Abstract
Introduction
Interleukin-6 (IL-6) can activate downstream signaling pathways in lung cancer cells, such as the STAT3 pathway, and is reported to be produced by tumor cells with activating EGFR mutations. We examined IL-6/STAT3 in lung cancer tumor tissues and the effects of siltuximab, a neutralizing antibody to human IL-6, in mouse models of lung cancer.
Methods
IL-6 and STAT3 activation levels were compared to tumor histology and presence of KRAS mutations in snap-frozen non-small cell lung cancer (NSCLC) tumors. The effects of siltuximab alone or in combination with erlotinib were examined in mouse xenograft models constructed using three cell line xenograft models and one primary explant mouse model. We examined the influence of cancer-associated fibroblasts (CAFs) on tumor growth and siltuximab effects.
Results
IL-6 levels were higher in tumors of squamous cell versus adenocarcinoma histology and were not associated with presence of KRAS mutations. Tyrosine phosphorylation status of STAT3 did not correlate with tumor IL-6 levels. Serine phosphorylation of STAT3 was correlated with KRAS mutation status. Both tumor and stromal cells contributed to total IL-6 within tumors. Siltuximab had minimal effect as a single agent in xenografts with tumor cells alone; however, in models co-administered with CAFs, siltuximab had more potent effects on tumor inhibition. We observed no effects of combined erlotinib and siltuximab.
Conclusions
IL-6 is elevated in subsets of human NSCLCs, especially with squamous cell histology. Tumors supported by stromal production of IL-6 appear to be the most vulnerable to tumor growth inhibition by siltuximab.
Keywords: IL-6, STAT3, siltuximab, KRAS, lung cancer
INTRODUCTION
Lung cancer continues to be the leading form of cancer death in both men and women in the United States. Given the inherent insensitivity of tumor cells to cytotoxic agents, identifying the drivers of lung cancer growth, survival, and metastasis is critical, as is the development of novel therapeutics. Members of the signal transducer and activator of transcription (STAT) family of transcription factors are potential targets in lung cancer and other cancers.1–3 STAT proteins are activated by upstream cytokines and tyrosine kinase signaling and control genes that regulate cancer hallmarks.4 Constitutive STAT3 activation is associated with cancers with poor prognosis via its anti-apoptotic and proliferative effects, as well as effects on immune evasion and tumor angiogenesis.5 STAT3 can become constitutively active in cancer cells via various cellular pathways, including upregulation of cytokines such as interleukin-6 (IL-6). In addition to cytokines, tyrosine kinases, such as receptor tyrosine kinases (epidermal growth factor receptor (EGFR), for example) and non-receptor tyrosine kinases (SRC family kinases) can also drive STAT3 signaling.6 Indirect or direct inhibition of STAT3 has been shown to affect tumor formation through inhibition of cell growth, induction of apoptosis, and/or inhibition of tumor angiogenesis. A number of recent studies have found activated STAT3 in lung cancer cell lines and tissues, thus suggesting a functional role for this target in subsets of lung cancer.7–11 Lung cancers with EGFR mutation have been reported to have more abundant IL-6 production, which drives STAT3 signaling and in some cases may drive resistance to EGFR tyrosine kinase inhibitors (TKIs).12, 13
Our previous studies examined STAT protein activation in lung cancer cell lines, including those with activating mutations in EGFR, and examined upstream kinases responsible for STAT3 phosphorylation and activation using small molecules, antibodies, and RNA interference. In these studies, we found more pronounced STAT3 activation in cells with activating EGFR mutations, yet inhibition of EGFR activity had no effect on STAT3 activation.9 Inhibition of JAK1 with small molecules or RNA interference resulted in loss of STAT3 tyrosine phosphorylation and inhibition of cell growth.10 Siltuximab (CNTO 328), an IL-6 neutralizing antibody, could inhibit STAT3 tyrosine phosphorylation in a cell-dependent manner. In H1650 cells harboring an activating EGFR mutation, siltuximab inhibited STAT3 tyrosine phosphorylation, resulting in inhibition of lung cancer cell growth in vivo. In addition, when siltuximab was combined with the EGFR inhibitor erlotinib, there was a more pronounced inhibition of STAT3 transcriptional activity. Our results suggest that indirect attacks on JAK1-STAT3 using an IL-6 neutralizing antibody with or without EGFR inhibition can inhibit lung cancer growth in lung cancer subsets.
Additional questions remain to be answered to fully realize the potential to translate these findings into early-phase clinical trials and to explore the utility of siltuximab in advanced lung cancer. In this study, we sought answers to the following questions. First, what is the incidence of tumors demonstrating constitutive IL-6 signaling, as this could represent an enriched group for future clinical trials. Second, how should siltuximab be combined with other molecular agents and in which tumor subtype? To explore these questions, we examined IL-6 and STAT3 signaling in 100 snap-frozen human non-small cell lung cancer (NSCLC) tumor tissues, as well as examined their histology and oncogene mutation status. We then explored efficacy of siltuximab with the EGFR-TKI erlotinib in three lung cancer cell line xenograft models. As part of these studies, we also studied the impact of cancer-associated fibroblasts (CAF) on tumor growth and sensitivity to siltuximab and erlotinib. CAF cells are known to be large producers of IL-6 in the cancer microenvironment, and we hypothesized that these heterotypic models may better recapitulate human lung cancers. Finally, we examined siltuximab in a primary explant mouse model of lung cancer. Our ultimate goal was to reveal the pattern of IL-6 to downstream STAT3 activation in NSCLC and to provide further evidence to guide future clinical cancer therapies.
MATERIALS AND METHODS
Materials
H1650 cells were obtained from ATCC (Manassas, VA), H157 cells were obtained from Dr. John Minna (University of Texas Southwestern University, Dallas, TX), and H322 cells were obtained from Dr. Paul Bunn (University of Colorado, Denver, CO). Cancer-associated fibroblasts (CAFS) (catalog #PC60199A1-CAF-P0) were obtained from Asterand (Detroit, MI) and were p0 fibroblasts isolated from tumor region of lung from a 77 year old Caucasian female with Stage I Squamous cell carcinoma of the lung. All cell lines have been maintained in a central repository at Moffitt since 2008. All cell lines had been authenticated by STR analysis (ACTG Inc, Wheeling, IL) as of September 2010, and all cells had been routinely tested and were negative for mycoplasma (PlasmoTest, InvivoGen, San Diego, CA). Erlotinib was purchased from ChemieTek (Indianapolis, IN). Siltuximab and murine anti-IL-6 antibody was supplied by Janssen R&D. Cells (5000) were grown in RPMI medium with 10% FBS and seeded in 96-well plates for cell viability assays, as previously described.10 We used the CellTiter-Glo® Luminescent Cell Viability Assay kit (Promega, Madison, WI) for cell viability assays, following the manufacturer's instructions. For Western blot analyses, cells were lysed in RIPA buffer with protease inhibitor cocktail tablets (Roche), as previously described.10 Total STAT3 antibodies were purchased from Santa Cruz Biotechnology, and tyrosine phosphorylated STAT3 (PY-STAT3; Tyr705) antibodies were from Cell Signaling Technologies (Cambridge, MA). Mouse rIL-6 was from R&D Systems (St Paul, MN).
