Abstract
Notch pathway plays an important role in the growth of high-grade serous ovarian (HGS-OvCa) and other cancers, but its clinical and biological mechanisms are not well understood. Here, we found that the Notch pathway alterations are prevalent and significantly related to poor clinical outcome in patients with ovarian cancer. Particularly, Notch3 alterations, including amplification and upregulation, were highly associated with poor patient survival. Targeting Notch3 inhibited OvCa growth and induced apoptosis. Importantly, we found that DNM-mediated endocytosis was required for selectively activating Jagged-1-mediated Notch3 signaling. Cleaved Notch3 expression was the critical determinant of response to Notch-targeted therapy. Collectively, these data identify previously unknown mechanisms underlying Notch3 signaling and identify new, biomarker-driven approaches for therapy.
Introduction
Notch signaling has been implicated in tumor angiogenesis processes such as vessel maturation; pericyte recruitment; branching; and cell differentiation, proliferation, and survival. In mammalian cells, this pathway comprises five transmembrane Notch ligands (Jagged-1, Jagged-2, Delta-like ligand [DLL] 1, DLL3, and DLL4) and four Notch receptors (Notch1–4). Mature Notch receptors are assembled as heterodimeric proteins, with each dimer comprised of a large extracellular ligand-binding domain, a single-pass transmembrane domain, and a smaller cytoplasmic subunit (Notch intracellular domain, NICD) (1). The activation of Notch signaling requires endocytosis and trafficking of both notch receptor and ligand. Upon ligand-receptor binding, it leads to cleavage of the Notch receptor via intramembrane proteolysis by gamma-secretase complex (including pesenilin, nicastin, APH1 and PEN2) and results in consequent release of NICD. The NICD fragment then enters the nucleus and interacts with nuclear DNA-binding factor, CSL (suppressor of hailess/LAG-1, RBPJK) to regulate transcription of the basic helix-loop-helix genes, hairy and Enhancer-of-split genes, and Notch target genes (2, 3).
However, the biological role of Notch pathway alterations in cancer growth and the clinical effects of these alterations are not well understood (4–7). In the present study, we performed an integrated and systematic analysis of the clinical relevance of the Notch pathway in high-grade serous ovarian cancer (HGS-OvCa) and identified novel mechanisms of Notch3 activation.
Materials and Methods
TCGA Clinical Analysis
Access to the TCGA database was approved by the National Cancer Institute. The University of Texas MD Anderson Cancer Center approved a waiver for performing our survival analysis with de-identified data. HGS-OvCa patients’ demographic characteristics and clinical data (histopathological information, treatment, and outcome parameters) were downloaded from the data portal for TCGA (http://tcga.cancer.gov) (Table S1).
The survival analysis output for the 316 study patients and complete information (OS and progression-free survival duration, expression, mutation, copy number) were downloaded from the cBio Cancer Portal for Genomics (http://www.cbioportal.org/public-portal/). Also, complete survival and gene expression information for our OS and progression-free survival analysis of 453 and 373 HGS-OvCa patients, respectively, was downloaded from TCGA. The patients’ mean age at diagnosis and tumor stage (as defined by the International Federation of Gynecology and Obstetrics), tumor grade, and surgical outcomes (residual tumor size) reflected those in individuals typically diagnosed with HGS-OvCa. The study patients’ tumor specimens had been resected before systemic treatment. All the patients had received a platinum agent, and 94% had received a taxane. The platforms used were described in the TCGA manuscript(4).
Copy-number alterations were analyzed using the Human Genome CGH Microarrays (244k, 415K, or 1M platforms; Agilent Technologies, Sugar Land, Texas), and focally amplified regions were identified using a modified method (4). Level 3 gene expression data were generated using three platforms: Agilent Technologies, GeneChip Human Exon ST Array (Affymetrix, Santa Clara, California), and GeneChip Human Genome U133A 2.0 Array (Affymetrix). We downloaded the Notch2–4 mutation data from TCGA (Table S1); these data were generated using the Genome Analyzer IIx platform (Illumina, San Diego, California) and the ABI SOLiD 3 System (Life Technologies/Applied Biosystems, Foster City, California).
Cell Lines and Cell Culture
OvCa cell lines (OVCAR3, OVCAR5, OVCAR420, SKOV3, SKOV3 TR, HeyA8, HeyA8 MDR, A2780, IGROV1, A2774, and HIO180) and uterine cancer cell line (Ishikawa) were obtained from the MD Anderson Characterized Cell Line Core Facility (Houston, Texas), which supplies authenticated cell lines. The cell lines were routinely tested to confirm the absence of mycoplasma, and all experiments were performed with cell lines at 60%–80% confluence.
OVCAR420, OVCAR3, SKOV3, SKOV3 TR, HeyA8, HeyA8 MDR, A2780, and IGROV1 cells were maintained and propagated in RPMI1640 medium supplemented with 10%–15% fetal bovine serum (FBS) and 0.1% gentamicin sulfate (Gemini Bio-Products, Sacramento, California). The medium used for the HeyA8 MDR and SKOV3 TR cells contained 100 nM docetaxel. OVCAR5 cells were maintained and propagated in Dulbecco’s modified Eagle’s medium/high-glucose medium supplemented with 15% FBS and 0.1% gentamicin sulfate. HIO180 and A2774 cell cultures were maintained in 10% cMEM.
Reagents and Antibodies
GSI was provided by Pfizer (New York, New York). Paclitaxel was purchased from the MD Anderson pharmacy. Notch3, Jagged-1, RPS6KB1, DNM1–3, control siRNAs, and Dynasore were purchased from Sigma-Aldrich (St. Louis, Missouri). Primers included PSEN1, APH1A, NCSTN, APH1B, PSENEN, RPS6KB1, DNM1–3, and 18s and were also purchased from Sigma-Aldrich. Antibodies used in this study included Jagged-1, Jagged-2, DLL1, DLL4, P70S6K (RPS6KB1), Notch1–4, and cleaved Notchs and were obtained from Cell Signaling Technology (Danvers, Massachusetts). Anti-DLL3 antibody was obtained from Abcam. β-actin was purchased from Sigma-Aldrich. CD31 was purchased from BD Biosciences (San Jose, California). Anti-Ki67 antibody was purchased from Neomarkers (Fremont, California). Antibody against cleaved caspase-3 was purchased from Cell Signaling Technology. Horseradish peroxidase-conjugated rat anti-mouse IgG2a was purchased from Harlan Bioproducts for Science (Indianapolis, Indiana).
Western Blot Analysis
Western blot analysis was performed to detect expression of Notch family members (Jagged-1, Jagged-2, DLL1, DLL3, DLL4, Notch1–4, and cleaved Notch (NICDs)) in a panel of OvCa cells. Cells were lysed with RIPA lysis buffer (50mM Tris-cl pH 7.4, 150mM NaCl, 1% NP40, 0.25% Na-deoxycholate, 1mM PMSF, 1x Roche complete mini protease inhibitor cocktail) and centrifuged for 15 minutes at 4°C. Protein concentrations were then measured using a Bio-Rad protein assay kit (Hercules, California). After loading the protein (25 μg/well), we separated bands on an 8%–10% gel using sodium dodecyl sulfate polyacrylamide gel electrophoresis. Bands were then transferred to nitrocellulose paper, blocked with 5% milk for 1 hour at room temperature, and incubated with primary antibodies against Notch1–4, Jagged-1, Jaged-2, and DLL4 (1:1000 dilution) and DLL3 (1:2000 dilution) overnight at 4°C. The samples were incubated with horseradish peroxidase-conjugated anti-mouse or anti-rabbit antibodies (GE Healthcare, Waukesha, Wisconsin) for 1 hour at room temperature. Blots were developed using an enhanced chemiluminescence detection kit (Pierce Biotechnology, Rockford, Illinois). Actin was used as a loading control, and all experiments were performed in duplicate. Densitometry was performed using the ImageJ software program (Bethesda, Maryland) to interpret differences in Western blot analysis results.
