epidermal cells (Epi). Actin and GAPDH were used as cytoplasmic markers, while COX IV was used as mitochondrial marker [lighter (low) and darker (high) exposures are shown]. WCE; whole cell extract, Cyto; cytoplasmic fraction, Mito; mitochondrial fraction.
Regulation and mitochondrial translocation of phosho-Stat3Ser727 in keratinocytes
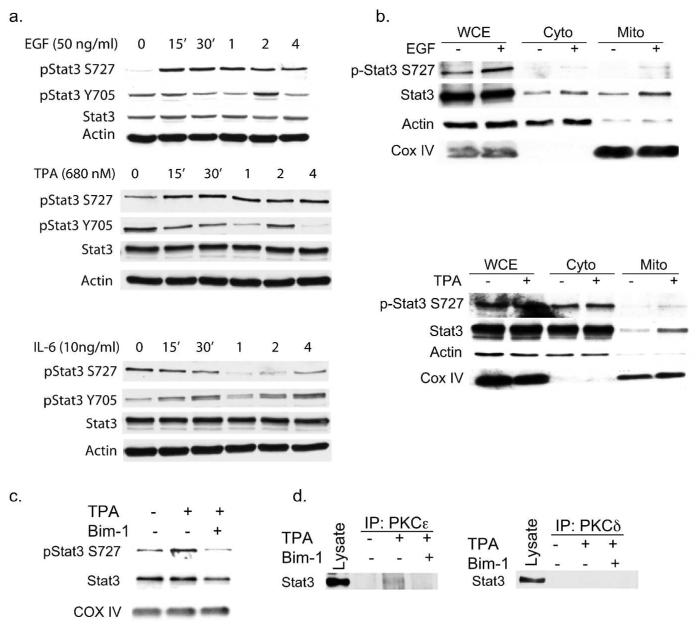
In light of the fact that phosphorylation of Stat3 on Ser727 (p-Stat3Ser727) is required for mitoStat3 localization, we examined the kinetics of Stat3Ser727 phosphorylation in keratinocytes. Western blot analyses of lysates from primary keratinocytes showed that p-Stat3Ser727 levels are induced within 15 min of treatment with either EGF or TPA, but not with IL-6 (Figure 2A). In contrast, we did not detect any immediate induction of p-Stat3Tyr705 levels with either EGF or TPA, as was the case with IL-6 treatment. Both human NCTC 2544 and mouse JB6 cells also showed comparable Ser727 phosphorylation kinetics (data not shown). To test whether Ser727 phosphorylation affected mitoStat3 levels, mitochondrial lysate fractions were isolated from primary keratinocytes 1 hr post-treatment with EGF and TPA and analyzed. As shown in Figure 2B, treatment with EGF or TPA increased the amount of total Stat3 and p-Stat3Ser727 levels in mitochondrial-enriched fractions in comparison to untreated controls.
Figure 2. Phosphorylation and translocation of mitoStat3.
(A) Kinetics of Stat3 phosphorylation. Primary keratinocytes were treated and harvested as indicated. Cell lysates were probed for phospho-Stat3Y705 and Stat3S727, Stat3 and actin as loading control.
(B) Mitochondrial translocation of Stat3 in mouse keratinocytes. Primary keratinocytes were treated as indicated for 1hr prior to mitochondrial cellular fractionation. Actin and COX IV were used as loading controls.
(C) Bim-1 inhibits mitochondrial serine phosphorylation of mitoStat3. Primary mouse keratinoctyes were treated with DMSO (control), TPA (680 nM) or pretreated for 1hr with Bim-1 and then treated with TPA for 1hr.
(D) PKCε association with mitoStat3. Mitochondrial lysates treated as above were immunoprecipitated with rabbit antibodies against PKCε or PKCδ and immunoblotted with a mouse anti-Stat3 antibody.
The phorbol ester tumor promoter, TPA, stimulates rapid activation of protein kinase C (PKC) (Weinstein 1985). PKCδ and PKCε are reported to bind and directly phosphorylate Stat3 Ser727 in mouse epidermal keratinocytes (Gartsbein et al. 2006; Aziz et al. 2007; Aziz et al. 2007). In addition, both PKCδ and PKCε have been previously shown to reside in and regulate mitochondria function (Li et al. 1999; Ardehali 2006; Budas and Mochly-Rosen 2007). Therefore, the possible role of PKCs in TPA-induced mitochondrial translocation of Stat3 was examined. To test this role, we first analyzed the effects of the pan PKC inhibitor Bisindolylmaleimide I (Bim-1) on Stat3 phosphorylation. As shown in Supplemental Figure 1A, pretreatment with Bim-1 inhibited the increase of p-Stat3Ser727 levels induced by TPA. In addition, pretreatment with Bim-1 reduced the basal level of p-Stat3Tyr705 and also reduced levels at 2 and 4 hrs following treatment with TPA. In contrast, phosphorylation of Stat3Tyr705 by IL-6 was not inhibited by Bim-1. Mitochondrial lysates from primary keratinocytes pretreated with Bim-1 followed by TPA treatment for 1 hr showed a decreased level of p-Stat3Ser727 and total Stat3 compared to mitochondrial fractions from cells treated only with TPA (Figure 2C). Consistent with previous reports, we detected an association of Stat3 with PKCδ and PKCε in whole cell extracts of cultured JB6 cells (Supplemental Figure S1). To test whether PKCs associated with Stat3 in mitochondria, we conducted immunoprecipitation analyses of mitochondrial lysates. As shown in Figure 2D, PKCε, but not PKCδ, was bound to mitoStat3 following treatment with TPA. The association of mitoStat3 and PKCε in mitochondrial-enriched fractions from TPA treated cells was inhibited by pretreatment with Bim-1.
