Abstract
Astrocytic tumors are the most common primary brain tumors in adults. ATRX mutations have been identified in gliomas and are correlated with its loss of expression, which causes alternative lengthening of telomeres (ALT) leading to genomic instability. In this study, we aimed to explore the role of ATRX mRNA expression alteration in the progression and subclassification of astrocytic tumors and examine its impact on clinical outcome. We investigated ATRX mRNA expression and its association with IDH1 and IDH2 mutations in 169 adult astrocytic tumors using whole transcriptome sequencing. In our cohort, low ATRX mRNA expression was detected in 68% of astrocytomas, 50% of anaplastic astrocytomas and 41.6% of glioblastomas. Low ATRX expression closely overlapped with mutations in IDH1/2 (P<0.0001) in astrocytic tumors across WHO grades II–IV. Significant association between low ATRX expression and longer overall survival was identified in our cohort (P<0.01). ATRX combined with IDH1/2 and Ki-67 was used to re-classify patients with astrocytic tumors: group A1 containing IDH1/2 mutations and low ATRX expression predicted a better prognostic outcome, whereas group A3 carrying wild-type IDH1/2 and high Ki-67 expression had the shortest overall survival; IDH-mutant tumors with low ATRX expression and IDH-wild-type tumors with high Ki-67 expression were grouped into group A2. In summary, our results showed that ATRX in cooperation with IDH1/2 and Ki-67 defines three subgroups of astrocytic tumors regardless of the conventional WHO grades consensus. The molecular stratification in astrocytic tumors may aid in treatment strategy selection, therapeutic trial design, and clinical prognosis evaluation.
Keywords: ATRX, IDH1/2, Ki-67, astrocytic tumors, whole transcriptome sequencing
INTRODUCTION
Glioma is the most common type of primary brain tumor among adults [1]. Tumors are graded on a WHO consensus-derived scale of I to IV according to their degree of malignancy, as judged by various histological features accompanied by genetic alterations [2]. Astrocytic tumors, which are the most common group of human gliomas, have an inherent tendency for recurrence and malignant progression, and usually cannot be cured by neurosurgical resection, radiotherapy and chemotherapy [3]. Malignant astrocytic tumors such as glioblastoma (GBM) are the most lethal intracranial tumors [4, 5]. The prognosis of astrocytic tumors depends on certain clinical factors, most notably age at diagnosis, clinical status as measured by the Karnofsky score, and extent of tumor resection, as well as the histological type, tumor grade and several molecular markers [3].
Over the past 30 years, recurrent chromosomal, cell biological, and genetic alterations have implicated a number of molecules in the different histological types and malignancy grades of astrocytic tumors [6]. Recently, various molecular markers have been reported to correlate with an improved group of adult gliomas. Somatic mutations in isocitrate dehydrogenase 1 and 2 (IDH1/2) were detected in approximately 80 % of diffuse and anaplastic astrocytomas as well as secondary GBMs. Tumors with IDH1/2 mutations had distinctive genetic and clinical characteristics, and patients with such mutations had a better outcome than those with wild-type (WT) IDH [7, 8]. We have previously reported that high expression levels of Ki-67 protein, which is associated with the cell cycle, mitosis and cell division, predict a shorter survival time for patients with malignant gliomas [9, 10]. Although several articles have delineated the molecular classification of astrocytic tumors, a more convenient method would be available for clinical application and would arouse the interest of medical practitioners [6, 11].
However, the clinical significance of molecular parameters for the diagnostic and prognostic prediction of astrocytic tumors is still limited. Recently, mutation/loss of ATRX was identified as a potent biomarker in lower-grade gliomas and was associated with recurrent gliomas [12-14]. Loss of ATRX-DAXX (death-domain associated protein) function impairs the heterochromatic state of telomeres, leading to telomere destabilization and thereby facilitating the development of alternative lengthening of telomeres (ALT) [15]. Recent studies characterized the molecular landscape of gliomas to better understand their molecular pathogenesis and to identify molecular subgroups of these tumors and aid in their classification for clinical management [13, 16, 17]. Identification of ATRX, CIC, and FUBP1 mutations refined the prognostic information provided by the known markers IDH, TP53, and 1p/19q loss of heterogeneity (LOH) [13, 18].
In this study, we sought to detect ATRX mRNA expression alterations that would enhance our understanding of the biology of astrocytic tumors and provide novel potential markers for prognosis. To achieve our goal, we applied RNA-seq to 169 astrocytic tumor samples in which three grades of distinct ATRX mRNA expression was demonstrated [19, 20]. Our approach highlighted the power of RNA-seq technology in identifying ATRX as a prognostic marker and characterizing three subgroups of astrocytic tumors (referred to as A1, A2 and A3).
RESULTS
Decreased ATRX mRNA expression was characteristically present in low-grade astrocytomas
In our dataset, all ATRX RPKM scores were classified into two categories, with the median expression of them as cutoff point. There were 84 samples, harboring lower ATRX RPKM score than the cutoff point, in the ATRX-low group, which included 30 As, 12 AAs and 42 GBMs. The ATRX-high group was comprised of 14 As, 12 AAs and 59 GBMs which bored higher ATRX RPKM score than the cutoff point. The frequency of ATRX mRNA low expression was higher in As (30/44, 68%) than in AAs (12/24, 50%), in pGBMs (35/81, 43%) and in sGBMs (7/20, 33%) (Table 1, Fig.1; P<0.05, Chi-Square test).. Consistent with the analysis above, ATRX mRNA expression (RPKM) was different in grade II-IV astrocytic tumors (Fig.2a; P<0.05) and decreased in As than in pGBMs and sGBM (Fig.2a; P<0.05, P<0.05, respectively). These findings confirmed a strong correlation between ATRX mRNA expression and malignancy in astrocytic tumors, suggesting low ATRX mRNA expression was characteristically present in low-grade astrocytomas.