RT-PCR
Relative levels of expression of the gamma-secretase complex (PSEN1, NCSTN, APH1A, APH1B, and PSENEN) were detected using qRT-PCR. DNM expression was detected using RT-PCR. Each RT-PCR was carried out with 500 ng of total RNA isolated from treated OvCa cells using an RNeasy Mini Kit (Qiagen, Hilden, Germany). Quantitative RT-PCR was performed using an ABI 7500 sequence detection system (Applied Biosystems) with a SensiMix SYBR Low-ROX kit (Bioline USA, Taunton, Massachusetts). Relative quantification of gene expression was performed using the 2−ΔΔCT method. The primer sequences are listed in Table S2.
In Vitro siRNA Transfection
All cell lines were transfected with Lipofectamine 2000 reagent (Invitrogen, Grand Island, New York) using either control or target siRNAs (Sigma-Aldrich) as specified in Table S2 according to the manufacturer’s protocol. Cells were plated in a 6-well plate 24 hours before incubation. For 1-well transfection, we added 2 μg of target siRNA (1 μg/μl) and 6 μl of Lipofectamine 2000 to 100 μl of serum-free medium, and the mixture was incubated for 30 minutes at room temperature. During transfection, we washed the 6-well plates once with 2 ml of phosphate-buffered saline (PBS), and then we added 100 μl of the mixture along with 1 ml of serum-free medium to the plates. After 4–6 hours of transfection, the medium was replaced with a medium containing FBS.
Apoptosis, Cell Cycle, and Anoikis Assays
Apoptosis in the OvCa cells was evaluated using an Annexin V apoptosis detection kit (BD Biosciences). Cells were incubated in trypsin-EDTA, and cell pellets were suspended in 1 ml of 1X Annexin V binding buffer. We incubated 100 μl of each cell suspension with 5 μl of Annexin V and 5 μl of 7-aminoactinomycin D at room temperature (25°C) in the dark for 30 minutes. Following this incubation, 400 μl of 1X binding buffer was added to each tube, and specimens were analyzed using an XL 4-color flow cytometer. Each experiment was repeated three times.
For cell-cycle analysis, we synchronized the treated cells by maintaining them in serum-free media for 24 hours. The cells were then trypsinized, washed with PBS, fixed in 70% cold ethanol, and stored overnight at 4°C. Cells were then centrifuged at 1200 revolutions per minute for 10 minutes at 4°C. After one wash with PBS, cells were suspended in propidium iodide (Roche Diagnostics, Indianapolis, Indiana) at 50 μg/ml and RNAse A (Qiagen) at 100 μg/ml and incubated in the dark at room temperature. We then determined the cell-cycle status using fluorescence-activated cell sorting.
For anoikis analysis, 2.0 × 105 OvCa cells were seeded on a 6-well plate. Twenty-four hours later, cells were transfected with siRNA. After 24 hours of transfection, 2.5 × 105 cells were lifted and plated on a 6-well poly(2-hydroxyethyl methacrylate)-treated plate in serum-free mito-PLUS medium and incubated for 72 hours at 37°C in a 5% carbon dioxide atmosphere. After incubation, detached and suspended cells were harvested in RPMI1640 medium and centrifuged at 500 × g for 10 minutes. Pellets were washed with PBS and fixed with ice-cold 75% (v/v) ethanol overnight at 4°C. After fixation, cells were washed with PBS and stained with 500 μl of a propidium iodide solution (50 mg/ml in PBS) containing 25 μg/ml RNase A. Cells were incubated at 37°C for 30 minutes and analyzed using fluorescence-activated cell sorting.
Cytotoxicity Assays
OvCa cells were plated in a 96-well plate and cultured overnight. Cells were then exposed to 0.25, 0.5, 1, 2, 4, 6, 8, or 10 μM of GSI for 72 hours. Control cells were treated with a vehicle at equal concentrations. To assess cell viability, we added 50 μl of 0.15% 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT; Sigma-Aldrich) to each well and incubated the well-plate for 2 hours at 37°C. The medium containing MTT was then removed, and 100 μl of dimethyl sulfoxide (Sigma-Aldrich) was added to each well. Cells were then incubated at room temperature for 10 minutes. The absorbance was read at 570 nm using a 96-well Synergy HT multi-mode microplate reader (Ceres UV 900C; BioTek, Winooski, Vermont). Cell viability was defined as the percentage of viable cells in the treatment group relative to viable cells in the control group. The experiments were repeated separately at least three times.
Microarray Analysis of Notch3 Downstream Genes
We analyzed the silencing of Notch3 with the Illumina cDNA microarray platform as described previously (8). Briefly, Notch3 in OVCAR3 cells was silenced in vitro using Notch3 siRNAs, and Notch3 silencing was confirmed via Western blot analysis. For microarray hybridization, total RNA was extracted from OvCa cells transfected with control siRNA or human Notch3 siRNA using a mirVana RNA isolation labeling kit (Ambion, Austin, Texas). We used 500 ng of total RNA to label and hybridize the cells according to Illumina’s protocols. After we scanned bead chips (Sentrix HumanHT-12 v3, Illumina) using an Illumina BeadArray reader, we normalized the microarray data using the quantile normalization method with the Linear Models for Microarray Data software program in the R language (Denton, Texas) (9). The level of expression of each gene was transformed into a log2 base before additional analysis was performed. Genes that were differentially expressed in control and Notch3-silenced cells were identified using a random-variance t-test; gene-expression differences were considered statistically significant if their p values were less than 0.001. A stringent significance threshold was used to limit the number of false-positive findings.
In addition, cluster analysis was performed using the Cluster and TreeView software programs (Berkeley, California) (10). The NetWalker software program was used for gene network analysis (11).
In Vivo Mouse Models
Female athymic nude mice (NCr-nu) were purchased from the Animal Production Area of the Frederick National Laboratory for Cancer Research (Frederick, Maryland). The animals were kept under specific pathogen free conditions in facilities approved by the Association for Assessment and Accreditation of Laboratory Animal Care International and in agreement with current regulations and standards of the United States Department of Health and Human Services, the United States Department of Agriculture, and the National Institutes of Health. Mice used in these experiments were 8–12 weeks old.
