Abstract
A new era of cancer immunotherapy has brought not only successful cancer vaccines but also immunomodulators, such as those that target checkpoint blockade in order to induce endogenous host immune responses. However, the immune system of cancer patients can be compromised through multiple means, including immune suppression by the tumor and by prior therapies such as chemotherapy and radiation. Therefore, a comprehensive means of assessing patient immunocompetence would seem helpful for determining whether patients are ready to benefit from immunotherapy, and perhaps even which immunotherapy might be most appropriate for them. Unfortunately, there are no standardized tests for immune competence, nor is there agreement on what to measure and what will be predictive of outcome. In this review, we will discuss the technologies and assays that might be most useful for this purpose. We argue for a comprehensive approach that should maximize the chances of developing predictive biomarkers for eventual clinical use.
Keywords: CIMT 2013, Immunocompetence, Immune monitoring, Mass cytometry
Introduction
In 1909, Paul Ehrlich hypothesized that the immune system played an important role in controlling the growth of cancer [1]. This hypothesis was supported by the classic experiments of Prehn and Main [2], which showed that mice could generate immune responses to methylcholanthrene-induced sarcomas. Yet for many years, surgery, chemotherapy, and radiation were used for the treatment of cancer, with little ability to successfully harness host anti-tumor immunity. Controversy about tumor immunogenicity certainly delayed progress in immunotherapy. Most cancers, unlike the chemically induced sarcomas and the relatively rare virus-associated tumors, were considered non-immunogenic or poorly immunogenic [3]. However, key discoveries in the 1980s showed that vaccination could change the immunogenic profile of naturally occurring tumors and that an effective immune response was required for this to occur [4, 5]. While tumors undoubtedly are selected to minimize their antigenic profile, it is now appreciated that most, if not all cancers induce some altered or overexpressed host proteins that can potentially function as antigens. In fact, the number of recognized tumor-associated antigens has grown tremendously [6], to include not only viral proteins (e.g., HPV) and chromosomal translocation products (e.g., bcr/abl), but also over-expressed normal proteins such as HER2/neu, telomerase, and mucin-1, aberrantly expressed germ line or differentiation antigens, and mutated normal proteins.
It is also now apparent that endogenous immune responses to these antigens can be detected, albeit at low levels, in many (perhaps most) individuals with cancer. This was elegantly demonstrated in the work of Lee et al. [7] in melanoma patients, using MHC-peptide tetramers. Our group has also shown that breast cancer patients often have detectable responses to tumor antigens such as HER2/neu, CEA, and MAGE-A3 [8], as demonstrated by intracellular cytokine staining (ICS). However, many groups have also demonstrated ways in which endogenous immune responses to tumors are suboptimal. These can range from poor avidity of the T cells [9] to incomplete differentiation and restricted cytokine profiles [8]. Thus, the current generation of cancer immunotherapy seeks to overcome the inadequacies of endogenous tumor responses. This can be accomplished by supplying monoclonal antibodies to tumor antigens, bispecific antibodies that direct T cells to tumors, or engineered T cells that themselves target tumor antigens. In these cases, the host immune system is itself not directly altered, but immune molecules or cells are provided as an anti-tumor therapy. However, another approach is to target the immune system directly, rather than the tumor, by vaccination to induce stronger and more appropriate responses, by monoclonal antibodies inducing costimulatory pathways or inhibiting immune regulatory checkpoints, or by specific cytokine therapy to enhance immune responses, among other strategies.
Within the past decade, many of these immunotherapy strategies have been proven effective, either alone or in combination with each other or with traditional therapeutic modalities. In terms of direct anti-tumor therapies, there are approved antibodies targeting receptors over-expressed in tumor cells (Cetuximab, or anti-EGFR, for colorectal and head and neck cancers [10], and Rituximab, or anti-CD20, for lymphoma [11]). Adoptive T-cell transfer has proven to have dramatic results in tumor eradication with metastatic melanoma patients [12]. Some of the most recent therapies to show promise in clinical trials are bispecific T-cell engagers (BiTEs) (e.g., blinatumomab, or anti-CD3/anti-CD19 antibody, for lymphoma and leukemia [13, 14]). In terms of “true” immunotherapies (that target the immune system and not the tumor directly), there are now approved cancer vaccines (Provenge for prostate cancer [15]) and monoclonal antibodies for checkpoint blockade (ipilimumab, or anti-CTLA-4, for melanoma [16]). Anti-PD-1 and anti-PD-L1, as well as other checkpoint antibodies, are in late-stage clinical testing. Several agonist antibodies to costimulatory receptors (CD40, CD137, and OX-40) are also in clinical trials. Yet the improvement in cancer patients treated with these immunotherapies, although dramatic, does not extend to the majority of patients. In general, many cancer clinical trials still fail to show clinical benefit, and this is in part related to heterogeneity in the host genetic background, the tumor, and the immune system.