Luminex Bio-Plex Assay and DNA Mutation Sequencing for Tumor Tissue
Frozen NSCLC specimens from 100 patients were obtained from the Moffitt Tissue Core repository. Collected samples were from patients who had provided written informed consent to participate in the Institutional Review Board-approved Moffitt Total Cancer Care (TCC®) protocol. We used the Multiplex Luminex assay to evaluate STAT3 activities and IL-6 expression levels using protein lysates from the frozen tissue. The Multiplex Luminex assay measured levels of PY-STAT3 (Tyr705), serine phosphorylated STAT3 (PS-STAT3; Ser727), and IL-6, with experiments run in triplicate as previously described.10 A standardized curve was used to calculate the IL-6 concentration using recombinant human IL-6 (BD Pharmagen) along a range from 0 to 10,000 pg (Supplemental Figure S1). Genomic DNA was examined for EGFR mutations on exons 18, 19, 20, and 21 and for KRAS mutations on exons 1 and 2. The sequencing results were analyzed using Lasergene (DNASTAR, Madison, WI), which measured the correlation between STAT3 activation and IL-6 expression in NSCLC cells.
Immunohistochemistry
Staining was performed on a Ventana Discovery XT automated system (Ventana Medical Systems, Tucson, AZ) as per manufacturer's protocol with proprietary reagents. Slides were deparaffinized with EZ Prep solution (Ventana) using an automated system. Ribo CC buffer (#760-107, Ventana) was used for heat-induced epitope retrieval. Murine monoclonal antibody to IL-6 (#ab9324, Abcam, Cambridge, MA) was used at a 1:100 concentration in Dako antibody diluent (Carpenteria, CA) for 32 minutes and detected with Ventana OmniMap anti-mouse secondary antibody for 16 minutes. Rabbit monoclonal antibody to PY-STAT3(Y705) (#9145, Cell Signaling Technologies, Danvers, MA) was used at a 1:100 concentration in PSS diluent (Carpenteria, CA) for 60 minutes and detected with Ventana UltraMap anti-rabbit secondary antibody for 20 minutes. Signal was detected with Ventana ChromoMap kit and slides were counterstained with hematoxylin. Slides were then dehydrated, coverslipped and imaged on an Aperio SlideScannerXT (Carlsbad, CA). For PY-STAT3(Y705), the Aperio Spectrum nuclear algorithm was used to quantify nuclear staining.
Xenograft Mouse Model
All animal protocols were approved by our Institutional Animal Care and Use Committee. Female CD-1 nu/nu mice were obtained from Charles River Laboratories (Wilmington, MA). The mice were injected with H1650 (adenocarcinoma), H322 (adenocarcinoma), or H157 (squamous) cell lines either alone or combined with CAFs in a 1:1 ratio. Cells were injected at a single subcutaneous site on the mouse flank (5 million cells per site mixed with 0.1 mL of Matrigel; BD Pharmingen, Franklin Lakes, NJ). After 10 to 14 days of tumor growth, mice were randomly divided into four groups with 15 mice in each treatment group: control, erlotinib only, anti-IL-6 antibodies only, and combination (erlotinib plus anti-IL-6 antibodies). Treatment was initiated when tumor size reached a volume of approximately 50–200 mm3. Siltuximab (10 mg/kg) was combined with anti-murine IL-6 monoclonal antibody (10 mg/kg) and administered three times per week intraperitoneally in PBS dilution. Erlotinib (50 mg/kg) was given daily by oral gavage in dilution of 0.5% of hydroxypropylmethylcellulose (Sigma, St. Louis, MO). Control animals received intraperitoneal injections of 100 µL PBS and 0.5% (w/v) hydroxypropylmethylcellulose by mouth, both as placebo. Tumor volume (in mm3), calculated as tumor length times width times width divided by 2, and body weight were measured two or three times per week.
Fresh lung adenocarcinoma tissue (EGFR wildtype, KRAS wildtype) was obtained from surgical specimens of patients undergoing tumor resection at the Moffitt Cancer Center and established as subcutaneous xenografts initially in nu/nu mice (F1 generation). In brief, primary tumor specimens were acquired at initial surgery from early-stage non-small cell lung cancer (NSCLC) patients, cut into small pieces and immediately subcutaneously transplanted in immunodeficient nu/nu mice. Consent for use of human tumor tissue was obtained from patients and the study was approved by the University of South Florida Institutional Review Board. The explanted tumor tissues from early tumor passages were maintained with ~2 or 3 passages. Frozen tumor xenografts from this initial passage (F3) were re-implanted subcutaneously in groups of five NOD/SCID mice (Taconic, Hudson, NY, USA) mice for each tumor sample, with two small pieces re-implanted per mouse (F4 generation). NOD/SCID mice were selected as tumors had better growth characteristics in our experience. When tumor size reached 1.5 cm, tumors were harvested, divided into small 3 × 3 × 3 mm pieces, and transplanted into another 18–22 mice, with one tumor per mouse (F5 generation). Thirty mice received subcutaneous implants of F5 tumor tissue and were observed for approximately 4–5 weeks for tumor volume growth. Treatment was started when tumor volume reached approximately 10–20 mm3. Siltuximab (10 mg/kg) was given twice per week intraperitoneally in PBS dilution while control animals received intraperitoneal injections of 100 µL PBS.
Statistical Methods
For tumor tissue experiments, statistical assays were designed to identify any significant relationships between profiles and relevant patient characteristics. Three continuous measures of IL-6, PY-STAT3, and PS-STAT3 were collected and then averaged. Correlations between two group variables were examined by Spearman non-parametric correlation with correlation coefficient (r). The unpaired t test with Welch's correction was used to examine significant differences between two study groups. Because of small, unequal, nonparametric sets with different variances and numbers, the Kruskal-Wallis one-way ANOVA, a nonparametric test, was used to compare three or more unpaired groups. Anderson-Darling statistics were used to examine whether log-transformed results were close to normal distribution. We used log2 transformation for tumor volume to allow volume measurements to approximate to normal distribution, since data were not normally distributed. Descriptive statistics indicated the mean, median, standard deviation, minimum, and maximum. Tukey’s multiple comparisons (compare all pairs of drug treatment group) were used to examine the one-way ANOVA for treatment groups. Significant differences are indicated with p values. For all statistical analyses and graphs, we used GraphPad version 5.0.