To generate tumors in the mice, we trypsinized subconfluent cultures of OVCAR5, A2780, and SKOV3 cells, mixed the cells with a medium containing 10% FBS, centrifuged the mixture at 1000 revolutions per minute for 5 minutes, and washed the cells in PBS. Mice were then injected intraperitoneally with 1 × 106 OVCAR5, A2780, or SKOV3 cells. In the OVCAR5 and A2780 models, mice were randomized into four treatment groups of 10 mice each: control siRNA, control siRNA with paclitaxel, Notch3 siRNA, and Notch3 siRNA with paclitaxel. In the SKOV3 model, mice were randomized into two treatment groups: control siRNA and Notch3 siRNA. Treatments were initiated 8 days after tumor-cell injection. Control and Notch3 siRNA were conjugated with CH (8), which we injected into the tail vein twice weekly (150 μg/kg per mouse). Paclitaxel was administered intraperitoneally once weekly (4 mg/kg per mouse). Mice were monitored daily and weighed weekly. All of the mice were subjected to necropsy when the control mice became moribund, which was approximately 28 days after A2780-cell injection, 35 days after OVCAR5-cell injection, and 38 days after SKOV3-cell injection. Each mouse’s total body weight and the location, weight, and number of tumors were recorded. Tumor specimens processed for further analysis were either preserved in optimum cutting temperature medium (Miles Laboratories, Elkhart, Indiana; for frozen sectioning) or fixed in formalin (for paraffin embedment).
To further validate target modulation (cleaved Notch3, Hes1 and RPS6KB1) in tumor tissues obtained from the in vivo A2780 model, ovarian cancer cells were injected intraperitoneally into the mice. Three weeks later, the mice were randomly allocated to two treatment groups (n = 5/group): 1) control, 2) Dynasore (60 mg/kg i.p. for two treatments, 72 hours apart). Tumor tissues were collected for Western blot analysis.
Immunohistochemical Staining
Paraffin-embedded orthotopic tumor specimens were used in immunohistochemical staining to detect cell proliferation (with Ki67), apoptosis (with cleaved caspase-3), and Notch3 expression. Tumor sections were deparaffinized, rehydrated, and transferred to PBS. After retrieving the antigens with citrate buffer (pH 6.0), we blocked the sections with 3% hydrogen peroxide in methanol and protein blocker at room temperature. The sections were then incubated with a monoclonal mouse anti-Ki67 antibody (1:200; Neomarkers), anti-Notch3 antibody (1:50; Santa Cruz Biotechnology, Santa Cruz, California), or anti-cleaved caspase-3 antibody (1:100; Biocare Medical, Concord, California) overnight at 4°C. After being washed with PBS, the sections were incubated with horseradish peroxidase-conjugated rat anti-mouse IgG2a (1:100; Harlan Bioproducts for Science).
For CD31 detection, frozen sections were fixed in cold acetone for 15 minutes, washed with PBS, blocked with protein blocker (4% fish gel), and then incubated with rat monoclonal anti-mouse CD31 (1:800, PharMingen, San Diego, CA) overnight at 4 °C. They were then washed with PBS and incubated with HRP-conjugated goat anti-rat IgG (1:200, Jackson ImmunoResearch Laboratories, West Grove, PA) for 1 hour. Reactive tissues were visualized using staining with 3,3′-diaminobenzidine (Research Genetics, Huntsville, AL) and followed by counterstaining with Gil’s hematoxylin (BioGenex Laboratories, San Ramon, CA).
To quantify Ki67, cleaved caspase-3 and Notch3, the percentage of cells positive for them was determined in five random 0.159-mm2 fields at 200× magnification. To quantify microvessel density (MVD) for each sample, the microvessels within five randomly selected 0.159-mm2 fields were counted at ×200 magnification. A single microvessel was defined as a discrete cluster for CD31 positivity.
Statistical Analysis
The Kaplan-Meier method was used to generate time-to-progression and survival curves. The statistical analysis was performed with the R statistical computing language (version 2.11.0; http://www.r-project.org/), and statistical significance was set at 0.05. For analysis of categorical data (tumor stage, tumor grade, and residual tumor size), the Fisher exact test was used to calculate p values. For in vivo data analysis, differences in continuous variables (mean body weight, tumor weight, tumor-cell proliferation, and apoptosis) were analyzed using the Mann-Whitney rank-sum test.
Results
Clinical Significance of Alterations of the Core Notch and Related Genes
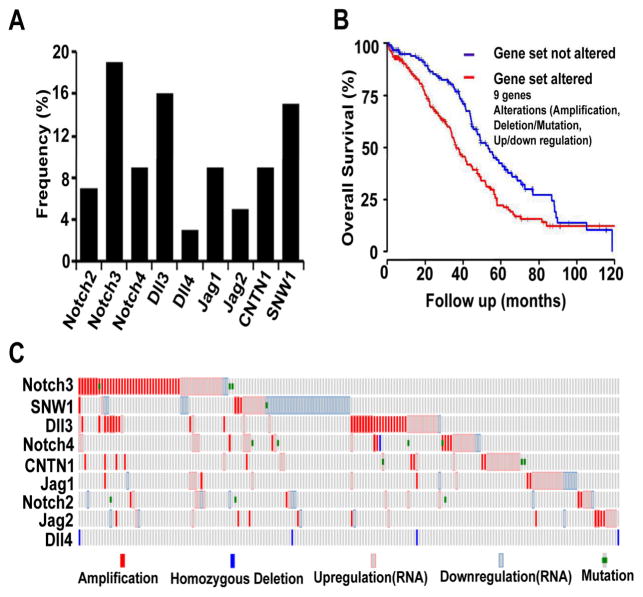
To determine the clinical relevance of alterations of the core Notch and related genes, including Notch ligands (Jagged-1, Jagged-2, DLL1, DLL3, and DLL4), Notch receptors (Notch1–4), and Notch-interacting genes (SNW1, CNTN1), we first examined the effect of these alterations on survival duration in 316 patients with HGS-OvCa from the TCGA data-set. We found that Notch3, DLL3, SNW1 and Jag1 were the genes altered (defined as amplification, upregulation or downregulation of expression, mutation, or homozygous deletion) most frequently (17%, 16%, 15% and 9% of cases, respectively) (Figure 1A). In total, 61% of the HGS-OvCa cases had alterations of at least one of the Notch pathway-related genes. Patients with these alterations had shorter overall survival (OS) durations than did those without the altered genes (median, 36.2 months versus 53.2 months; p = 0.001) (Figure 1B). The types of alterations for each of the nine genes are shown in Figure 1C. Notch pathway alterations were not significantly associated with other clinical parameters, including tumor stage (as defined by the International Federation of Gynecology and Obstetrics), grade, and residual tumor size (Tables S1a and S1c). Mutations of Notch2–4 did not occur frequently in HGS-OvCa cases (Table S1b), and there were no alterations in Notch1 or DLL1.
Figure 1. Alterations of Notch Genes in Patients with High-Grade Serous Ovarian Cancers (HGS-OvCa).
(A) Frequency of alterations of nine Notch-family genes and closely interacting genes. Among the alterations (amplification, upregulation and downregulation, mutation, and homozygous deletion), Notch3, delta-like ligand (DLL)3, SNW1 and Jagged1 were the most frequently altered genes (17%, 16%, 15%, and 9% of cases, respectively. (B) Overall survival (OS) durations in cases with and without gene alterations. The gene set consisted of Jagged-1, Jagged-2, DLL3, DLL4, CNTN1, Notch2, Notch3, Notch4, and SNW1. (C) Alteration patterns in the Notch signaling pathway. Each column represents an individual case with at least one alteration.
