Summary
Interchromosomal associations can regulate gene expression but little is known about the molecular basis of such associations. In response to antigen stimulation, naïve T cells can differentiate into Th1, Th2 and Th17 cells expressing IFN-γ, IL-4 and IL-17, respectively. We previously reported that in naïve T cells, the IFN-γ locus is associated with the Th2 cytokine locus. Here we show that the Th2 locus additionally associates with the IL-17 locus. This association requires a DNase I hypersensitive region (RHS6) at the Th2 locus. RHS6 and the IL-17 promoter both bear Oct-1 binding sites. Deletion of either of these sites or Oct-1 gene impairs the association. Oct-1 and CTCF bind their cognate sites cooperatively and CTCF-deficiency similarly impairs the association. Finally, defects in the association lead to enhanced IL-17 induction. Collectively, our data indicate Th17 lineage differentiation is restrained by the Th2 locus via interchromosomal associations organized by Oct-1 and CTCF.
Introduction
Naïve CD4+ T cells can differentiate into several lineages of T helper (Th) cells, including Th1, Th2 and Th17 cells, each defined by production of unique effector cytokines and expression of different master transcription factors (Kanno et al., 2012; Zhu et al., 2010). Th1 cells express interferon-γ (IFN-γ) and the transcription factor T-bet in response to intracellular pathogens (Kanno et al., 2012; Zhu et al., 2010). Th2 cells express interleukin-4 (IL-4), IL-5 and IL-13 and the transcription factor Gata-3 and function to eliminate helminthic infections (Kanno et al., 2012; Zhu et al., 2010). Th17 cells express IL-17A and IL-17F and the transcription factor RORγt and protect against extracellular bacteria and fungi (Kanno et al., 2012; Korn et al., 2009; Zhu et al., 2010). Deregulation of T cell differentiation and function is associated with different immunological disorders such as autoimmune disease and allergy.
A critical feature of T helper cell differentiation into a specific lineage is the suppression of other lineage fates and the silencing of lineage-inappropriate cytokine genes (Kanno et al., 2012; Zhu et al., 2010). For instance, in polarized Th1 cells expressing IFN-γ efficiently, the expression of IL-4 and IL-17 is repressed (Kanno et al., 2012). This is critically dependent upon the transcription factor T-bet, which promotes IFN-γ production, represses IL-4 transcription, and inhibits the function of Gata-3 and RORγt, thereby antagonizing Th2 and Th17 differentiation, respectively (Kanno et al., 2012). In Th2 cells, Gata-3 downregulates the expression of STAT4, which mediates IL-12 signaling and Th1 differentiation (Usui et al., 2003; Zhu et al., 2010). In addition, during Th2 differentiation, STAT5 activation, which is critical for the maintenance of Gata-3 expression (Guo et al., 2009), can also inhibit T-bet expression (Zhu et al., 2003; Zhu et al., 2010). However, it is still not fully understood how T helper cells suppress other lineage fates particularly in the early time period of differentiation before master transcription factors become expressed at high levels.
Much effort has been made to understand the cis-regulatory elements that control the expression of lineage-determining genes (Kanno et al., 2012). Such elements, which are often identified as conserved noncoding sequences (CNS) that exhibit DNase I hypersensitivity, not only contain binding sites for trans-acting factors but also constitute an integral part of gene structure and serve as centers of epigenetic changes (Kanno et al., 2012; Lee et al., 2006). Many cis-regulatory elements have been identified in the IFN-γ, IL-4/5/13 (Th2) and IL-17 loci in T cells. For instance, CNS-1, which contains three hypersensitive sites (HS), HSS1-HSS3, was identified in the intergenic region between the IL-4 and IL-13 loci (Takemoto et al., 1998). Two additional hypersensitive sites, HSIV and HSV are located at the 3′ end of the IL-4 gene (Agarwal and Rao, 1998). One of the most important cis-regulatory elements in the Th2 cytokine locus is the locus control region (LCR) (Lee et al., 2003). An LCR is a regulatory region with dual function. As an enhancer, the LCR promotes increased gene expression, while as an insulator it confers protection from the effects of neighboring chromatin on a transgene (Lee et al., 2006). LCRs are likely to be closely related to the recently described super enhancers (Whyte et al., 2013). The Th2 LCR is located at the 3′ end of the Rad50 gene and comprises several HS sites, RHS4-7, which are spaced over a 25kb region (Fields et al., 2004; Lee and Rao, 2004; Williams et al., 2013). In particular, the deletion of either RHS6 or RHS7 yields a dramatic reduction in Th2 cytokine expression, suggesting a critical role of the LCR in Th2 cytokine gene expression (Lee et al., 2005; Williams et al., 2013).
It is generally accepted that chromosomes are compartmentalized into discrete territories (Cremer and Cremer, 2001). Cell type-specific transcriptional regulation can be mediated in cis at the level of chromatin and genome organization through transcription factors and histone modifications (Hubner et al., 2012). Large scale intrachromosomal associations between distal cis-regulatory elements and cytokine gene promoters, which may serve as a platform to initiate transcription by recruiting trans-acting factors in close spatial proximity, have been reported at the Th2 cytokine locus during the differentiation of naïve T cells toward the Th2 lineage (Spilianakis and Flavell, 2004). The LCR of the Th2 cytokine locus plays a central role in mediating these intrachromosomal associations in Th2 cells (Spilianakis and Flavell, 2004). We have shown previously that the Th2 LCR on chromosome 11 associates with the IFN-γ gene on chromosome 10 at high frequencies in naïve T cells. Following differentiation of naïve T cells into either Th1 or Th2 effectors, the frequency of this interchromosomal association was strongly reduced in favor of interchromosomal enhancer-promoter associations (Spilianakis et al., 2005). This observation suggested that dynamic intra- and interchromosomal associations between specific loci regulate transcription activation or silencing during differentiation and cellular stimulation (Spilianakis et al., 2005). However, the underlying mechanism and the biological meaning of these intra- and interchromosomal associations remain unknown. Specifically, the occurrence of interchromosomal associations between other cytokine loci and the IFN-γ or the Th2 locus has not been examined. More importantly, it also remains unclear through which mechanisms these chromosomes associate in T cells. To date, very few factors are known to mediate or maintain the spatial organization of chromosomes.
In the present study, we report a novel interchromosomal association between the LCR of the Th2 cytokine locus on chromosome 11 and the IL-17 locus on chromosome 1 in naïve T cells. We demonstrate that the frequency of this association is significantly reduced after differentiation into either Th2 or Th17 cells and importantly that this association is mediated by a physical interaction between Oct-1 and CTCF. Deletion of Oct-1, CTCF, or the regulatory region of the Th2 locus resulted in loss of this association and in a concomitant reduction of binding of each factor to the cognate sites in naïve T cells. Our results suggest that these interchromosomal associations are coordinately regulated during the differentiation of naïve T cells into effector T cells. In addition, we identified two trans-acting factors that control the association. Lastly, we found that a reduction in the frequency of interchromosomal association led to enhanced IL-17 expression during the early period of differentiation of naïve T cells into Th17 cells. We propose that interchromosomal associations play a crucial role in gene activation or silencing during T helper differentiation.
