Abstract
Endophytic bacteria play a crucial role in plant life and are also drawing much attention for their capacity to produce bioactive compounds of relevant biotechnological interest. Here we present the characterisation of the cultivable endophytic bacteria of Lavandula angustifolia Mill.—a species used since antiquity for its therapeutic properties—since the production of bioactive metabolites from medical plants may reside also in the activity of bacterial endophytes through their direct production, PGPR activity on host, and/or elicitation of plant metabolism. Lavender tissues are inhabited by a tissue specific endophytic community dominated by Proteobacteria, highlighting also their difference from the rhizosphere environment where Actinobacteria and Firmicutes are also found. Leaves' endophytic community resulted as the most diverse from the other ecological niches. Overall, the findings reported here suggest: (i) the existence of different entry points for the endophytic community, (ii) its differentiation on the basis of the ecological niche variability, and (iii) a two-step colonization process for roots endophytes. Lastly, many isolates showed a strong inhibition potential against human pathogens and the molecular characterization demonstrated also the presence of not previously described isolates that may constitute a reservoir of bioactive compounds relevant in the field of pathogen control, phytoremediation, and human health.
1. Introduction
A diverse range of bacteria, including pathogens, mutualists, and commensals is supported by plants. These bacteria grow in and around roots, in the vasculature, and on aerial tissues [1, 2]. In particular, endophytic bacteria can be defined as those bacteria that colonize the internal tissue of the plants with no external sign of infection or negative effect on their host [2, 3]. It is increasingly evident that bacterial endophytes influence plant physiology, facilitating the uptake of nutrients such as nitrogen, phosphorus, sulphur, magnesium, and calcium [4] and showing plant growth-promoting activity (PGPR) related to the production of phytohormones involved in regulatory metabolism, such as ethylene [5], indole-3-acetic acid (IAA), and acetoin, 2,3-butanediol, [6–8]. Moreover, endophytic bacteria can also improve plant growth via nitrogen fixation [9]. Other relevant functions performed by endophytic bacteria are represented by the decrease or prevention of the pathogenic effects of certain parasitic microorganisms with the production of antimicrobial compounds or by increasing plant tolerance to pollution or stresses [3, 10]. Bacterial endophytes are drawing increasing interest as together with fungal endophytes they are reported to produce a number of bioactive metabolites relevant to human health, such as antibiotics [11], antitumor compounds [12, 13], and anti-inflammatory agents [14].
Endophytic bacteria can be further classified as “obligate” or “facultative” in accordance with their life strategies. Obligate endophytes are strictly dependent on the host plant for their growth and survival, besides, transmission to other plants could occur only by seeds or via vectors, while facultative endophytes could grow outside host plants [15]. Several studies have shown that facultative endophytes constitute the largest fraction of the endophytic bacterial communities [5]; in fact large plant-by-plant (both at interspecific and intraspecific level) differences in the bacterial communities composition have been found [16–20], supporting the idea that the ability to enter plant tissues is a widespread phenotype of soil and rhizosphere bacteria, and that plants exert only a limited selectivity on the colonizing bacterial communities [21], even if clues for a control over the bacterial colonizers operated by yet not completely clarified plant mechanism(s) are reported [5, 22]. Actually, a recent bioinformatic study has shown that inside the class of Alphaproteobacteria (which includes well known bacterial endophytes, such as rhizobia and methylobacteria) few genomic signatures only could distinguish endophytes from nonendophytes [23]. Another common feature of endophytic bacterial community is their strong temporal and spatial variability [16, 19, 24]; the community composition not only varies in the soil compartment between the rhizosphere and the root internal tissues in response to complex and not yet fully clarified biotic (plant species and genotype dependent) and abiotic (soil characteristics) driven processes [16, 24], but also among different aerial tissues (stems, leaves, flowers) of the same plant [25, 26]. Total endophytes are influenced not only by the location within the plant but also by the presence of the main components of the essential oil in the leaves of aromatic plants [27].
However, all the studies performed so far have analysed mainly the bacterial communities in terms of species composition (or higher taxonomic ranks), especially using cultivation-independent techniques, as 16S rRNA gene libraries or metagenetic sequencing [20, 28, 29]. While cultivation-independent techniques allow a deep coverage of taxonomic diversity of bacterial communities, they provide only partial information on community structure and clearly hamper the possibility to test the actual presence of PGPR activities and bioactive molecule production in the bacterial community.
Among crops, medicinal plants are stirring much attention due to the increasing demand for green chemistry approaches, sustainable practices, and especially in the quest for novel antibiotics able to tackle the increasing multidrug resistance of pathogenic bacteria. In spite of the high relevance of medicinal plants, to the best of our knowledge, very little, if nothing at all, is known about the endophytic bacterial communities isolated from medicinal plants. For this reason, in this work we isolated and preliminary characterized from a taxonomical viewpoint the aerobic heterotrophic cultivable endophytic bacterial community of lavender (Lavandula angustifolia Mill.syn. Lavandula officinalis Chaix, Lavandula vera DC, Lavandula spica L. var angustifolia Auct.). Lavender has a long history of medicinal use and is purported to possess relaxant, neurological, and antibacterial effects [30].
The essential oil of Lavender has been used since antiquity for a variety of medical application [31]; in particular anxiolytic activity of Lavender oil was confirmed [32] and its antimicrobial activity was demonstrated against different pathogens [33]. Moon et al. [34] reported that low (≤1%) concentrations of L. angustifolia and L. × intermedia oil can completely eliminate Giardia duodenalis, Trichomonas vaginalis, and Hexamita inflata in vitro. Preliminary results also highlighted the possibility for lavender essential oil to be used as antibiotic resistance modifying agent in microorganisms [35].
It has also been demonstrated that the production of bioactive compounds is significantly impacted by the genetic milieu of the plant and by environmental factors [31]; in this framework, it is increasingly evident that the collection of microorganism living in association with complex organisms, generally defined as microbiota, plays a fundamental role in shaping their phenotypic features. Since the qualiquantitative production of bioactive metabolites in medical plants may reside also in the activity of bacterial endophytes through the direct production, PGPR activity, and/or elicitation of plant metabolism [36], the purpose of this research was therefore to perform a cultivation-dependent study of the mesophilic aerobic heterotrophic bacterial endophytic community of the relevant medical and balsamic species L. angustifolia in order to (i) identify its composition, (ii) its diversity between different plant compartments (roots, stems, and leaves), and (iii) its diversity in relation to the rhizosphere bacterial community, and (iv) build a collection of isolates to be screened for the production of bioactive compounds, and (v) test a panel of randomly selected endophytic bacteria versus opportunistic human pathogens belonging to the Burkholderia cepacia complex (Bcc). We have chosen these bacteria since many strains of the complex are opportunistic human pathogens and represent a serious concern for cystic fibrosis (CF) patients and immunocompromised individuals [37], responsible for the “cepacia syndrome,” characterized by high fever, severe progressive respiratory failure, leukocytosis, and elevated erythrocyte sedimentation rate. Moreover, Bcc strains are (naturally) resistant to many antibiotics such as cephalosporin, β-lactams, polymyxins, and aminoglycosides; therefore, Bcc infections are very problematic to eradicate [37, 38]. In spite of this high degree of resistance to many antibiotics, it has been recently demonstrated that essential oils extracted from six medicinal plants are able to completely inhibit the growth of Bcc members, including those with a clinical origin and exhibiting resistance to many antibiotics [39].
2. Materials and Methods
2.1. Plant Sampling and Isolation of Mesophilic Cultivable Endophytic and Rhizosphere Bacteria
Five agamically propagated potted L. angustifolia plants were collected from the “Giardino delle Erbe,” located in Casola Valsenio, Italy, in June 2012. On the same day, plants were transported to the laboratory for processing. Pieces of 200 mg of fresh leaves, stems, and roots tissues were collected from each of the 5 plants and bulked (for a final sample of 1 g) to account for plants to plants variability. Roots and stems samples were divided into 1 cm long pieces and surface-sterilized for 40 s with 70% ethanol followed by 10 min with 2.5% sodium hypochlorite. Plant leaves were surface-sterilized for 20 s with 70% ethanol followed by 5 min with 2.5% sodium hypochlorite. To remove the disinfectant, sections were rinsed three times for 5 min in sterile distilled water. Samples were then dried with sterile filter paper and subsequently ground with 2 mL 0.9% sodium chloride with a sterile mortar. Aliquots (100 μL) of the last washing water were plated in triplicate as sterility controls. Samples (100 μL) of tissue extracts and their different dilutions were plated in triplicate. Aliquots of 200 mg of soil from each pot were collected and bulked for 1 g total. The soil samples were then resuspended in 5 mL of 10 mM Mg2SO4 and placed under stirring for 1 h at room temperature.
Endophytic and rhizospheric bacteria were grown in triplicate on solid 5% tryptone soya broth (TSB) medium (Oxoid Ltd., Basingstoke, Hampshire, UK) at 30°C for 72 h. The number of aerobic heterotrophic bacteria was determined as colony-forming units (CFUs). Each CFU determination was performed in triplicate and an average value of bacterial titre was determined. From each sample, colonies were randomly selected and singularly plated on 5% solid TSB Petri dishes. From each isolate, glycerol stock (25% final concentration) was prepared and stored at −80°C.
2.2. PCR Amplification and Sequencing of 16S rRNA Coding Genes from Bacterial Endophytes
Cell lysates were prepared by dissolving a bacterial colony in 100 μL sterile distilled water and incubation at 99°C for 10 min, followed by 5 min at 4°C. An aliquot of 2 μL lysate was used for the amplification reaction by polymerase chain reaction (PCR). Amplification of 16S rRNA genes was performed in a total volume of 20 μL containing 2 μL of 10X reaction buffer (Polymed, Firenze, Italy), 1.5 mM MgCl2, 10 pmol of each primer [27f, 5′-GAGAGTTTGATCCTGGCTCAG, and 1495r, 5′-CTACGGCTACCTTGTTACGA], 0.25 mM of dNTP mix, 2 U of Taq DNA polymerase (Polymed). PCR reaction conditions were as described by Mengoni et al. [40].
For sequencing reaction, amplified 16S rDNA fragments were excised from 1% agarose gels and purified using the QIAquick Gel Extraction (Qiagen) according to the manufacturer's instructions. Direct sequencing of amplicons was performed at the Genechron laboratory (Ylichron Srl, Italy) with primer 27f on an ABI3730 DNA analyser (Applied Biosystems, Foster City, CA, USA) using the Big Dye Terminator Kit.