Table 1. Clinicopathological Characteristics of the patients.
| Varible | ATRX-Low (n=84) | ATRX-High (n=85) | p value | |
|---|---|---|---|---|
| Median age | 42 | 45 | ||
| Age | ≥45 | 33 | 45 | >0.05 |
| <45 | 51 | 40 | ||
| Gender | Male | 43 | 63 | <0.01 |
| Female | 41 | 22 | ||
| Preoperative KPS score | ≥80 | 43 | 42 | >0.05 |
| <80 | 41 | 43 | ||
| Grade | II A | 30 | 14 | <0.05 |
| III AA | 12 | 12 | ||
| IV GBM | 42 | 59 | ||
| pGBM | 35 | 46 | ||
| sGBM | 7 | 13 | ||
| IDH1/2 status | Mutation | 44 | 22 | <0.0001 |
| Wild type | 34 | 60 | ||
| NA | 6 | 3 | ||
| Ki-67 expression | Low | 31 | 39 | >0.05 |
| High | 14 | 31 | ||
| NA | 39 | 15 | ||
| Extent of surgery | Total | 42 | 39 | >0.05 |
| Subtotal | 31 | 37 | ||
| NA | 11 | 9 | ||
| Radiotherapy | Yes | 53 | 51 | >0.05 |
| No | 26 | 28 | ||
| NA | 5 | 6 | ||
| Chemotherapy | Yes | 45 | 46 | >0.05 |
| No | 31 | 32 | ||
| NA | 8 | 7 |
Abbreviations: A, astrocytoma; AA, anaplastic astrocytoma; KPS, Karnofsky;pGBM, primary GBM; sGBM, secondary GBM.
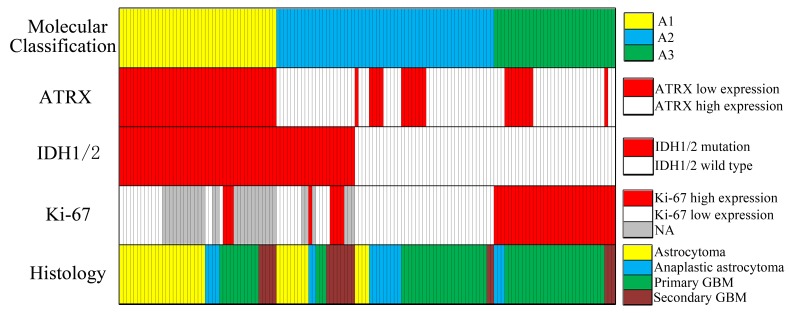
Figure 1. Overview of histology, molecular aberrations and molecular classification in the cohort (n=139).
Each column represents a patient. “Molecular Classification” is defined as A1 (IDH-mut astrocytic tumors with ATRX-low; n=44), A2 (IDH-mut tumors with ATRX-high and IDH-WT with low Ki-67 expression; n=61), A3 (IDH-WT tumors with high Ki-67 expression; n=34).
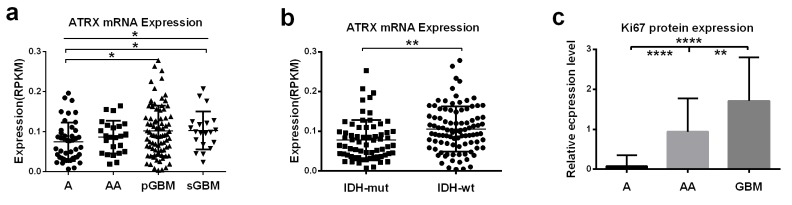
Figure 2. Correlation of ATRX mRNA expression and Ki-67 protein expression with histology and IDH1/2 mutational status.
a. ATRX mRNA expression (RPKM) was different in grade II-IV astrocytic tumors (P<0.05) and decreased in As than in pGBMs and sGBM (P<0.05, P<0.05, respectively). b. ATRX mRNA expression (RPKM) in tumors with IDH mutations was decreased compared with IDH-wild-type tumors (P<0.01). c. Ki-67 protein expression significantly differed in the three grades of astrocytic tumors (P<0.0001) and the expression levels in GBMs was significantly higher than that in astrocytomas and anaplastic astrocytomas (P<0.0001 and P<0.01, respectively). *P < 0.05, **P < 0.01, ****P < 0.0001.
ATRX mRNA expression alteration was strongly associated with IDH1/2 mutations in astrocytic tumors
To investigate the association between ATRX mRNA expression alteration and IDH1/2 mutations, we screened tumor samples in our cohort for IDH1/2 mutations using pyrosequencing. In accord with previous reports, we identified IDH1/2 mutations in 82.5% of As, 27.3% of AAs, 17.3% of pGBMs and 68.4% of sGBMs. Among the 84 tumor samples with low ATRX mRNA expression, 44 had mutations in either IDH1 or IDH2, while 34 were WT for both genes, indicating a strong association between ATRX mRNA expression and IDH1/2 mutations (Table 1, Fig.1; P<0.0001, Chi-Square test). In agreement with the analysis above, the expression of ATRX mRNA in tumors with IDH mutations was significantly decreased compared with WT IDH tumors (Fig. 2b; P<0.01).