Results
Interchromosomal associations between the Th2 and IL-17 loci in naïve T cells
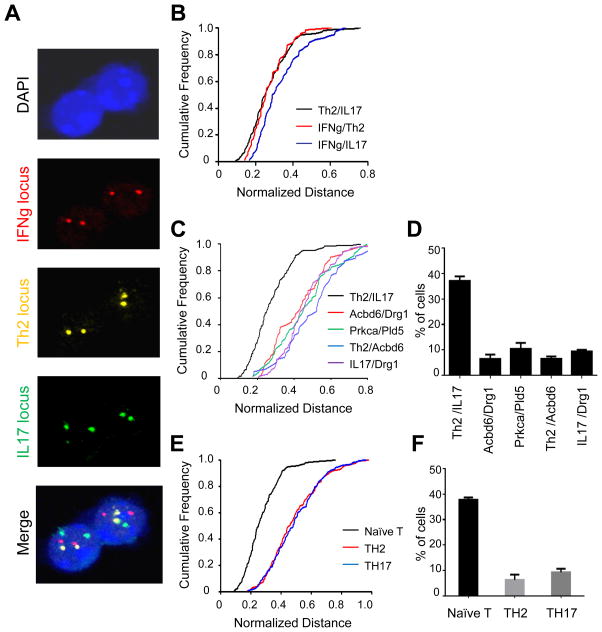
Th17 cells produce IL-17A and IL-17F as well as IL-22 and other cytokines and differentiate from naïve T cells through the action of TGF-β and inflammatory cytokines (Korn et al., 2009). Given that the IFN-γ locus (on chr. 10) and the Th2 cytokine locus (on chr. 11) associate monoallelically in naïve T cells (Spilianakis et al., 2005), we sought to determine whether the IL-17 locus on chromosome 1 similarly undergoes interchromosomal associations with other cytokine genes. We therefore performed three-color DNA FISH in naïve T cells and mapped the sub-nuclear positions of the IFN-γ, IL-4/13/5 and IL-17 loci (Figure 1A). We measured the closest distance between two loci in three-dimensions using Volocity software (Improvision) and then examined the distributions of interchromosomal associations between each pair of cytokine loci normalized for the nuclear diameter. Normalized distance values ranged between 0.1 and 0.8 in the population of naïve T cells examined. The Th2/IL-17 association was similar to the IFN-γ/Th2 association we have previously reported (Spilianakis et al., 2005) and somewhat closer than the IFN-γ/IL-17 association (Figures 1B and S1A). We next examined the distance between other combinations of either active or silent control genes which are located on the same respective chromosomes as the Th2 (e.g. Drg1 and Prkca), IL-17 (e.g. Acbd6 and Pld5) and IFN-γ loci (e.g. Fyn and Stx11). The frequency of Th2/IL-17 interchromosomal associations was higher than the frequency of interchromosomal associations between the control loci throughout the entire range of distances examined (Figures 1C and S1B). Using the Fisher’s Exact test, we determined that a cutoff normalized distance value of 0.2 detected most robustly a difference in association between the Th2 and IL-17 loci, as compared to the control genes (Figure S1C). A normalized distance value of 0.2 corresponds to an absolute separation distance of 1.5μm (Figure S1D). This criterion is consistent with our prior work (Spilianakis et al., 2005) and the literature on interchromosomal associations between the immunoglobulin loci in B cells (Hewitt et al., 2008). Approximately 40% of naïve T cells showed a distance equal to or less than 1.5μm between the Th2 and IL-17 loci, whereas only about 10% of naïve T cells showed interchromosomal associations between the control Acbd6/Drg1, Prkca/Pld5, Th2/Acbd6 and IL-17/Drg1 loci (Figure 1D). The IFN-γ/Th2 and IFN-γ/IL-17 interchromosomal distribution profiles also showed clear differences from the control gene distribution profiles (Figures S1E and S1F). Since the Th2/IL-17 association occurred at higher frequencies than the IFN-γ/IL-17 association, we focused further on the former association. In particular, we examined the fate of the Th2/IL-17 association in naïve and in differentiated effector Th2 and Th17 cells. We found that the Th2/IL-17 interchromosomal association, which predominates in naïve T cells, was dramatically reduced after differentiation into either Th2 or Th17 cells (Figures 1E, 1F and S1G).
Figure 1. An interchromosomal association between the Th2 locus and the IL-17 locus in naïve T cells.
(A) 3-D DNA FISH of naïve T cells for the IFN-γ locus (red), the Th2 cytokine locus (yellow) and the IL-17 locus (green). (B), (C) Cumulative frequency plots of normalized distances between the indicated loci in naïve T cells [N=200 for (B) and N=100 for (C)]. (D) Percentage of naïve T cells showing an interchromosomal distance equal to or below 1.5μm. (E) Cumulative frequency plots of normalized distances between the Th2 and IL-17 cytokine loci in naïve, Th2 and Th17 cells (N=200). (F) Percentage of cells with a Th2/IL-17 interchromosomal distance equal to or below 1.5μm. Data are representative of at least three independent experiments and represented as mean +/− SEM. The KS-test was performed and a P-value < 0.01 for (B) and a P-value < 0.001 for (C) and (E) was obtained. See also Figure S1.
RHS6 is the critical region that mediates interchromosomal associations between the Th2 and IL-17 loci in naïve T cells
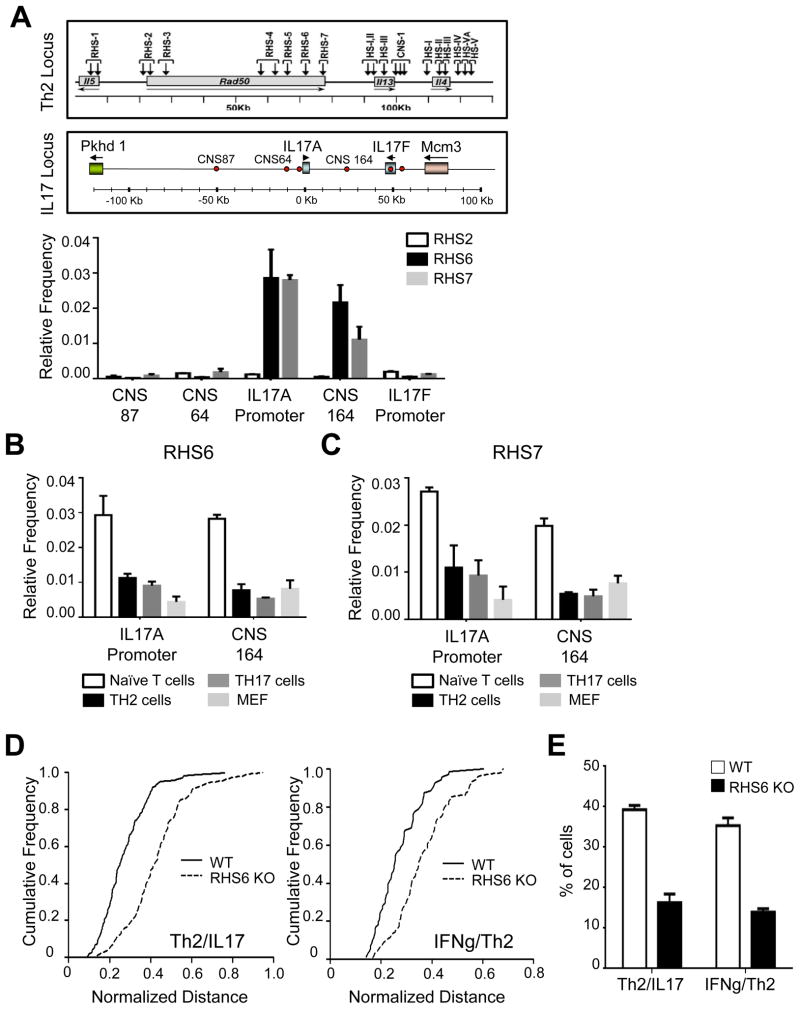
To identify the regions that mediate the Th2/IL-17 interchromosomal association, we performed chromosome conformation capture (3C) assays probing several hypersensitive regions within the Th2 locus and several conserved regions of the IL-17 locus. We detected reproducible 3C PCR chimeric products between either RHS6 or RHS7 of the Th2 locus and either the promoter of IL-17A or CNS164 of the IL-17 locus, suggesting that these regions mediate the association between the Th2 and IL-17 loci in naïve T cells (Figure 2A). When we cloned and sequenced all the 3C products, we confirmed that these were indeed chimeric products between the Th2 and IL-17 loci (Figure S2A and data not shown). We detected weak 3C PCR signals between the Th2 and IL-17 loci in effector Th2 and Th17 cells and in mouse embryonic fibroblasts (MEFs) (Figures 2B and 2C). These results suggested that several regulatory regions mediate the association between the Th2 and IL-17 loci in naïve T cells.
Figure 2. RHS6 is a critical region that mediates interchromosomal associations of the Th2 and IL-17 loci in naïve T cells.
(A) Frequency of 3C PCR chimeric products obtained with one primer annealing to a region of the Th2 locus (RHS2, RHS6 or RHS7) and the other primer annealing to a region of the IL-17 locus (CNS87, CNS64, the IL-17A promoter, CNS164, the IL-17F exon or the IL-17F promoter). For normalization, BAC DNA containing the Th2 locus or the IL-17 locus was digested, ligated and used as a 3C PCR template. (B) Frequency 3C PCR chimeric products between RHS6 and the IL-17A promoter or CNS164 in naïve T cells, Th2 cells, Th17 cells and MEFs. (C) Frequency 3C PCR chimeric products between RHS7 and the IL-17A promoter or CNS164 in naïve T cells, Th2 cells, Th17 cells and MEFs. (D) Cumulative frequency plots of normalized distances between the Th2 locus and the IL-17 locus (left) or the IFN-γ locus and Th2 locus (right) in WT and RHS6 KO naïve T cells (N=200). (E) Percentage of WT and RHS6 KO naïve T cells showing an interchromosomal separation equal to or below 1.5μm. Data are representative of at least three independent experiments and represented as mean +/− SEM. The KS-test was performed and P-value < 0.01 for (D) was obtained. See also Figure S2.
