Abstract
Background
Mycobacterium avium subsp. paratuberculosis (MAP) is an obligate intracellular pathogen that infects many ruminant species. The acquisition of foreign genes via horizontal gene transfer has been postulated to contribute to its pathogenesis, as these genetic elements are absent from its putative ancestor, M. avium subsp. hominissuis (MAH), an environmental organism with lesser pathogenicity. In this study, high-throughput sequencing of MAP transposon libraries were analyzed to qualitatively and quantitatively determine the contribution of individual genes to bacterial survival during infection.
Results
Out of 52384 TA dinucleotides present in the MAP K-10 genome, 12607 had a MycoMarT7 transposon in the input pool, interrupting 2443 of the 4350 genes in the MAP genome (56%). Of 96 genes situated in MAP-specific genomic islands, 82 were disrupted in the input pool, indicating that MAP-specific genomic regions are dispensable for in vitro growth (odds ratio = 0.21). Following 5 independent in vivo infections with this pool of mutants, the correlation between output pools was high for 4 of 5 (R = 0.49 to 0.61) enabling us to define genes whose disruption reproducibly reduced bacterial fitness in vivo. At three different thresholds for reduced fitness in vivo, MAP-specific genes were over-represented in the list of predicted essential genes. We also identified additional genes that were severely depleted after infection, and several of them have orthologues that are essential genes in M. tuberculosis.
Conclusions
This work indicates that the genetic elements required for the in vivo survival of MAP represent a combination of conserved mycobacterial virulence genes and MAP-specific genes acquired via horizontal gene transfer. In addition, the in vitro and in vivo essential genes identified in this study may be further characterized to offer a better understanding of MAP pathogenesis, and potentially contribute to the discovery of novel therapeutic and vaccine targets.
Electronic supplementary material
The online version of this article (doi:10.1186/1471-2164-15-415) contains supplementary material, which is available to authorized users.
Keywords: Mycobacterium avium, M. avium subsp. paratuberculosis, Transposon insertion sequencing, Horizontal gene transfer, Mycobacterial pathogenesis
Background
Mycobacterium avium subspecies paratuberculosis (MAP) is an intracellular pathogen that causes Johne’s disease, a chronic (2 to 5 years) intestinal inflammation in cattle, sheep, goats and other ruminants [1]. When MAP is shed into the environment from an infected host, its survival is finite, with no evidence of bacterial replication [2], indicating that the definitive host of MAP is the ruminant species in which it has co-evolved. In contrast, the closely-related organism, M. avium subspecies hominissuis (MAH), is considered an environmental generalist, as it can be isolated and propagated in a variety of reservoirs, including water sources and biofilms [3–5]. How MAP has evolved into a professional pathogen remains largely unknown.
In other bacterial pathogens such as Escherichia coli, Salmonella enterica, Shigella flexneri, and Yersinia enterocolitica, the transfer of DNA from one organism to another member of a different species has been shown to contribute to the emergence of virulent strains. Interestingly, in many cases the transferred DNA contains clusters of genes known as pathogenicity islands that enable the recipient strain to adapt to the host environment [6–11]. In the case of MAP, the completion of the genome sequences of MAP K10 and MAH 104 has greatly enabled the derivation of an evolutionary model for the emergence of MAP [12] along with the identification of MAP-specific genomic islands that are absent in MAH [13] (MAP: [GenBank:AE016958] [14] and revised version [GenBank: SRR060191] [15]; MAH: [GenBank:CP000479] provided by the J. Craig Venter Institute). Although horizontal gene transfer (HGT) has been detected in mycobacteria [16–18], the functional consequence of acquiring these novel genetic elements is currently unknown in this genus. In this study, we wished to examine whether MAP-specific genomic fragments contribute to the survival of MAP within the host.
Mutagenesis-mediated approaches have been employed extensively with great success for the determination of conditionally essential genes in a number of bacterial pathogens including Mycobacterium tuberculosis, Pseudomonas aeruginosa, Salmonella species, Vibrio cholerae, and Neisseria meningitidis[19–27]. Previous studies employing the transposon (Tn) mutagenesis strategy have identified MAP genes involved in metabolism and host adaptation using selected, genetically-defined mutants [28–31]. These studies indicate the feasibility of mutagenizing this organism with the purpose of conducting unbiased, genome-wide scale screens of conditionally essential genes of MAP. In this study, we have used high-throughput Illumina sequencing to characterize transposon libraries and identify genes whose disruption is deleterious for in vivo survival. In particular, we were interested in whether MAP-specific genes (i.e. genes absent from MAH strains) were over- or under-represented in genes predicted to contribute to survival in vitro or in vivo. Our data suggest that MAP-specific genes are dispensable for in vitro survival yet over-represented in genes required for MAP persistence in the mouse model. These findings present a methodology that can be readily applied to selected experimental conditions, including infection of natural mammalian hosts of MAP.
Results
Generation of M. avium subsp. paratuberculosis K-10 transposon library
Out of 52384 TA dinucleotides present in the MAP K-10 genome, 12607 were found to be targeted by the MycoMarT7 transposon in the input pool. We tallied TA positions that were aligned by ≥ 11 reads, resulting in 7784 unique disruptions. This corresponds to 2443 disrupted genes, or 56% of the 4350 genes in the MAP genome. The distribution of the mapped reads is shown in Figure 1.
Figure 1.
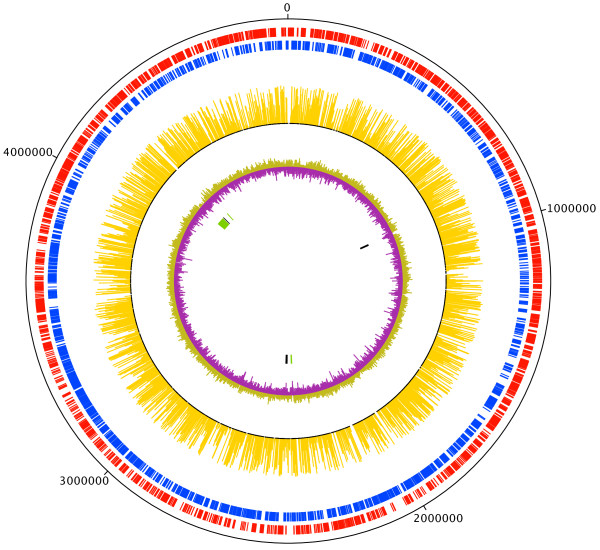
Input reads mapped onto the M. avium subsp. paratuberculosis K-10 genome. The amplitude of each peak corresponds to the number of sequence reads at a specified insertion site. The circles represent (from outer to inner): CDS on plus strand (red); CDS on minus strand (blue); transposon insertion reads (yellow);% GC plot (light brown = above average, purple = below average), and MAP-specific genomic islands in centre (LSPP4, 11, 12, 14, 15, 16 in alternating black and green blocks clockwise). CDS: coding DNA sequence.
