Abstract
Colon tumors are a major cause of cancer death, yet their molecular intricacies are not fully understood. We demonstrate that the histone demethylases JMJD2A, JMJD2B and JMJD2C are overexpressed in colon cancer cell lines, whereas another related protein, JMJD2D, is not. Interestingly, despite their high homology, the intracellular localization of JMJD2A-C is different in colon and other cancer cells, with JMJD2A being present comparably in the cytoplasm and nucleus, JMJD2B more prevalent in the nucleus and JMJD2C strongly associated with chromatin. This suggests that each of these three proteins performs different, non-redundant functions. Moreover, we show that JMJD2C (also called KDM4C) forms complexes with β-catenin, an oncoprotein whose overexpression is crucial for the development of most colonic tumors. In addition, JMJD2C downregulation reduced both growth and clonogenic capacity of HCT-116 colon cancer cells. Further, JMJD2C was required for efficient expression of the growth stimulatory proteins FRA1 and cyclin D1 as well as the survival factor BCL2. Lastly, we identified derivatives of curcumin as in vitro inhibitors of JMJD2 enzymes, suggesting that these curcuminoids could be useful for decreasing JMJD2 activity in vivo. In conclusion, our data highlight that overexpression of JMJD2C confers a pro-growth effect on colon cancer cells and, therefore, its inhibition by curcuminoids or other small molecules could be beneficial as an adjuvant therapy for colon cancer patients.
Keywords: β-catenin, colon cancer, curcumin, histone demethylase, JMJD2C, KDM4C
Introduction
Cancer is characterized by a combination of genetic mutations and epigenetic changes. Especially epigenetic regulators have recently attracted great attention in cancer research and several studies have shown how their dysregulation could lead to tumor formation. Accordingly, epigenetic processes have been identified as potential points of therapeutic interference [1].
Similar to lung, breast and prostate cancer, colon cancer is one of the major causes of death in developed countries. Overexpression of the β-catenin oncoprotein, due to loss of the adenomatous polyposis coli tumor suppressor or mutations in β-catenin preventing its destruction, is observed in the majority of colon cancer patients and an underlying cause of this disease [2]. In contrast, much less is known about epigenetic changes in colon cancer.
DNA methylation as well as the posttranslational modification of histones are major epigenetic events and often altered during tumorigenesis [3,4]. One of the prominent histone modifications is the methylation of lysine residues, which can become mono-, di- or trimethylated. This is a dynamic process that is governed by histone methyltransferases and demethylases [5]. Two classes of histone demethylases have been unveiled, the first one encompassing only two members (LSD1 and LSD2) while the second one is much larger and part of the Jumonji C domain (JMJD) protein family [6]. Within the JMJD protein family, the JMJD2A-D proteins, also called KDM4A-D, form a subfamily based on a high degree of overall sequence homology [7,8].
One of the JMJD2 subfamily members, JMJD2C/KDM4C/GASC1, was originally identified as having been amplified in esophageal cancer [9]. Similarly, JMJD2C gene amplification was subsequently observed in lung sarcomatoid carcinomas, desmoplastic medulloblastomas, breast cancer, primary mediastinal B-cell lymphoma and Hodgkin lymphoma [10-13]. In addition, translocation of the JMJD2C gene in mucosa-associated lymphoid tissue lymphoma resulted in its overexpression [14] and enhanced JMJD2C levels were also observed in breast tumors, medulloblastomas and prostate cancer [12,15-17]. All this suggests that JMJD2C overexpression is involved in tumorigenesis.
In mouse embryonic fibroblasts, overexpression of JMJD2C, but not its catalytic mutant, led to increased expression of the MDM2 oncoprotein, a key negative regulator of the p53 tumor suppressor, whereas JMJD2C siRNA reduced MDM2 levels. Accordingly, JMJD2C overexpression caused a decrease in p53 protein levels, which may represent one mechanism by which JMJD2C overexpression can contribute to cancer formation [18]. In breast cancer, JMJD2C may interact with the hypoxia-inducible factor 1α and thereby exert a pro-oncogenic role [19], while its physical interaction with and stimulation of the androgen receptor may be crucial in the development of prostate cancer [20]. Here, we have attempted to uncover if and how JMJD2C could be involved in colon cancer formation.
Materials and methods
Western blotting of cell extracts
Whole cell extracts were generated by collecting cells in Laemmli sample buffer and then boiling [21]. Alternatively, cells were dissolved from 10-cm dishes with 40 mM Hepes (pH 8.0)/10 mM EDTA/150 mM NaCl, pelleted by centrifugation [22] and then fractionated with the NE-PER nuclear and cytoplasmic extraction kit (Pierce Biotechnology) according to the manufacturer’s recommendation. Then, extracts were resolved by SDS polyacrylamide gel electrophoresis and proteins transferred to polyvinylidene difluoride membrane [23]. These membranes were incubated with primary rabbit antibodies followed by incubation with secondary anti-rabbit antibodies coupled to horseradish peroxidase [24]. After employing enhanced chemiluminescence, blots were exposed to film [25]. The following JMJD2 antibodies were used: JMJD2A (Bethyl A300-861A), JMJD2B (Bethyl A301-478A), JMJD2C (Bethyl A300-885A) and JMJD2D (Aviva System Biology ARP35946).
RNA interference
Two different JMJD2C shRNAs were cloned into the retroviral vector pSIREN-RetroQ [26]. The sequence targeted by shRNA #3 was 5’-CAUCAGUGGCAGAGAGUAA-3’ and by shRNA #5 5’-CCUAAGGAGUGGAAGCCAA-3’. These constructs were utilized to produce retrovirus in 293T cells as described before [27]. Then, HCT-116 cells were infected with retrovirus, selected with 1 μg/ml puromycin and expanded for further use [28].
Cell growth and clonogenic assays
1000 HCT-116 cells were seeded into 96-wells and grown in DMEM medium supplemented with 10% fetal calf serum [29]. At indicated days thereafter, cell growth was measured with the PrestoBlue cell viability kit (Invitrogen). Averages with standard deviations of triplicate experiments were determined. For clonogenic assays, 500, 1000 or 1500 HCT-116 cells were put into the wells of 6 cm-dishes and grown for three weeks. Then, cells were fixed in 10% methanol/10% acetic acid and stained with 0.4% crystal violet.
Coimmunoprecipitations
Human embryonic kidney 293T cells were grown in 6-cm dishes to ~25% confluency [30] and then transiently transfected by the calcium phosphate coprecipitation method [31,32]. 2 μg of empty vector pEV3S or Flag3-β-catenin, 2 μg of empty vector pCS3+-6Myc or 6Myc-JMJD2C, and 5 μg of KS+ (Stratagene) were cotransfected [33]. Cells were lysed in 50 mM Tris-HCl (pH 7.4), 50 mM NaF, 0.25 mM Na3VO4, 150 mM NaCl, 0.5% Igepal CA-630, 0.2 mM DTT, 2 μg/ml aprotinin, 10 μg/ml leupeptin, 1 μg/ml pepstatin A, 0.5 mM PMSF [34] and immunoprecipitations were performed with monoclonal antibodies (anti-Flag M2 or anti-Myc 9E10) as described [35]. Coprecipitated proteins were detected after Western blotting [36] employing enhanced chemiluminescence [37].