We next generated RHS6 KO mice to elucidate the role of this regulatory element in the expression of cytokines both in cis (Williams et al., 2013) and in trans via interchromosomal associations. We found no discernible differences in the development or in the nuclear size of naïve T cells isolated from WT and RHS6 KO mice (Figures S2B and S2C). Interestingly, we found that deletion of RHS6 dramatically reduced both the association frequency between the Th2 and IL-17 loci and the previously described association frequency between the IFN-γ and Th2 loci in naïve T cells (Figures 2D, 2E and S2D). To establish whether the reduction in the interchromosomal association frequency in RHS6 KO cells is T cell-intrinsic or extrinsic, for example as a consequence of alterations in the cytokine milieu, we performed bone marrow transplantations with a 50:50 mixture of WT (CD45.1) and RHS6 KO (CD45.2) cells into Rag-1 KO recipient mice. Two month after the transfer, we sorted naïve T cells and performed DNA FISH. We confirmed that the frequency of interchromosomal associations was reduced in CD45.2+ (RHS6 KO) naïve T cells compared to CD45.1+ WT naïve T cells isolated from these chimeric mice (Figure S2E). We conclude therefore that RHS6 is a critical region that mediates the interchromosomal association of the Th2 locus and the IL-17 locus in naïve T cells.
Oct-1 is bound at RHS6 and at the IL-17A promoter in naïve T cells
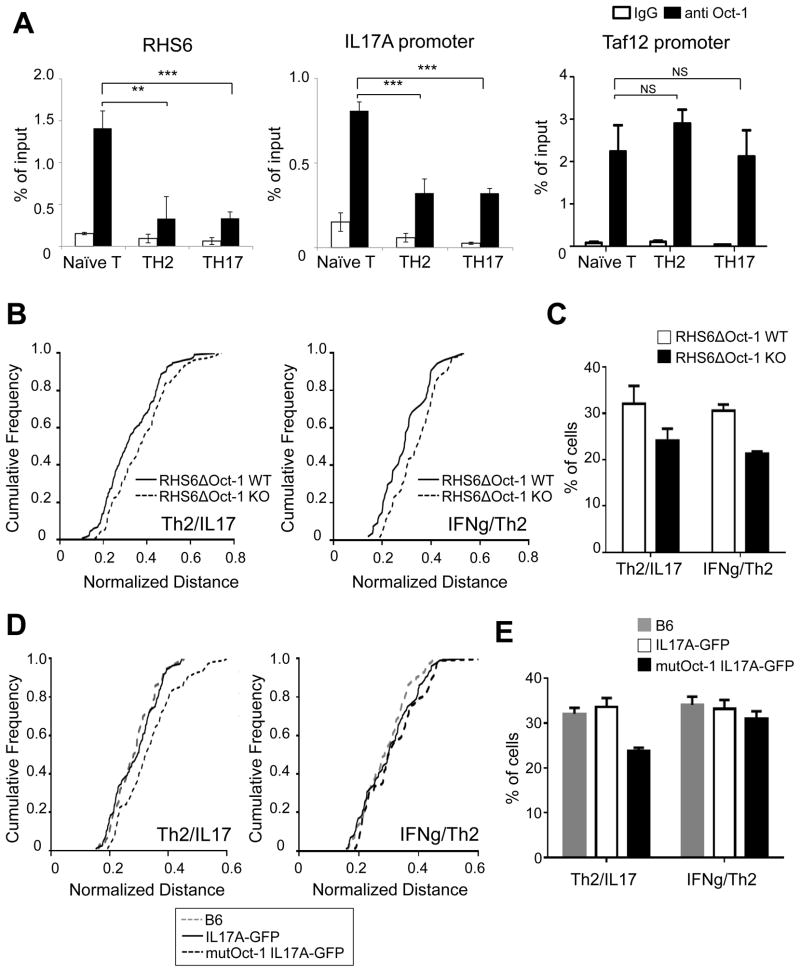
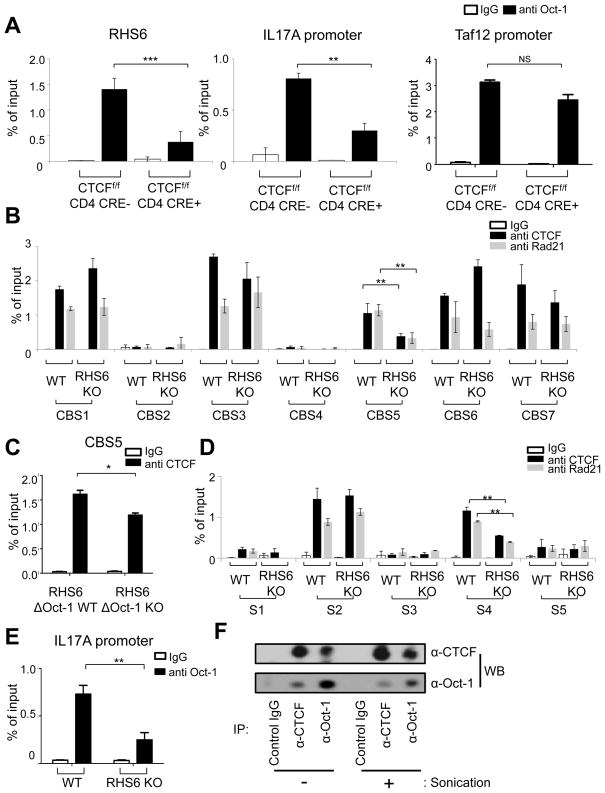
Next we investigated which factors bind to RHS6, RHS7, the promoter of IL-17A and to CNS164 of the IL-17 locus using the Biobase biological database and NCBI dcode.org. Among a few candidates, we selected Oct-1, since Oct-1 binding sites in RHS6 are evolutionarily conserved (Figure S3A) and Oct-1 is expressed in naïve T cells (data not shown). Using ChIP, we demonstrated that Oct-1 was bound to both RHS6 and to the promoter of IL-17A in the steady state in naïve T cells (Figure 3A). Interestingly, following differentiation into either Th2 or Th17 cells, Oct-1 binding to those regions was reduced, concomitant with the decrease in interchromosomal associations (Figure 3A). In contrast, we found that Oct-1 occupancy at the control Taf12 promoter was the same before and after differentiation of naïve T cells into effector cells (Kang et al., 2009) (Figure 3A). To directly demonstrate that the binding of Oct-1 to RHS6 mediates interchromosomal associations between the Th2 and IL-17 loci in naive T cells, we generated RHS6ΔOct-1 KO mice in which the two Oct-1 binding sites in RHS6 were deleted (Figures S3B and S3C). We found that deletion of the Oct-1 binding sites in RHS6 resulted in a significant reduction in the frequency of interchromosomal associations between the Th2 and IL-17 loci as well as between the IFN-γ and Th2 loci in naïve T cells (Figures 3B, 3C and S3D). In addition, we mutated the Oct-1 binding site within the IL-17A locus and investigated how this affects the associations. Since we carried out a site-directed mutagenesis within the construct used to generate IL-17A-GFP reporter mice (Esplugues et al., 2011) thereby yielding mutant reporter mice on the B6 background; and to ensure there were no background effects we compared the Oct-1 binding site mutant mice to wild type reporter mice and to C57BL/6 mice. First, we confirmed that the binding of Oct-1 to the promoter of IL-17A was reduced in the mutant cells (Figure S3E). Second, we found that a reduction in Oct-1 binding to the IL-17A promoter resulted in a reduction in the frequency of the Th2/IL-17 interchromosomal association but did not affect the frequency of the IFN-γ/Th2 association (Figures 3D, 3E and S3F). We could not find a significant difference between the association between loci of WT IL-17A-GFP reporter mice and C57BL/6 mice, which indicates that the insertion of GFP at the end of IL-17A gene does not affect the interchromosomal associations (Figures 3D, 3E and S3F). We propose that Oct-1 mediates the interchromosomal association between the Th2 and IL-17 loci by binding to RHS6 and to the IL-17A promoter.
Figure 3. Oct-1 is bound at RHS6 and at the IL-17 promoter in naïve T cells.
(A) ChIP-qPCR for Oct-1 on RHS6, the IL-17A promoter and the Taf12 promoter in naïve, Th2 and Th17 cells. The results were averaged for three independent experiments. The Student t-test was performed (**P < 0.01, ***P < 0.001). (B) Cumulative frequency plots of normalized distances between the Th2 and IL-17 loci (left) or between the IFN-γ and Th2 loci (right) in RHS6ΔOct-1 WT and RHS6ΔOct-1 KO naïve T cells (N=200). (C) Percentage of RHS6ΔOct-1 WT and RHS6ΔOct-1 KO naïve T cells showing an interchromosomal separation equal to or below 1.5μm. (D) Cumulative frequency plots of normalized distances between the Th2 and IL-17 loci (left) or between the IFN-γ and Th2 loci (right) in C57BL/6, IL-17A-GFP and IL-17A-GFP Oct-1 binding site mutant naïve T cells (N=100) (E) Percentage of C57BL/6, IL-17A-GFP and IL-17A-GFP Oct-1 binding site mutant naïve T cells showing an interchromosomal separation equal to or below 1.5μm. Data are representative of at least two independent experiments and represented as mean +/− SEM. The KS-test was performed and P-value < 0.05 for (B) and (D) was obtained. See also Figure S3.