MAP-specific genes within Large Sequence Polymorphisms (LSPPs) are dispensable for in vitro survival
Of the 96 genes situated within 6 previously defined MAP-specific genomic islands (also known as large sequence polymorphisms, LSPPs ([13]), 82 were disrupted. Based on the proportion of genes disrupted in the entire genome (56%), the expected number of surviving LSPP transposon mutants was 54. The difference between expected and observed surviving LSPP mutants was significant by chi-squared testing as indicated in Table 1. The odds ratio of LSPP disruption being associated with in vitro absence was calculated to be 0.21.
Table 1.
Summary of observed surviving LSP P mutants in the input pool
| Input | |||
|---|---|---|---|
| Observed | Survived Tn mutants | Did not grow in vitro | Total |
| LSPP | 82 | 14 | 96 |
| not LSPP | 2361 | 1893 | 4254 |
| Total | 2443 | 1907 | 4350 |
| Chi-square test | p = 5.1 × 10-9 |
The chi-squared test was used to compare the difference between the observed and the expected distribution of LSPP genes (expected values not shown). p value = 5.1 × 10-9.
Mutants with disruption in MAP-specific genes were depleted after animal infection
As described in the Methods section, we generated 5 independent sets of output:input ratios for all sites. When these ratios were compared across experiments, a correlation coefficient of ~ 0.5 to 0.6 was seen when comparing the ratios from outputs 1-4; in contrast, output 5 data revealed a much lower correlation coefficient when compared to each of the other 4 experiments (Table 2). As a result, output 5 was removed from downstream analysis. Each gene was assigned an output to input ratio, and the median of these ratios was 1.44, 1.50, 1.42, and 1.30 for the 4 output pools. To tease out genes important for in vivo fitness, we examined genes, that when disrupted, resulted in mutants with an output to input ratios less than 0.1 × average median of that experiment, reasoning that these genes would represent priority candidates for future targeted investigation. We identified a total of 415 depleted mutants at this threshold; while the expected number of depleted LSPP mutants was 14, we observed 26, demonstrating that the LSPP genes were over-represented in genes important for in vivo fitness. We tested two additional thresholds, 0.2 × median and 0.05 × median, and the results are summarized in Table 3. The odds ratio of LSPP gene disruption and in vivo depletion at 0.1 × median was 2.35. LSPP genes that were depleted in vivo at 0.1 × median are listed in Table 4. Lists of depleted genes at various median thresholds are presented in Additional file 1.
Table 2.
Correlation coefficient between each set of output:input ratio
| Ratio 1 | Ratio 2 | Ratio 3 | Ratio 4 | Ratio 5 | |
|---|---|---|---|---|---|
| Ratio 1 | n.a. | ||||
| Ratio 2 | 0.61 | n.a. | |||
| Ratio 3 | 0.54 | 0.59 | n.a. | ||
| Ratio 4 | 0.50 | 0.56 | 0.49 | n.a. | |
| Ratio 5 | 0.19 | 0.19 | 0.18 | 0.17 | n.a. |
n. a. = not applicable.
Table 3.
Summary of observed surviving LSP P mutants in the output pool
| Observed | Survived Tn | Depleted | Total |
|---|---|---|---|
| Output - 0.2 × median | |||
| LSPP | 49 | 33 | 82 |
| not LSPP | 1711 | 650 | 2361 |
| Total | 1760 | 683 | 2443 |
| Chi-square test | P = 0.01 | ||
| Output - 0.1 × median | |||
| LSPP | 56 | 26 | 82 |
| not LSPP | 1972 | 389 | 2361 |
| Total | 2028 | 415 | 2443 |
| Chi-square test | P = 0.0003 | ||
| Output - 0.05 × median | |||
| LSPP | 69 | 13 | 82 |
| not LSPP | 2137 | 224 | 2361 |
| Total | 2206 | 237 | 2443 |
| Chi-square test | P = 0.06 |
Summary of observed surviving LSPP mutants in the output pool at different thresholds compared to expected values (values not shown), and the p value of corresponding chi-square test.
Table 4.
LSP P genes depleted in vivo (as defined in [13])
| LSPP | Region description | Average ratio | Standard deviation | Predicted function |
|---|---|---|---|---|
| 4 | MAP0856c (MAPK_2912) | 0.0843 | 0.0720 | H.P. |
| 4 | MAP0862 (MAPK_2906) | 0.0189 | 0.0378 | H.P. |
| 4 | MAP0865 (MAPK_2903) | 0.0000 | 0.0000 | Cell division protein |
| 11 | MAP2148 (MAPK_1620) | 0.0653 | 0.0640 | Phage integrase |
| 11 | MAP2150 (MAPK_1618) | 0.0827 | 0.0087 | Transposase |
| 11 | MAP2154c (MAPK_1614) | 0.1038 | 0.0153 | H.P. |
| 11 | MAP2157 (MAPK_1611) | 0.1216 | 0.0910 | Transposase |
| 11 | MAP2158 (MAPK_1610) | 0.1255 | 0.0333 | H.P. |
| 12 | MAP2185c (MAPK_1583) | 0.1410 | 0.1087 | Amidohydrolase |
| 12 | MAP2194 (MAPK_1574) | 0.0787 | 0.0703 | Mce family protein |
| 14 | MAP3731c (MAPK_0037) | 0.0302 | 0.0509 | ABC transporter ATP-binding Protein |
| 14 | MAP3733c (MAPK_0035) | 0.1041 | 0.0890 | H.P. |
| 14 | MAP3735c (MAPK_0033) | 0.0557 | 0.0533 | ABC transporter ATP-binding Protein |
| 14 | MAP3741 (MAPK_0027) | 0.0039 | 0.0005 | H.P. |
| 14 | MAP3742 (MAPK_0026) | 0.0265 | 0.0178 | H.P. |
| 14 | MAP3745 (MAPK_0023) | 0.0104 | 0.0154 | H.P. |
| 14 | MAP3750 (MAPK_0018) | 0.0111 | 0.0057 | MmpS1 family protein |
| 14 | MAP3751 (MAPK_0017) | 0.0746 | 0.0970 | transmembrane transport protein, MmpL4_5 |
| 14 | MAP3757c (MAPK_0011) | 0.0923 | 0.0679 | H.P. |
| 14 | MAP3759c (MAPK_3761) | 0.0836 | 0.0106 | Tranposase |
| 14 | MAP3760c (n. a.) | 0.0400 | 0.0065 | H.P. |
| 14 | MAP3763c (MAPK_3765) | 0.0372 | 0.0267 | PapA2_3 |
| 14 | MAP3764c (MAPK_3766) | 0.1181 | 0.1928 | Pks2 |
| 15 | MAP3776c (MAPK_3778) | 0.1353 | 0.2561 | ABC transporter periplasmic solute binding protein |
| 16 | MAP3816 (MAPK_3818) | 0.0701 | 0.0757 | Phage integrase |
| 16 | MAP3817c (MAPK_3819) | 0.1251 | 0.0114 | H.P. |
LSPP genes depleted in vivo with an average ratio of less than 0.1 × median (0.153). Gene annotation by Li et al shown in second column with Wynne et al annotation in brackets. H.P. = Hypothetical protein, Mce = Mammalian cell entry, Mmp = Membrane protein, Pap = Polyketide associated protein, Pks = Polyketide synthase, n. a. = not annotated.