2.3. Sequence Analysis and Phylogenetic Tree Construction
The sequences presented in this work have been deposited in GeneBank database under the accession numbers KF202531-KF202915. Partial 16S rDNA sequences were matched against nucleotide sequences available in GenBank database using the BLASTn program [41].
MUSCLE [42] (http://www.drive5.com/muscle) was used to align the 16S rDNA sequences obtained with the most similar orthologous sequences retrieved from the Ribosomal Database Project (RDP) database (http://rdp.cme.msu.edu/). Alignments were trimmed to eliminate poorly aligned region and used to build Bayesian, Maximum Parsimony (MP), and the Neighbor joining (NJ) dendrograms.
Bayesian dendrograms were obtained with MrBayes 3.2 [43] with GTR substitution model with gamma-distributed rate variation across sites with 1000000 generations. MP dendrograms were obtained with MEGA 5 [44] (http://www.megasoftware.net/) using the Tree-Bisection-Regrafting (TBR) algorithm with search level 3 in which the initial trees were obtained by the random addition of sequences (500 replicates); the robustness of the inferred trees was evaluated by 1000 bootstrap resamplings; consistency and retention indexes (CI and RI) were calculated with Mesquite software 2.75 (http://mesquiteproject.org). NJ dendrograms were obtained after calculation of a Kimura two-parameter distance matrix with the software Mega 5. The robustness of the inferred trees was evaluated by 1000 bootstrap resamplings.
2.4. Statistical Analysis
Pairwise sequence identity values (not taking deletion into account) were calculated using the stand-alone Clustal Omega [45]. Genetic distances among OTU were calculated using the Kimura 2 parameter model and 1000 bootstraps replications in the MEGA 5.2.1 software [44]. Diversity indexes were calculated with the PAST3 software [46]. Pairwise differentiation (Fst) values were calculated inside the GenoDive v 2.0b25 software (http://www.patrickmeirmans.com/software/GenoDive.html), using vector of presence/absence of the bacterial genera in the different samples with 999 permutations.
2.5. Cross-Streaking Experiments
Antibacterial activity was determined by using the cross-streak method [47, 48]. Hereinafter, endophytic bacterial isolates to be tested for inhibitory activity will be termed “tester” strains, whereas Bcc strains used as a target will be called “target” strains. Cross-streaking experiments were carried out as previously described [47] by using Petri dishes. Tester strains were streaked across one-half of an agar plate with PCA medium and incubated at 30°C. After 2 days of incubation, target strains were streaked on PCA medium perpendicular to the initial streak and plates were further incubated at 30°C for 2 days. The antagonistic effect was indicated by the failure of the target strains to grow. The list of target Bcc strains used in this work is reported in Table 3.
Table 3.
The 40 Burkholderia cepacia complex strains used as targets in the cross streak experiments.
| Target strain | ||
|---|---|---|
| Strain | Species | Origin |
| FCF1 | B. cepacia | CF |
| FCF3 | ||
|
| ||
| LMG17588 | B. multivorans | ENV |
|
| ||
| FCF16 | B. cenocepacia (IIIA) | CF |
| J2315 | ||
|
| ||
| FCF18 | B. cenocepacia (IIIB) | CF |
| FCF20 | ||
| FCF23 | ||
| FCF24 | ||
| FCF27 | ||
| FCF29 | ||
| FCF30 | ||
| LMG16654 | ||
| C5424 | ||
| CEP511 | ||
| MVPC1/16 | ENV | |
| MVPC1/73 | ||
|
| ||
| LMG19230 | B. cenocepacia (IIIC) | ENV |
| LMG19240 | ||
|
| ||
| FCF38 | B. cenocepacia (IIID) | CF |
| LMG21462 | ||
|
| ||
| FCF41 | B. stabilis | CF |
|
| ||
| FCF42 | B. vietnamiensis | CF |
| TVV75 | ENV | |
|
| ||
| LMG18941 | B. dolosa | CF |
| LMG18942 | ||
| LMG18943 | ||
|
| ||
| MCI7 | B. ambifaria | ENV |
| LMG19467 | CF | |
| LMG19182 | ENV | |
|
| ||
| LMG16670 | B. anthina | ENV |
|
| ||
| FCF43 | B. pyrrocina | CF |
|
| ||
| LSED4 | B. lata | CF |
|
| ||
| LMG24064 | B. latens | CF |
|
| ||
| LMG24065 | B. diffusa | CF |
|
| ||
| LMG23361 | B. contaminans | AI |
|
| ||
| LMG24067 | B. seminals | CF |
|
| ||
| LMG24068 | B. metallica | CF |
|
| ||
| LMG24066 | B. arboris | ENV |
|
| ||
| LMG24263 | B. ubonensis | NI |
Abbreviations: CF: strains isolated from cystic fibrosis patients; AI: strains isolated from animal infection; NI: strains isolated from nosocomial infection; ENV: environmental strain.
3. Results
3.1. Composition of Endophytic Bacterial Communities Isolated from L. Angustifolia
Aerobic heterotrophic culturable bacteria were isolated from leaves, stems, and roots of 5 plants of Lavandula angustifolia. Leaves samples had the highest number of CFU/g fresh weight (6.8 × 105), whereas roots had the lowest values (1.6 × 104), the difference being anyway of one order of magnitude only (stem and rhizospheric soil showed titers of 6.5 × 104 and 3.4 × 105, resp.). Plates were visually inspected and no increase in colony number was observed with an extended incubation time of up to 7 days.
On the triplicate plates of the same plant portion, 100 colonies were randomly selected and isolated on TSA agar plates. A collection of 400 colonies was then prepared with isolates from the four samples types, namely, rhizospheric soil, roots, stems, and leaves.
In order to determine the taxonomic composition of the bacterial communities isolated from the different compartments of L. angustifolia plants and from rhizospheric soil, 16S rRNA genes were PCR-amplified from the isolates of the collection as described in Materials and Methods. An amplicon of the expected size was obtained from each bacterial isolate (data not shown) and the nucleotide sequence of the amplicons was then determined. In this way we obtained 395 sequences, each of which was used as seed to probe databases.
Table 1 reports the percentage distribution of the identified bacterial phyla in the different plant compartments and in the rhizospheric soil, along with standard diversity indexes calculated on genera distribution; the results show a relative peak of diversity in the leaf and a minimum in the stem (Shannon index 2.29 and 1.06, resp.). Figure 1 depicts the taxonomic composition of the total lavender aerobic heterotrophic cultivable endophytic community made up by a total of 11 genera; the majority of the isolated strains (88%) belonged to proteobacteria, with a dominance of gammaproteobacteria (74%), Pseudomonas being the most abundant genus accounting for the 51% of total community, followed by Stenotrophomonas (13%) and Pantoea (9%). Rhizobium is also frequently found in the internal tissue of lavender, representing 14% of the total collection. Actinomycetales are also represented (8%) with the genus Microbacterium being the most abundant (6%). Other genera were found, namely, Bacillus, Plantibacter, Psychrobacter, Sanguibacter, Salinibacterium, and Jeotgalibacillus representing collectively 6% of the endophytic collection.
Table 1.
Percentage distribution of bacterial phyla isolated from L. angustifolia roots, stem, leaves, and rhizospheric soil; standard diversity indexes built on genera distribution are also presented.
| Soil | Roots | Stem | Leaves | |
|---|---|---|---|---|
| Proteobacteria | 40 | 94 | 95 | 93 |
| Firmicutes | 44 | 6 | 3 | — |
| Actinobacteria | 16 | — | 2 | 7 |
|
| ||||
| Richness | 7 | 6 | 5 | 8 |
| Evenness | 0.73 | 0.72 | 0.45 | 0.76 |
| Shannon index | 2.06 | 1.88 | 1.06 | 2.29 |
Figure 1.
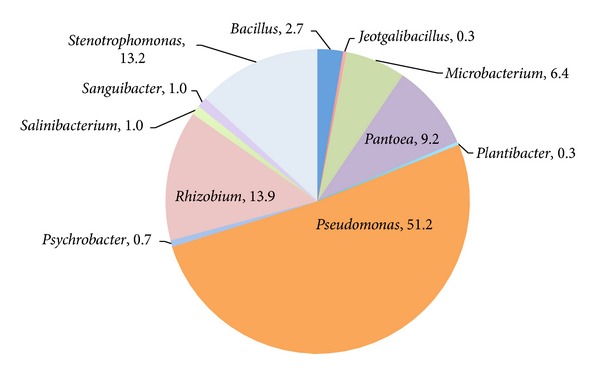
Composition of the aerobic heterotrophic endophytic (sensu stricto) bacterial community in L. angustifolia tissues. The composition is reported as percentages of the total number of characterized isolates (n = 395).
Bacteria isolated from the rhizospheric soil showed a quite different taxonomic composition (Figure 2(a)) when compared to the overall endophytic community; in the rhizosphere, Bacillus is the most abundant genus (44%), followed by Pseudomonas (30%), Microbacterium (10%), Rhizobium (7%), and Arthrobacter (6%), a genus that is not detected in the endophytic community. Figure 2 shows also the composition of the endophytic communities isolated from the different tissues (panels (b), (c), and (d), for roots, stem, and leaves, resp.); the three compartments are characterized by strikingly different communities, for the differential presence of genera (e.g., Bacillus is found in roots and stem, but absent in the leaves, Sanguibacter and Plantibacter are detected in the leaves only, Jeotgalibacillus is found only in the roots) and the overall diversity (8 genera are found in the leaves and only 5 genera are in the stem) and the different relative presence of the other most abundant genera.
Figure 2.