Stat3 binds to mitochondrial DNA
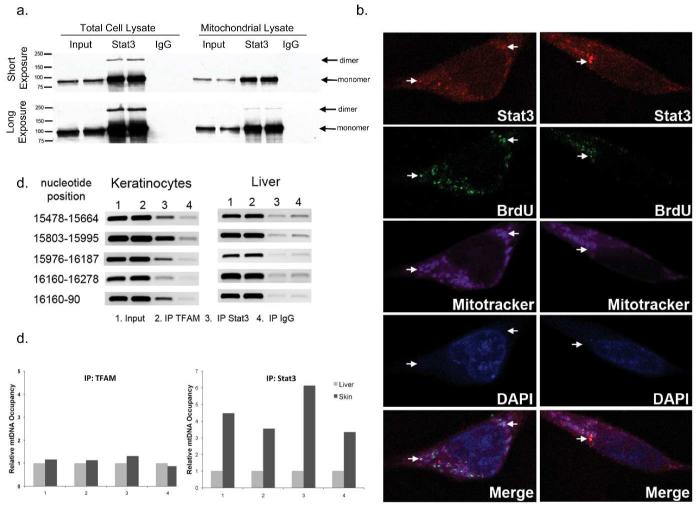
To determine if mitoStat3 exists in a monomer and/or dimer state, immunoprecipitations of whole cell and mitochondrial lysates from NCTC 2544 cells were performed with a mouse anti-Stat3 antibody, followed by immunoblotting with a rabbit anti-Stat3 antibody. As shown in Figure 3A, protein bands at approximately 90 kDa and 180 kDa were detected in Stat3 pull downs from whole cell lysates as well as mitochondrial-enriched fractions following a longer exposure suggesting Stat3 dimer formation in keratinocyte mitochondria. Furthermore, examination of mouse and human (not shown) mitochondrial DNA (mtDNA) sequence revealed multiple generic Stat or GAS (TTN4-6AA) binding sequences within the regulatory d-loop region (Supplemental Figure S2). Given these data, we examined whether mitoStat3 binds to mtDNA. To test this, a double labeling immunocytochemical experiment was conducted, probing for endogenous Stat3 and BrdU labeled mtDNA in NCTC 2544 keratinocytes. As shown in Figure 3B, Stat3 colocalized with BrdU labeled mtDNA. To further substantiate mitoStat3-mtDNA binding, a chromatin immunoprecipitation (ChIP) assay was conducted using purified mitochondrial fractions obtained from mouse epidermal keratinocytes. In accordance with the double labeling experiment, PCR analyses of Stat3-mtDNA pull downs using primers spanning potential Stat binding/GAS sequences in the regulatory d-loop region showed binding of Stat3 (Figure 3C). Binding of a constitutively active/dimerized Stat3 mutant to mtDNA was also observed in mitochondrial ChIP analyses of mitochondria from keratinocytes of BK5.Stat3C transgenic mice (Supplemental Figure S3). Interestingly, Stat3 bound weakly, at background levels, in Stat3-mtDNA pull downs from liver mitochondrial lysates. In fact, RT-PCR analyses showed that mitoStat3 in keratinocytes occupied the mtDNA d-loop region approximately 3-6 fold higher compared to mitoStat3 from liver (Figure 3D). In contrast, the mitochondrial transcription factor A (TFAM), which was used as a positive control, was bound at similar levels to mtDNA from both liver and epidermal keratinocytes.
Figure 3. mitoStat3 binds to mtDNA in keratinocytes.
(A) Whole cell and mitochondrial lysates were immunoprecipitated with mouse anti-Stat3 and immunoblotted with rabbit anti-Stat3. Arrows denote Stat3 monomers and dimers.
(B) mitoStat3 colocalizes with mtDNA. Immunostaining of BrdU (green) labeled mitochondria and Stat3 (Cy5, pseudocolor red). Mitotracker red staining changed to pseudocolor magenta.
(C) Stat3 binds to mtDNA. Mitochondrial ChIP assay was conducted from epidermal keratinocytes and liver mitochondria. Immunoprecipitations conducted with Stat3, TFAM and rabbit IgG. PCR carried out with primers spanning STAT binding sequences at indicated nucleotide positions.
(D) Quantification of Stat3 mtDNA occupancy via Real-Time PCR. Input was used to normalize and liver pulldowns were used as reference. Primer sets 1-4 correspond to amplicons at nucleotide positions, 15478-15664, 15803-15995, 15976-16187 and 16160-90 respectively.
mitoStat3 binds to TFAM and throughout the mitochondrial genome
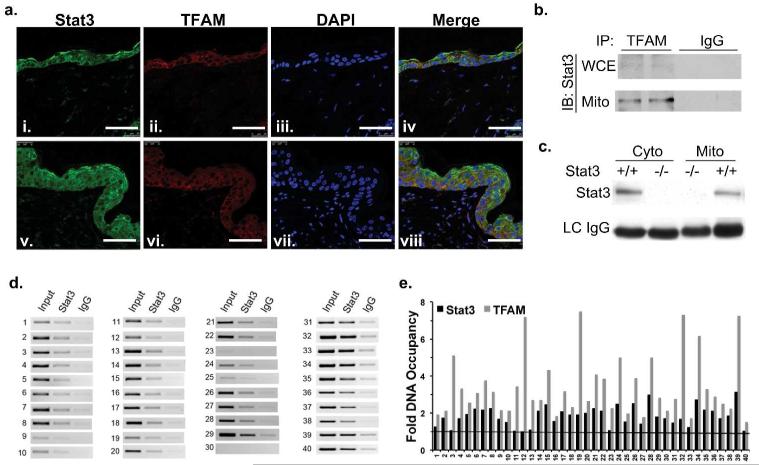
Given the data in Figure 3 showing that Stat3 binds to the mtDNA d-loop regulatory region, the association of Stat3 with TFAM, which is known to bind and regulate mtDNA replication and transcription, was evaluated (Campbell et al. 2012). TFAM and Stat3 colocalized in epidermal keratinocytes of both control (acetone treated, i-iv) and hyperplastic (TPA treated, v-viii) skin sections (Figure 4A). Colocalization of TFAM and Stat3 was observed in the basal cell layer of the epidermis (proliferative compartment), which was increased in the hyperplastic epidermis. Immunoprecipitation experiments of whole cell lysates and mitochondrial enriched fractions from NCTC 2544 cells with an anti-TFAM antibody showed association with Stat3 (Figure 4B). Additionally, similar pull down experiments of cytoplasmic and mitochondrial enriched lysates, from Stat3fl/fl and HK5Cre.Stat3fl/fl primary keratinocytes (negative control) showed clear co-immunoprecipitation of Stat3 (Figure 4C). While TFAM binds to sequence-specific sites, it also has non-specific DNA binding activity and is bound randomly to the majority of mitochondrial genomic DNA (Kanki et al. 2004; Campbell et al. 2012). Given our observed TFAM-Stat3 interaction, we conducted a mitochondrial genome wide ChIP assay for Stat3-mDNA complexes. Primer pairs spanning the entire 16.3 kb mouse mitochondrial genome were designed to amplify approximately every 300-400 bp of mtDNA (Supplemental Table S1). As shown in Figure 4D, PCR analyses demonstrated that Stat3 binds various sites throughout the mitochondrial genome. In addition, we compared Stat3 and TFAM mtDNA binding throughout the mitochondrial genome by qPCR. While TFAM bound to a greater extent to mtDNA, mitoStat3 binding to mtDNA followed a similar pattern (Figure 4E).