MAP “in vivo” essential genes and comparison with M. tuberculosis conditionally attenuated mutants
Among the 415 depleted mutants at 0.1 × median, 29 were undetectable in the output pool in all 4 mice analyzed. Compared to Tuberculist (http://tuberculist.epfl.ch/) [32] in which in vitro and in vivo M. tuberculosis essential genes are compiled [19–21], many orthologues of these MAP genes are also essential, either in vitro or in vivo, in M. tuberculosis. A list of these genes is presented in Table 5.
Table 5.
Genes absent from all 4 output pools
| Zero output | Mtb orthologue | Essentiality in H37Rv | Protein function in M. tuberculosis |
|---|---|---|---|
| MAP0298 (MAPK_3470) | Rv1129c | Essential for growth of H37Rv on cholesterol in vitro | Probable transcriptional regulator protein |
| MAP0704 (MAPK_3064) | Rv3121 | Essential for in vitro growth | Probable cytochrome P450 141 Cyp141 |
| MAP0865 (MAPK_2903) | Rv0284 | Essential for in vitro growth | ESX conserved component EccC3 ESX-3 type VII |
| MAP0908c (MAPK_2860) | Rv0966c | Non-essential for in vitro growth | Hypothetical protein |
| MAP0977 (MAPK_2791) | Rv1013 | Required for growth in C57BL/6 J mouse spleen | Putative polyketide synthase Pks16 |
| MAP1031c (MAPK_2737) | Rv2601 | Non-essential for in vitro growth | Probable spermidine synthase SpeE |
| MAP1082c (MAPK_2686) | Rv1936 | Non-essential for in vitro growth | Possible monooxygenase |
| MAP1195c (MAPK_2573) | Rv1467c | Non-essential for in vitro growth | Probable acyl-CoA dehydrogenase FadE15 |
| MAP1236c (MAPK_2532) | Rv2938 | Required for growth in C57BL/6 J mouse spleen | Probable daunorubicin-dim-transport integral membrane protein ABC transporter DrrC |
| MAP1576 (MAPK_2192) | Rv1866 | Non-essential for in vitro growth | Hypothetical protein |
| MAP1584c (MAPK_2184) | Rv2897c | Essential for in vitro growth | Hypothetical protein |
| MAP1601 (MAPK_2167) | n.a. | ||
| MAP1605c (MAPK_2163) | Rv1882c | Non-essential for in vitro growth | Probable short-chain type dehydrogenase/reductase |
| MAP1822c (MAPK_1946) | Rv3903c | Non-essential for in vitro growth | Hypothetical alanine and proline rich protein |
| MAP1835c (MAPK_1933) | Rv2110c | Essential for in vitro growth and encoded proteasome required for persistence in mice | Proteasome beta subunit PrcB |
| MAP1914 (MAPK_1854) | Rv2176 | Non-essential for in vitro growth | Probable transmembrane serine/threonine-protein kinase L PknL (protein kinase L) |
| MAP2008 (MAPK_1760) | Rv2259 | n.d. | S-nitrosomycothiol reductase MscR |
| MAP2385c (MAPK_1383) | Rv3543c | Essential for growth of H37Rv on cholesterol in vitro | Probable acyl-CoA dehydrogenase FadE29 |
| MAP2439c (MAPK_1329) | Rv1321 | Non-essential for in vitro growth | Hypothetical protein |
| MAP2582c (MAPK_1186) | Rv1866 | Non-essential for in vitro growth | Hypothetical protein |
| MAP2964c (MAPK_0804) | Rv1701 | Essential for in vitro growth | Probable integrase/recombinase |
| MAP3070 (MAPK_0698) | Rv3829c | Non-essential for in vitro growth | Probable dehydrogenase |
| MAP3131 (MAPK_0637) | Rv0450c | Essential for in vitro growth | Probable conserved transmembrane transport protein MmpL4 |
| MAP3327c (MAPK_0441) | Rv3529c | Non-essential for in vitro growth | Hypothetical protein |
| MAP3352c (MAPK_0416) | Rv1358 | n.d. | Probable transcriptional regulatory protein |
| MAP3420c (MAPK_0348) | Rv1705c | Non-essential for in vitro growth | PPE family protein PPE22 |
| MAP3699c (MAPK_0069) | Rv0249c | Required for growth in C57BL/6 J mouse spleen | Probable succinate dehydrogenase [membrane anchor subunit] |
| MAP3951c (MAPK_3953) | Rv0457c | Non-essential for in vitro growth | Probable peptidase |
| MAP4117c (MAPK_4119) | Rv0645c | Non-essential for in vitro growth | Methoxy mycolic acid synthase 1 |
M. tuberculosis orthologues are assigned based on the KEGG database (Kyoto Encyclopedia Genes and Genomes; http://www.kegg.jp/[56]) and essentiality information is found on Tuberculist. Gene annotation by Li et al shown in first column with Wynne et al annotation in brackets. n.a. = not applicable, n.d. = no data, PPE = Pro-Pro-Glu.