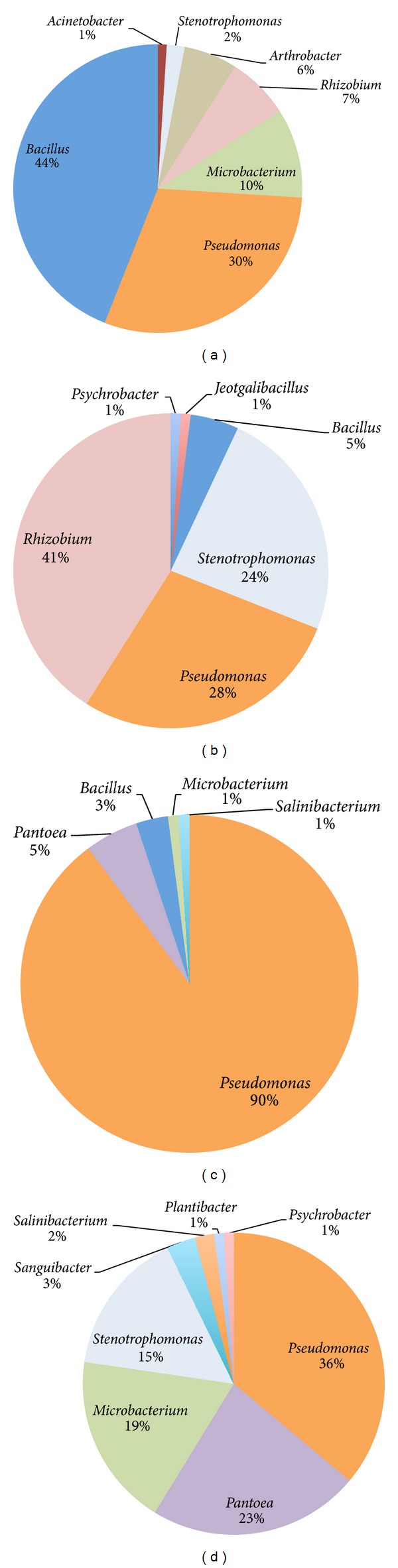
Composition of the aerobic heterotrophic bacterial community of L. angustifolia rhizospheric soil (a), roots (b), stem (c), and leaves (d). The composition is reported as percentages of the total number of characterized isolates for each sample.
Figure 3 shows the distribution of main genera (occurring in >5% of total isolates) in the 3 lavender tissues and in its rhizosphere. Isolates belonging to the genus Bacillus decrease their abundance from the rhizosphere to the stem and disappear in the leaves, while Pantoea and Microbacterium show a similar distribution in stems and leaves, but are both absent in the roots. Isolates from the Pseudomonas genus dominate in the stem community and are similarly occurring (ca. 30% of total) in the other 3 samples. Rhizobium spp. was found in soil and represented 40% of roots isolates. Lastly, Stenotrophomonas spp. is abundant in the roots and leaves, but absent in the stem. To further analyse the diversity of the bacterial communities isolated, pairwise differentiation (Fst) values were calculated on vector of presence/absence of bacterial genera; the lowest differentiation was registered among roots and rhizospheric soil (Fst = 0.198) communities, while the highest among stems and leaves (Fst = 0.512). Overall, the stem endophytic community resulted as the most differentiated from the others (Fst = 0.428 and 0.394 versus roots and rhizospheric soil, resp.) while the leaves community showed similar differentiation values when compared to rhizospheric soil (Fst = 0.245) and roots (Fst = 0.239).
Figure 3.
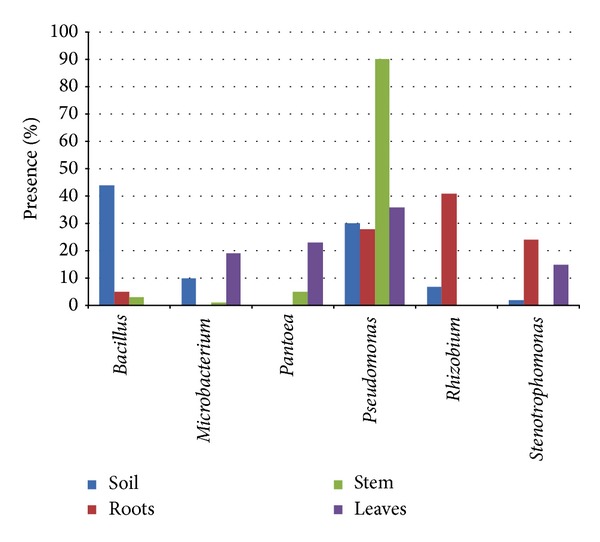
Comparison of the relative abundance of bacterial genera in the different samples; only genera with percentages >5% in at least one of the samples are reported.
Considering the low number of bacterial phyla found in the endophytic community (>95% of isolates were assigned to only 6 genera), we analysed the intrageneric level of diversity by means of 16S rRNA gene sequence identity values. Data obtained are shown in Table 2. From the 16S rRNA gene sequence diversity indices reported that the isolates assigned to the genus Rhizobium possessed the lowest overall diversity, suggesting the presence of a low number of species belonging to the genus Rhizobium. On the other extreme, the Pantoea isolates showed a stronger differentiation supporting the idea of the presence of different species (sequence identity < 97%). To further characterize the isolated endophytic and rhizosphere bacteria, Bayesian, Maximum Parsimony and Neighbor Joining trees based on 16S rRNA genes were constructed for the most abundant genera, namely, Stenotrophomonas (Figure 4 and Suppl. Figures 3 and 9 in Supplementary Material available online at http://dx.doi.org/10.1155/2014/650905), Rhizobium (Figure 5 and Suppl. Figures 4 and 10), Pantoea (Figure 6 and Suppl. Figures 5 and 11), Microbacterium (Figure 7 and Suppl. Figures 6 and 12), Bacillus (Suppl. Figures 1, 7, and 13), and Pseudomonas (Suppl. Figures 2, 8, and 14). Overall, Bayesian and NJ dendrograms showed a stronger support (and consistency of the results) than the MP ones (see also Suppl. Table 4). The analysis of the Bayesian phylogenetic trees revealed the following.
Endophytic bacteria isolated from the leaves and roots of lavender and assigned to the genus Stenotrophomonas (Figure 4) are clearly split in two groups. One group contains isolates clustering with Stenotrophomonas chelatiphaga; interestingly this cluster comprises isolates from both roots and leaves (no Stenotrophomonas were isolated from the stem), suggesting that two compartments might share bacteria belonging to the same species, even though the polymorphism shown by the aligned partial 16S sequences might suggest a diversification at the strain level. The second cluster embeds roots isolates (with the exception of leaves isolate LL44), clustering with Stenotrophomonas rizophila.
Concerning rhizobia (Figure 5), a grouping in one main cluster of low differentiate sequences is present. These groups might contain new clades of plant-associated rhizobia, which deserve more investigation in the future.
From the phylogenetic tree of Figure 6, Pantoea isolated endophytes (from lavender stem and leaves) cluster with already described Pantoea spp.; another group of isolates (all from the leaves) are diverging and thus may represent none yet described strains are found to be associated with plants.
Figure 7 depicts the phylogenetic position of Microbacterium isolates (mainly from leaves and rhizosphere) in the context of the genus reference strains; noticeably many leaves isolates cluster with M. hydrocarbonoxydans and M. oleivorans, species whose members are able to perform crude oil degradation [49].
Concerning Bacillus isolates (Suppl. Figure 1), a cluster of soil isolates including Bacillus mojavensis was disclosed. Another group, again comprising soil isolates, showed similarities with the stress resistant B. safensis.
Lastly, more than the half of the bacterial isolates were affiliated to the genus Pseudomonas; the phylogenetic tree reported in Supplemental Figure 2 is quite complex. Regardless of the overall low support of the tree, (typical of Pseudomonas phylogenies), some observations may be drawn; the presence of large unresolved clusters shows clearly a low divergence of the isolated colonies, implying the existence of clonal populations; Pseudomonas spp. isolated from the different compartments are intermixed, suggesting the presence of a continuum rhizosphere-leaves (Pseudomonas is the only genus found in all the 4 sample analysed).
Table 2.
Intragenus diversity analysis based on 16S rRNA gene sequence identity; mean identity of all versus all 16S rRNA gene sequences (for each genus); mean distance (Kimura 2 parameter); number of pairwise comparisons of 16S sequences with identity >97%, considered as threshold of species identity (in brackets the total number of comparisons and the percentage); number of OTU with at least 1 pairwise comparison with identity >97%.
| Genus | Number of isolates | Mean identity among OTU (%) | Mean distance among OTU | Number of pairwise comparisons with identity >97% | Number of OTU with identity >97% |
|---|---|---|---|---|---|
| Pseudomonas | 172 | 94.25 | 0.013 | 3840 (14878, 25.8%) | 170 |
| Bacillus | 47 | 93.04 | 0.027 | 329 (1128, 29%) | 46 |
| Rhizobia | 46 | 98.61 | 0.016 | 952 (1081, 88%) | 46 |
| Stenotrophomonas | 37 | 95.34 | 0.023 | 415 (703, 59%) | 37 |
| Microbacterium | 30 | 95.00 | 0.059 | 120 (465, 25%) | 29 |
| Pantoea | 26 | 86.95 | 0.126 | 24 (351, 6.8%) | 7 |
Figure 4.
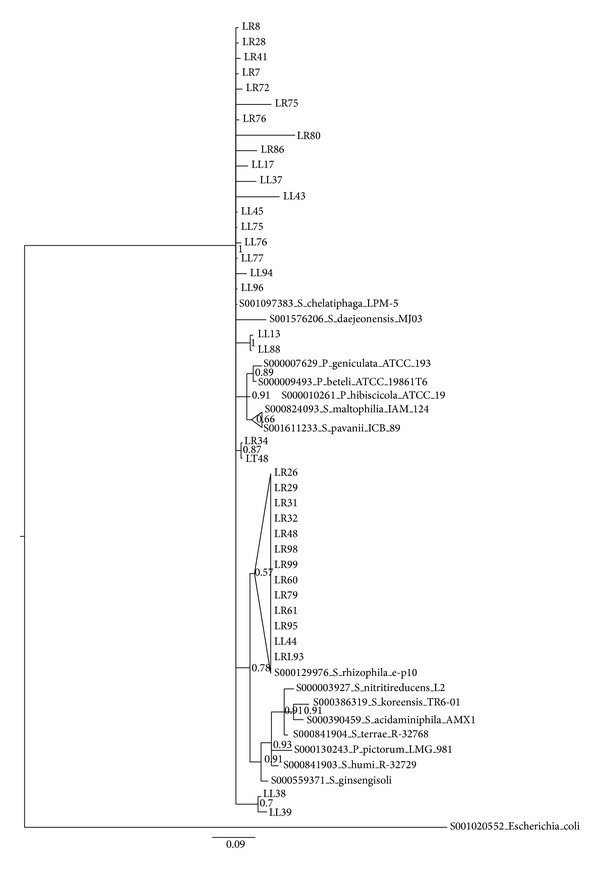
Bayesian dendrogram showing the relationships among the 16S rDNA sequences of 37 isolates belonging to the genus Stenotrophomonas and those of reference type strains. Posterior probability values are indicated at the node. Nodes are collapsed at 70% probability. LL = bacteria isolated from the leaves, LR = bacteria isolated from the roots (see Section 2 for details).
Figure 5.