RESULTS
Patterns of IL-6 and STAT3 Activation in NSCLC
We studied 100 NSCLC human specimens for IL-6 and downstream STAT3 activation and correlated these results with tumor histology and presence of KRAS mutations. We also examined whether IL-6 levels were associated with patterns of STAT3 activation. Tumor tissue characteristics are shown in Table 1. All stages of disease were represented, although most were from earlier stages amenable to surgical resection. Overall, nearly 50% were characterized as adenocarcinoma, 25% were characterized as squamous cell carcinoma, with the remaining tissues characterized as NSCLC, not otherwise specified (NOS). Our sequencing analyses indicated that 5% of our tissues were EGFR-mutant and 25% were KRAS-mutant. Because of the low number of EGFR mutants, we were only able to compare IL-6 and STAT3 signaling between the KRAS-mutant and wild-type tumors.
TABLE 1.
Characterization of the tumor tissues
| Total Samples (N = 100) | |
|---|---|
| TNM Path Stage Group, n | |
| I | 33 |
| II | 22 |
| III | 16 |
| IV | 6 |
| Uncharacterized | 23 |
| Histology, n | |
| Adenocarcinoma | 50 |
| Squamous cell carcinoma | 25 |
| NSCLC (uncharacterized) | 25 |
| Mutations in EGFR and KRAS, n | |
| KRAS | 25 |
| EGFR | 5 |
| Neither | 70 |
100 frozen non-small cell lung carcinoma specimens were obtained from our Institution’s Tissue Core Facility and evaluated for the tumor type and disease stage. The mutations of EGFR and KRAS were identified by genomic PCR.
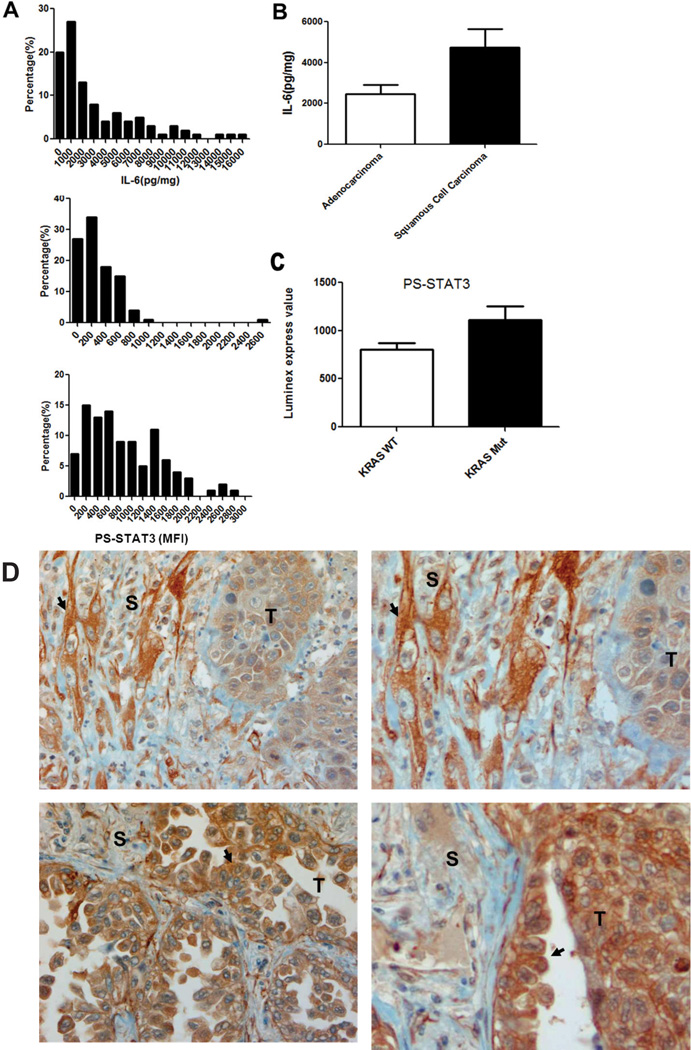
As shown in Figure 1A, overall, over 70% of the tumors demonstrated a higher level of PY-STAT3 (Tyr705) and IL-6 than a prototypical NSCLC cell line, such as A549, which has more modest expression of IL-6 and PY-STAT3 (Tyr705) (data not shown). When we measured the correlation between IL-6 and PY-STAT3 and IL-6 and PS-STAT3, we did not find a strong correlation between PY-STAT3 and IL-6 (r = −0.1365, p > 0.05) and PS-STAT3 and IL-6 (r = 0.219, p > 0.05). Similar analyses within adenocarcinoma, squamous cell lung cancer and NSCLC NOS also failed to demonstrate any significant correlations (data not shown). These data suggest that STAT3 phosphorylation is not a good surrogate marker of total tumor IL-6 levels. As shown in Figure 1B, IL-6 levels in the squamous cell carcinoma group were nearly two-fold higher than in the adenocarcinoma group. No significant differences of PY-STAT3 and PS-STAT3 were found between these histological groups. However, as shown in Figure 1C, we did find that PS-STAT3 levels in the KRAS mutation group were higher than those in wild-type tumors, suggesting that high PS-STAT3 levels may contribute to tumorigenesis in K-RAS mutation setting (p < 0.05).
Figure 1. IL-6 and STAT3 activation patterns in NSCLC specimens.
(A) Histogram indicating distribution of IL-6, PY-STAT3, and PS-STAT3 levels in frozen non-small cell lung carcinoma (NSCLC) specimens from 100 patients by Luminex multiplex assay. (B) Bar graph indicating differences in IL-6 levels in adenocarcinoma and squamous cell carcinoma tissue based on tumor histology. IL-6 levels (pg/mg) were calculated by standardized regression linear curve and normalized with the protein extracts from tissue (mg). P values indicate comparisons versus controls: *p < 0.05, **p < 0.01, and ***p < 0.005. (C) Bar graph indicating differences in PS-STAT3 wild-type and KRAS tumor via PCR sequencing results. P values indicate comparisons versus controls: *p < 0.05, **p < 0.01, and ***p < 0.005. (D) Immunohistochemistry for IL-6 in 2 high expressing cells. Top: stromal cell (S) expressing IL-6 (arrow); bottom: tumor cells (T) expressing IL-6 (arrow).
We used IL-6 immunohistochemistry to examine the cell source of the IL-6 found in our tumors and performed a focused analysis on 10 cases that had high levels of IL-6 detected in tumor lysates. Interestingly, we identified two generalized patterns. First, within the tumor, we observed tumor cells that expressed little IL-6 and stromal cells that produced abundant levels of IL-6 (Figure 1D, top panel). Second, within other tumors, we observed (Figure 1D, bottom panel) tumor cells that expressed abundant levels of IL-6. Data on these 10 cases is summarized in Table 2. Therefore, IL-6 production in the tumors can be produced either in an autocrine fashion by the tumor cells or by stromal cells, such as CAFs.