Discussion
To unambiguously investigate the essentiality of individual genes on a genome-wide scale, the present study generated a large transposon mutant pool (input) that was subjected to in vivo selection (output). High-throughput Illumina sequencing technology was used to determine the exact position of transposon insertion site, and the number of reads at each insertion site in the input and output pools were then analyzed to identify the gene set important for MAP survival inside a mammalian host. Our data indicate that MAP-specific genes were under-represented in genes required for survival in vitro but over-represented among those predicted to contribute to survival in vivo, with both results highly statistically significant. Furthermore, our data identified MAP genes that are conserved across other mycobacterial species whose disruption resulted in an inability to survive in vivo, potentially offering candidate genes for the generation of live, attenuated vaccines.
Of the 4350 genes in the MAP genome, 2443 (56%) genes were disrupted by the transposon, indicating that we did not achieve 100% saturation in our input pool. While some of genes are presumably essential in vitro, and cannot be disrupted, our result fell short of expectations and suggested that we had incomplete disruption coverage of the genome. Although we harvested ~ 90000 clones for the input pool, we only achieved ~ 12000 unique transposon insertion mutants. This phenomenon was likely due to a bottlenecking effect during sample preparation or sequencing stage. This sparse disruption frequency prevented us from calling essential domains within a gene with statistical confidence. To address this issue in future studies, we will generate more independent libraries to maximally saturate the number of transposon insertions, which would allow us to study not only at the gene level but also intergenic regions and domains required for optimal growth under different conditions. Nonetheless, the assessment of these 2443 genes, including 82 MAP-specific genes, provides the first comprehensive portrait of genes required for survival of MAP, in vitro and in vivo. This method can now be readily applied to defined culture conditions that are deemed representative of the life cycle of MAP or to in vivo infections of the natural host, to test whether there are host-specific essential genes in the MAP genome.
MAP-specific genes are distributed on 6 genomic islands known as large sequence polymorphisms (LSPPs) [13]. These gene clusters are absent in M. avium subsp. hominissuis (MAH), the putative ancestor of MAP and a generally non-pathogenic strain [13], thus we were particularly interested in assessing whether the presence of these genes have increased MAP’s fitness as a professional pathogen. Indeed, we observed nearly twice as many LSPP mutants to be depleted than expected after animal infection and our results indicated that these findings were not clustered to 1 island, but rather pertained to each of these 6 genome islands. Of note, different groups, using different comparison strains and technical platforms, have estimated the precise number of MAP-specific genes differently. Using the set of MAP-specific genes described by Castellanos et al.[33] which comprises 200 MAP-specific genes including the 96 LSPP genes identified by Alexander et al. [13], we observed 160 disrupted genes, and 42 were depleted after the in vivo challenge. The enrichment was higher than expected (p value = 0.0012) and the odds ratio of MAP-specific gene disruption and in vivo attenuation was 1.82 in this case, showing a similar trend as our previous analysis.
Within LSPP4, MAP0856c shares no homology with any known protein; the closest orthologue of MAP0862 is found in Acidothermus cellulolyticus, a cellolytic thermophilic actinobacterium [34]. Of particular interest, the disruption of MAP0865 led to complete absence of mutants carrying this mutation in all outputs. MAP0865 is conserved in the cell division protein FtsK in Streptomyces violaceusniger. In M. tuberculosis, ftsK (Rv2748c) is essential for in vitro growth [19, 21] and has been predicted to be involved in cell division [32].
Genes found to be depleted in LSPP11 include: MAP2148, with a phage integrase orthologue in Geodermatophilus obscurus, a bacterium often found in stressful environments [35]; MAP2150 and MAP2157, each likely encodes a transposase; MAP2154c and MAP2158 have no known function or orthologue in another organism.
Within LSPP12, MAP2185c was found to be important for in vivo growth; it shares homology with an amidohydrolase found in Frankia, a genus of bacteria that are nitrogen-fixing and often plant symbionts [36]. Another gene, MAP2194 is part of the mammalian cell entry (mce) operon. In M. tuberculosis, the mce genes are known to facilitate mycobacterial cell entry and thus virulence factors [19, 37, 38]. The mce gene clusters are predicted to function as ATP-binding cassette (ABC) transporters for cholesterol [39–41], a substrate implicated in MAP pathogenesis [42].
LSPP14 constitutes the largest MAP-specific genomic island, and contains several blocks predicted to mediate functions such as metal acquisition and synthesis of metabolic and transport proteins [13]. In this study, MAP3731c, MAP3733c, and MAP3735c were found to be depleted in the output pool. They are part of an inorganic metal uptake functional unit that spans MAP3731c to MAP3736c. Strikingly the attenuated vaccine strain 316 F has been reported to have a deletion spanning MAP3714-MAP3735c[43]; this region has therefore been independently linked to in vivo survival by both gene deletion and Tn-induced gene disruption. In addition, MAP3734c-3736c have been found to be upregulated during bovine epithelial cells and macrophages [44] while a transcriptomic study found that MAP3731-3736c were downregulated in infected bovine tissues [45]. MAP3740 to MAP3746 has been predicted to be involved in siderophore biosynthesis; disruption in MAP3741, MAP3742, MAP3745 all resulted in reduction in the output pool. As the first gene involved in mycobacterial siderophore (mycobactin) biosynthesis is truncated in MAP K-10 [14], it is of great interest to elucidate the function of this genetic element. Another set of depleted genes consisted of MAP3750 and MAP3751, encoding membrane protein MmpS1 and MmpL4. Other depleted genes include MAP3757c, a probable leucyl-tRNA synthetase, MAP3759 a transposase, MAP3760c a predicted methylase and two adjacent genes, MAP3763c, and MAP3764c, predicted to code for proteins involved in polyketide synthesis (PapA3 and Pks2 respectively) [46].
LSPP15 contains a putative metal uptake operon with a ferric uptake regulator (Fur)-like transcriptional regulator. In our study we identified disruption in MAP3776c, the first gene in this genomic region led to depletion in the output. MAP3776c encodes the solute-binding portion of an ABC transporter and is found to be downregulated in infected tissue [45]. The functional characterization of this operon is currently underway in our laboratory. Finally, LSPP16 contained two depleted genes, MAP3816 which encodes a phage integrase, and MAP3817c which encodes a protein possibly involved in thiamine biosynthesis [47].
