Abstract
We previously identified a cross-tick species conserved tick feeding stimuli responsive Amblyomma americanum (Aam) AV422 gene. This study demonstrates that AamAV422 belongs to a novel group of arthropod proteins that is characterized by 14 cysteine amino acid residues: C23-X7/9-C33-X23/24-C58-C8-C67X7-X75-X23-C99-X15-C115-X10-C126X24/25/33-C150C151-X7-C159-X8-X168-X23/24-C192-X9/10-C202 predicted to form seven disulfide bonds. We show that AamAV422 protein is a ubiquitously expressed protein that is injected into the host within the first 24 h of the tick attaching onto the host as revealed by western blotting analyses of recombinant (r)AamAV422, tick saliva and dissected tick organ protein extracts using antibodies to 24 h and 48 h tick saliva proteins (TSPs). Native AamAV422 is apparently involved with mediating tick anti-hemostasis and anti-complement functions in that rAamAV422 delayed plasma clotting time in a dose responsive manner by up to ~160 s, prevented platelet aggregation by up to ~16% and caused ~24% reduction in production of terminal complement complexes. Target validation analysis revealed that rAamAV422 is a potential candidate for a cocktail or multivalent tick vaccine preparation in that RNA interference (RNAi)-mediated silencing of AamAV422 mRNA caused a statistically significant (~44%) reduction in tick engorgement weights, which is proxy for amounts of ingested blood. We speculate that AamAV422 is a potential target antigen for development of the highly desired universal tick vaccine in that consistent with high conservation among ticks, antibodies to 24 h Ixodes scapularis TSPs specifically bound rAamAV422. We discuss data in this study in the context of advancing the biology of tick feeding physiology and discovery of potential target antigens for tick vaccine development.
Keywords: Amblyomma americanum tick feeding biology, Highly conserved tick saliva protein, Anticoagulant function
1. Introduction
Ticks represent the most successful arthropod vectors of animal and human disease agents in terms of diversity and they are second only to mosquitoes in terms of impact. The molecular basis of hard tick feeding is emerging. It is known that ticks continuously change their protein profiles throughout their feeding cycle. The differentially expressed tick proteins are thought to regulate specialized biological events that are unique to different phases of the tick feeding process (Wang et al., 1999; Mulenga et al., 2007). Within 5 - 30 min of inserting their hyposthome into the skin of the host, ticks secrete a mixture of substances into the host (Sonenshine, 1993). These substances are thought to regulate the initial stages of tick feeding: an adhesive substance called cement anchors the tick onto host skin, with other substances involved in creating the tick-feeding site. In a related study, Wang et al. (1999) observed that if Rhipicephalus appendiculatus ticks were disrupted from feeding at day 6 and then incubated at 25°C for 14 days, their re-attachment to the animal to start feeding was accompanied by reprogramming of the salivary gland protein profiles.
The tick feeding cycle includes the preparatory feeding phase (PFP) (attachment and creation of feeding lesion) during the first 24 – 36 h, the slow feeding phase (moderate blood meal uptake, pathogen transmission, increase in physical size) that may last up to 7 days and the rapid feeding phase (feeding to repletion) that may occur within 24 h after 7 days or more of feeding (Sonenshine, 1993). Studies that have attempted to identify tick proteins which regulate tick feeding have mostly targeted ticks that were attached to the host for periods of 3-5 days (Mulenga et al., 2001, 2003a, b; Valenzuela et al., 2000, 2002; Ribeiro et al., 2006). These approaches have led to the discovery of several tick bioactive enzymes with the properties of anticoagulants, anti-complement, anti-inflammatory, kinase activity, anti-oxidant and other bioactive activities (Ribeiro et al., 2006). This approach has potential to miss those genes that regulate events at the start of the tick feeding process. The goal in our research is to identify tick saliva proteins (TSPs) that regulate the beginning stages of the tick feeding process. Our assumption is that these genes play essential role(s) in regulating the PFP which precedes key facets of tick parasitism: blood meal feeding, disease agent acquisition and transmission (Sonenshine, 1993). Except for some viruses such as the Powassan virus and tick-borne encephalitis virus that are transmitted to the animal within minutes of the tick attaching onto the animal (Ebel and Kramer, 2004), the majority of tick-borne disease agents such Borrelia, Theileria, Babesia are not transmitted until after ticks have been feeding for 2-3 days (Piesman and Spielman, 1980; Piesman et al., 1987; Katavolos et al., 1998; Konnai et al., 2007). Thus tick proteins regulating the PFP represent important target antigens for development of novel tick control methods assuming that disruption of the PFP may protect animals against the key facets of tick parasitism.
A number of studies have reported observations that newly molted ticks less than 7-21 days old show a reluctance to engage the host and feed (Gladney, 1970; Tukahirwa, 1976; Davey, 1987) and they do not respond to CO2 stimuli, which is a proxy for host breathing. In contrast they walk away from the source, indicating they are not ready to initiate feeding (Anderson et al., 1998). However, ticks older than 21 days show a strong response to CO2 stimuli; these ticks move toward the source indicating physiological readiness to engage the host. This observation suggests that attainment of physiological readiness to feed is accompanied by differential gene expression. Previously, we used subtractive hybridization methodology to subtract the cDNA library of newly molted (~0 to 1 day) adult Amblyomma americanum female ticks that were not stimulated to start feeding, against the cDNA library of 5 week old unfed ticks that were exposed to feeding stimuli for ~ 7 h (Mulenga et al., 2007). This approach allowed us to identify 40 genes that were differentially upregulated or induced in response to attainment of appetence and/or exposure to feeding stimuli (Mulenga et al., 2007). Since our previous study (Mulenga et al., 2007), two similar studies that used new generation sequencing approaches to identify genes associated with regulating the initial phases of Rhipicephalus microplus tick feeding have been described (Lew-Tabor et al., 2010; Rodriguez-Valle et al., 2010).
Advances in sequencing technologies are leading to unprecedented amounts of sequence data that provide immense opportunities for in-depth understanding of the molecular basis of tick physiology. For ticks and indeed other parasitic organisms the main limitation is that the majority of genes coming through the discovery pipelines are of unknown function or orphan genes (Mulenga et al., 2007; Wang et al., 2007). While orphan genes pose a challenge in designing follow-up functional analyses experiments, they provide interesting opportunities for discovery of druggable or vaccine targets against parasites including ticks. From our experience the majority of orphan genes do not have mammalian homologs, which is highly desirable as this reduces the potential for candidate antigens to be unsuitable immunogens because they are recognized as self by the host or anti-parasitic drugs cross-reacting with host mammalian proteins. As part of our prioritization strategy, we selected candidate orphan genes on the basis of showing conservation in other tick species, which signals that the candidate gene is potentially associated with an important pathway that is evolutionarily conserved in the parasitic organism but not the mammalian host. Implementation of this strategy was made possible due to the availability of multiple expressed sequence tag (EST) and RNA sequencing research outputs for Ambylomma tick spp. (Nene et al., 2002; Aljamali et al., 2009; Karim et al., 2011), other tick species (Hill and Guttierrez, 2000; Valenzuella et al., 2002; Nene et al., 2004; Santos et al., 2004; Guerrero etal., 2005; Ribeiro et al., 2006; Wang et al., 2007; Anatriello et al., 2010; Lees et al., 2010; Lew-Tabor et al., 2010; Rodriguez-Valle et al., 2010; Francischetti et al., 2011) and the Ixodes scapularis genome sequence data (Pagel et al., 2007), that allowed us to identify some of tick orphan genes that are cross-tick species conserved (Mulenga et al., 2007). In the current study the goal was to deorphanize and gain insight into the significance and role(s) of the A. americanum (Aam) AV422 gene in tick feeding regulation. AamAV422 was identified among tick feeding stimuli responsive genes and was found to be expressed in multiple tick tissues with the transcript level increasing particularly in the salivary gland, as the tick continued to feed (Mulenga et al., 2007). This study shows that AamV422 is a TSP that is injected into the host from the first 24 h of the tick feeding process and is apparently involved in mediating tick anti-hemostatic and anti-complement functions. Data in this study also show that AamAV422 apparently belongs to an exclusive novel family of arthropod hypothetical proteins that is characterized by seven disulfide bonds. The data are discussed in the context of advances in tick feeding regulation.
2. Materials and methods
2.1. Tick feeding, dissections and total protein extraction
Amblyomma americanum ticks used in this study were purchased from the laboratory of Dr. Pete Teel in the Entomology Department of Texas A&M University, College Station, Texas, USA. The ticks were fed on rabbit ears or on the back of a calf under conditions described in animal use protocols approved by Texas A & M University Institutional Animal Care and Use Committee. Ticks were placed in a cotton orthopedic stockinet containment cell on top of the rabbit ear to avoid ticks getting into the ear canal of the rabbits as previously described (Chalaire et al., 2011). On rabbits, the cotton stockinet was attached to the rabbit ear using Kamar adhesive (Jeffers Inc, Dothan, AL, USA). Likewise, on the calf, the cotton stockinet was attached to the back of the calf using Kamar adhesive. Fifteen, 10 and eight ticks that were, respectively, partially fed for 24, 72 and 120 h were manually detached and dissected as previously described (Mulenga et al., 2003a). Dissected tick organs, salivary glands, (SG), midguts (MG), carcass (CA, tick remnant after removal of SG and MG) were homogenized in lysis buffer (25 mM Tris-HCl pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% NP-40 and 5% glycerol) containing Halt protease inhibitor cocktail (Thermo Fisher Scientific, Rockford, IL, USA) using a Sonic dismembranator (Thermo Fisher Scientific). The disrupted samples were centrifuged at 18,000 g to clarify the supernatant that was then stored at −80°C until use.
2.2. Tick saliva and haemolymph collection
Tick saliva was collected using methods initially described by Ribeiro et al., (2004) with slight modifications (Chalaire et al., 2011). Tick saliva was collected from 20 female ticks per time point: 48, 96 and 120 h. Ticks were rinsed in PBS and dried between paper towels. Subsequently, ticks were secured dorsal side up using double-sided tape affixed to a glass slide. To induce salivation, 3-5 μL (depending on size of the tick) of pilocarpine hydrochloride (50 mg/mL in 95% ethanol) (Sigma-Aldrich, St. Louis, MO, USA) was applied to the scutum. Ticks were left at room temperature until the ethanol had dried and then individually placed onto a matrix of a DNA binding spin column insert (Agilent technologies (formerly Strategene), Santa Clara, CA, USA). The spin column containing the tick was placed into a 1.5 ml collection tube, partially capped and incubated at 35°C for 3 h to induce salivation under humidified conditions. Humid conditions were provided by placing wet paper towels at the bottom of a covered container that was holding tubes containing ticks. Following incubation, 10 μL of PBS (pH 7.4) was placed onto tick mouthparts and then the column was spun at 2,000 g to elute the saliva. The eluted saliva were pooled and stored at −80°C
Tick haemolymph was harvested from 20 ticks per time point at 24, 72 and 120 h. Within the first hour of detaching ticks, they were washed in sterile PBS, dried between paper towels and sliced with a razor blade along the leg line of the alloscutum. Sliced ticks were then placed in well slides immersed in 50 μL of PBS and gentle pressure applied to squeeze out haemolymph into PBS. Harvested haemolymph with added protease inhibitor cocktail was stored at −80°C until used.