TABLE 2.
IL-6 Immunohistochemistry Summary
| Tumor | Histology | Tumor IL-6 | Stroma IL-6 |
|---|---|---|---|
| 1 | Adeno | Negative | Negative |
| 2 | Adeno | Negative | Positive |
| 3 | Squamous | Negative | Positive |
| 4 | Squamous | Negative | Positive |
| 5 | Adeno | Negative | Negative |
| 6 | Adeno | Positive | Negative |
| 7 | NOS | Negative | Positive |
| 8 | Adeno | Negative | Negative |
| 9 | Adeno | Positive | Positive |
| 10 | Adeno | Negative | Negative |
Ten NSCLC tissues were examined for tumor, stromal, or both IL-6 presence using IL-6 immunohistochemistry. Positive = positive for the presence of IL-6 by immunohistochemistry; Negative = lack of IL-6 signal by immunohistochemistry.
Effects of Combined Siltuximab and Erlotinib on NSCLC in vivo
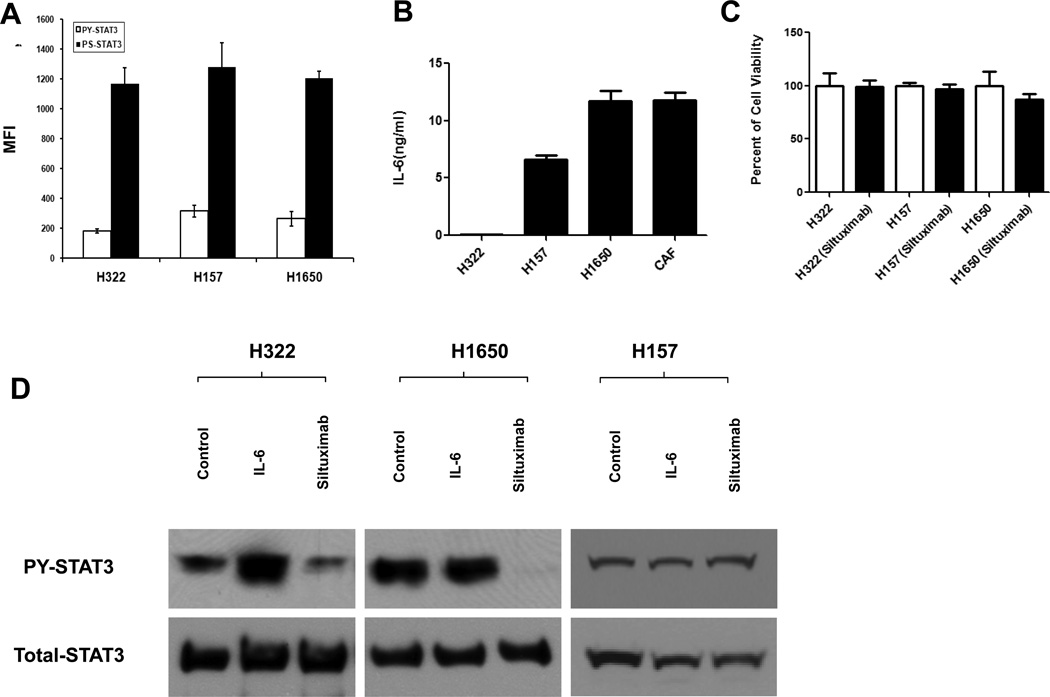
We next examined how siltuximab affects tumor growth in vivo using human lung cancer xenograft models. Using two adenocarcinoma cell lines (H1650, H322) and a squamous cell carcinoma cell line (H157), we characterized production of IL-6 in these cells, their expression of phosphorylated STAT3, and the in vitro effects of siltuximab on PY-STAT3. Luminex results confirmed basal expression of PY-STAT3 and PS-STAT3 in all three cell lines (Figure 2A). As shown in Figure 2B, the H157 and H1650 cells produced high amounts of autocrine IL-6, whereas H322 cells produced negligible amounts of IL-6. We also examined CAF cells and confirmed that CAF produced high levels of IL-6 compared with the other cell lines. Regarding the in vitro effects of siltuximab on lung cancer cell viability, except for very modest effects on H1650 growth, we observed no differences with siltuximab treatment versus control treatment (Figure 2C). Next, we examined basal PY-STAT3 and responses to IL-6 and siltuximab by Western blotting. As shown in the Figure 2D, PY-STAT3 levels were stimulated by IL-6 in H322 cells and this could be repressed by siltuximab. In H1650 and H157 cells, the levels of PY-STAT3 were not further increased by exogenous levels of IL-6. Siltuximab repressed PY-STAT3 levels in H1650 cells, confirming our previous studies.10
Figure 2. Patterns of in vitro IL-6 and STAT3 activation in lung cancer cells.
H322, H157, and H1650 cells were cultured in 10 mL of RPMI 1640 with 10% FBS, and CAF was cultured in 10 mL of DMEM with 10% FBS for 48 hours in 10-cm dishes. Protein and culture medium were collected for Luminex or immunoblot assay. (A) Luminex results show the endogenous levels of PY-STAT3 and PS-STAT3. (B) Luminex results indicate IL-6 levels from cell culture medium which normalized by cell numbers. (C) CellTiter-GLO assay results performed after 72 hours following incubation with siltuximab or PBS as vehicle control. (D) Immunoblot results indicate PY-STAT3 levels change via stimulation with IL-6 or treated with siltuximab. Cells were collected after 10 minutes of IL-6 stimulation or 16 hours of siltuximab.