Compared to two previous studies that screened MAP transposon libraries for attenuated mutants, we also observed depletion (below 0.2 × median) of MAP1694, MAP2231, MAP2232, MAP3963, MAP2205c, MAP3212, and MAP3607 in the output pool. Both studies used ATCC strain 19698 and different infection models (Balb/c mice and bovine kidney epithelial cells) [30, 31], and the consistency between these studies and our data suggests that these genes are very likely to be important for the survival of MAP in a mammalian host. Table 5 lists genes in which disruption resulted in complete absence in the output pool. A closer examination of these genes revealed that some of them are essential for in vitro growth in M. tuberculosis. A possible explanation is that our input pool was only passaged in rich growth medium (7H9) once, thus mutants with disruption in these essential genes were not completely eliminated but potentially growing poorly prior to infection.
To our knowledge, the present study is the first report that describes the assessment of conditionally important genes in MAP at a genome-wide scale. As MAP is a very slow-growing and fastidious microorganism, this transposon-mediated screen offers a powerful and unbiased tool for identifying the genetic basis for survival of MAP within a mammalian host. Further functional characterization of these promising candidates will undoubtedly shed light on the metabolism, genetic regulation, and virulence of MAP.
Conclusions
The present study demonstrates that MAP-specific genes are over-represented in genes required for MAP to survive in vivo, but under-represented for its growth in vitro. Our finding provides support for the notion that horizontally transferred genetic elements specific to MAP contributed to its emergence as a professional pathogen. In addition, genes identified as essential for growth of MAP in vitro and in vivo present as potential targets for therapeutic development.
Methods
Bacteria and growth conditions
Mycobacterium avium subsp. paratuberculosis K-10 was used as the parental strain for transposon mutant library construction. Bacteria were grown with rotation at 37°C in Middlebrook 7H9 medium (Difco Laboratories, Detroit, MI) containing 0.2% glycerol, 0.1% Tween 80 (Sigma-Aldrich, St. Louis, MO), 10% albumin-dextrose-catalase (Becton Dickinson and Co., Sparks, MD), and 2 μg/ml of mycobactin J (Allied Monitor, IN). Transduction mutants were selected on Middlebrook 7H10 solid medium supplemented with 10% oleic acid-albumin-dextrose-catalase (Becton Dickinson and Co., Sparks, MD) and 50 μg/ml of kanamycin.
Transposon insertion mutant library construction
Transposon library was generated as described [48]. Briefly, the MycoMarT7 phagemid was titered and amplified using M. smegmatis at 30°C. The phagemid contains the kanamycin-marked MycoMarT7 transposon that can be integrated into a TA dinucleotide site in the host DNA and has been extensively used to create high-density mutagenesis in mycobacteria [49]. Mycobacterium avium subsp. paratuberculosis at an OD600 of ~0.6 were transduced with ~3 × 109 phages in MP buffer (50 mM Tris-HCl [pH 7.6], 150 mM NaCl, 2 mM CaCl2) for 4 hours at 37°C, transferred to 7H9 medium for 24 hours with rotation at 37°C, and subsequently plated on selective 7H10 medium. Kanamycin-resistant colonies (~8.8 × 104) were evenly resuspended in 7H9 containing 25% glycerol and kanamycin, aliquoted and stored at -80°C until further use.
Animals
C57BL/6 mice were purchased from Jackson Laboratories and maintained in a pathogen-free environment at the McGill University Health Centre. All animal experiments were in compliance with the regulations of the Canadian Council of Animal care and approved by the McGill University Animal Committee. Five mice were intraperitoneally injected with 0.74 × 108 colony-forming units (CFUs) of transposon mutants. The inoculum was plated on 7H10 agar media for colony quantification as well as to study the input pool. One month after infection, the mice were sacrificed, and their spleens were aseptically removed, homogenized, and plated onto 7H10 kanaymcin plates to harvest surviving mutants (5 output pools).
Genomic library preparation
High quality genomic DNA was extracted from input and output plates as described [50]. Subsequent DNA partial digestion, ligation to asymmetric adapters, transposon junction amplification, addition of Illumina sequencing sites by nested PCR were performed according to [21]. Amplified fragments between 250 – 400 base pairs were gel-purified and sequenced with generic Illumina primer (5′ACACTCTTTCCCTACACGACGCTCTTCCGATCT) using an Illumina HiSeq2000 system at the McGill University and Génome Québec Innovation Centre, and 100 base pair reads were generated.
Sequence mapping and analysis
Transposon sequence up to the TA insertion site and regions of lower quality bases were trimmed off in all sequenced reads using a custom Python script. The sequences were aligned to the M. avium subsp. paratuberculosis K-10 reference genome [14] using Bowtie2 alignment software [51]. Reads aligned to multiple sites are assigned randomly to a mapped site. Aligned Sequence Alignment/Map (SAM) files were converted into binary BAM files using SAMtools [52]. Reads were then parsed and mapped to genomic coordinates of the TA sites using MATLAB® with custom scripts. For each TA insertion site, the number of reads detected and strand orientation were determined. Each insertion site coordinate was mapped to a protein coding gene or an intergenic region annotated in RefSeq file NC_002944.2.ptt (ftp://ftp.ncbi.nlm.nih.gov/genomes/Bacteria/Mycobacterium_avium_paratuberculosis_K_10_uid57699/NC_002944.gff). Insertion sites with ≤ 10 reads in the input pool were not considered in further analyses as we wished to test for relative depletion in the output compared to the input and needed a robust denominator as the basis for this comparison. The relative representation of each mutant after in vivo challenge was determined by calculating the ratio present in the output pool compared to the ratio present in the input pool (reads at each insertion/total reads in output divided by reads at each insertion/total reads in input). Read position was visualized by either Integrative Genomics Viewer (http://www.broadinstitute.org/igv/) [53, 54] or DNAPlotter (http://www.sanger.ac.uk/resources/software/dnaplotter/) [55] and multiple sites within a gene were then assessed together to generate estimates of essentiality as a function of genes. The output:input ratio of all disrupted insertion sites with more than 10 reads are listed in the Additional file 1. In addition, in all tables and supplemental data we also provided gene annotation generated by Li et al.[14] as well as the revised version by Wynne et al.[15] to improve accuracy as well as consistency for other researchers. Genes depleted in output pools are listed in Table 5 along with ortholog essentiality in M. tuberculosis and their putative functions [32, 56].
