Abstract
Despite increased implementation of screening colonoscopy, interval cancers in the proximal colon remain a major public health concern. This fact underscores the limitations of current screening paradigms and the need for developing advanced endoscopic techniques. The density of aberrant crypt foci (ACF), the earliest identifiable mucosal abnormality, may serve as a surrogate marker for colon cancer risk, but has rarely been studied in the proximal colon. To this end, high-definition (HD) chromoendoscopy was conducted to define the relevance of ACF in the proximal colon. In addition, due to limited ACF size, the development of a combinatorial approach was required to maximize data acquisition obtained from individual biopsy samples. Proximal and distal ACF samples were characterized for a total of 105 mutations across 22 known tumor suppressor and proto-oncogenes using high-throughput Sequenom MASSarray analysis. From this profiling, a discrete number of somatic mutations were identified, including APCR876* and FLT3I836M, as well as a deletion within the EGFR gene. Combined, these data highlight the significance of ACF within the context of colon cancer pathogenesis, particularly in the proximal colon.
Implications
The identification of cancer-related mutations in commonly overlooked mucosal lesions underscores the preventive benefit of implementing advanced endoscopic screening to larger patient populations, particularly in the proximal colon.
Keywords: Premalignant lesions, gastrointestinal cancers: colorectal, aberrant crypt foci, laser capture microdissection, DNA mass spectrometry
Introduction
Screening colonoscopy has been established as an effective strategy to reduce the risk of colorectal cancer (1). However, recent evidence suggests that protection may be most successful in the distal colorectum since the incidence of proximal colon cancer has not been significantly reduced by the implementation of widespread screening colonoscopy (2). The frequency of interval colon cancers, or those cancers developing between screening procedures, raises the possibility that proximal tumors in particular may have accelerated growth characteristics that limits the effectiveness of colonoscopy(3). This lack of protection afforded by colonoscopy, in particular within the proximal colon, underscores the need for advanced endoscopic techniques that may be used to identify small precursor lesions that would otherwise be missed by traditional screening approaches.
The multi-stage colon cancer model provides a paradigm for understanding the role of early mutations in cancer progression through defined histological stages (4). Aberrant crypt foci (ACF) are morphologically identifiable mucosal abnormalities, a subset of which may be precancerous and contribute to a 'field defect' within the mucosa(5). With the use of dye-spray (e.g. indigo carmine or methylene blue) and high-definition magnifying colonoscopy, the in situ identification of ACF has become relatively straightforward(6, 7). Although their utility as a surrogate marker of colon cancer has been challenged(8, 9), we recently reported that elevated numbers of distal ACF observed during index colonoscopy predicts the development of advanced neoplasia within a five-year screening interval(10).
The following study was undertaken to more accurately define the molecular alterations that are present within colonic ACF. Using DNA mass spectrometry (DNA-MS) combined with laser capture microdissection (LCM), we report a highly sensitive method to interrogate the mutational spectrum of microscopic biopsies following their removal from the human colon. A limited number of somatic mutations have been identified, including mutations to APCR876* and FLT3I836M, as well as an insertion/deletion within the EGFR gene, underscoring the biological significance of ACF within the context of CRC and in particular within the proximal colon.
Materials and Methods
Subject Selection
All subjects underwent a total screening colonoscopy at the University of Connecticut Health Center (UCHC) in accordance with Institutional policies. Patients who met the Amsterdam criteria for familial adenomatous polyposis (FAP) or hereditary non-polyposis CRC (HNPCC) were excluded from the study. This study was performed only following Institutional Review Board approval and receipt of written informed consent from the subjects.
ACF Collection and Characterization
ACF were identified in situ and biopsied from grossly normal-appearing colonic mucosa during high-definition, close-focus magnifying chromoendsocopy. The proximal colon from the cecum to the right hepatic flexure, in addition to the distal 20-cm of the colorectum, were sprayed with a freshly prepared solution of 1% indigo carmine. ACF were visualized and photographed using a high-definition colonoscope (Olympus, PCF-190; Olympus Corp., Center Valley, PA) with visualization from 2–100 mm at 60× magnification. A finding was accepted as an ACF if 5 or more crypts have an increased lumen diameter (1.5–2×), thick crypt walls, or abnormally shaped lumens relative to the surrounding mucosa. In addition, the lesion must be less than 5 mm in diameter to be considered an ACF. Biopsies were immediately embedded in freezing medium (OCT), flash-frozen, and stored at -80°C (11).