Figure 4. Stat3 associates with TFAM and is bound throughout the mitochondrial genome.
(A) Paraffin embedded normal skin (i-iv) and TPA treated hyperplastic skin (v-viii) were immunostained for Stat3 (green) and TFAM (red). Scale bar = 50 m.
(B) NCTC 2544 whole cell and mitochondrial lysates were immunoprecipitated with TFAM and blotted for Stat3.
(C) Cytoplasmic and mitochondrial fractions from Stat3fl/fl (+/+) and HK5Cre.Stat3fl/fl (−/−) keratinoctyes were immunoprecipitated with mouse anti-TFAM and blotted for Stat3.
(D) Stat3 association with mitochondrial genome. PCR reactions were carried out on DNA fragments from mtDNA-Stat3 immunoprecipitates. Forty primers sets (Supplemental Table S1) spanning the mitochondrial genome amplifying every 300-400 bp were used.
(E) qPCR was conducted to quantify the amount of TFAM and Stat3 binding throughout the mitochondrial genome using the primer sets as above. Relative fluorescence units were used to normalize to input and then fold binding relative to the IgG negative control was calculated.
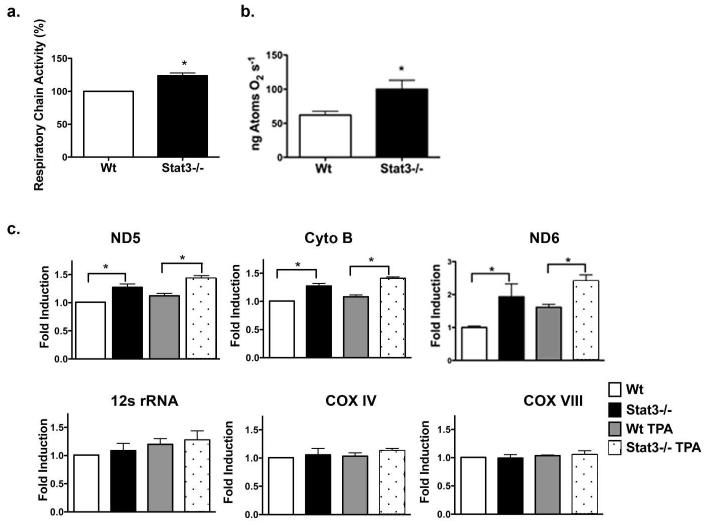
Stat3 regulation of mitochondrial respiration and mitochondrial-encoded genes
To further evaluate the function of mitoStat3, experiments were performed to determine whether association of mitoStat3 with mtDNA influences mitochondrial respiration and transcription of mitochondrial-encoded genes. To test this, Stat3−/− primary keratinocytes from HK5Cre.Stat3fl/fl mice were utilized and compared with keratinocytes from wild-type mice. Utilizing a resazurin dye based method (Figure 5A) and a Clarke-type oxygen polarographic method (Figure 5B), mitochondrial respiratory chain activity was found to be elevated in Stat3−/− primary keratinocytes compared to wild-type controls. Furthermore, we observed a significant induction of mitochondrial-encoded cytochrome b (Cyto B) and NADH dehydrogenase 5 (ND5) mRNA transcripts in Stat3−/− primary keratinoctyes, along with various other mitochondrial encoded genes (Supplemental Figure S4A), compared to wild-type controls (Figure 5C). In addition, NADH dehydrogenase 6 (ND6), which is encoded on the light strand (reverse strand) of the mitochondrial genome, was also significantly upregulated in Stat3−/− primary keratinocytes (Figure 5C). TPA treatment further induced the transcription of mitochondrial-encoded genes Cyto B, ND5 and ND6 in Stat3−/− primary keratinocytes. In further experiments the expression of nuclear encoded genes that could influence mitochondrial biogenesis was analyzed (i.e. mTERF, PGC-1α and NRF-1). Interestingly, the expression of these genes was also induced by the loss of Stat3 in primary keratinocytes (Supplemental Figure S4A). However, no differences in the expression of the nuclear-encoded mitochondrial proteins cytochrome c oxidiase subunit VIIIa (Cox8a) or cytochrome c oxidase subunit IV (Cox4), were observed in Stat3−/− keratinocytes compared to wild-type keratinocytes (Figure 5B). Furthermore, no difference in mtDNA copy number was observed in Stat3−/− keratinocytes compared to wild-type keratinocytes (Supplemental figure S4B). Collectively, these results suggest that mitoStat3 may regulate transcription of mitochondrial-encoded genes.
Figure 5. Stat3 regulates mitochondrial-encoded genes.
(A) Stat3 regulation of mitochondrial respiratory chain activity. Primary adult keratinocytes from Stat3fl/fl (Wt) and HK5.Cre;Stat3fl/fl (Stat3−/−) were assayed for mitochondrial activity by reduction of resazurin. Values represent mean ± SEM (n=4). (*, P = 0.0265 calculated by Mann-Whitney U test).
(B) Oxygen consumption measured using Clarke-type electrode from Stat3fl/fl (Wt) and HK5.Cre;Stat3fl/fl (Stat3−/−) single cell epidermal keratinocytes in Krebs-Ringer solution. Values represent mean ± SEM (n=3). (*, P = 0.0480 calculated by Mann-Whitney U test).