ATRX mRNA expression alteration was a potent prognostic factor and could subclassify astrocytic tumors in combination with IDH1/2 mutations and Ki-67 expression
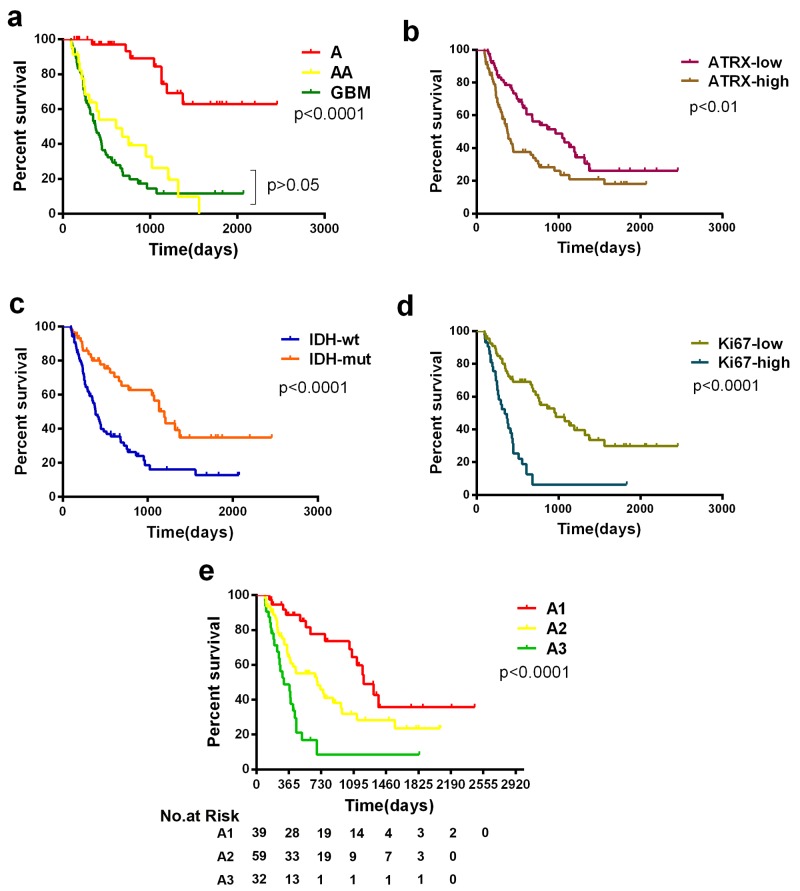
The clinical characteristics of 169 patients in our cohort were described between the two groups with low or high ATRX mRNA expression levels (Table.1). Notably, in the present study, the prognostic value of histological grading was not significant in high-grade astrocytic tumors (Table 2, Fig.3a; AAs vs. GBMs: P>0.05). However, patients in the ATRX-low group displayed significantly longer overall survival than patients in the ATRX-high group (Table 2, Fig.3b; Median OS=965 vs. 381 days; log-rank test, P<0.01). Our results also illustrated that high Ki-67 expression was dominant in WT IDH1/2 astrocytic tumors (Fig.1, P<0.05, Chi-Square test), and its relative expression level was significantly different in the three malignancy grades(Fig.2c). Therefore, to further refine the molecular classification of astrocytic tumors, we incorporated the other two prognostic markers into the model, IDH1/2 mutational status and Ki-67 protein expression, which were also associated with the clinical outcome of patients with astrocytic tumors.(Fig.3c, Fig.3d, Table 2). Based on these findings, we first classified astrocytic tumors into IDH-mut and IDH-WT tumors and then defined IDH-mut tumors with ATRX-low as A1, IDH-WT tumors with high Ki-67 expression as A3, and grouped IDH-mut tumors with ATRX-high and IDH-WT tumors with low Ki-67 expression into A2 (Fig.4). The A1 subgroup was correlated with a better clinical outcome (median OS, 4 years). In contrast, the A3 subgroup was associated with a poorer clinical outcome (median OS, 1 year). Correlation of the A2 subgroup with respect to clinical outcome fell between the A1 and A3 subgroups (median OS, 2 years). As showed in Figure 3e, survival analysis of the new classification also demonstrated a remarkable separation of the clinical course in the three molecular subgroups (log-rank test, P<0.0001). To study the influence of the three molecular markers (IDH1/2, ATRX and Ki-67) we used in the our classidication schema, multivariate Cox regression analyses were used for the adjustment of these factors (Table 3). We confirmed that IDH1/2 mutation status, ATRX mRNA expression and Ki-67 expression were three independent of each other prognostic factors in our cohort and the new classification scheme was dependent on these three factors to predict survival value. Furthermore, upon incorporation of only our classification and the WHO grading scheme (Table 3), the prognostic value of our classification was still significant, independent of the WHO grades, and served as an addition to the latter.
Table 2. Prognostic factors associated with OS in the univariate analysis for astrocytic tumors.
| Variable | Hazard ratio | 95% CI | p value | |
|---|---|---|---|---|
| Age | ≥45 vs. <45 | 1.744 | 1.146-2.635 | <0.01 |
| Gender | Male vs. Female | 0.756 | 0.486-1.177 | >0.05 |
| Preoperative KPS score | ≥80 vs. <80 | 0.169 | 0.105-0.270 | <0.0001 |
| New Classfication | A1 vs. A2 | 2.103 | 1.143-3.868 | <0.05 |
| A2 vs. A3 | 2.251 | 1.309-3.872 | <0.01 | |
| Grade | A vs. AA | 6.997 | 2.981-16.420 | <0.0001 |
| AA vs. GBM | 1.345 | 0.789-2.291 | >0.05 | |
| IDH1/2 mutation status | Mut vs. Wt | 2.552 | 1.596-4.082 | <0.0001 |
| ATRX mRNA expression | Low vs. High | 1.872 | 1.228-2.855 | <0.01 |
| Ki-67 expression | Low vs. High | 3.552 | 2.115-5.966 | <0.0001 |
| Extent of resection | Total vs. Subtotal | 1.473 | 0.943-2.301 | 0.089 |
| Radiotherapy | Yes vs. No | 1.674 | 1.067-2.626 | <0.05 |
| Chemotherapy | Yes vs. No | 0.791 | 0.504-1.240 | >0.05 |
Abbreviations: OS,overall survival; A, astrocytoma; AA, anaplastic astrocytoma; KPS, Karnofsky performance status; Mut, mutation; Wt, wild type.
Figure 3. Kaplan-Meier estimates of survival for astrocytic tumor patients.
a. Among patients with astrocytic tumors of all grades, there was no significant difference in survival between anaplastic astrocytomas and GBMs when patients were only stratified by the histopathological diagnosis of their tumors. b, c, d. ATRX, IDH1/2 status and Ki-67 are biomarkers associated with survival of patients in astrocytic tumors. e. Among all patients with astrocytic tumors (n=139, except non-classifiable), there was a significant difference in overall survival between the new three subgroups, and the A1 group and A2 group survived significantly longer (median OS, 1208 and 689 days, respectively) than the A3 group (309 days; P<0.0001).