Frozen sections were stained with hematoxylin and eosin (H&E) and routine histological analyses were performed by two independent,, board-certified human gastrointestinal pathologists blinded to clinical findings according to our previously established criteria (11). Dysplastic ACF are characterized histologically by enlarged upper cryptal regions of irregular shape with stratified, elongated nuclei and a general dysplastic appearance. Hyperplastic ACF are characterized according to the same criteria applied to hyperplastic polyps(12). These hyperplastic ACF are subclassified into serrated and distended (non-serrated) pathologies as previously described (11). Briefly, serrated ACF are defined as ACF that show stellate luminal shape upon cross-section with a prominent component of columnar crypts with microvessicular cytoplasm. Distended ACF lack serration, prominently feature goblet cells, and frequently exhibit tufting of the surface epithelium.
Laser capture microdissection and DNA purification
A Veritas microdissector was used to capture ~5,000 cells (or the equivalent of approximately one mm2 of collected tissue area) from 12-µm thick frozen serial sections of ACF prepared on PEN membrane glass slides (Applied Biosystems, Foster City, CA). Genomic DNA was extracted from aberrant crypts (isolated from surrounding stroma and normal mucosa by LCM) using the PicoPure DNA extraction kit (Applied Biosystems). Samples were then cleaned using the DNA Wizard protocol (Promega, Madison, WI)
Somatic mutation screening using DNA mass-spectrometry
Mutation screening was performed at the Yale Center for Genome Analysis using the Sequenom MASSArray DNA-MS approach. This method uses a matrix assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) platform to detect single base mutations with increased sensitivity(13). Extracted DNA was amplified using multiplexed OncoCarta PCR primers targeting 105 mutations across 22 known tumor suppressor and proto-oncogenes (v.3.0, Sequenom Inc., San Diego, CA; Supplemental Table 1). The DNA concentration per reaction approached 1 ng of gDNA, a concentration that is within the limits of detection for the Sequenom platform(13). DNA mass-spectrometry was carried out as previously described (13). Briefly, target regions are amplified through multiplex PCR reactions. Shrimp alkaline phosphatase is added to the reaction to dephosphorylate unincorporated dNTPs and an extension reaction mix is added. A post-PCR primer extension reaction creates a single nucleotide extension using mass-modified terminators and the reactions are desalted using 6 mg of CLEAN Resin. Reaction products are then printed onto a SpectroCHIP to be read by MALDI-TOF MS. Mass spectrometry data is then analyzed using Typer4.0 software to identify sample genotype with respect to the assayed mutation.
Somatic mutations were confirmed by extracting DNA from whole blood using the DNAeasy Blood and Tissue Kit standard protocol (Qiagen; Venlo, Netherlands) and sequenced by GeneWiz, Inc. (South Plainfield, NJ). To determine mutation status in adenomas, DNA was extracted from two 10-micron FFPE curls using the QIAamp DNA FFPE Tissue Kit standard protocol and sequenced (GeneWiz, Inc.) Primer sequences for PCR amplification and Sanger sequencing were: EGFR F: 5’-CCCCAGCAATATCAGCCTTA-3’ R: 5’-ATGAGGTACTCGTCGGCATC-3’ ; APC F: 5’-GCATGCTCAAGAACCTCATTC-3’ R: 5’-TAGGTCGGCTGGGTATTGAC-3’ ; FLT3 F: 5’-AGAACTGCAGCCACCATAGC-3’ R: 5’-ACCTCATGATCTGCCCTCCT-3’ (Integrated DNA Technologies, Coralville, IA). Sanger sequencing for BRAF and KRAS mutations was performed as previously described (14).
Immunohistochemistry
Immunohistochemistry (IHC) was performed on frozen ACF prepared at 7-µm thickness. Sections were treated with 3% hydrogen peroxide, blocked and incubated in anti-β-catenin primary antibody (1:2,000; Sigma-Aldrich Co, St. Louis, MO). Sections were incubated in ImmPRESS anti-mouse Ig (Vector Laboratories, Burlingame, CA). Signal detection was achieved using a 3,3’-diaminobenzidine solution (Vector Laboratories) and tissues were counterstained with methyl green nuclear stain.