The binding of CTCF to RHS7 of the Th2 locus and to CNS164 of the IL-17 locus is reduced after differentiation of naïve T cells into Th2 or Th17 cells
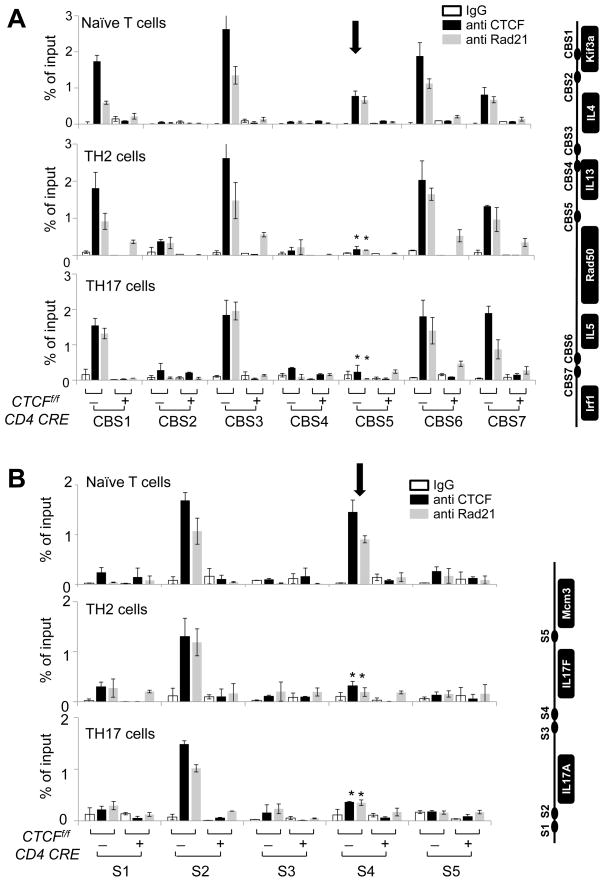
Since CTCF and cohesin have been implicated in mediating long-range chromosomal associations (Merkenschlager and Odom, 2013), and CTCF binding sites within the Th2 locus have been reported previously (Ribeiro de Almeida et al., 2009), we sought to determine the binding profiles of these factors to the regulatory regions of the Th2 and IL-17 loci. Indeed, we confirmed by ChIP that CTCF and cohesin were bound at CBS1, CBS3, CBS6 and CBS7 in Th2 cells as previously reported (Ribeiro de Almeida et al., 2009) (Figure 4A). Moreover, CTCF and cohesin were bound to these four sites in naïve T cells and in effector Th17 cells (Figure 4A). Interestingly, in naïve T cells, CTCF and cohesin were also bound at CBS5, which is located within RHS7 but unusually for a CTCF site, this binding was decreased upon differentiation of naïve cells into either effector Th2 or Th17 cells (Figure 4A). Next we examined the binding profile of CTCF and cohesin in the IL-17 locus and found five putative CTCF binding sites (Dr. Tae Hoon Kim, personal communication; Figure 4B, right panel). Using ChIP, we confirmed that CTCF and cohesin were indeed bound at the S2 and S4 sites of the IL-17 locus in naïve T cells; however upon differentiation of naïve T cells into Th17 cells, binding of both factors was reduced at the S4 site, which is located within CNS164 (Figure 4B). Thus, we conclude that CTCF and cohesin bind to both RHS7 in the Th2 locus and to CNS164 in the IL-17 locus in naïve T cells. Binding of both factors to these sites was diminished upon differentiation, concomitant with the decrease in the frequency of the interchromosomal Th2/IL-17 associations.
Figure 4. CTCF and cohesin bind to the Th2 locus and the IL-17 locus in T cells.
ChIP-qPCR for control IgG, anti-CTCF and anti-Rad21 was performed using SYBR Green and the designated primers within the Th2 locus (A) and the IL-17 locus (B). The location of each binding site is indicated on the right of the graphs. The results were averaged for three independent experiments and represented as mean +/− SEM. Statistical significance was calculated using the Student t-test comparing naïve and effector T cells (*P < 0.05).
The binding of Oct-1 or CTCF to their cognate sites is reduced in the absence of either CTCF or RHS6 in naïve T cells
We next sought to determine the consequence of deletion of CTCF on the occupancy of Oct-1 within RHS6 of the Th2 locus and the IL-17A promoter. There was no significant difference in the development of CTCF WT and CTCF KO naïve T cells or in the expression of Oct-1 in CTCF WT and KO T cells (Figures S4A and S4B). Interestingly, however, binding of Oct-1 to RHS6 or to the IL-17A promoter was significantly reduced in the absence of CTCF in naïve T cells (Figure 5A). Next we checked the binding of CTCF and cohesin at their respective sites in RHS6 WT and KO T cells and found that the binding of both proteins to CBS5 of RHS7 in the Th2 locus was reduced in the absence of the adjacent RHS6 site, which is 6kb upstream (Figure 5B). In addition, we confirmed that binding of CTCF to CBS5 was also reduced by deletion of the Oct-1 binding sites in RHS6, suggesting that the reduction in CTCF binding to CBS5 is due to the absence of Oct-1 binding at RHS6 (Figure 5C). To check the possibility that the absence of Oct-1 binding at RHS6 caused epigenetic changes at RHS7, thereby indirectly influencing CTCF binding to CBS5 of RHS7, we performed ChIP-qPCR for H3K4me3 and H3K27me3. We could not detect any changes in the histone methylation pattern, and we therefore conclude that epigenetic changes secondary to Oct-1 binding are unlikely to influence binding of CTCF to CBS5 in the absence of Oct-1 binding at RHS6 (Figure S4C). Notably, binding of CTCF and cohesin to the S4 site of the IL-17 locus in naïve T cells was also reduced upon deletion of RHS6 in the Th2 locus despite the fact that these sites are located on different chromosomes (Figure 5D). Furthermore, we observed that binding of Oct-1 to the IL-17A promoter was reduced in the absence of RHS6 (Figure 5E). The fact that Oct-1 and CTCF reciprocally affect each other’s binding to their respective sites led us to consider the possibility of a physical interaction between these two factors. To address this, we performed co-immunoprecipitation experiments with or without sonication. We used sonication to eliminate the possibility that Oct-1 and CTCF were bound to the same DNA fragment and interacted in cis. Using both conditions, we observed that CTCF and Oct-1 are associated in naïve T cells (Figure 5F). Additionally, in RHS6ΔOct-1 KO naïve T cells, binding of Oct-1 to the IL-17A promoter and binding of CTCF to S4 of CNS164 was not changed (data not shown). This suggests other factors may be involved in Th2/IL-17 association. Oct-1 and CTCF could be part of a larger complex of proteins that mediates the association.
Figure 5. The binding of Oct-1 and CTCF to their cognate sites is reduced in the absence of either CTCF or RHS6 in naïve T cells.
(A) ChIP-qPCR for Oct-1 on RHS6, the IL-17A promoter and the Taf12 promoter in CTCFf/f CD4cre- and CTCFf/f CD4cre+ naïve T cells. (B) ChIP-qPCR for CTCF or Rad21 on the Th2 locus in WT and RHS6 KO naïve T cells. (C) ChIP-qPCR for CTCF on CBS5 of the Th2 locus in RHS6ΔOct-1 WT and RHS6ΔOct-1 KO mice naïve T cells. (D) ChIP-qPCR for CTCF or Rad21 on the IL-17 locus in WT and RHS6 KO naïve T cells. (E) ChIP-qPCR for Oct-1 on the IL-17 promoter in WT and RHS6 KO mice naïve T cells. The results were averaged for three independent experiments and represented as mean +/− SEM. The Student t-test was performed (*P < 0.05, **P < 0.01, ***P < 0.001). (F) Co-immunoprecipitation of Oct-1 with CTCF and vice versa. Intact or sonicated cell lysates were incubated with control IgG, anti-CTCF or anti-Oct-1. Co-IP samples were subjected to Western blotting with either anti-CTCF or anti-Oct-1 as indicated. See also Figure S4.