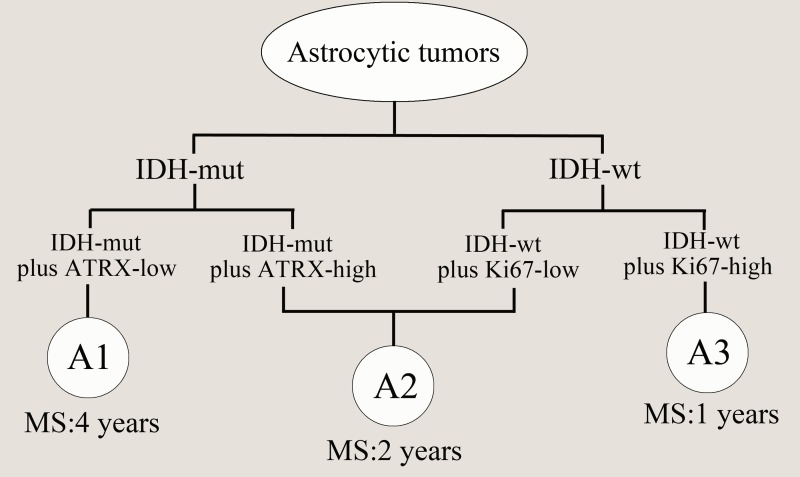
Figure 4. Model for classification of astrocytic tumors based on molecular markers.
IDH-mut tumors with ATRX-low was defined as A1 (median OS=4 years), IDH-WT tumors with high Ki-67 expression as A3 (median OS=1 years), and IDH-mut tumors with ATRX-high and IDH-WT with low Ki-67 expression were termed as A2 (median OS=2 years). MS: median OS.
Table 3. Multivariate COX regression models for overall survival.
| Variable | Hazard ratio | 95% CI | p value |
|---|---|---|---|
| COX model of Classification, IDH1/2, ATRX and Ki-67 | |||
| Classification(A1 vs. A2 vs. A3) | 0.464 | 0.193-1.115 | 0.086 |
| IDH1/2 (Mut vs. Wt) | 2.884 | 1.137-7.313 | 0.026 |
| ATRX (Low vs. High) | 2.665 | 1.386-5.123 | 0.003 |
| Ki-67 (Low vs. High) | 5.626 | 2.337-13.545 | <0.0001 |
| COX model of Classification and Grade | |||
| Classification(A1 vs. A2 vs. A3) | 1.541 | 1.078-2.204 | 0.018 |
| Grade (A vs. AA vs. GBM) | 2.095 | 1.468-2.990 | <0.0001 |
Abbreviations: A, astrocytoma; AA, anaplastic astrocytoma; Mut, mutation; Wt, wild type.
DISCUSSION
Mutations and loss of expression of alpha-thalassemia/mental retardation syndrome X-linked (ATRX) was first reported in pancreatic neuroendocrine tumors [21]. The protein encoded by ATRX plays multiple cellular roles, including chromatin remodeling at telomeres where it is required for the incorporation of the histone variant H3.3 [15]. Additionally, multiple reports have demonstrated that ATRX mutation or loss of expression results in ALT and genomic instability [13, 15, 22], and many ALT cancers harbor mutations in ATRX or DAXX genes encoding proteins that interact with each other at telomeres [15, 21].
In several previous studies, ATRX mutation was reported to be strongly correlated with its loss of expression, and may predict better prognosis in astrocytic tumors with IDH mutations [13, 15, 21, 22]. However, whether ATRX mRNA expression alteration could affect overall survival of patients with astrocytic tumors remained unclear. In our present work, we first examined ATRX mRNA expression level using whole transcriptome sequencing, because RNA-seq can add benefits for gene expression analysis such as quantitation of transcripts, improved dynamic range, and additional capabilities for detecting expressed single nucleotide variants (SNVs), translocations, and transcript isoform switches compared with microarray and immunochemistry [27]; and from the dataset of 169 astrocytic tumors we found that patients with low ATRX expression had a significantly longer median overall survival than those with high ATRX expression (965 vs. 381 days). This suggests that ATRX mRNA expression alteration plays an important role in the clinical course of astrocytic tumors. This may further promote the development of clinical substratification for astrocytic tumors based on this molecular aberration. According to Jiao's study, a large frequency of ATRX mutation and loss was reported in grade II (67%) and III astroctyomas (73%) by using exon sequencing[13]. In Wiestler's report, the frequency of ATRX loss was 45% (29/65) in anaplastic astrocytomas, detected by immunohistochemistry, which was also much lower than that in Jiao's report (AA, 73%, 32/44)[28]. Another article delineated that the frequency of ATRX alteration, tested by both exon sequencing and immunohistochemistry, was 100% (4/4) and 53% (16/30) respectively in anaplastic astrocytomas[29]. In the present study, for the first time to our knowledge, we detected ATRX mRNA expression using whole transcriptome sequencing in a large sample size. 68% of grade II astrocytomas harbored low ATRX expression, similar to the Jiao's result from exon sequencing data, whereas the incidence of low ATRX expression was only 50% in anaplastic astocytomas and 33% in secondary glioblastomas, and up to 43% of primary glioblastomas contained low ATRX expression. We thought that the difference in the above articles mainly resulted from the number of samples and the different levels detected by different testing methods, both of which could impact the frequency of ATRX alteration.
IDH1/2 mutations typically occur in low-grade gliomas and anaplastic astrocytic, oligodendroglial and mixed oligoastroglial tumors and in secondary GBMs, but rarely in primary GBMs [7, 30]. Numerous studies have identified IDH1/2 mutations as a more powerful prognostic marker in glioma patients, and found that IDH mutational status delineates molecularly and clinically distinct subclasses of gliomas[31]. In 2009, Yan's study indicated that mutations of IDH1 and IDH2 occurred in majority of several types of malignant gliomas and reported the incidence of IDH1/2 mutation in diffuse astrocytoma (90%), anaplastic astrocytoma (73%) primary GBM (5%) and secondary GBM (85%). In the present study, the frequency of IDH1/2 gene mutations in astrocytoma, anaplastic astrocytoma, primary glioblastoma and secondary glioblastoma was 82.5% (33/40), 27.3% (6/22), 17.3% (14/81) and 68.4% (13/19), respectively. Our team previous reports delineated that the incidence of IDH1/2 mutation in Chinese population with anaplastic astrocytomas and primary glioblastoma was separately 42.9% (24/56) and 16.1% (19/118) [32, 33] and the reasons why there was somewhat difference in the frequency of IDH mutations from the result of American and European reports might be explained as follows: (1) there was a much larger sample size in our study which could reveal much detailed overview of IDH1/2 gene mutation in anaplastic astrocytomas; (2) the distinct proportion of histopathological subtypes; (3) the ethnic differences[7, 32-34]And recent articles have demonstrated that the ATRX, IDH1/2 and TP53 mutant phenotype is important in the early development and progression of astrocytic tumors [12, 13, 25, 28]. In our study, we also showed that decreased ATRX mRNA expression was characteristically enriched in low-grade astrocytomas and this ATRX alteration significantly overlapped with IDH1/2 mutations (P<0.0001). Interestingly, the relationship between high Ki-67 expression levels and WT IDH was further confirmed in this study. Our previous reports showed that IDH1 mutations were associated with low Ki-67 expression in primary GBMs [32], and that Ki-67 protein expression is an independent prognostic marker in GBM patients[10]. In contrast with IDH1/2 mutations and decreased ATRX expression being widely considered as key aberrations in the early stage of astrocytic tumors [7, 35], increasing Ki-67 expression may be the final event in the progression of these tumors [9, 10, 33].