Results and Discussion
High-definition chromoendoscopy positively identifies ACF in the proximal colon
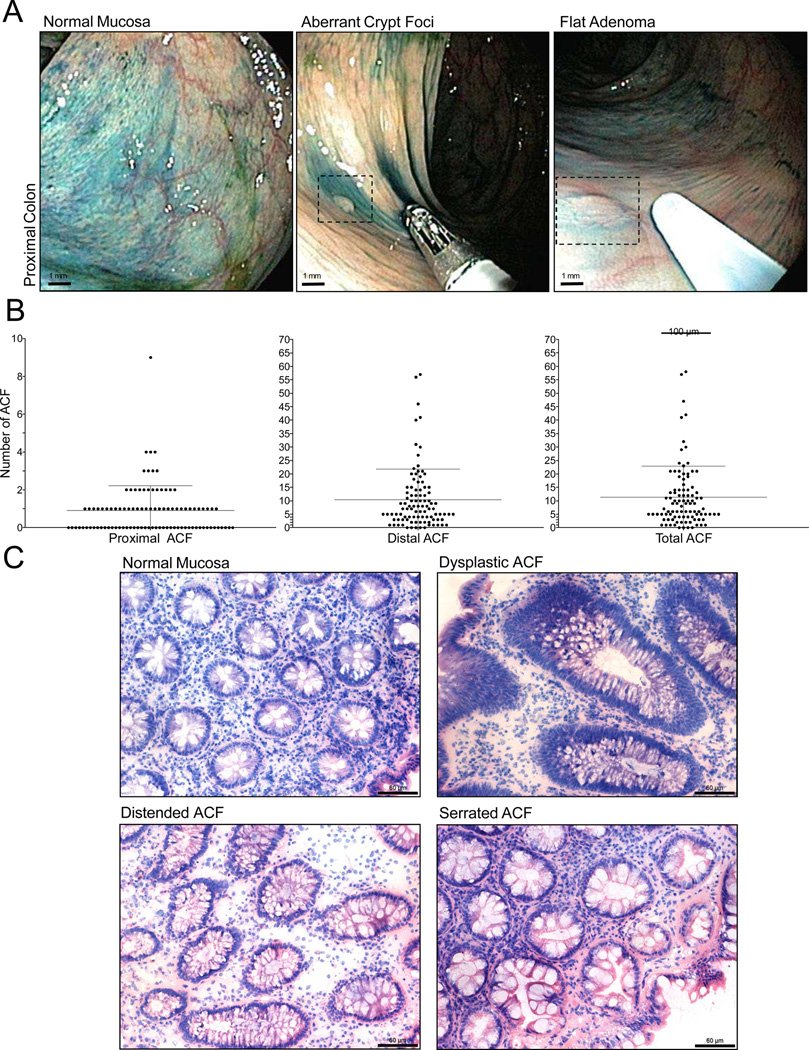
Using high-definition chromoendoscopy, we and others have previously demonstrated the ability to identify ACF within the distal colon(6, 7). We have recently expanded our analysis to include the identification of ACF within the proximal colon, a region of the colon that includes the cecum to the splenic flexure (Fig. 1A). A total of 96 patients have been screened and 88 proximal ACF and 1,010 distal ACF have been identified (Fig. 1B). The majority of the subjects were undergoing a screening colonoscopy (n=70/96; 73%) and approximately half of the subjects (n = 47/96; 49%) had at least one polyp identified and removed during the procedure. Histologic evaluation reveals that proximal ACF harbor the full spectrum of established histologic abnormalities present within distal ACF, including hyperplasia (serrated or distended) and dysplasia (Fig. 1C).
Figure 1. Endoscopic detection of proximal ACF.
A, High-definition magnifying colonoscopy combined with dye-spray reveals diminutive flat lesions of the proximal colorectum, including ACF and flat adenomas. B, Frequency of ACF according to colonic location identified during screening chromoendoscopy. C, Histologic appearance of normal colonic mucosa and ACF procured from the proximal colon. Proximal lesions display the same type of morphologies commonly associated with distal colon ACF, including dysplasia and hyperplasia (distended and serrated) (200× magnification; bar = 60 µm).