2.3. Full-length AamAV422 cDNA by rapid amplification of cDNA ends (RACE), bioinformatics and phylogenetic analyses
We had previously shown that the AamAV422 cDNA was strongly expressed in ticks that had fed for 120-168 h (Mulenga et al., 2007). To clone full-length cDNA, ~5 μg of total RNA from 120 h fed whole A. americanum ticks, which was extracted using TRIZOL reagent (Life Technologies, Carlsbad, CA, USA), was used to synthesize 3′ and 5′ RACE cDNA templates using the SMART RACE cDNA Synthesis Kit according to the manufacturer’s instructions (Clontech, Mountain View, CA, USA). After sequencing the 3′ and 5′ AamAV422 cDNA fragments, a PCR primer at the 5′ was designed to amplify the full-length AamAV422 cDNA in a 3′ RACE approach. Routinely, PCR fragments were cloned into the pGEMT vectors (Promega, Madison, WI, USA) and sequenced with SP6 and T7 promoter primers. Sequence analysis was performed using the MacVector DNA sequence analysis software (Accelrys, San Diego, CA, USA).
To gain insight into the probable function and properties of AamAV422, the AamAV422 protein sequence was subjected to amino acid motif scanning and physico-chemical property analyses using ProtParam and ScanProsite software (Expasy web server, http://ca.expasy.org/). Visual inspection of the AamAV422 sequence revealed multiple cysteine amino acid residues in the predicted AamAV422 protein. To assess whether the cysteine amino acid residues may be involved in disulfide bond formation, the AamAV422 amino acid sequence was scanned with the DiAminoacid Neural Network Application (DiANNA 1.1 web server, http://clavius.bc.edu/~clotelab/DiANNA/, Ferre and Clote 2005a,b, 2006). To examine the features that characterize AamAV422 protein, AamAV422 and the closest matches from GenBank were subjected to multiple sequence alignment analyses using MacVector software.
To determine the phylogenetic relationship of Aam422 protein sequence to its homologs in GenBank, a guide phylogenetic tree was developed based on MacVector Neighbor joining method. Sequences used in the analysis included I. scapularis (Accession numbers EEC13894, EEC18638, EEC18639, EEC18204, EEC18201, EEC18200, EEC19641, EEC12857, EEC19641 and Y66605), Amblyomma maculatum, (AEO35804, AEO36190 and AEO33794) (Karim et al., 2011), R. appendiculatus (CD782378), Rhipicephalus microplus (CK189031) (Guerrero et al., 2005) and Amblyomma variegatum (BM293119 and DAA34157) (Nene et al., 2002). Non-tick sequences included the fruit fly, Drosophila yoruba (EDW92254), scorpions, Mesobuthus eupus (ABR21053) and Hottentotta judaicus (ADY39539) (Morgenstern et al., 2011) and Buff-tailed bumblebee Bombus terrestris (XP_003396818).
2.4. Expression of rAamAV422 in Pichia pastoris
To construct the rAamAV422 expression plasmid, the mature AamAV422 protein-coding domain was subcloned into the pPICZαA (Life Technologies, USA), EcoRI/NotI sites using forward (5′GAGGCATGCAACTGCCACCTG3′) and reverse (5′TTGCCCAGGTAGGCGGAGAAG3′) primers respectively. The pPICZαA-AamAV422 expression plasmid was used to transform X-33 Pichia pastoris strain (Invitrogen, USA) by electroporation using a ECM 630 electroporator with parameters set to ~1,500 kv, 25 μF and 200 Ω. Transformed X-33 cells were then selected for methanol utilization. Selected recombinant transformants were cultured at 28°C to A600 of 1 before inducing protein expression by daily feeding of yeast cultures with 5% methanol for 48 h. During the preliminary experiment, it was observed that rAamAV422 starts to auto-degrade after 3 days in culture. The pPICZαA plasmid secreted the recombinant protein into the media. To purify rAamAV422, it was first precipitated by adding 525 g of ammonium sulfate per liter of culture supernatant and the protein allowed to precipitate overnight at 4°C with stirring. Following overnight incubation, the protein precipitate was centrifuged and the pellet dissolved in PBS, pH 7.4. For affinity purification, the recombinant protein in PBS was diluted in 2X native column binding buffer and then affinity purified under native conditions using the NiCl2 charged HiTrap™ Chelating HP column (GE Healthcare Bio-Sciences Corp, Piscataway, NJ, USA). The affinity-purified rAamAV422 was dialyzed against PBS or normal saline (0.9% NaCl in sterile distilled water) and stored at −80°C until required.
The expression of rAamAV422 in both bacteria and yeast was routinely confirmed by standard SDS-PAGE with Coomassie blue or silver staining, as well as western blotting analysis using antibodies (Abs) to N- and C-terminal histidine tags, respectively. Affinity purified rAamAV422 was dialyzed into three different buffers: normal saline (0.9% sodium chloride), 10 mM HEPES with 150 mM sodium chloride, pH 7.4 or PBS, pH 7.4.
Primary sequence analysis revealed putative N-glycosylation sites in AamAV422. To check whether rAamAV422 was glycosylated, affinity purified rAamAV422 was treated with the PNGase F enzyme according to instructions from the manufacturer (New England Biolabs, Ipswich, MA, USA). Deglycosylated rAamAV422 was detected by western blotting analysis using antibodies to the C-terminal histidine-tag as described above.
2.5. Production of rabbit Abs to 24 h A. americanum and I. scapularis TSPs
Production of Abs to 48 h A. americanum TSPs by repeated 48 h infestations of rabbits has been previously reported (Chalaire et al., 2011). In the current study, these protocols were repeated to produce Abs to 24 h A. americanum and I. scapularis TSPs by repeated 24 h infestations of rabbits with 10 female A. americanum or I. scapularis female ticks. Infestations were done four times per week every other week and repeated three times.
2.6. Validating secretion into the host, temporal and spatial expression analyses of AamAV422
To validate whether native AamAV422 was secreted into the host during tick feeding, affinity purified rAamAV422 was subjected to routine western blotting analysis, using Abs to 24 and 48 h A. americanum TSPs. Our preliminary analysis showed that rAamAV422 specifically reacted with Ab to 24 and 48 h A. americanum TSPs. To validate whether the observed specific reactivity of rAamAV422 with Ab to 24 h and 48 h TSP was consistent, 48, 96 and 120 h TSPs and 24, 72 and 120 h haemolymph, protein extracts of dissected tick organs (SG, MG and CA) and whole ticks were subjected to western blotting analyses using mono-specific Abs to rAamAV422.
Mono-specific Abs to rAamAV422 were produced as previously described (Mulenga et al., 1999) with slight modifications. Briefly rAamAV422 PVDF (a polymer of vinylidene fluoride) membrane strips were incubated in excess amounts of Abs to 48 h A. americanum TSP for 4 h at room temperature. Following appropriate washing, the PVDF strip was incubated in 500 μL of 100 mM glycine buffer, pH 2.8, for 5 min at room termperature to elute the mono-specific Ab to rAamAV422. Immediately after eluting, the Ab was neutralized by adding 0.1 volume of 10X PBS (pH 7.4) and 1 M Tris base. We made three elutions per strip. The eluted Ab was stored at −80°C until used.
2.7. Validating cross-reactivity of rAamAV422 with Abs to I. scapularis TSPs
We previously reported that the AamAV422 amino acid sequence was 87% identical to its I. scapularis homolog (Mulenga et al., 2007). To determine whether, consistent with sequence identity levels, AamAV422 cross-reacted with Abs to other tick species homologs, rAamAV422 was subjected to routine western blotting analysis using Abs to 24 h I. scapularis TSPs.
2.8. Antimicrobial, anti-platelet aggregation, anti-blood clotting and anti-complement functions
Antimicrobial function was assayed by co-incubating rAamAV422 with DH5α Escherichia coli K-12 strain used for routine gene cloning at 37°C for 10, 20 and 30 min in PBS, pH 7.4. An overnight DH5α culture was washed in 1 ml of PBS three times and then resuspended in 500 μL of PBS. DH5α aliquots were co-incubated with varying amounts of rAamAV422, ~0.16, 0.8 and 1.6 μg, which corresponded to ~0.032, 0.16, 0.32 μM of the affinity-purified protein in 200 μL of reaction volume. Subsequently 108 fold dilutions were plated onto Luria agar overnight, incubated at 37°C and colony forming units manually counted.
Anti-platelet aggregation function of rAamAV422 was investigated using the whole blood approach. Cattle blood collected from the Texas A&M University slaughterhouse was mixed with sodium citrate in a 9:1 ratio to prevent clotting. Citrated cattle blood was then used in the assays within 3 h of collection. Citrated whole cattle blood (500 μL) was diluted in a 1:1 ratio with normal saline (0.9% NaCl) and pre-incubated for 5 min at 37°C with or without 5 and 20 μg or 0.2 and 0.8 μ M of affinity-purified rAamAV422. Following pre-incubation, platelet aggregation was induced by the addition of ADP to 20 μM final concentration. Platelet aggregation as a function of increased appendance in Ω was then monitored and recorded using the Whole blood platelet aggregometer (Chono-Log Coorporation, Harvetown, PA, USA).
The effect of rAamAV422 on blood clotting function was investigated using routine blood coagulation assays, recalcification time (RCT), Activated Partial Thromboplastin Time (APPT), Prothrombin Time (PT), and Thrombin Time (TT) as previously published (Spillert and Lazaro, 1995; Nazareth et al., 2006; Derem et al., 2009; Liao et al., 2009; Gao et al., 2011). The RCT assay, which measures the integrity of the blood clotting system in its entirety, was done according to previously published methods (Liao et al., 2009; Gao et al., 2011). Briefly, 20μL of citrated human plasma was pre-incubated at 37°C for 10 min without or with serially diluted affinity purified rAamAV422, 6.6, 3.3, 0.825 and 0.415 μg or 3.77, 1.89, 0.471 and 0.237 μM in 70 μL of 10 mM HEPES buffer, pH 7.4. Adding 10 μl of 25 mM CaCl2 triggered clotting, which was subsequently monitored at A650 at 20 s intervals for 10 min using a VersaMax microplate reader (Molecular Devices, Sunnyvale, CA, USA) set to 37°C. The interpretation of data was that formation and firming of the clot was directly proportional increase in A650.
The PT assay, which measures the integrity of the extrinsic blood clotting activation pathway, is triggered by injury to blood vessels as occurs during tick feeding (Davie et al., 1991; Lefkowitz, 2006). To investigate whether AamAV422 interferes with the extrinsic blood-clotting activation pathway, 100 μL of citrated plasma was pre-incubated for 10 min at 37°C with an excess amount (6.0 μM or 16.5 μg) of affinity purified rAamAV422. Subsequently, adding 200 μL of the PT reagent triggered the clotting reaction. Clotting time was immediately monitored and determined using the KC1 DELTA coagulometer (Trinity Biotech, NJ, USA).
