Abstract
Membrane protein folding and topogenesis are tuned to a given lipid profile since lipids and proteins have co-evolved to follow a set of interdependent rules governing final protein topological organization. Transmembrane domain (TMD) topology is determined via a dynamic process in which topogenic signals in the nascent protein are recognized and interpreted initially by the translocon followed by a given lipid profile in accordance with the Positive Inside Rule. The net zero charged phospholipid phosphatidylethanolamine and other neutral lipids dampen the translocation potential of negatively charged residues in favor of the cytoplasmic retention potential of positively charged residues (Charge Balance Rule). This explains why positively charged residues are more potent topological signals than negatively charged residues. Dynamic changes in orientation of TMDs during or after membrane insertion are attributed to non-sequential cooperative and collective lipid–protein charge interactions as well as long-term interactions within a protein. The proportion of dual topological conformers of a membrane protein varies in a dose responsive manner with changes in the membrane lipid composition not only in vivo but also in vitro and therefore is determined by the membrane lipid composition. Switching between two opposite TMD topologies can occur in either direction in vivo and also in liposomes (designated as fliposomes) independent of any other cellular factors. Such lipid-dependent post-insertional reversibility of TMD orientation indicates a thermodynamically driven process that can occur at any time and in any cell membrane driven by changes in the lipid composition. This dynamic view of protein topological organization influenced by the lipid environment reveals previously unrecognized possibilities for cellular regulation and understanding of disease states resulting from mis-folded proteins. This article is part of a Special Issue entitled: Protein Trafficking & Secretion.
Keywords: Topogenesis, Membrane protein topology, Charge Balance Rule, Phosphatidylethanolamine, Positive Inside Rule, Dual topology
1. Introduction
A common architectural feature of polytopic α-helical membrane proteins is their membrane topology, i.e. the number of transmembrane domains (TMDs) and their orientation with respect to the plane of the membrane lipid bilayer. A central question in the generation of membrane protein topology (topogenesis) is how a given protein sequence will orient itself and fold in a given lipid environment. This problem is a fundamental aspect of membrane protein biogenesis. The orientation of transmembrane α-helices is a prerequisite for correct three-dimensional assembly of bundles of TMDs with proper tertiary contacts and exposure of extramembrane domains (EMDs) on the physiologically relevant side of the membrane.
The topology of a polytopic membrane protein is determined by a complex interplay between the topogenic signals residing within the protein sequence, the interaction of the protein with the translocon and insertion machinery, short-range and long-range interactions within the protein and the final environment of the protein largely determined by membrane lipid composition. The nascent polypeptide chain is first directed to and inserted into the membrane by the targeting and translocation machinery. Final protein organization is determined by interaction of the protein with itself, the aqueous extra-membrane environment, the charged membrane surface, and specific hydrophilic (head groups) and hydrophobic (fatty acids) domains of the lipids that make up the membrane bilayer. Although some topogenic signals within polytopic membrane proteins have been identified, the interpretation of these signals by the membrane insertion machinery and the lipid bilayer are not fully understood. For the vast majority of proteins a unique and stable topological organization is established during initial TMD insertion. However, some topological decisions in response to the local lipid environment appear to be made after initial bilayer insertion, during late folding events or even after initial folding into a compact structure. Also an increasing number of proteins have been found to display dual topological organization either in the same membrane or for the same protein in different membrane locations within the same cell. How such dual topology is generated and where such topological decisions are made are largely unknown.
After a brief review of membrane protein biogenesis, the focus will be on the role of the lipid bilayer in membrane protein topogenesis wherein we propose the Charge Balance Rule as an extension of the Positive Inside Rule.
2. Membrane protein biogenesis
The initial topological decision of how to orient a TMD appears to be made by the translocon (Fig. 1A). Overall hydrophobicity of individual protein domains is the primary driving force for membrane integration efficiency (Fig. 1B, #1) due to the energetically favorable partitioning of TMDs into the lipid bilayer [1,2]. The correlation between the free energy value for such partitioning of model peptides measured in isolated cell membranes and liposomes provided the evidence that membrane protein insertion is thermodynamically rather than kinetically controlled. Such insertion is driven by protein–lipid rather than protein–protein interactions [3–5]. However, the orientation of TMDs in the lipid bilayer is the result of several other factors described below.
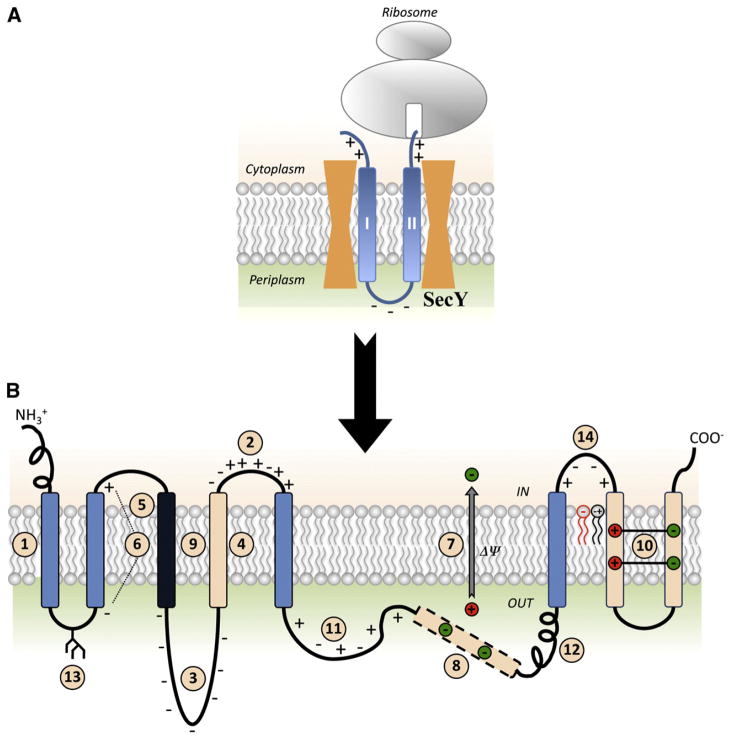
Fig. 1.
Summary of factors determining membrane protein topogenesis and folding. (A) Initial orientation according to the Positive Inside Rule of a nascent two-TMD hairpin is shown in the SecY translocon channel. (B) The features of polytopic membrane proteins and biological membranes that influence folding and final TMD topology. TMDs are color coded for relative hydrophobicity: Light Blue indicates average hydrophobicity; Black indicates above average hydrophobicity; Beige indicates low hydrophobicity. Insertion efficiency of a TMD is driven by overall hydrophobicity with initial orientation determined within the translocon channel (#1) by the Positive Inside Rule (#2) or Charge Difference Rule (#6). Orientation and final topology are influenced by negatively charged residues present in high numbers (#3), flanking a marginally hydrophobic TMD (#4) or that lie at the end of a highly hydrophobic domain (#5) as well as the membrane potential (#7). After exit from the translocon into the lipid bilayer, topology is subject to interactions within the protein where TMDs of low hydrophobicity may become EMDs (#8), highly hydrophobic TMDs stabilize neighboring TMDs of low hydrophobicity (#9), or charged TMDs form salt bridges in the lipid bilayer (#10). Domains with conflicting signals can provide dynamic molecular hinges between independently folding domains (#8 and #11). Rapid, stable folding of an EMD (#12) or co-translational glycosylation of EMDs in eukaryotic cells (#13) can prevent transmembrane shuffling by the translocon. Finally, lipid–protein interactions dictated by the Charge Balance Rule (#14) determine topological organization during initial membrane insertion as well as after folding of membrane proteins.
2.1. Positive Inside Rule
Membrane protein topology appears to be primarily determined by charged residues in the EMDs flanking hydrophobic TMDs and can be described in most cases by the statistically derived and experimentally confirmed Positive Inside Rule (Fig. 1B, #2) [6,7]. This rule is based on the enrichment in positively charged residues in EMDs not translocated across the membrane as compared to those translocated across the membrane. Indeed positively charged residues are 4-times more abundant on the cytoplasmic side of membranes versus the trans side [8]. In most cases the precise position of the charge is unimportant with the contribution of charges being additive while the density of the charges within a net positively charged domain does matter [9].
Although the Positive Inside Rule discounts the importance of negatively charged residues, the rule is not absolute since positively and negatively charged amino acids are found on both sides of membranes [8]. EMDs with a net negative charge are found on the cytoplasmic side of the membrane [10]. Negatively charged residues appear to be topologically active as translocation signals if they are present in high numbers (Fig. 1B, #3) [11], flank a marginally hydrophobic TMD (Fig. 1B, #4) [12] or lie within an uninterrupted window of seven flanking residues from the end of a highly hydrophobic TMD (Fig. 1B, #5) [13]. Several negatively charged residues are required to translocate a cytoplasmic domain with even a single positively charged residue [11]. However, the topological effect of positively charged residues can still be attenuated [14] or even overridden by negatively charged residues in a position-specific manner for bacterial [9,13] and eukaryotic membrane proteins [15].
Although there are exceptions to the Positive Inside Rule, the orientation of most TMDs can be simply determined by the presence of positively charged residues in EMDs. However, it is not clear when and how positively charged residues exert their effect on topology and why they are retained in the cytosol. What determines the relative potency of positive and negative charges and their effectiveness in TMD orientation are not completely understood. It is also not clear why positively charged residues are generally dominant retention signals over negatively charged residues (acting as translocation signals) under physiological conditions.
2.2. Charge Difference Rule
Despite the fact that positive-inside bias also appears to be true for most eukaryotic membrane proteins, the net electrical charge difference between EMDs flanking a TMD (more positive on the cytosolic side) has been shown statistically to correlate with domain orientation [16]. This Charge Difference Rule (Fig. 1B, #6) differs from the Positive Inside Rule by giving positive and negative charges equal topological strength. If this hypothesis is correct, the orientation of TMDs should be affected by altering the charge balance surrounding a TMD. Indeed this effect was demonstrated for single spanning hybrid chimeric constructs in yeast [17] and Escherichia coli [18]. These results clearly demonstrate that a monotopic transmembrane protein can be engineered to adopt either of the two opposite TMD orientations and that charge balance around the TMD is a determinant of orientation. However, for a polytopic membrane protein the charge difference across the first TMD only dictates its initial orientation and does not affect the topology of downstream regions [19] demonstrating that the subsequent downstream TMDs and EMDs possess essential topological information. Therefore, direct application of this rule to polytopic membrane proteins is complicated by the collective interactions between multiple TMDs and EMDs.
2.3. Sequential versus non-sequential mode of topogenesis
According to the sequential model of TMD topogenesis, the most N-terminal sequence defines its own orientation as well as the orientations of all subsequent TMDs in alternating orientations [16]. However, deletion of one TMD of the E. coli lactose permease (LacY) [20] or maltose transporter (MalF) [21] and perturbation of the orientation of N-terminal TMD of the tetracycline/H+ antiporter (TetA) [22] did not affect the topology of downstream TMDs. The inversion of the charge difference between EMDs flanking the first TMD of the glucose transporter (Glut1) prevented its insertion but did not affect the topology of the rest of the molecule [19]. Therefore the insertion of TMDs does not proceed sequentially in a strictly linear manner from the N- to the C-terminus. This opens the possibility for non-sequential insertion mechanisms, where interactions between neighboring and distant TMDs or re-orientation of TMDs after the insertion process determine the final topology.
Indeed, posttranslational reorientation of eukaryotic, bacterial and viral membrane proteins is not unprecedented. A TMD is not committed to a final topology after initiation of insertion and may reorient as protein synthesis continues [23] or until further reorientation is blocked by glycosylation [24]. Viral hepatitis C protein NS4B [25] and human aquaporin 1 [23] TMDs are initially inserted in one orientation and then flip following synthesis of the remainder of the protein or during post-translational maturation.
2.4. Topological determinants
2.4.1. Translocon
The translocon (Fig. 1A) provides the permissive hydrophilic environment required for concurrent TMD insertion and distribution of the flanking EMDs with different net charges to the extramembrane space [26]. The conserved charged residues in Sec61p, a subunit of the yeast Sec-translocon, are involved in the initial orientation of a TMD according to the Positive Inside Rule [27]; a negatively charged residue (Glu382) lies near the cytoplasmic end of the channel while positively charged residues (Arg67 and Arg74) lie closer to the trans end of the channel. The appropriate presentation of oppositely charged residues to the translocon channel according the Positive Inside Rule may in part be explained by the relative time for completion of translocation of TMDs with different flanking charges [28]. Reduction in the rate of translocation of TMDs with a high content of positively charged residues in flanking EMDs allows for a longer time for TMDs to properly orient before release from the translocon. Negatively charged residues have minor effects on translocation delay.