Syntheses of FLLL compounds
The syntheses of FLLL-7 [38], FLLL-8 [38], FLLL-24 [39], FLLL-32 [40] and FLLL-60 [41] have been reported previously. To synthesize FLLL-59, sodium hydroxide (12.5 ml of a 5 M solution) was added to 125 μl of aliquat and the mixture was heated to reflux. 2,6-Dime-thylpyrimidine (296 μl, 2.5 mmol) was added to the mixture and stirred for 5 min followed by addition of 3,4-dimethoxybenzaldehyde (5 mmol). The resultant mixture was stirred for 1 h. After the reflux, the mixture was cooled to room temperature and a yellow precipitate formed. This precipitate was filtered, washed with ethanol and recrystallized from ethanol. The melting point was 127-129°C; 1H NMR (CDCl3, 300 MHz) δ 9.05 (d, J = 0.9 Hz, 1H), 7.84 (d, J = 15.9 Hz, 2H), 7.28 (s, 1H), 7.19 (s, 1H), 7.16 (s, 3H), 6.94 (d, J = 15.9 Hz, 2H), 6.90 (d, J = 7.8 Hz, 2H), 3.95 (s, 6H), 3.92 (s, 6H); 13C NMR (CDCl3, 400 MHz) δ 162.7, 158.5, 150.3, 149.1, 136.7, 128.6, 121.7, 115.6, 111.1, 109.3, 56.8, 56.7; HRMS-TOF m/z (M + Na)+ calculated for C24H24N2O4Na 427.1634, found 427.1622.
Demethylation assay
Glutathione S-transferase (GST) proteins fused to amino acids 2-350 of human JMJD2A, 2-352 of human JMJD2C and 2-523 of human JMJD2D were produced in Escherichia coli and purified with the help of glutathione agarose [42]. In vitro demethylation assays were then performed essentially as described before [43]. FLLL compounds were dissolved in DMSO and used at 1 mM concentration. Reaction mixtures were separated by SDS polyacrylamide gel electrophoresis and trimethylated H3K9 revealed by standard Western blotting [44].
Results
Expression of JMJD2 proteins in colon cancer cells
To assess whether JMJD2C might be overexpressed in colon cancer, we studied its expression in five different colon cancer cell lines compared to untransformed CCD-841-CoN colon cells (Figure 1). JMJD2C was overexpressed in all five colon cancer cell lines, similar to another protein, the RNA helicase DDX5, which was previously shown to be overexpressed in colon tumors [45,46]. Likewise, with the exception of RKO cells, β-catenin was also overexpressed in the colon cancer cells studied. We additionally explored the expression of the three close relatives of JMJD2C. Like JMJD2C, JMJD2A and JMJD2B were overexpressed in all five colon cancer cell lines, whereas JMJD2D was not (Figure 1). These data suggest that overexpression of JMJD2C occurs jointly with an increase in JMJD2A and JMJD2B protein levels in colon tumors.
Figure 1.
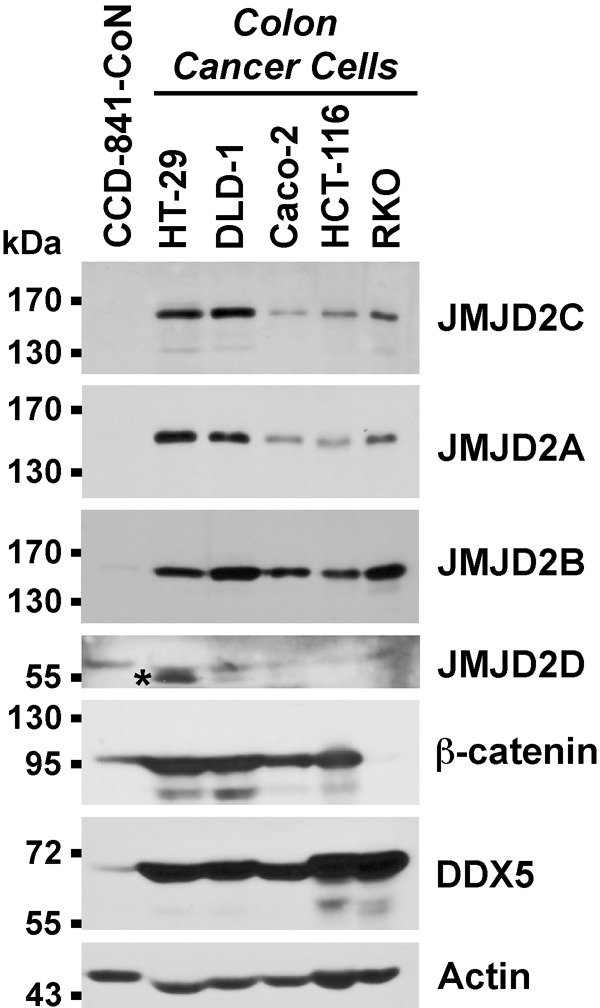
Western blots of whole cell extracts. The asterisk marks an unspecific band recognized by JMJD2D antibodies in HT-29 colon cancer cells.
Intracellular distribution of JMJD2C
Next, we were interested in which intracellular compartments JMJD2C would reside. To this end, we fractionated several cell lines into cytoplasm, nucleus and an insoluble fraction, which largely consisted of the nuclear matrix and attached chromatin. We observed that JMJD2C was prominently present in the insoluble fraction and also resident in the nucleus and cytoplasm of colon cancer and other cells (Figure 2). In contrast, none of the other three JMJD2 proteins was appreciably present in the insoluble fraction. Moreover, while JMJD2B and JMJD2D were primarily in the cell nucleus, JMJD2A often displayed comparable levels in the cytoplasm and nucleus. These data implicate that JMJD2A-C, which are all overexpressed in colon cancer cells, behave differently and may thus perform non-overlapping functions.
Figure 2.
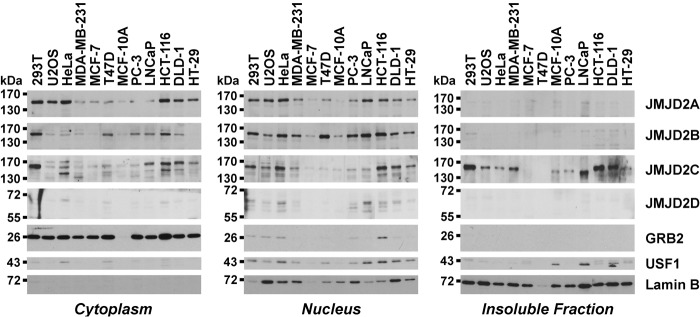
Intracellular localization of JMJD2 proteins in colon cancer cells (HCT-116, DLD-1, HT-29), transformed human embryonal kidney cells (293T), osteosarcoma cells (U2OS), cancerous (MDA-MB-231, MCF-7, T47D) and untransformed (MCF-10A) breast cells, or prostate tumor cells (PC-3, LNCaP). Cells were fractionated into cytosolic, nuclear and insoluble parts and the presence of JMJD2 proteins probed by Western blotting. The growth factor-receptor bound 2 (GRB2) adaptor protein is a marker for cytoplasm, whereas lamin B marks the cell nucleus and the insoluble nuclear matrix. Upstream transcription factor 1 (USF1), which is present primarily in nuclei and the insoluble chromatin fraction, was employed as another control.