ACF within the distal colon share many genetic and histologic abnormalities associated with more advanced colonic neoplasia. For example, hyperplastic ACF commonly harbor BRAF or KRAS mutations(14), resulting in increased ERK activation(11). The presence of ACF with dysplastic histological features, however, is uncommon within the distal colon, although in one case a dysplastic ACF was found to carry a novel somatic mutation to the APC gene(14). A high frequency of APC mutations has been reported previously in dysplastic ACF(15), but the inclusion of patients with FAP disease in this study may have exaggerated the prevalence of APC mutations. In an earlier study, we found that, a significant percentage of ACF (~ 40%) do not harbor mutations to BRAF or KRAS(11). Thus in the present study, a broader genetic screen was undertaken to identify additional mutations that may contribute to early neoplastic changes in the colon.
Application of DNA-MS for high-throughput genotyping of ACF
Comprehensive mutation profiling of early neoplasia may provide insight into future colon cancer risk(16). However, there are a number of technical challenges associated with the application of high-throughput screening strategies to the analysis of small mucosal biopsy specimens. In addition, the number of morphologically abnormal colonic crypts within an ACF biopsy is limited, necessitating the use of LCM to enrich for subpopulations of aberrant and normal-appearing colonocytes. Recent advances in DNA-MS technology using MALDI-TOF have provided a high-throughput, multiplexed approach to allow somatic mutational profiling with as little as 1 ng of input genomic DNA(13). To examine the feasibility of performing DNA-MS analyses on micro-dissected ACF samples, DNA was extracted from approximately 5,000 colonocytes and subjected to the MASSArray PCR standard protocol. Amplification of genomic loci was achieved with less than 1 ng of input genomic DNA (Supplemental Fig. 1). A total of twenty-six micro-dissected ACF were then subjected to DNA-MS analyses. This group included seven proximal ACF, nineteen distal ACF, and four normal colon biopsies.
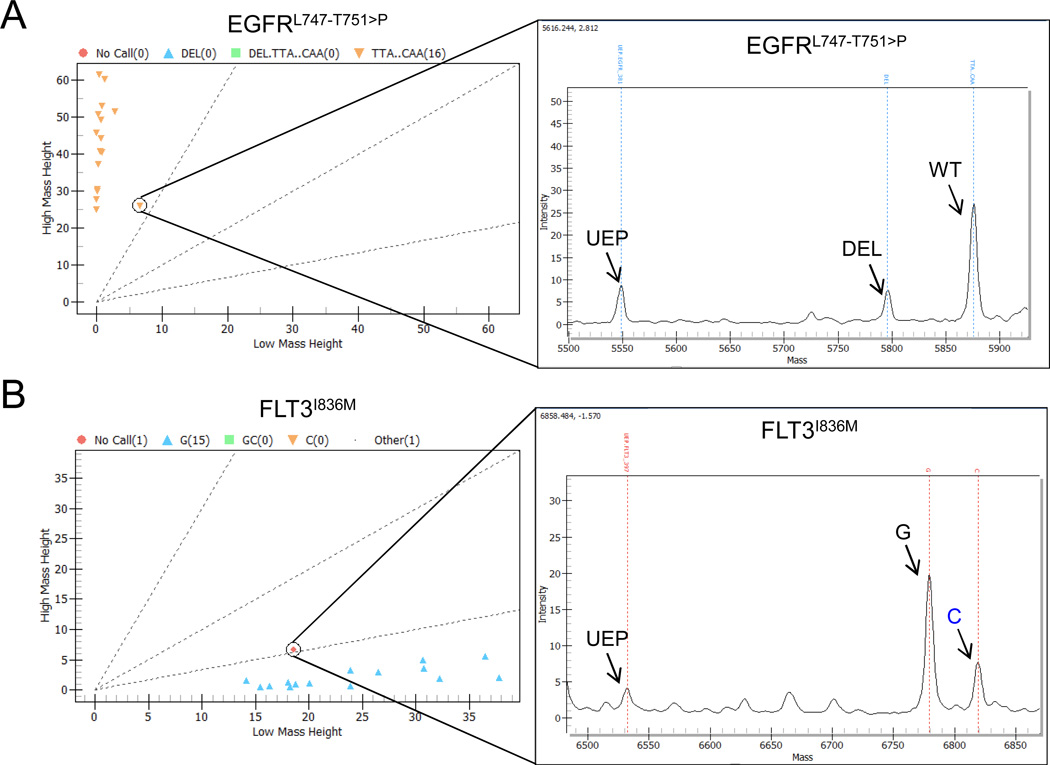
As shown in Fig. 2A, a proximal, serrated hyperplastic ACF was positive for the in-frame insertion/deletion, c.2239_2251delTTAAGAGAAGCAAinsC (AA:L747_T751>P) in the EGFR gene (19.6% deletion: 80.4% wild-type) as detected by MASSArray This mutation was not present in the subject's blood, confirming that it was somatically acquired While EGFR mutations occur in approximately 6% of colorectal cancers (COSMIC, n = 3,102), they are much more frequent in lung adenocarcinomas (25.2%, COSMIC, n = 26,293). This specific mutation has not been previously detected in colorectal cancer and accounts for <1% of lung EGFR mutations, but is associated with a gain of EGFR function (17).