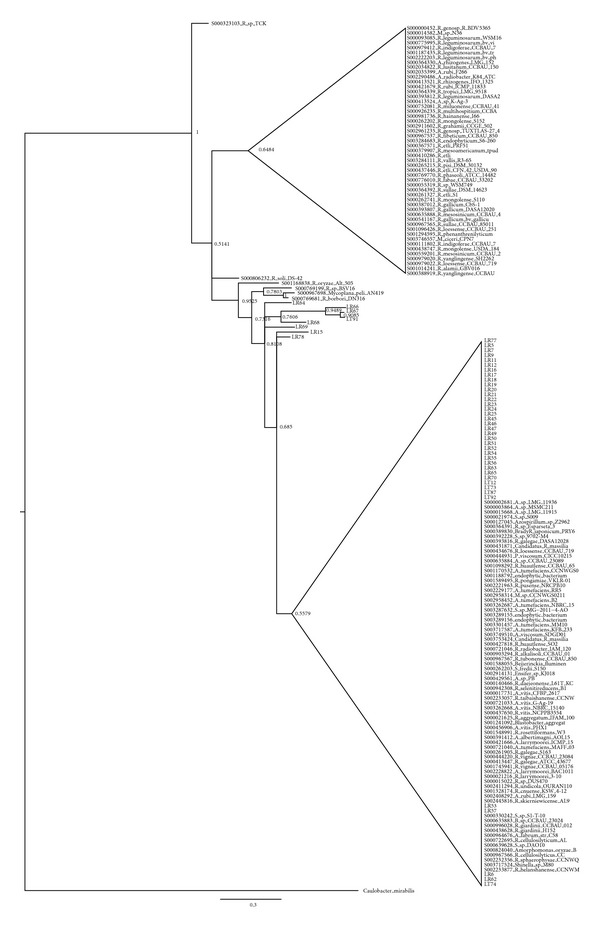
Bayesian dendrogram showing the relationships among the 16S rRNA gene sequences of 46 isolates belonging to the genus Rhizobium and those of reference type strains. Posterior probability values are indicated at each node. Nodes are collapsed at 70% probability. LT = bacteria isolated from the rhizosphere, LR = bacteria isolated from the roots (see Section 2 for details).
Figure 6.
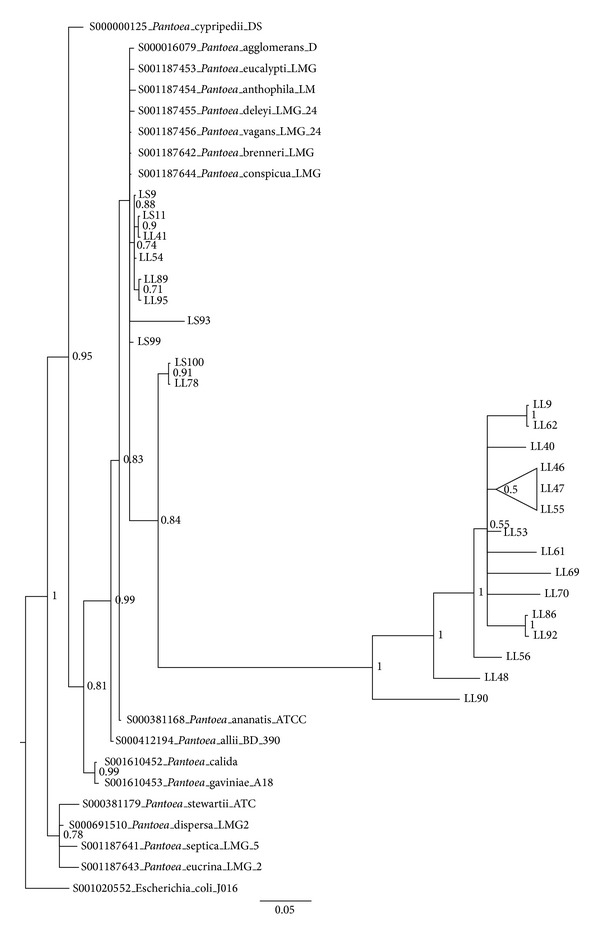
Bayesian dendrogram showing the relationships among the 16S rRNA gene sequences of 25 isolates belonging to the genus Pantoea and those of reference type strains. Posterior probability values are indicated at the node. Nodes are collapsed at 70% probability. LS = bacteria isolated from the stem, LL = bacteria isolated from the leaves (see Section 2 for details).
Figure 7.
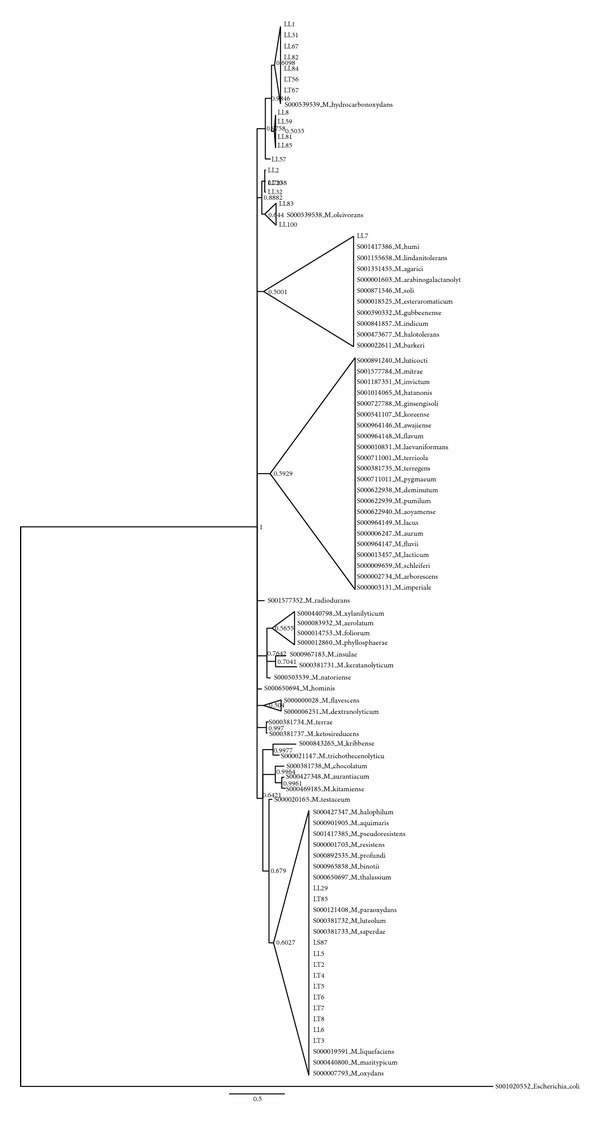
Bayesian dendrogram showing the relationships among the 16S rRNA gene sequences of 30 isolates belonging to the genus Microbacterium and those of reference type strains. Posterior probability values are indicated at the node. Nodes are collapsed at 70% probability. LT = bacteria isolated from the rhizosphere, LS = bacteria isolated from the stem, LL = bacteria isolated from the leaves (see Section 2 for details).
3.2. Inhibition of Burkholderia Cepacia Complex Strains Growth by L. angustifolia Endophytic Bacteria
In order to check the ability of L. angustifolia bacterial endophytes to antagonize the growth of (opportunistic) human pathogenic bacteria, cross-streak experiments were carried out as described in Section 2 using a panel of the 19 randomly chosen different endophytic strains isolated from soil, roots, or leafs and phylogenetically assigned to six different genera (Bacillus, Microbacterium, Pantoea, Plantibacter, Pseudomonas, and Rhizobium), as testers versus 40 Bcc strains representative of seventeen species (out of eighteen) and with either environmental or clinical origin, with some of which being multidrug-resistant. Data obtained are shown in Table 4. The analysis of these data revealed that all the tester strains were able to completely inhibit the growth of Bcc strains, including those with a clinical source and that exhibited multidrug resistance; this finding suggested that lavender endophytic bacteria are able to synthesize (strong) antimicrobial compounds. However, tester strains showed a different pattern of Bcc growth inhibition, even those affiliated to the same genus. In addition to this, there is no apparent difference in the antimicrobial potential between strains belonging to different plant compartments. Concerning the sensitivity of Bcc strains to the antimicrobial activity of lavender endophytes, strains belonging to different species exhibited a different sensitivity spectrum. However, it is quite interesting that strains with clinical origin appeared to be more sensitive to the antimicrobials synthesized by endophytic bacteria than their environmental counterparts (see for instance strains belonging to the species Burkholderia cenocepacia IIIB) (Supplementary Table 3).
Table 4.
Growth of 40 Burkholderia cepacia complex strains in the presence/absence of endophytic bacterial strains isolated from lavender.