The frequency of interchromosomal associations between the Th2 and the IL-17 loci is reduced in the absence of either Oct-1 or CTCF in naïve T cells
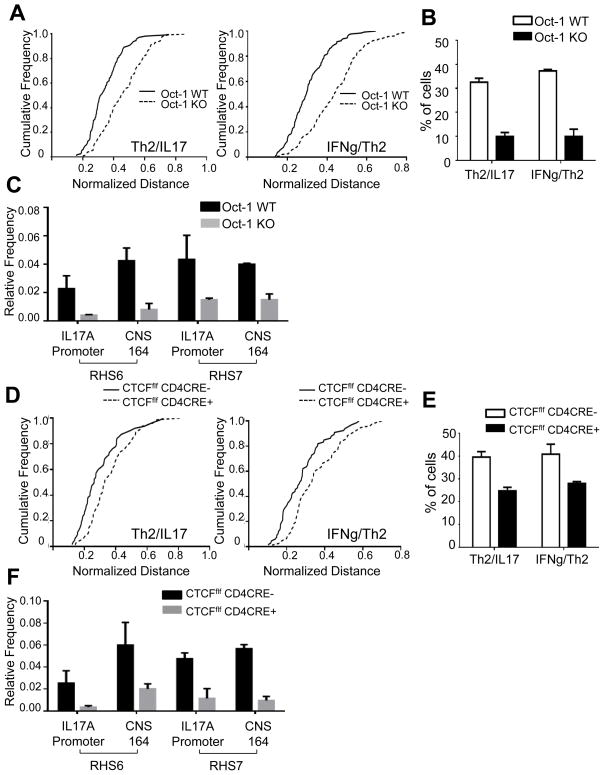
We next examined the roles of Oct-1 and CTCF in mediating interchromosomal associations in naïve T cells. Because knock-out of Oct-1 confers a prenatal lethal phenotype, we performed fetal liver transplantations into Rag-1 KO mice. Naïve T cells isolated from Rag-1 KO mice transplanted with either Oct-1 WT or Oct-1 KO fetal liver, were developmentally indistinguishable (Figure S5A). We examined the frequency of interchromosomal associations in naïve T cells in the presence or absence of Oct-1. Interestingly, the Th2/IL-17 associations were greatly reduced in the absence of Oct-1 (Figures 6A, 6B and S5B). We also confirmed that the frequency of associations between the Th2 and IL-17 loci in Oct-1 KO T cells was reduced compared to Oct-1 WT T cells by 3C assays (Figure 6C). In addition, CTCF KO T cells likewise showed a reduction in interchromosomal associations by all parameters examined (Figures 6D, 6E, 6F and S5C). There was no detectable difference in the association frequency between the control Drg1 and Acbd6 loci in Oct-1 WT and Oct-1 KO T cells or between CTCF WT and CTCF KO T cells (data not shown). As we found previously that IFN-γ/Th2 associations were dependent on RHS7 (Spilianakis et al., 2005), we examined its role in the Th2/IL-17 association. Thus, we also found that the frequency of Th2 and IL-17 associations was likewise reduced in RHS7 KO naïve T cells (Figure S5D). Overall, we conclude that Oct-1 and CTCF play a role in the juxtaposition of the Th2/IL-17 loci in naïve T cells.
Figure 6. The frequency of interchromosomal associations between the Th2 locus and the IL-17 locus is reduced in the absence of either Oct-1 or CTCF in naïve T cells.
(A) Cumulative frequency plots of normalized distances between the Th2 and the IL-17 loci (left panel), and between the IFN-γ and IL-17 loci (right panel) in Oct-1 WT and Oct-1 KO recipient mice (N=100). (B) Percentage of naïve T cells from Oct-1 WT recipient and Oct-1 KO recipient mice showing an interchromosomal distance equal to or below 1.5μm. (C) Frequency of 3C chimeric products between the Th2 locus (RHS6 or RHS7) and the IL-17 locus (the IL-17A promoter or CNS164) in naïve T cells from Oct-1 WT and Oct-1 KO recipient mice. (D) Cumulative frequency plots of normalized distances between the Th2 and the IL-17 loci (left panel), and between the IFN-γ and the IL-17 loci (right panel) in CTCFf/f CD4 cre+ and CTCFf/f CD4 cre- naïve T cells (N=100). (E) Percentage of CTCFf/f CD4cre- and CTCFf/f CD4cre+ naïve T cells showing an interchromosomal distance equal to or below 1.5μm. (F) Frequency 3C PCR chimeric products between the Th2 locus (RHS6 or RHS7) and the IL-17 locus (the IL-17A promoter or CNS164) in naïve T cells from CTCFf/f CD4 cre+ and CTCFf/f CD4 cre- mice. Data are representative of at least three independent experiments and represented as mean +/− SEM. The KS-test was performed for each of the cumulative frequency plots. P-value < 0.001 for (A) and P-value < 0.05 for (D). See also Figure S5.
A reduction in the frequency of interchromosomal associations leads to increased IL-17A expression at early time points of differentiation of naïve T cells towards the Th17 lineage
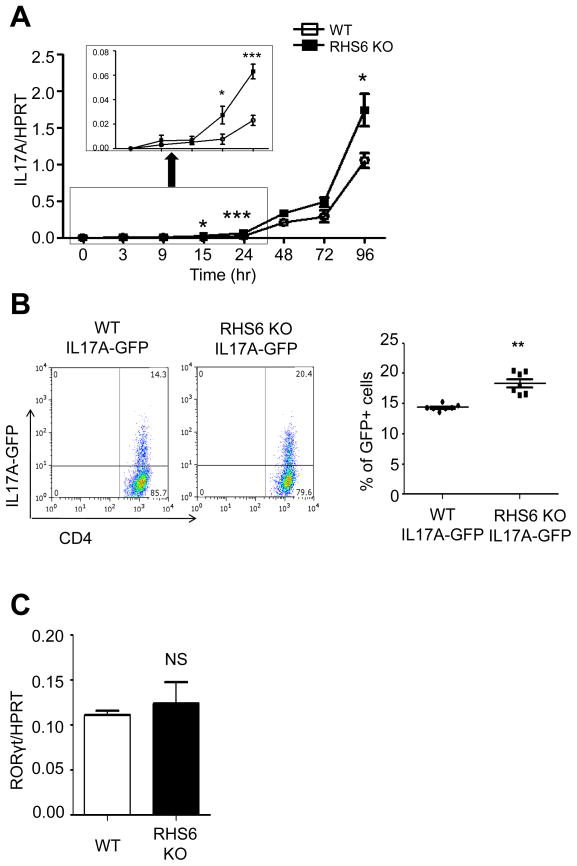
Lastly, we examined the functional role of interchromosomal associations in the expression of IL-17A in RHS6 WT and KO T cells. Since RHS6 is a critical regulatory component of the Th2 locus, deletion of RHS6 results in a dramatic reduction in Th2 cytokine expression (Williams et al., 2013). Interestingly, although RHS6 is located on a different chromosome, deletion of RHS6 resulted in increased IL-17A expression at early time points of activation (Figure 7A), as well as in an increased frequency of IL-17A producing cells (Figure 7B). There was no detectable difference in RORγt expression between WT and RHS6 KO cells (Figure 7C). Since the lymphoid organ environment of RHS6 KO mice might differ from that of WT mice as a consequence of the altered ability to produce Th2 cytokines, we sought to minimize such differences by allowing WT and KO cells to develop in the same environment. We therefore performed bone marrow transplantations with mixtures of WT (CD45.1) and RHS6 KO (CD45.2) cells into Rag-1 KO mice. Five mice were analyzed and showed an approximately 2-fold increase in IL-17A expression in RHS6 KO cells compared to WT cells (Figure S6A). This result suggests that the increase in IL-17A expression observed in RHS6 KO mice is T cell intrinsic and not a consequence of an alteration in the immune status of the mice. In similar experiments, Th17 cells isolated from either Oct-1 KO or CTCF KO mice showed an increase in IL-17A expression compared to the WT both at the mRNA and at the protein levels at early time points of activation (Figures S6B and S6C). Thus, the increased IL-17A production in either Oct-1 KO or CTCF KO T cells compared to the WT, correlated with the loss of Th2/IL-17 interchromosomal associations, although we cannot exclude an indirect effect of Oct-1 and CTCF. However, the fact that both the RHS6ΔOct-1 KO and the IL-17A Oct-1 binding site mutant alleles phenocopy to some extent the Oct-1 KO interchromosomal phenotype suggests that some effects at least must be local.
Figure 7. Analysis of IL-17 expression in RHS6 WT and KO T cells.
(A) Quantitative PCR of IL-17 mRNA expression. Naïve T cells from WT and RHS6 KO mice were treated with plate-bound anti-CD3, anti-CD28, TGF-β, IL-6 and IL-23 for the indicated time. The Student t-test was performed (*P < 0.05. ***P < 0.001). (B) MACS purified CD4+ cells from IL-17A-IRES-GFP reporter mice and RHS6 KO IL-17A-IRES-GFP reporter mice were cultured in Th17 polarizing conditions for 2 days. The statistical significance was calculated using the Student t-test (**P < 0.01). (C) Quantitative PCR for RORγt mRNA expression at 24hrs of differentiation. Data are representative of at least three independent experiments and represented as mean +/− SEM. The Student t-test was performed. See also Figure S6.