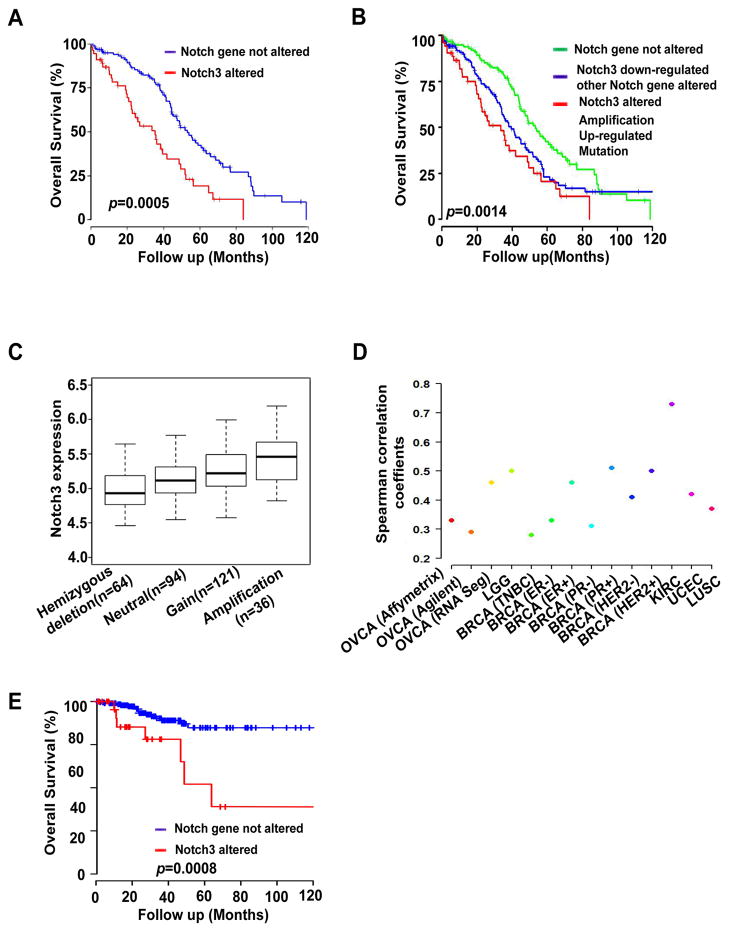
We next focused specifically on Notch3 due to its frequent alterations. Patients with Notch3 alterations had significantly shorter OS durations than did patients without these alterations (median, 35.3 months versus 52.2 months; p = 0.0005; Figure 2A). Furthermore, patients with amplification, increased expression, and mutation of Notch3 genes had significantly shorter OS durations than did those without these alterations (median OS duration, 33.6 months versus 53.2 months; p = 0.0014; Figure 2B). Notch3 amplification alone was not associated with OS duration, but there was a trend of shorter OS duration in patients with amplification (Figure S1A). Notch3 expression was correlated with its amplification (p < 0.001; Figure 2C), according to Spearman correlation analysis. Notch3 overexpression was also significantly correlated with the GATA2 and C/EBP transcriptional factors based on Spearman correlation analysis (Supplementary Table 3). Importantly, we identified a significant correlation between Jagged-1 and Notch3 expression in HGS-OvCa samples as well as in breast, uterine, and kidney cancers (Figure 2D; Figure S1B, Table 1). Notch3 amplification and upregulation were also significantly correlated with shorter survival in patients with uterine cancer (median OS duration, 56 months); patients without these alterations were still alive at the end of this study (p = 0.0008; Figure 2E).
Figure 2. Overall Survival (OS) Durations in Patients with High-Grade Serous Ovarian Cancers (HGS-OvCa) with Notch3 Alterations.
(A) OS durations in cases with and without Notch3 alterations. (B) OS durations in cases with amplified, upregulated, or mutated Notch3 genes; with downregulated Notch3 expression and alterations of other Notch genes; and without Notch gene alterations. (C) Correlation between Notch3 expression in patients with Notch amplification and that in patients with gain, neutral, or hemizygous deletion of Notch3. (D) Correlation between Notch3 and Jag1 expression in human cancers. HGS-OvCa =Ovarian serous adenocarcinoma; HNCS=Head and neck squamous cell carcinoma; LGG=Brain low grade glioma; BRCA=Breast invasive carcinoma; CRC=Colon and rectum adenocarcinoma; KIRC=Kidney renal clear cell carcinoma; UCEC=Uterine corpus endometrioid carcinoma; LUSC=Lung squamous cell carcinoma; TNBC=Triple-negative breast cancer. (E) OS durations in patients with uterine cancer with Notch3 alterations and without Notch alterations (amplification and upregulation).
Table 1.
Correlation between Notch3 and Jagged-1 expression in Human cancers
| Cancer type | Coefficient | p | Size |
|---|---|---|---|
| HGS-OvCa (Affymetrix microarray) | 0.33 | 3.28E-16 | 569 |
| HGS-OvCa (Agilent microarray) | 0.29 | 3.48E-12 | 571 |
| HGS-OvCa (IlluminaRNASeq) | 0.46 | 2.09E-22 | 411 |
| HNSC | 0.08 | 0.16677 | 303 |
| LGG | 0.50 | <0.0000001 | 174 |
| BRCA-TNBC | 0.28 | 0.002831 | 112 |
| BRCA-ER− | 0.33 | 4.8E-05 | 149 |
| BRCA-ER+ | 0.46 | <0.000001 | 511 |
| BRCA-PR− | 0.31 | 4.65E-06 | 215 |
| BRCA-PR+ | 0.51 | <0.0000001 | 443 |
| BRCA-HER2− | 0.41 | <0.0000001 | 582 |
| BRCA-HER2+ | 0.5 | 4.54E-07 | 95 |
| CRC | 0.13 | 0.035451 | 264 |
| KIRC | 0.73 | 3.17E-78 | 470 |
| UCEC | 0.42 | 0.000001 | 333 |
| LUSC | 0.37 | 2.27-08 | 223 |
HGS-OvCa =Ovarian serous adenocarcinoma; HNCS=Head and neck squamous cell carcinoma; LGG=Brain low grade glioma; BRCA=Breast invasive carcinoma; CRC=Colon and rectum adenocarcinoma; KIRC=Kidney renal clear cell carcinoma; UCEC=Uterine corpus endometrioid carcinoma; LUSC=Lung squamous cell carcinoma; TNBC=Triple-negative breast cancer.