(C) Quantitative analysis of mitochondrial and nuclear encoded genes. Adult primary keratinocytes from Stat3fl/fl (Wt) and HK5.Cre;Stat3fl/fl (Stat3−/−) were treated with DMSO (control) or 680 nM TPA for 1hr. Real time quantitative PCR was carried out using Sybr green on an ABI Viia 7 RT-PCR system using comparative Ct method. Triplicate samples were run in each experiment and averaged in individual experiments. Values represent mean ± SEM (n=4). (*, P = 0.0268 calculated by Mann-Whitney U test.)
DISCUSSION
As noted in the introduction, recent data suggest that Stat3 functions in mitochondria, possibly as an interacting protein with ETC components (Gough et al. 2009; Wegrzyn et al. 2009). However, this reported mitochondrial function remains controversial since there is a large disparity between the molecular ratios of Stat3 and ETC components in some cell types (Phillips et al. 2010). In this study, we provide evidence for a previously unreported role of Stat3 in mitochondria in keratinocytes. In this regard, we have shown that mitoStat3 binds to mtDNA, associates with TFAM and appears to regulate the expression of mitochondrial-encoded genes.
The data presented in this report demonstrate that mitochondria of epidermal keratinocytes (both in vivo and in primary cell culture) contain a pool of Stat3. In addition, mitoStat3 was detected in SCCs derived from a two-stage skin carcinogenesis protocol. Compared to liver and heart, epidermal keratinocytes contain a relatively low mitochondrial load, as shown by levels of the mitochondrial marker, COX IV (Figure 1C). However, western blot analyses revealed that mitoStat3 protein levels were similar between mitochondrial protein lysates obtained from these three tissues. Consequently, keratinocytes contain greater amounts of Stat3 per mitochondria as compared to cells that are more abundant for mitochondria. In addition, treatment of primary keratinocytes with TPA or EGF increased levels of p-Stat3Ser727 and induced mitochondrial translocation of Stat3. Increased p-Stat3Ser727 levels seen following treatment with TPA were dependent on PKC activation, as illustrated by inhibition using the pan PKC inhibitor, Bim-1. PKCε, but not PKCδ was found to interact with Stat3 in mitochondrial fractions. In addition, PKC inhibition reduced Stat3 mitochondrial localization, p-Stat3Ser727 levels and abrogated Stat3-PKCε association in mitochondria. These data are consistent with previous publications showing interaction of Stat3 with PKCε and PKCε mediated phosphorylation of Stat3 at Ser727 in mouse keratinoctyes treated with either TPA or UVB irradiation (Aziz et al. 2007; Aziz et al. 2007). Like mitoStat3, PKCε has also been shown to interact with components of the ETC in cardiomyocytes (Ardehali 2006; Budas et al. 2007; Szczepanek et al. 2012). These observations suggest a possible mitoStat3-PKC dependent mitochondrial import mechanism in response to TPA treatment. However, it is also possible that Stat3 is imported via a PKC independent mechanism and is phosphorylated by PKCε within mitochondria, as PKCs maintain kinase activity in mitochondria (Backer et al. 1986).
As shown in Figure 3, endogenous Stat3 homodimers as well as monomers were observed by immunoprecipitation of Stat3 from mitochondrial-enriched protein lysates, which suggested that mitoStat3 is capable of binding mtDNA. Consistent with this observation, a constitutively active/dimerized Stat3 mutant (Stat3C) has recently been reported to localize in mitochondria of Stat3C knockin mouse embryonic fibroblasts (Demaria et al. 2010). In addition, mitoStat3 colocalized labeled mtDNA in (Figure 3B) and mitochondrial ChIP analyses showed binding of mitoStat3 to the mitochondrial d-loop, which is the regulatory region for transcription of mtDNA (Scarpulla 2008). Similarly, the constitutively active/dimerized Stat3C mutant bound mtDNA in mitochondria from BK5.Stat3C transgenic mice (Supplemental Figure S3). These data are consistent with an earlier publication showing that activated Stat3 binds to mtDNA in human platelet mitochondria (Vassilev et al. 2002). More recently, an mtDNA deoxyribonuclease I footprinting/protection assay showed that protected mtDNA fragments aligned with the Stat3 consensus recognition sequence in various cell lines (Mercer et al. 2011). Collectively, these data show that mitoStat3 binds to mtDNA.
TFAM has both sequence-dependent and sequence-independent DNA binding activity and its abundance allows it to cover the whole mitochondrial genome (Scarpulla 2008). In this regard, TFAM regulates mitochondrial DNA copy number and mitochondrial chromosome stability (Scarpulla 2008). This feature of TFAM may explain the observed binding of Stat3 throughout the mitochondrial genome. Another interesting observation was that interaction between TFAM and Stat3 was also detected in cytoplasmic fractions from keratinocytes in addition to mitochondrial fractions. Although TFAM is predominantly a mitochondrial localized protein, it is also found outside of mitochondria (Pastukh et al. 2007). To our knowledge, this interaction has not been previously reported, however TFAM has recently been shown to bind the promoter region and regulate known Stat3 nuclear target genes, including Survivin and Bcl-2 (Han et al. 2011; Kurita et al. 2012). Whether or not TFAM associates with nuclear Stat3 is undetermined. Nevertheless, our data demonstrates that mitoStat3 binds to TFAM as well as throughout the mitochondrial genome, in a manner similar to TFAM.
Lastly, our data shows that mitoStat3 binding to TFAM and mtDNA appears to have a functional role in keratinocyte mitochondria. In this regard, loss of Stat3 resulted in an increased level of mitochondrial respiratory chain activity in primary keratinocytes. In addition, we observed a statistically significant increase in mRNA levels of several mitochondrial-encoded genes (e.g., Cyto b, ND5 and ND6) in Stat3−/− keratinocytes. The mitochondrial genome contains three transcriptional start sites within the d-loop regulatory region, HSP1, HSP2 and LSP (reverse strand) (Peralta et al. 2012). Transcription from the HSP2 start site results in a polycistronic mRNA that encodes the two rRNAs and 12 of 13 mitochondrial encoded genes. Therefore, a uniform induction of mitochondrial encoded genes and rRNAs is expected. However, despite their polycistronic origin, we did not observe a uniform increase in mitochondrial transcripts. By deep sequencing of mitochondrial RNA, Mercer and colleagues have also recently observed a wide variation in mitochondrial transcript abundance, and have shown that this is due to transcript processing and mRNA maturation events (Mercer et al. 2011). We also observed an increase in nuclear encoded mTERF, PGC-1α and NRF-1 in Stat3−/− keratinocytes, which could induce mitochondrial biogenesis and transcription. However, the lack of induction of nuclear encoded ETC component mRNAs (e.g., COX IV and COXVIII) coupled with no changes in mtDNA copy number in Stat3−/− keratinocytes supports the conclusion that the increase in mitochondrial transcripts is due to loss of mitoStat3 and not an increase in nuclear encoded genes, PGC-1α or NRF-1.