Interaction of JMJD2C with β-catenin
In the vast majority of sporadic colonic tumors, overexpression of β-catenin is crucial for their development [47]. This causes a profound change of the transcriptome, since the β-catenin oncoprotein is a transcriptional cofactor [48]. Thus, we hypothesized that the transcriptional cofactor JMJD2C might interact with β-catenin. To test this, we coexpressed Flag-tagged β-catenin with Myc-tagged JMJD2C and assessed whether JMJD2C would coprecipitate with β-catenin. Indeed, after immunoprecipitation with Flag antibodies, JMJD2C was observed in the immunoprecipitate upon probing with Myc antibodies (Figure 3A). We then performed a reverse immunoprecipitation and consistently found that β-catenin coprecipitated with JMJD2C (Figure 3B). These results strongly suggested that JMJD2C may act as a transcriptional regulator in cooperation with β-catenin.
Figure 3.
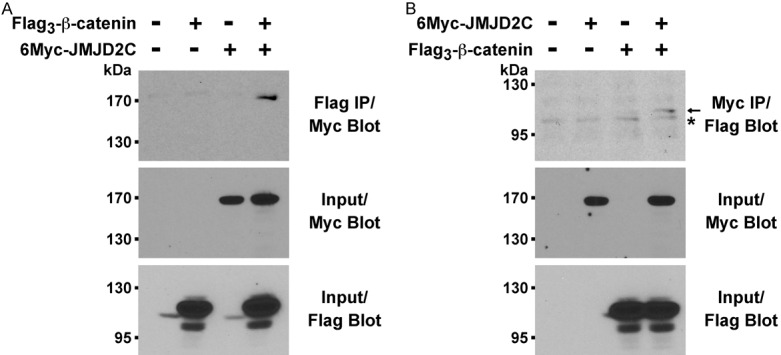
Binding of JMJD2C to β-catenin. A. Flag-tagged β-catenin was coexpressed with Myc-tagged JMJD2C in 293T cells. After anti-Flag immunoprecipitation (IP), coprecipitated JMJD2C was detected by anti-Myc Western blotting (top panel). The bottom two panels reveal input levels of 6Myc-JMJD2C and Flag3-β-catenin. B. Reverse order coimmunoprecipitation assay. Asterisk marks an unspecific band; arrow points at Flag-tagged β-catenin.
Consequences of JMJD2C ablation in HCT-116 cells
We wished to explore how JMJD2C might affect the physiology of colon cancer cells. Thus, we expressed two different JMJD2C shRNAs in HCT-116 colon cancer cells. Both shRNAs led to a robust depletion of JMJD2C compared to control shRNA (Figure 4). Consistent with a putative role as a β-catenin cofactor, JMJD2C ablation led to reduced expression of the oncogenic transcription factor FRA1 and the cell cycle regulator cyclin D1, both of which are regulated by β-catenin at the transcriptional level [49-51]. In contrast, JMJD2C downregulation had no impact on the expression levels of β-catenin itself or actin (Figure 4).
Figure 4.
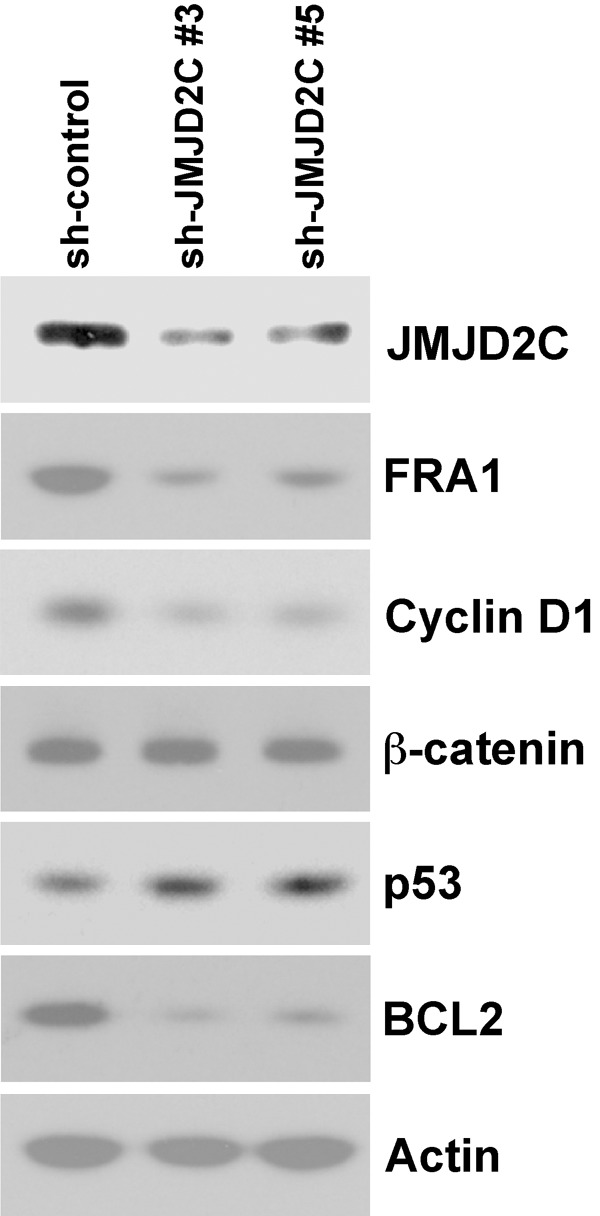
Impact of JMJD2C downregulation on the expression of oncoproteins (FRA1, cyclin D1, β-catenin, BCL2), the tumor suppressor p53 or actin in HCT-116 cells.
Next, we assessed how JMJD2C shRNA would affect the expression of the p53 tumor suppressor, which has been shown to interact with JMJD2C [52] and whose levels were reported to be negatively affected by JMJD2C overexpression [18]. Accordingly, downregulation of JMJD2C led to a slight increase of p53 protein levels (Figure 4). Further, expression of the anti-apoptotic BCL2 protein is negatively regulated by p53 [53,54]. Therefore, we also tested the impact of JMJD2C downregulation on BCL2 expression and found it drastically reduced (Figure 4). Currently, we do not know whether this is a consequence of the observed increase of p53 protein levels and/or other causes such as the ability of JMJD2C to support p53 in repressing the BCL2 gene promoter. Regardless, our results suggest that JMJD2C might promote survival of HCT-116 colon cancer cells via a BCL2-dependent mechanism and stimulate proliferation via upregulation of FRA1 and cyclin D1.
And indeed, when we tested the growth of HCT-116 colon cancer cells in the presence of JMJD2C shRNA, we found a significant reduction in cell growth with both shRNAs utilized (Figure 5A). Similarly, the clonogenic activity of HCT-116 cells was drastically reduced upon downregulation of JMJD2C (Figure 5B). These in vivo data implicate that JMJD2C promotes the oncogenic potential of colon cancer cells.
Figure 5.