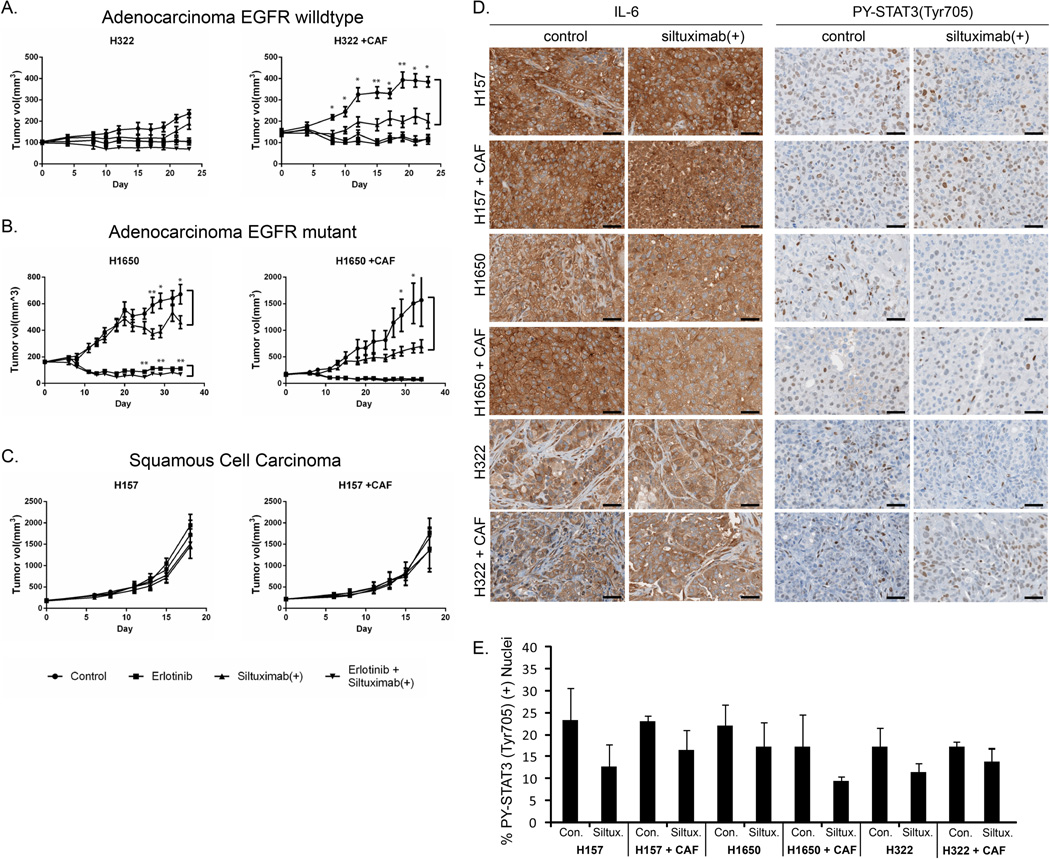
Three cell lines were used in xenograft models to study the in vivo effects of siltuximab: 1) H322, representing cells that produce no autocrine IL-6 but can be stimulated by IL-6 from the environment; 2) H1650, representing cells that produce their own autocrine IL-6 that drives STAT3 signaling; and 3) H157, representing cells with high autocrine IL-6 yet inhibition of IL-6 by siltuximab has no effect on downstream signaling. Siltuximab alone and combination treatment with siltuximab and erlotinib were examined in these mouse xenograft models. To target both human IL-6 from human cell lines and mouse IL-6 produced by murine stromal cells, we combined anti-human IL-6 (siltuximab) with anti-mouse IL-6 antibody. This is referred to as “siltuximab(+)” in the subsequent discussion and figures. We also tested these treatment combinations in a co-culture model with CAF cells based on our results suggesting that stromal cells may be producers of IL-6 and this could be important in tumor growth and sensitivity to siltuximab (Figure 3A–C). Levels of PY-STAT3(Tyr705) and IL-6 were examined by immunohistochemistry in a subset of these xenograft models and we observed strong evidence of both IL-6 and PY-STAT3 (Tyr705) in all tumors examined (Figure 3D). We found that all xenograft models produced similarly abundant IL-6 and we did not observe the intra-tumoral heterogeneity seen in patient specimens. We also observe IL-6 staining in both tumor and stromal cells. Surprisingly, levels of IL-6 were largely independent of cell line histology or presence of CAFs. Although a modest reduction of nuclear PY-STAT3 (Tyr705) was detected in response to siltuximab(+) across all models, these differences were not statistically significant (Figure 3E). Because our previous data showed that combined siltuximab plus erlotinib can reduce STAT3 activation and phosphorylation of serine and tyrosine in H1650 cell line in vitro, the EGFR inhibitor erlotinib was selected for combination strategy in the present study.10
Figure 3. Effects of siltuximab(+) in mouse xenograft models of lung cancer.
CD-1 nu/nu mice were divided into 4 treatment groups of 15 mice of each for H322, H1650, and H157 xenograft. models The single-agent treated group was exposed to 10mg/kg of each anti-human IL-6 and anti-mouse IL-6 antibodies (siltuximab +) 3 times per week or 50 mg/kg of erlotinib daily; the combination group was exposed to both siltuximab(+) and erlotinib. The control group was treated identically as the single-agent and combined groups, but with control vehicle (PBS) intraperitoneally and 0.5% of hydroxypropylmethylcellulose orally. Tumor growth (in mm3) was assessed every 2–3 days. Error bars indicate variation of tumor volume, and p values indicate the significant comparison in two groups: *p < 0.05, **p < 0.01, and ***p < 0.005. (A) Tumor growth in H322 and H322 + CAF xenografts, with significant differences shown in control and siltuximab(+) groups. (B) Tumor growth in H1650 and H1650 + CAF xenografts, with significant differences observed in control versus siltuximab(+) and in erlotinib versus combination group. (C) Tumor growth in H157 and H157 + CAF xenograft. No significant differences were found. (D) Representative images of tumor cells in cell line xenograft models, demonstrating IL-6 and PY-STAT3 (Tyr705) staining. Black scale bars represent 40µM. E) Quantification of nuclear PY-STAT3(Tyr705) using Spectrum nuclear algorithm (Aperio). Graphs show mean percentage of nuclei staining positive per treatment (N=3), with error bars representing s.d. All p-values >.05 for control vs siltuximab(+).
H322 model
Erlotinib alone had a strong inhibitory effect on tumor growth, consistent with previously published studies.14 Compared to the other models studied below (H1650, H157), tumors grew slowly in the H322 model system and siltuximab(+) therapy had little effect on the tumors. No additional effect of combined erlotinib and siltuximab(+) was observed. Addition of CAF cells clearly increased the growth of tumors compared with H322 cells alone, and erlotinib alone again had a strong effect on tumor growth. However, compared to the results with H322 tumors alone, tumors with H322 and CAF cells demonstrated strong inhibition of growth following single-agent siltuximab(+) therapy. No additional effect of combined erlotinib and siltuximab(+) was observed in the CAF + H322 model.
H1650 model
Erlotinib, as expected, had a strong inhibitory effect on tumor growth, in this EGFR-mutant lung cancer model.12 The tumors grew slowly in the H1650 model system. Siltuximab therapy had modest effects on the H1650 tumors compared to effects with erlotinib. Combined erlotinib and siltuximab(+) had a statistically significant but very modest reduction in tumor growth compared to single agents alone. Similar to the observations with the H322 tumors, addition of CAF cells clearly increased tumor growth compared with H1650 tumors alone, and erlotinib again had strong effects on growth of these tumors. Compared to the results with H1650 tumors alone, tumors produced with H1650 and CAF cells demonstrated stronger inhibition of growth following single-agent siltuximab(+) therapy. No additional effect of combined erlotinib and siltuximab(+) was observed in the CAF + H1650 model.