The concept of a nuclear transcription factor having a non-canonical role in the mitochondria is not entirely surprising (e.g., p53, IRF3, NF-κB and STATs) [reviewed in (Szczepanek et al. 2012)]. Evidence includes data showing binding of mtDNA and gene regulation by nuclear transcription factors such as NF-κB, CREB and MEF2D (Lee et al. 2005; Benavides et al. 2011; Johnson et al. 2011). In fact, similar to mitoStat3, NF-κB was shown to negatively regulate mitochondrial-encoded genes (e.g., cyotochrome c oxidase subunit III (CoxIII) and Cyto B) (Cogswell et al. 2003; Johnson et al. 2011). In addition, reduction of NF-κB resulted in increased mitochondrial respiration, similar to our results seen with loss of Stat3 (Johnson et al. 2011). Stat5 also translocates to leukemic T cell mitochondria and is able to bind mtDNA in vitro upon cytokine stimulation (Chueh et al. 2010). Both Stat5 and NF-κB are known to associate with Stat3, thus, it would be interesting to determine whether these proteins associate with mitoStat3 and function cooperatively in regulation of gene expression in mitochondria (Novak et al. 1996; Lee et al. 2008; Lee et al. 2009).
In closing, the current data demonstrate, to our knowledge, a previously unreported role for mitoStat3 in keratinocytes. We hypothesize that mitoStat3 is able to bind to and potentially regulate mtDNA transcription, presumably through the previously undescribed association with the mitochondrial transcription factor TFAM although we cannot rule out a more direct mtDNA binding mechanism at the present time. While the majority of mitochondrial proteins are encoded in the nucleus, this pathway appears to represent a localized transcriptional regulatory mechanism that can alter mitochondrial biogenesis and respiration. Both EGF and TPA induced Stat3 mitochondrial translocation. Furthermore, TPA-induced mitochondrial translocation of Stat3 is mediated by PKCε. These data suggest a possible role for mitoStat3 in epidermal proliferation and/or differentiation. Whether this function of mitoStat3 plays a significant role in epithelial tumorigenesis remains to be further investigated.
MATERIALS AND METHODS
Primary keratinocyte isolation and cell culture
Primary mouse keratinocytes were isolated from 6-8 week old FVB or HK5.Cre;Stat3fl/fl mice as previously described (Dlugosz et al. 1995). Primary keratinocytes were plated on collagen coated plates and maintained in complete keratinocyte growth medium consisting of Eagle’s minimal essential base medium without Ca2+ supplemented as previously described (Dlugosz et al. 1995). Human NCTC 2544 cells were grown in RPMI-1640 medium supplemented with 10% FBS. Mouse epidermal JB6 (P+) epidermal cells were grown in MEM supplemented with 5% FBS.
Western blot and co-immunoprecipitations
For straight western blot experiments, keratinocytes were lysed with RIPA lysis buffer (50 mM Tris, 150 mM NaCl, 1% NP-40, 0.5% DOCS, 0.1% SDS). For co-immunoprecipitation experiments cells were lysed with 0.5% NP-40 lysis buffer (50 mM Tris, 150 mM NaCl, 50 mM NaF, and NP-40). All lysis buffers were supplemented with protease inhibitor cocktail, phosphatase inhibitor cocktail and 1 mM DTT just prior to use (Sigma Aldrich). For antibodies used see Supplemental Materials and Methods.
Subcellular fractionation and mitochondrial purification
A differential centrifugation subcellular mitochondrial fractionation experimental procedure was adapted and modified from a previously described method (Vander Heiden et al. 1997). For more details see Supplemental Materials and Methods.
Mitochondrial respiratory chain activity
Adult primary keratinocytes were seeded on 12 well plates at equal densities between genotypes. Stock solution of resazurin (0.01% w/v) was added to cell culture media and incubated for 4 hrs. Activity was measured by absorbance taken at 570 nm minus the reference at 600 nm and adjusted to background levels. Values were normalized to total mg of protein. Oxygen consumption was also measured with a YSI 5300 oxygen monitor equipped with a Clarke-type oxygen electrode (Yellow Springs Instruments, Yellow Springs, OH) following the manufacturer’s instructions.
Immunofluorescence staining and confocal microscopy
Mitochondrial DNA labeling was facilitated by inhibiting nuclear DNA synthesis with 20 μM aphidicolin, followed by incubation with 15 mM BrdU for 3 hrs (Davis and Clayton 1996). For more details on immunofluorescence staining see Supplemental Materials and Methods.
Mitochondrial ChIP assay
Pierce Agarose ChIP kit was used following manufacturer’s protocol modified for mitochondrial ChIP assay. In brief, mtDNA and mitochondrial proteins from intact mitochondrial pellets were cross-linked with 1% formaldehyde. DNA fragmentation was achieved enzymatically with micrococcal nuclease at 37°C for 15 min according to manufacturer’s instructions. Cross-linked mitochondria were then lysed with 0.5% NP-40 lysis buffer and approximately 100 μg of protein lysate was used for immunoprecipitation with indicated antibodies at 4°C overnight. ChIP grade protein A beads were subsequently added for 1.5 hrs followed by wash steps with wash buffers provided in kit. Immunoprecipitations were eluted with 150 μl IP elution buffer, followed by reverse cross-linking by heating at 65°C in 200 mM NaCl and proteinase K digestion (4 μg). DNA was recovered by DNA clean-up column. PCR analysis was conducted using an ABI thermocycler for 28 cycles and products resolved by agarose gel.