Thus, we speculated that IDH1/2 mutations accompanied by ATRX or Ki-67 may represent a distinct biological process during the development of astrocytic tumors from the original tumor cells. Excitingly, regardless of histological grading, our new molecular classification model on the basis of IDH1/2 mutational status and ATRX/Ki-67 expression could exactly reflect the biological properties of the three subgroups of astrocytic tumors with distinct clinical prognosis. A1 tumors are clearly defined with low ATRX expresion, and IDH-mut, likely representing low grade II astroctyomas with the best prognosis. A3 tumors mainly harbor IDH-wt, high Ki-67 expression with the poorest prognosis, like glioblastoma. A2 subgroup included patients with or without IDH1/2 mutations, and also included cases with low and high ATRX expression. This phenomenon could be explained by tumor heterogeneity within this subgroup. As we all know, malignant gilomas are hardly composed of a single homogeneous population, but rather by a heterogeneous ensemble of cells that differ in many biological features, such as morphology, proliferation rate, invasive behavior, metastatic potential, drug resistance, gene expression and genetic abnormalities[36, 37]. The molecular basis of heterogeneity in gliomas was evidenced by previous studies that found markedly different genetic instability phenotypes from clinical specimens, and even intratumoral samples [17, 38-40]. A growing number of genetic aberrations and their frequency were identified in different types of human gliomas, some of which also displayed outstanding significance in molecular subclassification. Primary glioblastomas are characterized by EGFR amplification, PTEN mutation and absence of IDH1/2 mutations, while secondary glioblastomas are characterized by TP53 mutations, IDH1/2 mutations and lack of EGFR amplification[41]. However, we didn't introduce these markers into our analyses because of the much lower frequency of EGFR amplification or PTEN mutation in our dataset than that from Western researches. Co-deletion of 1p/19q was recently shown to be associated with CIC and FUBP1 mutations, mainly seen in oligodendroglioma and mutually exclusive with ATRX mutation [13, 42]. Integrating IDH–CIC–FUBP1 has served as a good definition to characterize oligodendroglioma[13]. Thus, in our work, we analyzed neither LOH of 1p/19q nor the mutational status of CIC/FUBP1 in astrocytomas. In 2013, TERT promoter mutations were reported in glioma, and the frequency of these mutations was particularly high among primary glioblastomas (65/78, 83%) and pure oligodendroglial tumors (35/45, 78%), while relatively low in astrocytomas, including grade II astrocytomas, grade III astrocytomas and secondary GBM (4/40, 11%), suggesting that primary GBMs with IDH1/2- wt and TERT promoter mutation could be a heterogenous subtype [23, 24, 26]. However, there was still controversies in the prognostic value of TERT promoter mutation[43]. This is the critical reason why we defined astrocytic tumors using IDH1/2 mutational status plus ATRX or Ki67 expression, a common proliferation marker in clinical practice [10]. Undoubtedly, the introduction of additional genetic markers may help clear up some of the heterogeneity in A2 subgroup. As mentioned above, we still need to take advantage of classical and emerging biomarker to interpret the heterogeneity so that the molecular classification can further improve, providing new clue for prognosis, treatment selection.In conclusion, our results revealed that ATRX mRNA expression alteration plays a critical role in astrocytic tumors and a new molecular stratification can serve as a useful addition to conventional glioma classification. Compared with Jiao's classification[13], our scheme further classified three subtypes (referred as A1, A2 and A3) of I-A and I-X gliomas. A1 tumor was similar to I-A glioma, and A3 tumor was a branch of I-X glioma; A2 tumor was the transformation stage between A1 and A3. Anyway, new molecular stratification above could serve as a useful addition to conventional glioma classification.
METHODS
Clinical characteristics of samples
One hundred sixty-nine samples from the Chinese Glioma Genome Atlas (CGGA) were included in our study, including 44 astrocytomas (WHO II, As), 24 anaplastic astrocytomas (WHO III, AAs), 81 primary glioblastomas (WHO IV, pGBMs) and 20 secondary glioblastomas (WHO IV, sGBMs). All of the patients (age range: 18–81 years) underwent surgical resection. Patients were eligible for the study if their diagnosis was established histologically by two neuropathologists according to the 2007 WHO classification guidelines [2]. Tumor tissue samples were obtained by surgical resection. All patients provided written informed consent, and the study was approved by the ethics committees of the participating hospitals. Survival data were collected by clinics during patient visits and/or phone interviews. Patients who underwent biopsy alone were not followed up at our center and were therefore excluded from the survival analysis.
Whole transcriptome sequencing
Total RNA was isolated using a RNeasy Mini Kit (Qiagen) according to the manufacturer's instructions. A pestle and a QIAshredder (Qiagen) were used to disrupt and homogenize frozen tissue. RNA intensity was checked using 2100 Bioanalyzer (Agilent Technologies) and only high quality samples with an RNA Integrity Number (RIN) value greater than or equal to 7.0 were used to construct the sequencing library. The subsequent steps included end repair, adapter ligation, size selection and polymerase chain reaction enrichment. The length of DNA fragment was measured using a 2100 Bioanalyzer, with median insert sizes of 200 nucleotides. The libraries were sequenced on the Illumina HiSeq 2000 platform using the 101-bp pair-end sequencing strategy. Short sequence reads were aligned to the human reference genome (Hg 19 Refseq) using the Burrows-Wheeler Aligner (BWA, Version 0.6.2-r126) [44].