Figure 2. DNA-MS identifies FLT3I836M and a deletion to EGFR gene in hyperplastic ACF.
A, DNA-MS identification of an insertion/deletion to the EGFR gene in a serrated hyperplastic proximal colon ACF. B, DNA-MS detection of a G>C missense mutation resulting in FLT3I836M somatic mutation in a distended hyperplastic distal colon ACF. UEP=unextended primer
Additionally, a distal, distended hyperplastic ACF was positive for a FLT3 c.2508C>G (I836M) mutation (24.0% mutant: 76.0% wild-type) and confirmed by Sanger-sequencing to be somatic (Fig. 2B). FLT3 mutations have widely been implicated in cases of acute myeloid leukemia (23.5%, COSMIC, n = 63,213) and mutations to FLT3 occur in 4.8% of colorectal cancers (COSMIC, n = 1,003). To our knowledge, the c.2508C>G mutation found in this study has never before been reported in colorectal cancer. The I836M missense mutation has been shown previously to cause constitutive activation of the receptor tyrosine kinase and subsequent downstream signaling(18). Constitutive activation of these two receptor tyrosine kinases, not previously implicated in ACF formation, may contribute to the aberrant activation of MAPK signaling reported earlier in hyperplastic lesions(11).
While the commercially available panel used in this study does not include KRAS or BRAF, we have previously reported a high frequency of mutations to these proto-oncogenes in distal hyperplastic ACF using conventional PCR-based sequencing (11, 14). Thus to establish the presence of BRAFV600E and KRAS (codons 12 and 13) mutations, Sanger-sequencing was performed on seven randomly selected ACF that had no other somatic mutations detected by the OncoCarta panel. Four of the seven ACF were positive for KRAS mutations and none of the ACF had BRAFV600E mutations.
DNA mass spectrometry identifies a rare somatic APC mutation in a proximal colon ACF
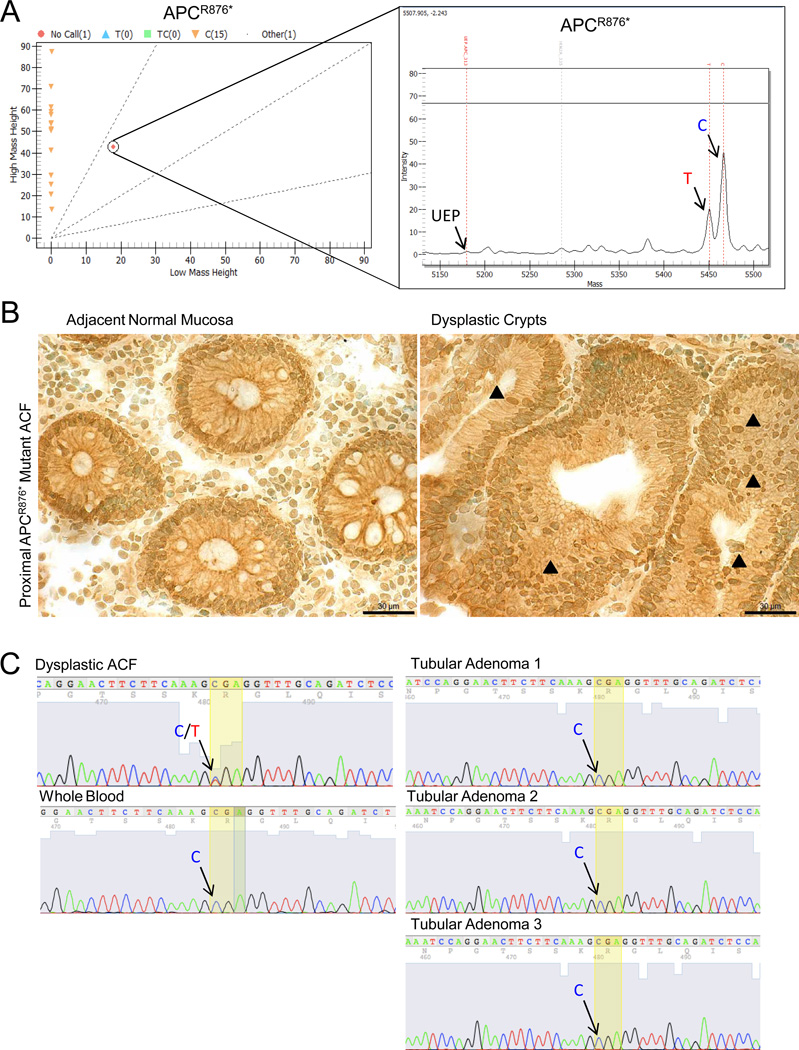
An APCR876* mutation, a c.2626C>T (30.4% mutant T: 69.6% wild-type C), was identified in a proximal dysplastic ACF (Fig. 3A) In order to determine the function of this rare mutation, immunolocalization of the Wnt target protein, β-catenin, was performed on the dysplastic ACF. As shown in Figure 3B, there was an increase in nuclear accumulation of β-catenin in the dysplastic colonocytes compared to adjacent normal crypts. While mutations to APC are frequently detected in colorectal cancers (42%; COSMIC, n=3,982), the APCR876* mutation is relatively rare, accounting for approximately 2.5% of all APC mutations. The c.2626C>T non-sense mutation is most commonly somatically acquired(19), although it has occasionally been detected in FAP pedigrees (19–21). In the present study, however, this mutation was confirmed to be somatic, as it was not detected in a matched blood sample (Fig. 3C), thereby ruling out the possibility of underlying FAP disease. Interestingly, this mutation was not detected in any of the three synchronous tubular adenomas removed during the chromoendoscopy procedure (Fig. 3C).