Biological Effects of Notch3 and cleaved Notch3 in Cancer Cells
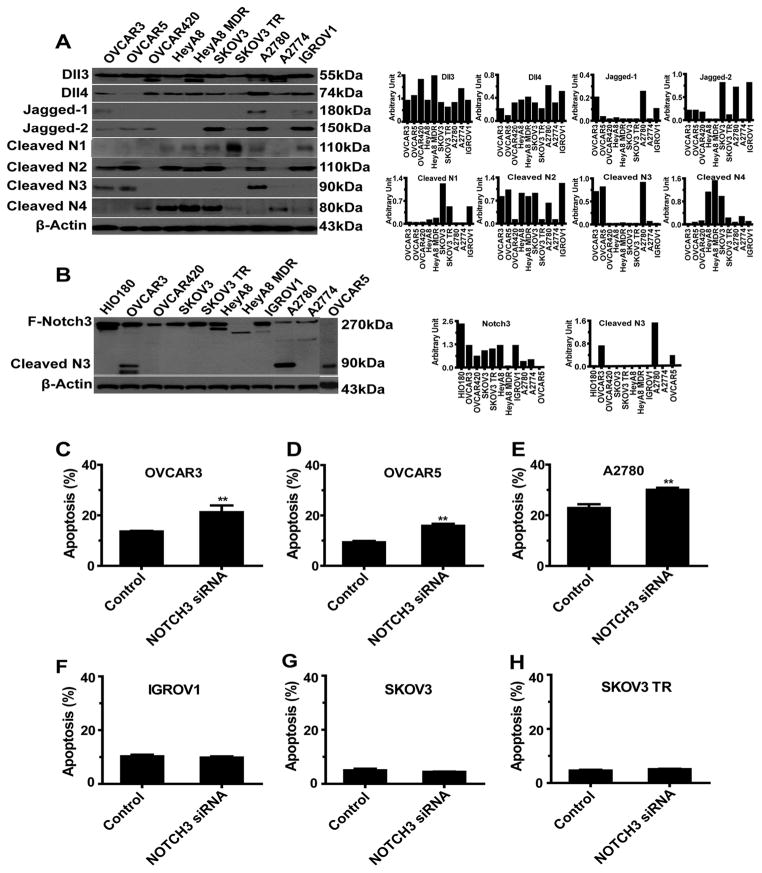
We first used Western blot analysis to screen a panel of OvCa cell lines for expression of Notch family members (Figures 3A and 3B). We selected both cleaved Notch3 (NICD3)-positive (OVCAR3, OVCAR5, and A2780) and NICD3-negative (SKOV3, SKOV3 TR, and IGROV1) OvCa cells for additional in vitro and in vivo studies. After transfecting cancer cells with Notch3 siRNA (Figure S2A), we noticed high levels of apoptosis in the NICD3-positive cells (Figures 3C–3E), but not in the NICD3-negative cells (Figures 3F–3H); these results suggest that NICD3 expression is an important determinant of the biological functions of Notch3 in cancer cells. Furthermore, compared with levels in control cells, levels of anoikis increased greatly in NICD3-positive cells transfected with Notch3 siRNA (Figure S2B). Notch3 siRNA also induced cell-cycle arrest in the G2/M phase in NICD3-positive cells, but not in NICD3-negative cells (Figure S2C).
Figure 3. In Vitro Functional Studies of Notch3 Silencing.
(A) Western blot analysis of expression of 8 Notch-family proteins in a panel of 10 ovarian cancer (OvCa) cell lines. (B) Western blot analysis of expression of full-length Notch3 and cleaved Notch3 (NICD) in a panel of 10 OvCa cell lines. (C–H) Apoptosis in OvCa cells (NICD3-positive: OVCAR5, OVCAR3, and A2780; NICD3-negative: IGROV1, SKOV3, and SKOV3 TR) after exposure to control siRNA or Notch3 siRNA for 72 hours. **p < 0.01.
To determine whether the cleaved Notch3 is a determinant of sensitivity to Notch-targeted therapy, we treated human epithelial OvCa cells (OVCAR5, OVCAR3, SKOV3, HeyA8, and A2780) with a gamma-secretase inhibitor (GSI) at various concentrations for 72 hours and assessed cell viability. The half-maximal inhibitory concentrations of GSI ranged from 3.06 to 7.73 μM. The NICD3-positive cells (A2780, OVCAR3, and OVCAR5) were more sensitive to treatment with GSI than were the NICD3-negative cells (SKOV3 and HeyA8; Figure S2D). GSI-based treatment induced substantially more apoptosis in the NICD3-positive cells than in the NICD3-negative cells (Figure S2E).
Because Notch3 siRNA arrested cells in the G2/M phase, we examined whether Notch3 siRNA could enhance sensitivity to paclitaxel. We found that silencing Notch3 significantly enhanced the sensitivity of OVCAR3 cells to treatment with paclitaxel (p<0.01, Figures S2F–G)
Dynamin-Dependent Endocytosis Is Required for Jag1-Mediated Notch3 Activation
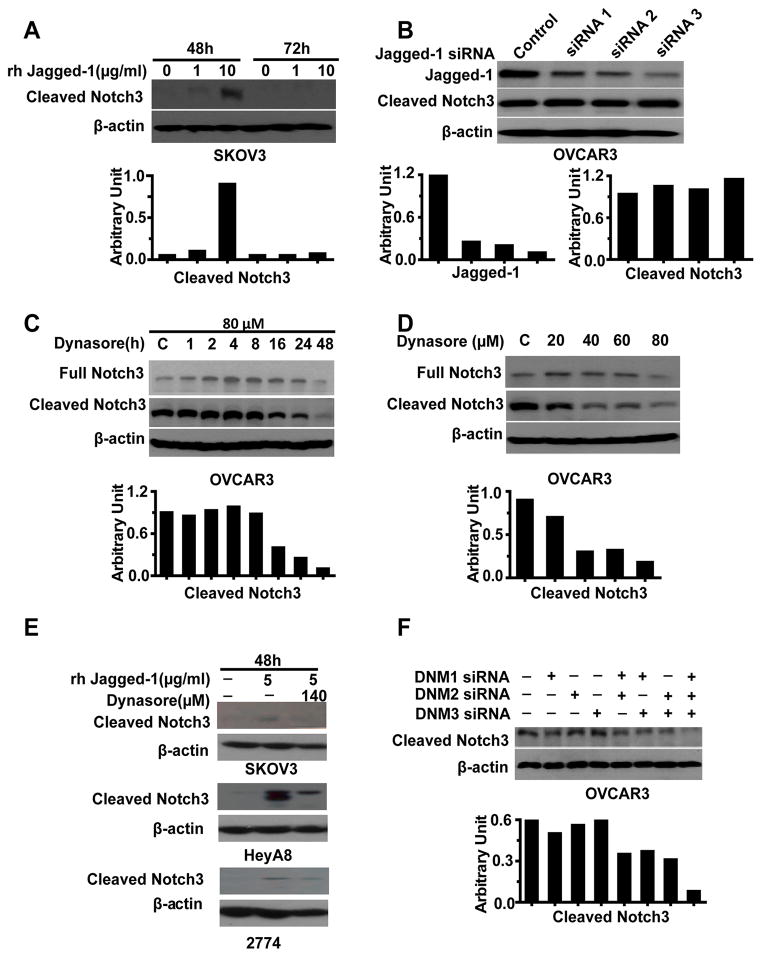
Next, we considered potential explanations for selective cleaved Notch3 expression in cancer cell lines. Given the role of the gamma-secretase complex (PSEN1, APH1A, NCSTN, APH1B, and PSENEN) in Notch proteolysis, we first used quantitative reverse transcription polymerase chain reaction (qRT-PCR) to examine the expression of the gamma-secretase complex in the panel of OvCa cells. These five genes were not differentially expressed between the NICD3-positive or -negative cells (Figure S3). Since there was no association between the gamma-secretase complex and cleaved Notch3 expression, we next used TCGA data and the Spearman’s rank correlation coefficient to analyze the relationship between Notch3 and its potential ligands in HGS-OvCa patients. There was a significant correlation between Notch3 and Jagged-1 expression in HGS-OvCa as well as other cancers, including cancers of the uterus, breast, and kidney (p < 0.0001; Table 1). Therefore, we next evaluated whether Jagged-1 stimulation could induce Notch3 proteolysis in NICD3-negative SKOV3 cells. Whereas treatment with recombinant human Jagged-1 (rhJagged-1) (10 μg/ml) at 48 hours increased cleaved Notch3 expression in these cells compared with controls (Figure 4A), Jagged-1 silencing did not decrease cleaved Notch3 levels in NICD3-positive cells (Figure 4B).These findings prompted us to consider whether endocytosis may be involved in Notch3 activation.