| Species | Strain | Origin | LL1 | LL2 | LL3 | LL4 | LL6 | LL7 | LL9 | LL10 | LS1 | LS2 | LS3 | LS4 | LS5 | LS6 | LR1 | LR2 | LR3 | LR4 | LR5 | C- |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| B. cepacia | FCF 1 | CF | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + |
| B. cepacia | FCF 3 | CF | +/− | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + |
| B. multivorans | LMG 17588 | Env | + | − | − | + | +/− | +/− | − | +/− | + | − | + | +/− | +/− | − | + | − | + | − | − | + |
| B. cenocepacia (IIIA) | FCF 16 | CF | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + |
| B. cenocepacia (IIIA) | J2315 | CF | + | − | − | +/− | +/− | − | − | +/− | − | − | + | − | − | − | − | − | − | − | − | + |
| B. cenocepacia (IIIB) | FCF 18 | CF | + | − | − | − | − | − | − | − | − | − | − | − | − | − | +/− | − | − | − | − | + |
| B. cenocepacia (IIIB) | FCF 20 | CF | + | + | + | + | + | +/− | + | +/− | +/− | +/− | +/− | + | +/− | + | +/− | + | + | + | + | + |
| B. cenocepacia (IIIB) | FCF 23 | CF | + | − | − | − | − | − | − | − | +/− − | − | + | +/− | − | − | + | − | − | − | − | + |
| B. cenocepacia (IIIB) | FCF 24 | CF | + | − | − | − | − | − | − | − | +/− | − | +/− | − | − | − | + | − | − | − | − | + |
| B. cenocepacia (IIIB) | FCF 27 | CF | + | − | − | − | − | − | − | − | +/− | − | − | − | − | − | − | − | − | − | − | + |
| B. cenocepacia (IIIB) | FCF 29 | CF | + | − | − | +/− | +/− | − | − | +/− | − | − | − | − | − | − | + | − | + | − | − | + |
| B. cenocepacia (IIIB) | FCF 30 | CF | + | − | − | +/− | +/− | − | − | +/− | +/− | − | − | − | − | − | + | − | + | − | − | + |
| B. cenocepacia (IIIB) | LMG 16654 | CF | + | +/− | − | + | +/− | + | − | + | + | − | + | + | − | − | + | − | + | − | − | + |
| B. cenocepacia (IIIB) | C5424 | CF | + | +/− | − | +/− | − | − | − | +/− | − | − | +/− | − | − | − | +/− | − | + | − | − | + |
| B. cenocepacia (IIIB) | CEP511 | CF | + | − | − | +/− | − | − | − | +/− | +/− | − | +/− | +/− | − | − | + | − | + | − | − | + |
| B. cenocepacia (IIIB) | MVPC 1/16 | Env | + | +/− | − | +/− | + | +/− | − | +/− | +/− | +/− | +/− | − | +/− | − | +/− | − | + | − | +/− | + |
| B. cenocepacia (IIIB) | MVPC 1/73 | Env | + | + | + | +/− | + | + | +/− | + | + | + | + | + | + | + | + | + | + | + | +/− | + |
| B. cenocepacia (IIIIC) | LMG 19230 | Env | + | + | + | + | + | + | +/− | + | + | + | + | + | + | + | + | + | + | + | + | + |
| B. cenocepacia (IIIC) | LMG 19240 | Env | + | + | + | + | +/− | + | − | + | + | +/− | + | + | + | +/− | + | + | + | + | +/− | + |
| B. cenocepacia (IIID) | FCF 38 | CF | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + |
| B. cenocepacia (IIID) | LMG 21462 | CF | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + |
| B. stabilis | FCF 41 | CF | + | +/− | + | + | +/− | +/− | − | + | + | + | − | + | +/− | − | + | + | + | +/− | − | + |
| B. vietnamiensis | FCF 42 | CF | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + |
| B. vietnamiensis | TVV 75 | Env | + | +/− | − | +/− | − | − | − | +/− | − | − | + | +/− | − | − | +/− | − | + | − | − | + |
| B. dolosa | LMG 18941 | CF | + | − | − | +/− | +/− | +/− | − | +/− | − | − | + | +/− | − | − | +/− | +/− | + | − | − | + |
| B. dolosa | LMG 18942 | CF | + | + | − | − | +/− | +/− | − | + | + | +/− | + | +/− | − | − | + | +/− | + | +/− | − | + |
| B. dolosa | LMG 18943 | CF | + | +/− | − | + | +/− | +/− | − | +/− | +/− | − | + | +/− | − | − | + | +/− | + | − | − | + |
| B. ambifaria | MCI 7 | Env | + | + | − | +/− | +/− | +/− | − | + | +/− | − | + | +/− | − | − | + | − | + | − | − | + |
| B. ambifaria | LMG 19467 | CF | + | + | − | +/− | +/− | − | − | + | +/− | − | + | +/− | − | − | + | − | +/− | − | − | + |
| B. ambifaria | LMG 19182 | Env | + | + | − | +/− | +/− | − | − | + | − | − | + | − | − | − | + | − | +/− | − | − | + |
| B. anthina | LMG 16670 | Env | + | − | − | − | − | + | − | + | − | − | + | +/− | − | − | + | − | +/− | − | − | + |
| B. pyrrocinia | FCF 43 | CF | +/− | − | − | − | − | − | − | − | − | − | +/− | − | − | − | − | − | +/− | − | − | + |
| B. lata | LSED 4 | CF | + | − | + | − | − | +/− | − | + | − | − | + | +/− | − | − | +/− | +/− | +/− | − | − | + |
| B. latens | LMG 24064 | CF | + | − | + | +/− | − | +/− | − | + | + | − | + | + | − | + | + | + | + | + | − | + |
| B. diffusa | LMG 24065 | CF | + | +/− | + | +/− | +/− | + | − | + | + | − | + | + | − | +/− | + | + | + | + | − | + |
| B. contaminans | LMG 23361 | AI | + | +/− | +/− | +/− | +/− | + | − | + | + | + | + | + | + | +/− | + | + | + | + | − | + |
| B. seminalis | LMG 24067 | CF | + | +/− | + | +/− | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | + |
| B. metallica | LMG 24068 | CF | + | + | + | +/− | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | + |
| B. arboris | LMG 24066 | Env | + | +/− | + | +/− | + | + | +/− | + | + | + | + | + | +/− | +/− | + | + | + | +/− | − | + |
| B. ubonensis | LMG 24263 | NI | + | − | − | − | − | +/− | − | +/− | + | − | + | + | − | − | +/− | − | +/− | − | − | + |
Abbreviations: CF: strains isolated from cystic fibrosis patients; AI: strains isolated from animal infection; NI: strains isolated from nosocomial infection; ENV: environmental strain; L: strains isolated from the leaf; S: strains isolated from the stem; R: strains isolated from the root.
Symbols: +: growth; +/−: reduced growth; +/− −: very reduced growth; −: no growth; C-: Petri dishes containing only the target strains.
4. Discussion
Medicinal plants are stirring the attention of many researchers, due to the presence of compounds that constitute a large fraction of the current pharmacopoeias. Natural products have been the source of most of the active ingredients of medicines and more than 80% of drug substances are natural products or inspired by a natural compound, including most of the of anticancer and anti-infective agents [50]. Lavender plants are grown mainly for the production of essential oil, which has antiseptic and anti-inflammatory properties. In spite of the relative harsh environment for bacterial colonization, L. angustifolia tissues were found to be rich in bacterial diversity. However, the endophytic community isolated from different plant compartments showed different levels of diversity, with the leaf showing the highest level and the stem the lowest (Table 1). It is noteworthy that the same level of diversity (regardless of community composition) showed by the leaf and roots community is already reported in other species [26] even if there are few direct comparisons of roots and phyllosphere bacterial communities, especially comparisons using material from the same plants. The microbial community residing in the phyllosphere is faced with a nutrient poor and variable environment that is characterized by fluctuating temperature, humidity, and UV radiation and so it is predicted to be more diverse than that within the rhizosphere, a more homeostatic environment. These findings, supported also by the similar bacterial titre, are in agreement with the idea that the source of inoculum for the leaf community (except for the vertically transferred via seeds) may be exclusive (e.g., aerosol, even if the process is not yet clarified, Bulgarelli et al. 2013) and not related to transmission from the roots. Moreover, the low diversity of the community isolated in the stem, dominated by clonally (Table 2) populations of Pseudomonas, could be related to the highly specific main tissue (phloem) present there, which could provide an homeostatic environment, rich in nutrient (sucrose contained in the phloem tissues) but poorer (if compared to leaf and roots) in secondary metabolites that may promote ecological niche differentiation.
Concerning the taxonomic community composition, different taxonomic profiles were found for endosphere and rhizosphere. More in detail the rhizosphere community showed quite similar diversity indexes if compared to the roots one, but with a different composition; as an example, members of the phylum Actinobacteria are completely absent from the root internal tissues that are largely dominated by Proteobacteria, while they are present in the soil. To explain this finding a two-step model has been proposed [16]: soil abiotic properties determine the structure of the initial bacterial communities that is then modified by rhizodeposits with plant roots cell wall features and released metabolites promoting the growth of organotrophic bacteria, thereby initiating a soil community shift. In the second step, convergent host genotype-dependent selection [51, 52] fine-tunes community profiles thriving on the rhizoplane and within plant roots.
Moreover, endosphere communities were different between plant organs (roots, stems, and leaves), even if the internal tissues are dominated, as already reported [25, 52–54] by members of the phylum Proteobacteria; an example, while rhizobia were isolated from root internal tissues, they are completely absent from stems and leaves (Figures 2 and 3); isolates belonging to the genus Pantoea, on the contrary, were isolated from aerial parts but not from root tissues. Hence, it is possible that plant might “select” specific taxa for entering inside its compartment. Such hypothesis is supported also by the pairwise differentiation values; the communities isolated from the different tissues, even those in physical continuity, are in fact strongly differentiated with the stem, representing a low diversity compartment separating the two high diversity organs roots and leaves. In this view it is noteworthy that in lavender only Pseudomonas represents a ubiquitous and abundant genus of both rhizosphere and endosphere (Figure 3). The analysis of intrageneric diversity pointed out also that Pseudomonas isolates belong to a low differentiated clonal population (Table 2 and Suppl. Figure 2), suggesting also that this component of the community either diffused inside the organs from a radical or foliar entry point or established during plants development, putatively from the seed inoculum. A similar consideration can be proposed for isolates of rhizobia but for the rhizosphere-root internal tissues continuum: a low taxonomically differentiated community from the soil entered (increasing its relative abundance) inside the plant organ. An entry point from the leaves is on the contrary consistent with the distribution of Pantoea isolates with their strong diversity (Table 2) suggesting a diversification in response to the high variable leaf environment. The isolates belonging to the genus Stenotrophomonas show an interesting distribution pattern (Figure 3); they are rare in the rhizosphere, abundant in the roots and in the leaves, and absent in the stem; this finding suggests a different entry point or a negative selection in the stem; the second explanation is supported by admixture of root and leaves isolates (Figure 4).
The detailed phylogenetic analyses of the isolates enabled us also to putatively ascribe some isolates to already described endophytic strains that can be targeted for functional characterisation; many Microbacterium leaves isolates cluster with M. hydrocarbonoxydans and M. oleivorans, two species with crude oil degrading activity [49] being this metabolic activity that is found frequently associated with a phyllosphere adapted lifestyle [55]. Several isolates cluster with bacteria with interesting biotechnological or ecological applications, such as S. chelatiphaga, a remarkable species with biotechnological potential in phytoremediation [56] and S. rizophila a plant associated bacterium with antifungal activity [57] or Bacillus mojavensis a bacteriumclosely related to B. subtilis and already reported having an endophytic lifestyle and showing an antagonistic role against plant pathogenic fungi [58]. On the contrary, many lavender Pseudomonas isolates cluster with known plant pathogen strains highlighting the sometimes subtle difference among pathogenic and endophytic lifestyle and with Pantoea agglomerans (and also with lavender isolates clustering withinthe B. licheniformis and B. soronensis groups), a known plant associated bacterium, and an opportunistic pathogen in immune-compromised human individuals drawing attention to the possible pathogenic action of endophytic bacteria in humans [59, 60].
The overall analysis of the dendrograms points out that the characterization of isolates from endophytic communities can lead to the identification (as an example for Pantoea and rhizobia) of not yet described strains that may represent untapped resources of bioactive compounds of agricultural or medical interest.