Discussion
In this study, we identified and characterized a novel interchromosomal association in T cells between the IL-17 gene on chromosome 1 and the regulatory regions of the Th2 cytokine locus on chromosome 11 in naïve CD4 T cells. As we reported previously for the IFN-γ/Th2 association (Spilianakis et al., 2005), the frequency of the Th2/IL-17 association was higher in naïve T cells than in effector T cells. We identified the regions that mediate this interchromosomal association, as hypersensitive sites within the Th2 LCR and the IL-17 promoter region and determined that these are binding sites for Oct-1 and CTCF. Deletion of Oct-1, CTCF or the RHS6 of the Th2 locus resulted in a reduced frequency of interchromosomal associations and in a concomitant reduction of binding of each factor to the cognate sites in naïve T cells. Finally, we demonstrated that defects in the Th2/IL-17 association led to enhanced IL-17 expression, paralleled by an increased frequency of IL-17 producing cells early during differentiation of naïve T cells toward the Th17 lineage.
Although Oct-1 has been widely studied as a regulator of transcription in many cell types (Herr and Cleary, 1995; Rosenfeld, 1991), its precise role is still unclear. Oct-1 is known to have both activating (Duncliffe et al., 1997; Fletcher et al., 1987; Zhou et al., 2007) and silencing activity (dela Paz et al., 2007; Kakizawa et al., 2001; Schwachtgen et al., 1998). Moreover, Oct-1 has been suggested to be a regulatable stabilizer of repressed and inducible transcriptional states (Shakya et al., 2011). Notably, Oct-1 acts at hypersensitive sites that are distant from the promoters of target genes in several cases. For example, Oct-1 regulates the IL-3 gene in activated T cells via a T cell-specific enhancer, which is located 14kb upstream of the gene (Duncliffe et al., 1997). In another case, Oct-1 was found at a macrophage-specific hypersensitive site approximately 10kb upstream of the IL-12b gene, where it played a role in nucleosome remodeling (Zhou et al., 2007). In addition, a common single polymorphism, which creates a binding site for Oct-1 within a hypersensitive site found 1.5kb upstream of the IL-13 gene, was strongly associated with high IL-13 expression and total serum IgE levels (Kiesler et al., 2009). We believe these reports correlate well with our hypothesis that Oct-1 does not only act as an activator or repressor, but also functions as a global regulator, which mediates or maintains the spatial organization of chromosomes.
CTCF is well known as an insulator (Bell et al., 1999) and a central player in regulating long-range interactions in conjunction with cohesin (Wendt et al., 2008; Hadjur et al., 2009). Numerous reports have explored the functional cooperation between CTCF and Oct-1. For example, in the IGF2/H19 imprinting control region (ICR), CTCF selectively binds the unmethylated maternal allele and blocks the expression of maternal IGF2 (Merkenschlager and Odom, 2013). Surprisingly, the Oct-1 binding site in the IGF2/H19 ICR functions as a maintenance sequence for the unmethylated state (Hori et al., 2002). Moreover, mutations of the Oct-1 binding sites were discovered in a patient with Beckwith-Wiedemann syndrome, a growth disorder that results from methylation defects in the IGF2/H19 ICR (Demars et al., 2010; Poole et al., 2012). In addition, Oct-1 binding motifs occur preferentially within nuclear lamina associated domains, which provide structural support by anchoring chromatin to the nuclear envelope and whose borders are demarcated by CTCF (Guelen et al., 2008; Tang et al., 2008). We, therefore, believe that these reports are consistent with our findings that Oct-1 and CTCF regulate the spatial arrangement of chromatin by mediating interchromosomal associations of cytokines loci in T cells. Another Oct family member, Oct-4 has recently been shown to interact with CTCF and to control homologous X-chromosome pairing and counting (Donohoe et al., 2009). We speculate that Oct family proteins act as mediators of spatial organization of chromosomes in multiple cell types in cooperation with CTCF.
We have identified Oct-1 and CTCF as mediators of interchromosomal associations in naïve T cells, but the dynamic aspects of this process remain to. Our analysis only captures snapshots of gene positions at different time points of T cell differentiation and activation. It is possible that these represent stochastic gene encounters in the interchromatin domain (Williams et al., 2010). However, our analysis clearly demonstrates that the associations of the Th2 and IL-17 loci occurs at much higher frequencies than that of randomly selected genes located on same chromosomes in naïve T cells. In addition, the frequency of cytokine gene associations are dramatically reduced in response to biologic stimuli that lead to the differentiation of naïve T cells into effector T cells. Moreover, we demonstrated that the frequency of cytokine interchromosomal associations is reduced by depletion of two factors, Oct-1 and CTCF, involved in T cell differentiation. Collectively, our data suggest that the association of the IFN-γ, IL-4/5/13 and IL-17 loci in naïve T cells is regulated phenomenon, which has functional consequences on cytokine expression.
Interestingly, deletion of the DNase I hypersensitive site of Th2 cytokine locus (RHS6), which mediates interchromosomal associations, led to increased expression of IL-17 despite being located on a different chromosome. The fact that deficiency of Oct-1 or CTCF, both impaired interchromosomal associations and led to enhanced IL-17 expression upon T cell activation, strongly suggests that interchromosomal associations functionally affect gene expression, although we cannot exclude the possibility of indirect effects caused by deletion of hypersensitive sites and relevant proteins.
Recently, OBF.1, a transcriptional coactivator of Oct-1, was reported to promote Th17 differentiation (Yosef et al., 2013). Although OBF.1 is known to be constitutively expressed in B cells, OBF.1 expression and function is induced with phorbol esters and ionomycin in Jurkat T cells and in primary murine thymocytes (Zwilling et al., 1997). Moreover, posttranslational modifications, such as phosphorylation at Ser184, are required for the inducible activation of OBF.1 (Zwilling et al., 1997). We speculate that Oct-1 may have a dual function in T cells: it may control (1) the kinetics of IL-17 induction via interchromosomal associations during the early period of differentiation of naïve T cells into Th17 effectors and (2) the threshold of IL-17 induction in activated Th17 effectors in cooperation with OBF.1. Therefore, the transcriptional role of Oct-1 in the absence (e.g. in naïve T cells) or presence (e.g. in activated effector T cells) of OBF.1 should be separately considered.
In conclusion, we have shown that interchromosomal associations between the Th2/IL-17 alternatively expressed cytokine genes, which predominate in naïve T cells, diminish in frequency upon differentiation and lineage commitment. We propose that Oct-1 and CTCF, bound to the RHS6 site of the Th2 locus and to the IL-17 promoter, mediate these trans associations. Cooperatively these factors promote the formation of a three-dimensional interchromosomal conformation, which governs the timing and level of cytokine gene expression. We propose that Oct-1 and CTCF play a crucial role in the transcriptional activation of lineage-specific cytokine genes and in the repression of alternative cell fates.
Experimental Procedures
Mice
C57BL/6 mice were purchased from Jackson Laboratories or NCI. RHS6 KO mice were generated in our lab (Williams et al., 2013). CTCF conditional KO mice were previously described (Ribeiro de Almeida et al., 2009). Oct-1 KO mice were provided by Dr. D. Tantin (University of Utah, USA) and IL-17A--GFP reporter mice were generated in our lab (Esplugues et al., 2011). All animals were maintained in accordance with Yale University Institutional Animal Care and Use Committee.
Cell sorting and cell culture conditions
CD4+ T cells were isolated from the spleen and lymph nodes of WT and KO mice and enriched with CD4 microbeads (Miltenyi Biotech). Naïve T cells (CD4+ CD25- NK1.1- CD44low CD62Lhigh) were sorted by flow cytometry and activated with plate-bound anti-CD3, anti-CD28, cytokines and neutralizing antibodies. IL-4, IL-2 and anti-IFN-γ were used for Th2 cell differentiation and TGF-β, IL-23, IL-6, anti-IL-4 and anti-IFN-γ were used for Th17 cell differentiation. For CTCF WT and CTCF KO T cell cultures, PMA and ionomycin stimulation was used as described previously (Ribeiro de Almeida et al., 2009).