Electronic supplementary material
Additional file 1: Tn insertion data from input and output pools. First tab: “All data” – region description, genomic position, total reads at each site with ≥ 11 reads aligned, proportion of each site relative to sequenced library, Output:Input ratio, and median value of ratio 1-4. Second tab: “0.2 × median” – insertion sites, genes (derived from insertion sites), and LSPP genes depleted at 0.2 × median. Third tab: “0.1 × median” – insertion sites, genes (derived from insertion sites), and LSPP genes depleted at 0.1 × median. Fourth tab: “0.05 × median” – insertion sites, genes (derived from insertion sites), and LSPP genes depleted at 0.05 × median. For tabs 2-4, data were analyzed using LSPP genes determined by Alexander et al[13]. For the fifth tab: “0.1 × median Castellanos et al” – insertion sites, genes (derived from insertion sites) and MAP-specific genes depleted at 0.1 × median, data were analyzed using MAP-specific genes identified by Castellanos et al[33]. (XLSX 2 MB)
Acknowledgements
We thank Dr. Eric Rubin (Harvard University, Cambridge, MA) for MycomarT7 phagemid, Génome Québec for optimizing Illumina sequencing conditions and Dr. James Wynne (CSIRO Australian Animal Health Laboratory, Geelong, Australia) for providing resequenced MAP K-10 files to facilitate analysis. We also thank members of the Behr, Reed and Schurr labs for input and suggestions. This work was supported by an operating grant MOP-97813 from the Canadian Institutes of Health Research (CIHR) to MB. MB is Chercheur National of the FRSQ and a William Dawson Scholar of McGill University.
Footnotes
Competing interests
The authors claim no competing interests.
Authors’ contributions
MAB conceived and designed the study with JW. JW and LK performed the experiments. AM optimized and performed Illumina sequencing. JW and JRP performed the bioinformatics analyses, and MAB and JW analyzed the data. MAB and JW wrote the manuscript. All authors read and approved the final manuscript.
Contributor Information
Joyce Wang, Email: joyce.wang2@mail.mcgill.ca.
Justin R Pritchard, Email: justin.pritchard@gmail.com.
Louis Kreitmann, Email: louis.kreitmann@gmail.com.
Alexandre Montpetit, Email: alexandre.montpetit@mail.mcgill.ca.
Marcel A Behr, Email: marcel.behr@mcgill.ca.
References
- 1.Chacon O, Bermudez LE, Barletta RG. Johne’s disease, inflammatory bowel disease, and Mycobacterium paratuberculosis. Annu Rev Microbiol. 2004;58:329–363. doi: 10.1146/annurev.micro.58.030603.123726. [DOI] [PubMed] [Google Scholar]
- 2.Whittington RJ, Marshall DJ, Nicholls PJ, Marsh IB, Reddacliff LA. Survival and dormancy of Mycobacterium avium subsp. paratuberculosis in the environment. Appl Environ Microbiol. 2004;70(5):2989–3004. doi: 10.1128/AEM.70.5.2989-3004.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johansen TB, Agdestein A, Olsen I, Nilsen SF, Holstad G, Djonne B. Biofilm formation by Mycobacterium avium isolates originating from humans, swine and birds. BMC Microbiol. 2009;9:159. doi: 10.1186/1471-2180-9-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feazel LM, Baumgartner LK, Peterson KL, Frank DN, Harris JK, Pace NR. Opportunistic pathogens enriched in showerhead biofilms. Proc Natl Acad Sci U S A. 2009;106(38):16393–16399. doi: 10.1073/pnas.0908446106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Castillo-Rodal AI, Mazari-Hiriart M, Lloret-Sanchez LT, Sachman-Ruiz B, Vinuesa P, Lopez-Vidal Y. Potentially pathogenic nontuberculous mycobacteria found in aquatic systems. Analysis from a reclaimed water and water distribution system in Mexico City. Eur J Clin Microbiol Infect Dis. 2012;31(5):683–694. doi: 10.1007/s10096-011-1359-y. [DOI] [PubMed] [Google Scholar]
- 6.Ochman H, Lawrence JG, Groisman EA. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405(6784):299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- 7.Lacher DW, Steinsland H, Blank TE, Donnenberg MS, Whittam TS. Molecular evolution of typical enteropathogenic Escherichia coli: clonal analysis by multilocus sequence typing and virulence gene allelic profiling. J Bacteriol. 2007;189(2):342–350. doi: 10.1128/JB.01472-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tomljenovic-Berube AM, Henriksbo B, Porwollik S, Cooper CA, Tuinema BR, McClelland M, Coombes BK. Mapping and regulation of genes within Salmonella pathogenicity island 12 that contribute to in vivo fitness of Salmonella enterica Serovar Typhimurium. Infect Immun. 2013;81(7):2394–2404. doi: 10.1128/IAI.00067-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sasakawa C, Kamata K, Sakai T, Makino S, Yamada M, Okada N, Yoshikawa M. Virulence-associated genetic regions comprising 31 kilobases of the 230-kilobase plasmid in Shigella flexneri 2a. J Bacteriol. 1988;170(6):2480–2484. doi: 10.1128/jb.170.6.2480-2484.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu P, Elliott J, McCready P, Skowronski E, Garnes J, Kobayashi A, Brubaker RR, Garcia E. Structural organization of virulence-associated plasmids of Yersinia pestis. J Bacteriol. 1998;180(19):5192–5202. doi: 10.1128/jb.180.19.5192-5202.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Juhas M, van der Meer JR, Gaillard M, Harding RM, Hood DW, Crook DW. Genomic islands: tools of bacterial horizontal gene transfer and evolution. FEMS Microbiol Rev. 2009;33(2):376–393. doi: 10.1111/j.1574-6976.2008.00136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Turenne CY, Collins DM, Alexander DC, Behr MA. Mycobacterium avium subsp. paratuberculosis and M. avium subsp. avium are independently evolved pathogenic clones of a much broader group of M. avium organisms. J Bacteriol. 2008;190(7):2479–2487. doi: 10.1128/JB.01691-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alexander DC, Turenne CY, Behr MA. Insertion and deletion events that define the pathogen Mycobacterium avium subsp. paratuberculosis. J Bacteriol. 2009;191(3):1018–1025. doi: 10.1128/JB.01340-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li L, Bannantine JP, Zhang Q, Amonsin A, May BJ, Alt D, Banerji N, Kanjilal S, Kapur V. The complete genome sequence of Mycobacterium avium subspecies paratuberculosis. Proc Natl Acad Sci U S A. 2005;102(35):12344–12349. doi: 10.1073/pnas.0505662102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wynne JW, Seemann T, Bulach DM, Coutts SA, Talaat AM, Michalski WP. Resequencing the Mycobacterium avium subsp. paratuberculosis K10 genome: improved annotation and revised genome sequence. J Bacteriol. 