Monitoring immune competence
Most of the current immunotherapies work at least in part by activating the immune system to target the tumor. But without knowing the immune competence of the patient, such strategies may fail, or work only in a fraction of patients. Tumors are known to induce immune suppression; over a decade ago, CD4+ T-cell anergy was reported as a common early event in tumor progression [17]. Immune suppression after radiation [18] and chemotherapy [19] has long been documented in cancer patients. These findings help explain tumor progression in spite of the presence of tumor-specific T-cell populations. They also argue for investigating immune competence in cancer patients as a prerequisite to initiating immunotherapy.
The argument has been made that pre-selection of patients with favorable immune profiles could be beneficial to trials of cancer vaccines [20]. In fact, the concept of the “immunoscore” as a fourth axis of tumor staging (after tumor size, nodal involvement, and metastases) has been widely publicized [21–23]. As put forth by Galon et al., the “immunoscore” is derived by immunohistochemical quantitation of infiltrating CD4+ and CD8+ T cells in tumor biopsies. This is undoubtedly an important development for the field, as the number and ratio of these cells appear to be prognostic of clinical outcomes.
However, despite the development and recognition received for the immunoscore, much more is possible with regard to immune competence measurement. For one thing, many specific phenotypes of CD4+ and CD8+ T cells can participate in either enhancing or blocking tumor immunity. These include cytotoxic T cells (CTL), helper T cells (Th1, Th2, or Th17), and regulatory T cells (Treg). For each of these classes, there are of course many phenotypic and functional variants [24]. Expression levels of specific activating or suppressing receptor/ligand combinations associated with T-cell activation, such as PD1/PDL-1 [25], CD137/CD137L [26], or CTLA-4/CD80 [27, 28] can be important for immune competence. There is also growing appreciation for the role of NK cells [29], as well as macrophages [30], in anti-tumor immunity induced by specific immunotherapies. Dendritic cells, B cells, and antibody production also have the potential to play an important role in anti-tumor immunity, and myeloid-derived suppressor cells (MDSC) have received recent attention as correlating with clinical outcome [31, 32].
Thus, a more comprehensive approach to measuring immune competence is needed. One way to assess the immune system on a relatively global level is to perform in vitro stimulation with a mitogen (such as PMA + ionomycin) and quantitatively analyze the cellular phenotypes that produce key cytokines in response to the stimulation. A variation of this approach was used in an early study from our group, in which the frequency of CD4+ T cells producing IFNγ in response to the superantigen SEB was assessed [33]. Despite the small sample size and heterogeneity of responsiveness to SEB in healthy controls, patients with multiple myeloma had generally much lower responses than controls, suggesting they were mostly immune suppressed.
Another functional readout that could be used to measure immune competence is intracellular phosphosignaling [34]. Recent work from our group (Shen-Orr, submitted for publication) has found variable levels of defective pSTAT signaling in response to in vitro cytokine stimulation, in a subset of elderly individuals. These defects appear to be related to chronic inflammation.
The technology of choice: mass cytometry
Today, the readout of either cytokines or phosphoprotein signaling, in combination with cell phenotyping, is routinely done by multiparameter flow cytometry [34, 35], among other methods. However, the number of markers required to comprehensively subset immune cells, even at a fairly granular level, is on the order of 20 or so. Adding in a variety of either cytokine or phosphoepitope antibodies would bring the panel upwards of 30 markers or more. This is clearly beyond the ability of traditional multicolor flow cytometry.