2.4.2. Membrane potential
Another factor, which may be involved in interpretation of the Positive Inside Rule, is the electrochemical membrane potential (Fig. 1B, #7). The positive outward potential in most bacteria introduces an asymmetry that may act electrophoretically to translocate negatively charged EMDs [29,30] and retain positively charged EMDs [31] during membrane integration especially when the hydrophobicity of an associated TMD is low [12]. In E. coli the dissipation of the membrane potential by a protonophore renders negatively and positively charged residues topologically equivalent [29]. However, the retention of positive charges in the cytoplasm cannot be determined solely by membrane potential since the Positive Inside Rule is still obeyed in the obligate acidophilic bacterium Sulfolobus acidocaldarius, which has a negative outward membrane potential [32]. In addition the endoplasmic reticulum membrane potential is nearly zero [17].
In principle TMD orientation could be achieved either by the active electrophoretic movement of negatively charged loops across the membrane or by passive prevention of translocation of positively charged EMDs through electrostatic interactions with the negative charge of the membrane surface due to phospholipid head groups [17,32–34]. Therefore, the topogenic strength of charged residues may be due to a combination of both electrostatic and electrophoretic effects acting on oppositely charged amino acid residues. The role of lipid–protein charge interactions and membrane potential in determining final protein topology appears to be more complex than originally proposed as will be discussed below.
2.4.3. Making topological decisions: how, where and when?
It is not clear how (what cellular topological determinants are involved), where (locally or globally within a protein itself or either within or outside of the translocon) and when (co- or post-translationally, during late folding events or after final folding) positively charged residues exert their effect on topology. The vast majority of proteins are not inserted directly and spontaneously into the membrane, but their insertion is guided by the protein-conducting channel of the translocon aided by insertase complexes (YidC or membrane protein insertase (MPIase)) [35,36]. As TMDs enter the lipid bilayer after lateral release from the translocon, short-range protein–protein interactions (with at least two known insertases), short-range lipid–protein interactions and long-range interhelical interactions within inserted proteins collectively govern topological and folding events resulting in a final compact native structure.
Early dogma assumed that the final topology of integral membrane proteins is determined at the time of insertion of TMDs into membrane. However, the nascent polypeptide is not committed to a final topology even after initiation of membrane insertion and can transiently adopt alternative or mixed topologies in order to be properly exposed to the lipid bilayer and extramembrane space [37]. A newly synthesized TMD can dynamically reorient within the translocon pore in search of the most energetically favorable organization. Although perhaps flexible and able to oligomerize [38], the single protein-conducting channel would be too small to accommodate more than a single α-helix domain or possibly one TMD hairpin [39]. Protein segments of 80 or more residues would be sterically unable to reorient themselves within the translocon [40]. Therefore, the contribution of the translocon in making a topological decision is limited by time [37], the number of TMDs accommodated by the pore and the diameter of the translocation pore [41,42], which is still a matter of debate [38]. Since post-translational rearrangement of TMD topology or reorientation of several TMDs is not unprecedented [23,25], a nascent domain may be released from the translocon into the surrounding lipid bilayer prior to any final commitment of TMD orientation.
2.4.4. Conflicting topogenic sequences and violation of topological rules
Competition between opposing topogenic signals and dynamic short- and long-range interactions between nascent TMDs and their adjacent EMDs may influence either initial or final topology resulting in unexpected topological organizations. The topogenic effect of positively charged residues can be bypassed by closely spaced negatively charged residues [13], negatively charged residues present in high numbers [11], the presence of an unusually long TMD [43] or highly hydrophobic TMDs [44].
Although hydrophobicity is the predominant factor determining insertion efficiency, the application of the TMD tendency scale to whole genomic data revealed an overlap of TMDs and EMDs in the “semihydrophobic” range [45]. This raises the possibility that a significant number of proteins have domains that can be either TMDs or EMDs depending on the properties of the remaining protein. TMDs with low hydrophobicity do not exert strong stop-transfer function and may end up as an EMD (Fig. 1B, #8) [46]. Alternatively, a less hydrophobic domain can be forced to adopt a transmembrane orientation due to dominant insertion of neighboring hydrophobic domains (Fig. 1B, #9) or by formation of salt bridges with other TMDs (Fig. 1B, #10). Even hydrophobic domains can be forced to form mini-loops [47] that insert into but do not traverse the membrane due to changes in the lipid environment [48] or the properties of neighboring domains [42]. Such conflicting topogenic sequences can provide a “molecular hinge” between independently folding multiple membrane spanning domains (Fig. 1B #8 and #11), which exist as multiple topological conformers or interconvert between two topological conformers in response to changes in the lipid environment as will be discussed later.
3. Systematic alteration of membrane lipid composition
As extensively reviewed [49,50], the combined inner and outer membranes of Gram-negative bacteria such as E. coli are composed of three major phospholipids: zwitterionic phosphatidylethanolamine (PE, 70–80%) and anionic phosphatidylglycerol (PG, 20–25%) plus cardiolipin (CL, 5–10%). Given that the inner leaflet of the outer membrane of E. coli is about 90% PE, the inner membrane contains near equal amounts of zwitterionic and anionic phospholipids. The phospholipid composition and the ratio of zwitterionic to anionic phospholipids remain quite constant over a wide range of growth conditions. Other bacteria each have their own unique lipid composition including a wide spectrum of zwitterionic, anionic, uncharged and even cationic lipids [51]. Eukaryotic cells from single cell organisms to mammals display a much more complex mixture of membrane lipids with the added variable of cell and individual internal organelle membranes with significantly different lipid compositions [52]. In each membrane of the latter, dynamic temporal changes occur in local lipid environments. Except for mitochondrial membrane proteins, the majority of the remaining membrane proteins are first inserted into the endoplasmic reticulum membrane and then transported to their resident membrane by vesicular transport processes. Thus eukaryotic membrane proteins are exposed to a dynamically changing lipid environment during intracellular trafficking.
In order to uncover the role of lipids in membrane protein assembly, structure and function, methods for systematically changing lipid composition were developed [50,53]. Although genetic manipulation of eukaryotic lipid composition is possible, the presence of most lipids in all organelles, except the unique localization of CL to the mitochondria, results in complex pleiotropic effects and cell death. Bacteria are much more accommodating to large changes in membrane lipid composition generally resulting in compromised but viable mutants with altered membrane lipid composition (Fig. 2A). However, even in the latter case changes in lipid composition results in pleiotropic effects so that any direct involvement of a lipid in a cellular process must be verified by in vitro reconstitution studies. Given the vast information on lipid synthesis coupled with extensive studies on membrane protein synthesis and assembly, E. coli is an ideal vehicle to study the role of lipids in membrane protein synthesis, structure and function [50,53].
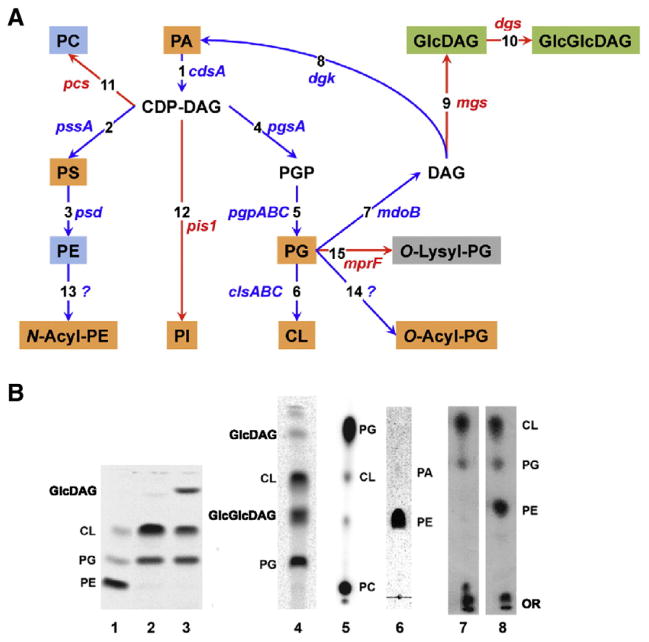
Fig. 2.
Manipulating membrane phospholipid composition in vivo using E. coli. (A) Pathways native to E. coli are noted with blue arrows, and pathways resulting from foreign genes introduced into E. coli are noted with red arrows [50]. Lipids are color coded as zwitterionic (blue), neutral (green), anionic (orange) or cationic (gray). The genes encoding the following enzymes and associated with each biosynthetic step are listed next to the arrows: (1) CDP-diacylglycerol synthase; (2) PS synthase; (3) PS decarboxylase; (4) PGP synthase; (5) PGP phos-phatases; (6) CL synthases; (7) PG: membrane derived oligosaccharide sn-glycerol-1-P transferase; (8) diacylglycerol kinase; (9) GlcDAG synthase (Acholeplasma laidlawii); (10) GlcGlcDAG synthase (A. laidlawii); (11) PC synthase (Legionella pneumophila); (12) PI synthase (Saccharomyces cerevisiae); (13) N-Acyl PE synthase; (14) O-Acyl PG synthase; and (15) O-Lysyl PG synthase (Staphylococcus aureus) utilizing Lysyl-tRNA as the lysine donor. (B) Lipid profiles displayed by thin layer chromatography of 32PO4-labeled E. coli mutants with altered lipid compositions. Lane 1. AL95 (ΔpssA) has wild type phospholipid composition (80 mol% PE and 20 mol% PG plus CL) due to complementation by a plasmid (pDD72) copy of the pssA gene that encodes the committed step to PE biosynthesis. Lane 2. AL95 is PE-lacking due to the null allele of the pssA gene and contains mainly CL and PG. Lane 3. Introduction of the mgs gene from A. laidlawii into strain AL95 results in 35 mol% GlcDAG. The remaining lipids are primarily PG (35 mol%) and CL (25 mol%). Lane 4. Introduction into AL95 of the mgs and dgs genes from A. laidlawii results in about 30–40 mol% GlcGlcDAG with less than 1 mol% GlcDAG. Lane 5. Introduction into AL95 of the pcs gene from L. pneumophila results in about 70 mol% PC with the remainder being PG (26 mol%) plus CL (2.5 mol%). Lane 6. UE54 carries a null allele of the pgsA gene encoding the committed step to PG and CL biosynthesis making it devoid of PG and CL and containing about 90 mol% PE, 4 mol% PA and 3.2 mol% CDP-diacylglycerol. Lane 7. Introduction into AL95 of the mprF gene from S. aureus results in about 60 mol% of O-Lysyl-PG (LPG) with the remainder being PG (9 mol%) plus CL (30 mol%) and other minor lipids. Lane 8. Introduction of the mprF gene into “wild-type” AL95/pDD72 results in about 23 mol% of LPG, 44 mol% of PE, 11 mol% of PG and 22 mol% of CL.
The phospholipid biosynthetic pathway and the genes responsible for encoding the respective biosynthetic enzymes are shown in Fig. 2A. Interestingly, overproduction of any of these enzymes has little effect on lipid composition. However, mutations in any step result in the expected downstream loss of lipids (Fig. 2B). Null mutations prior to the formation of CDP-diacylglycerol are lethal while null mutations in downstream steps are viable under a set of defined growth conditions [50]. Complete elimination of CL (ΔclsABC) with an increase in PG levels was only possible recently [54]. Such mutants are quite healthy but do display some phenotypes that have not been studied in detail. Complete lack of PG and CL (ΔpgsA) requires a genetic background with multiple suppressor mutations [55]. Such mutants also still contain about 10% anionic phospholipids mainly composed of phosphatidic acid (PA), CDP-diacylglycerol and N-acyl-PE [56], which complicates defining a role for anionic lipids via whole cell studies. Null mutants in PE synthesis (Δpsd) lack PE but accumulate large amounts of phosphatidylserine (PS), which appears to substitute for several functions of PE [57]. However, ΔpssA mutants lack all amino-containing and zwitterionic phospholipids with the remainder being only anionic lipids primarily composed of PG and CL. For viability these mutants require medium containing mmolar levels of divalent cations (calcium, magnesium or strontium). They also lack the ability to grow on μmolar levels of lactose as a carbon source or in the absence of amino acid supplemented minimal medium [58,59]. This latter defect is due to the inability to effect energy dependent uptake of several sugars and amino acids, as will be discussed later.