Figure 4. Notch3 Activation by Dynamin (DNM)-Dependent Endocytosis.
(A) Effect of Jagged-1 stimulation on Notch3 activation in SKOV3 cells. (B) Effect of Jagged-1 silencing on Notch3 activation in OVCAR3 cells. (C and D) Effect of treatment with dynasore on Notch3 activation in OVCAR3 cells at different time points and doses. (E) Effect of treatment with dynasore on Jagged-1-mediated Notch3 activation in NICD3-negative ovarian cancer cells (SKOV3, 2774 and HeyA8). (F) Effect of DNM silencing on Notch3 activation in OVCAR3 cells.
To determine whether endocytosis is involved in Notch3 proteolysis, OVCAR3 cells were treated with dynasore, a cell-permeable inhibitor of dynamin (DNM). Cleaved Notch3 expression was decreased in a time- and dose-dependent manner (Figures 4C and 4D). Dynasore treatment indeed increased the expression of full Notch3 expression at lower doses and at 1 - 24 hours, likely due to its rapid effect on inhibiting Notch3 receptor degradation, but this treatment decreased full Notch3 and Jagged-1 at higher doses and 48 hours (Figures 4C and 4D, and Figure S4A). Importantly, dynasore prevented Jagged-1-stimulated cleaved Notch3 expression in the NICD3-negative cells (SKOV3 cells, HeyA8 and 2774), suggesting that endocytosis was required for Jagged1-mediated Notch3 activation (Figure 4E, Figure S4B). Next, to identify which DNMs are involved in this endocytotic process to activate Notch3, we screened a panel of OvCa cells for expression of DNM1, DNM2, and DNM3. Among these, DNM1 and DNM2 were expressed in all the cells tested, but DNM3 was present only in some cells (Figure S4C). We then treated the OVCAR3 cells with DNM1, DNM2, or DNM3 siRNA or pools of two or three of these DNM siRNAs (Table S4, Figure S4D). Treatment with the pools of three DNM siRNAs resulted in much lower cleaved expression in the OVCAR3 cells than did treatment with the individual siRNA duplexes (Figure 4F), demonstrating that DNM-dependent endocytosis was a critical determinant of Notch3 activation.
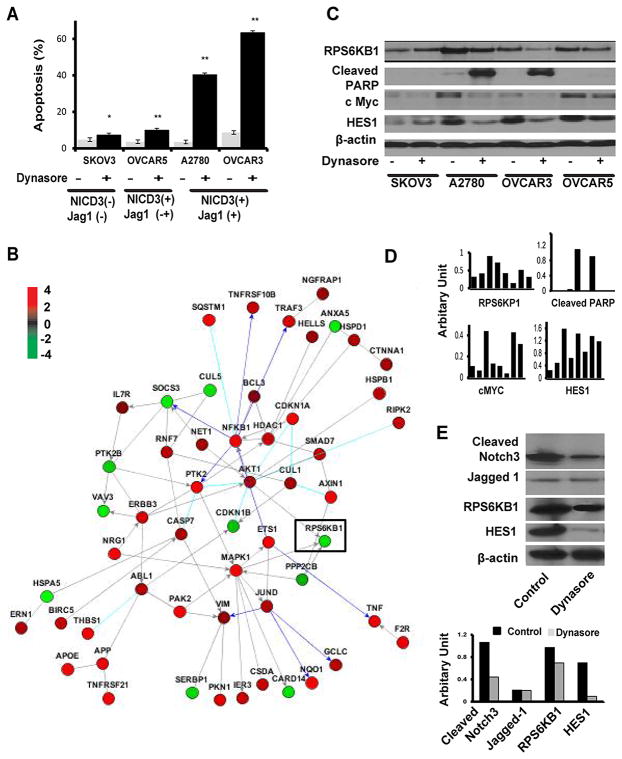
To determine whether inhibiting DNM-mediated endocytosis via dynasore could affect apoptosis, NICD3-positive cells (OVCAR5, OVCAR3, and A2780) and NICD3-negative cells (SKOV3) were treated with dynasore (60 μM) for 72 hours. Induction of apoptosis was significantly higher in the Jag1/NICD3-positive cells (A2780 and OVCAR3) than in the Jagged-1/NICD3-negative cells (SKOV3; Figure 5A; p<0.001). Apoptosis was only slightly increased in the NICD3-positive OVCAR5 cells treated with Dynasore, likely because of the weak expression of Jagged-1 in these cells (Figure 5A). Induction of apoptosis by Dynasore treatment was also noted in uterine cancer cells (Ishikawa), which express Notch3 and Jagged-1 (Figure S4E–F). Dynasore treatment increased Jagged-1 expression in uterine cancer cells (Figure S4E), which may occur due to a possible feedback loop of Notch inhibition or cross-talk with other pathways such as Wnt/Beta catenin.
Figure 5. Microarray Analysis of Notch3-targeted Genes in OvCa Cells.
(A) Analysis of apoptosis in NICD3-positive and NICD3-negative cells after exposure to dynasore for 72 hours. *p < 0.05;**p < 0.01(B) Pathway analysis of apoptosis-related genes in OVCAR3 cells treated with Notch3 siRNA or control siRNA. (C, D) Western blot and image analysis of RPS6KP1, cleaved PARP, cMyc, and Hes-1 in OvCa cells treated with dynasore. (E) Western blot analysis of target modulation (Cleaved Notch 3, HES1, Jagged-1 and RPS6KB1) in tumor tissues obtained from the in vivo A2780 model. Ovarian cancer cells were injected intraperitoneally into the mice. Three weeks later, the mice were randomly allocated to two treatment groups (n = 5/group): 1) control, 2) Dynasore.
Next, we searched for potential Notch3-targeted genes that could be involved in apoptosis in OvCa cells by hierarchical clustering analysis following Notch3 silencing. There were 461 genes differentially expressed in the OVCAR3 cells following Notch3 silencing, when defined as a fold change > 0.7 (Table S4). Given the robust induction of apoptosis in NICD3-positive cells, a pathway analysis focusing on apoptosis was performed. Among the 56 genes in this pathway, RPS6KB1 expression was particularly downregulated in response to Notch3 silencing (Figures 5B). Moreover, dynasore treatment decreased RPS6KB1 expression by 3.0 and 1.3 fold in OVCAR3 and A2780 cells respectively (Figures 5C); we found that cleaved poly ADP ribose polymerase dramatically increased in OVCAR3 and A2780 cells, but not in SKOV3 cells, suggesting that RPS6KB1 was a dominant downstream target of the Notch3 pathway involved in apoptosis. Since it has been reported that PI3K/AKT/mTOR signaling could be modulated by direct repression of PTEN via the Notch target gene Hes1 or cMyc (12), we tested cMyc and HES1 in these cells following dynasore treatment. Dynasore treatment decreased cMyc and HES1 levels by 2-fold in the OVCAR3 and A2780 cells, but not in the NICD3-negative cells (Figure 5C, 5D).