Fortunately, a new generation of flow cytometry instrumentation, based on mass spectrometry readout of heavy metal ion-labeled probes, has been developed [36–38]. Termed “mass cytometry,” and supported by a commercial instrument (CyTOF, DVS Sciences, Toronto, CA), this technology has a twofold benefit over traditional fluorescence-based flow cytometry. First, the number of labels that can be measured per sample is greatly enhanced (presently about 40, but increasing regularly). Second, the “spillover” of signal from one detector channel to another is dramatically reduced, eliminating the need for calculating compensation matrices and reducing the noise associated with such spillover.
The first application of mass cytometry to large-scale immune profiling was carried out by Bendall et al. [39] using a panel of 34 cell surface and phospho-specific antibodies. Additional studies have used ICS, along with cell surface phenotyping, on this platform as well [40]. Extensive discussion of the platform’s benefits and caveats have been published elsewhere [38, 41] and will not be repeated here. However, it suffices to state that this platform is ideally suited, and in fact already proven, for readout of highly multiparameter phosphoepitope and intracellular cytokine analysis.
Our laboratory has recently built 31–39 parameter CyTOF panels for each of these types of applications (phosphoepitope and intracellular analysis). The antibodies in these panels are shown in Table 1. The intracellular cytokine panel is currently being used with PBMCs from cancer patients prior to and after immunotherapy. The cells are stimulated for 4 h with PMA + ionomycin, and cell phenotypes and functions are analyzed.
Table 1.
Antibody panels for mass cytometry using ICS and phosphoepitope analysis (phospho-CyTOF)
| CyTOF ICS | Phospho-CyTOF | ||||||
|---|---|---|---|---|---|---|---|
| Metal label | Specificity | Clone | Source | Metal label | Specificity | Clone | Source |
| 102–110Pd | barcoding (opt.) | 102-110Pd | barcoding (opt.) | ||||
| 113Cd | CD57 | HCD57, BioLegend | In house | 113Cd | |||
| 115In | Live/dead | 115In | Live/dead | ||||
| 139La | CD49d | 9F10, BioLegend | In house | 139La | |||
| 141Pr | CD45RA | HI100, BioLegend | In house | 141Pr | |||
| 142Nd | CD19 | SJ25C1, Southern Biotech | In house | 142Nd | CD19 | HIB19 | DVS |
| 143Nd | CD8 | SK1, BioLegend | In house | 143Nd | |||
| 144Nd | CD69 | MCA 2806, AbD Serotec | In house | 144Nd | |||
| 145Nd | CD4 | RPA-T4 | DVS | 145Nd | CD4 | RPA-T4 | DVS |
| 146Nd | Granzyme B | GB11, Abcam | In house | 146Nd | |||
| 147Sm | CD20 | 2H7 | DVS | 147Sm | CD20 | H1, BD | In house |
| 148Nd | MIP1ß | D21-1351, BD | In house | 148Nd | |||
| 149Sm | CD85j | 292319, R&D Systems | In house | 149Sm | CD7 | CD7-687, BioLegend | In house |
| 150Nd | CD45RO | UCHL1, BioLegend | In house | 150Nd | CD3 | UCHT1, BD | In house |
| 151Eu | CD38 | HB-7, BD | In house | 151Eu | CD123 | 9F5 | DVS |