Therefore, ΔpssA mutants are an ideal vehicle to study the role of PE and the ratio of zwitterionic to anionic lipids in cell function. For the latter case, strains were developed in which the chromosomal pssA gene was placed under exogenous expression control using the arabinose synthesis (ParaB) or tetracycline resistance (Ptet) promoter [48,60,61]. Comparison of PE-containing and PE-lacking cells was used to analyze cells at the two extremes of steady state lipid composition. Cells in which PE levels are regulated were used in three types of experiments: growth of cells at different inducer levels to study the effects at steady state on membrane proteins of varying the ratio of PE to PG plus CL; growth first in the absence of inducer followed by addition of inducer to study the effect of introduction of PE on fully assembled membrane proteins; and growth in the presence of inducer followed by removal of inducer to study the effect of PE depletion on mature membrane proteins. In addition, E. coli strains were developed, which express enzymes from other bacteria that synthesize lipids not present in E. coli (Fig. 2A). Since large changes in membrane lipid composition affect multiple processes (some direct and some indirect), it is important to carry out studies in reconstituted in vitro systems to isolate the point at which lipids are directly involved in cell processes. For membrane protein synthesis, assembly and function it is important to differentiate effects on the protein insertion and assembly machinery from direct effects on membrane proteins.
4. Lipids as topological determinants
Since proteins and lipids have co-evolved to guarantee assembly of functional membrane proteins, a lipid involvement in topological decisions only becomes evident when membrane lipid composition is changed. Use of reagent strains of E. coli cells to systematically regulate membrane lipid composition [58,60,62–64] coupled with methods for determining protein topology revealed that membrane lipid composition is a critical determinant of TMD orientation with respect to the plane of the membrane bilayer. The substituted cysteine accessibility method as applied to TMD topology (SCAMTMD) [65,66] was refined to assess the topological organization of membrane proteins in whole cells [61,67,68], in inside out and right side out cell membrane vesicles [61] and after reconstitution into liposomes of defined lipid composition [69,70]. When assembled in membranes lacking PE, the orientation of the N-terminal six-TMD α-helical bundle of lactose permease (LacY) [48,61] and the N-terminal two-TMD hairpin of phenylalanine permease (PheP) [68] and γ-aminobutyrate permease (GabP) [71] are inverted with respect to the plane of the membrane bilayer and the C-terminal domains of each respective protein (Fig. 3B and D).
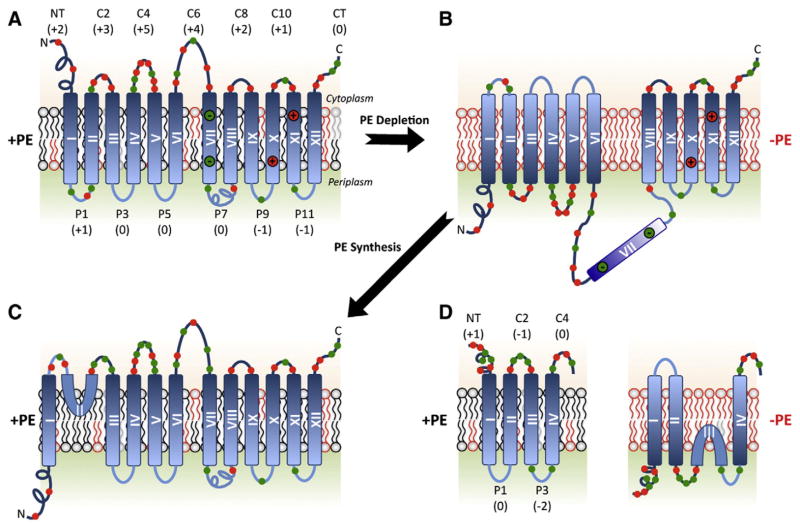
Fig. 3.
Topological organization of LacY, PheP, and GabP as a function of membrane lipid composition. TMDs (Roman numerals) and EMDs (Arabic numerals) are sequentially numbered from the N-terminus to C-terminus with EMDs exposed to the periplasm (P) or cytoplasm (C) as in wild type cells. Net charge of EMDs and distribution of positive (red) and negative (green) charges are shown. Topology of LacY is shown after initial assembly in PE-containing cells (A), after either initial assembly in PE-lacking cells or after dilution of PE following initial assembly in PE-containing cells (B), or after initiation of PE synthesis post-assembly of LacY in PE-lacking cells (C). EMD P7 is recognized by monoclonal antibody 4B1 in PE-containing (specific conformation) but not in PE-lacking (loss of native conformation) cells. Charges in TMD VII salt bridge with charges in TMDs X and XI in native LacY. The interconversion of topological conformers (A, B and C) is reversible in both directions. Topology and EMD charge distribution of the lipid sensitive domains of PheP and GabP are shown in PE-containing and PE-lacking cells (D).
Structural features, individual chemical properties, and the collective physical properties of lipids in association with each other must be considered when assessing protein–lipid interactions. The charge properties of PE are dampened by the formation of an internal charge paired ring between the phosphate and amine moieties [72]. Such a ring is not formed by the zwitterionic phospholipid PC due to the trimethylated amine. However, both PE and PC have no net charge and are both capable of countering the high negative charge density contributed to the lipid bilayer surface by net negatively charged phospholipids such as CL, PA, PG or phosphatidylinositol (PI). The glycolipids, monoglucosyl diacylglycerol (GlcDAG) and diglucosyl diacylglycerol (GlcGlcDAG), have no charge character and therefore are also effective diluents of negative charge (Fig. 2A). If collective charge properties of the membrane surface are important for proper membrane topogenesis, all these lipids should share the same capability to support proper topogenesis of polytopic membrane proteins in spite of the marked differences in their physical and chemical properties.
To address this question under physiological conditions, foreign lipids with net zero charge were introduced into a PE-lacking strain of E. coli (Fig. 2B). Introduction of the gene (mgs) encoding the A. laidlawii GlcDAG synthase or the mgs gene and the gene (dgs) encoding the A. laidlawii GlcGlcDAG synthase resulted in GlcDAG [59] or GlcGlcDAG [73], respectively, replacing the normal 70% PE with about 30–40% of the neutral glycolipid (Fig. 2B). Introduction of the L. pneumophila pcsA gene (encoding PC synthase) into the PE-lacking mutant coupled with supplementation of the growth medium with choline resulted in 70% PC with the remainder being mostly PG and CL [62] (Fig. 2B). Strikingly, the replacement of PE in vivo by the foreign lipids PC [62], GlcDAG [64] or GlcGlcDAG [73] restored wild type TMD topology of the N-terminal helical bundle of LacY. Moreover, LacY reconstituted into proteoliposomes containing in vivo equivalent amounts of PE, PC, GlcDAG, or GlcGlcDAG plus anionic PG and CL displayed native topology; the inverted topology for the N-terminal six-TMD bundle was observed in proteoliposomes made from only negatively charged PG and CL [74]. The faithful mimicking in vitro of in vivo effects of the absence or presence of net zero charged lipids strongly indicates that direct lipid–LacY interactions govern membrane protein TMD organization.
Therefore, TMD topology is sensitive to the charge density on the membrane surface i.e. neutral GlcDAG and GlcGlcDAG and zwitterionic PE and PC all dilute the negative charge of the membrane surface created by PG and CL. At the same time these net neutral lipids support native LacY topology. Physical properties such as imposing lateral stress or membrane curvature (properties of only PE and GlcDAG) appear not to be important determinants of topological organization. Although it is surprising that E. coli is tolerant to such major changes in membrane lipid composition, these results emphasize the importance of similar collective charge properties of lipids in determining membrane protein topology rather than a strict structural or physical requirement, as observed for proteins.
Another topologically important feature of polytopic membrane proteins is the folding of EMDs [75]. Since an EMD needs to be unfolded for translocation or reversal of orientation by the translocon, rapid and stable co-translational folding or glycosylation (Fig. 1, #12 or #13, respectively) can ensure the location of an EMD. In the case of LacY proper folding of EMD P7 depends on PE acting as a non-protein molecular chaperone (termed lipochaperone) [76–78]. As will be discussed in the next section, proper folding of P7 is necessary for wild type LacY transport activity and does not occur in cells or liposomes containing only anionic lipids. Thus, membrane lipid composition influences both the topological organization and subtler local domain folding of membrane proteins.
5. Lipid-dependent structure–function relationships
The secondary transporters like LacY carry out energy dependent transport to concentrate substrate by coupling uphill accumulation of substrate to downhill movement of protons generated by the electrochemical membrane potential. In the absence of a membrane potential these transporters facilitate downhill movement of substrate and thus equilibrate substrate across the membrane. In the absence of PE or other net neutral lipids LacY, PheP and GabP [68,71,79] and possibly many other secondary transporters only carry out downhill transport even though the cell maintains a robust membrane potential [79]. The retention of partial function indicates a compact folded structure even in the absence of PE. In the case of LacY there is a strong correlation between the proper folding of EMD P7 and uphill transport function [80]. Native folding of this domain can be assessed by recognition of P7 by the conformationally sensitive monoclonal antibody 4B1 [80]. Mis-folding of P7 alters the packing of TMDs in the C-terminal half of LacY. This significantly lowers the abnormally high pKa of a glutamate in TMD X thus disrupting the proton wire involved in symport of the substrate and a proton [81]. Proper folding of P7 and uphill transport is supported by PE [77], GlcDAG and PC in vivo but not by GlcGlcDAG [62]. The head group of GlcGlcDAG may be too large to allow complete acquisition of P7 domain structure because of steric clashes with the bulky disaccharide group. Reconstitution of purified LacY (from PE-containing or PE-lacking cells) in liposomes made of total lipid extracts from cells containing PE, PC or GlcDAG (but not GlcGlcDAG) also supported uphill transport and recognition by monoclonal antibody 4B1 indicating a direct effect of these lipids on the proper folding of the P7 domain [74]. Previous reconstitution studies of LacY into liposomes containing PCs with only unsaturated fatty acids did not result in uphill transport [82]. However, PE and PC containing at least one saturated fatty acid or those derived from E. coli cells (primarily one saturated and one unsaturated fatty acid) did support uphill transport and recognition of P7 by monoclonal antibody 4B1 after reconstitution [74]. Therefore, proper folding of P7 and support for uphill transport does not strictly depend on the chemical properties of the net neutral head group but rather on the combined physical and chemical properties of the head group and the fatty acids.
6. What property of proteins makes them sensitive to membrane lipid composition?
Inspection of the distribution of charged residues in the cytoplasmic EMDs of LacY, PheP and GabP (that are mis-oriented in PE-lacking cells) shows a pattern of positively charged residues mixed with negatively charged residues (Fig. 3). In fact NT, C2 and C4 of PheP are net +1, −1 and neutral, respectively. This suggests that the translocation potential of negatively charged amino acids, which is known to be weak relative to the retention potential of positively charged residues [68], is enhanced in the absence of PE. In order to investigate whether the presence of both negatively and positively charged residues is the basis for topological sensitivity to the lipid environment, charged residues within the cytoplasmic domains of the N-terminal bundle of LacY and N-terminal hairpin of PheP were altered and topological organization was assessed in whole cells as a function of lipid composition using SCAMTMD [48,67].
Increasing the net positive charge of the cytoplasmic face of the N-terminal six-TMD bundle of LacY by one in a position independent manner prevented inversion in PE-lacking cells [48]. A similar result was found for the N-terminal hairpin of PheP [67]. However, inversion of this LacY domain in PE-containing cells required a change in net charge from +6 to −6 for the whole-bundle cytoplasmic surface. This result strongly indicates that the weak translocation signal provided by negatively charged residues is due to the presence of PE. Therefore, PE dampens the translocation potential of negatively charged residues in favor of positively charged residues acting as retention signals. Also the net charge of the cytoplasmic surface of the whole N-terminal six-TMD bundle responds as an independently folding unit.
These results support (i) the dominance of the positively charged residues as retention signals over negatively charged residue as translocation signals in PE-containing cells, (ii) the attenuating effect of PE on the translocation potential of negatively charged residues and (iii) suggest that negatively charged residues can play an active role as topogenic signals dependent on the membrane lipid composition.