Three-dimensional (3-D) FISH
Three-dimensional FISH was done as described previously (Spilianakis et al., 2005). BAC clones were purchased from Genome systems (B172 for Th2 locus) or CHORI (RPCI.24-352N22 for IFN-γ, RP24-305203 for IL-17, RP24-368F20 for Stx11, RP24-235K8 for Fyn, RP24-217E8 for Drg1, RP24-371K6 for Prkca, RP23-357C20 for Pld5 and RP23-30B13 for Acbd6). Two micrograms of BAC DNA was labeled by nick translation (Abbott) with ChromoTide Alexa Fluor 647-12-OBEA-dCTP (Molecular Probes), Spectrum Green (Vysis) or Spectrum Orange (Vysis), according to the manufacturer’s instructions. The images were acquired using a Leica TCS SP5 confocal microscope. The distance between two loci in 3-D was measured using the Volocity software (Improvision). Some scans were scored in a blinded manner.
Chromosome Conformation Capture (3C) analysis
The 3C analysis was done as described previously (Spilianakis and Flavell, 2004; Spilianakis et al., 2005). The precipitated 3C products were quantitated using the Quantitect SYBR Green PCR Kit (Qiagen) in a 7500 Fast Real Time PCR System (Applied Biosystems). The sequencing of 3C PCR products was performed in two ways. First, the 3C products, which were obtained after 35 cycles of PCR, were isolated from agarose gels using the Qiagen PCR gel extraction kit (Qiagen) and sequenced directly. Second, the 3C products, which were obtained after 23 cycles of PCR, were cloned into the TA vector (Invitrogen) and sequenced.
Chromatin Immunoprecipitation (ChIP)
ChIP was performed as described previously (Kim et al., 2007; Lerner et al., 2003). Antibodies were purchased from Cell Signaling for anti-CTCF (#3418), from Abcam for anti-Rad21 (ab992) and from Santa Cruz for Oct-1 (sc-232X). The precipitated chromatin fragments were quantitated with a Quantitect SYBR Green PCR Kit (Qiagen) in a 7500 Fast Real Time PCR System (Applied Biosystems).
Co-immunoprecipitation (Co-IP) Assay
The cells were lysed in 50mM Tris (pH 8.0), 150mM NaCl, 1% NP-40, 0.5% sodium deoxycholate and 0.1% SDS, 1mM dithiothreitol (DTT) and 1x complete protease inhibitor cocktail (Roche Diagnostics) after crosslinking with dimethyl adipimidate 2HCl (Pierce) for 90 min. The cell lysates were sonicated for 0 min or 10 min. Control IgG, anti-CTCF or anti-Oct-1 were added and incubated at 4°C. Eluates were used for Western blotting.
Real time PCR
The expression of mRNA was measured using Taqman probes (Applied Biosystems) in a 7500 Fast Real Time PCR System (Applied Biosystems).
Adoptive transplantation
Adoptive transfer of fetal liver or bone marrow was performed as described previously (Wang et al., 2004). Briefly, the fetal liver cells were harvested from 12.5 day embryos of Oct-1 WT or Oct-1 KO mice. Bone marrow cells were harvested from WT and RHS6 KO mice. Rag-1 KO recipient mice (5–8 weeks old) were sub-lethally irradiated and at least one million fetal liver cells were retro-orbitally injected. The recipient mice were analyzed 8–10 weeks after transfer.
Statistical analysis
An unpaired Student t-test was performed for statistical analysis for all studies using the GraphPad Prism software. Kolmogorov-Smirnow test was performed using the Minitab 16 statistical software. Fisher’s Exact test was performed using the program R. A P value < 0.05 was considered statistically significant.
Supplementary Material
Highlights.
There is an interchromosomal association of the Th2 and IL-17 loci in naïve T cells
The association require DNase I hypersensitive regions at the Th2 locus
Oct-1 mediates the association by binding these sites in cooperation with CTCF
Defect in the association leads to enhanced IL-17 induction
Acknowledgments
We thank D. Tantin for providing the Oct-1 KO mice, T.H. Kim for providing information on CTCF binding and F. Parisi and Y. Kluger for statistical analysis. We also thank D. Schatz, T. Chi and A. Williams for critical reading of the manuscript and C. Lieber for assistance with the manuscript preparation. L.K.K. was supported by the Korea Research Foundation Grant funded by Korean Government (KRF-2008-357-C00106). C.E.Z. was the recipient of an NIH predoctoral training grant (T32-GM07499). L.K.K., C.E.Z. and R.A.F. wrote the manuscript. This work was supported in part by the Howard Hughes Medical Institute (L.K.K., R.A.F.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agarwal S, Rao A. Modulation of chromatin structure regulates cytokine gene expression during T cell differentiation. Immunity. 1998;9:765–775. doi: 10.1016/s1074-7613(00)80642-1. [DOI] [PubMed] [Google Scholar]
- Bell AC, West AG, Felsenfeld G. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell. 1999;98:387–396. doi: 10.1016/s0092-8674(00)81967-4. [DOI] [PubMed] [Google Scholar]
- Cremer T, Cremer C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nature reviews Genetics. 2001;2:292–301. doi: 10.1038/35066075. [DOI] [PubMed] [Google Scholar]
- dela Paz NG, Simeonidis S, Leo C, Rose DW, Collins T. Regulation of NF-kappaB-dependent gene expression by the POU domain transcription factor Oct-1. J Biol Chem. 2007;282:8424–8434. doi: 10.1074/jbc.M606923200. [DOI] [PubMed] [Google Scholar]
- Demars J, Shmela ME, Rossignol S, Okabe J, Netchine I, Azzi S, Cabrol S, Le Caignec C, David A, Le Bouc Y, et al. Analysis of the IGF2/H19 imprinting control region uncovers new genetic defects, including mutations of OCT-binding sequences, in patients with 11p15 fetal growth disorders. Hum Mol Genet. 2010;19:803–814. doi: 10.1093/hmg/ddp549. [DOI] [PubMed] [Google Scholar]
- Donohoe ME, Silva SS, Pinter SF, Xu N, Lee JT. The pluripotency factor Oct4 interacts with Ctcf and also controls X-chromosome pairing and counting. Nature. 2009;460:128–132. doi: 10.1038/nature08098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncliffe KN, Bert AG, Vadas MA, Cockerill PN. A T cell-specific enhancer in the interleukin-3 locus is activated cooperatively by Oct and NFAT elements within a DNase I-hypersensitive site. Immunity. 1997;6:175–185. doi: 10.1016/s1074-7613(00)80424-0. [DOI] [PubMed] [Google Scholar]
- Esplugues E, Huber S, Gagliani N, Hauser AE, Town T, Wan YY, O’Connor W, Jr, Rongvaux A, Van Rooijen N, Haberman AM, et al. Control of TH17 cells occurs in the small intestine. Nature. 2011;475:514–518. doi: 10.1038/nature10228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields PE, Lee GR, Kim ST, Bartsevich VV, Flavell RA. Th2-specific chromatin remodeling and enhancer activity in the Th2 cytokine locus control region. Immunity. 2004;21:865–876. doi: 10.1016/j.immuni.2004.10.015. [DOI] [PubMed] [Google Scholar]
- Fletcher C, Heintz N, Roeder RG. Purification and characterization of OTF-1, a transcription factor regulating cell cycle expression of a human histone H2b gene. Cell. 1987;51:773–781. doi: 10.1016/0092-8674(87)90100-0. [DOI] [PubMed] [Google Scholar]
- Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, Eussen BH, de Klein A, Wessels L, de Laat W, et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–951. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- Guo L, Wei G, Zhu J, Liao W, Leonard WJ, Zhao K, Paul W. IL-1 family members and STAT activators induce cytokine production by Th2, Th17, and Th1 cells. Proc Natl Acad Sci U S A. 2009;106:13463–13468. doi: 10.1073/pnas.0906988106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjur S, Williams LM, Ryan NK, Cobb BS, Sexton T, Fraser P, Fisher AG, Merkenschlager M. Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature. 2009;460:410–413. doi: 10.1038/nature08079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herr W, Cleary MA. The POU domain: versatility in transcriptional regulation by a flexible two-in-one DNA-binding domain. Genes Dev. 1995;9:1679–1693. doi: 10.1101/gad.9.14.1679. [DOI] [PubMed] [Google Scholar]
- Hewitt SL, Farmer D, Marszalek K, Cadera E, Liang HE, Xu Y, Schlissel MS, Skok JA. Association between the Igk and Igh immunoglobulin loci mediated by the 3′ Igk enhancer induces ‘decontraction’ of the Igh locus in pre-B cells. Nat Immunol. 2008;9:396–404. doi: 10.1038/ni1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori N, Nakano H, Takeuchi T, Kato H, Hamaguchi S, Oshimura M, Sato K. A dyad oct-binding sequence functions as a maintenance sequence for the unmethylated state within the H19/Igf2-imprinted control region. J Biol Chem. 2002;277:27960–27967. doi: 10.1074/jbc.M202280200. [DOI] [PubMed] [Google Scholar]