The APPT assay, which measures the integrity of the intrinsic blood clotting activation pathway, can be triggered by multiple factors such as internal blood vessel injury and bacterial infection (Davie et al., 1991; Lefkowitz, 2006). To determine whether AamAV422 interferes with functioning of the intrinsic blood clotting activation pathway, 100 μL of human plasma was pre-incubated for 10 min at 37°C with an amount (6.0 μM or 16.5 μg) of affinity purified rAamAV422. Subsequently, adding 100 μL of the APPT reagent and continuing to incubate at 37°C for an additional 3 min activated the reaction. Addition of 100 μL of 25mM CaCl2+ to the reaction of plasma and APPT reagent mixture triggered clotting. APPT clotting time was determined using the KC1 DELTA coagulometer.
The extrinsic and intrinsic blood clotting pathways converge to the common pathway, when activated thrombin converts fibrinogen to fibrin (Davie et al., 1991; Lefkowitz, 2006). The TT assay measures normal function of activated thrombin to convert fibrinogen to fibrin, the final step that is necessary to produce sufficient fibrin needed to produce the blood clot (Davie et al., 1991; Lefkowitz, 2006). To investigate whether AamAV422 interfered with thrombin conversion of fibrinogen to fibrin, 100 μL of plasma was pre-incubated at 37°C for 10 min with 6.0 μM or 16.5 μg of affinity purified rAamAV422. Adding the 200 μL of the TT reagent triggered conversion of fibrinogen to fibrin followed by clot formation. Clotting time was determined using the KC1 DELTA coagulometer as described above.
To gauge the potential of AamAV422 to interfere with the complement activation pathway, the anti-complement function of rAamAV422 was evaluated using the MicroVue CH50 ELISA kit (Quidel, San Diego, CA, USA). This kit quantifies the amount of terminal complement complexes (TCC) that are formed when the complement system is activated via the classical pathway as a function of integrity of the system. Thus, to assess whether native AamAV422 affected the function of the complement activation pathway, reference human serum (20 μL) was co-incubated with 20 μL of PBS, pH 7.4 (3.3 μg or 3 μM of affinity purified rAamAV422) for 10 min at 37°C in a 40 μL reaction volume. The serum-rAamAV422 reaction mixture was then incubated with ~123 μL of the complement activator solution for 1 h at 37°C. To quantify the amount of formed TCC, the reaction mix (diluted 1:200) was bound to human anti-TCC Abs coated onto micro-wells. Following appropriate washing, the bound TCC was reacted with horseradish peroxidase (HRP)-conjugated ligand. Subsequently the chromogenic HRP substrate was added to the wells to quantify TCC that bound onto Abs. The intensity of color development, which directly represented the amount of formed TCCs, was quantified at 450 nm.
2.9. Target validation by RNA mediated gene silencing of AamAV422 mRNA
RNA interference (RNAi)-mediated silencing of AamAV422 mRNA was performed as previously described (Mulenga et al., 2008; Mulenga and Khumthong, 2010a,b). A ~600 bp cDNA template used for dsRNA synthesis was amplified with primers (For: 5′TAATACGACTCACTATAGGGGACCTTCGCCCTGTTCGCC3′, Rev: 5′TAATACGACTCACTATAGGGCCTGGCGTTGGCCTCGCCG3′) with an added T7 promoter (underlined) using the plasmid template. The amplified cDNA template was selected on the basis that it had not have long regions that showed identity to other genes to avoid non-specific effects of RNAi silencing. Using a 10 μL gastight syringe (Hamilton, Reno, NV, USA), 25 female A. americanum ticks were microinjected with 0.5-1μL (2-3 μg/μL) of ~600 bp of AamAV422 or GFP (control) dsRNA in Tris-EDTA buffer. Microinjected ticks were allowed to recover overnight at 22°C before placing them on a calf to feed as previously described (Mulenga and Khumthong, 2010a). To validate whether AamAV422 mRNA was disrupted, three ticks per treatment were sampled at 48 h post-attachment and then individually processed for total RNA extraction using TRIZOL. Extracted RNA was subjected to two-step qualitative reverse transcriptase (RT)-PCR using AamAV422 open reading frame (ORF) primers, which were placed outside the cDNA region that was use for dsRNA synthesis. Ticks used in validation of RNAi silencing were sampled at 48 h to ensure that the ticks were alive and could attempt to feed. Tick actin PCR primers (Mulenga et al., 2009) were used as an internal control.
The effect of silencing AamAV422 mRNA was determined by assessing tick feeding and reproductive efficiency parameters including attachment rates, tick mortality during and after feeding, engorgement weight (EW) as an index of imbibed blood, and egg mass conversion ratio (EMCR) as a measure of the engorged female tick to convert its blood meal to egg mass. EMCR was determined as a fraction of egg mass over EW. Attachment rates were determined by subtracting the number of unattached ticks from the total number of ticks that were placed on the animal. Mortality was determined by counting dead ticks found in the cell 48 h post-attachment. The time to spontaneous detachment was also recorded. Spontaneously detached ticks were incubated at 25°C to allow them to lay eggs. Statistical significance between treatment groups was confirmed using one-way ANOVA and post-hoc pair-wise comparisons using Tukey’s honestly significant difference (HSD) hosted by the Website for Statistical Computation at Vassar University, USA (http://faculty.vassar.edu/lowry/anova1u.html). For feeding of ticks on calves, ticks were placed in cotton orthopedic stockinet containment cells on the back of the calf.
3. Results
3.1. AamAV422 belongs to an exclusive arthropod protein family characterized by seven disulfide bonds
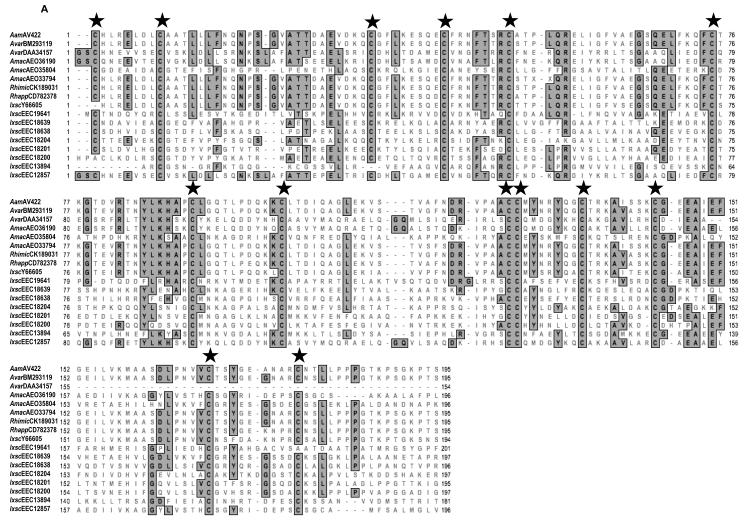
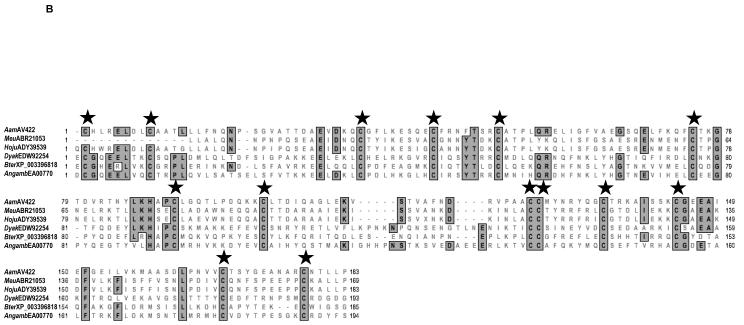
RACE (3′ and 5′) was successfully used to clone a full-length 1,400 bp AamAV422 cDNA (NCBI Accession no. KC222016). The AamAV422 cDNA contained a 696 bp ORF that encodes for a putatively secreted protein that is 231 amino acid residues long. Amino acid motif scanning analysis was used to identity the probable biological functions of AamAV422. Except for the putative N-glycosylation site at position 43 (not shown), amino acid motif scanning analysis of the AamAV422 protein did not retrieve any known amino acid motifs. BLASTP scanning of AamAV422 exclusively retrieved arthropod protein sequences (results not shown) indicating that AamAV422 does not have any known homologs in non-arthropod organisms. Visual inspection of the AamAV422 amino acid sequence revealed that the AamAV422 protein contained 15 cysteine amino acid residues at positions 23, 25, 33, 58, 67, 75, 99, 115, 126, 150, 151, 159, 168, 192 and 202 (numbering of cysteine amino acid residues is based on AamAV422 preprotein amino sequence). Scanning of AamAV422 amino acid sequence on DiANNA web server (Ferre and Clote 2005a,b, 2006) predicted that 14 of the 15 cysteine amino acid residues will form seven disulfide bonds involving C23 and C115, C33 and C58, C67 and C150, C75 and C151, C99 and C126, C159 and C192, C168 and C202 as summarized in Fig. 1. We previously reported that AamAV422 amino acid sequence was ~87% homologous to I. scapularis (Y66605) and ~94-97% identical to its homologs in R. microplus (CK189031), A. variegatum (BM293119) and R. appendiculatus (CD782378) (Mulenga et al., 2007). Here we update our sequence analysis to include sequences such as A. maculatum (AEO33794) that show 96% amino acid identity to AamAV422 and other arthropod sequences that show an apparent structural similarity to AamAV422 as indicated by consensus cysteine amino acid residues (Fig. 2). Multiple sequence alignment of the AamAV422 amino acid sequence with AamAV422-like sequences in other tick species (Fig. 2A) and non-tick species sequences (Fig. 2B), revealed that with exception of C25, 14 of the 15 cysteine amino acid residues were well conserved displaying a distinct pattern, “C23-X7/9-C33-X23/24-C58-X8-C67-X7-C75-X23-C99-X15-C115-X10-C126-X24/25/33-C150C151-X7-C159-X8-C168-X23/24-C192-X9/10-C202”.
Fig. 1.
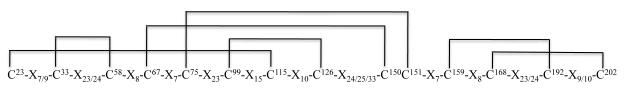
Predicted disulfide bonds and zinc binding sites in Amblyomma americanum (Aam)AV422 predicted protein sequence. The translated AamAV422 protein was scanned on the DiAminoacid Neural Network Application (DiANNA 1.1 web server, http://clavius.bc.edu/~clotelab/DiANNA/). Numbering of cysteine residues forming disulfide bonds starts from the first methionine of AamAV422 pre-protein. Predicted disulfide bond pairings, C23-C115, C33-C58, C67-C150, C75-C151, C99-C126, C159-C192, C168-C202, are marked with solid lines.
Fig. 2.
Multiple sequence alignment of Amblyomma americanum (Aam)AV422 amino acid sequence with homologs in other ticks (A) and non-tick arthropods (B). Amino acid sequences were aligned using the CustalWsequence alignment tool in MacVector DNA analysis software. Conserved amino acid residues are shaded gray. Stars (★) denotes conserved cysteine amino acid residues. Sequences are denoted by an abbreviation of their species name followed by their GenBank accession number. AamAV422, A. americanum (KC222016); Rhimic, Rhipicephalus microplus (CK189031); Rhapp, Rhipicephalus appendiculatus (CD782378); Amac, Amblyomma maculatum (AE033794, AE035804, AE036190); Avar, Amblyomma variegatum (DAA34157 and BM293119); Ixsc, Ixodes scapularis (Y66605, EEC1287, EEC13894, EEC18200, EEC18201, EEC18204, EEC18638, EEC18639 and EEC19641); Angamb, Anopheles gambiae (EAA00770); Meu, Mesobuthus eupeus (ABR21053); Hoju, Hottentotta judaica (ADY39539); Bter, Bombus terrestris (XP_003396818); Dyak, Drosophila yakuba (EDW92254).