Even though it has not been still completely demonstrated, it cannot be excluded that the possibility that the qualiquantitative spectrum of bioactive metabolites in medical plants and their essential oils may be related also on the activity of bacterial endophytes as they may promote plant health and growth, elicit plant metabolism, or directly produce biotechnologically relevant compounds [36]. In this context, the characterization of cultivable bacterial communities is a fundamental step to this purpose since it paves the way to the possibility to build collections of isolates that may be phenotypically typed for important traits. In our opinion, data obtained in this work (Table 4 and Supplementary Tables 2 and 3) offers a preliminary but very promising example of the biotechnological potential of bacteria isolated from medicinal plants. In this pilot study in fact the majority of the tested endophytes showed to inhibit the growth of several (in some case all) human pathogenic strains belonging to the Bcc complexes that are known for their resistance to traditional antibiotics. These data are particularly interesting if correlated with the recent finding that essential oils from different medicinal plants are able to strongly (or completely) interfere with the growth of the same Bcc strains [61] and that bacterial endophytes isolated from plants of the same species exhibited the same bioactivity (Maida et al., in preparation). It is therefore possible that the therapeutic properties of the essential oil might reside also on bioactive compounds of microbial origin. We are completely aware that the determination of the correlation (eventually) existing between the composition of bacterial endophytic communities and the bioactivity of essential oils extracted from a medicinal plant would require the parallel analysis of the bacterial community and the essential oil activity extracted from the same plant. However, data obtained in this work and those from Maida et al. [62] support this idea.
It is also particularly intriguing, the idea that the antimicrobial activity of essential oils may reside on the action of multiple compounds, some of them produced or elicited by the microbiota, limiting the observed rapid evolution of human pathogenic bacteria resistant to single molecule antimicrobials.
The characterization of the heterotrophic aerobic endophytic bacterial community of L. angustifolia highlighted the existence of a diversified community between different organs/tissues that may be related to the coexistence of different sources of inoculum and/or a selection of the communities promoted by nutrient sand metabolites availability and antagonistic forces. Several not previously characterized strains have been isolated and molecularly typed, confirming that the analysis of the bacterial communities inhabiting extreme or unconventional environments may represent a proper strategy for the discovery of untapped sources of functional biodiversity and bioactive molecule production.
Supplementary Material
Supplementary Figure 1: Bayesian dendrogram showing the relationships among the 16S rDNA sequences of 47 isolates belonging to the genus Bacillus and those of reference type strains. Posterior probability values are indicated at the node. Nodes are collapsed at 70% probability. LT = bacteria isolated from the rhizosphere, LR = bacteria isolated from the roots, LS = bacteria isolated from the stem.
Supplementary Figure 2: Bayesian dendrogram showing the relationships among the 16S rDNA sequences of isolates belonging to the genus Pseudomonas and those of reference type strains. Posterior probability values are indicated at the node. Nodes are collapsed at 70% probability. LT= bacteria isolated from the rhizosphere, LR = bacteria isolated from the roots, LS = bacteria isolated from the stem, LL = bacteria isolated from the leaves.
Supplementary Figure 3: Maximum parsimony dendrogram showing the relationships among the 16S rDNA sequences of 37 isolates belonging to the genus Stenotrophomonas and those of reference type strains. Bootstrap values are indicated at the node. LL= bacteria isolated from the leaves, LR = bacteria isolated from the roots (see Material and Methods and Suppl. Table 4 for details).
Supplementary Figure 4: Maximum parsimony dendrogram showing the relationships among the 16S rDNA sequences of 46 isolates belonging to the genus Rhizobium and those of reference type strains. Bootstrap values are indicated at each node. Nodes are collapsed at 70% probability. LT= bacteria isolated from the rhizosphere, LR = bacteria isolated from the roots (see Material and Methods and Suppl. Table 4 for details).
Supplementary Figure 5: Maximum parsimony dendrogram showing the relationships among the 16S rDNA sequences of 25 isolates belonging to the genus Pantoea and those of reference type strains. Bootstrap values are indicated at the node. Nodes are collapsed at 70% probability. LS = bacteria isolated from the stem, LL = bacteria isolated from the leaves (see Material and Methods and Suppl. Table 4 for details).
Supplementary Figure 6: Maximum parsimony dendrogram showing the relationships among the 16S rDNA sequences of 30 isolates belonging to the genus Microbacterium and those of reference type strains. Bootstrap values are indicated at the node. Nodes are collapsed at 70% probability. LT = bacteria isolated from the rhizosphere, LS = bacteria isolated from the stem, LL = bacteria isolated from the leaves (see Material and Methods and Suppl. Table 4 for details).
Supplementary Figure 7: Maximum parsimony dendrogram showing the relationships among the 16S rDNA sequences of 47 isolates belonging to the genus Bacillus and those of reference type strains. Bootstrap values are indicated at the node. Nodes are collapsed at 70% probability. LT = bacteria isolated from the rhizosphere, LR = bacteria isolated from the roots, LS = bacteria isolated from the stem (see Material and Methods and Suppl. Table 4 for details).
Supplementary Figure 8: Maximum parsimony dendrogram showing the relationships among the 16S rDNA sequences of isolates belonging to the genus Pseudomonas and those of reference type strains. Bootstrap values are indicated at the node. Nodes are collapsed at 70% probability. LT= bacteria isolated from the rhizosphere, LR = bacteria isolated from the roots, LS = bacteria isolated from the stem, LL = bacteria isolated from the leaves (see Material and Methods and Suppl. Table 4 for details).
Supplementary Figure 9: NJ dendrogram showing the relationships among the 16S rDNA sequences of 37 isolates belonging to the genus Stenotrophomans and those of reference type strains. Scale bars represent the Kimura-2 distance. Bootstrap values are indicated at the node. Nodes are collapsed at 70% probability. LL= bacteria isolated from the leaves, LR = bacteria isolated from the roots.
Supplementary Figure 10: NJ dendrogram showing the relationships among the 16S rDNA sequences of 46 isolates belonging to the genus Rhizobium and those of reference type strains. Scale bars represent the Kimura-2 distance. Bootstrap values are indicated at each node. Nodes are collapsed at 70% probability. LT= bacteria isolated from the rhizosphere, LR = bacteria isolated from the roots.
Supplementary Figure 11: NJ dendrogram showing the relationships among the 16S rDNA sequences of 25 isolates belonging to the genus Pantoea and those of reference type strains. Scale bars represent the Kimura-2 distance. Bootstrap values are indicated at the node. Nodes are collapsed at 70% probability. LS = bacteria isolated from the stem, LL = bacteria isolated from the leaves.
Supplementary Figure 12: NJ dendrogram showing the relationships among the 16S rDNA sequences of 30 isolates belonging to the genus Microbacterium and those of reference type strains. Scale bars represent the Kimura-2 distance. Bootstrap values are indicated at the node. Nodes are collapsed at 70% probability. LT = bacteria isolated from the rhizosphere, LS = bacteria isolated from the stem, LL = bacteria isolated from the leaves.
Supplementary Figure 13: NJ dendrogram showing the relationships among the 16S rDNA sequences of 47 isolates belonging to the genus Bacillus and those of reference type strains. Scale bars represent the Kimura-2 distance. Bootstrap values are indicated at the node. Nodes are collapsed at 70% probability. LT = bacteria isolated from the rhizosphere, LR = bacteria isolated from the roots, LS = bacteria isolated from the stem.
Supplementary Figure 14: NJ dendrogram showing the relationships among the 16S rDNA sequences of isolates belonging to the genus Pseudomonas and those of reference type strains. Scale bars represent the Kimura-2 distance. Bootstrap values are indicated at the node. Nodes are collapsed at 70% probability. LT= bacteria isolated from the rhizosphere, LR = bacteria isolated from the roots, LS = bacteria isolated from the stem, LL = bacteria isolated from the leaves.
Supplementary Table 1: List of bacterial endophytic strains used in this work as tester in the crossstreak experiments.
Supplementary Table 2: Number of Bcc target strains whose growth is inhibited by lavender bacterial endophytes.
Supplementary Table 3: Sensitivity spectrum exhibited by the 40 Bcc strains in the cross streak experiment vs a panel of lavender endophytic bacterial isolates.
Supplementary Table 4: Length of aligned matrices (bp), Variable and conserved characters number, Parsimony-informative characters, Consistency Index (CI), Retention Index (RI) and species used as outgroup of dendrograms obtained with Maximun Parsimony (see Materials and Methods for details).
Acknowledgments
This work was supported by grants from the Ente Cassa di Risparmio di Firenze (Grant 2013.0657, “Herbiome: new antimicrobial compounds from endophytic bacteria isolated from medicinal plants”). Giovanni Emiliani is financially supported by the Regione Toscana POR CRO FSE 2007-2013 Asse IV Grant “Sysbiofor” (CNR-9). Marco Fondi and Elena Perrin are financially supported by the FEMS Advanced Fellowship (FAF2012) and the “Buzzati-Traverso” Foundation, respectively.