| 152Sm | TNF | Mab11 | DVS | 152Sm | CD27 | 0323, BioLegend | In house |
| 153Eu | CD3 | UCHT1, BD | In house | 153Eu | CD45RA | H100 | DVS |
| 154Sm | CD107a | H4A3, BD | In house | 154Sm | CD45 (opt.) | HI30 | DVS |
| 155Gd | GMCSF | BVD2-21C11, BD | In house | 155Gd | |||
| 156Gd | CD94 | HP-3D9, BD | In house | 156Gd | pp38 | D3F9 | DVS |
| 157Gd | IL-2 | MQ1-17h12, eBioscience | In house | 157Gd | CD24 | ML5, BioLegend | In house |
| 158Gd | IFNγ | 4S.B3, BioLegend | In house | 158Gd | pSTAT3 | 4/P-Stat3 | DVS |
| 159 Tb | HLA-DR | G46-6, BD | In house | 159 Tb | CD11c | Bu15 | DVS |
| 160Gd | CD14 | M5E2 | DVS | 160Gd | CD14 | M5E2 | DVS |
| 161Dy | CD137 | 4B4-1, BD | In house | 161Dy | IgD | IA6-2, BioLegend | In house |
| 162Dy | IL-10 (indirect) | JES3-12G8, BioLegend | In house | 162Dy | pErk1/2 | 20A, BD | In house |
| 163Dy | CD137L | C65-485, BD | In house | 163Dy | IkBtot | L35A5, CST | In house |
| 164Dy | IL-17 | N49-653 | DVS | 164Dy | CD25 | M-A251, BD | In house |
| 165Ho | CD127 | A019D5, BioLegend | In house | 165Ho | pS6 | N7-548, BD | In house |
| 166Er | CD33 | P67.8, BD | In house | 166Er | CD16 | B73.1, BD | In house |
| 167Er | CD27 | L128, BD | In house | 167Er | CD38 | HIT-2 | DVS |
| 168Er | FcER1 | 9E1, Abcam | In house | 168Er | CD8 | SK1 | DVS |
| 169Tm | CCR7 | 150503, R&D Systems | In house | 169Tm | pSTAT1 | 4a, BD | In house |
| 170Er | PDL-1 | 10F.9G2, BioLegend | In house | 170Er | CD3 | UCHT1 | DVS |
| 171Yb | PDL-2 | TY25, BioLegend | In house | 171Yb | CD66 (opt.) | CD66a-B1.1 | DVS |
| 172Yb | PD1 | EH12.1, BioLegend | In house | 172Yb | pSTAT5 | 47/Stat5 (pY694), BD | In house |
| 173Yb | Perforin | B-D48, Abcam | In house | 173Yb | pPLCg2 | K86-68937, BD | In house |
| 174Yb | CD16 | 3G8, BioLegend | In house | 174Yb | HLA-DR | L243 | DVS |
| 175Lu | CD56 | NCAM16.2, BD | In house | 175Lu | CD56 | HCD56, BioLegend | In house |
| 176Yb | CD25 | M-A251, BD | In house | 176Yb | CD127 | A019D5 | DVS |
Analyzing the data for biomarkers
A complexity of the mass cytometry approach is how to analyze 40-parameter single-cell data. While one can certainly perform directed gating using bivariate dot plots, this soon becomes overwhelming and is likely to miss potentially key populations or responses. More global visualization approaches have been published, including SPADE [42], viSNE [43], and heat maps [44]. While SPADE performs clustering of related cells, and viSNE does not, both SPADE and viSNE display the data of a single file in a two-dimensional representation of relatedness. Cells (in viSNE) or clusters (in SPADE) are displayed as circles in a nearest neighbor analysis, where those most related to each other are grouped closest together.
Another analytical approach that has been applied to CyTOF data is principal components analysis (PCA) [40]. This is useful for showing differences between particular cell populations, e.g., influenza-specific versus CMV-specific T cells.