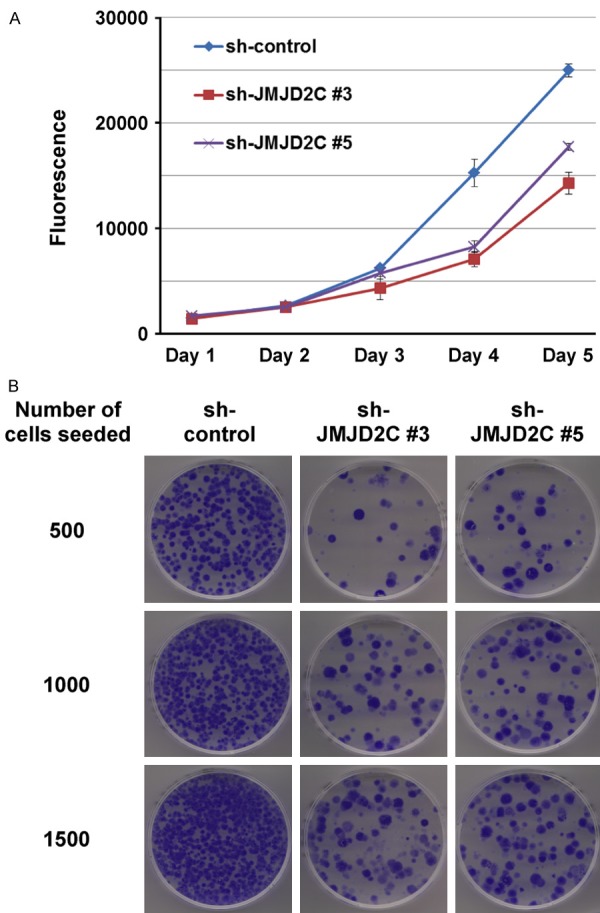
A. Reduced cell growth in HCT-116 cells expressing JMJD2C shRNA. B. Likewise, decreased ability to form colonies upon JMJD2C downregulation.
JMJD2 inhibition by curcuminoids
Curcumin is a yellow natural phenol found in the spice turmeric, which has received wide attention as a potential chemopreventive and chemotherapeutic agent [55]. Notably, curcumin has been shown to induce apoptosis in HCT-116 cells [56,57]. We speculated that a component of curcumin’s growth repressive role in colon cancer cells may include the inhibition of JMJD2 histone demethylases. To increase its solubility and/or bioavailability, many derivatives of curcumin, the so called curcuminoids, have been chemically synthesized [58]. Therefore, we focused on such curcuminoids. Specifically, we studied FLLL-7 and FLLL-8, which were shown to inhibit prostate and breast cancer cell growth [38], FLLL-32 that inhibits the function of the STAT3 transcription factor [40], and FLLL-24 [39] and FLLL-60 [41] that have not been studied as potential cancer drugs (Figure 6A). In addition, we synthesized a novel curcuminoid, FLLL-59 (Figure 6B).
Figure 6.
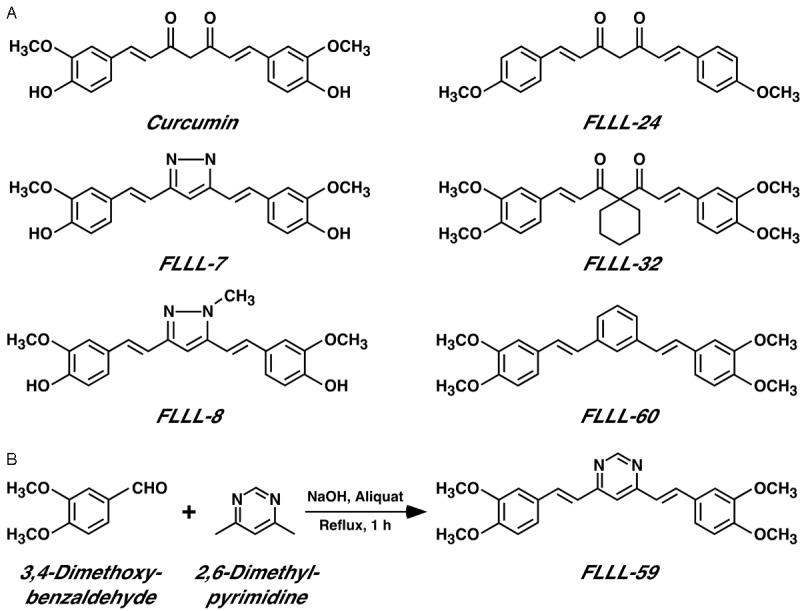
A. Structure of curcumin (keto tautomer) and five previously described FLLL curcuminoids. B. Synthesis scheme for FLLL-59.
We then tested these FLLL compounds in an in vitro demethylation assay. To this end, GST-JMJD2 fusion proteins were incubated with histones in vitro in the presence of the carrier DMSO or FLLL compounds dissolved in DMSO. Efficient demethylation of histone H3 trimethylated on lysine 9 was observed in the presence of GST-JMJD2C (compare lanes 1 and 2 in Figure 7). Notably, FLLL-8 and more so FLLL-24 significantly inhibited this enzymatic activity of JMJD2C, while other FLLL compounds had little to no effect. FLLL-8 and FLLL-24 were also effective in inhibiting the catalytic activity of JMJD2A and JMJD2D (Figure 7). Moreover, other FLLL compounds showed differences in their ability to suppress JMJD2 catalytic activity. For instance, FLLL-60 inhibited JMJD2A and JMJD2D, but not JMJD2C, while FLLL-32 robustly inhibited only JMJD2D. Thus, some curcuminoids do not inhibit all JMJD2 proteins at the same time, but rather display a preference for specific members of the JMJD2 histone demethylase family.
Figure 7.
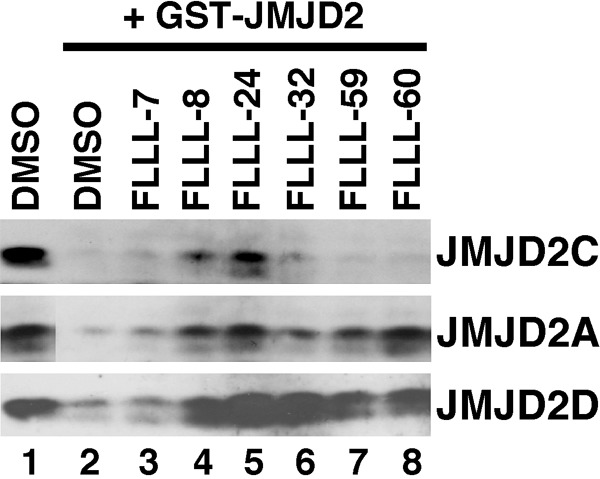
Inhibition of JMJD2 catalytic activity by FLLL curcuminoids. Histones were incubated in vitro without (lane 1) or with (lanes 2-8) GST-JMJD2C (top panel), GST-JMJD2A (middle panel) or GST-JMJD2D (bottom panel). Shown are Western blots revealing trimethylation of lysine 9 on histone H3.