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
References
- 1.Reinhold-Hurek B, Hurek T. Living inside plants: bacterial endophytes. Current Opinion in Plant Biology. 2011;14(4):435–443. doi: 10.1016/j.pbi.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 2.Rosenblueth M, Martínez-Romero E. Bacterial endophytes and their interactions with hosts. Molecular Plant-Microbe Interactions. 2006;19(8):827–837. doi: 10.1094/MPMI-19-0827. [DOI] [PubMed] [Google Scholar]
- 3.Ryan RP, Germaine K, Franks A, Ryan DJ, Dowling DN. Bacterial endophytes: recent developments and applications. FEMS Microbiology Letters. 2008;278(1):1–9. doi: 10.1111/j.1574-6968.2007.00918.x. [DOI] [PubMed] [Google Scholar]
- 4.Çakmakçi R, Dönmez F, Aydın A, Şahin F. Growth promotion of plants by plant growth-promoting rhizobacteria under greenhouse and two different field soil conditions. Soil Biology and Biochemistry. 2006;38:1482–1487. [Google Scholar]
- 5.Hardoim PR, van Overbeek LS, Elsas JDV. Properties of bacterial endophytes and their proposed role in plant growth. Trends in Microbiology. 2008;16(10):463–471. doi: 10.1016/j.tim.2008.07.008. [DOI] [PubMed] [Google Scholar]
- 6.Compant S, Clément C, Sessitsch A. Plant growth-promoting bacteria in the rhizo- and endosphere of plants: their role, colonization, mechanisms involved and prospects for utilization. Soil Biology and Biochemistry. 2010;42(5):669–678. [Google Scholar]
- 7.Glick BR. Bacterial ACC deaminase and the alleviation of plant stress. Advances in Applied Microbiology. 2004;56:291–312. doi: 10.1016/S0065-2164(04)56009-4. [DOI] [PubMed] [Google Scholar]
- 8.Glick BR, Todorovic B, Czarny J, Cheng Z, Duan J, McConkey B. Promotion of plant growth by bacterial ACC deaminase. Critical Reviews in Plant Sciences. 2007;26(5-6):227–242. [Google Scholar]
- 9.Doty SL, Oakley B, Xin G, et al. Diazotrophic endophytes of native black cottonwood and willow. Symbiosis. 2009;47(1):23–33. [Google Scholar]
- 10.Lugtenberg B, Kamilova F. Plant-growth-promoting rhizobacteria. Annual Review of Microbiology. 2009;63:541–556. doi: 10.1146/annurev.micro.62.081307.162918. [DOI] [PubMed] [Google Scholar]
- 11.Castillo UF, Strobel GA, Ford EJ, et al. Munumbicins, wide-spectrum antibiotics produced by Streptomyces NRRL 30562, endophytic on Kennedia nigriscans . Microbiology. 2002;148(9):2675–2685. doi: 10.1099/00221287-148-9-2675. [DOI] [PubMed] [Google Scholar]
- 12.Igarashi Y, Trujillo ME, Martínez-Molina E, et al. Antitumor anthraquinones from an endophytic actinomycete Micromonospora lupini sp. nov. Bioorganic & Medicinal Chemistry Letters. 2007;17(13):3702–3705. doi: 10.1016/j.bmcl.2007.04.039. [DOI] [PubMed] [Google Scholar]
- 13.Shweta S, Bindu JH, Raghu J, et al. Isolation of endophytic bacteria producing the anti-cancer alkaloid camptothecine from Miquelia dentata Bedd. (Icacinaceae) Phytomedicine. 2013;20:913–917. doi: 10.1016/j.phymed.2013.04.004. [DOI] [PubMed] [Google Scholar]
- 14.Taechowisan T, Wanbanjob A, Tuntiwachwuttikul P, Liu J. Anti-inflammatory activity of lansais from endophytic Streptomyces sp. SUC1 in LPS-induced RAW 264.7 cells. Food and Agricultural Immunology. 2009;20(1):67–77. [Google Scholar]
- 15.Hardoim PR, Hardoim CCP, van Overbeek LS, van Elsas JD. Dynamics of seed-borne rice endophytes on early plant growth stages. PLoS ONE. 2012;7(2) doi: 10.1371/journal.pone.0030438.e30438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bulgarelli D, Schlaeppi K, Spaepen S, van Themaat EVL, Schulze-Lefert P. Structure and functions of the bacterial microbiota of plants. Annual Review of Plant Biology. 2013;64:807–838. doi: 10.1146/annurev-arplant-050312-120106. [DOI] [PubMed] [Google Scholar]
- 17.Mengoni A, Mocali S, Surico G, Tegli S, Fani R. Fluctuation of endophytic bacteria and phytoplasmosis in elm trees. Microbiological Research. 2003;158(4):363–369. doi: 10.1078/0944-5013-00216. [DOI] [PubMed] [Google Scholar]
- 18.Mengoni A, Pini F, Huang L-N, Shu W-S, Bazzicalupo M. Plant-by-plant variations of bacterial communities associated with leaves of the nickel hyperaccumulator Alyssum bertolonii desv. Microbial Ecology. 2009;58(3):660–667. doi: 10.1007/s00248-009-9537-5. [DOI] [PubMed] [Google Scholar]
- 19.Mocali S, Bertelli E, di Cello F, et al. Fluctuation of bacteria isolated from elm tissues during different seasons and from different plant organs. Research in Microbiology. 2003;154(2):105–114. doi: 10.1016/S0923-2508(03)00031-7. [DOI] [PubMed] [Google Scholar]
- 20.Pini F, Frascella A, Santopolo L, et al. Exploring the plant-associated bacterial communities in Medicago sativa L. BMC Microbiology. 2012;12, article 78 doi: 10.1186/1471-2180-12-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mengoni A, Cecchi L, Gonnelli C. Nickel hyperaccumulating plants and Alyssum bertolonii: model systems for studying biogeochemical interactions in serpentine soils. In: Kothe E, Varma A, editors. Bio-Geo Interactions in Metal-Contaminated Soils. Berlin, Germany: Springer; 2012. pp. 279–296. [Google Scholar]
- 22.Ikeda S, Okubo T, Anda M, et al. Community- and genome-based views of plant-associated bacteria: plant-bacterial interactions in soybean and rice. Plant and Cell Physiology. 2010;51(9):1398–1410. doi: 10.1093/pcp/pcq119. [DOI] [PubMed] [Google Scholar]
- 23.Pini F, Galardini M, Bazzicalupo M, Mengoni A. Plant-bacteria association and symbiosis: are there common genomic traits in alphaproteobacteria? Genes. 2011;2(4):1017–1032. doi: 10.3390/genes2041017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aleklett K, Hart M. The root microbiota‚ a fingerprint in the soil? Plant Soil. 2013;370:671–686. [Google Scholar]
- 25.Beneduzi A, Moreira F, Costa PB, et al. Diversity and plant growth promoting evaluation abilities of bacteria isolated from sugarcane cultivated in the South of Brazil. Applied Soil Ecology. 2013;63:94–104. [Google Scholar]
- 26.Bodenhausen N, Horton MW, Bergelson J. Bacterial communities associated with the leaves and the roots of Arabidopsis thaliana . PLoS ONE. 2013;8(2) doi: 10.1371/journal.pone.0056329.e56329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.da Silva TF, Vollú RE, Jurelevicius D, et al. Does the essential oil of Lippia sidoides Cham. (pepper-rosmarin) affect its endophytic microbial community? BMC Microbiology. 2013;13(1, article 29) doi: 10.1186/1471-2180-13-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lucero ME, Unc A, Cooke P, Dowd S, Sun S. Endophyte microbiome diversity in micropropagated Atriplex canescens and Atriplex torreyi var griffithsii . PLoS ONE. 2011;6(3) doi: 10.1371/journal.pone.0017693.e17693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lundberg DS, Lebeis SL, Paredes SH, et al. Defining the core Arabidopsis thaliana root microbiome. Nature. 2012;487(7409):86–90. doi: 10.1038/nature11237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cavanagh HM, Wilkinson JM. Biological activities of lavender essential oil. Phytotherapy Research. 2002;16(4):301–308. doi: 10.1002/ptr.1103. [DOI] [PubMed] [Google Scholar]
- 31.Woronuk G, Demissie Z, Rheault M, Mahmoud S. Biosynthesis and therapeutic properties of Lavandula essential oil constituents. Planta Medica. 2011;77(1):7–15. doi: 10.1055/s-0030-1250136. [DOI] [PubMed] [Google Scholar]
- 32.Perry R, Terry R, Watson LK, Ernst E. Is lavender an anxiolytic drug? A systematic review of randomised clinical trials. Phytomedicine. 2012;19:825–835. doi: 10.1016/j.phymed.2012.02.013. [DOI] [PubMed] [Google Scholar]
- 33.de Rapper S, Kamatou G, Viljoen A, van Vuuren S. The in vitro antimicrobial activity of Lavandula angustifolia essential oil in combination with other aroma-therapeutic oils. Evidence-Based Complementary and Alternative Medicine. 2013;2013:10 pages. doi: 10.1155/2013/852049.852049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moon T, Wilkinson JM, Cavanagh HM. Antiparasitic activity of two Lavandula essential oils against Giardia duodenalis, Trichomonas vaginalis and Hexamita inflata . Parasitology Research. 2006;99(6):722–728. doi: 10.1007/s00436-006-0234-8. [DOI] [PubMed] [Google Scholar]
- 35.Yap PS, Lim SH, Hu CP, Yiap BC. Combination of essential oils and antibiotics reduce antibiotic resistance in plasmid-conferred multidrug resistant bacteria. Phytomedicine. 2013;20:710–713. doi: 10.1016/j.phymed.2013.02.013. [DOI] [PubMed] [Google Scholar]
- 36.Brader G, Compant S, Mitter B, Trognitz F, Sessitsch A. Metabolic potential of endophytic bacteria. Current Opinion in Biotechnology. 2014;27:30–37. doi: 10.1016/j.copbio.2013.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Drevinek P, Mahenthiralingam E. Burkholderia cenocepacia in cystic fibrosis: epidemiology and molecular mechanisms of virulence. Clinical Microbiology and Infection. 2010;16(7):821–830. doi: 10.1111/j.1469-0691.2010.03237.x. [DOI] [PubMed] [Google Scholar]
- 38.Bazzini S, Udine C, Riccardi G. Molecular approaches to pathogenesis study of Burkholderia cenocepacia, an important cystic fibrosis opportunistic bacterium. Applied Microbiology and Biotechnology. 2011;92(5):887–895. doi: 10.1007/s00253-011-3616-5. [DOI] [PubMed] [Google Scholar]
- 39.Maida I, lo Nostro A, Pesavento G, et al. Exploring the anti-Burkholderia cepacia complex activity of essential oils: a preliminary analysis. Evidence-Based Complementary and Alternative Medicine. 2014;2014:10 pages. doi: 10.1155/2014/573518.573518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mengoni A, Barzanti R, Gonnelli C, Gabbrielli R, Bazzicalupo M. Characterization of nickel-resistant bacteria isolated from serpentine soil. Environmental Microbiology. 2001;3(11):691–698. doi: 10.1046/j.1462-2920.2001.00243.x. [DOI] [PubMed] [Google Scholar]
- 41.Altschul SF, Madden TL, Schaffer AA, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ronquist F, Teslenko M, van der Mark P, et al. Mrbayes 3.2: efficient bayesian phylogenetic inference and model choice across a large model space. Systematic Biology. 2012;61(3):539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sievers F, Wilm A, Dineen D, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Molecular Systems Biology. 2011;7, article 539 doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hammer Ø, Harper DAT, Ryan PD. PAST: paleontological statistics software package for education and data analysis. Palaeontologia Electronica. 2001;4(1):1–9. [Google Scholar]
- 47.Papaleo MC, Fondi M, Maida I, et al. Sponge-associated microbial Antarctic communities exhibiting antimicrobial activity against Burkholderia cepacia complex bacteria. Biotechnology Advances. 2012;30(1):272–293. doi: 10.1016/j.biotechadv.2011.06.011. [DOI] [PubMed] [Google Scholar]