2010;192(23):6319–6320. doi: 10.1128/JB.00972-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Veyrier F, Pletzer D, Turenne C, Behr MA. Phylogenetic detection of horizontal gene transfer during the step-wise genesis of Mycobacterium tuberculosis. BMC Evol Biol. 2009;9:196. doi: 10.1186/1471-2148-9-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Becq J, Churlaud C, Deschavanne P. A benchmark of parametric methods for horizontal transfers detection. PLoS One. 2010;5(4):e9989. doi: 10.1371/journal.pone.0009989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Supply P, Marceau M, Mangenot S, Roche D, Rouanet C, Khanna V, Majlessi L, Criscuolo A, Tap J, Pawlik A, Fiette L, Orgeur M, Fabre M, Parmentier C, Frigui W, Simeone R, Boritsch EC, Debrie AS, Willery E, Walker D, Quail MA, Ma L, Bouchier C, Salvignol G, Sayes F, Cascioferro A, Seemann T, Barbe V, Locht C, Gutierrez MC, et al. Genomic analysis of smooth tubercle bacilli provides insights into ancestry and pathoadaptation of Mycobacterium tuberculosis. Nat Genet. 2013;45(2):172–179. doi: 10.1038/ng.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sassetti CM, Boyd DH, Rubin EJ. Comprehensive identification of conditionally essential genes in mycobacteria. Proc Natl Acad Sci U S A. 2001;98(22):12712–12717. doi: 10.1073/pnas.231275498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rengarajan J, Bloom BR, Rubin EJ. Genome-wide requirements for Mycobacterium tuberculosis adaptation and survival in macrophages. Proc Natl Acad Sci U S A. 2005;102(23):8327–8332. doi: 10.1073/pnas.0503272102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Griffin JE, Gawronski JD, Dejesus MA, Ioerger TR, Akerley BJ, Sassetti CM. High-resolution phenotypic profiling defines genes essential for mycobacterial growth and cholesterol catabolism. PLoS Pathog. 2011;7(9):e1002251. doi: 10.1371/journal.ppat.1002251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang YJ, Reddy MC, Ioerger TR, Rothchild AC, Dartois V, Schuster BM, Trauner A, Wallis D, Galaviz S, Huttenhower C, Sacchettini JC, Behar SM, Rubin EJ. Tryptophan biosynthesis protects mycobacteria from CD4 T-cell-mediated killing. Cell. 2013;155(6):1296–1308. doi: 10.1016/j.cell.2013.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Skurnik D, Roux D, Aschard H, Cattoir V, Yoder-Himes D, Lory S, Pier GB. A comprehensive analysis of in vitro and in vivo genetic fitness of Pseudomonas aeruginosa using high-throughput sequencing of transposon libraries. PLoS Pathog. 2013;9(9):e1003582. doi: 10.1371/journal.ppat.1003582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barquist L, Langridge GC, Turner DJ, Phan MD, Turner AK, Bateman A, Parkhill J, Wain J, Gardner PP. A comparison of dense transposon insertion libraries in the Salmonella serovars Typhi and Typhimurium. Nucleic Acids Res. 2013;41(8):4549–4564. doi: 10.1093/nar/gkt148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chao MC, Pritchard JR, Zhang YJ, Rubin EJ, Livny J, Davis BM, Waldor MK. High-resolution definition of the Vibrio cholerae essential gene set with hidden Markov model-based analyses of transposon-insertion sequencing data. Nucleic Acids Res. 2013;41(19):9033–9048. doi: 10.1093/nar/gkt654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang YJ, Ioerger TR, Huttenhower C, Long JE, Sassetti CM, Sacchettini JC, Rubin EJ. Global assessment of genomic regions required for growth in Mycobacterium tuberculosis. PLoS Pathog. 2012;8(9):e1002946. doi: 10.1371/journal.ppat.1002946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mendum TA, Newcombe J, Mannan AA, Kierzek AM, McFadden J. Interrogation of global mutagenesis data with a genome scale model of Neisseria meningitidis to assess gene fitness in vitro and in sera. Genome Biol. 2011;12(12):R127. doi: 10.1186/gb-2011-12-12-r127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cavaignac SM, White SJ, de Lisle GW, Collins DM. Construction and screening of Mycobacterium paratuberculosis insertional mutant libraries. Arch Microbiol. 2000;173(3):229–231. doi: 10.1007/s002039900132. [DOI] [PubMed] [Google Scholar]
- 29.Harris NB, Feng Z, Liu X, Cirillo SL, Cirillo JD, Barletta RG. Development of a transposon mutagenesis system for Mycobacterium avium subsp. paratuberculosis. FEMS Microbiol Lett. 1999;175(1):21–26. doi: 10.1111/j.1574-6968.1999.tb13597.x. [DOI] [PubMed] [Google Scholar]
- 30.Shin SJ, Wu CW, Steinberg H, Talaat AM. Identification of novel virulence determinants in Mycobacterium paratuberculosis by screening a library of insertional mutants. Infect Immun. 2006;74(7):3825–3833. doi: 10.1128/IAI.01742-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alonso-Hearn M, Patel D, Danelishvili L, Meunier-Goddik L, Bermudez LE. The Mycobacterium avium subsp. paratuberculosis MAP3464 gene encodes an oxidoreductase involved in invasion of bovine epithelial cells through the activation of host cell Cdc42. Infect Immun. 2008;76(1):170–178. doi: 10.1128/IAI.01913-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lew JM, Kapopoulou A, Jones LM, Cole ST. TubercuList–10 years after. Tuberculosis. 2011;91(1):1–7. doi: 10.1016/j.tube.2010.09.008. [DOI] [PubMed] [Google Scholar]