Molecular analyses
IDH mutations. Genomic DNA was extracted from frozen tissues with a QIAamp DNA Mini Kit (Qiagen) according to the manufacturer's protocol. DNA concentration and quality were measured using a Nano-Drop ND-1000 spectrophotometer (NanoDrop Technologies, Houston, TX). Pyrosequencing of IDH1/2 mutations was supported by Gene-tech (Shanghai, China) and performed on a Pyro-Mark Q96 ID System (Qiagen, Valencia, Calif). The primers 5'-GCTTGTGAGTGGATGGGTAAAAC-3', 5'-Biotin-TTGCC AACATGACTTACTTGATC- 3' for IDH1 and 5'-ATCCTGGGGGGGACTGTCTT-3', 5'- Biotin-CTCTCCACCCTGGCCT ACCT-3' for IDH2 were used for PCR amplification, and the primers 5'-TG GA TGGGT AAAACCT-3' for IDH1 and 5'-AGCCCATCACCATTG-3' for IDH2 were used for pyrosequencing [32, 33].
Ki-67 protein expression. Immunohistochemistry was performed to detect Ki-67 protein expression according to the manufacturer's protocol. Anti-Ki-67 antibody (Santa Cruz Biotechnology, Santa Cruz, CA) was used at a dilution of 1:100. Each slide stained was individually reviewed and scored by two independent neuropathologists. Staining was scored using a 4-point scale from 0 to 3, with 0=no or rare occurrence of staining, 1 = 10% of cells positively stained, 2 = 10–30 % of cells positively stained, 3 = >30 % of cells positively stained. Score 2 and 3 were defined as strong nuclear staining in at least 10 % of the tumor cells. Score 0 and 1 were defined as positive staining of <30 % of cells. Controls without primary antibody and positive control tissues were included in all experiments to ensure the quality of the staining [9, 34].
Statistical analysis
RNA sequencing data was downloaded from CGGA [1, 45]. Gene expression was calculated using the RPKM method (reads per kilobase transcriptome per million reads) [46, 47]. The RPKM method is able to remove the influence of varying gene lengths and sequencing discrepancies from the calculation of gene expression. Therefore, the calculated gene expression can be directly used to compare the differences in gene expression among samples. We defined the median value of ATRX mRNA expression RPKM score as the cutoff point in distinguishing the expression level of each sample. Certain a sample, harboring higher ATRX RPKM score than the cutoff point, fell into ATRX-high, and in contrast, certain a sample with lower ATRX RPKM than the cutoff was termed as ATRX-low.
Overall survival was estimated from the date of diagnosis to the date of either death or last follow-up. Patients were censored at the time they were last known to be alive. Overall survival was assessed using the Kaplan–Meier method and the log-rank test was used for comparison between groups. Student's t-test and analysis of variance (ANOVA) were used to determine significant differences in GraphPad Prism Version 6.01. Clinical and pathological characteristics between cohorts were compared using the Chi-Square test in SPSS, version 13.0, software for Windows (SPSS). All differences were considered statistically significant at P<0.05.
Acknowledgments
We thank Yuling Yang for tissue sample collection and clinical data retrieval. This work was supported by the following grants: 1. National High Technology Research and Development Program (No. 2012AA02A508); 2. International Science and Technology Cooperation Program (No.2012DFA30470); 3. National Natural Science Foundation of China (No. 91229121, No. 81201993); 4. Project of Ministry of Health P. R. China (No.w2011bx059); 5. National 973 program (No.2011CB707804).
Reference
- 1.Yan W, Zhang W, You G, Zhang JX, Han L, Bao ZS, Wang YZ, Liu YW, Jiang CL, Kang CS, You YP, Jiang T. Molecular classification of gliomas based on whole genome gene expression: a systematic report of 225 samples from the Chinese Glioma Cooperative Group. Neuro-Oncology. 2012;14(12):1432–1440. doi: 10.1093/neuonc/nos263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathologica. 2007;114(2):97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reifenberger G, Collins VP. Pathology and molecular genetics of astrocytic gliomas. Journal of molecular medicine. 2004;82(10):656–670. doi: 10.1007/s00109-004-0564-x. [DOI] [PubMed] [Google Scholar]
- 4.Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A, Hahn WC, Ligon KL, Louis DN, Brennan C, Chin L, DePinho RA, Cavenee WK. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007;21(21):2683–2710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- 5.Ohgaki H, Dessen P, Jourde B, Horstmann S, Nishikawa T, Di Patre PL, Burkhard C, Schuler D, Probst-Hensch NM, Maiorka PC, Baeza N, Pisani P, Yonekawa Y, Yasargil MG, Lutolf UM, Kleihues P. Genetic pathways to glioblastoma: a population-based study. Cancer Res. 2004;64(19):6892–6899. doi: 10.1158/0008-5472.CAN-04-1337. [DOI] [PubMed] [Google Scholar]
- 6.Gorovets D, Kannan K, Shen R, Kastenhuber ER, Islamdoust N, Campos C, Pentsova E, Heguy A, Jhanwar SC, Mellinghoff IK, Chan TA, Huse JT. IDH mutation and neuroglial developmental features define clinically distinct subclasses of lower grade diffuse astrocytic glioma. Clinical cancer research : an official journal of the American Association for Cancer Research. 2012;18(9):2490–2501. doi: 10.1158/1078-0432.CCR-11-2977. [DOI] [PubMed] [Google Scholar]
- 7.Yan H, Parsons DW, Jin GL, McLendon R, Rasheed BA, Yuan WS, Kos I, Batinic-Haberle I, Jones S, Riggins GJ, Friedman H, Friedman A, Reardon D, Herndon J, Kinzler KW, Velculescu VE, et al. IDH1 and IDH2 Mutations in Gliomas. New England Journal of Medicine. 2009;360(8):765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wen PY, Kesari S. Malignant gliomas in adults. New England Journal of Medicine. 2008;359(5):492–507. doi: 10.1056/NEJMra0708126. [DOI] [PubMed] [Google Scholar]