Real Time PCR
Reverse transcription of mRNA was carried according to manufacturer’s suggestions with 1 μg of total RNA using SuperScript III First-Strand cDNA synthesis kit (Life Technologies). Real time PCR was carried out on an Applied Biosystems ViiA 7 Real-Time PCR system using Sybr green dye and analyzed using comparative CT method. For more details see Supplemental Materials and Methods.
Statistical analyses
For comparison of mitochondrial respiratory chain activity and analyses of gene induction by Real-Time PCR, Mann-Whitney U test was used. Statistical analyses were conducted using GraphPad Prism 4 software (La Jolla, CA).
Supplementary Material
Acknowledgements
We thank Lauren Pascale for help in the preparation and submission of this manuscript. We also thank Dr. Ted Mills for critical reading of the manuscript and helpful suggestions related to this work.
Footnotes
Conflict of Interest
The authors state no conflict of interest.
References
- Ardehali H. Signaling mechanisms in ischemic preconditioning: interaction of PKCepsilon and MitoK(ATP) in the inner membrane of mitochondria. Circ Res. 2006;99(8):798–800. doi: 10.1161/01.RES.0000247029.31997.a4. [DOI] [PubMed] [Google Scholar]
- Aziz MH, Manoharan HT, Sand JM, Verma AK. Protein kinase Cepsilon interacts with Stat3 and regulates its activation that is essential for the development of skin cancer. Mol Carcinog. 2007;46(8):646–653. doi: 10.1002/mc.20356. [DOI] [PubMed] [Google Scholar]
- Aziz MH, Manoharan HT, Verma AK. Protein kinase C epsilon, which sensitizes skin to sun’s UV radiation-induced cutaneous damage and development of squamous cell carcinomas, associates with Stat3. Cancer Res. 2007;67(3):1385–1394. doi: 10.1158/0008-5472.CAN-06-3350. [DOI] [PubMed] [Google Scholar]
- Backer JM, Arcoleo JP, Weinstein IB. Protein phosphorylation in isolated mitochondria and the effects of protein kinase C. FEBS Lett. 1986;200(1):161–164. doi: 10.1016/0014-5793(86)80530-0. [DOI] [PubMed] [Google Scholar]
- Benavides F, Blando J, Perez CJ, Garg R, Conti CJ, DiGiovanni J, Kazanietz MG. Transgenic overexpression of PKCepsilon in the mouse prostate induces preneoplastic lesions. Cell Cycle. 2011;10(2):268–277. doi: 10.4161/cc.10.2.14469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boengler K, Hilfiker-Kleiner D, Heusch G, Schulz R. Inhibition of permeability transition pore opening by mitochondrial STAT3 and its role in myocardial ischemia/reperfusion. Basic Res Cardiol. 2010;105(6):771–785. doi: 10.1007/s00395-010-0124-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman T, Garcia R, Turkson J, Jove R. STATs in oncogenesis. Oncogene. 2000;19(21):2474–2488. doi: 10.1038/sj.onc.1203527. [DOI] [PubMed] [Google Scholar]
- Bromberg J, Darnell JE., Jr. The role of STATs in transcriptional control and their impact on cellular function. Oncogene. 2000;19(21):2468–2473. doi: 10.1038/sj.onc.1203476. [DOI] [PubMed] [Google Scholar]
- Budas GR, Churchill EN, Mochly-Rosen D. Cardioprotective mechanisms of PKC isozyme-selective activators and inhibitors in the treatment of ischemia-reperfusion injury. Pharmacol Res. 2007;55(6):523–536. doi: 10.1016/j.phrs.2007.04.005. [DOI] [PubMed] [Google Scholar]
- Budas GR, Mochly-Rosen D. Mitochondrial protein kinase Cepsilon (PKCepsilon): emerging role in cardiac protection from ischaemic damage. Biochem Soc Trans. 2007;35(Pt 5):1052–1054. doi: 10.1042/BST0351052. [DOI] [PubMed] [Google Scholar]
- Campbell CT, Kolesar JE, Kaufman BA. Mitochondrial transcription factor A regulates mitochondrial transcription initiation, DNA packaging, and genome copy number. Biochim Biophys Acta. 2012;1819(9-10):921–929. doi: 10.1016/j.bbagrm.2012.03.002. [DOI] [PubMed] [Google Scholar]
- Chan KS, Carbajal S, Kiguchi K, Clifford J, Sano S, DiGiovanni J. Epidermal growth factor receptor-mediated activation of Stat3 during multistage skin carcinogenesis. Cancer Res. 2004;64(7):2382–2389. doi: 10.1158/0008-5472.can-03-3197. [DOI] [PubMed] [Google Scholar]
- Chan KS, Sano S, Kataoka K, Abel E, Carbajal S, Beltran L, Clifford J, Peavey M, Shen J, Digiovanni J. Forced expression of a constitutively active form of Stat3 in mouse epidermis enhances malignant progression of skin tumors induced by two-stage carcinogenesis. Oncogene. 2008;27(8):1087–1094. doi: 10.1038/sj.onc.1210726. [DOI] [PubMed] [Google Scholar]
- Chan KS, Sano S, Kiguchi K, Anders J, Komazawa N, Takeda J, DiGiovanni J. Disruption of Stat3 reveals a critical role in both the initiation and the promotion stages of epithelial carcinogenesis. J Clin Invest. 2004;114(5):720–728. doi: 10.1172/JCI21032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chueh FY, Leong KF, Yu CL. Mitochondrial translocation of signal transducer and activator of transcription 5 (STAT5) in leukemic T cells and cytokine-stimulated cells. Biochem Biophys Res Commun. 2010;402(4):778–783. doi: 10.1016/j.bbrc.2010.10.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogswell PC, Kashatus DF, Keifer JA, Guttridge DC, Reuther JY, Bristow C, Roy S, Nicholson DW, Baldwin AS., Jr. NF-kappa B and I kappa B alpha are found in the mitochondria. Evidence for regulation of mitochondrial gene expression by NF-kappa B. J Biol Chem. 2003;278(5):2963–2968. doi: 10.1074/jbc.M209995200. [DOI] [PubMed] [Google Scholar]