Discussion
In this study, we uncovered that the histone demethylase JMJD2C is overexpressed in colon cancer cells and required for maximal HCT-116 cell growth. Moreover, we demonstrated that JMJD2C can be in a complex with β-catenin and thereby regulate the expression of many crucial β-catenin downstream effectors. Interestingly, JMJD2C appeared to be co-overexpressed with JMJD2A and JMJD2B, but not JMJD2D, in colon cancer cells. Although we cannot rule out that this co-overexpression is due to simultaneous transcriptional upregulation of the JMJD2A-C genes, it is possible that a common mechanism applies to enhance the stability of JMJD2A-C proteins. In particular, since JMJD2D is lacking the C-terminal half of JMJD2A-C [8], one may envision that post-translational modification of JMJD2A-C within their C-termini could be responsible for a stabilization of these proteins in colon cancer cells. Since JMJD2A and JMJD2C (and possibly JMJD2B) also form heteromers with each other, whereas JMJD2D does not [59], conceivably even modification of just one out of the three JMJD2A-C proteins may protect the other two by sequestering them away in heteromeric complexes. Further, JMJD2A-C displayed a distinct pattern of intracellular localization, suggesting that each protein performs at least some different functions and therefore overexpression of JMJD2A, JMJD2B and JMJD2C is non-redundantly contributing to colon tumorigenesis.
JMJD2C is capable of demethylating histone H3 on lysines 9 and 36 and histone H1.4 on lysine 26 [17,60,61]. In particular, trimethylated H3K9 and H1.4K26 confer a repressed chromatin status onto gene promoters [62,63], which may explain why JMJD2C can activate transcription at targets such as FRA1 or cyclin D1. In contrast, trimethylated H3K36 has often been associated with stimulation of gene expression [64], indicating that JMJD2C may also exert repressive functions as observed at the promoter of the p21 cell cycle inhibitor [52]. Lastly, JMJD2C is potentially also capable of demethylating non-histone proteins. In fact, it can demethylate polycomb 2 protein, thereby facilitating the activation of growth control genes [65]. All these activities of JMJD2C may be relevant in tumor cells. Interestingly, JMJD2C has also been shown to be a positive regulator of Nanog transcription and thereby promote self-renewal of embryonic stem cells [66], suggesting that overexpression of JMJD2C may as well stimulate the renewal of cancer stem cells.
FRA1 is a DNA-binding transcription factor and cyclin D1 is a prominent cell cycle regulator. Both are encoded by oncogenes that are transcriptionally regulated by β-catenin [49-51]. Moreover, FRA1 and cyclin D1 are upregulated in colonic tumors [67,68]. Together with our observation that JMJD2C and β-catenin form complexes, this suggests a scenario whereby JMJD2C activates the transcriptional potential of β-catenin leading to the expression of pro-growth genes such as FRA1 and cyclin D1. On top of this, JMJD2C and β-catenin may be involved in a feed-forward mechanism, because β-catenin may bind and thereby activate the JMJD2C promoter [69]. Additionally, we found that another oncoprotein, the survival molecule BCL2, was downregulated when JMJD2C expression was suppressed by shRNAs. Notably, BCL2 has also been reported to be overexpressed in adenomas and carcinomas of the colon [70,71]. Since BCL2 is negatively regulated by the p53 tumor suppressor [53,54] and JMJD2C appears to repress p53 expression, at least part of the JMJD2C-dependent BCL2 expression could be due to JMJD2C’s impact on p53 levels. This would accord with BCL2 expression being inversely correlated with that of the p53 tumor suppressor during colon tumor formation [72].
Several JMJD2C inhibitors have been identified before. These include N-oxalylglycine and hydroxamate analogs bearing a tertiary amine [43,73], 8-hydroxyquinoline chemotypes [74], catechols [75], compounds with a 4-hydroxypyrazole scaffold [76] and JIB-04 [77]. Here, we identified curcuminoids as other promising JMJD2C inhibitors. In particular, curcumin itself has been shown to have essentially no toxicity [55], implying that curcuminoids may be safe to use in the clinic. Notably, a previous study showed that FLLL-32 suppressed growth of colon cancer cells, including HCT-116 [78], suggesting that FLLL compounds might be useful in the treatment of colon cancer. The fact that some FLLL compounds inhibit multiple JMJD2 proteins may even be advantageous, since JMJD2A-C appear to be often co-overexpressed and thus could be inhibited with just one drug.
Altogether, our data support the notion that JMJD2C is an oncogenic protein in colon cancer, in part due to its ability to augment β-catenin in regulating gene transcription. Further, the identification of curcuminoids as JMJD2 inhibitors has provided novel lead compounds that could be further developed to effectively inhibit JMJD2C and its relatives in vivo and thus help to ameliorate clinical management of colon cancer patients.
Acknowledgements
This study was supported by grants from the Peggy and Charles Stephenson Cancer Center (to M. A. I. and R. J.), the National Cancer Institute (R01 CA154745; to R. J.) and the Basic Science Research Program of the National Research Foundation of Korea/Ministry of Education (357-2010-1-E00006; to T.-D. K.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the granting agencies.
Disclosure of conflict of interest
None.
References
- 1.Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150:12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 2.Clevers H, Nusse R. Wnt/beta-catenin signaling and disease. Cell. 2012;149:1192–1205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 3.Kulis M, Esteller M. DNA methylation and cancer. Adv Genet. 2010;70:27–56. doi: 10.1016/B978-0-12-380866-0.60002-2. [DOI] [PubMed] [Google Scholar]
- 4.Chi P, Allis CD, Wang GG. Covalent histone modifications--miswritten, misinterpreted and mis-erased in human cancers. Nat Rev Cancer. 2010;10:457–469. doi: 10.1038/nrc2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Black JC, Van Rechem C, Whetstine JR. Histone lysine methylation dynamics: establishment, regulation, and biological impact. Mol Cell. 2012;48:491–507. doi: 10.1016/j.molcel.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kooistra SM, Helin K. Molecular mechanisms and potential functions of histone demethylases. Nat Rev Mol Cell Biol. 2012;13:297–311. doi: 10.1038/nrm3327. [DOI] [PubMed] [Google Scholar]