We also validated the target modulation in vivo by examining the effects of Dynasore on expression of Jagged-1, cleaved Notch3, HES1 and RP6SKB1 in the A2780 model. After two i.p. injections of Dynasore (60mg/kg), cleaved Notch3, HES1 and RP6SKB1 levels, but not Jagged-1, were decreased in tumors treated with Dynasore (Figure 5E).
In Vivo Targeting of Notch3 with siRNAs
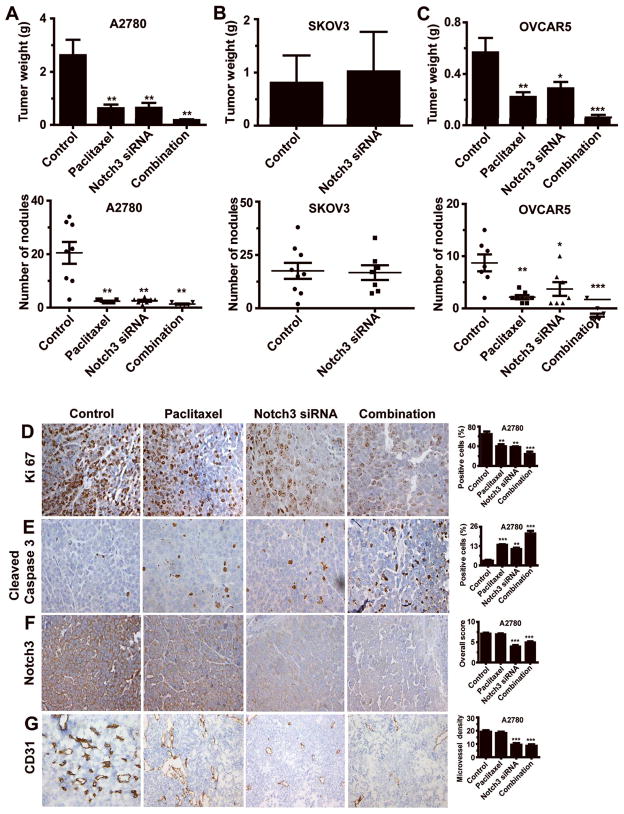
On the basis of our in vitro findings, we next tested whether silencing Notch3 could induce apoptosis in vivo and enhance sensitivity of cancer cells to chemotherapy. For in vivo testing, the A2780 (NICD3/Jagged1-positive) and SKOV3 (NICD3/Jagged-1-negative) models were used. For systemic delivery of siRNA, the well-characterized chitosan (CH) delivery system (8) was used. In the A2780 model, tumors treated with Notch3 siRNA-CH or paclitaxel alone weighed 74.1% (p = 0.0093) and 75.7% (p = 0.0081) less, respectively, than control siRNA-CH treated tumors (Figure 6A). Combining Notch3 siRNA-CH and paclitaxel reduced tumor weight by 99.3% (p = 0.003, compared with tumor weight in control mice). Mice receiving this combination treatment also had significantly fewer tumor nodules on average than did control siRNA-CH treated mice (p = 0.001). In the SKOV3 model, Notch3 silencing did not affect tumor weight or the number of tumor nodules (Figure 6B). Treatment with Notch3 siRNA-CH did not significantly affect body weight (Figure S5A). In the OVCAR5 (NICD3-positive) model, tumors treated with Notch3 siRNA-CH or paclitaxel alone weighed 49.1% (p = 0.0432) and 60.7% (p = 0.013) less, respectively, than did the control siRNA-CH tumors (Figure 6C). Also, combining Notch3 siRNA-CH and paclitaxel decreased tumor weights by 78.7% and 72.3%, respectively, from weights with either treatment alone (p = 0.001 and 0.003, respectively). Body weight was not significantly affected by treatment (Figure S5A).
Figure 6. In Vivo Study of Notch3 siRNA in Mouse Models of Ovarian Cancer (OvCa).
(A–C) Effect of treatment with Notch3 siRNA and/or paclitaxel on tumor weight in Notch3 intracellular domain (NICD3)-positive models (OVCAR5 and A2780) and NICD3-negative model (SKOV3). The error bars represent standard error of the mean (SEM). *p < 0.05; **p < 0.01; ***p < 0.001. (D–G) Effect of treatment with Notch3 siRNA and/or paclitaxel on biological endpoints: cell proliferation (Ki67 expression), apoptosis (cleaved caspase-3 expression), Notch3 expression and microvessel density (CD31). Original magnification, 200×. The error bars represent standard error of the mean (SEM). *p < 0.05; **p < 0.01; ***p < 0.001.
Notch3 levels were significantly decreased in the Notch3 siRNA-CH-treated groups compared with the levels in the untreated mice (p<0.01). In the A2780 model, apoptosis was significantly higher in the tumors treated with Notch3 silencing than in untreated cells (Figure 6D–6G, p<0.01). Compared with the controls, proliferation and angiogenesis were significantly decreased in the tumors treated with the combination therapy. Similar effects were noted in the OVCAR5 model (p<0.01; Figure S5B–S5E).
Discussion
The key findings from our work are that Notch pathway alterations are prevalent and significantly related to poor clinical outcome in patients with ovarian cancer. Particularly, Notch3 alterations, including amplification and upregulation, were highly associated with poor survival. Targeting Notch3 inhibited the growth of OvCa and induced apoptosis. Importantly, we found that DNM-mediated endocytosis was required for selectively activating Jag1-mediated Notch3 signaling. Cleaved Notch3 expression was the critical determinant of response to Notch-targeted therapy. Collectively, these data identify previously unknown mechanisms underlying Notch3 signaling and identify new, biomarker-driven approaches for therapy.
Why Notch3 expression is preferentially upregulated in patients with OvCa is not clear. Several mechanisms may contribute to its upregulation. Notch3 upregulation was partially correlated with its amplification; Notch3 expression may be caused by transcriptional activation or loss of negative regulators such as SNW1, since the ability of NICD3 to recruit co-activators and co-repressors and undergo conformational changes is different from that of Notch1 and Notch2 (13). However, we cannot rule out a role for SNW1 in the negative regulation of Notch3 pathway activation in such cases (14). Unlike the proteolysis findings with Notch1, Notch2, and Notch4, we found that Notch3 proteolysis was selectively activated in OvCa cells. We considered potential explanations for selective cleaved Notch3 expression in OvCa cells, such as differential expression of gamma secretase complex in NICD3-positive and NICD3-negative OvCa cells, preferential pairing of ligands and Notch3, and ligand endocytosis for signaling activation and receptor degradation. Our data did not support an association between gamma secretase complex and cleaved Notch3 expression in NICD3-positive versus negative OvCa cells. It has been suggested that preferential pairing of Notch3 and Jagged-1 or DLL4 can occur in colon cancer cells (13). We found that Notch3 proteolysis was selectively activated in OvCa cells in a Jagged-1-dependent manner, which highlights a critical role for Jagged-1 in activating Notch3 signaling. Jagged-1 expression has also been reported to be regulated directly by Notch 3 and Wnt/beta catenin in ovarian cancer, which might contribute to maintaining long-term Notch3 signaling activation in cancer cells (15).