- Davis AF, Clayton DA. In situ localization of mitochondrial DNA replication in intact mammalian cells. J Cell Biol. 1996;135(4):883–893. doi: 10.1083/jcb.135.4.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demaria M, Giorgi C, Lebiedzinska M, Esposito G, D’Angeli L, Bartoli A, Gough DJ, Turkson J, Levy DE, Watson CJ, Wieckowski MR, Provero P, Pinton P, Poli V. A STAT3-mediated metabolic switch is involved in tumour transformation and STAT3 addiction. Aging (Albany NY) 2010;2(11):823–842. doi: 10.18632/aging.100232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dlugosz AA, Glick AB, Tennenbaum T, Weinberg WC, Yuspa SH. Isolation and utilization of epidermal keratinocytes for oncogene research. Methods Enzymol. 1995;254:3–20. doi: 10.1016/0076-6879(95)54003-2. [DOI] [PubMed] [Google Scholar]
- Gartsbein M, Alt A, Hashimoto K, Nakajima K, Kuroki T, Tennenbaum T. The role of protein kinase C delta activation and STAT3 Ser727 phosphorylation in insulin-induced keratinocyte proliferation. J Cell Sci. 2006;119(Pt 3):470–481. doi: 10.1242/jcs.02744. [DOI] [PubMed] [Google Scholar]
- Gough DJ, Corlett A, Schlessinger K, Wegrzyn J, Larner AC, Levy DE. Mitochondrial STAT3 supports Ras-dependent oncogenic transformation. Science. 2009;324(5935):1713–1716. doi: 10.1126/science.1171721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han B, Izumi H, Yasuniwa Y, Akiyama M, Yamaguchi T, Fujimoto N, Matsumoto T, Wu B, Tanimoto A, Sasaguri Y, Kohno K. Human mitochondrial transcription factor A functions in both nuclei and mitochondria and regulates cancer cell growth. Biochem Biophys Res Commun. 2011;408(1):45–51. doi: 10.1016/j.bbrc.2011.03.114. [DOI] [PubMed] [Google Scholar]
- Hazan-Halevy I, Harris D, Liu Z, Liu J, Li P, Chen X, Shanker S, Ferrajoli A, Keating MJ, Estrov Z. STAT3 is constitutively phosphorylated on serine 727 residues, binds DNA, and activates transcription in CLL cells. Blood. 2010;115(14):2852–2863. doi: 10.1182/blood-2009-10-230060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RF, Witzel, Perkins ND. p53-dependent regulation of mitochondrial energy production by the RelA subunit of NF-kappaB. Cancer Res. 2011;71(16):5588–5597. doi: 10.1158/0008-5472.CAN-10-4252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki T, Ohgaki K, Gaspari M, Gustafsson CM, Fukuoh A, Sasaki N, Hamasaki N, Kang D. Architectural role of mitochondrial transcription factor A in maintenance of human mitochondrial DNA. Mol Cell Biol. 2004;24(22):9823–9834. doi: 10.1128/MCB.24.22.9823-9834.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DJ, Angel JM, Sano S, DiGiovanni J. Constitutive activation and targeted disruption of signal transducer and activator of transcription 3 (Stat3) in mouse epidermis reveal its critical role in UVB-induced skin carcinogenesis. Oncogene. 2009;28(7):950–960. doi: 10.1038/onc.2008.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DJ, Chan KS, Sano S, Digiovanni J. Signal transducer and activator of transcription 3 (Stat3) in epithelial carcinogenesis. Mol Carcinog. 2007;46(8):725–731. doi: 10.1002/mc.20342. [DOI] [PubMed] [Google Scholar]
- Kim DJ, Kataoka K, Rao D, Kiguchi K, Cotsarelis G, Digiovanni J. Targeted disruption of stat3 reveals a major role for follicular stem cells in skin tumor initiation. Cancer Res. 2009;69(19):7587–7594. doi: 10.1158/0008-5472.CAN-09-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurita T, Izumi H, Kagami S, Kawagoe T, Toki N, Matsuura Y, Hachisuga T, Kohno K. Mitochondrial transcription factor A regulates BCL2L1 gene expression and is a prognostic factor in serous ovarian cancer. Cancer Sci. 2012;103(2):239–444. doi: 10.1111/j.1349-7006.2011.02156.x. [DOI] [PubMed] [Google Scholar]
- Lee H, Herrmann A, Deng JH, Kujawski M, Niu G, Li Z, Forman S, Jove R, Pardoll DM, Yu H. Persistently activated Stat3 maintains constitutive NF-kappaB activity in tumors. Cancer Cell. 2009;15(4):283–293. doi: 10.1016/j.ccr.2009.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Kim CH, Simon DK, Aminova LR, Andreyev AY, Kushnareva YE, Murphy AN, Lonze BE, Kim KS, Ginty DD, Ferrante RJ, Ryu H, Ratan RR. Mitochondrial cyclic AMP response element-binding protein (CREB) mediates mitochondrial gene expression and neuronal survival. J Biol Chem. 2005;280(49):40398–40401. doi: 10.1074/jbc.C500140200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TL, Yeh J, Friedman J, Yan B, Yang X, Yeh NT, Van Waes C, Chen Z. A signal network involving coactivated NF-kappaB and STAT3 and altered p53 modulates BAX/BCL-XL expression and promotes cell survival of head and neck squamous cell carcinomas. Int J Cancer. 2008;122(9):1987–1998. doi: 10.1002/ijc.23324. [DOI] [PubMed] [Google Scholar]
- Levy DE, Darnell JE., Jr. Stats: transcriptional control and biological impact. Nat Rev Mol Cell Biol. 2002;3(9):651–662. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- Li L, Lorenzo PS, Bogi K, Blumberg PM, Yuspa SH. Protein kinase Cdelta targets mitochondria, alters mitochondrial membrane potential, and induces apoptosis in normal and neoplastic keratinocytes when overexpressed by an adenoviral vector. Mol Cell Biol. 1999;19(12):8547–8558. doi: 10.1128/mcb.19.12.8547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Neph S, Dinger ME, Crawford J, Smith MA, Shearwood AM, Haugen E, Bracken CP, Rackham O, Stamatoyannopoulos JA, Filipovska A, Mattick JS. The human mitochondrial transcriptome. Cell. 2011;146(4):645–658. doi: 10.1016/j.cell.2011.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak U, Mui A, Miyajima A, Paradiso L. Formation of STAT5-containing DNA binding complexes in response to colony-stimulating factor-1 and platelet-derived growth factor. J Biol Chem. 1996;271(31):18350–18354. doi: 10.1074/jbc.271.31.18350. [DOI] [PubMed] [Google Scholar]