- 7.Katoh M, Katoh M. Identification and characterization of JMJD2 family genes in silico. Int J Oncol. 2004;24:1623–1628. [PubMed] [Google Scholar]
- 8.Berry WL, Janknecht R. KDM4/JMJD2 histone demethylases: epigenetic regulators in cancer cells. Cancer Res. 2013;73:2936–2942. doi: 10.1158/0008-5472.CAN-12-4300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang ZQ, Imoto I, Fukuda Y, Pimkhaokham A, Shimada Y, Imamura M, Sugano S, Nakamura Y, Inazawa J. Identification of a novel gene, GASC1, within an amplicon at 9p23-24 frequently detected in esophageal cancer cell lines. Cancer Res. 2000;60:4735–4739. [PubMed] [Google Scholar]
- 10.Italiano A, Attias R, Aurias A, Perot G, Burel-Vandenbos F, Otto J, Venissac N, Pedeutour F. Molecular cytogenetic characterization of a metastatic lung sarcomatoid carcinoma: 9p23 neocentromere and 9p23 approximately p24 amplification including JAK2 and JMJD2C. Cancer Genet Cytogenet. 2006;167:122–130. doi: 10.1016/j.cancergencyto.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 11.Ehrbrecht A, Müller U, Wolter M, Hoischen A, Koch A, Radlwimmer B, Actor B, Mincheva A, Pietsch T, Lichter P, Reifenberger G, Weber RG. Comprehensive genomic analysis of desmoplastic medulloblastomas: identification of novel amplified genes and separate evaluation of the different histological components. J Pathol. 2006;208:554–563. doi: 10.1002/path.1925. [DOI] [PubMed] [Google Scholar]
- 12.Liu G, Bollig-Fischer A, Kreike B, van de Vijver MJ, Abrams J, Ethier SP, Yang ZQ. Genomic amplification and oncogenic properties of the GASC1 histone demethylase gene in breast cancer. Oncogene. 2009;28:4491–4500. doi: 10.1038/onc.2009.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rui L, Emre NC, Kruhlak MJ, Chung HJ, Steidl C, Slack G, Wright GW, Lenz G, Ngo VN, Shaffer AL, Xu W, Zhao H, Yang Y, Lamy L, Davis RE, Xiao W, Powell J, Maloney D, Thomas CJ, Moller P, Rosenwald A, Ott G, Muller-Hermelink HK, Savage K, Connors JM, Rimsza LM, Campo E, Jaffe ES, Delabie J, Smeland EB, Weisenburger DD, Chan WC, Gascoyne RD, Levens D, Staudt LM. Cooperative epigenetic modulation by cancer amplicon genes. Cancer Cell. 2010;18:590–605. doi: 10.1016/j.ccr.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vinatzer U, Gollinger M, Mullauer L, Raderer M, Chott A, Streubel B. Mucosa-associated lymphoid tissue lymphoma: novel translocations including rearrangements of ODZ2, JMJD2C, and CNN3. Clin Cancer Res. 2008;14:6426–6431. doi: 10.1158/1078-0432.CCR-08-0702. [DOI] [PubMed] [Google Scholar]
- 15.Berdel B, Nieminen K, Soini Y, Tengstrom M, Malinen M, Kosma VM, Palvimo J, Mannermaa A. Histone demethylase GASC1 - a potential prognostic and predictive marker in invasive breast cancer. BMC Cancer. 2012;12:516. doi: 10.1186/1471-2407-12-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Northcott PA, Nakahara Y, Wu X, Feuk L, Ellison DW, Croul S, Mack S, Kongkham PN, Peacock J, Dubuc A, Ra YS, Zilberberg K, McLeod J, Scherer SW, Sunil Rao J, Eberhart CG, Grajkowska W, Gillespie Y, Lach B, Grundy R, Pollack IF, Hamilton RL, Van Meter T, Carlotti CG, Boop F, Bigner D, Gilbertson RJ, Rutka JT, Taylor MD. Multiple recurrent genetic events converge on control of histone lysine methylation in medulloblastoma. Nat Genet. 2009;41:465–472. doi: 10.1038/ng.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cloos PA, Christensen J, Agger K, Maiolica A, Rappsilber J, Antal T, Hansen KH, Helin K. The putative oncogene GASC1 demethylates tri- and dimethylated lysine 9 on histone H3. Nature. 2006;442:307–311. doi: 10.1038/nature04837. [DOI] [PubMed] [Google Scholar]
- 18.Ishimura A, Terashima M, Kimura H, Akagi K, Suzuki Y, Sugano S, Suzuki T. Jmjd2c histone demethylase enhances the expression of Mdm2 oncogene. Biochem Biophys Res Commun. 2009;389:366–371. doi: 10.1016/j.bbrc.2009.08.155. [DOI] [PubMed] [Google Scholar]
- 19.Luo W, Chang R, Zhong J, Pandey A, Semenza GL. Histone demethylase JMJD2C is a coactivator for hypoxia-inducible factor 1 that is required for breast cancer progression. Proc Natl Acad Sci U S A. 2012;109:E3367–E3376. doi: 10.1073/pnas.1217394109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wissmann M, Yin N, Muller JM, Greschik H, Fodor BD, Jenuwein T, Vogler C, Schneider R, Gunther T, Buettner R, Metzger E, Schule R. Cooperative demethylation by JMJD2C and LSD1 promotes androgen receptor-dependent gene expression. Nat Cell Biol. 2007;9:347–353. doi: 10.1038/ncb1546. [DOI] [PubMed] [Google Scholar]
- 21.Kim TD, Shin S, Janknecht R. Repression of Smad3 activity by histone demethylase SMCX/JARID1C. Biochem Biophys Res Commun. 2008;366:563–567. doi: 10.1016/j.bbrc.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 22.Shin S, Janknecht R. Concerted activation of the Mdm2 promoter by p72 RNA helicase and the coactivators p300 and P/CAF. J Cell Biochem. 2007;101:1252–1265. doi: 10.1002/jcb.21250. [DOI] [PubMed] [Google Scholar]
- 23.Papoutsopoulou S, Janknecht R. Phosphorylation of ETS transcription factor ER81 in a complex with its coactivators CREB-binding protein and p300. Mol Cell Biol. 2000;20:7300–7310. doi: 10.1128/mcb.20.19.7300-7310.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goel A, Janknecht R. Acetylation-mediated transcriptional activation of the ETS protein ER81 by p300, P/CAF, and HER2/Neu. Mol Cell Biol. 2003;23:6243–6254. doi: 10.1128/MCB.23.17.6243-6254.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rossow KL, Janknecht R. The Ewing’s sarcoma gene product functions as a transcriptional activator. Cancer Res. 2001;61:2690–2695. [PubMed] [Google Scholar]
- 26.Berry WL, Shin S, Lightfoot SA, Janknecht R. Oncogenic features of the JMJD2A histone demethylase in breast cancer. Int J Oncol. 2012;41:1701–1706. doi: 10.3892/ijo.2012.1618. [DOI] [PubMed] [Google Scholar]