3.2. AamAV422-like protein family segregates into two clusters
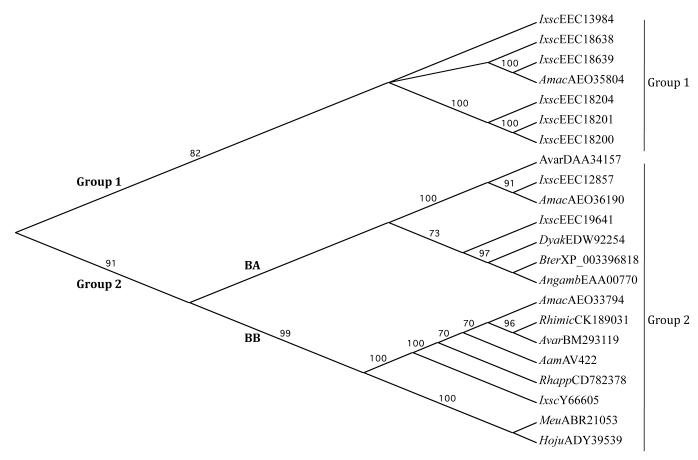
The midpoint rooted guide phylogenetic tree analysis in Fig. 3 showed that AamAV422-like protein sequences in Fig. 2A and B segregated into two groups supported by 82 and 91% bootstrap values. Group 1 contains exclusive AamAV422-like tick sequences. Except for one sequence, A. maculatum (AEO35804), the first group of sequences contains a majority of I. scapularis AamAV422-like sequences (EEC18200, EEC18201, EEC18204, EEC18638 and EEC18639). The identity levels between sequences in Group 1 ranged from 24-47%, with the exception of A. maculatum (AEO35804) and I. scapularis (EEC18639) which show amino acid identity levels at 58%, (not shown). Group 2 has the majority of sequences that further segregates into two subgroups, BA and BB. In the BB subgroup, AamAV422 sequence clusters with other tick sequences, I. scapularis (Y66605), R. appendiculatus (CD782378), R. microplus (CK189031), A. maculatum (AEO33794), A. variegatum (BM2931119), and two non-tick sequences, the Lesser Asian scorpion, Mesobuthus eupeus (ABR21053) and Scorpion Hottentotta judaica (ADY39539). AamAV422 shows 87-99% amino acid identity to the other tick sequences in subgroup BB, while the identity level drops to ~32-36% compared with non-tick sequences. Tick AamAV422-like sequences in BA subgroup, I. scapularis (EEC12857 and EEC19641), A. maculatum (AEO36190) and A. variegatum (DAA34157) segregate together with Anopheles gambiae (EAA00770), buff-tailed bumblebee, Bombus terrestris (XP_003396818) and Drosophila yakuba (EDW92254). Except for EEC19641 which shows ~20-30% amino acid identity to other tick sequences in subgroup BA, the amino acid identity levels jump to between 56-70% among other tick sequences (not shown). It is particularly interesting to note that the highest amino acid identity level, 70%, was observed between A. maculatum (AEO36190) and I. scapularis (EEC12857) sequences.
Fig. 3.
Neighbor joining guide phylogenetic tree showing the relationship of Amblyomma americanum AamAv422 putative protein to AamAV422-like proteins from ticks (n = 15) and non-tick arthropods (n = 6). The predicted AamAV422 protein sequence was aligned with AamAV422-like sequences: Rhimic, Rhipicephalus microplus (CK189031); Rhapp, Rhipicephalus appendiculatus (CD782378); Amac, Amblyomma maculatum (AE033794, AE035804, AE036190); Avar, Amblyomma variegatum (DAA34157 and BM293119); Ixsc, Ixodes scapularis (Y66605, EEC1287, EEC13894, EEC18200, EEC18201, EEC18204, EEC18638, EEC18639 and EEC19641); Angamb, Anopheles gambiae (EAA00770); Meu, Mesobuthus eupeus (ABR21053); Hoju, Hottentotta judaica (ADY39539); Bter, Bombus terrestris (XP_003396818) and Dyak, Drosophila yakuba (EDW92254), using ClustalW. The guide phylogenetic tree was developed using the MacVector Neighbor joining method default parameters as described in Section 2.3. Groups 1 and 2, and subgroups BA and BB are indicated. Numbers on each branch represent bootstrap values that signify the level of confidence in the branch.
3.3. Expression and affinity purification of rAamAV422 in P. pastoris
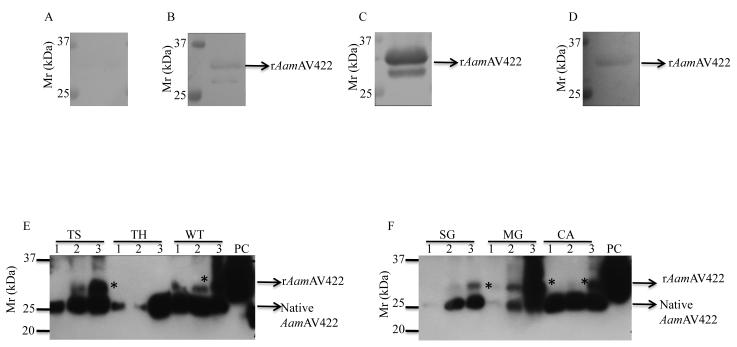
Recombinant AamAV422 was successfully expressed in Pichia pastoris (Fig. 4). The pilot expression analysis over 5 days (Fig. 4A) revealed that expression of rAamAV422 peaked between 24 and 48 h time points but started to autodegrade from day 3. A combination of overnight ammonium sulfate precipitation at 4°C and affinity purification using NiCl2+ charged columns under native conditions were successfully used to purify rAamAV422 from yeast-spent media (Fig. 4B). The calculated molecular size of rAamAV422 is ~27 kDa: sum of ~24 kDa molecular weight of mature AamAV422 protein with an added ~3.5 kDa C-terminus fusion comprising the hexahistidine tag. However, the observed molecular mass of rAamAV422 is ~30 kDa, which may suggest possible post-translational modification (Fig. 4B). When treated with the PNGase F N-deglycosylation enzyme, a reduction of the molecular mass of rAamAV422 from ~30 to ~27 kDa was observed (Fig. 4C). This observation suggests that N-glycosylation contributed ~3 kDa to the molecular mass of rAamAV422 (Fig. 4C).
Fig. 4.
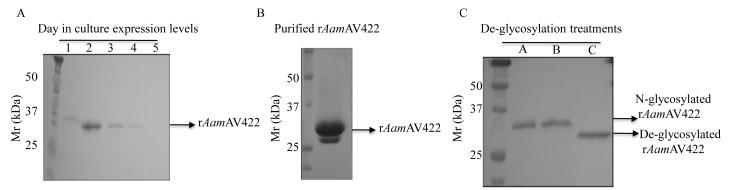
Expression and affinity purification of recombinant (r) Amblyomma americanum AamAV422 expression in Pichia pastoris. (A) Cumulative daily expression levels of rAamAV422 through 5 days of culture. Construction of expression plasmid, induction and validation of expression levels using the antibody to the C-terminus hexahistidine tag and affinity purification of rAamAV422 were performed as described in Section 2.4. (B) Validation of affinity purification of rAamAV422 using Coommassie blue staining was accomplished by a combination of ammonium sulfate precipitation and affinity purification on NiCl2+ charged columns as described in Section 2.4. (C) Validation of N-glycosylation posttranslational modification of rAamAV422: 20 μg of affinity-purified rAamAV422 was treated with the PNGase F enzyme as described in Section 2.4. De-glycosylated rAamAV422 was detected by western blotting analysis using antibodies to the C-terminus hexahistidine tag. Lane A, Non-treated rAamAV422; lane B, rAamAV422 treated with PNGase F buffer only, lane C, deglycosylated rAamAV422.
3.4. AamAV422 protein is ubiquitously expressed, injected into the host from 24 h of tick feeding and cross-reacts with Abs to I. scapularis TSPs
The possibility of AamAV422 being secreted into the host during tick feeding was investigated by subjecting rAamAV422 to western blotting analysis using Abs raised to 24 and 48 h A. americanum TSPs. As summarized in Fig. 5, rAamAV422 did not bind pre-immune serum (Fig. 5A), but specifically bound with Abs to 24 h (Fig. 5B) and 48 h (Fig. 5C) A. americanum tick saliva, indicating that native AamAV422 protein is injected into the host within the first 24 h of tick feeding. It was also interesting to note that consistent with our reported comparative sequence analysis data that showed 87% amino acid residue identity between AamAV422 and its I. scapularis homolog (Mulenga et al., 2007), Abs to 24 h I. scapularis TSPs specifically reacted with rAamAV422 (Fig. 5D).
Fig. 5.
Western blotting analyses. Validation of Amblyomma americanum AamAV422 protein secretion into the host, cross-reactivity of recombinant (r)AamAV422 with antibodies to other ticks, temporal and spatial expression analyses of AamAV422 were done by routine western blotting analyses as described in section 2.6. rAamAV422 western blots were screened with rabbit pre-immune serum (A), antibodies to 24 h (B) and 48 h (C) A. americanum tick saliva proteins (TSPs) and antibodies to Ixodes scapularis (D) TSPs. Mono-specific antibodies were eluted as described and used to screen western blots of tick saliva (TS) proteins, tick hemolymph (TH) and protein extracts of whole ticks (WT) (E), and dissected tick organs, salivary glands (SG), Midgut (MG) and carcass (CA) (F). TS lanes 1, 2 and 3 are TSPs harvested from 48, 96 and 120 h fed ticks, respectively. TH, WT, SG, MG and CA lanes 1, 2 and 3 are hemolymph and protein extracts of 24, 72 and 120 h fed ticks. PC, rAamAV422 used as positive control; *, a potentially glycosylated native AamAV422 form.
To test whether our observations in Fig. 5B and C were physiologically consistent and not an artifact, we successfully eluted mono-specific Abs to rAamAV422 and used them to screen western blots of 48, 96 and 120 h TSPs, haemolymph, protein extracts of whole ticks (Fig. 5E) and dissected tick organs, SG, MG and CA (Fig. 5F). The expectation was that, if data shown in Fig. 5B and C were physiologically consistent, mono-specific Abs to rAamAV422 would specifically bind at the very least the expected ~24 kDa mature AamAV422 protein band and/or other high molecular weight due to possible post translational modifications. In agreement with this expectation mono-specific Abs to rAamAV42 specifically reacted the expected ~24 kDa Aam AV422 mature protein bands on blots shown in Fig. 5E (tick saliva, haemolymph and whole tick) and Fig. 5F (SG, MG and CA). In addition to the expected ~24 kDa AamAV422 protein band, mono-specific Ab also bound an ~ 28 kDa protein from the 96 h tick feeding time point. We suspect that the 28 kDa protein band is the glycosylated form of native AamAV422. It is also notable that in haemolymph (Fig. 5E), SG and MG (Fig. 5F), the AamAV422 protein band is weak.