- 9.Liu Y, Tang K, Yan W, Wang Y, You G, Kang C, Jiang T, Zhang W. Identifying Ki-67 specific miRNA-mRNA interactions in malignant astrocytomas. Neurosci Lett. 2013 doi: 10.1016/j.neulet.2013.04.030. [DOI] [PubMed] [Google Scholar]
- 10.Jin Q, Zhang W, Qiu XG, Yan W, You G, Liu YW, Jiang T, Wang L. Gene expression profiling reveals Ki-67 associated proliferation signature in human glioblastoma. Chinese medical journal. 2011;124(17):2584–2588. [PubMed] [Google Scholar]
- 11.Toedt G, Barbus S, Wolter M, Felsberg J, Tews B, Blond F, Sabel MC, Hofmann S, Becker N, Hartmann C, Ohgaki H, von Deimling A, Wiestler OD, Hahn M, Lichter P, Reifenberger G, et al. Molecular signatures classify astrocytic gliomas by IDH1 mutation status. International journal of cancer Journal international du cancer. 2011;128(5):1095–1103. doi: 10.1002/ijc.25448. [DOI] [PubMed] [Google Scholar]
- 12.Kannan K, Inagaki A, Silber J, Gorovets D, Zhang J, Kastenhuber ER, Heguy A, Petrini JH, Chan TA, Huse JT. Whole-exome sequencing identifies ATRX mutation as a key molecular determinant in lower-grade glioma. Oncotarget. 2012;3(10):1194–1203. doi: 10.18632/oncotarget.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiao Y, Killela PJ, Reitman ZJ, Rasheed AB, Heaphy CM, de Wilde RF, Rodriguez FJ, Rosemberg S, Oba-Shinjo SM, Nagahashi Marie SK, Bettegowda C, Agrawal N, Lipp E, Pirozzi C, Lopez G, He Y, et al. Frequent ATRX, CIC, FUBP1 and IDH1 mutations refine the classification of malignant gliomas. Oncotarget. 2012;3(7):709–722. doi: 10.18632/oncotarget.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.E B. Mutational Analysis Reveals the Origin and Therapy-Driven Evolution of Recurrent Glioma. Science. 2013 doi: 10.1126/science.1239947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heaphy CM, de Wilde RF, Jiao Y, Klein AP, Edil BH, Shi C, Bettegowda C, Rodriguez FJ, Eberhart CG, Hebbar S, Offerhaus GJ, McLendon R, Rasheed BA, He Y, Yan H, Bigner DD, et al. Altered telomeres in tumors with ATRX and DAXX mutations. Science. 2011;333(6041):425. doi: 10.1126/science.1207313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, Alexe G, Lawrence M, O'Kelly M, Tamayo P, Weir BA, Gabriel S, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brennan CW, Verhaak RG, McKenna A, Campos B, Noushmehr H, Salama SR, Zheng S, Chakravarty D, Sanborn JZ, Berman SH, Beroukhim R, Bernard B, Wu CJ, Genovese G, Shmulevich I, Barnholtz-Sloan J, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155(2):462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nobusawa S, Watanabe T, Kleihues P, Ohgaki H. IDH1 Mutations as Molecular Signature and Predictive Factor of Secondary Glioblastomas. Clinical Cancer Research. 2009;15(19):6002–6007. doi: 10.1158/1078-0432.CCR-09-0715. [DOI] [PubMed] [Google Scholar]
- 19.Kridel R, Meissner B, Rogic S, Boyle M, Telenius A, Woolcock B, Gunawardana J, Jenkins C, Cochrane C, Ben-Neriah S, Tan K, Morin RD, Opat S, Sehn LH, Connors JM, Marra MA, et al. Whole transcriptome sequencing reveals recurrent NOTCH1 mutations in mantle cell lymphoma. Blood. 2012;119(9):1963–1971. doi: 10.1182/blood-2011-11-391474. [DOI] [PubMed] [Google Scholar]
- 20.Greif PA, Eck SH, Konstandin NP, Benet-Pages A, Ksienzyk B, Dufour A, Vetter AT, Popp HD, Lorenz-Depiereux B, Meitinger T, Bohlander SK, Strom TM. Identification of recurring tumor-specific somatic mutations in acute myeloid leukemia by transcriptome sequencing. Leukemia. 2011;25(5):821–827. doi: 10.1038/leu.2011.19. [DOI] [PubMed] [Google Scholar]
- 21.Jiao Y, Shi C, Edil BH, de Wilde RF, Klimstra DS, Maitra A, Schulick RD, Tang LH, Wolfgang CL, Choti MA, Velculescu VE, Diaz LA, Jr, Vogelstein B, Kinzler KW, Hruban RH, Papadopoulos N. DAXX/ATRX, MEN1, and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumors. Science. 2011;331(6021):1199–1203. doi: 10.1126/science.1200609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schwartzentruber J, Korshunov A, Liu XY, Jones DT, Pfaff E, Jacob K, Sturm D, Fontebasso AM, Quang DA, Tonjes M, Hovestadt V, Albrecht S, Kool M, Nantel A, Konermann C, Lindroth A, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482(7384):226–231. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- 23.Killela PJ, Reitman ZJ, Jiao YC, Bettegowda C, Agrawal N, Diaz LA, Friedman AH, Friedman H, Gallia GL, Giovanella BC, Grollman AP, He TC, He YP, Hruban RH, Jallo GI, Mandahl N, et al. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(15):6021–6026. doi: 10.1073/pnas.1303607110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horn S, Figl A, Rachakonda PS, Fischer C, Sucker A, Gast A, Kadel S, Moll I, Nagore E, Hemminki K, Schadendorf D, Kumar R. TERT promoter mutations in familial and sporadic melanoma. Science. 2013;339(6122):959–961. doi: 10.1126/science.1230062. [DOI] [PubMed] [Google Scholar]
- 25.Liu XY, Gerges N, Korshunov A, Sabha N, Khuong-Quang DA, Fontebasso AM, Fleming A, Hadjadj D, Schwartzentruber J, Majewski J, Dong Z, Siegel P, Albrecht S, Croul S, Jones DT, Kool M, et al. Frequent ATRX mutations and loss of expression in adult diffuse astrocytic tumors carrying IDH1/IDH2 and TP53 mutations. Acta Neuropathol. 2012;124(5):615–625. doi: 10.1007/s00401-012-1031-3. [DOI] [PubMed] [Google Scholar]