7. Long- and short-range lipid–protein interactions
What is necessary and sufficient for a change in TMD orientation within a membrane in response to changes in membrane lipid composition? A primary feature would be the presence of cytoplasmic domains containing a mixture of negatively and positively charged residues flanking a TMD. However, such domains must be free of restrictions preventing different orientations relative to other parts of the protein. In order for the N- and C-terminal six-TMD bundles of LacY to respond to the lipid environment independent of each other there must exist a flexible hinge region (or topogenic conflicting domain, Fig. 1B, #10) between the independently folding domains. Indeed the inversion of N-terminal bundle of LacY in PE-lacking cells coincides with periplasmic exposure (Fig. 3B) of the marginally hydrophobic (ΔG = −1.4 kcal/mol) TMD VII [48]. The hydrophilic nature of TMD VII appears to directly influence the operation of the molecular hinge within LacY. An increase in TMD VII hydrophobicity by replacement of one negatively charged residue by a hydrophobic residue (Asp240Ile mutation) prevents TMD VII exposure to the periplasm and simultaneously blocks the inversion of the N-terminal helical bundle. Consistent with this conclusion is that in the case of PheP and GabP, TMD III becomes a mini-loop (Fig. 3D) that no longer spans the membrane to allow the N-terminal hairpin to invert in PE-lacking cells [67,68,71].
The thermodynamically favorable inversion of the N-terminal bundle brought about by lipid–protein interactions is countered by the thermodynamically unfavorable aqueous exposure of this TMD when its hydrophobicity is increased [48]. Therefore, the positive driving force for inversion must balance the negative energy cost of exposing a TMD to an aqueous environment. Since the translocation potential of negatively charged residues is greatly enhanced in the absence of PE, it would be expected that increasing the translocation potential of neighboring EMDs could overcome this thermodynamic barrier to inversion of topology. This proved to be the case. An increase in the net negative charge of the cytoplasmic surface of the N-terminal bundle from +6 to −6 overcame the block to inversion in PE-lacking cells caused by an increase in hydrophobicity of TMD VII [67].
In support of these conclusions the higher hydrophobicity of TMD VII in the E. coli sucrose permease (CscB) relative to LacY appears to desensitize CscB to topological inversion in PE-lacking cells [67]. However, a progressive increase in the net negative charge of the cytoplasmic surface of the six-TMD N-terminal helical bundle of CscB resulted in a complete inversion of TMDs N-terminal to TMD VII first in PE-lacking cells (from +6 to −6) and then in PE-containing cells (from +6 to −10).
The results with LacY, PheP and CscB demonstrate that PE enhances the retention potential of positively charged residues and reduces the translocation potential of negatively charged residues thereby providing a molecular basis for the operation of the Positive Inside Rule for domains containing a mixture of positively and negatively charged residues particularly when the latter are in excess. The thermodynamic block to exposure of a hydrophobic TMD to an aqueous environment can be overridden by the increased translocation potential of negatively charged residues in PE-lacking cells. The results with LacY and CscB illustrate how long-range interactions can override local electrostatic and hydrophobic forces.
8. How is the Positive Inside Rule executed?
The native topological orientation of LacY in PE-lacking cells was maintained by the elimination in a position-independent manner of anyone of three negative charges within otherwise positively charged EMDs (either C2, C4 or C6) on the cytoplasmic surface of the N-terminal bundle. Similarly, introduction of a single positively charged residue was sufficient to prevent mis-orientation [48]. Therefore, positively charged residues exert their topogenic effect globally and can be potent even in a retrograde manner, i.e. a change in the charge of EMD C6 (located between TMDs VI and VII) or a change in the hydrophobicity of TMD VII results in a topological change in TMDs that were already synthesized and most likely residing outside of the translocon. This finding was further supported in the case of EmrE where placement of a single positively charged amino acid at the C-terminus was potent enough to reorient the preceding four- or five-TMD helical bundle [83] post-translationally.
Therefore either LacY [48], the EmrE monomer [83] or CscB (Vitrac, Bogdanov and Dowhan, unpublished data) can be forced to obey the Positive Inside Rule either by elimination of negatively charged residues or by introduction of positively charged residues within any of the cytoplasmic domains thus demonstrating the high potency of positively charged residues. These results also provided the evidence that interpretation of Positive Inside Rule could be started from anywhere within a nascent sequence even in a retrograde manner (i.e. from the C-end to N-end). Thus the final topology is determined by cooperative short-range and long-range lipid–protein and intra-protein interactions that occur well after the nascent polypeptide exits the translocon.
9. The Charge Balance Rule
If interactions in the membrane–aqueous interfacial region between charged EMDs and the collective charge of the membrane surface are a determinant of TMD orientation, then alterations in the charge nature of either the lipid head groups or the protein domains should affect final protein topology. As was described in previous sections, the effects of net charge were the same whether these resulted from the lipid or the protein thus supporting charge interaction in a complementary manner between the cytoplasmic domains and the bilayer surface as a determinant of topological organization. Therefore, the proposed Charge Balance Rule (Fig. 1B #14) is an extension of the Positive Inside Rule with incorporation of the effects of lipid–protein interactions [84]. A byproduct of these effects allows the presence of negatively charged amino acids in cytoplasmic domains necessary to support protein function without affecting protein topology [48,85]. Several of the negatively charged residues on the cytoplasmic side of LacY are important in its function but would be counterproductive if allowed to be strong topogenic signals. Furthermore, the principle of the Charge Balance Rule can be extended to a balance between long-range interactions, i.e. positive thermodynamic driving force derived from lipid–protein interactions can balance with the loss of hydrophobic forces resulting from the exposure of a TMD to an aqueous environment.
Earlier studies indicated that the interaction between positively charged amino acids and negatively charged lipid head groups may provide the molecular basis for the Positive Inside Rule [34]. Increasing the net positive charge of EMDs or increasing membrane anionic lipid content prevented translocation of EMDs containing only positively charged and neutral residues. These results initially appear to be in conflict with the Charge Balance Rule. However, the role of negatively charged amino acids was not considered in these experiments. For EMDs containing negatively charged residues, increasing the membrane surface negative charge density increases the translocation potential of negatively charged residues in opposition to the retention potential of positively charged residues [48,67]. This is consistent with requiring a large number of negatively charged residues to induce inversion in PE-containing cells while requiring an increase in number of positively charged residues to prevent inversion in PE-lacking cells. Therefore, interactions of positively charged residues with anionic lipid head groups may still explain execution of the Positive Inside Rule but the attenuating effect of negatively charged residues must be considered as the collective negative charge density of the membrane surface is increased.
How might membrane lipid composition affect the translocation potential of negatively charged (acidic) amino acids in EMDs? The pKa of negatively charged amino acids of the multidrug transporter LmrP from Lactococcus lactis was determined as a function of the lipid composition after reconstitution in proteoliposomes [86]. In proteoliposomes composed of PE, PG and CL, pKa values of acidic residues were unexpectedly raised to 6–7 making these residues less prone to ionization. However, in the presence of only PG and CL, the pKa values of acidic residues were 4–5, which is also unexpected due to the high negative charge density of the interfacial region that should hinder ionization [87]. Therefore, the pKa upshift caused by PE appears to selectively neutralize negatively charged residues. Transbilayer asymmetry of PE in the inner membrane might play a role if E. coli asymmetry (unknown) mirrors that of Bacillus megaterium where the inner leaflet contains twice the PE of the outer leaflet [88]. Such a distribution would dampen negative charges at the cytoplasmic surface with a reduced effect at the periplasmic surface. The positive outward membrane potential, which was demonstrated to drive negatively charged EMDs outward [29,30], could also provide an asymmetric component. For EMDs containing a mixture of negative and positive charges the presence of PE would increase cytoplasmic retention of the EMD by neutralizing negatively charged residues. This would result in an increase in the positive charge of the EMD. Conversely, the absence of PE would potentiate ionization of acidic amino acids in favor of domain translocation (Fig. 4).
Fig. 4.
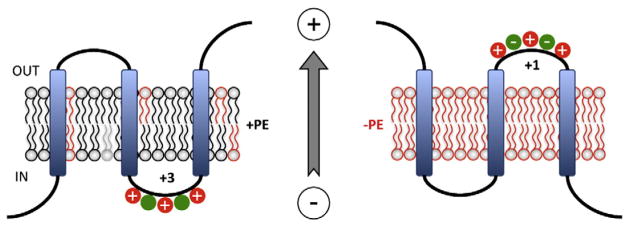
PE and the Charge Balance Rule. A cytoplasmic EMD is shown containing a mixture of negatively and positively charged amino acids. Left Panel. According to the Charge Balance Rule PE (black) would raise the pKa and suppress the translocation potential of negatively charged residues (green), which increases the effective positive charge potential of the EMD (+3) thus favoring its retention on the cytoplasmic side of the membrane. Right Panel. In the absence of PE (red) negatively charged residues exert their full translocation potential and result in translocation of the domain that now exhibits a lower effective net positive charge (+1). Even though the charge is still net +1, negatively charged residues may exhibit stronger signals than positively charged residues in the absence of PE. The membrane potential (positive outward) determines EMD directionality. This figure was modified from the original figure published in [85] © Annual Reviews of Biochemistry.
The Charge Balance Rule was recently tested in vitro with a eubacterial proteorhodopsin [89]. This seven TMD membrane protein has an inherent asymmetric surface charge density across the membrane. The net negative N-terminal EMD and the net positive C-terminal EMD domain are oriented to opposite sides of the membrane. The orientation of the protein reconstituted into liposomes was monitored as a function of the surface charge density (zwitterionic, anionic or cationic) of the lipid bilayer. The protein assumed an N-terminal out and C-terminal in orientation in negatively charged liposomes and an opposite orientation in net neutral or positively charged liposomes. The effective charge of the EMDs and the lipid bilayer surface charge cooperatively governed preferential orientation of this polytopic membrane protein. Therefore, the Charge Balance Rule appears to be a general assembly rule for membrane proteins.
10. Post-assembly reversibility of topology
Since lipid–protein interactions apparently independent and outside of the translocon are determinants of final protein topology, can changes in the topology and structure of a fully folded and functional membrane protein occur in response to changes in the lipid environment? Such a result would be in conflict with dogma that assumes stable orientation of highly hydrophobic TMDs due to the assumed large free energy barrier to passage of hydrophilic flanking EMDs through the lipid bilayer. The development of E. coli strains in which lipid composition can be controlled dynamically as a function of added inducers for the expression of lipid biosynthetic enzymes provided the means to answer this question in vivo [48,60,61]. LacY synthesis in initially PE-lacking cells was first induced by isopropyl β-D-thiogalactoside from its native promoter followed by removal of the inducer (ceases new LacY synthesis). Addition of an inducer for the pssA gene resulted in synthesize of PE post-assembly of LacY into its inverted topology. Analysis of the topological organization of LacY by SCAMTMD showed a dramatic reorientation of TMD III–TMD VI to their native orientation and insertion of the solvent exposed TMD VII into the membrane (Fig. 3C). The N-terminal EMD and TMD I of LacY remained in their non-native orientation, and TMD II became a mini-loop or new hinge domain that no longer spanned the membrane [48]. This post-assembly reorientation of TMDs was shown to be fully reversible by first synthesis of LacY in PE-containing cells followed by removal of both inducers. Dilution of PE to low levels resulted in the inverted topological organization of LacY (Fig. 3B) [60]. Therefore, the topological re-organization post-assembly of proteins is bi-directional. Although surprising, these results are still consistent with the Charge Balance Rule. The increase in membrane PE content after complete folding of LacY would be expected to increase the probability of the N-terminal helical bundle to adopt the correct orientation due to strengthening of the Positive Inside Rule. The depletion of PE post-assembly of LacY in PE-containing membranes would increase translocation potential of negatively charged residues (Fig. 4) resulting in inversion of topology.
E. coli LacY and PheP so far are the only native polytopic membrane proteins for which lipid-dependent post-insertional TMD re-orientation has been definitively demonstrated [48,61,68]. However post-insertional re-orientation is not restricted to the secondary transporters. SecG, an integral membrane subunit of the E. coli translocon, undergoes reversible re-orientation coupled to its function in ATP- and SecA-dependent translocation of pre-proteins [90]. In the resting state SecG is a helical hairpin with the N- and C-termini exposed to the periplasmic space. However, SecG undergoes transmembrane topology inversion during the protein translocation process. It is quite interesting that the N-terminal domain of SecG is net negative and does not possess a positive charge consistent with its periplasmic exposure. Positively and negatively charged residues exist close to each other in equal amounts among the cytoplasmic EMD and the long C-terminal EMD. Close encounter of SecG with a novel glycolipid is required for reversible inversion of the SecG hairpin. The glycolipid is composed of diacylglycerol and a glycan chain of three acetylated aminosugars linked through pyrophosphate [36].