3.5. rAamAV422 delays plasma clotting, inhibits platelet aggregation and complement activation
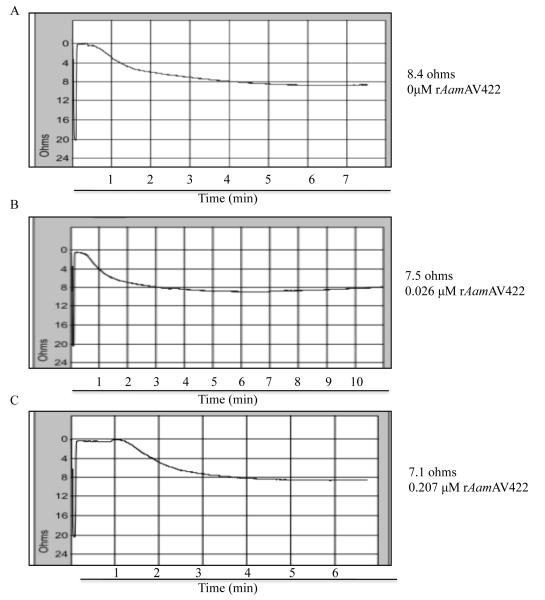
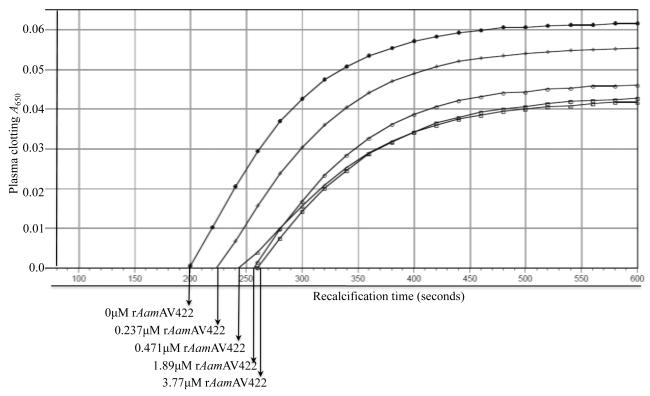
Given the data from Fig. 5B and C demonstrating that native AamAV422 was injected into the host during tick feeding, biological functions of this protein at the tick-host interface were investigated. To do this rAamAV422 was screened against a battery of assays to gauge whether this protein had antimicrobial, anti-platelet aggregation, anti-plasma clotting and anti-complement activation functions. The data summarized in Figs. 6 - 8 show that, except for antimicrobial activity, rAamAV422 affected platelet aggregation, plasma clotting and complement activation. In whole blood platelet aggregation approach summarized in Fig. 6, the level of platelet aggregation was correlated with an increase in electrical resistance. Thus it is interesting to note that in the presence of 5 and 20 μg or 0.2 and 0.8 μM of affinity purified rAamAV422, platelet aggregation was reduced in a dose responsive manner by ~11% (7.5/8.4 Ω, Fig. 6B) and 15% (7.1/8.4 Ω, Fig. 6C), respectively, when compared with platelet aggregation in the absence of rAamAV422 (Fig. 6A). Although no effect was observed in assays to measure the effect of rAamAV422 on plasma clotting time in the extrinsic (PT assay), intrinsic (APPT assay) and the common pathway (TT assay) (result not shown), it delayed plasma clotting in a dose-responsive manner when subjected to the RCT assay, which measures the functional integrity of the blood clotting system in its entirety (Fig. 7). As summarized in Fig. 7, plasma clotting was delayed by ~170, 165, 135 and 75 s in the presence of 3.77, 1.89, 0.471 and 0.237 μM rAamAV422. In the RCT assay, A650 levels correlate with how firm the plasma clot is. It is interesting to note that, in addition to delaying plasma clotting time, the plasma clot firmness was reduced by ~13-35% (not shown). Likewise, pre-incubation of serum with 3 μM rAamAV422 reduced production of terminal complement complexes by ~24% (Fig. 8).
Fig. 6.
Anti-platelet aggregation function assay. The effect of recombinant (r) Amblyomma americanum AamAV422 protein (rAamAV422) on platelet aggregation was done using the whole blood approach as described in Section 2.8. Five hundred μL of cattle whole blood mixed 9:1 with sodium citrate diluted with equal volume normal saline (0.9% sodium chloride) was pre-incubated with indicated various amounts of rAamAV422 for 10 min at 37°C. Adding 20 μM ADP triggered platelet aggregation, which was subsequently monitored as a function of electrical resistance using the Chrono-Log aggregometer.
Fig. 8.
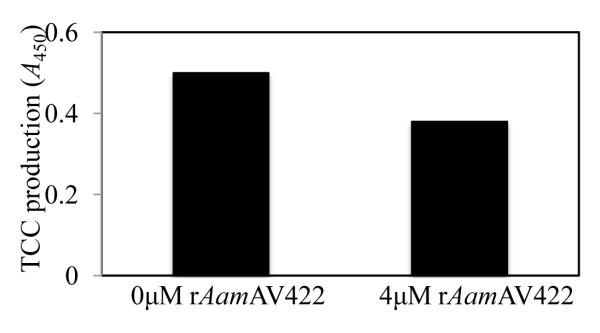
The effect of recombinant (r) Amblyomma americanum AamAV422 protein (rAamAV422) against classical complement activation pathway. Indicated amounts of rAamAV422 protein were pre-incubated with serum for 10 min at 37°C and subjected to the classical complement activation assay as described in Section 2.8.
Fig. 7.
The effect of recombinant (r) Amblyomma americanum AamAV422 protein (rAamAV422) against plasma clotting in the recalcification time assay. Twenty μL of citrated human plasma diluted up to 90 μL with 10 mM HEPES buffer plus 150 mM NaCl, pH 7.4, was pre-incubated at 37°C for 5 and 10 min with indicated amounts of affinity purified rAamAV422. In the microplate, clotting time was triggered by adding 10 μL of 25 mM CaCl2+ and then monitored as described in Section 2.8.
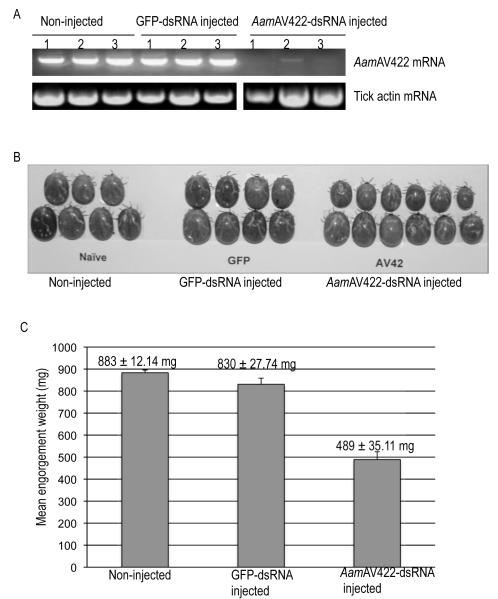
3.6. AamAV422 silencing significantly reduces the tick blood meal size
To characterize the significance of AamAV422 in tick feeding biology, the effect of disrupting AamAV422 mRNA using the RNAi silencing approach was investigated (Fig. 9). Two step RT-PCR using cDNA synthesized from naïve (not injected with dsRNA), GFP-dsRNA and AamAV422-dsRNA injected animals validated that injection of AamAV422-dsRNA into unfed female ticks caused partial disruption of AamAV422 mRNA by 48 h post-attachment (Fig. 9A). Disruption of the AamAV422 mRNA appeared not to negatively affect the tick’s ability to attach to the animal, start and complete feeding, in that no differences between AamAV422-dsRNA injected and controls (GFP dsRNA injected and non-injected) were observed (not shown). Similarly, disruption of AamAV422 mRNA did not have any lethal effect on ticks in that mortality rates of ticks between treated and control ticks were not apparently different (not shown). Apart from AamAV422-dsRNA injected ticks being apparently smaller than control ticks, there was no other visible physical phenotype that was observed (Fig. 9B). EW, an index of the amount of blood taken in by ticks, ranged from 260-660 mg for AamAV422-dsRNA injected ticks compared with EWs of control ticks, non-injected (750-940 mg) and GFP-dsRNA injected (750-1,070 mg). At the P<0.05 level of significance, one-way ANOVA analysis of the mean EW revealed that disruption of AamAV422 mRNA caused a significant reduction in the amount of blood taken in treated ticks (F (2, 32) = 65.36, P<0.0001). Tukey’s HSD post-hoc comparison of mean EWs revealed that the mean EW of AamAV422-dsRNA injected ticks (M = 489 mg ± S.D. (35.11 mg) was significantly smaller than both control ticks, non-injected (M = 883 ± 12.14 mg, GFP-dsRNA injected, M = 830 ± 27.74 mg) (Fig. 9C). AamAV422 mRNA disruption did not affect the tick fecundity in that 91% (11/12) were able to lay eggs (not shown). Additionally, a one-way ANOVA revealed that mean EMCRs of AamAV422-dsRNA injected ticks were not significantly different from control ticks (not shown).
Fig. 9.
Validating the significance of Amblyomma americanum AamAV422 protein in tick feeding success. (A) Validating the disruption of AamAV422 mRNA in AamAV422-double stranded (ds) RNA injected ticks. Approximately 25-30 ticks were microinjected with 0.5-1 μL (2-3 μg/μL) of AamAV422 or GFP (control) dsRNA in Tris-EDTA buffer. At 48 h post-attachment, three ticks per treatment, non-injected and GFP-dsRNA injected control ticks, together with the AamAV422-dsRNA injected ticks, were manually detached. Ticks were individually processed for total RNA extraction, DNase treated to eliminate genomic DNA contamination and then subjected to two-step reverse transcriptase (RT)-PCR using AamAV422 PCR primers. Equivalent amounts, ~15 μL of PCR products, were resolved on 2% agarose gels containing 1 μg/ml of ethidium bromide. (B) Physical phenotype of AamAV422-dsRNA injected ticks. Ticks were photographed after spontaneous detachment. (C) Effect of RNA interference (RNAi)-mediated silencing of the AamAV422 mRNA disruption on Amblyomma americanum female tick feeding success. After spontaneously detaching from the host, ticks were individually weighed to determine engorgement weights (EWs) as indices for amounts of blood imbibed by ticks. EWs were subjected to one-way ANOVA as described in Section 2.9 to determine statistical significance. Mean (M) EWs (MEW ± S.D.) were plotted as bar graphs. *, significant differences in MEWs between AamAV422-dsRNA injected ticks and both control groups. P < 0.0001.