Figure 3. DNA-MS reveals APCR876* somatic mutation in a proximal dysplastic ACF.
A, DNA-MS reveals a c.2626C>T substitution resulting in an APCR876* mutation in a proximal dysplastic ACF. UEP = unextended primer. B, IHC demonstrates an accumulation of nuclear β-catenin (black arrowheads) in dysplastic crypts positive for the APCR876* mutation compared to adjacent normal crypts. (400× magnification; bar = 30 µm) C, Sanger sequencing confirms that the APCR876* mutation is somatically acquired. Blood and synchronous tubular adenomas are wild-type at codon 876 of APC.
Sporadic forms of CRC are most likely the result of early somatic mutations resulting in the clonal expansion of initiated progenitor cells. ACF have been shown to be monoclonal and as such are likely the direct consequence of this initial clonal expansion (22). Polyps and adenomas are presumed to arise from these lesions following the accumulation of multiple genetic alterations within this initiated cell population. Clearly, additional somatic mutations must occur to drive neoplastic progression (4). Additional changes have been identified in early stages of CRC that contribute to carcinogenesis, including the acquisition of aberrant promoter methylation (7) and microsatellite instability (23). Therefore, it is reasonable to anticipate that only a small percentage of ACF may sustain multiple oncogenic mutations at this early precancerous stage. In fact, in the present study there was no evidence for the presence of multiple somatic mutations, at least within the cancer panel that was tested.
The in situ identification of colonic ACF, and in particular those from the proximal colon, is dependent upon the use of dye-spray and high-definition magnifying endoscopy and we believe that in some cases this additional clinical effort is warranted. For example, the identification of the APC mutation, APCR876*, associated with the formation of aggressive, invasive carcinomas(24), in a commonly missed mucosal lesion underscores the potential preventive benefit of implementing this clinical approach to larger patient populations. It is possible that ACF that harbor this and other significant somatic mutations have the capacity to rapidly progress to a more advanced neoplasia, perhaps representing a subset of “missed lesions” that may be responsible for interval colon cancers. Expanded molecular classification of proximal ACF with a DNA-MS panel designed specifically for CRC, as well as establishing detailed associations of proximal ACF with known CRC risk factors, will be necessary to firmly establish their pre-malignant potential and utility as a surrogate marker for future cancer risk.