- 48.lo Giudice A, Bruni V, Michaud L. Characterization of Antarctic psychrotrophic bacteria with antibacterial activities against terrestrial microorganisms. Journal of Basic Microbiology. 2007;47(6):496–505. doi: 10.1002/jobm.200700227. [DOI] [PubMed] [Google Scholar]
- 49.Schippers A, Bosecker K, Spröer C, Schumann P. Microbacterium oleivorans sp. nov. and Microbacterium hydrocarbonoxydans sp. nov., novel crude-oil-degrading Gram-positive bacteria. International Journal of Systematic and Evolutionary Microbiology. 2005;55(2):655–660. doi: 10.1099/ijs.0.63305-0. [DOI] [PubMed] [Google Scholar]
- 50.McChesney JD, Venkataraman SK, Henri JT. Plant natural products: back to the future or into extinction? Phytochemistry. 2007;68(14):2015–2022. doi: 10.1016/j.phytochem.2007.04.032. [DOI] [PubMed] [Google Scholar]
- 51.Manter DK, Delgado JA, Holm DG, Stong RA. Pyrosequencing reveals a highly diverse and cultivar-specific bacterial endophyte community in potato roots. Microbial Ecology. 2010;60(1):157–166. doi: 10.1007/s00248-010-9658-x. [DOI] [PubMed] [Google Scholar]
- 52.Ulrich K, Ulrich A, Ewald D. Diversity of endophytic bacterial communities in poplar grown under field conditions. FEMS Microbiology Ecology. 2008;63(2):169–180. doi: 10.1111/j.1574-6941.2007.00419.x. [DOI] [PubMed] [Google Scholar]
- 53.Gottel NR, Castro HF, Kerley M, et al. Distinct microbial communities within the endosphere and rhizosphere of Populus deltoides roots across contrasting soil types. Applied and Environmental Microbiology. 2011;77(17):5934–5944. doi: 10.1128/AEM.05255-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ma B, Lv X, Warren A, Gong J. Shifts in diversity and community structure of endophytic bacteria and archaea across root, stem and leaf tissues in the common reed, Phragmites australis, along a salinity gradient in a marine tidal wetland of northern China. Antonie van Leeuwenhoek. 2013;104(5):759–768. doi: 10.1007/s10482-013-9984-3. [DOI] [PubMed] [Google Scholar]
- 55.Vorholt JA. Microbial life in the phyllosphere. Nature Reviews Microbiology. 2012;10(12):828–840. doi: 10.1038/nrmicro2910. [DOI] [PubMed] [Google Scholar]
- 56.Ryan RP, Monchy S, Cardinale M, et al. The versatility and adaptation of bacteria from the genus Stenotrophomonas . Nature Reviews Microbiology. 2009;7(7):514–525. doi: 10.1038/nrmicro2163. [DOI] [PubMed] [Google Scholar]
- 57.Wolf A, Fritze A, Hagemann M, Berg G. Stenotrophomonas rhizophila sp. nov., a novel plant-associated bacterium with antifungal properties. International Journal of Systematic and Evolutionary Microbiology. 2002;52(6):1937–1944. doi: 10.1099/00207713-52-6-1937. [DOI] [PubMed] [Google Scholar]
- 58.Bacon CW, Hinton DM. Endophytic and biological control potential of Bacillus mojavensis and related species. Biological Control. 2002;23(3):274–284. [Google Scholar]
- 59.Berg G, Eberl L, Hartmann A. The rhizosphere as a reservoir for opportunistic human pathogenic bacteria. Environmental Microbiology. 2005;7(11):1673–1685. doi: 10.1111/j.1462-2920.2005.00891.x. [DOI] [PubMed] [Google Scholar]
- 60.Tyler HL, Triplett EW. Plants as a habitat for beneficial and/or human pathogenic bacteria. Annual Review of Phytopathology. 2008;46:53–73. doi: 10.1146/annurev.phyto.011708.103102. [DOI] [PubMed] [Google Scholar]
- 61.Perry R, Terry R, Watson LK, Ernst E. Is lavender an anxiolytic drug? A systematic review of randomised clinical trials. Phytomedicine. 2012;19(8-9):825–835. doi: 10.1016/j.phymed.2012.02.013. [DOI] [PubMed] [Google Scholar]
- 62.Maida I, lo Nostro A, Pesavento G, et al. Exploring the anti-Burkholderia cepacia complex activity of essential oils: a preliminary analysis. Evidence-Based Complementary and Alternative Medicine. 2014;2014:10 pages. doi: 10.1155/2014/573518.573518 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1: Bayesian dendrogram showing the relationships among the 16S rDNA sequences of 47 isolates belonging to the genus Bacillus and those of reference type strains. Posterior probability values are indicated at the node. Nodes are collapsed at 70% probability. LT = bacteria isolated from the rhizosphere, LR = bacteria isolated from the roots, LS = bacteria isolated from the stem.
Supplementary Figure 2: Bayesian dendrogram showing the relationships among the 16S rDNA sequences of isolates belonging to the genus Pseudomonas and those of reference type strains. Posterior probability values are indicated at the node. Nodes are collapsed at 70% probability. LT= bacteria isolated from the rhizosphere, LR = bacteria isolated from the roots, LS = bacteria isolated from the stem, LL = bacteria isolated from the leaves.
Supplementary Figure 3: Maximum parsimony dendrogram showing the relationships among the 16S rDNA sequences of 37 isolates belonging to the genus Stenotrophomonas and those of reference type strains. Bootstrap values are indicated at the node. LL= bacteria isolated from the leaves, LR = bacteria isolated from the roots (see Material and Methods and Suppl. Table 4 for details).
Supplementary Figure 4: Maximum parsimony dendrogram showing the relationships among the 16S rDNA sequences of 46 isolates belonging to the genus Rhizobium and those of reference type strains. Bootstrap values are indicated at each node. Nodes are collapsed at 70% probability. LT= bacteria isolated from the rhizosphere, LR = bacteria isolated from the roots (see Material and Methods and Suppl. Table 4 for details).
Supplementary Figure 5: Maximum parsimony dendrogram showing the relationships among the 16S rDNA sequences of 25 isolates belonging to the genus Pantoea and those of reference type strains. Bootstrap values are indicated at the node. Nodes are collapsed at 70% probability. LS = bacteria isolated from the stem, LL = bacteria isolated from the leaves (see Material and Methods and Suppl. Table 4 for details).
Supplementary Figure 6: Maximum parsimony dendrogram showing the relationships among the 16S rDNA sequences of 30 isolates belonging to the genus Microbacterium and those of reference type strains. Bootstrap values are indicated at the node. Nodes are collapsed at 70% probability. LT = bacteria isolated from the rhizosphere, LS = bacteria isolated from the stem, LL = bacteria isolated from the leaves (see Material and Methods and Suppl. Table 4 for details).
Supplementary Figure 7: Maximum parsimony dendrogram showing the relationships among the 16S rDNA sequences of 47 isolates belonging to the genus Bacillus and those of reference type strains. Bootstrap values are indicated at the node. Nodes are collapsed at 70% probability. LT = bacteria isolated from the rhizosphere, LR = bacteria isolated from the roots, LS = bacteria isolated from the stem (see Material and Methods and Suppl. Table 4 for details).
Supplementary Figure 8: Maximum parsimony dendrogram showing the relationships among the 16S rDNA sequences of isolates belonging to the genus Pseudomonas and those of reference type strains. Bootstrap values are indicated at the node. Nodes are collapsed at 70% probability. LT= bacteria isolated from the rhizosphere, LR = bacteria isolated from the roots, LS = bacteria isolated from the stem, LL = bacteria isolated from the leaves (see Material and Methods and Suppl. Table 4 for details).
Supplementary Figure 9: NJ dendrogram showing the relationships among the 16S rDNA sequences of 37 isolates belonging to the genus Stenotrophomans and those of reference type strains. Scale bars represent the Kimura-2 distance. Bootstrap values are indicated at the node. Nodes are collapsed at 70% probability. LL= bacteria isolated from the leaves, LR = bacteria isolated from the roots.
Supplementary Figure 10: NJ dendrogram showing the relationships among the 16S rDNA sequences of 46 isolates belonging to the genus Rhizobium and those of reference type strains. Scale bars represent the Kimura-2 distance. Bootstrap values are indicated at each node. Nodes are collapsed at 70% probability. LT= bacteria isolated from the rhizosphere, LR = bacteria isolated from the roots.
Supplementary Figure 11: NJ dendrogram showing the relationships among the 16S rDNA sequences of 25 isolates belonging to the genus Pantoea and those of reference type strains. Scale bars represent the Kimura-2 distance. Bootstrap values are indicated at the node. Nodes are collapsed at 70% probability. LS = bacteria isolated from the stem, LL = bacteria isolated from the leaves.
Supplementary Figure 12: NJ dendrogram showing the relationships among the 16S rDNA sequences of 30 isolates belonging to the genus Microbacterium and those of reference type strains. Scale bars represent the Kimura-2 distance. Bootstrap values are indicated at the node. Nodes are collapsed at 70% probability. LT = bacteria isolated from the rhizosphere, LS = bacteria isolated from the stem, LL = bacteria isolated from the leaves.
Supplementary Figure 13: NJ dendrogram showing the relationships among the 16S rDNA sequences of 47 isolates belonging to the genus Bacillus and those of reference type strains. Scale bars represent the Kimura-2 distance. Bootstrap values are indicated at the node. Nodes are collapsed at 70% probability. LT = bacteria isolated from the rhizosphere, LR = bacteria isolated from the roots, LS = bacteria isolated from the stem.
Supplementary Figure 14: NJ dendrogram showing the relationships among the 16S rDNA sequences of isolates belonging to the genus Pseudomonas and those of reference type strains. Scale bars represent the Kimura-2 distance. Bootstrap values are indicated at the node. Nodes are collapsed at 70% probability. LT= bacteria isolated from the rhizosphere, LR = bacteria isolated from the roots, LS = bacteria isolated from the stem, LL = bacteria isolated from the leaves.
Supplementary Table 1: List of bacterial endophytic strains used in this work as tester in the crossstreak experiments.
Supplementary Table 2: Number of Bcc target strains whose growth is inhibited by lavender bacterial endophytes.
Supplementary Table 3: Sensitivity spectrum exhibited by the 40 Bcc strains in the cross streak experiment vs a panel of lavender endophytic bacterial isolates.
Supplementary Table 4: Length of aligned matrices (bp), Variable and conserved characters number, Parsimony-informative characters, Consistency Index (CI), Retention Index (RI) and species used as outgroup of dendrograms obtained with Maximun Parsimony (see Materials and Methods for details).