The exact function of ligand endocytosis in intracellular signaling is unknown. Two possible models have been reported: one that involves ligand-induced Notch signaling (16–18) and another that involves alternative endocytotic adaptors such as dynamin, epsins and actin (17). Prior to our work, role of endocytotic adaptors in Jagged-1-mediated endocytosis in activating Notch3 in ovarian cancer was unknown. Three different dynamin genes have been identified in mammals, including DNM1, DNM2 and DNM3. Dynamin has been reported as an endocytotic adaptor that functions in membrane tabulation and fission of budding vesiculo-tubular structures and regulates ligand internalization (19). We demonstrated that selective Notch3 activation is dependent on DNM-mediated endocytosis and blocking DNMs with dynamin inhibitor significantly increased apoptosis in OvCa, which represents an innovative strategy for inhibiting Notch3 activation.
Taken together, our findings demonstrate that Notch pathway alterations, especially in Notch3 (amplification or upregulation of expression), are prevalent in HGS-OvCa cases and shorten OS durations. Cleaved Notch3 is a key determinant of Notch3 biology, and DNM-mediated endocytosis is required for Jagged1-mediated Notch3 activation. Targeting Notch3 pathway activation with DNM inhibitors (20) combined with paclitaxel could be considered for future clinical investigation.
Supplementary Material
Acknowledgments
Portions of this work were supported by the National Institutes of Health (grants P50 CA083639, CA109298, P50 CA098258, UH2 TR000943, CA128797, and U54 CA151668), the Ovarian Cancer Research Fund (a Program Project Development Grant), the United States Department of Defense (grants OC120547 and OC093146), the Zarrow Foundation, the Marcus Foundation, the Betty Anne Asche Murray Distinguished Professorship, MD Anderson Cancer Center Support Grant CA016672, the Gilder Foundation, and the RGK Foundation. J.B.-M., B.Z., R.A.P., and H.D. were supported by a National Cancer Institute/United States Department of Health and Human Services/National Institutes of Health Training Grant (T32 CA101642). W.H. was partially supported by the Gynecologic Cancer Foundation/Florence and Marshall Schwid Ovarian Cancer Award.
We thank Nicholas B. Jennings for technical support and Diane Hackett and Jill Delsigne, Department of Scientific Publications, for editing the manuscript.
Footnotes
The authors report no conflicts of interest.
References
- 1.Chan YM, Jan YN. Roles for proteolysis and trafficking in notch maturation and signal transduction. Cell. 1998;94:423–6. doi: 10.1016/s0092-8674(00)81583-4. [DOI] [PubMed] [Google Scholar]
- 2.Artavanis-Tsakonas S, Muskavitch MA. Notch: the past, the present, and the future. Curr Top Dev Biol. 2010;92:1–29. doi: 10.1016/S0070-2153(10)92001-2. [DOI] [PubMed] [Google Scholar]
- 3.Wang Z, Li Y, Ahmad A, Azmi AS, Banerjee S, Kong D, et al. Targeting Notch signaling pathway to overcome drug resistance for cancer therapy. Biochim Biophys Acta. 2010;1806:258–67. doi: 10.1016/j.bbcan.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.TCGA Research Group. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–15. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cooke SL, Brenton JD. Evolution of platinum resistance in high-grade serous ovarian cancer. Lancet Oncol. 2011;12:1169–74. doi: 10.1016/S1470-2045(11)70123-1. [DOI] [PubMed] [Google Scholar]
- 6.Landen CN, Jr, Birrer MJ, Sood AK. Early events in the pathogenesis of epithelial ovarian cancer. J Clin Oncol. 2008;26:995–1005. doi: 10.1200/JCO.2006.07.9970. [DOI] [PubMed] [Google Scholar]
- 7.Saad AF, Hu W, Sood AK. Microenvironment and pathogenesis of epithelial ovarian cancer. Horm Cancer. 2010;1:277–90. doi: 10.1007/s12672-010-0054-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu C, Han HD, Mangala LS, Ali-Fehmi R, Newton CS, Ozbun L, et al. Regulation of tumor angiogenesis by EZH2. Cancer Cell. 2010;18:185–97. doi: 10.1016/j.ccr.2010.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smyth GK. Bioinformatics and Computational Biology: Solutions using R and Bioconductor. New York: Springer; 2005. Limma: linear models for microarray data. [Google Scholar]
- 10.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–8. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Komurov K, Dursun S, Erdin S, Ram PT. NetWalker: a contextual network analysis tool for functional genomics. BMC Genomics. 2012;13:282. doi: 10.1186/1471-2164-13-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wong GW, Knowles GC, Mak TW, Ferrando AA, Zuniga-Pflucker JC. HES1 opposes a PTEN-dependent check on survival, differentiation, and proliferation of TCRbeta-selected mouse thymocytes. Blood. 2012;120:1439–48. doi: 10.1182/blood-2011-12-395319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Serafin V, Persano L, Moserle L, Esposito G, Ghisi M, Curtarello M, et al. Notch3 signalling promotes tumour growth in colorectal cancer. J Pathol. 2011;224:448–60. doi: 10.1002/path.2895. [DOI] [PubMed] [Google Scholar]
- 14.Bellavia D, Checquolo S, Campese AF, Felli MP, Gulino A, Screpanti I. Notch3: from subtle structural differences to functional diversity. Oncogene. 2008;27:5092–8. doi: 10.1038/onc.2008.230. [DOI] [PubMed] [Google Scholar]
- 15.Chen X, Stoeck A, Lee SJ, Shih Ie M, Wang MM, Wang TL. Jagged1 expression regulated by Notch3 and Wnt/beta-catenin signaling pathways in ovarian cancer. Oncotarget. 2010;1:210–8. doi: 10.18632/oncotarget.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamamoto S, Charng WL, Bellen HJ. Endocytosis and intracellular trafficking of Notch and its ligands. Curr Top Dev Biol. 2010;92:165–200. doi: 10.1016/S0070-2153(10)92005-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meloty-Kapella L, Shergill B, Kuon J, Botvinick E, Weinmaster G. Notch ligand endocytosis generates mechanical pulling force dependent on dynamin, epsins, and actin. Dev Cell. 2012;22:1299–312. doi: 10.1016/j.devcel.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Le Borgne R, Bardin A, Schweisguth F. The roles of receptor and ligand endocytosis in regulating Notch signaling. Development. 2005;132:1751–62. doi: 10.1242/dev.01789. [DOI] [PubMed] [Google Scholar]
- 19.Kirchhausen T, Macia E, Pelish HE. Use of dynasore, the small molecule inhibitor of dynamin, in the regulation of endocytosis. Methods Enzymol. 2008;438:77–93. doi: 10.1016/S0076-6879(07)38006-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Masaike Y, Takagi T, Hirota M, Yamada J, Ishihara S, Yung TM, et al. Identification of dynamin-2-mediated endocytosis as a new target of osteoporosis drugs, bisphosphonates. Mol Pharmacol. 2010;77:262–9. doi: 10.1124/mol.109.059006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.