- Pastukh V, Shokolenko I, Wang B, Wilson G, Alexeyev M. Human mitochondrial transcription factor A possesses multiple subcellular targeting signals. FEBS J. 2007;274(24):6488–6499. doi: 10.1111/j.1742-4658.2007.06167.x. [DOI] [PubMed] [Google Scholar]
- Peralta S, Wang X, Moraes CT. Mitochondrial transcription: lessons from mouse models. Biochim Biophys Acta. 2012;1819(9-10):961–969. doi: 10.1016/j.bbagrm.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips D, Reilley MJ, Aponte AM, Wang G, Boja E, Gucek M, Balaban RS. Stoichiometry of STAT3 and mitochondrial proteins: Implications for the regulation of oxidative phosphorylation by protein-protein interactions. J Biol Chem. 2010;285(31):23532–23536. doi: 10.1074/jbc.C110.152652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin HR, Kim HJ, Kim JY, Hurt EM, Klarmann GJ, Kawasaki BT, Duhagon Serrat MA, Farrar WL. Activation of signal transducer and activator of transcription 3 through a phosphomimetic serine 727 promotes prostate tumorigenesis independent of tyrosine 705 phosphorylation. Cancer Res. 2008;68(19):7736–7741. doi: 10.1158/0008-5472.CAN-08-1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano S, Chan KS, DiGiovanni J. Impact of Stat3 activation upon skin biology: a dichotomy of its role between homeostasis and diseases. J Dermatol Sci. 2008;50(1):1–14. doi: 10.1016/j.jdermsci.2007.05.016. [DOI] [PubMed] [Google Scholar]
- Sarafian TA, Montes C, Imura T, Qi J, Coppola G, Geschwind DH, Sofroniew MV. Disruption of astrocyte STAT3 signaling decreases mitochondrial function and increases oxidative stress in vitro. PLoS One. 2010;5(3):e9532. doi: 10.1371/journal.pone.0009532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpulla RC. Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol Rev. 2008;88(2):611–638. doi: 10.1152/physrev.00025.2007. [DOI] [PubMed] [Google Scholar]
- Shen Y, Schlessinger K, Zhu X, Meffre E, Quimby F, Levy DE, Darnell JE., Jr. Essential role of STAT3 in postnatal survival and growth revealed by mice lacking STAT3 serine 727 phosphorylation. Mol Cell Biol. 2004;24(1):407–419. doi: 10.1128/MCB.24.1.407-419.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczepanek K, Chen Q, Larner AC, Lesnefsky EJ. Cytoprotection by the modulation of mitochondrial electron transport chain: the emerging role of mitochondrial STAT3. Mitochondrion. 2012;12(2):180–189. doi: 10.1016/j.mito.2011.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczepanek K, Lesnefsky EJ, Larner AC. Multi-tasking: nuclear transcription factors with novel roles in the mitochondria. Trends Cell Biol. 2012;22(8):429–437. doi: 10.1016/j.tcb.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turkson J, Jove R. STAT proteins: molecular targets for cancer drug discovery. Oncogene. 2000;19(56):6613–6626. doi: 10.1038/sj.onc.1204086. [DOI] [PubMed] [Google Scholar]
- Vander Heiden MG, Chandel NS, Williamson EK, Schumacker PT, Thompson CB. Bcl-xL regulates the membrane potential and volume homeostasis of mitochondria. Cell. 1997;91(5):627–637. doi: 10.1016/s0092-8674(00)80450-x. [DOI] [PubMed] [Google Scholar]
- Vassilev AO, Lorenz DR, Tibbles HE, Uckun FM. Role of the leukemia-associated transcription factor STAT3 in platelet physiology. Leuk Lymphoma. 2002;43(7):1461–1467. doi: 10.1080/1042819022386716. [DOI] [PubMed] [Google Scholar]
- Wegrzyn J, Potla R, Chwae YJ, Sepuri NB, Zhang Q, Koeck T, Derecka M, Szczepanek K, Szelag M, Gornicka A, Moh A, Moghaddas S, Chen Q, Bobbili S, Cichy J, Dulak J, Baker DP, Wolfman A, Stuehr D, Hassan MO, Fu XY, Avadhani N, Drake JI, Fawcett P, Lesnefsky EJ, Larner AC. Function of mitochondrial Stat3 in cellular respiration. Science. 2009;323(5915):793–797. doi: 10.1126/science.1164551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein IB. Cell culture studies on the mechanism of action of chemical carcinogens and tumor promoters. Carcinog Compr Surv. 1985;10:177–187. [PubMed] [Google Scholar]
- Wen Z, Zhong Z, Darnell JE., Jr. Maximal activation of transcription by Stat1 and Stat3 requires both tyrosine and serine phosphorylation. Cell. 1995;82(2):241–250. doi: 10.1016/0092-8674(95)90311-9. [DOI] [PubMed] [Google Scholar]
- Yang SF, Yuan SS, Yeh YT, Wu MT, Su JH, Hung SC, Chai CY. The role of p-STAT3 (ser727) revealed by its association with Ki-67 in cervical intraepithelial neoplasia. Gynecol Oncol. 2005;98(3):446–452. doi: 10.1016/j.ygyno.2005.05.032. [DOI] [PubMed] [Google Scholar]
- Yeh YT, Ou-Yang F, Chen IF, Yang SF, Wang YY, Chuang HY, Su JH, Hou MF, Yuan SS. STAT3 ser727 phosphorylation and its association with negative estrogen receptor status in breast infiltrating ductal carcinoma. Int J Cancer. 2006;118(12):2943–2947. doi: 10.1002/ijc.21771. [DOI] [PubMed] [Google Scholar]
- Zhong Z, Wen Z, Darnell JE., Jr. Stat3: a STAT family member activated by tyrosine phosphorylation in response to epidermal growth factor and interleukin-6. Science. 1994;264(5155):95–98. doi: 10.1126/science.8140422. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.