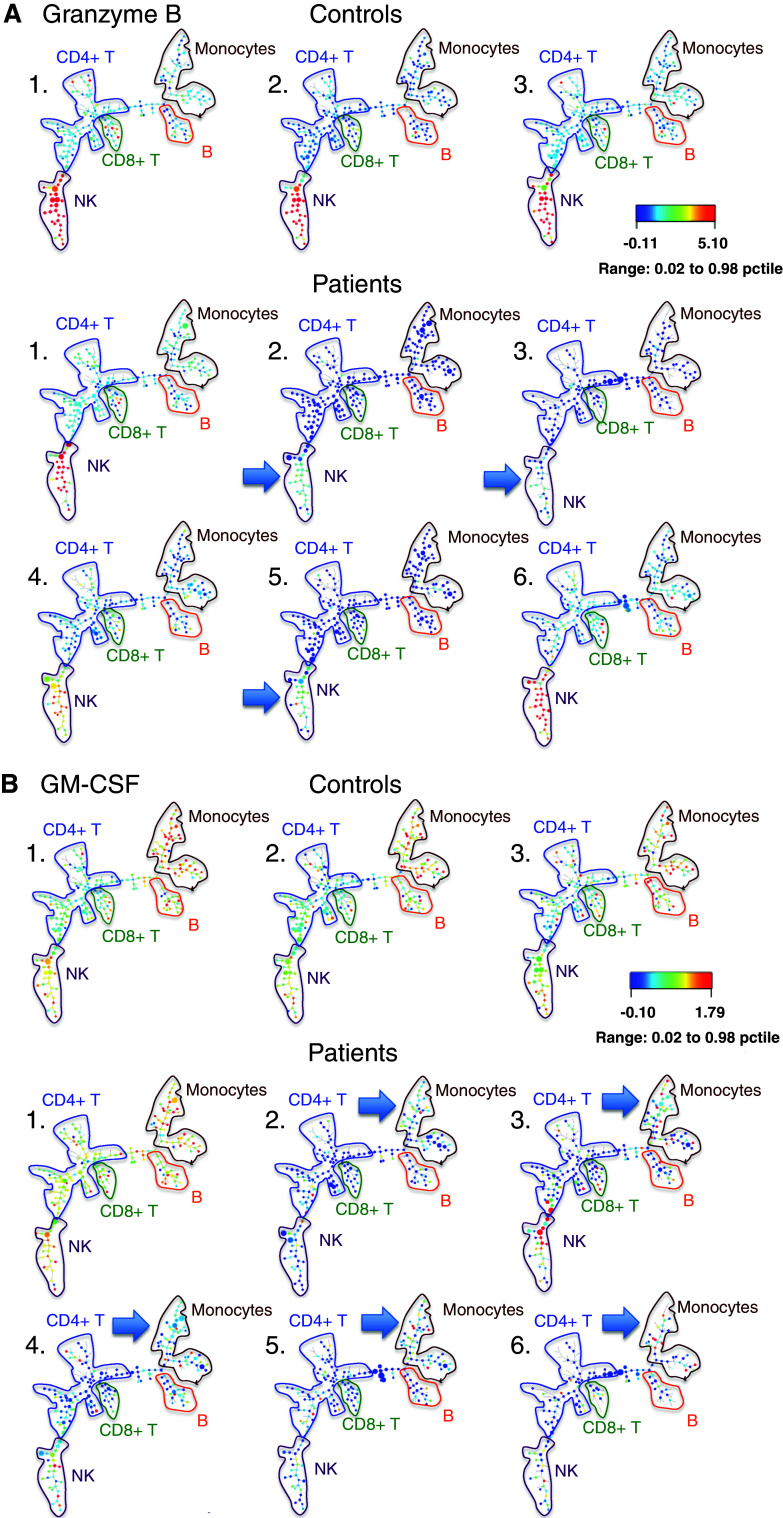
We have begun to use SPADE to compare the mass cytometry profiles of PMA + ionomycin-stimulated PBMCs of cancer patients and healthy controls (Fig. 1). A number of cytokines in stimulated baseline cancer patient samples (IL-17, IL-10, granzyme B, GM-CSF, CD107a) showed diverse maps compared to controls that had very homogeneous maps. This type of analysis suggests that the cancer patients are more heterogeneous in their immune profiles than controls, sometimes showing increases and sometimes decreases in specific cytokine responses. We therefore have some hope that there will be immune phenotypes that correlate with therapy outcome.
Fig. 1.
SPADE analysis of PMA + ionomycin-stimulated PBMCs from three healthy controls (top row) and six head and neck cancer patients (bottom row) prior to immunotherapy. Major cell lineages are annotated based on lineage marker expression. Cell clusters are colored for granzyme B (a), or for GM-CSF (b) median intensity (arbitrary units), and cluster size is proportional to cell number. Note the lack of granzyme B expression in NK cells and lack of GM-CSF expression in monocytes, in certain patients compared to controls (blue arrows)
A variety of informatics approaches will likely be needed to mine data such as the CyTOF profiles now being generated. Eventually, these should lead us to refine our assays, perhaps focusing in more detail on subsets and functions that are most promising. Finally, simpler assays than the CyTOF will undoubtedly be possible to design, and these simpler assays will likely be the ones that make it into clinical diagnostics, if there are to be any diagnostics of this kind. But at present we are still casting a wide net: we do not yet know which, if any, immune competence markers will correlate with clinical outcomes in particular cancer types; and we do not know how immune competence might best be modulated to achieve more successful cancer therapies.