Supplementary Material
Acknowledgements
We thank Dr. T.V. Rajan for providing a detailed pathologic analysis of ACF. We also would like to thank Dr. Shi Yang at Boston Medical Center for performing KRAS and BRAF sequencing services.
Grant Support: This work was supported by the State of Connecticut Department of Public Health, Biomedical Research Application #2012-0913 (D.W.R.) and the National Institutes of Health 1RO1CA159976 (D.W.R.)
Footnotes
Disclosure of Potential Conflicts of Interest: We have no potential conflicts of interest to disclose.
Authors’ Contributions:
Conception and design: D.A. Drew, D.W. Rosenberg
Development of methodology: D.A. Drew, T.J. Devers, D.W. Rosenberg
Acquisition of data: D.A. Drew, T.J. Devers, M.J. O’Brien, N. Horelik, J. Levine
Analysis and interpretation of data: D.A. Drew, T.J. Devers, M.J. O’Brien, D.W. Rosenberg
Writing, review, and/or revision of the manuscript: D.A. Drew, T.J. Devers, D.W. Rosenberg
Study Supervision: D.W. Rosenberg
References
- 1.Rougier P, Mitry E. Epidemiology, treatment and chemoprevention in colorectal cancer. Annals of oncology : official journal of the European Society for Medical Oncology / ESMO. 2003;14(Suppl 2):ii3–ii5. doi: 10.1093/annonc/mdg722. [DOI] [PubMed] [Google Scholar]
- 2.Baxter NN, Goldwasser MA, Paszat LF, Saskin R, Urbach DR, Rabeneck L. Association of colonoscopy and death from colorectal cancer. Annals of internal medicine. 2009;150:1–8. doi: 10.7326/0003-4819-150-1-200901060-00306. [DOI] [PubMed] [Google Scholar]
- 3.Cooper GS, Xu F, Barnholtz Sloan JS, Schluchter MD, Koroukian SM. Prevalence and predictors of interval colorectal cancers in medicare beneficiaries. Cancer. 2012;118:3044–3052. doi: 10.1002/cncr.26602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vogelstein B, Fearon ER, Hamilton SR, Kern SE, Preisinger AC, Leppert M, et al. Genetic alterations during colorectal-tumor development. The New England journal of medicine. 1988;319:525–532. doi: 10.1056/NEJM198809013190901. [DOI] [PubMed] [Google Scholar]
- 5.Bird RP. Observation and quantification of aberrant crypts in the murine colon treated with a colon carcinogen: preliminary findings. Cancer letters. 1987;37:147–151. doi: 10.1016/0304-3835(87)90157-1. [DOI] [PubMed] [Google Scholar]
- 6.Yokota T, Sugano K, Kondo H, Saito D, Sugihara K, Fukayama N, et al. Detection of aberrant crypt foci by magnifying colonoscopy. Gastrointestinal endoscopy. 1997;46:61–65. doi: 10.1016/s0016-5107(97)70212-8. [DOI] [PubMed] [Google Scholar]
- 7.Greenspan EJ, Jablonski MA, Rajan TV, Levine J, Belinsky GS, Rosenberg DW. Epigenetic alterations in RASSF1A in human aberrant crypt foci. Carcinogenesis. 2006;27:1316–1322. doi: 10.1093/carcin/bgi373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lance P, Hamilton SR. Sporadic aberrant crypt foci are not a surrogate endpoint for colorectal adenoma prevention. Cancer Prev Res (Phila) 2008;1:4–8. doi: 10.1158/1940-6207.CAPR-08-0043. [DOI] [PubMed] [Google Scholar]
- 9.Stevens RG, Pretlow TP, Hurlstone DP, Giardina C, Rosenberg DW. Comment re:"Sporadic aberrant crypt foci are not a surrogate endpoint for colorectal adenoma prevention" and "Aberrant crypt foci in the adenoma prevention with celecoxib trial". Cancer Prev Res (Phila) 2008;1:215–216. doi: 10.1158/1940-6207.CAPR-08-0094. author reply 6. [DOI] [PubMed] [Google Scholar]
- 10.Anderson JC, Swede H, Rustagi T, Protiva P, Pleau D, Brenner BM, et al. Aberrant crypt foci as predictors of colorectal neoplasia on repeat colonoscopy. Cancer causes & control : CCC. 2012;23:355–361. doi: 10.1007/s10552-011-9884-7. [DOI] [PubMed] [Google Scholar]