- 27.Shin S, Oh S, An S, Janknecht R. ETS variant 1 regulates matrix metalloproteinase-7 transcription in LNCaP prostate cancer cells. Oncol Rep. 2013;29:306–314. doi: 10.3892/or.2012.2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oh S, Shin S, Lightfoot SA, Janknecht R. 14-3-3 proteins modulate the ETS transcription factor ETV1 in prostate cancer. Cancer Res. 2013;73:5110–5119. doi: 10.1158/0008-5472.CAN-13-0578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim TD, Oh S, Shin S, Janknecht R. Regulation of tumor suppressor p53 and HCT116 cell physiology by histone demethylase JMJD2D/KDM4D. PLoS One. 2012;7:e34618. doi: 10.1371/journal.pone.0034618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mooney SM, Goel A, D’Assoro AB, Salisbury JL, Janknecht R. Pleiotropic effects of p300-mediated acetylation on p68 and p72 RNA helicase. J Biol Chem. 2010;285:30443–30452. doi: 10.1074/jbc.M110.143792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dowdy SC, Mariani A, Janknecht R. HER2/Neu- and TAK1-mediated up-regulation of the transforming growth factor beta inhibitor Smad7 via the ETS protein ER81. J Biol Chem. 2003;278:44377–44384. doi: 10.1074/jbc.M307202200. [DOI] [PubMed] [Google Scholar]
- 32.DiTacchio L, Bowles J, Shin S, Lim DS, Koopman P, Janknecht R. Transcription factors ER71/ETV2 and SOX9 participate in a positive feedback loop in fetal and adult mouse testis. J Biol Chem. 2012;287:23657–23666. doi: 10.1074/jbc.M111.320101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mooney SM, Grande JP, Salisbury JL, Janknecht R. Sumoylation of p68 and p72 RNA helicases affects protein stability and transactivation potential. Biochemistry. 2010;49:1–10. doi: 10.1021/bi901263m. [DOI] [PubMed] [Google Scholar]
- 34.Knebel J, De Haro L, Janknecht R. Repression of transcription by TSGA/Jmjd1a, a novel interaction partner of the ETS protein ER71. J Cell Biochem. 2006;99:319–329. doi: 10.1002/jcb.20945. [DOI] [PubMed] [Google Scholar]
- 35.Janknecht R. Regulation of the ER81 transcription factor and its coactivators by mitogen- and stress-activated protein kinase 1 (MSK1) Oncogene. 2003;22:746–755. doi: 10.1038/sj.onc.1206185. [DOI] [PubMed] [Google Scholar]
- 36.Goueli BS, Janknecht R. Upregulation of the catalytic telomerase subunit by the transcription factor ER81 and oncogenic HER2/Neu, Ras, or Raf. Mol Cell Biol. 2004;24:25–35. doi: 10.1128/MCB.24.1.25-35.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goel A, Janknecht R. Concerted activation of ETS protein ER81 by p160 coactivators, the acetyltransferase p300 and the receptor tyrosine kinase HER2/Neu. J Biol Chem. 2004;279:14909–14916. doi: 10.1074/jbc.M400036200. [DOI] [PubMed] [Google Scholar]
- 38.Fuchs JR, Pandit B, Bhasin D, Etter JP, Regan N, Abdelhamid D, Li C, Lin J, Li PK. Structure-activity relationship studies of curcumin analogues. Bioorg Med Chem Lett. 2009;19:2065–2069. doi: 10.1016/j.bmcl.2009.01.104. [DOI] [PubMed] [Google Scholar]
- 39.Khan MA, El-Khatib R, Rainsford KD, Whitehouse MW. Synthesis and anti-inflammatory properties of some aromatic and heterocyclic aromatic curcuminoids. Bioorg Chem. 2012;40:30–38. doi: 10.1016/j.bioorg.2011.11.004. [DOI] [PubMed] [Google Scholar]
- 40.Lin L, Hutzen B, Zuo M, Ball S, Deangelis S, Foust E, Pandit B, Ihnat MA, Shenoy SS, Kulp S, Li PK, Li C, Fuchs J, Lin J. Novel STAT3 phosphorylation inhibitors exhibit potent growth-suppressive activity in pancreatic and breast cancer cells. Cancer Res. 2010;70:2445–2454. doi: 10.1158/0008-5472.CAN-09-2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reinke AA, Gestwicki JE. Structure-activity relationships of amyloid beta-aggregation inhibitors based on curcumin: influence of linker length and flexibility. Chem Biol Drug Des. 2007;70:206–215. doi: 10.1111/j.1747-0285.2007.00557.x. [DOI] [PubMed] [Google Scholar]
- 42.Shin S, Kim TD, Jin F, van Deursen JM, Dehm SM, Tindall DJ, Grande JP, Munz JM, Vasmatzis G, Janknecht R. Induction of prostatic intraepithelial neoplasia and modulation of androgen receptor by ETS variant 1/ETS-related protein 81. Cancer Res. 2009;69:8102–8110. doi: 10.1158/0008-5472.CAN-09-0941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hamada S, Kim TD, Suzuki T, Itoh Y, Tsumoto H, Nakagawa H, Janknecht R, Miyata N. Synthesis and activity of N-oxalylglycine and its derivatives as Jumonji C-domain-containing histone lysine demethylase inhibitors. Bioorg Med Chem Lett. 2009;19:2852–2855. doi: 10.1016/j.bmcl.2009.03.098. [DOI] [PubMed] [Google Scholar]
- 44.Wu J, Janknecht R. Regulation of the ETS transcription factor ER81 by the 90-kDa ribosomal S6 kinase 1 and protein kinase A. J Biol Chem. 2002;277:42669–42679. doi: 10.1074/jbc.M205501200. [DOI] [PubMed] [Google Scholar]
- 45.Shin S, Rossow KL, Grande JP, Janknecht R. Involvement of RNA helicases p68 and p72 in colon cancer. Cancer Res. 2007;67:7572–7578. doi: 10.1158/0008-5472.CAN-06-4652. [DOI] [PubMed] [Google Scholar]
- 46.Janknecht R. Multi-talented DEAD-box proteins and potential tumor promoters: p68 RNA helicase (DDX5) and its paralog, p72 RNA helicase (DDX17) Am J Transl Res. 2010;2:223–234. [PMC free article] [PubMed] [Google Scholar]
- 47.Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 48.Mosimann C, Hausmann G, Basler K. Beta-catenin hits chromatin: regulation of Wnt target gene activation. Nat Rev Mol Cell Biol. 2009;10:276–286. doi: 10.1038/nrm2654. [DOI] [PubMed] [Google Scholar]
- 49.Mann B, Gelos M, Siedow A, Hanski ML, Gratchev A, Ilyas M, Bodmer WF, Moyer MP, Riecken EO, Buhr HJ, Hanski C. Target genes of beta-catenin-T cell-factor/lymphoid-enhancer-factor signaling in human colorectal carcinomas. Proc Natl Acad Sci U S A. 1999;96:1603–1608. doi: 10.1073/pnas.96.4.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shtutman M, Zhurinsky J, Simcha I, Albanese C, D’Amico M, Pestell R, Ben-Ze’ev A. The cyclin D1 gene is a target of the beta-catenin/LEF-1 pathway. Proc Natl Acad Sci U S A. 1999;96:5522–5527. doi: 10.1073/pnas.96.10.5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tetsu O, McCormick F. Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature. 1999;398:422–426. doi: 10.1038/18884. [DOI] [PubMed] [Google Scholar]
- 52.Kim TD, Shin S, Berry WL, Oh S, Janknecht R. The JMJD2A demethylase regulates apoptosis and proliferation in colon cancer cells. J Cell Biochem. 2012;113:1368–1376. doi: 10.1002/jcb.24009. [DOI] [PubMed] [Google Scholar]
- 53.Miyashita T, Harigai M, Hanada M, Reed JC. Identification of a p53-dependent negative response element in the bcl-2 gene. Cancer Res. 1994;54:3131–3135. [PubMed] [Google Scholar]
- 54.Budhram-Mahadeo V, Morris PJ, Smith MD, Midgley CA, Boxer LM, Latchman DS. p53 suppresses the activation of the Bcl-2 promoter by the Brn-3a POU family transcription factor. J Biol Chem. 1999;274:15237–15244. doi: 10.1074/jbc.274.21.15237. [DOI] [PubMed] [Google Scholar]