Conclusions
We are clearly only at the beginning of developing the knowledge required to do effective patient prognosis on the basis of immune competence measurements. However, we have argued here that the new wave of cancer immunotherapies should greatly benefit from comprehensive immune competence measurement and that the platform of mass cytometry provides an ideal discovery tool for doing such measurements. With the first steps already being taken by the development of the immunoscore, we look forward to seeing more sophisticated tests of immune competence for cancer patients being applied in the near future. It then remains to be seen what biomarkers will emerge for prognosis of outcome or choice of immunotherapy modality.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
This paper is a Focussed Research Review based on a presentation given at the Eleventh Annual Meeting of the Association for Cancer Immunotherapy (CIMT), held in Mainz, Germany, 14–16 May, 2013. It is part of a CII series of Focussed Research Reviews and meeting report.
References
- 1.Ehrlich P. Uber den jetzigen Stand der Karzinomforschung. Ned Tijdschr Gneneeskd. 1909;53:273–290. [Google Scholar]
- 2.Prehn RT, Main JM. Immunity to methylcholanthrene-induced sarcomas. J Natl Cancer Inst. 1957;18:769–778. [PubMed] [Google Scholar]
- 3.Hewitt HB, Blake ER, Walder AS. A critique of the evidence for active host defence against cancer, based on personal studies of 27 murine tumours of spontaneous origin. Br J Cancer. 1976;33:241–259. doi: 10.1038/bjc.1976.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Van Pel A, Boon T. Protection against a nonimmunogenic mouse leukemia by an immunogenic variant obtained by mutagenesis. Proc Natl Acad Sci USA. 1982;79:4718–4722. doi: 10.1073/pnas.79.15.4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vessiere F, Georlette M, Warnier G, et al. Immunogenic variants obtained by mutagenesis of mouse Lewis lung carcinoma. Recognition of variant-specific antigens by cytolytic T lymphocytes. Eur J Cancer Clin Oncol. 1982;18:867–874. doi: 10.1016/0277-5379(82)90197-3. [DOI] [PubMed] [Google Scholar]
- 6.Vigneron N, Stroobant V, Van den Eynde BJ, van der Bruggen P. Database of T cell-defined human tumor antigens: the 2013 update. Cancer Immun. 2013;13:15. [PMC free article] [PubMed] [Google Scholar]
- 7.Lee PP, Yee C, Savage PA, et al. Characterization of circulating T cells specific for tumor-associated antigens in melanoma patients. Nat Med. 1999;5:677–685. doi: 10.1038/9525. [DOI] [PubMed] [Google Scholar]
- 8.Inokuma M, Rosa dela C, Schmitt C, et al. Functional T cell responses to tumor antigens in breast cancer patients have a distinct phenotype and cytokine signature. J Immunol. 2007;179:2627–2633. doi: 10.4049/jimmunol.179.4.2627. [DOI] [PubMed] [Google Scholar]
- 9.Rubio V, Stuge TB, Singh N, et al. Ex vivo identification, isolation and analysis of tumor-cytolytic T cells. Nat Med. 2003;9:1377–1382. doi: 10.1038/nm942. [DOI] [PubMed] [Google Scholar]
- 10.(2006) Cetuximab approved by FDA for treatment of head and neck squamous cell cancer. Cancer Biol Ther 5:340–342 [PubMed]
- 11.Reff ME, Carner K, Chambers KS, et al. Depletion of B cells in vivo by a chimeric mouse human monoclonal antibody to CD20. Blood. 1994;83:435–445. [PubMed] [Google Scholar]
- 12.Rosenberg SA, Yang JC, Sherry RM, et al. Durable complete responses in heavily pretreated patients with metastatic melanoma using T-cell transfer immunotherapy. Clin Cancer Res. 2011;17:4550–4557. doi: 10.1158/1078-0432.CCR-11-0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dreier T, Baeuerle PA, Fichtner I, et al. T cell costimulus-independent and very efficacious inhibition of tumor growth in mice bearing subcutaneous or leukemic human B cell lymphoma xenografts by a CD19-/CD3- bispecific single-chain antibody construct. J Immunol. 2003;170:4397–4402. doi: 10.4049/jimmunol.170.8.4397. [DOI] [PubMed] [Google Scholar]