- 33.Castellanos E, Aranaz A, Gould KA, Linedale R, Stevenson K, Alvarez J, Dominguez L, de Juan L, Hinds J, Bull TJ. Discovery of stable and variable differences in the Mycobacterium avium subsp. paratuberculosis type I, II, and III genomes by pan-genome microarray analysis. Appl Environ Microbiol. 2009;75(3):676–686. doi: 10.1128/AEM.01683-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barabote RD, Xie G, Leu DH, Normand P, Necsulea A, Daubin V, Médigue C, Adney WS, Xu XC, Lapidus A, Parales RE, Detter C, Pujic P, Bruce D, Lavire C, Challacombe JF, Brettin TS, Berry AM. Complete genome of the cellulolytic thermophile Acidothermus cellulolyticus 11B provides insights into its ecophysiological and evolutionary adaptations. Genome Res. 2009;19(6):1033–1043. doi: 10.1101/gr.084848.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ivanova N, Sikorski J, Jando M, Munk C, Lapidus A, Glavina Del Rio T, Copeland A, Tice H, Cheng JF, Lucas S, Chen F, Nolan M, Bruce D, Goodwin L, Pitluck S, Mavromatis K, Mikhailova N, Pati A, Chen A, Palaniappan K, Land M, Hauser L, Chang YJ, Jeffries CD, Meincke L, Brettin T, Detter JC, Rohde M, Göker M, Bristow J, et al. Complete genome sequence of Geodermatophilus obscurus type strain (G-20) Stand Genomic Sci. 2010;2(2):158–167. doi: 10.4056/sigs.711311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Benson DR, Silvester WB. Biology of Frankia strains, actinomycete symbionts of actinorhizal plants. Microbiol Rev. 1993;57(2):293–319. doi: 10.1128/mr.57.2.293-319.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arruda S, Bomfim G, Knights R, Huima-Byron T, Riley LW. Cloning of an M. tuberculosis DNA fragment associated with entry and survival inside cells. Science. 1993;261(5127):1454–1457. doi: 10.1126/science.8367727. [DOI] [PubMed] [Google Scholar]
- 38.Gioffré A, Infante E, Aguilar D, Santangelo MP, Klepp L, Amadio A, Meikle V, Etchechoury I, Romano MI, Cataldi A, Hernández RP, Bigi F. Mutation in mce operons attenuates Mycobacterium tuberculosis virulence. Microbes Infect. 2005;7(3):325–334. doi: 10.1016/j.micinf.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 39.Joshi SM, Pandey AK, Capite N, Fortune SM, Rubin EJ, Sassetti CM. Characterization of mycobacterial virulence genes through genetic interaction mapping. Proc Natl Acad Sci U S A. 2006;103(31):11760–11765. doi: 10.1073/pnas.0603179103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van der Geize R, Yam K, Heuser T, Wilbrink MH, Hara H, Anderton MC, Sim E, Dijkhuizen L, Davies JE, Mohn WW, Eltis LD. A gene cluster encoding cholesterol catabolism in a soil actinomycete provides insight into Mycobacterium tuberculosis survival in macrophages. Proc Natl Acad Sci U S A. 2007;104(6):1947–1952. doi: 10.1073/pnas.0605728104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Casali N, Riley LW. A phylogenomic analysis of the Actinomycetales mce operons. BMC Genomics. 2007;8:60. doi: 10.1186/1471-2164-8-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Keown DA, Collings DA, Keenan JI. Uptake and persistence of Mycobacterium avium subsp. paratuberculosis in human monocytes. Infect Immun. 2012;80(11):3768–3775. doi: 10.1128/IAI.00534-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bull TJ, Schock A, Sharp JM, Greene M, McKendrick IJ, Sales J, Linedale R, Stevenson K. Genomic variations associated with attenuation in Mycobacterium avium subsp. paratuberculosis vaccine strains. BMC Microbiol. 2013;13:11. doi: 10.1186/1471-2180-13-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lamont EA, Xu WW, Sreevatsan S. Host-Mycobacterium avium subsp. paratuberculosis interactome reveals a novel iron assimilation mechanism linked to nitric oxide stress during early infection. BMC Genomics. 2013;14:694. doi: 10.1186/1471-2164-14-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Janagama HK, Lamont EA, George S, Bannantine JP, Xu WW, Tu ZJ, Wells SJ, Schefers J, Sreevatsan S. Primary transcriptomes of Mycobacterium avium subsp. paratuberculosis reveal proprietary pathways in tissue and macrophages. BMC Genomics. 2010;11:561. doi: 10.1186/1471-2164-11-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hatzios SK, Schelle MW, Holsclaw CM, Behrens CR, Botyanszki Z, Lin FL, Carlson BL, Kumar P, Leary JA, Bertozzi CR. PapA3 is an acyltransferase required for polyacyltrehalose biosynthesis in Mycobacterium tuberculosis. J Biol Chem. 2009;284(19):12745–12751. doi: 10.1074/jbc.M809088200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paustian ML, Amonsin A, Kapur V, Bannantine JP. Characterization of novel coding sequences specific to Mycobacterium avium subsp. paratuberculosis: implications for diagnosis of Johne’s Disease. J Clin Microbiol. 2004;42(6):2675–2681. doi: 10.1128/JCM.42.6.2675-2681.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Murry JP, Sassetti CM, Lane JM, Xie Z, Rubin EJ. Transposon site hybridization in Mycobacterium tuberculosis. Methods Mol Biol. 2008;416:45–59. doi: 10.1007/978-1-59745-321-9_4. [DOI] [PubMed] [Google Scholar]
- 49.Rubin EJ, Akerley BJ, Novik VN, Lampe DJ, Husson RN, Mekalanos JJ. In vivo transposition of mariner-based elements in enteric bacteria and mycobacteria. Proc Natl Acad Sci U S A. 1999;96(4):1645–1650. doi: 10.1073/pnas.96.4.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Radomski N, Kreitmann L, McIntosh F, Behr MA. The critical role of DNA extraction for detection of mycobacteria in tissues. PLoS One. 2013;8(10):e78749. doi: 10.1371/journal.pone.0078749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. Integrative genomics viewer. Nat Biotechnol. 2011;29(1):24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14(2):178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Carver T, Thomson N, Bleasby A, Berriman M, Parkhill J. DNAPlotter: circular and linear interactive genome visualization. Bioinformatics. 2009;25(1):119–120. doi: 10.1093/bioinformatics/btn578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40(Database issue):D109–D114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Tn insertion data from input and output pools. First tab: “All data” – region description, genomic position, total reads at each site with ≥ 11 reads aligned, proportion of each site relative to sequenced library, Output:Input ratio, and median value of ratio 1-4. Second tab: “0.2 × median” – insertion sites, genes (derived from insertion sites), and LSPP genes depleted at 0.2 × median. Third tab: “0.1 × median” – insertion sites, genes (derived from insertion sites), and LSPP genes depleted at 0.1 × median. Fourth tab: “0.05 × median” – insertion sites, genes (derived from insertion sites), and LSPP genes depleted at 0.05 × median. For tabs 2-4, data were analyzed using LSPP genes determined by Alexander et al[13]. For the fifth tab: “0.1 × median Castellanos et al” – insertion sites, genes (derived from insertion sites) and MAP-specific genes depleted at 0.1 × median, data were analyzed using MAP-specific genes identified by Castellanos et al[33]. (XLSX 2 MB)