- Hubner MR, Eckersley-Maslin MA, Spector DL. Chromatin organization and transcriptional regulation. Current opinion in genetics & development. 2012 doi: 10.1016/j.gde.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakizawa T, Miyamoto T, Ichikawa K, Takeda T, Suzuki S, Mori J, Kumagai M, Yamashita K, Hashizume K. Silencing mediator for retinoid and thyroid hormone receptors interacts with octamer transcription factor-1 and acts as a transcriptional repressor. J Biol Chem. 2001;276:9720–9725. doi: 10.1074/jbc.M008531200. [DOI] [PubMed] [Google Scholar]
- Kang J, Gemberling M, Nakamura M, Whitby FG, Handa H, Fairbrother WG, Tantin D. A general mechanism for transcription regulation by Oct1 and Oct4 in response to genotoxic and oxidative stress. Genes Dev. 2009;23:208–222. doi: 10.1101/gad.1750709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno Y, Vahedi G, Hirahara K, Singleton K, O’Shea JJ. Transcriptional and epigenetic control of T helper cell specification: molecular mechanisms underlying commitment and plasticity. Annu Rev Immunol. 2012;30:707–731. doi: 10.1146/annurev-immunol-020711-075058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiesler P, Shakya A, Tantin D, Vercelli D. An allergy-associated polymorphism in a novel regulatory element enhances IL13 expression. Hum Mol Genet. 2009;18:4513–4520. doi: 10.1093/hmg/ddp411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim LK, Choi UY, Cho HS, Lee JS, Lee WB, Kim J, Jeong K, Shim J, Kim-Ha J, Kim YJ. Down-regulation of NF-kappaB target genes by the AP-1 and STAT complex during the innate immune response in Drosophila. PLoS Biol. 2007;5:e238. doi: 10.1371/journal.pbio.0050238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korn T, Bettelli E, Oukka M, Kuchroo VK. IL-17 and Th17 Cells. Annu Rev Immunol. 2009;27:485–517. doi: 10.1146/annurev.immunol.021908.132710. [DOI] [PubMed] [Google Scholar]
- Lee DU, Rao A. Molecular analysis of a locus control region in the T helper 2 cytokine gene cluster: a target for STAT6 but not GATA3. Proc Natl Acad Sci U S A. 2004;101:16010–16015. doi: 10.1073/pnas.0407031101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee GR, Fields PE, Griffin TJ, Flavell RA. Regulation of the Th2 cytokine locus by a locus control region. Immunity. 2003;19:145–153. doi: 10.1016/s1074-7613(03)00179-1. [DOI] [PubMed] [Google Scholar]
- Lee GR, Kim ST, Spilianakis CG, Fields PE, Flavell RA. T helper cell differentiation: regulation by cis elements and epigenetics. Immunity. 2006;24:369–379. doi: 10.1016/j.immuni.2006.03.007. [DOI] [PubMed] [Google Scholar]
- Lee GR, Spilianakis CG, Flavell RA. Hypersensitive site 7 of the TH2 locus control region is essential for expressing TH2 cytokine genes and for long-range intrachromosomal interactions. Nat Immunol. 2005;6:42–48. doi: 10.1038/ni1148. [DOI] [PubMed] [Google Scholar]
- Lerner L, Henriksen MA, Zhang X, Darnell JE., Jr STAT3-dependent enhanceosome assembly and disassembly: synergy with GR for full transcriptional increase of the alpha 2-macroglobulin gene. Genes Dev. 2003;17:2564–2577. doi: 10.1101/gad.1135003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkenschlager M, Odom DT. CTCF and cohesin: linking gene regulatory elements with their targets. Cell. 2013;152:1285–1297. doi: 10.1016/j.cell.2013.02.029. [DOI] [PubMed] [Google Scholar]
- Poole RL, Leith DJ, Docherty LE, Shmela ME, Gicquel C, Splitt M, Temple IK, Mackay DJ. Beckwith-Wiedemann syndrome caused by maternally inherited mutation of an OCT-binding motif in the IGF2/H19-imprinting control region, ICR1. European journal of human genetics : EJHG. 2012;20:240–243. doi: 10.1038/ejhg.2011.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro de Almeida C, Heath H, Krpic S, Dingjan GM, van Hamburg JP, Bergen I, van de Nobelen S, Sleutels F, Grosveld F, Galjart N, et al. Critical role for the transcription regulator CCCTC-binding factor in the control of Th2 cytokine expression. J Immunol. 2009;182:999–1010. doi: 10.4049/jimmunol.182.2.999. [DOI] [PubMed] [Google Scholar]
- Rosenfeld MG. POU-domain transcription factors: pou-er-ful developmental regulators. Genes Dev. 1991;5:897–907. doi: 10.1101/gad.5.6.897. [DOI] [PubMed] [Google Scholar]
- Schwachtgen JL, Remacle JE, Janel N, Brys R, Huylebroeck D, Meyer D, Kerbiriou-Nabias D. Oct-1 is involved in the transcriptional repression of the von willebrand factor gene promoter. Blood. 1998;92:1247–1258. [PubMed] [Google Scholar]
- Shakya A, Kang J, Chumley J, Williams MA, Tantin D. Oct1 is a switchable, bipotential stabilizer of repressed and inducible transcriptional states. J Biol Chem. 2011;286:450–459. doi: 10.1074/jbc.M110.174045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spilianakis CG, Flavell RA. Long-range intrachromosomal interactions in the T helper type 2 cytokine locus. Nat Immunol. 2004;5:1017–1027. doi: 10.1038/ni1115. [DOI] [PubMed] [Google Scholar]
- Spilianakis CG, Lalioti MD, Town T, Lee GR, Flavell RA. Interchromosomal associations between alternatively expressed loci. Nature. 2005;435:637–645. doi: 10.1038/nature03574. [DOI] [PubMed] [Google Scholar]
- Takemoto N, Koyano-Nakagawa N, Yokota T, Arai N, Miyatake S, Arai K. Th2-specific DNase I-hypersensitive sites in the murine IL-13 and IL-4 intergenic region. Int Immunol. 1998;10:1981–1985. doi: 10.1093/intimm/10.12.1981. [DOI] [PubMed] [Google Scholar]
- Tang CW, Maya-Mendoza A, Martin C, Zeng K, Chen S, Feret D, Wilson SA, Jackson DA. The integrity of a lamin-B1-dependent nucleoskeleton is a fundamental determinant of RNA synthesis in human cells. Journal of cell science. 2008;121:1014–1024. doi: 10.1242/jcs.020982. [DOI] [PubMed] [Google Scholar]
- Usui T, Nishikomori R, Kitani A, Strober W. GATA-3 suppresses Th1 development by downregulation of Stat4 and not through effects on IL-12Rbeta2 chain or T-bet. Immunity. 2003;18:415–428. doi: 10.1016/s1074-7613(03)00057-8. [DOI] [PubMed] [Google Scholar]
- Wang VE, Tantin D, Chen J, Sharp PA. B cell development and immunoglobulin transcription in Oct-1-deficient mice. Proc Natl Acad Sci U S A. 2004;101:2005–2010. doi: 10.1073/pnas.0307304101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, Tsutsumi S, Nagae G, Ishihara K, Mishiro T, et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams A, Lee GR, Spilianakis CG, Hwang SS, Eisenbarth SC, Flavell RA. Hypersensitive site 6 of the Th2 locus control region is essential for Th2 cytokine expression. Proc Natl Acad Sci U S A. 2013 doi: 10.1073/pnas.1304720110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams A, Spilianakis CG, Flavell RA. Interchromosomal association and gene regulation in trans. Trends Genet. 2010;26:188–197. doi: 10.1016/j.tig.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yosef N, Shalek AK, Gaublomme JT, Jin H, Lee Y, Awasthi A, Wu C, Karwacz K, Xiao S, Jorgolli M, et al. Dynamic regulatory network controlling TH17 cell differentiation. Nature. 2013;496:461–468. doi: 10.1038/nature11981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Nazarian AA, Xu J, Tantin D, Corcoran LM, Smale ST. An inducible enhancer required for Il12b promoter activity in an insulated chromatin environment. Mol Cell Biol. 2007;27:2698–2712. doi: 10.1128/MCB.00788-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Cote-Sierra J, Guo L, Paul WE. Stat5 activation plays a critical role in Th2 differentiation. Immunity. 2003;19:739–748. doi: 10.1016/s1074-7613(03)00292-9. [DOI] [PubMed] [Google Scholar]
- Zhu J, Yamane H, Paul WE. Differentiation of effector CD4 T cell populations (*) Annu Rev Immunol. 2010;28:445–489. doi: 10.1146/annurev-immunol-030409-101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwilling S, Dieckmann A, Pfisterer P, Angel P, Wirth T. Inducible expression and phosphorylation of coactivator BOB.1/OBF.1 in T cells. Science. 1997;277:221–225. doi: 10.1126/science.277.5323.221. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.