4. Discussion
Cross-tick species conservation, predominant expression in tick salivary glands and increased amounts of transcript as ticks continue to feed (Mulenga et al., 2007) prompted our interest to explore the biology of AamAV422 in tick feeding regulation. The specificity of Abs to both 24 h and 48 h TSPs as well as the reactivity of mono-specific Abs to rAamAV422 on western blots of rAamAV422, tick saliva, haemolymph and dissected tick organs led us to conclude that this protein was ubiquitously expressed and injected into the host within 24 h of tick feeding. In the literature two other cross-tick species conserved TSPs, tick calreticulin (Sanders et al., 1998, 1999; Ferreira et al., 2002; Xu et al., 2004, 2005; Alarcon-Chaidez et al., 2006; Gao et al., 2008; Kaewhom et al., 2008; Parizi et al., 2009) and histamine release factor (Mulenga et al., 2003a Mulenga and Azad, 2005; Dai et al., 2010) have been described. A common feature between tick calreticulin and histamine release factor is that they both have homologs in vertebrates (Mulenga et al., 2003b, Mulenga and Azad, 2005; Coe and Michalak, 2009; Martins et al., 2010). From this perspective, AamAV422 is a unique cross-tick species conserved TSP in that it apparently belongs to an exclusive family of arthropod proteins that has no vertebrate homologs. For lack of a conventional term, we are referring to this family of proteins as the “AamAV422-like protein family”. From the perspective of discovery of druggable and/or tick vaccine target antigens, the observation that AamAV422 does not have vertebrate homologs is advantageous. It may mean that targeting AamAV422, as a druggable and/or vaccine target antigen for tick control, will have no unintended consequences on the vertebrate host. From the analyses in this study it is apparent that ticks encode multiple AamAV422-like proteins that segregate into three distinct groups. The observation that within each of the three clusters on the phylogenetic tree, AamAV422-like protein sequences were highly homologous, is suggestive of the possibility that this protein family regulates biological functions that are evolutionarily conserved in all ticks. It is interesting to note that tick AamAV422-like protein sequences from different clusters on the phylogenetic tree shared very low amino acid identities which may signal that biological functions regulated by members of this protein family may be related but not redundant.
It is apparent from the phylogenetic analysis data that the AamAV422-like group in ticks has several members that cluster in three unique clusters. On the basis of sequence similarity that shows low inter-cluster but high intra-cluster amino acid sequence identity levels, it is possible that the three groupings of AamAV422-like proteins in ticks represent three uniquely conserved but related pathways. Our long-term interest is to explore the role(s) of AamAV422 and its homologs in regulating early stages of the tick feeding process, attachment onto host skin, creation of the tick feeding site and conditioning of the host for tick-borne disease agents to colonize the hosts. Thus our data validating that native AamAV422 protein was part of the TSP complex that is injected into the host from 24 h through 120 h of the tick feeding process is significant. Consistent with previous findings that Aam AV422 mRNA levels increased with tick feeding (Mulenga et al., 2007), the observation that the AamAV422 protein band was weaker on 24 h SG and MG blots, but increased at subsequent time points, indicates that AamAV422 protein expression increases with tick feeding. Ticks differentially inject multiple proteins into the host to regulate the different phases of the tick feeding process (Wang et al., 1999; Mulenga et al., 2007). Proteins such as AamAV422 that are injected into the host within the first 24 h of tick feeding may potentially regulate events associated with the PFP when the tick attaches to host skin, creates the feeding site for subsequent blood meal feeding, and starts to condition the host to allow for transmitted tick-borne disease agents to colonize the host with minimal impediments. Thus, successfully blocking functions of proteins such as AamAV422 that regulate the PFP represent attractive target antigens for development of tick vaccines, in that such a vaccine, if successful, will block the key facets of tick parasitism that are preceded by PFP.
A major drawback in tick molecular biological research is that the majority of candidate genes identified though transcriptome projects are of unknown function (Ribeiro et al., 2006; Mulenga et al., 2007; Wang et al., 2007; Karim et al., 2011). This presents a challenge when designing experiments to understand the role(s) of these candidate genes in tick feeding physiology. In our laboratory, we have developed a deorphanization strategy to understand the role(s) of candidate proteins at the tick-host interface. Our battery of assays were designed on the understanding that for a tick to successfully feed, it must keep blood in a fluid state at both the feeding site and in the gut. Additionally the tick must condition the host by modulating innate immune responses such as complement activation so that tick-borne disease agents can colonize the host with minimal impediment (Valenzuella et al., 2002; Daix et al., 2007). On the basis of data from our deorphanization assays, it is apparent that native AamAV422 may be involved in mediating the tick’s anti-haemostatic and anti-complement activation functions. Blood clotting can be initiated via the extrinsic pathway, which is triggered by injury to blood vessels as occurs during tick feeding or via the intrinsic pathway that can be initiated by multiple triggers such as internal injury, hyperlipidemia or bacteriaal infection (Davie et al., 1991). Either one of the two blood clotting pathways culminates in the common pathway, when activated thrombin converts fibrinogen to fibrin. Our efforts to home in on which of the three pathways was being affected by rAamAV422 to exert the observed delayed plasma clotting time were inconclusive. Normal functioning of the intrinsic blood clotting activation pathway requires two metal ion co-factors, calcium and zinc (Davie et al., 1991; Kluszynski et al., 1997; Tubek et al., 2008a,b). It is interesting to note that in addition to being secreted into the host, AamAV422 protein is also expressed in multiple tick organs including the MG. There is a possibility that AamAV422 expressed in the tick MG potentially participates in preventing blood from clotting in the MG. In addition to ensuring that host blood is in a fluid state at the tick-feeding site, the tick has to maintain host blood in a fluid state in the MG in order for ticks to digest and assimilate nutrients and acquire tick-borne disease agents. Tick anticoagulant (Ibrahim et al., 2001; Ehebauer et al., 2002; Narasimhan et al., 2002; Lai et al., 2004; Nazareth et al., 2006; Mans et al., 2008; Ricci et al., 2007; Decrem et al., 2009; Gao et al., 2011,) anti-platelet aggregation (Chmelar et al., 2011) and those with anti-complement (Valenzuella et al., 2002; Daix et al, 2007) functions have been reported from multiple tick species. The uniqueness of AamAV422 is that it shows high cross-tick species conservation, which signals that it regulates functions that are central to all ticks.
Another important goal of this study was to investigate the significance of AamAV422 in tick feeding physiology. Consistent with our assumption that cross-tick conservation signaled an important role for this molecule in tick feeding regulation, RNAi-mediated silencing of AamAV422 mRNA caused a significant reduction in the amount of blood that was ingested by ticks, demonstrating that AamAV422 protein was important to the tick feeding success. Despite the reduced blood meal sizes, AamAV422 mRNA silencing did not affect the ability of ticks to convert their blood meal to eggs, indicating that AamAV422 may not play a role in tick reproduction. It is also interesting to note that, while AamAV422 was injected into the host from 24 h of tick feeding, RNAi silencing did not affect the tick’s ability to start and complete feeding. This may be explained by the fact that AamAV422 may not be the sole molecule that is important in regulating the tick feeding success. The goal of RNAi silencing is to lower or eliminate the expression of the target protein and in so doing affect the biological function of the cognate protein. The weakness of the RNAi silencing experiment conducted in this study is that we did not validate whether or not disruption of the AamAV422 mRNA resulted in reductions or elimination of the AamAV422 protein. Given that we did not achieve complete disruption of the AamAV422 mRNA, it’s more than likely that the AamAV422 protein was expressed, albeit at low levels. Whether or not this was enough to affect other tick feeding parameters other than blood meal uptake cannot be determined from results from this study. Chalaire et al. (2011) have also recently observed that despite complete silencing of target mRNA, proteins that were expressed prior to RNAi silencing remain and are potentially functional. Another limitation of the RNA silencing experiment in this study is that due to the lack of genomic sequence data for A. americanum, we did not exhaustively screen for gene targets that may have sequence regions that are identical to the dsRNA target in this study. There is a possibility that if there are genes in the A. americanum genome that have regions that are identical to AamAV422, the small interfering RNA that results from dicing the AamAV422 dsRNA may trigger degradation of other genes and thus the observed RNAi silencing effects may not be specific to AamAV422. However, we are confident that this was not the case, as scanning AamAV422 against an extensive number of tick ESTs in GenBank retrieved genes that showed similarity on the basis of conserved cysteine amino acid residues and not strings of identical sequence regions.
Another interesting finding in this study is that consistent with being cross-tick species conserved, Abs to I. scapularis TSPs cross-reacted with rAamAV422. This feature represents an important opportunity to target AamAV422 as a candidate for a universal tick vaccine. In nature, animals are infested by different tick species. Thus, an ideal vaccine that will target different tick species has been advocated (Fragoso et al., 1998; Parizi et al., 2012). Experiments to investigate the vaccination effect of AamAV422 are currently underway.
Highlights.