The mechanism by which CD38 catalyzes the formation and hydrolysis of cyclic ADP-ribose with the former acting as an intracellular Ca2+-mobilizing messenger remains controversial due to exposure of the C-terminal catalytic domain to the extracellular surface. Recently, it was demonstrated that CD38 exists as a mixture of two potentially in-terconvertible topologically forms [91]. A TMD is followed by the catalytic C-terminal domain, which is either oriented to the exterior of the cell (type II orientation) or the interior of the cell (type III orientation). The number of positive charged amino acids is about the same in the EMDs flanking this TMD. However, TMD orientation of CD38 was converted from a mixture of type II and type III orientations to uniform type III by changing four positively charged amino acids to aspartic acid in the EMD N-terminal to the TMD. This conversion increased intra-cellular levels of cyclic ADP-ribose. Phosphorylation/dephosphorylation of three native serines within this same EMD was suggested as a physiological post-assembly mechanism to regulate the ratio between the type II and type III orientation and thus cyclic ADP-ribose levels.
The ability of membrane proteins to undergo large topological rearrangements post-assembly into functional proteins challenges the dogma that once transmembrane orientation is established in a fully mature protein, topology is stable and static. Clearly membrane protein structure is dynamic rather than static and can respond to changes in membrane lipid composition in the local environment, which occurs during cell division, membrane fission and fusion, movement of proteins in and out of lipid rafts, and metabolic changes in polyphosphorylated PI pools. As proteins traffic through the secretory system of eukaryotic cells, membrane proteins encounter dramatic changes in the lipid environment. Topological changes in membrane proteins post-assembly are a possible means of regulating protein function or coding proteins for turnover. Such changes could occur by transfer of the protein to a different cellular membrane, dynamic changes in local lipid composition within the same membrane, or post-translational charge modification of EMD charges of a membrane protein such as by phosphorylation/dephosphorylation cycles [91]. The freely reversible interconversion of topological conformers in response to changes in lipid environment can explain the different topological organizations of the same protein in different membranes in the same cell. Therefore, the effects of changes in lipid environment must be considered as a possible regulatory mechanism.
11. How are lipid-dependent transmembrane topological inversions governed?
Phospholipids might exert their effect on membrane protein topology either directly by interacting with the topogenic signals or indirectly by influencing the assembly machinery. The second possibility represents a reasonable alternative, since many of the components of the assembly machinery are known to be lipid-dependent [92]. The former possibility is reasonable based on simple equilibrium and the lowest energy considerations within a given lipid environment.
Several lines of in vitro evidence strongly indicate that polytopic membrane protein topogenesis is driven thermodynamically by direct lipid–protein interactions. LacY purified from either PE-lacking or PE-containing cells adopts inverted or proper topology after reconstitution in PE-lacking or PE-containing liposomes, respectively. This demonstrates that the topogenic influence of lipids is largely independent of other protein factors and from the in vivo folding history of the protein [70]. These experiments leave little doubt that direct lipid–protein interactions outside and independent of the translocon machinery are determinants of TMD orientation even after full protein maturation. These in vitro results support conclusions drawn from in vivo experiments where N-terminal topological organization after release from the translocon is influenced by decoding of downstream [48] or even C-terminal topogenic signals [83].
Further confirmation has been obtained that TMD re-orientation post-assembly requires no other cellular factors other than a change in lipid environment. The topological organization of LacY was analyzed in an in vitro proteoliposome system in which lipid composition can be systematically controlled before (liposomes) and after (fliposomes) reconstitution [69] using a methyl-β-cyclodextrin-mediated lipid exchange technique [93]. Methyl-β-cyclodextrin facilitates the rapid exchange of phospholipids between the outer monolayers of donor multilamellar lipid vesicles and recipient small unilamellar vesicles (liposomes) without bilayer fusion or disruption of bilayer integrity [93]. This new fliposome technology allows determination of the minimum and sufficient requirements for a protein to flip between topologically distinct states. The approach also provides a means to establish whether this interconversion is thermodynamically driven by the properties of the protein interacting with its lipid environment independent of other cellular factors. The in vivo interconversion between topological conformers of LacY was faithfully reproduced in a PE dose-dependent manner by either increasing or decreasing PE levels in pro-teoliposomes after reconstitution of LacY purified from PE-containing or PE-lacking cells.
Therefore, post-assembly bi-directional changes in TMD orientation are thermodynamically driven and determined by direct lipid–protein interactions, which are governed by the inherent properties of the protein and its lipid environment. Initial orientations and post-assembly reorientations may still require unknown molecular chaperones or other factors to kinetically aid in response to the lipid environment. However, in the fliposome system the rates of protein flipping in both directions occurred on a second scale following the changing lipid composition of the proteoliposomes (Vitrac, Bogdanov and Dowhan, unpublished result). This result is consistent with the low energy barrier observed in vivo for transmembrane rearrangements driven primarily by direct lipid–protein interactions rather than an indirect involvement of other factors affected by changes in lipid composition. The apparent low activation energy in vivo and in vitro for protein re-organization in the lipid bilayer strongly suggests a thermodynamically driven process, which can occur at any time and in any membrane. Therefore, either most membrane proteins must be designed to prevent such topological changes while a subset of proteins may be designed to take advantage of topological dynamics to regulate or change their function.
Interestingly, reconstitution of LacY [69] and other membrane proteins [94] by rapid dilution in the presence of preformed liposomes containing native lipid composition results in a uniform topological orientation. However, overall orientation is opposite that observed in cells with normally cytoplasmic domains oriented outward and normally periplasmic domains oriented inward. In such experiments there is no membrane potential and lipid bilayer asymmetry is not a factor. In this case the effect of net neutral lipids like PE in raising the pKa of acidic amino acids (see Section 9) might provide an explanation. The presence of PE would make EMDs containing both negatively and positively charged amino acids highly positively charged. EMDs containing mostly negatively charged amino acids would be essentially neutral. This would favor the retention of the former positively charged EMDs and translocation of the latter more neutral EMDs through the low-dielectric hydrophobic lipid bilayer in accordance with a physical mechanism involving electrically neutral residue transfer [87]. Conversely, in the absence of PE the EMDs enriched in acidic residues would become negatively charged, which would hinder their translocation. EMDs containing both negatively and positively charged residues would become more neutral thus facilitating translocation through the lipid bilayer. The result would be an overall opposite, as compared to proteoliposomes containing PE, but still unidirectional orientation. The energetically favorable partitioning of a membrane protein into the lipid bilayer would provide the driving force for translocation of neutral EMDs across the membrane.
However, what might be the driving force for dynamic changes in protein topological organization in response to a change in proteoliposome lipid composition is yet unknown. In the lipid exchange process lipids enter the outer leaflet of proteoliposomes, but lipid bilayer asymmetry appears not to be a factor [69]. In the case of LacY a consideration of the effects of a change in lipid composition on the properties of the two aspartate residues in TM VII may provide the driving force for reorientation. In the absence of PE the low hydrophobicity of TM VII (now an EMD in close association with the surface of the lipid bilayer [48]) would be maintained by ionization of the aspartate residues. Introduction of PE would promote selective protonation of these residues due to the experimentally demonstrated effect of this lipid on the pKa of acidic amino acids [86]. The result would be a drastic increase in the hydrophobicity of TM VII favoring its insertion into hydrophobic core of the membrane. The free energy gained from insertion of TM VII would thermodynamically drive reorientation of the N-terminal helical bundle as was demonstrated in whole cells [48]. Dilution of PE would have the opposite effect. Further experimental verification of the effects of membrane lipid composition on the properties of charged amino acids remains to be carried out.
12. Lipid-dependent generation of dual topologies
Dogma assumes that all copies of an integral membrane protein have the same orientation relative to the membrane bilayer and such orientation once attained does not change. Yet a number of membrane proteins have been shown to exist in more than one topological organization with respect to their TMDs either in the same membrane or in different membranes within the same cell [60,95,96]. The best documented cases for membrane proteins with dual topologies are bi-functional enzymes or proteins, which have been reviewed elsewhere [95]. The generation of equal amounts of oppositely oriented proteins within the same membrane is well beyond the control of the translocon [97]. Therefore, existence of dual and multiple topologies of the same membrane protein raises intriguing questions concerning the mechanism of membrane protein topogenesis. So far there is no mechanistic understanding of how dual topologies are generated and maintained within one membrane or between different membranes in the same cell. How might the ratio of topological conformers be controlled? Are topological conformers in rapid equilibrium with each other or stably expressed? If multiple conformers are interconvertible, what factors are involved? Is topological organization highly cooperative resulting in a sharp transition between conformers or of low cooperativity resulting in mixtures of conformers?
By using the tet-regulated control of pssA gene expression, the level of PE can be regulated in a dose responsive manner between 5% and 75% of total phospholipid uniformly throughout the cell culture [60]. This allows the titration of the topological effect of lipids in vivo. In vivo LacY displays a mixture of topological conformations ranging from complete inversion of the N-terminal helical bundle to mixed topology and then to completely native topology as PE is increased from near zero to 75% of membrane phospholipids at the time of initial synthesis. At intermediate levels of PE (about 30%), LacY co-exists in living cells as two topologically distinct and stable conformers in about equal amounts (i.e. the wild type conformation and the non-native inverted conformation normally seen in cells lacking PE, see Fig. 3A and B). There was no threshold level of PE determining a sharp transition from one topological conformer to the other. Co-existing conformers were thermodynamically locked and not in rapid equilibrium at a steady-state lipid composition. Therefore, mixtures of topological conformers in the same membrane are not only dependent on the protein sequence as previously demonstrated [46,67] but are also regulated by membrane lipid composition in a dose-dependent manner. However, increasing or decreasing PE content post-assembly resulted in the expected increase or decrease of the native conformer, respectively.
The fliposome system [69] was utilized to titrate in vitro topological effects of lipids in the same manner as was done in vivo to determine the minimum and sufficient requirements for a protein to adopt a dual topology. Reconstitution of LacY in liposomes with varied PE content also resulted in the same ratio of topological conformers as observed in vivo at varying PE content. Therefore, LacY can insert simultaneously into the liposomal bilayer in two opposite orientations. The ratio between these multiple conformers is determined by the mol% of PE [69]. The in vitro experiments also demonstrated that the multiple topological conformers were not in rapid equilibrium with each other. The ratio of conformers can also change in response to post-assembly changes in lipid composition as observed in living cells [60]. Most proposed mechanisms for generation of multiple topologies rely on proteins with weak topogenic signals [98,99]. However, LacY possesses strong topological signals and highly hydrophobic membrane domains. Therefore, in vivo and in vitro experiments with LacY indicate that membrane lipid composition in concert with protein sequence can be the basis for the generation and stable co-existence of dual topologies for a single protein in the same membrane.
These in vitro results further confirm that the balance of electrostatic and hydrophobic forces and the collective properties of both the protein and lipid environment are important decision-making factors in the thermodynamically driven process of membrane protein topogenesis. The results support conclusions that could not be made based solely on in vivo observations. The formation and co-existence of multiple topological conformers is an inherent property of some membrane proteins and their lipid environment. The membrane protein insertion/ assembly machinery or any other cellular factors are not necessarily required to orchestrate a mixed topological organization within the same membrane. Since examples exist in nature of proteins that exhibit topological and functional duality, these results provide a potential molecular mechanism by which the same gene product can exist in multiple structural forms.
The bi-directionality and reversibility of topogenesis of transmembrane protein structure observed in vivo and in vitro can be used to build a thermodynamic based and lipid-dependent model for shifting the equilibrium between different conformational states of a membrane protein (Fig. 5). At the extremes of lipid composition, a nascent protein folds into its single respective lowest energy state. At an intermediate lipid composition, there is a point during the folding process where multiple conformer precursors are in rapid equilibrium with the proportion of each conformer dictated by the lipid composition. As each precursor continues to fold to its lowest energy state, the final conformers are separated by a high activation energy. This provides a thermodynamic block to interconversion between the two conformations resulting in stable dual conformations within the same membrane. The simplest interpretation of the bi-directional reorganization process is that raising or lowering of PE levels would destabilize the folded state of the inverted or native conformer, respectively. Increasing the energy state of the destabilized conformer would allow reorientation of TMDs in spontaneous response to the new lipid environment.
Fig. 5.