- 26.Arita H, Narita Y, Fukushima S, Tateishi K, Matsushita Y, Yoshida A, Miyakita Y, Ohno M, Collins VP, Kawahara N, Shibui S, Ichimura K. Upregulating mutations in the TERT promoter commonly occur in adult malignant gliomas and are strongly associated with total 1p19q loss. Acta Neuropathologica. 2013;126(2):267–276. doi: 10.1007/s00401-013-1141-6. [DOI] [PubMed] [Google Scholar]
- 27.Ozsolak F, Milos PM. RNA sequencing: advances, challenges and opportunities. Nature Reviews Genetics. 2011;12(2):87–98. doi: 10.1038/nrg2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wiestler B, Capper D, Holland-Letz T, Korshunov A, von Deimling A, Pfister SM, Platten M, Weller M, Wick W. ATRX loss refines the classification of anaplastic gliomas and identifies a subgroup of IDH mutant astrocytic tumors with better prognosis. Acta Neuropathol. 2013;126(3):443–451. doi: 10.1007/s00401-013-1156-z. [DOI] [PubMed] [Google Scholar]
- 29.Liu XY, Gerges N, Korshunov A, Sabha N, Khuong-Quang DA, Fontebasso AM, Fleming A, Hadjadj D, Schwartzentruber J, Majewski J, Dong ZF, Siegel P, Albrecht S, Croul S, Jones DTW, Kool M, et al. Frequent ATRX mutations and loss of expression in adult diffuse astrocytic tumors carrying IDH1/IDH2 and TP53 mutations. Acta Neuropathologica. 2012;124(5):615–625. doi: 10.1007/s00401-012-1031-3. [DOI] [PubMed] [Google Scholar]
- 30.Balss J, Meyer J, Mueller W, Korshunov A, Hartmann C, von Deimling A. Analysis of the IDH1 codon 132 mutation in brain tumors. Acta Neuropathol. 2008;116(6):597–602. doi: 10.1007/s00401-008-0455-2. [DOI] [PubMed] [Google Scholar]
- 31.Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E, Campos C, Fabius AW, Lu C, Ward PS, Thompson CB, Kaufman A, Guryanova O, Levine R, Heguy A, Viale A, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483(7390):479–483. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yan W, Zhang W, You G, Bao ZS, Wang YZ, Liu YW, Kang CS, You YP, Wang L, Jiang T. Correlation of IDH1 Mutation with Clinicopathologic Factors and Prognosis in Primary Glioblastoma: A Report of 118 Patients from China. Plos One. 2012;7(1) doi: 10.1371/journal.pone.0030339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang CB, Bao ZS, Wang HJ, Yan W, Liu YW, Li MY, Zhang W, Chen L, Jiang T. Correlation of IDH1/2 mutation with clinicopathologic factors and prognosis in anaplastic gliomas: a report of 203 patients from China. J Cancer Res Clin Oncol. 2013 doi: 10.1007/s00432-013-1519-9. [DOI] [PubMed] [Google Scholar]
- 34.Yang P, Wang Y, Peng X, You G, Zhang W, Yan W, Bao Z, Wang Y, Qiu X, Jiang T. Management and survival rates in patients with glioma in China (2004-2010): a retrospective study from a single-institution. J Neurooncol. 2013 doi: 10.1007/s11060-013-1103-9. [DOI] [PubMed] [Google Scholar]
- 35.Goze C, Mansour L, Rigau V, Duffau H. Distinct IDH1/IDH2 mutation profiles in purely insular versus paralimbic WHO Grade II gliomas Laboratory investigation. Journal of neurosurgery. 2013;118(4):866–872. doi: 10.3171/2012.11.JNS121100. [DOI] [PubMed] [Google Scholar]
- 36.Bonavia R, Inda MM, Cavenee WK, Furnari FB. Heterogeneity maintenance in glioblastoma: a social network. Cancer Res. 2011;71(12):4055–4060. doi: 10.1158/0008-5472.CAN-11-0153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Olar A, Aldape KD. Using the molecular classification of glioblastoma to inform personalized treatment. The Journal of pathology. 2014;232(2):165–177. doi: 10.1002/path.4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shapiro JR, Yung WK, Shapiro WR. Isolation, karyotype, and clonal growth of heterogeneous subpopulations of human malignant gliomas. Cancer Res. 1981;41(6):2349–2359. [PubMed] [Google Scholar]
- 39.Sottoriva A, Spiteri I, Piccirillo SG, Touloumis A, Collins VP, Marioni JC, Curtis C, Watts C, Tavare S. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc Natl Acad Sci U S A. 2013;110(10):4009–4014. doi: 10.1073/pnas.1219747110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chin L. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parsons DW, Jones S, Zhang XS, Lin JCH, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, Olivi A, McLendon R, Rasheed BA, Keir S, Nikolskaya T, Nikolsky Y, et al. An integrated genomic analysis of human glioblastoma Multiforme. Science. 2008;321(5897):1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bettegowda C, Agrawal N, Jiao Y, Sausen M, Wood LD, Hruban RH, Rodriguez FJ, Cahill DP, McLendon R, Riggins G, Velculescu VE, Oba-Shinjo SM, Marie SK, Vogelstein B, Bigner D, Yan H, et al. Mutations in CIC and FUBP1 contribute to human oligodendroglioma. Science. 2011;333(6048):1453–1455. doi: 10.1126/science.1210557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nonoguchi N, Ohta T, Oh JE, Kim YH, Kleihues P, Ohgaki H. TERT promoter mutations in primary and secondary glioblastomas. Acta Neuropathol. 2013 doi: 10.1007/s00401-013-1163-0. [DOI] [PubMed] [Google Scholar]
- 44.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang W, Zhang J, Hoadley K, Kushwaha D, Ramakrishnan V, Li S, Kang C, You Y, Jiang C, Song SW, Jiang T, Chen CC. miR-181d: a predictive glioblastoma biomarker that downregulates MGMT expression. Neuro Oncol. 2012;14(6):712–719. doi: 10.1093/neuonc/nos089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5(7):621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 47.Audic S, Claverie JM. The significance of digital gene expression profiles. Genome research. 1997;7(10):986–995. doi: 10.1101/gr.7.10.986. [DOI] [PubMed] [Google Scholar]