- 55.Gupta SC, Patchva S, Koh W, Aggarwal BB. Discovery of curcumin, a component of golden spice, and its miraculous biological activities. Clin Exp Pharmacol Physiol. 2012;39:283–299. doi: 10.1111/j.1440-1681.2011.05648.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Collett GP, Campbell FC. Curcumin induces c-jun N-terminal kinase-dependent apoptosis in HCT116 human colon cancer cells. Carcinogenesis. 2004;25:2183–2189. doi: 10.1093/carcin/bgh233. [DOI] [PubMed] [Google Scholar]
- 57.Watson JL, Hill R, Lee PW, Giacomantonio CA, Hoskin DW. Curcumin induces apoptosis in HCT-116 human colon cancer cells in a p21-independent manner. Exp Mol Pathol. 2008;84:230–233. doi: 10.1016/j.yexmp.2008.02.002. [DOI] [PubMed] [Google Scholar]
- 58.Esatbeyoglu T, Huebbe P, Ernst IM, Chin D, Wagner AE, Rimbach G. Curcumin--from molecule to biological function. Angew Chem Int Ed Engl. 2012;51:5308–5332. doi: 10.1002/anie.201107724. [DOI] [PubMed] [Google Scholar]
- 59.Shin S, Janknecht R. Diversity within the JMJD2 histone demethylase family. Biochem Biophys Res Commun. 2007;353:973–977. doi: 10.1016/j.bbrc.2006.12.147. [DOI] [PubMed] [Google Scholar]
- 60.Whetstine JR, Nottke A, Lan F, Huarte M, Smolikov S, Chen Z, Spooner E, Li E, Zhang G, Colaiacovo M, Shi Y. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell. 2006;125:467–481. doi: 10.1016/j.cell.2006.03.028. [DOI] [PubMed] [Google Scholar]
- 61.Trojer P, Zhang J, Yonezawa M, Schmidt A, Zheng H, Jenuwein T, Reinberg D. Dynamic Histone H1 Isotype 4 Methylation and Demethylation by Histone Lysine Methyltransferase G9a/KMT1C and the Jumonji Domain-containing JMJD2/KDM4 Proteins. J Biol Chem. 2009;284:8395–8405. doi: 10.1074/jbc.M807818200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 63.Daujat S, Zeissler U, Waldmann T, Happel N, Schneider R. HP1 binds specifically to Lys26-methylated histone H1.4, whereas simultaneous Ser27 phosphorylation blocks HP1 binding. J Biol Chem. 2005;280:38090–38095. doi: 10.1074/jbc.C500229200. [DOI] [PubMed] [Google Scholar]
- 64.Wagner EJ, Carpenter PB. Understanding the language of Lys36 methylation at histone H3. Nat Rev Mol Cell Biol. 2012;13:115–126. doi: 10.1038/nrm3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang L, Lin C, Liu W, Zhang J, Ohgi KA, Grinstein JD, Dorrestein PC, Rosenfeld MG. ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell. 2011;147:773–788. doi: 10.1016/j.cell.2011.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Loh YH, Zhang W, Chen X, George J, Ng HH. Jmjd1a and Jmjd2c histone H3 Lys 9 demethylases regulate self-renewal in embryonic stem cells. Genes Dev. 2007;21:2545–2557. doi: 10.1101/gad.1588207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang HL, Wang J, Xiao SY, Haydon R, Stoiber D, He TC, Bissonnette M, Hart J. Elevated protein expression of cyclin D1 and Fra-1 but decreased expression of c-Myc in human colorectal adenocarcinomas overexpressing beta-catenin. Int J Cancer. 2002;101:301–310. doi: 10.1002/ijc.10630. [DOI] [PubMed] [Google Scholar]
- 68.Bartkova J, Lukas J, Strauss M, Bartek J. The PRAD-1/cyclin D1 oncogene product accumulates aberrantly in a subset of colorectal carcinomas. Int J Cancer. 1994;58:568–573. doi: 10.1002/ijc.2910580420. [DOI] [PubMed] [Google Scholar]
- 69.Yamamoto S, Tateishi K, Kudo Y, Yamamoto K, Isagawa T, Nagae G, Nakatsuka T, Asaoka Y, Ijichi H, Hirata Y, Otsuka M, Ikenoue T, Aburatani H, Omata M, Koike K. Histone demethylase KDM4C regulates sphere formation by mediating the cross talk between Wnt and Notch pathways in colonic cancer cells. Carcinogenesis. 2013;34:2380–2388. doi: 10.1093/carcin/bgt174. [DOI] [PubMed] [Google Scholar]
- 70.Hague A, Moorghen M, Hicks D, Chapman M, Paraskeva C. BCL-2 expression in human colorectal adenomas and carcinomas. Oncogene. 1994;9:3367–3370. [PubMed] [Google Scholar]
- 71.Bosari S, Moneghini L, Graziani D, Lee AK, Murray JJ, Coggi G, Viale G. bcl-2 oncoprotein in colorectal hyperplastic polyps, adenomas, and adenocarcinomas. Hum Pathol. 1995;26:534–540. doi: 10.1016/0046-8177(95)90250-3. [DOI] [PubMed] [Google Scholar]
- 72.Sinicrope FA, Ruan SB, Cleary KR, Stephens LC, Lee JJ, Levin B. bcl-2 and p53 oncoprotein expression during colorectal tumorigenesis. Cancer Res. 1995;55:237–241. [PubMed] [Google Scholar]
- 73.Hamada S, Suzuki T, Mino K, Koseki K, Oehme F, Flamme I, Ozasa H, Itoh Y, Ogasawara D, Komaarashi H, Kato A, Tsumoto H, Nakagawa H, Hasegawa M, Sasaki R, Mizukami T, Miyata N. Design, synthesis, enzyme-inhibitory activity, and effect on human cancer cells of a novel series of jumonji domain-containing protein 2 histone demethylase inhibitors. J Med Chem. 2010;53:5629–5638. doi: 10.1021/jm1003655. [DOI] [PubMed] [Google Scholar]
- 74.Hutchinson SE, Leveridge MV, Heathcote ML, Francis P, Williams L, Gee M, Munoz-Muriedas J, Leavens B, Shillings A, Jones E, Homes P, Baddeley S, Chung CW, Bridges A, Argyrou A. Enabling lead discovery for histone lysine demethylases by high-throughput RapidFire mass spectrometry. J Biomol Screen. 2012;17:39–48. doi: 10.1177/1087057111416660. [DOI] [PubMed] [Google Scholar]
- 75.Nielsen AL, Kristensen LH, Stephansen KB, Kristensen JB, Helgstrand C, Lees M, Cloos P, Helin K, Gajhede M, Olsen L. Identification of catechols as histone-lysine demethylase inhibitors. FEBS Lett. 2012;586:1190–1194. doi: 10.1016/j.febslet.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 76.Leurs U, Clausen RP, Kristensen JL, Lohse B. Inhibitor scaffold for the histone lysine demethylase KDM4C (JMJD2C) Bioorg Med Chem Lett. 2012;22:5811–5813. doi: 10.1016/j.bmcl.2012.07.091. [DOI] [PubMed] [Google Scholar]
- 77.Wang L, Chang J, Varghese D, Dellinger M, Kumar S, Best AM, Ruiz J, Bruick R, Pena-Llopis S, Xu J, Babinski DJ, Frantz DE, Brekken RA, Quinn AM, Simeonov A, Easmon J, Martinez ED. A small molecule modulates Jumonji histone demethylase activity and selectively inhibits cancer growth. Nat Commun. 2013;4:2035. doi: 10.1038/ncomms3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lin L, Deangelis S, Foust E, Fuchs J, Li C, Li PK, Schwartz EB, Lesinski GB, Benson D, Lu J, Hoyt D, Lin J. A novel small molecule inhibits STAT3 phosphorylation and DNA binding activity and exhibits potent growth suppressive activity in human cancer cells. Mol Cancer. 2010;9:217. doi: 10.1186/1476-4598-9-217. [DOI] [PMC free article] [PubMed] [Google Scholar]