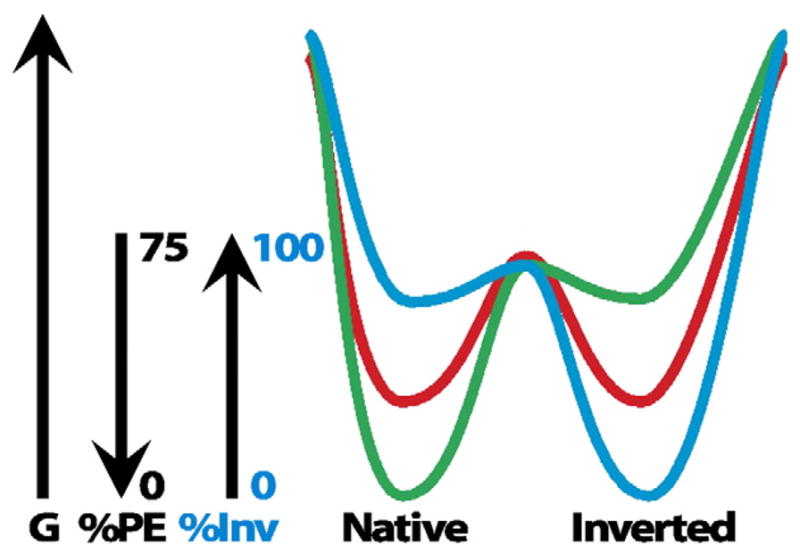
Dual minima energy folding funnel for LacY as a function of membrane lipid composition. The schematic depicts a two-dimensional section through the three-dimensional protein folding funnel. The folding of LacY to its lowest free energy state (G) proceeds via a funnel-shaped energy landscape whose shape is defined by the physicochemical properties of the lipid environment (green, 75% PE; red, intermediate % PE; blue, 0% PE). The topology of the N-terminal bundle of LacY is responsive to the membrane PE content. During initial folding of LacY multiple topological conformers are in rapid equilibrium, but subsequent folding events result in a stable mixture of conformers determined by the percent PE and separated by a high activation energy. A change in the PE content raises the free energy of the conformers resulting in a redistribution of conformers governed by the new PE content. This figure was originally published in [60] © the American Society for Biochemistry and Molecular Biology.
Since such interconversions are solely thermodynamically driven, a change in lipid composition within any cell membrane need not require other cellular factors to initiate a change in protein organization. Membrane protein topogenesis can be re-directed to a new TMD organization by either post-translational (phosphorylation) modifications of proteins [91] or by different lipid compositions. As noted earlier, the latter changes along the intracellular protein trafficking pathways. Thus proteins can either organize into multiple conformers at the time of synthesis in the endoplasmic reticulum or bacterial membrane or once transported to their final resident membrane undergo a change in topological organization. The fact that protein sequence defines topological organization but is written for a specific lipid environment explains improper transmembrane organization of many membrane proteins when expressed in a foreign host [100]. This thermodynamic model of dynamic membrane protein topogenesis explains how both uniform and dual topological organization of different membrane proteins can co-exist within the same membrane. The model also provides a physiologically plausible mechanism and theoretical basis for further investigation of proteins in more complex eukaryotic systems where multiple protein conformers exist.
Several reports demonstrated that the changes in cholesterol levels in the endoplasmic reticulum membrane might affect topogenesis and result in either aberrant topology or topological heterogeneity of membrane proteins. Because cholesterol does not possess any charge, it can act to dilute the negatively charged membrane surface as dictated by the Charge Balance Rule. The large (L) envelope protein of hepatitis B virus exists in two functionally distinct topological conformers [101,102] whose distribution varies with the endoplasmic reticulum cholesterol content. Progressive lowering of cholesterol levels in host cells converts the dual topology of viral L protein into a uniform one [103].
Spermatozoa become capable of fertilization through a series of membrane changes including a change in lipid composition resulting in a drastic reorganization of both lipids and proteins leading to uniform raft formation. Remarkably the topological organization of a bull sperm Na+/K+ ATPase, a protein involved in cell signaling in sperm during capacitation (depletion of sperm coat cholesterol and glycoproteins), was found to be critically dependent on cholesterol levels after reconstitution into proteoliposomes with lipid compositions that mirrored in vivo raft compositions. Cholesterol directly affected orientation of this protein. After incorporation of the β subunit of Na+/K+ ATPase into liposomes, the orientation of the protein was right side out in the presence of cholesterol and inside out in the presence of drastically reduced cholesterol. These observations mimic events, which could be associated with movement of the protein in and out of lipid rafts, raft break down or cholesterol efflux from sperm membranes during capacitation [104]. Thus the results suggest an interesting mechanism for regulation of the function of a membrane protein through sensing of cholesterol levels or movement in and out of lipid rafts.
13. Membrane protein topological bias and membrane lipid composition
The ratio of net negatively charged phospholipids to net zero charged lipids (zwitterionic or uncharged) is highly regulated and kept constant [51] in Gram-negative and Gram-positive bacteria although there is a broad range of ratios when comparing different bacteria. If the Charge Balance Rule applies generally over a wide range of organisms, then the net positive charge of cytoplasmic EMDs of homologous membranes protein over a broad number of bacteria should increase as the proportion of anionic lipids increases in membranes in order to maintain the same protein topological orientation.
A bioinformatic approach was used to compare the sequences of bacterial small multidrug resistance (SMR) protein homologues and the lipid profiles of their respective host organisms [51]. The results were as predicted by the Charge Balance Rule. As the ratio of anionic to net neutral lipids in the host membranes increased, the inside positive bias of EMDs of homologous SMR proteins increased presumably to maintain the same topological organization. Therefore, evolution cannot only impart different membrane topologies on related and homologous proteins by shuffling positively charged residues between EMDs, as has been proposed [99], but also through changes in membrane lipid composition without introducing mutations in the protein [60]. These results suggest that during the course of evolution both proteins and lipids co-evolved in the context of the lipid environment of the membrane in which both are mutually dependent on each other.
14. Conclusions
Multiple assembly rules, topogenic sequences and topological determinants (translocon, phospholipids, membrane potential) ensure proper membrane protein topogenesis. At the same time these same factors allow for dynamic post-translational TMD orientation resulting in uniform topology, alternative topologies or dual topology depending on the membrane lipid composition or physiological state of the cell. By taking advantage of the well-characterized E. coli system and in vitro membrane protein reconstitution techniques, the lipid composition in genetically altered strains or in proteoliposomes can be manipulated temporally or in a dose-dependent manner. Using these systems the role of individual lipids in the complex processes of membrane protein assembly and protein structural dynamics can be determined. The close agreement between in vivo and in vitro results indicates that the general principles governing these processes in bacteria can be extrapolated to more complex biological systems.
TMD orientation is determined by the sum of responses of the translocation machinery, membrane potential and membrane lipid composition to independent signals provided by TMD hydrophobicity and flanking positive and negative charges. All must act in concert initially at the time of presentation to the translocon channel and then to the lipid bilayer after clearance from the translocon. The translocation machinery initially influences TMD orientation, but final topology is determined outside the translocon in the accordance with Charge Balance Rule by short-range interactions between the protein and the lipid environment and by long-range interactions within the protein as independently folding domains assume their native structure. Lipid–protein interactions affect the potency of charged residues as topological signals and thus provide a physiological basis for Positive Inside Rule. According to the Charge Balance Rule the translocation potential of negatively charged amino acids working in opposition to the Positive Inside Rule is largely attenuated by the presence of PE and other net neutral lipids. This explains the dominance of positively charged residues as retention signals in wild type cells. Topological organization of a membrane protein, once established, is not static but can be dynamic in response to changes in lipid environment. Membrane protein topogenesis is a primarily thermodynamically driven process with lipid–protein interactions as a major determinant of which topological organization is energetically favored. Other factors such as the hydrophobicity of a TMD to potentially act as a hinge region between independently folding domains also determines the existence of dual topological states or the possibility to change topology in response to changes in the lipid environment. These dynamic TMD rearrangements constitute an entirely novel lipid-dependent biological switch for generation of functional and structural heterogeneity for a single polypeptide chain within the same membrane or different subcellular membranes. This proof of principle observation has important implication for membrane proteins in eukaryotic cells. Lipid-dependent TMD conformational dependence and switches illustrate an elegant way to regulate a membrane protein function between different membranes or within the same membrane where lipid composition is not uniform either spatially or temporally. Therefore, differences in lipid composition between membranes encountered by proteins during intracellular trafficking and sorting may play an important role in determining eukaryotic membrane protein organization and function.
At the moment alteration of lipid composition in mammalian cells or whole animals poses significant barriers as an experimental approach to validate extrapolation of folding and assembly rules to more complex biological systems. Limited studies may be possible in yeast since viable mutants with altered lipid composition [105] are available. However, there is close correlation in E. coli between the effects of lipid alteration from in vivo studies and in vitro proteoliposomes studies. Therefore, the fliposome reconstitution systems described here should be directly applicable to mammalian proteins to determine if changes in local membrane lipid composition have similar effects as observed in bacteria.
Acknowledgments
The work of the authors was supported by funds awarded to W.D. from the National Institutes of General Medical Sciences grant GM 20478 and the John Dunn Research Foundation.
Abbreviations
- PA
phosphatidic acid
- PE
phosphatidylethanolamine
- PG
phosphatidylglycerol
- PGP
phosphatidylglycerophosphate
- DAG
diacylglycerol
- CL
cardiolipin
- PA
phosphatidic acid
- PS
phosphatidylserine
- PC
phosphatidylcholine
- PI
phosphatidylinositol
- GlcDAG
monoglucosyl diacylglycerol
- GlcGlcDAG
diglucosyl diacylglycerol
- LacY
lactose permease
- PheP
phenylalanine permease
- GabP
γ-aminobutyrate permease
- CscB
sucrose permease
Footnotes
This article is part of a Special Issue entitled: Protein Trafficking & Secretion.
Contributor Information
Mikhail Bogdanov, Email: Mikhail.V.Bogdanov@uth.tmc.edu.
William Dowhan, Email: William.Dowhan@uth.tmc.edu.