H157 model
Erlotinib had no effect on tumor growth in this squamous cell line with wild-type EGFR, consistent with previously published results.14 The cells grew briskly and aggressively using this model system. Siltuximab(+) therapy had little effect on these tumors. This was somewhat expected because, although IL-6 levels were observed to be produced by the H157 cells, siltuximab had no effect on downstream STAT3 signaling in cell culture (Figure 3C). No additional effect of combined erlotinib and siltuximab(+) was observed. Results with CAF were identical to those observed with H157 cells alone, with no increased tumor growth observed with co-administration of CAF cells. H157 tumors, with or without CAF, did not respond to siltuximab(+), suggesting that H157 cells are not dependent on the IL-6/STAT3 signaling pathway, even with a high expression of IL-6.
Primary Explant Model
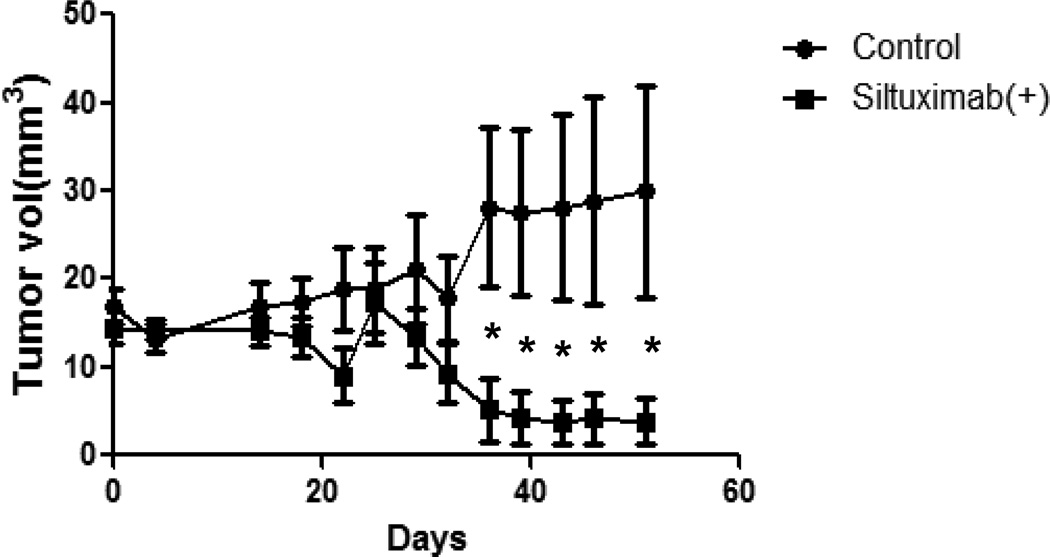
The primary xenograft was an adenocarcinoma with wild-type K-RAS and EGFR. This primary human NSCLC xenograft model closely recapitulates the properties of the parental tumors.15 One of the advantages of using direct xenografting of human cancers is that there is no clonal selection, and all cellular fractions including stem cell populations existing in a tumor are transplanted. We initially attempted to propagate three primary xenograft models that had initially grown in immunodeficient mice. However, two of the initial models failed to propagate and we abandoned studying these further. The third model grew extremely slowly, requiring nearly 50 days of growth before robust measurements could be obtained and drug treatment experiments (control treatment and siltuximab(+) alone) were started at 5th generation from a single primary tumor. We had originally planed to study the combination of erlotinib and siltuximab in this model but the slow pace resulted in less animals available for study. As shown in Figure 4, although both treatment groups responded with a slow tumor growth at first, after 36 days mice receiving siltuximab(+) had significantly less tumor size than control treated mice.
Figure 4. Effects of siltuximab(+) on NSCLC primary explant model.
NOD/SCID mice were divided into two groups with 6 pairs of mice for each primary xenograft, as described in Materials and Methods. The single-agent treatment group was given 10 mg/kg of siltuximab(+) (injected intraperitoneally) 3 times per week; controls were given the same volume of PBS. Tumor growth (in mm3) was assessed every 2–3 days. Error bars indicate variation of tumor volume, and p values indicate the significant comparison in two groups: *p < 0.05, **p < 0.01, ***p < 0.005.
DISCUSSION
The purpose of this study was to examine patterns of IL-6 signaling in NSCLC and to determine the effects of neutralizing IL-6 antibodies (siltuximab) on tumor growth. First, when we examined patterns of IL-6 expression in NSCLC tissues and correlated these results with tumor histology, EGFR and KRAS mutation status, and downstream activation of STAT3 signaling, we found IL-6 to be twice as abundant in squamous cell histology versus adenocarcinoma histology. We observed no differences in IL-6 production in KRAS-mutant adenocarcinoma versus wild-type KRAS adenocarcinoma cells. Based on studies that found that KRAS transformation requires IL-6, we had hypothesized that KRAS mutant cancers would have more IL-6, but this was not the case.16, 17 However, given the small sample size, we were unable to determine whether IL-6 is higher in tumors with activating EGFR mutations, as has been observed in lung cancer cell lines.13 We had hypothesized that high levels of IL-6 in the tumor would translate to high levels of activated PY-STAT3 in the tumor, but this result was not observed. This finding may suggest additional complexities in regulation of STAT3 phosphorylation beyond simply reflecting intra-tumor IL-6 levels. In addition, this result suggests that PY-STAT3 in tumor cells, which can be assayed by immunohistochemistry, may not be accurate enough to be used as a predictive marker to identify tumors driven by high IL-6 levels and therefore most appropriate for siltuximab therapy. Interestingly, KRAS mutation was related to serine phosphorylation of STAT3, suggesting that downstream MAP kinase activation by KRAS mutations may signal to STAT3, as has been suggested in cell culture experiments.18, 19
Our second aim was to determine the effects of IL-6 neutralizing antibodies on xenograft lung cancer model systems and to determine combination effects with EGFR inhibition by erlotinib and effects of stromal production of IL-6 by CAF cells. We used CAF cells along with tumor cells (H1650, H322, and H157) in our in vivo models since we observed some tumors with abundant production of IL-6 by stromal cells (Figure 1D). IL6 can be produced by both tumor cells and stromal cells in the tumor microenvironment. We and others previously demonstrated that lung cancer cells can express IL6 as well as IL6 receptor subunits.11, 20 Conversely, hepatic cells, mesenchymal stem cells, cancer associated fibroblasts, and osteoclasts have been reported in various contexts to produce IL6.21–25 In the H322 model, we observed increased tumor growth by co-administration of CAF cells that could be inhibited by IL-6 blockade (siltuximab(+)). We interpret these results to mean that some subsets of lung cancer tumors may be driven by stromal IL-6 that can be neutralized by siltuximab(+), resulting in inhibition of tumor growth. In this model, as well as in the H1650 and H157, we observed no additional benefit of co-targeting EGFR and IL-6. Similar findings were observed in the H1650 cell, where CAF cells increased tumor volume that could be blocked with siltuximab. We also observed some modest reduction in tumor growth in H1650 tumors alone, consistent with the autocrine signaling of IL-6 to downstream STAT3, although the effect was less pronounced compared to erlotinib monotherapy. Finally, the H157 squamous cell line was completely resistant to IL-6 neutralizing antibody therapy either with or without CAF cells. This cell line appears to have IL-6-independent growth. Lastly, we examined one primary explant model with wild-type EGFR and KRAS status and found modest effects of tumor growth later in the experimental timeline. This experiment was somewhat provocative as some tumors stopped growing and regressed toward later days of treatment. More experiments will be necessary to fully determine whether similar results would be observed in a larger number of animal tumor models.