- 11.Drew DA, Devers T, Horelik N, Yang S, O'Brien M, Wu R, et al. Nanoproteomic analysis of extracellular receptor kinase-1/2 post-translational activation in microdissected human hyperplastic colon lesions. Proteomics. 2013;13:1428–1436. doi: 10.1002/pmic.201200430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang S, Farraye FA, Mack C, Posnik O, O'Brien MJ. BRAF and KRAS Mutations in hyperplastic polyps and serrated adenomas of the colorectum: relationship to histology and CpG island methylation status. The American journal of surgical pathology. 2004;28:1452–1459. doi: 10.1097/01.pas.0000141404.56839.6a. [DOI] [PubMed] [Google Scholar]
- 13.Fumagalli D, Gavin PG, Taniyama Y, Kim SI, Choi HJ, Paik S, et al. A rapid, sensitive, reproducible and cost-effective method for mutation profiling of colon cancer and metastatic lymph nodes. BMC Cancer. 2010;10:101. doi: 10.1186/1471-2407-10-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rosenberg DW, Yang S, Pleau DC, Greenspan EJ, Stevens RG, Rajan TV, et al. Mutations in BRAF and KRAS differentially distinguish serrated versus non-serrated hyperplastic aberrant crypt foci in humans. Cancer research. 2007;67:3551–3554. doi: 10.1158/0008-5472.CAN-07-0343. [DOI] [PubMed] [Google Scholar]
- 15.Takayama T, Ohi M, Hayashi T, Miyanishi K, Nobuoka A, Nakajima T, et al. Analysis of K-ras APC, beta-catenin in aberrant crypt foci in sporadic adenoma, cancer, and familial adenomatous polyposis. Gastroenterology. 2001;121:599–611. doi: 10.1053/gast.2001.27203. [DOI] [PubMed] [Google Scholar]
- 16.Network TCGA. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kosaka T, Yatabe Y, Endoh H, Kuwano H, Takahashi T, Mitsudomi T. Mutations of the epidermal growth factor receptor gene in lung cancer: biological and clinical implications. Cancer research. 2004;64:8919–8923. doi: 10.1158/0008-5472.CAN-04-2818. [DOI] [PubMed] [Google Scholar]
- 18.Grundler R, Thiede C, Miething C, Steudel C, Peschel C, Duyster J. Sensitivity toward tyrosine kinase inhibitors varies between different activating mutations of the FLT3 receptor. Blood. 2003;102:646–651. doi: 10.1182/blood-2002-11-3441. [DOI] [PubMed] [Google Scholar]
- 19.Miyaki M, Konishi M, Kikuchi-Yanoshita R, Enomoto M, Igari T, Tanaka K, et al. Characteristics of somatic mutation of the adenomatous polyposis coli gene in colorectal tumors. Cancer research. 1994;54:3011–3020. [PubMed] [Google Scholar]
- 20.Aretz S, Stienen D, Uhlhaas S, Pagenstecher C, Mangold E, Caspari R, et al. Large submicroscopic genomic APC deletions are a common cause of typical familial adenomatous polyposis. Journal of medical genetics. 2005;42:185–192. doi: 10.1136/jmg.2004.022822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kanter-Smoler G, Fritzell K, Rohlin A, Engwall Y, Hallberg B, Bergman A, et al. Clinical characterization and the mutation spectrum in Swedish adenomatous polyposis families. BMC medicine. 2008;6:10. doi: 10.1186/1741-7015-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Siu IM, Robinson DR, Schwartz S, Kung HJ, Pretlow TG, Petersen RB, et al. The identification of monoclonality in human aberrant crypt foci. Cancer research. 1999;59:63–66. [PubMed] [Google Scholar]
- 23.Greenspan EJ, Cyr JL, Pleau DC, Levine J, Rajan TV, Rosenberg DW, et al. Microsatellite instability in aberrant crypt foci from patients without concurrent colon cancer. Carcinogenesis. 2007;28:769–776. doi: 10.1093/carcin/bgl209. [DOI] [PubMed] [Google Scholar]
- 24.De Rosa M, Scarano MI, Panariello L, Morelli G, Riegler G, Rossi GB, et al. The mutation spectrum of the APC gene in FAP patients from southern Italy: detection of known and four novel mutations. Human mutation. 2003;21:655–656. doi: 10.1002/humu.9151. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.