AamAV422 belongs to an exclusive group of arthropod proteins
AamAV422-like group of proteins characterized by seven putative disulfide bonds
AamAV422 protein is injected into the host within 24h of tick feeding
AamAV422 cross-reacts with antibodies to 24 h Ixodes scapularis tick saliva proteins
Native AamAV422 associated modulates hemostasis and complement activation
Acknowledgements
This work was supported by grant support from the National Institute of Allergy and Infectious Diseases National Institutes of Health (NIAID/NIH), USA grant (AI081093) to AM. AI was a visiting PhD student from Brazil supported by a scholarship from the Brazilian National Council for Scientific and Technological Development (CNPq).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alarcon-Chaidez F, Ryan R, Wikel S, Dardick K, Lawler C, Foppa IM, Tomas P, Cushman A, Hsieh A, Spielman A, Bouchard KR, Dias F, Aslanzadeh J, Krause PJ. Confirmation of tick bite by detection of antibody to Ixodes calreticulin salivary protein. Clin. Vaccine Immunol. 2006;13:1217–1222. doi: 10.1128/CVI.00201-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aljamali MN, Ramakrishnan VG, Weng H, Tucker JS, Sauer JR, Essenberg RC. Microarray analysis of gene expression changes in feeding female and male lone star ticks, Amblyomma americanum (L) Arch Insect Biochem Physiol. 2009;71:236–253. doi: 10.1002/arch.20318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anatriello E, Ribeiro JM, de Miranda-Santos IK, Brandão LG, Anderson JM, Valenzuela JG, Maruyama SR, Silva JS, Ferreira BR. An insight into the sialotranscriptome of the brown dog tick, Rhipicephalus sanguineus. BMC Genomics. 2010;11:450. doi: 10.1186/1471-2164-11-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson RB, Scrimgeour GJ, Reuben K. Responses of the ixodid tick, Amblyomma hebraeum (Acari: Ixodidae), to carbon dioxide. Exp. Appl. Acarol. 1998;22:667–681. [Google Scholar]
- Chalaire KC, Kim TK, Garcia-Rodriguez H, Mulenga A. Amblyomma americanum (L.) (Acari: Ixodidae) tick salivary gland serine protease inhibitor (serpin) 6 is secreted into tick saliva during tick feeding. J. Exp. Biol. 2011;214:665–6673. doi: 10.1242/jeb.052076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Chen B, Yao XQ, Gui BS, Ou Y, Ouyang JM. Anticoagulation of Diethyl Citrate and Its Comparison with Sodium Citrate in an Animal Model. Blood Purif. 2011;33:30–36. doi: 10.1159/000330891. [DOI] [PubMed] [Google Scholar]
- Chmelar J, Oliveira CJ, Rezacova P, Francischetti IM, Kovarova Z, Pejler G, Kopacek P, Ribeiro JM, Mares M, Kopecky J, Kotsyfakis M. A tick salivary protein targets cathepsin G and chymase and inhibits host inflammation and platelet aggregation. Blood. 2011;117:736–744. doi: 10.1182/blood-2010-06-293241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coe H, Michalak M. Calcium binding chaperones of the endoplasmic reticulum. Gen. Physiol. Biophys. 2009:F96–F103. [PubMed] [Google Scholar]
- Dai J, Narasimhan S, Zhang L, Liu L, Wang P, Fikrig E. Tick histamine release factor is critical for Ixodes scapularis engorgement and transmission of the lyme disease agent. PLoS Pathog. 2010;6:e1001205. doi: 10.1371/journal.ppat.1001205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daix V, Schroeder H, Praet N, Georgin JP, Chiappino I, Gillet L, de Fays K, Decrem Y, Leboulle G, Godfroid E, Bollen A, Pastoret PP, Gern L, Sharp PM, Vanderplasschen A. Ixodes ticks belonging to the Ixodes ricinus complex encode a family of anticomplement proteins. Insect Mol. Biol. 2007;16:155–166. doi: 10.1111/j.1365-2583.2006.00710.x. [DOI] [PubMed] [Google Scholar]
- Davey RB. Effect of age of Boophilus microplus larvae (Aacri: Ixodidae) on attachment to cattle. J. Med. Entomol. 1987;24:118–120. doi: 10.1093/jmedent/24.1.118. [DOI] [PubMed] [Google Scholar]
- Davie EW, Fujikawa K, Kisiel W. The coagulation cascade: initiation, maintenance, and regulation. Biochemistry. 1991;30:10363–10370. doi: 10.1021/bi00107a001. [DOI] [PubMed] [Google Scholar]
- Decrem Y, Rath G, Blasioli V, Cauchie P, Robert S, Beaufays J, Frère JM, Feron O, Dogné JM, Dessy C, Vanhamme L, Godfroid E. Ir-CPI, a coagulation contact phase inhibitor from the tick Ixodes ricinus, inhibits thrombus formation without impairing hemostasis. J. Exp. Med. 2009;206:2381–2395. doi: 10.1084/jem.20091007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebel GD, Kramer LD. Short report: duration of tick attachment required for transmission of powassan virus by deer ticks. Am. J. Trop. Med. Hyg. 2004;71:268–271. [PubMed] [Google Scholar]
- Ehebauer MT, Mans BJ, Gaspar AR, Neitz AW. Identification of extrinsic blood coagulation pathway inhibitors from the tick Ornithodoros savignyi (Acari: Argasidae) Exp. Parasitol. 2002;101:138–148. doi: 10.1016/s0014-4894(02)00102-9. [DOI] [PubMed] [Google Scholar]
- Ferre F, Clote P. Disulfide connectivity prediction using secondary structure information and diresidue frequencies. Bioinformatics. 2005a;21:2336–2346. doi: 10.1093/bioinformatics/bti328. [DOI] [PubMed] [Google Scholar]
- Ferre F, Clote P. DiANNA: a web server for disulfide connectivity prediction. Nucleic Acids Res. 2005b;33(Web Server issue):W230–232. doi: 10.1093/nar/gki412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferre F, Clote P. DiANNA 1.1: an extension of the DiANNA web server for ternary cysteine classification. Accepted for publication in Nucl. Acids Res. 2006;34(Web Server issue):W182–185. doi: 10.1093/nar/gkl189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira CA, Da Silva, Vaz I, da Silva SS, Haag KL, Valenzuela JG, Masuda A. Cloning and partial characterization of a Boophilus microplus (Acari: Ixodidae) calreticulin. Exp. Parasitol. 2002;101:25–34. doi: 10.1016/s0014-4894(02)00032-2. [DOI] [PubMed] [Google Scholar]
- Fragoso H, Rad PH, Ortiz M, Rodriguez M, Redondo M, Herrera L, de la Fuente J. Protection against Boophilus annulatus infestations in cattle vaccinated with the B. microplus Bm86-containing vaccine Gavac. Vaccine. 1998;16:990–1992. doi: 10.1016/s0264-410x(98)00116-9. [DOI] [PubMed] [Google Scholar]
- Francischetti IM, Anderson JM, Manoukis N, Pham VM, Ribeiro JM. An insight into the sialotranscriptome and proteome of the coarse bontlegged tick, Hyalomma marginatum rufipes. J Proteomics. 2011;74:2892–2908. doi: 10.1016/j.jprot.2011.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Luo J, Fan R, Fingerle V, Guan G, Liu Z, Li Y, Zhao H, Ma M, Liu J, Liu A, Ren Q, Dang Z, Sugimoto C, Yin H. Cloning and characterization of a cDNA clone encoding calreticulin from Haemaphysalis qinghaiensis (Acari: Ixodidae) Parasitol. Res. 2008;102:737–746. doi: 10.1007/s00436-007-0826-y. [DOI] [PubMed] [Google Scholar]
- Gao X, Shi L, Zhou Y, Cao J, Zhang H, Zhou J. Characterization of the anticoagulant protein Rhipilin-1 from the Rhipicephalus haemaphysaloides tick. J. Insect Physiol. 2011;57:339–343. doi: 10.1016/j.jinsphys.2010.12.001. [DOI] [PubMed] [Google Scholar]
- Gladney WJ, Drummond RO, Whetstone TM, Ernst SE. Effect of age on the attachment rate the parasitic stage of the Lone Star tick, Amblyomma americanum (Linnaeus) (Aacrina: Ixodidae), in the laboratory. J. Med. Ento. 1970;7:92–95. doi: 10.1093/jmedent/7.1.92. [DOI] [PubMed] [Google Scholar]
- Guerrero FD, Miller RJ, Rousseau ME, Sunkara S, Quackenbush J, Lee Y, Nene V. BmiGI: a database of cDNAs expressed in Boophilus microplus, the tropical/southern cattle tick. Insect Biochem Mol Biol. 2005;35:585–595. doi: 10.1016/j.ibmb.2005.01.020. [DOI] [PubMed] [Google Scholar]
- Hill CA, Gutierrez JA. Analysis of the expressed genome of the lone star tick, Amblyomma americanum (Acari: Ixodidae) using an expressed sequence tag approach. Microb Comp Genomics. 2000;5:89–101. doi: 10.1089/10906590050179774. [DOI] [PubMed] [Google Scholar]
- Ibrahim MA, Ghazy AH, Maharem TM, Khalil MI. Factor Xa (FXa) inhibitor from the nymphs of the camel tick Hyalomma dromedarii. Comp. Biochem. Physiol. B. 2001;130:501–512. doi: 10.1016/s1096-4959(01)00459-6. [DOI] [PubMed] [Google Scholar]
- Kaewhom P, Stich RW, Needham GR, Jittapalapong S. Molecular analysis of calreticulin expressed in salivary glands of Rhipicephalus (Boophilus) microplus indigenous to Thailand. Ann. N.Y. Acad .Sci. 2008;1149:53–57. doi: 10.1196/annals.1428.033. [DOI] [PubMed] [Google Scholar]
- Karim S, Singh P, Ribeiro JM. A deep insight into the sialotranscriptome of the gulf coast tick, Amblyomma maculatum. PLoS One. 2011;6(12) doi: 10.1371/journal.pone.0028525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katavolos P, Armstrong PM, Dawson JE, Telford SR., III Duration of tick attachment required for transmission of granulocytic ehrlichiosis. J. Infect. Dis. 1998;177:1422–1425. doi: 10.1086/517829. [DOI] [PubMed] [Google Scholar]
- Kluszynski BA, Kim C, Faulk WP. Zinc as a cofactor for heparin neutralization by histidine-rich glycoprotein. J. Biol. Chem. 1997;272:13541–1357. doi: 10.1074/jbc.272.21.13541. [DOI] [PubMed] [Google Scholar]
- Konnai S, Yamada S, Imamura S, Simuunza M, Chembens M, Chota A, Nambota A, Ohashi K, Onuma M. Attachment duration required for Rhipicephalus appendiculatus to transmit Theileria parva to the host. Vector Borne Zoonotic Dis. 2007;7:241–248. doi: 10.1089/vbz.2006.0616. [DOI] [PubMed] [Google Scholar]
- Lai R, Takeuchi H, Jonczy J, Rees HH, Turner PC. A thrombin inhibitor from the ixodid tick, Amblyomma hebraeum. Gene. 2004;342:243–249. doi: 10.1016/j.gene.2004.07.012. [DOI] [PubMed] [Google Scholar]
- Lees K, Woods DJ, Bowman AS. Transcriptome analysis of the synganglion from the brown dog tick, Rhipicephalus sanguineus. Insect Mol Biol. 2010;19:273–282. doi: 10.1111/j.1365-2583.2009.00968.x. [DOI] [PubMed] [Google Scholar]
- Lefkowitz JB. Coagulation Pathway and Physiology. JB Lippincott Co; Philadelphia, PA: 2006. [Google Scholar]
- Levi M, Schultz M, van der Poll T. Disseminated intravascular coagulation in infectious disease. Semin. Thromb. Hemost. 2010;36:367–377. doi: 10.1055/s-0030-1254046. [DOI] [PubMed] [Google Scholar]
- Lew-Tabor AE, Moolhuijzen PM, Vance ME, Kurscheid S, Valle MR, Jarrett S, Minchin CM, Jackson LA, Jonsson NN, Bellgard MI, Guerrero FD. Suppressive subtractive hybridization analysis of Rhipicephalus (Boophilus) microplus larval and adult transcript expression during attachment and feeding. Vet Parasitol. 2010;167:304–320. doi: 10.1016/j.vetpar.2009.09.033. [DOI] [PubMed] [Google Scholar]