This study has implications for potential use of IL-6 neutralizing antibodies for lung cancer. First, we observed more IL-6 production in squamous cell tumors; thus, further experiments in additional squamous cell models, both cell line xenograft and primary explant, should be considered. While some targetable lesions have been observed in squamous cell lung cancer (DDR2 mutations, FGFR1 gene amplification, mutations), the tumor type has lacked success in targeted agent efficacy in clinic compared to adenocarcinoma.26–30 Our single H157 model should not dissuade further experiments in squamous cell models. Second, our animal xenograft experiments suggest that IL-6 neutralizing antibodies may work best in tumors that have stromal production of IL-6 that is driving tumor cell growth in a paracrine manner. Further studies examining CAF cells in human tumor samples, their production of IL-6, and measurements of IL-6-driven signaling in the corresponding tumor cells (perhaps with IL-6 immunohistochemistry to look for stromal cells expressing IL-6) may identify subsets of tumors that benefit from IL-6-neutralizing antibodies such as siltuximab. Third, we observed no clinically relevant improvement in tumor growth with combined erlotinib and siltuximab compared to erlotinib alone, at least in the models studied. Nonetheless, in both the H1650 and H322 cell xenograft models, siltuximab did have some degree of single agent activity, especially in co-culture models with CAF cells.
We acknowledge that our study may have some limitations. Tumors with EGFR mutations should be further studied for IL-6 levels and STAT3 activation. Previous studies have suggested that tumor cells with activating EGFR mutations drive STAT3 signaling through IL-6 production and targeting IL-6 impaired tumorigenesis.13 We had inadequate numbers of EGFR-mutant tumors to make any conclusions about IL-6 production in these tumors. This is also important for further study given a potential role of IL-6 in driving resistance to EGFR-TKIs.12 However, both of these studies, along with our own results, suggest that IL-6 targeting using siltuximab may have benefit in NSCLC subtypes. Our results are limited in squamous cell models since we only tested the H157 model that grows well in xenografts. Further studies should be considered in squamous cell models, as our tumor studies suggested more IL6 production in these tumor types. Use of squamous cell lines from conditionally reprogrammed cell models may expand the ability to study more squamous cell models. Likewise, our use of primary xenograft models was limited and further studies in these systems could be more informative. We used simple subcutaneous xenograft models, and more striking results may be obtained in orthotopic models, especially in organs that produce higher levels of IL-6, such as bone.31 It would be interesting and important to consider these additional types of models, as bone metastasis is relatively common in lung cancer and other solid tumors. We limited our studies to tumor growth and therefore may have missed some important effects on tumor invasion and metastasis. Some studies have suggested that IL-6 may be important in lung cancer stem cells, as opposed to the bulk tumor cells.32 Beyond direct targeting of tumor cells, targeting IL-6-related inflammation may have beneficial effects on patient quality of life through improvement in cachexia and lung cancer-related anemia.33 In conclusion, our results and those of others suggest that further study of IL-6 targeting with siltuximab should be considered in human clinical trials. Our studies would suggest that IL-6 production in the tumor stroma may be a surrogate marker for siltuximab activity and should be examined in patient samples along with EGFR mutation status and prior response to EGFR TKI therapy.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Rasa Hamilton for editorial assistance, Fumi Kinose for assistance with cell culture, Noel Clarke for immunohistochemistry, Linda Ley and Carol Ulge for administrative assistance, and Tiffany Razabdouski for assistance with animal studies.
Funding support: Support for these studies was provided by a research agreement through Centocor Ortho Biotech Oncology Research & Development, Radnor, PA 19087 and through NCI 1R01 CA121182-01A1.
DISCLOSURE OF POTENTIAL CONFLICTS OF INTEREST. Dr. Haura has received research support for these studies from Centocor. Dr. Doshi and Dr. Sasser are employees of Janssen R&D.
REFERENCES
- 1.Yu H, Jove R. The STATs of cancer--new molecular targets come of age. Nat Rev Cancer. 2004;4:97–105. doi: 10.1038/nrc1275. [DOI] [PubMed] [Google Scholar]
- 2.Haura EB, Turkson J, Jove R. Mechanisms of disease: Insights into the emerging role of signal transducers and activators of transcription in cancer. Nature clinical practice Oncology. 2005;2:315–324. doi: 10.1038/ncponc0195. [DOI] [PubMed] [Google Scholar]
- 3.Bromberg JF, Wrzeszczynska MH, Devgan G, et al. Stat3 as an oncogene. Cell. 1999;98:295–303. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- 4.Yu CY, Wang L, Khaletskiy A, et al. STAT3 activation is required for interleukin-6 induced transformation in tumor-promotion sensitive mouse skin epithelial cells. Oncogene. 2002;21:3949–3960. doi: 10.1038/sj.onc.1205499. [DOI] [PubMed] [Google Scholar]
- 5.Niu G, Wright KL, Huang M, et al. Constitutive Stat3 activity up-regulates VEGF expression and tumor angiogenesis. Oncogene. 2002;21:2000–2008. doi: 10.1038/sj.onc.1205260. [DOI] [PubMed] [Google Scholar]
- 6.Berishaj M, Gao SP, Ahmed S, et al. Stat3 is tyrosine-phosphorylated through the interleukin-6/glycoprotein 130/Janus kinase pathway in breast cancer. Breast Cancer Res. 2007;9:R32. doi: 10.1186/bcr1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Song L, Turkson J, Karras JG, et al. Activation of Stat3 by receptor tyrosine kinases and cytokines regulates survival in human non-small cell carcinoma cells. Oncogene. 2003;22:4150–4165. doi: 10.1038/sj.onc.1206479. [DOI] [PubMed] [Google Scholar]
- 8.Dauer DJ, Ferraro B, Song L, et al. Stat3 regulates genes common to both wound healing and cancer. Oncogene. 2005;24:3397–3408. doi: 10.1038/sj.onc.1208469. [DOI] [PubMed] [Google Scholar]
- 9.Haura EB, Zheng Z, Song L, et al. Activated epidermal growth factor receptor-Stat-3 signaling promotes tumor survival in vivo in non-small cell lung cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2005;11:8288–8294. doi: 10.1158/1078-0432.CCR-05-0827. [DOI] [PubMed] [Google Scholar]