- 14.Nagorsen D, Baeuerle PA. Immunomodulatory therapy of cancer with T cell-engaging BiTE antibody blinatumomab. Exp Cell Res. 2011;317:1255–1260. doi: 10.1016/j.yexcr.2011.03.010. [DOI] [PubMed] [Google Scholar]
- 15.Cheever MA, Higano CS. PROVENGE (Sipuleucel-T) in prostate cancer: the first FDA-approved therapeutic cancer vaccine. Clin Cancer Res. 2011;17:3520–3526. doi: 10.1158/1078-0432.CCR-10-3126. [DOI] [PubMed] [Google Scholar]
- 16.Wolchok JD, Hodi FS, Weber JS, et al. Development of ipilimumab: a novel immunotherapeutic approach for the treatment of advanced melanoma. Ann N Y Acad Sci. 2013;1291:1–13. doi: 10.1111/nyas.12180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Staveley-O’Carroll K, Sotomayor E, Montgomery J, et al. Induction of antigen-specific T cell anergy: an early event in the course of tumor progression. Proc Natl Acad Sci USA. 1998;95:1178–1183. doi: 10.1073/pnas.95.3.1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cosimi AB, Brunstetter FH, Kemmerer WT, Miller BN. Cellular immune competence of breast cancer patients receiving radiotherapy. Arch Surg. 1973;107:531–535. doi: 10.1001/archsurg.1973.01350220015005. [DOI] [PubMed] [Google Scholar]
- 19.Harris J, Sengar D, Stewart T, Hyslop D. The effect of immunosuppressive chemotherapy on immune function in patients with malignant disease. Cancer. 1976;37:1058–1069. doi: 10.1002/1097-0142(197602)37:2+<1058::AID-CNCR2820370813>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 20.Ogi C, Aruga A. Immunological monitoring of anticancer vaccines in clinical trials. Oncoimmunology. 2013;2:e26012. doi: 10.4161/onci.26012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ascierto PA, Capone M, Urba WJ, et al. The additional facet of immunoscore: immunoprofiling as a possible predictive tool for cancer treatment. J Transl Med. 2013;11:54. doi: 10.1186/1479-5876-11-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Galon J, Franck P, Marincola FM, et al. Cancer classification using the immunoscore: a worldwide task force. J Transl Med. 2012;10:205. doi: 10.1186/1479-5876-10-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Galon J, Pagès F, Marincola FM, et al. The immune score as a new possible approach for the classification of cancer. J Transl Med. 2012;10:1. doi: 10.1186/1479-5876-10-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mahnke YD, Brodie TM, Sallusto F, et al. The who’s who of T-cell differentiation: human memory T-cell subsets. Eur J Immunol. 2013;43:2797–2809. doi: 10.1002/eji.201343751. [DOI] [PubMed] [Google Scholar]
- 25.Badoual C, Hans S, Merillon N, et al. PD-1-expressing tumor-infiltrating T cells are a favorable prognostic biomarker in HPV-associated head and neck cancer. Cancer Res. 2013;73:128–138. doi: 10.1158/0008-5472.CAN-12-2606. [DOI] [PubMed] [Google Scholar]
- 26.Yan X, Johnson BD, Orentas RJ. Murine CD8 lymphocyte expansion in vitro by artificial antigen-presenting cells expressing CD137L (4-1BBL) is superior to CD28, and CD137L expressed on neuroblastoma expands CD8 tumour-reactive effector cells in vivo. Immunology. 2004;112:105–116. doi: 10.1111/j.1365-2567.2004.01853.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang W, Yu D, Sarnaik AA, et al. Biomarkers on melanoma patient T cells associated with ipilimumab treatment. J Transl Med. 2012;10:146. doi: 10.1186/1479-5876-10-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu L, Liu J, Chen X, et al. CTLA-4 gene polymorphism +49 A/G contributes to genetic susceptibility to two infection-related cancers—hepatocellular carcinoma and cervical cancer. Hum Immunol. 2010;71:888–891. doi: 10.1016/j.humimm.2010.05.023. [DOI] [PubMed] [Google Scholar]