References
- 1.Enquist K, Fransson M, Boekel C, Bengtsson I, Geiger K, Lang L, Pettersson A, Johansson S, von Heijne G, Nilsson I. Membrane-integration characteristics of two ABC transporters, CFTR and P-glycoprotein. J Mol Biol. 2009;387:1153–1164. doi: 10.1016/j.jmb.2009.02.035. [DOI] [PubMed] [Google Scholar]
- 2.Lee E, Manoil C. Mutations eliminating the protein export function of a membrane-spanning sequence. J Biol Chem. 1994;269:28822–28828. [PubMed] [Google Scholar]
- 3.Hessa T, Kim H, Bihlmaier K, Lundin C, Boekel J, Andersson H, Nilsson I, White SH, von Heijne G. Recognition of transmembrane helices by the endoplasmic reticulum translocon. Nature. 2005;433:377–381. doi: 10.1038/nature03216. [DOI] [PubMed] [Google Scholar]
- 4.Mackenzie KR. Folding and stability of alpha-helical integral membrane proteins. Chem Rev. 2006;106:1931–1977. doi: 10.1021/cr0404388. [DOI] [PubMed] [Google Scholar]
- 5.White SH, von Heijne G. Do protein–lipid interactions determine the recognition of transmembrane helices at the ER translocon? Biochem Soc Trans. 2005;33:1012–1015. doi: 10.1042/BST20051012. [DOI] [PubMed] [Google Scholar]
- 6.Nilsson J, Persson B, von Heijne G. Comparative analysis of amino acid distributions in integral membrane proteins from 107 genomes. Proteins. 2005;60:606–616. doi: 10.1002/prot.20583. [DOI] [PubMed] [Google Scholar]
- 7.von Heijne G. Control of topology and mode of assembly of a polytopic membrane protein by positively charged residues. Nature. 1989;341:456–458. doi: 10.1038/341456a0. [DOI] [PubMed] [Google Scholar]
- 8.von Heijne G. The distribution of positively charged residues in bacterial inner membrane proteins correlates with the trans-membrane topology. EMBO J. 1986;5:3021–3027. doi: 10.1002/j.1460-2075.1986.tb04601.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andersson H, Bakker E, von Heijne G. Different positively charged amino acids have similar effects on the topology of a polytopic transmembrane protein in Escherichia coli. J Biol Chem. 1992;267:1491–1495. [PubMed] [Google Scholar]
- 10.Pi J, Chow H, Pittard AJ. Study of second-site suppression in the pheP gene for the phenylalanine transporter of Escherichia coli. J Bacteriol. 2002;184:5842–5847. doi: 10.1128/JB.184.21.5842-5847.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nilsson I, von Heijne G. Fine-tuning the topology of a polytopic membrane protein: role of positively and negatively charged amino acids. Cell. 1990;62:1135–1141. doi: 10.1016/0092-8674(90)90390-z. [DOI] [PubMed] [Google Scholar]
- 12.Delgado-Partin VM, Dalbey RE. The proton motive force, acting on acidic residues, promotes translocation of amino-terminal domains of membrane proteins when the hydrophobicity of the translocation signal is low. J Biol Chem. 1998;273:9927–9934. doi: 10.1074/jbc.273.16.9927. [DOI] [PubMed] [Google Scholar]
- 13.Rutz C, Rosenthal W, Schulein R. A single negatively charged residue affects the orientation of a membrane protein in the inner membrane of Escherichia coli only when it is located adjacent to a transmembrane domain. J Biol Chem. 1999;274:33757–33763. doi: 10.1074/jbc.274.47.33757. [DOI] [PubMed] [Google Scholar]
- 14.Andersson H, von Heijne G. Position-specific Asp-Lys pairing can affect signal sequence function and membrane protein topology. J Biol Chem. 1993;268:21389–21393. [PubMed] [Google Scholar]
- 15.Parks GD, Lamb RA. Role of NH2-terminal positively charged residues in estab-lishing membrane protein topology. J Biol Chem. 1993;268:19101–19109. [PubMed] [Google Scholar]
- 16.Hartmann E, Rapoport TA, Lodish HF. Predicting the orientation of eukaryotic membrane-spanning proteins. Proc Natl Acad Sci U S A. 1989;86:5786–5790. doi: 10.1073/pnas.86.15.5786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harley CA, Holt JA, Turner R, Tipper DJ. Transmembrane protein insertion orientation in yeast depends on the charge difference across transmembrane segments, their total hydrophobicity, and its distribution. J Biol Chem. 1998;273:24963–24971. doi: 10.1074/jbc.273.38.24963. [DOI] [PubMed] [Google Scholar]
- 18.Kim H, Paul S, Gennity J, Jennity J, Inouye M. Reversible topology of a bifunction-al transmembrane protein depends upon the charge balance around its transmembrane domain. Mol Microbiol. 1994;11:819–831. doi: 10.1111/j.1365-2958.1994.tb00360.x. [DOI] [PubMed] [Google Scholar]
- 19.Sato M, Hresko R, Mueckler M. Testing the charge difference hypothesis for the assembly of a eucaryotic multispanning membrane protein. J Biol Chem. 1998;273:25203–25208. doi: 10.1074/jbc.273.39.25203. [DOI] [PubMed] [Google Scholar]
- 20.Bibi E, Stearns SM, Kaback HR. The N-terminal 22 amino acid residues in the lactose permease of Escherichia coli are not obligatory for membrane insertion or transport activity. Proc Natl Acad Sci U S A. 1992;89:3180–3184. doi: 10.1073/pnas.89.8.3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ehrmann M, Beckwith J. Proper insertion of a complex membrane protein in the absence of its amino-terminal export signal. J Biol Chem. 1991;266:16530–16533. [PubMed] [Google Scholar]
- 22.Guo D, Liu J, Motlagh A, Jewell J, Miller KW. Efficient insertion of odd-numbered transmembrane segments of the tetracycline resistance protein requires even-numbered segments. J Biol Chem. 1996;271:30829–30834. doi: 10.1074/jbc.271.48.30829. [DOI] [PubMed] [Google Scholar]
- 23.Lu Y, Turnbull IR, Bragin A, Carveth K, Verkman AS, Skach WR. Reorientation of aquaporin-1 topology during maturation in the endoplasmic reticulum. Mol Biol Cell. 2000;11:2973–2985. doi: 10.1091/mbc.11.9.2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goder V, Bieri C, Spiess M. Glycosylation can influence topogenesis of membrane proteins and reveals dynamic reorientation of nascent polypeptides within the translocon. J Cell Biol. 1999;147:257–266. doi: 10.1083/jcb.147.2.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lundin M, Monne M, Widell A, Von Heijne G, Persson MA. Topology of the membrane-associated hepatitis C virus protein NS4B. J Virol. 2003;77:5428–5438. doi: 10.1128/JVI.77.9.5428-5438.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Higy M, Junne T, Spiess M. Topogenesis of membrane proteins at the endoplasmic reticulum. Biochemistry. 2004;43:12716–12722. doi: 10.1021/bi048368m. [DOI] [PubMed] [Google Scholar]
- 27.Goder V, Junne T, Spiess M. Sec61p contributes to signal sequence orientation according to the positive-inside rule. Mol Biol Cell. 2004;15:1470–1478. doi: 10.1091/mbc.E03-08-0599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Monne M, Hessa T, Thissen L, von Heijne G. Competition between neighboring topogenic signals during membrane protein insertion into the ER. FEBS J. 2005;272:28–36. doi: 10.1111/j.1742-4658.2004.04394.x. [DOI] [PubMed] [Google Scholar]
- 29.Andersson H, von Heijne G. Membrane protein topology: effects of ΔμH+ on the translocation of charged residues explain the ‘positive inside’ rule. EMBO J. 1994;13:2267–2272. doi: 10.1002/j.1460-2075.1994.tb06508.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cao G, Kuhn A, Dalbey RE. The translocation of negatively charged residues across the membrane is driven by the electrochemical potential: evidence for an electrophoresis-like membrane transfer mechanism. EMBO J. 1995;14:866–875. doi: 10.1002/j.1460-2075.1995.tb07068.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cao G, Dalbey RE. Translocation of N-terminal tails across the plasma membrane. EMBO J. 1994;13:4662–4669. doi: 10.1002/j.1460-2075.1994.tb06789.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van de Vossenberg JL, Albers SV, van der Does C, Driessen AJ, van Klompenburg W. The positive inside rule is not determined by the polarity of the delta psi (transmembrane electrical potential) Mol Microbiol. 1998;29:1125–1127. doi: 10.1046/j.1365-2958.1998.01001.x. [DOI] [PubMed] [Google Scholar]
- 33.Gallusser A, Kuhn A. Initial steps in protein membrane insertion. Bacteriophage M13 procoat protein binds to the membrane surface by electrostatic interaction. EMBO J. 1990;9:2723–2729. doi: 10.1002/j.1460-2075.1990.tb07459.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Klompenburg W, Nilsson I, von Heijne G, de Kruijff B. Anionic phospholipids are determinants of membrane protein topology. EMBO J. 1997;16:4261–4266. doi: 10.1093/emboj/16.14.4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luirink J, von Heijne G, Houben E, de Gier JW. Biogenesis of inner membrane proteins in Escherichia coli. Annu Rev Microbiol. 2005;59:329–355. doi: 10.1146/annurev.micro.59.030804.121246. [DOI] [PubMed] [Google Scholar]
- 36.Moser M, Nagamori S, Huber M, Tokuda H, Nishiyama K. Glycolipozyme MPIase is essential for topology inversion of SecG during preprotein translocation. Proc Natl Acad Sci U S A. 2013;110:9734–9739. doi: 10.1073/pnas.1303160110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goder V, Spiess M. Molecular mechanism of signal sequence orientation in the endoplasmic reticulum. EMBO J. 2003;22:3645–3653. doi: 10.1093/emboj/cdg361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Skach WR. Cellular mechanisms of membrane protein folding. Nat Struct Mol Biol. 2009;16:606–612. doi: 10.1038/nsmb.1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kida Y, Morimoto F, Sakaguchi M. Two translocating hydrophilic segments of a nascent chain span the ER membrane during multispanning protein topogenesis. J Cell Biol. 2007;179:1441–1452. doi: 10.1083/jcb.200707050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ismail N, Crawshaw SG, Cross BC, Haagsma AC, High S. Specific transmembrane segments are selectively delayed at the ER translocon during opsin biogenesis. Biochem J. 2008;411:495–506. doi: 10.1042/BJ20071597. [DOI] [PubMed] [Google Scholar]
- 41.Sadlish H, Pitonzo D, Johnson AE, Skach WR. Sequential triage of transmembrane segments by Sec61alpha during biogenesis of a native multispanning membrane protein. Nat Struct Mol Biol. 2005;12:870–878. doi: 10.1038/nsmb994. [DOI] [PubMed] [Google Scholar]
- 42.Van den Berg B, Clemons WM, Jr, Collinson I, Modis Y, Hartmann E, Harrison SC, Rapoport TA. X-ray structure of a protein-conducting channel. Nature. 2004;427:36–44. doi: 10.1038/nature02218. [DOI] [PubMed] [Google Scholar]
- 43.Sakaguchi M, Tomiyoshi R, Kuroiwa T, Mihara K, Omura T. Functions of signal and signal-anchor sequences are determined by the balance between the hydrophobic segment and the N-terminal charge. Proc Natl Acad Sci U S A. 1992;89:16–19. doi: 10.1073/pnas.89.1.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kuroiwa T, Sakaguchi M, Mihara K, Omura T. Structural requirements for interruption of protein translocation across rough endoplasmic reticulum membrane. J Biochem. 1990;108:829–834. doi: 10.1093/oxfordjournals.jbchem.a123288. [DOI] [PubMed] [Google Scholar]
- 45.Zhao G, London E. An amino acid “transmembrane tendency” scale that approaches the theoretical limit to accuracy for prediction of transmembrane helices: relationship to biological hydrophobicity. Protein Sci. 2006;15:1987–2001. doi: 10.1110/ps.062286306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gafvelin G, von Heijne G. Topological “frustration” in multispanning E. coli inner membrane proteins. Cell. 1994;77:401–412. doi: 10.1016/0092-8674(94)90155-4. [DOI] [PubMed] [Google Scholar]
- 47.Lasso G, Antoniw JF, Mullins JG. A combinatorial pattern discovery approach for the prediction of membrane dipping (re-entrant) loops. Bioinformatics. 2006;22:e290–e297. doi: 10.1093/bioinformatics/btl209. [DOI] [PubMed] [Google Scholar]
- 48.Bogdanov M, Xie J, Heacock P, Dowhan W. To flip or not to flip: lipid–protein charge interactions are a determinant of final membrane protein topology. J Cell Biol. 2008;182:925–935. doi: 10.1083/jcb.200803097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dowhan W. Molecular basis for membrane phospholipid diversity: why are there so many lipids? Annu Rev Biochem. 1997;66:199–232. doi: 10.1146/annurev.biochem.66.1.199. [DOI] [PubMed] [Google Scholar]
- 50.Dowhan W. A retrospective: use of Escherichia coli as a vehicle to study phospholipid synthesis and function. Biochim Biophys Acta. 2013;1831:471–494. doi: 10.1016/j.bbalip.2012.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bay DC, Turner RJ. Membrane composition influences the topology bias of bacterial integral membrane proteins. Biochim Biophys Acta. 2013;1828:260–270. doi: 10.1016/j.bbamem.2012.09.003. [DOI] [PubMed] [Google Scholar]
- 52.Dowhan W, Bogdanov M, Mileykovskaya E. Functional Roles of Lipids in Membranes. In: Vance DE, Vance JE, editors. Biochemistry of Lipids Lipoproteins and Membranes. Elsevier Press; Amsterdam: 2008. pp. 1–37. [Google Scholar]