- 10.Song L, Rawal B, Nemeth JA, et al. JAK1 activates STAT3 activity in non-small-cell lung cancer cells and IL-6 neutralizing antibodies can suppress JAK1-STAT3 signaling. Molecular cancer therapeutics. 2011;10:481–494. doi: 10.1158/1535-7163.MCT-10-0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao SP, Mark KG, Leslie K, et al. Mutations in the EGFR kinase domain mediate STAT3 activation via IL-6 production in human lung adenocarcinomas. The Journal of clinical investigation. 2007;117:3846–3856. doi: 10.1172/JCI31871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yao Z, Fenoglio S, Gao DC, et al. TGF-beta IL-6 axis mediates selective and adaptive mechanisms of resistance to molecular targeted therapy in lung cancer. Proc Natl Acad Sci U S A. 107:15535–15540. doi: 10.1073/pnas.1009472107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao SP, Mark KG, Leslie K, et al. Mutations in the EGFR kinase domain mediate STAT3 activation via IL-6 production in human lung adenocarcinomas. J Clin Invest. 2007;117:3846–3856. doi: 10.1172/JCI31871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Helfrich BA, Raben D, Varella-Garcia M, et al. Antitumor activity of the epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor gefitinib (ZD1839, Iressa) in non-small cell lung cancer cell lines correlates with gene copy number and EGFR mutations but not EGFR protein levels. Clinical cancer research : an official journal of the American Association for Cancer Research. 2006;12:7117–7125. doi: 10.1158/1078-0432.CCR-06-0760. [DOI] [PubMed] [Google Scholar]
- 15.Fichtner I, Rolff J, Soong R, et al. Establishment of patient-derived non-small cell lung cancer xenografts as models for the identification of predictive biomarkers. Clin Cancer Res. 2008;14:6456–6468. doi: 10.1158/1078-0432.CCR-08-0138. [DOI] [PubMed] [Google Scholar]
- 16.Ancrile B, Lim KH, Counter CM. Oncogenic Ras-induced secretion of IL6 is required for tumorigenesis. Genes & development. 2007;21:1714–1719. doi: 10.1101/gad.1549407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ancrile BB, O'Hayer KM, Counter CM. Oncogenic ras-induced expression of cytokines: a new target of anti-cancer therapeutics. Mol Interv. 2008;8:22–27. doi: 10.1124/mi.8.1.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Turkson J, Bowman T, Adnane J, et al. Requirement for Ras/Rac1-mediated p38 and c-Jun N-terminal kinase signaling in Stat3 transcriptional activity induced by the Src oncoprotein. Mol Cell Biol. 1999;19:7519–7528. doi: 10.1128/mcb.19.11.7519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wen Z, Zhong Z, Darnell JE., Jr Maximal activation of transcription by Stat1 and Stat3 requires both tyrosine and serine phosphorylation. Cell. 1995;82:241–250. doi: 10.1016/0092-8674(95)90311-9. [DOI] [PubMed] [Google Scholar]
- 20.Haura EB, Livingston S, Coppola D. Autocrine Interleukin-6/Interleukin-6 Receptor Stimulation in Non-Small-Cell Lung Cancer. Clin Lung Cancer. 2006;7:273–275. doi: 10.3816/CLC.2006.n.006. [DOI] [PubMed] [Google Scholar]
- 21.Kim Y, Fiel MI, Albanis E, et al. Anti-fibrotic activity and enhanced interleukin-6 production by hepatic stellate cells in response to imatinib mesylate. Liver Int. 2012;32:1008–1017. doi: 10.1111/j.1478-3231.2012.02806.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kerpedjieva SS, Kim DS, Barbeau DJ, et al. EGFR ligands drive multipotential stromal cells to produce multiple growth factors and cytokines via early growth response-1. Stem Cells Dev. 2012;21:2541–2551. doi: 10.1089/scd.2011.0711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spaeth EL, Dembinski JL, Sasser AK, et al. Mesenchymal stem cell transition to tumor-associated fibroblasts contributes to fibrovascular network expansion and tumor progression. PLoS One. 2009;4:e4992. doi: 10.1371/journal.pone.0004992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Erez N, Glanz S, Raz Y, et al. Cancer associated fibroblasts express pro-inflammatory factors in human breast and ovarian tumors. Biochemical and Biophysical Research Communications. 2013;437:397–402. doi: 10.1016/j.bbrc.2013.06.089. [DOI] [PubMed] [Google Scholar]
- 25.Kayamori K, Sakamoto K, Nakashima T, et al. Roles of interleukin-6 and parathyroid hormone-related peptide in osteoclast formation associated with oral cancers: significance of interleukin-6 synthesized by stromal cells in response to cancer cells. The American journal of pathology. 2010;176:968–980. doi: 10.2353/ajpath.2010.090299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hammerman PS, Sos ML, Ramos AH, et al. Mutations in the DDR2 kinase gene identify a novel therapeutic target in squamous cell lung cancer. Cancer Discovery. 2011;1:78–89. doi: 10.1158/2159-8274.CD-11-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weiss J, Sos ML, Seidel D, et al. Frequent and focal FGFR1 amplification associates with therapeutically tractable FGFR1 dependency in squamous cell lung cancer. Sci Transl Med. 2010;2:62ra93. doi: 10.1126/scitranslmed.3001451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sos ML, Thomas RK. Genetic insight and therapeutic targets in squamous-cell lung cancer. Oncogene. 2012;31:4811–4814. doi: 10.1038/onc.2011.640. [DOI] [PubMed] [Google Scholar]
- 29.Hammerman PS, Hayes DN, Wilkerson MD, et al. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dutt A, Ramos AH, Hammerman PS, et al. Inhibitor-sensitive FGFR1 amplification in human non-small cell lung cancer. PLoS One. 2011;6:e20351. doi: 10.1371/journal.pone.0020351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schafer ZT, Brugge JS. IL-6 involvement in epithelial cancers. The Journal of clinical investigation. 2007;117:3660–3663. doi: 10.1172/JCI34237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yi H, Cho HJ, Cho SM, et al. Blockade of interleukin-6 receptor suppresses the proliferation of H460 lung cancer stem cells. International journal of oncology. 2012;41:310–316. doi: 10.3892/ijo.2012.1447. [DOI] [PubMed] [Google Scholar]
- 33.Bayliss TJ, Smith JT, Schuster M, et al. A humanized anti-IL-6 antibody (ALD518) in non-small cell lung cancer. Expert opinion on biological therapy. 2011;11:1663–1668. doi: 10.1517/14712598.2011.627850. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.