- Liao M, Zhou J, Gong H, Boldbaatar D, Shirafuji R, Battur B, Nishikawa Y, Fujisaki K. Hemalin, a thrombin inhibitor isolated from a midgut cDNA library from the hard tick Haemaphysalis longicornis. J. Insect Physiol. 2009;55:164–173. doi: 10.1016/j.jinsphys.2008.11.004. [DOI] [PubMed] [Google Scholar]
- Mans BJ, Andersen JF, Schwan TG, Ribeiro JM. Characterization of anti-hemostatic factors in the argasid, Argas monolakensis: implications for the evolution of blood-feeding in the soft tick family. Insect. Biochem. Mol. Biol. 2008;38:22–41. doi: 10.1016/j.ibmb.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins I, Kepp O, Galluzzi L, Senovilla L, Schlemmer F, Adjemian S, Menger L, Michaud M, Zitvogel L, Kroemer G. Surface-exposed calreticulin in the interaction between dying cells and phagocytes. Ann. N.Y. Acad. Sci. 2010;1209:77–82. doi: 10.1111/j.1749-6632.2010.05740.x. [DOI] [PubMed] [Google Scholar]
- Morgenstern D, Rohde BH, King GF, Tal T, Sher D, Zlotkin E. The tale of a resting gland: transcriptome of a replete venom gland from the scorpion Hottentotta judaicus. Toxicon. 2011;57:695–703. doi: 10.1016/j.toxicon.2011.02.001. [DOI] [PubMed] [Google Scholar]
- Mulenga A, Sugimoto C, Sako Y, Ohashi K, Musoke A, Shubash M, Onuma M. Molecular characterization of a Haemaphysalis longicornis tick salivary gland-associated 29-kilodalton protein and its effect as a vaccine against tick infestation in rabbits. Infect. Immun. 1999;67:1652–1658. doi: 10.1128/iai.67.4.1652-1658.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulenga A, Sugimoto C, Ingram G, Ohashi K, Misao O. Characterization of two cDNAs encoding serine proteinases from the hard tick Haemaphysalis longicornis. Insect Biochem. Mol. Biol. 2001;31:817–825. doi: 10.1016/s0965-1748(00)00187-9. [DOI] [PubMed] [Google Scholar]
- Mulenga A, Macaluso KR, Simser JA, Azad AF. The American dog tick, Dermacentor variabilis, encodes a functional histamine release factor homolog. Insect Biochem. Mol. Biol. 2003a;33:911–919. doi: 10.1016/s0965-1748(03)00097-3. [DOI] [PubMed] [Google Scholar]
- Mulenga A, Misao O, Sugimoto C. Three serine proteinases from midguts of the hard tick Rhipicephalus appendiculatus; cDNA cloning and preliminary characterization. Exp. Appl. Acarol. 2003b;29:151–164. doi: 10.1023/a:1024278402288. [DOI] [PubMed] [Google Scholar]
- Mulenga A, Azad AF. The molecular and biological analysis of ixodid ticks histamine release factors. Exp. Appl. Acarol. 2005;37:215–229. doi: 10.1007/s10493-005-3261-8. [DOI] [PubMed] [Google Scholar]
- Mulenga A, Blandon M, Khumthong R. The molecular basis of the Amblyomma americanum tick attachment phase. Exp. Appl. Acarol. 2007;41:267–287. doi: 10.1007/s10493-007-9064-3. [DOI] [PubMed] [Google Scholar]
- Mulenga A, Khumthong R, Chalaire KC, Strey O, Teel P. Molecular and biological characterization of the Amblyomma americanum organic anion transporter polypeptide. J. Exp. Biol. 2008;211:3401–3408. doi: 10.1242/jeb.022376. [DOI] [PubMed] [Google Scholar]
- Mulenga A, Erikson K. A snapshot of the Ixodes scapularis degradome. Gene. 2009;482:78–93. doi: 10.1016/j.gene.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulenga A, Khumthong R. Disrupting the Amblyomma americanum (L.) CD147 receptor homolog prevents ticks from feeding to repletion and blocks spontaneous detachment of ticks from their host. Insect. Biochem. Mol. Biol. 2010a;40:524–532. doi: 10.1016/j.ibmb.2010.04.012. [DOI] [PubMed] [Google Scholar]
- Mulenga A, Khumthong R. Silencing of three Amblyomma americanum (L.) insulin-like growth factor binding protein-related proteins prevents ticks from feeding to repletion. J. Exp. Biol. 2010b;213:1153–1161. doi: 10.1242/jeb.035204. [DOI] [PubMed] [Google Scholar]
- Narasimhan S, Koski RA, Beaulieu B, Anderson JF, Ramamoorthi N, Kantor F, Cappello M, Fikrig E. A novel family of anticoagulants from the saliva of Ixodes scapularis. Insect Mol. Biol. 2002;11:641–650. doi: 10.1046/j.1365-2583.2002.00375.x. [DOI] [PubMed] [Google Scholar]
- Nazareth RA, Tomaz LS, Ortiz-Costa S, Atella GC, Ribeiro JM, Francischetti IM, Monteiro RQ. Antithrombotic properties of Ixolaris, a potent inhibitor of the extrinsic pathway of the coagulation cascade. Thromb. Haemost. 2006;96:7–13. doi: 10.1160/TH06-02-0105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nene V, Lee D, Quackenbush J, Skilton R, Mwaura S, Gardner MJ, Bishop R. AvGI, an index of genes transcribed in the salivary glands of the ixodid tick Amblyomma variegatum. Int J Parasitol. 2002;32:1447–1456. doi: 10.1016/s0020-7519(02)00159-5. [DOI] [PubMed] [Google Scholar]
- Nene V, Lee D, Kang’a S, Skilton R, Shah T, de Villiers E, Mwaura S, Taylor D, Quackenbush J, Bishop R. Genes transcribed in the salivary glands of female Rhipicephalus appendiculatus ticks infected with Theileria parva. Insect Biochem Mol Biol. 2004;34:1117–1128. doi: 10.1016/j.ibmb.2004.07.002. [DOI] [PubMed] [Google Scholar]
- Pagel VZJ, Geraci NS, Guerrero FD, Wikel SK, Stuart JJ, Nene VM, Hill CA. Tick genomics: the Ixodes genome project and beyond. Int J Parasitol. 2007;37:1297–305. doi: 10.1016/j.ijpara.2007.05.011. [DOI] [PubMed] [Google Scholar]
- Parizi LF, Rech H, Ferreira CA, Imamura S, Ohashi K, Onuma M, Masuda A, Vaz Ida S., Jr. Comparative immunogenicity of Haemaphysalis longicornis and Rhipicephalus (Boophilus) microplus calreticulins. Vet. Parasitol. 2009;164:282–290. doi: 10.1016/j.vetpar.2009.05.023. [DOI] [PubMed] [Google Scholar]
- Parizi LF, Githaka NW, Logullo C, Konnai S, Masuda A, Ohashi K, da Silva Vaz I., Jr. The quest for a universal vaccine against ticks: Cross-immunity insights. Vet J. 2012;194:158–165. doi: 10.1016/j.tvjl.2012.05.023. [DOI] [PubMed] [Google Scholar]
- Piesman J, Spielman A. Human babesiosis on Nantucket Island: prevalence of Babesia microti in ticks. Am J Trop Med Hyg. 1980;29:742–746. doi: 10.4269/ajtmh.1980.29.742. [DOI] [PubMed] [Google Scholar]
- Piesman J, Mather TN, Sinsky RJ, Spielman A. Duration of tick attachment and Borrelia burgdorferi transmission. J. Clin. Microbiol. 1987;25:557–558. doi: 10.1128/jcm.25.3.557-558.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro JM, Alarcon-Chaidez F, Mans BJ, Mather TN, Wikel SK. An annotated catalog of salivary gland transcripts from Ixodes scapularis ticks. Insect Biochem. Mol. Biol. 2006;36:111–129. doi: 10.1016/j.ibmb.2005.11.005. [DOI] [PubMed] [Google Scholar]
- Ribeiro JM, Zeidner NS, Ledin K, Dolan MC, Mather TN. How much pilocarpine contaminates pilocarpine-induced tick saliva? Med Vet Entomol. 2004;18:20–24. doi: 10.1111/j.0269-283x.2003.0469.x. [DOI] [PubMed] [Google Scholar]
- Ricci CG, Pinto AF, Berger M, Termignoni C. A thrombin inhibitor from the gut of Boophilus microplus ticks. Exp. Appl. Acarol. 2007;42:291–300. doi: 10.1007/s10493-007-9097-7. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Valle M, Lew-Tabor A, Gondro C, Moolhuijzen P, Vance M, Guerrero FD, Bellgard M, Jorgensen W. Comparative microarray analysis of Rhipicephalus (Boophilus) microplus expression profiles of larvae pre-attachment and feeding adult female stages on Bos indicus and Bos taurus cattle. BMC Genomics. 2010;11:437. doi: 10.1186/1471-2164-11-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders ML, Jaworski DC, Sanchez JL, DeFraites RF, Glass GE, Scott AL, Raha S, Ritchie BC, Needham GR, Schwartz BS. Antibody to a cDNA-derived calreticulin protein from Amblyomma americanum as a biomarker of tick exposure in humans. Am. J. Trop. Med. Hyg. 1998;59:279–285. doi: 10.4269/ajtmh.1998.59.279. [DOI] [PubMed] [Google Scholar]
- Sanders ML, Glass GE, Nadelman RB, Wormser GP, Scott AL, Raha S, Ritchie BC, Jaworski DC, Schwartz BS. Antibody levels to recombinant tick calreticulin increase in humans after exposure to Ixodes scapularis (Say) and are correlated with tick engorgement indices. Am. J. Epidemiol. 1999;149:777–784. doi: 10.1093/oxfordjournals.aje.a009887. [DOI] [PubMed] [Google Scholar]
- Santos IK, Valenzuela JG, Ribeiro JM, de Castro M, Costa JN, Costa AM, da Silva ER, Neto OB, Rocha C, Daffre S, Ferreira BR, da Silva JS, Szabó MP, Bechara GH. Gene discovery in Boophilus microplus, the cattle tick: the transcriptomes of ovaries, salivary glands, and hemocytes. Ann N Y Acad Sci. 2004;1026:242–246. doi: 10.1196/annals.1307.037. [DOI] [PubMed] [Google Scholar]
- Sonenshine DE. Biology of ticks. Oxford University Press; Oxford: 1993. [Google Scholar]
- Spillert CR, Lazaro EJ. Modified recalcification time (MRT): a sensitive cancer test? Review of the evidence. J. Natl. Med. Assoc. 1995;87:687–692. [PMC free article] [PubMed] [Google Scholar]
- Tubek S, Grzanka P, Tubek I. Role of zinc in hemostasis: a review. Biol. Trace Elem. Res. 2008a;121:1–8. doi: 10.1007/s12011-007-8038-y. [DOI] [PubMed] [Google Scholar]
- Tubek S, Grzanka P, Tubek I. The role of zinc in thrombosis and pulmonary embolism in the course of antiphospholipid syndrome (APS)--short review. Biol. Trace Elem. Res. 2008b;122:193–196. doi: 10.1007/s12011-007-8077-4. [DOI] [PubMed] [Google Scholar]
- Tukahirwa EM. The feeding behaviour of larvae, nymphs and adults of Rhipicephalus appendiculatus. Parasitology. 1976;72:65–74. doi: 10.1017/s0031182000043195. [DOI] [PubMed] [Google Scholar]
- Valenzuela JG, Charlab R, Mather TN, Ribeiro JM. Purification, cloning, and expression of a novel salivary anticomplement protein from the tick, Ixodes scapularis. J. Biol. Chem. 2000;275:18717–18723. doi: 10.1074/jbc.M001486200. [DOI] [PubMed] [Google Scholar]
- Valenzuela JG, Francischetti IM, Pham VM, Garfield MK, Mather TN, Ribeiro JM. Exploring the sialome of the tick Ixodes scapularis. J Exp Biol. 2002;205:2843–2864. doi: 10.1242/jeb.205.18.2843. [DOI] [PubMed] [Google Scholar]
- Wang H, Henbest PJ, Nuttall PA. Successful interrupted feeding of adult Rhipicephalus appendiculatus (Ixodidae) is accompanied by reprogramming of salivary gland protein expression. Parasitology. 1999;119:143–149. doi: 10.1017/s0031182099004540. [DOI] [PubMed] [Google Scholar]
- Wang M, Guerrero FD, Pertea G, Nene VM. Global comparative analysis of ESTs from the southern cattle tick, Rhipicephalus (Boophilus) microplus. BMC Genomics. 2007;12:368. doi: 10.1186/1471-2164-8-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G, Fang QQ, Keirans JE, Durden LA. Cloning and sequencing of putative calreticulin complementary DNAs from four hard tick species. J. Parasitol. 2004;90:73–78. doi: 10.1645/GE-157R. [DOI] [PubMed] [Google Scholar]
- Xu G, Fang QQ, Sun Y, Keirans JE, Durden LA. Hard tick calreticulin (CRT) gene coding regions have only one intron with conserved positions and variable sizes. J Parasitol. 2005;1:1326–1331. doi: 10.1645/GE-344R1.1. [DOI] [PubMed] [Google Scholar]