- 53.Dowhan W, Bogdanov M. Molecular genetic and biochemical approaches for defining lipid-dependent membrane protein folding. Biochim Biophys Acta. 2012;1818:1097–1107. doi: 10.1016/j.bbamem.2011.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tan BK, Bogdanov M, Zhao J, Dowhan W, Raetz CR, Guan Z. Discovery of a cardiolipin synthase utilizing phosphatidylethanolamine and phosphatidylglycerol as substrates. Proc Natl Acad Sci U S A. 2012;109:16504–16509. doi: 10.1073/pnas.1212797109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shiba Y, Yokoyama Y, Aono Y, Kiuchi T, Kusaka J, Matsumoto K, Hara H. Activation of the Rcs signal transduction system is responsible for the thermosensitive growth defect of an Escherichia coli mutant lacking phosphatidylglycerol and cardiolipin. J Bacteriol. 2004;186:6526–6535. doi: 10.1128/JB.186.19.6526-6535.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mileykovskaya E, Ryan AC, Mo X, Lin CC, Khalaf KI, Dowhan W, Garrett TA. Phosphatidic acid and N-acyl phosphatidylethanolamine form membrane domains in Escherichia coli mutant lacking cardiolipin and phosphatidylglycerol. J Biol Chem. 2009;284:2990–3000. doi: 10.1074/jbc.M805189200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hawrot E, Kennedy EP. Phospholipid composition and membrane function in phosphatidylserine decarboxylase mutants of Escherichia coli. J Biol Chem. 1978;253:8213–8220. [PubMed] [Google Scholar]
- 58.DeChavigny A, Heacock PN, Dowhan W. Phosphatidylethanolamine may not be essential for the viability of Escherichia coli. J Biol Chem. 1991;266:5323–5332. [PubMed] [Google Scholar]
- 59.Wikstrom M, Xie J, Bogdanov M, Mileykovskaya E, Heacock P, Wieslander A, Dowhan W. Monoglucosyldiacylglycerol, a foreign lipid, can substitute for phosphati-dylethanolamine in essential membrane-associated functions in Escherichia coli. J Biol Chem. 2004;279:10484–10493. doi: 10.1074/jbc.M310183200. [DOI] [PubMed] [Google Scholar]
- 60.Bogdanov M, Dowhan W. Lipid-dependent generation of a dual topology for a membrane protein. J Biol Chem. 2012;287:37939–37948. doi: 10.1074/jbc.M112.404103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bogdanov M, Heacock PN, Dowhan W. A polytopic membrane protein displays a reversible topology dependent on membrane lipid composition. EMBO J. 2002;21:2107–2116. doi: 10.1093/emboj/21.9.2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bogdanov M, Heacock P, Guan Z, Dowhan W. Plasticity of lipid–protein interactions in the function and topogenesis of the membrane protein lactose permease from Escherichia coli. Proc Natl Acad Sci U S A. 2010;107:15057–15062. doi: 10.1073/pnas.1006286107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bogdanov M, Mileykovskaya E, Dowhan W. Lipids in the assembly of membrane proteins and organization of protein supercomplexes: implications for lipid-linked disorders. Subcell Biochem. 2008;49:197–239. doi: 10.1007/978-1-4020-8831-5_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xie J, Bogdanov M, Heacock P, Dowhan W. Phosphatidylethanolamine and monoglucosyldiacylglycerol are interchangeable in supporting topogenesis and function of the polytopic membrane protein lactose permease. J Biol Chem. 2006;281:19172–19178. doi: 10.1074/jbc.M602565200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bogdanov M, Heacock PN, Dowhan W. Study of polytopic membrane protein topological organization as a function of membrane lipid composition. Methods Mol Biol. 2010;619:79–101. doi: 10.1007/978-1-60327-412-8_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bogdanov M, Zhang W, Xie J, Dowhan W. Transmembrane protein topology mapping by the substituted cysteine accessibility method (SCAM™): application to lipid-specific membrane protein topogenesis. Methods. 2005;36:148–171. doi: 10.1016/j.ymeth.2004.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vitrac H, Bogdanov M, Heacock P, Dowhan W. Lipids and topological rules of membrane protein assembly: balance between long- and short-range lipid–protein interactions. J Biol Chem. 2011;286:15182–15194. doi: 10.1074/jbc.M110.214387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang W, Bogdanov M, Pi J, Pittard AJ, Dowhan W. Reversible topological organization within a polytopic membrane protein is governed by a change in membrane phospholipid composition. J Biol Chem. 2003;278:50128–50135. doi: 10.1074/jbc.M309840200. [DOI] [PubMed] [Google Scholar]
- 69.Vitrac H, Bogdanov M, Dowhan W. In vitro reconstitution of lipid-dependent dual topology and postassembly topological switching of a membrane protein. Proc Natl Acad Sci U S A. 2013;110:9338–9343. doi: 10.1073/pnas.1304375110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang X, Bogdanov M, Dowhan W. Topology of polytopic membrane protein subdomains is dictated by membrane phospholipid composition. EMBO J. 2002;21:5673–5681. doi: 10.1093/emboj/cdf571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang W, Campbell HA, King SC, Dowhan W. Phospholipids as determinants of membrane protein topology. Phosphatidylethanolamine is required for the proper topological organization of the gamma-aminobutyric acid permease (GabP) of Escherichia coli. J Biol Chem. 2005;280:26032–26038. doi: 10.1074/jbc.M504929200. [DOI] [PubMed] [Google Scholar]
- 72.Seimiya T, Ashida M, Hayashi M, Muramatsu T, Hara I. Hydrogen bond among the ionic groups of ampholytic phospholipid. Chem Phys Lipids. 1978;21:69–76. doi: 10.1016/0009-3084(78)90028-2. [DOI] [PubMed] [Google Scholar]
- 73.Wikstrom M, Kelly AA, Georgiev A, Eriksson HM, Klement MR, Bogdanov M, Dowhan W, Wieslander A. Lipid-engineered Escherichia coli membranes reveal critical lipid headgroup size for protein function. J Biol Chem. 2009;284:954–965. doi: 10.1074/jbc.M804482200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vitrac H, Bogdanov M, Dowhan W. Proper fatty acid composition rather than an ionizable lipid amine is required for full transport function of lactose permease from Escherichia coli. J Biol Chem. 2013;288:5873–5885. doi: 10.1074/jbc.M112.442988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Granseth E, von Heijne G, Elofsson A. A study of the membrane–water interface region of membrane proteins. J Mol Biol. 2005;346:377–385. doi: 10.1016/j.jmb.2004.11.036. [DOI] [PubMed] [Google Scholar]
- 76.Bogdanov M, Dowhan W. Lipid-assisted protein folding. J Biol Chem. 1999;274:36827–36830. doi: 10.1074/jbc.274.52.36827. [DOI] [PubMed] [Google Scholar]
- 77.Bogdanov M, Sun J, Kaback HR, Dowhan W. A phospholipid acts as a chaperone in assembly of a membrane transport protein. J Biol Chem. 1996;271:11615–11618. doi: 10.1074/jbc.271.20.11615. [DOI] [PubMed] [Google Scholar]
- 78.Bogdanov M, Umeda M, Dowhan W. Phospholipid-assisted refolding of an integral membrane protein. Minimum structural features for phosphatidylethanol-amine to act as a molecular chaperone. J Biol Chem. 1999;274:12339–12345. doi: 10.1074/jbc.274.18.12339. [DOI] [PubMed] [Google Scholar]
- 79.Bogdanov M, Dowhan W. Phosphatidylethanolamine is required for in vivo function of the membrane-associated lactose permease of Escherichia coli. J Biol Chem. 1995;270:732–739. doi: 10.1074/jbc.270.2.732. [DOI] [PubMed] [Google Scholar]
- 80.Sun J, Wu J, Carrasco N, Kaback HR. Identification of the epitope for monoclonal antibody 4B1 which uncouples lactose and proton translocation in the lactose permease of Escherichia coli. Biochemistry. 1996;35:990–998. doi: 10.1021/bi952166w. [DOI] [PubMed] [Google Scholar]
- 81.Frillingos S, Kaback HR. Monoclonal antibody 4B1 alters the pKa of a carboxylic acid at position 325 (helix X) of the lactose permease of Escherichia coli. Biochemistry. 1996;35:10166–10171. doi: 10.1021/bi960995r. [DOI] [PubMed] [Google Scholar]
- 82.Chen CC, Wilson TH. The phospholipid requirement for activity of the lactose carrier of Escherichia coli. J Biol Chem. 1984;259:10150–10158. [PubMed] [Google Scholar]
- 83.Seppälä S, Slusky JS, Lloris-Garcera P, Rapp M, von Heijne G. Control of membrane protein topology by a single C-terminal residue. Science. 2010;328:1698–1700. doi: 10.1126/science.1188950. [DOI] [PubMed] [Google Scholar]
- 84.Bogdanov M, Xie J, Dowhan W. Lipid–protein interactions drive membrane protein topogenesis in accordance with the positive inside rule. J Biol Chem. 2009;284:9637–9641. doi: 10.1074/jbc.R800081200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dowhan W, Bogdanov M. Lipid-dependent membrane protein topogenesis. Annu Rev Biochem. 2009;78:515–540. doi: 10.1146/annurev.biochem.77.060806.091251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gbaguidi B, Hakizimana P, Vandenbussche G, Ruysschaert JM. Conformational changes in a bacterial multidrug transporter are phosphatidylethanolamine-dependent. Cell Mol Life Sci. 2007;64:1571–1582. doi: 10.1007/s00018-007-7031-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Krishtalik LI, Cramer WA. On the physical basis for the cis-positive rule describing protein orientation in biological membranes. FEBS Lett. 1995;369:140–143. doi: 10.1016/0014-5793(95)00756-y. [DOI] [PubMed] [Google Scholar]
- 88.Rothman JE, Kennedy EP. Asymmetrical distribution of phospholipids in the membrane of Bacillus megaterium. J Mol Biol. 1977;110:603–618. doi: 10.1016/s0022-2836(77)80114-9. [DOI] [PubMed] [Google Scholar]
- 89.Tunuguntla R, Bangar M, Kim K, Stroeve P, Ajo-Franklin CM, Noy A. Lipid bilayer composition can influence the orientation of proteorhodopsin in artificial membranes. Biophys J. 2013;105:1388–1396. doi: 10.1016/j.bpj.2013.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sugai R, Takemae K, Tokuda H, Nishiyama K. Topology inversion of SecG is essential for cytosolic SecA-dependent stimulation of protein translocation. J Biol Chem. 2007;282:29540–29548. doi: 10.1074/jbc.M704716200. [DOI] [PubMed] [Google Scholar]
- 91.Zhao YJ, Lam CM, Lee HC. The membrane-bound enzyme CD38 exists in two opposing orientations. Sci Signal. 2012;5:ra67. doi: 10.1126/scisignal.2002700. [DOI] [PubMed] [Google Scholar]
- 92.Van Voorst F, De Kruijff B. Role of lipids in the translocation of proteins across membranes. Biochem J. 2000;347(Pt 3):601–612. doi: 10.1042/0264-6021:3470601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cheng HT, Megha E. London, preparation and properties of asymmetric vesicles that mimic cell membranes: effect upon lipid raft formation and transmembrane helix orientation. J Biol Chem. 2009;284:6079–6092. doi: 10.1074/jbc.M806077200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.de Lima Santos H, Lopes ML, Maggio B, Ciancaglini P. Na, K-ATPase reconstituted in liposomes: effects of lipid composition on hydrolytic activity and enzyme orientation. Colloids Surf B: Biointerfaces. 2005;41:239–248. doi: 10.1016/j.colsurfb.2004.12.013. [DOI] [PubMed] [Google Scholar]
- 95.Levy D. Membrane proteins which exhibit multiple topological orientations. Essays Biochem. 1996;31:49–60. [PubMed] [Google Scholar]
- 96.von Heijne G. Membrane-protein topology. Nat Rev Mol Cell Biol. 2006;7:909–918. doi: 10.1038/nrm2063. [DOI] [PubMed] [Google Scholar]
- 97.Schuldiner S. Controversy over EmrE structure. Science. 2007;317:748–751. doi: 10.1126/science.317.5839.748d. [DOI] [PubMed] [Google Scholar]
- 98.Bowie JU. Flip-flopping membrane proteins. Nat Struct Mol Biol. 2006;13:94–96. doi: 10.1038/nsmb0206-94. [DOI] [PubMed] [Google Scholar]
- 99.Rapp M, Granseth E, Seppälä S, von Heijne G. Identification and evolution of dual-topology membrane proteins. Nat Struct Mol Biol. 2006;13:112–116. doi: 10.1038/nsmb1057. [DOI] [PubMed] [Google Scholar]
- 100.Linton KJ, Higgins CF. P-glycoprotein misfolds in Escherichia coli: evidence against alternating-topology models of the transport cycle. Mol Membr Biol. 2002;19:51–58. doi: 10.1080/09687680110103622. [DOI] [PubMed] [Google Scholar]
- 101.Lambert C, Mann S, Prange R. Assessment of determinants affecting the dual topology of hepadnaviral large envelope proteins. J Gen Virol. 2004;85:1221–1225. doi: 10.1099/vir.0.19737-0. [DOI] [PubMed] [Google Scholar]
- 102.Lambert C, Prange R. Chaperone action in the posttranslational topological reorientation of the hepatitis B virus large envelope protein: implications for translocational regulation. Proc Natl Acad Sci U S A. 2003;100:5199–5204. doi: 10.1073/pnas.0930813100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dorobantu C, Macovei A, Lazar C, Dwek RA, Zitzmann N, Branza-Nichita N. Cholesterol depletion of hepatoma cells impairs hepatitis B virus envelopment by altering the topology of the large envelope protein. J Virol. 2011;85:13373–13383. doi: 10.1128/JVI.05423-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hickey KD, Buhr MM. Lipid bilayer composition affects transmembrane protein orientation and function. J Lipids. 2011;2011:208457. doi: 10.1155/2011/208457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Henry SA, Kohlwein SD, Carman GM. Metabolism and regulation of glycerolipids in the yeast Saccharomyces cerevisiae. Genetics. 2012;190:317–349. doi: 10.1534/genetics.111.130286. [DOI] [PMC free article] [PubMed] [Google Scholar]