- 29.Grobárová V, Benson V, Rozbeský D, et al. Re-evaluation of the involvement of NK cells and C-type lectin-like NK receptors in modulation of immune responses by multivalent GlcNAc-terminated oligosaccharides. Immunol Lett. 2013;156:110–117. doi: 10.1016/j.imlet.2013.09.009. [DOI] [PubMed] [Google Scholar]
- 30.Beatty GL, Chiorean EG, Fishman MP, et al. CD40 agonists alter tumor stroma and show efficacy against pancreatic carcinoma in mice and humans. Science. 2011;331:1612–1616. doi: 10.1126/science.1198443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walter S, Weinschenk T, Stenzl A, et al. Multipeptide immune response to cancer vaccine IMA901 after single-dose cyclophosphamide associates with longer patient survival. Nat Med. 2012;18:1254–1261. doi: 10.1038/nm.2883. [DOI] [PubMed] [Google Scholar]
- 32.Meyer C, Cagnon L, Costa-Nunes CM, et al. Frequencies of circulating MDSC correlate with clinical outcome of melanoma patients treated with ipilimumab. Cancer Immunol Immunother. 2013 doi: 10.1007/s00262-013-1508-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maecker HT, Auffermann-Gretzinger S, Nomura LE, et al. Detection of CD4 T-cell responses to a tumor vaccine by cytokine flow cytometry. Clin Cancer Res. 2001;7:902s–908s. [PubMed] [Google Scholar]
- 34.Perez OD, Mitchell D, Campos R et al (2005) Multiparameter analysis of intracellular phosphoepitopes in immunophenotyped cell populations by flow cytometry. Curr Protoc Cytom. Chapter 6: Unit 6.20. doi:10.1002/0471142956.cy0620s32 [DOI] [PubMed]
- 35.Maecker HT. Multiparameter flow cytometry monitoring of T cell responses. Methods Mol Biol. 2009;485:375–391. doi: 10.1007/978-1-59745-170-3_25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bandura DR, Baranov VI, Ornatsky OI, et al. Mass cytometry: technique for real time single cell multitarget immunoassay based on inductively coupled plasma time-of-flight mass spectrometry. Anal Chem. 2009;81:6813–6822. doi: 10.1021/ac901049w. [DOI] [PubMed] [Google Scholar]
- 37.Ornatsky O, Bandura D, Baranov V, et al. Highly multiparametric analysis by mass cytometry. J Immunol Methods. 2010;361:1–20. doi: 10.1016/j.jim.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 38.Tanner SD, Baranov VI, Ornatsky OI, et al. An introduction to mass cytometry: fundamentals and applications. Cancer Immunol Immunother. 2013;62:955–965. doi: 10.1007/s00262-013-1416-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bendall SC, Simonds EF, Qiu P, et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science. 2011;332:687–696. doi: 10.1126/science.1198704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Newell EW, Sigal N, Bendall SC, et al. Cytometry by time-of-flight shows combinatorial cytokine expression and virus-specific cell niches within a continuum of CD8+ T cell phenotypes. Immunity. 2012;36:142–152. doi: 10.1016/j.immuni.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bendall SC, Nolan GP, Roederer M, Chattopadhyay PK. A deep profiler’s guide to cytometry. Trends Immunol. 2012;33:323–332. doi: 10.1016/j.it.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simonds EF, Bendall SC, Gibbs KD et al (2011) Extracting a cellular hierarchy from high-dimensional cytometry data with SPADE. Nat Biotechnol 1–8. doi:10.1038/nbt.1991 [DOI] [PMC free article] [PubMed]
- 43.Amir E-AD, Davis KL, Tadmor MD, et al. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat Biotechnol. 2013;31:545–552. doi: 10.1038/nbt.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Newell EW, Sigal N, Nair N, et al. Combinatorial tetramer staining and mass cytometry analysis facilitate T-cell epitope mapping and characterization. Nat Biotechnol. 2013;31:623–629. doi: 10.1038/nbt.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]