Abstract
Hantaviruses have tri-segmented negative sense RNA genome. The viral M-segment RNA encodes a glycoprotein precursor (GPC), which is cleaved into two glycoprotein molecules Gn and Gc that form spikes on the viral envelope. We previously reported that Gn is degraded shortly after synthesis by the host autophagy machinery. However, Gn being an important integral component of the virion, must escape degradation during the packaging and assembly stage of virus replication cycle. The mechanism regulating the intrinsic steady-state levels of Gn during the course of virus replication cycle is not clear. We transfected cells with plasmids expressing viral S-segment RNA, nucleocapsid protein and glycoproteins Gn and Gc and monitored their expression levels over time. These studies revealed that accumulation of nucleocapsid protein, glycoprotein Gc and viral S-segment RNA helped to stabilize Gn. These observations suggest that initiation of virus assembly may help Gn to escape autophagic degradation by yet unknown mechanism
1. Introduction
Autophagy, originally considered as cellular response to starvation, is an intercellular mechanism required for both the maintenance of cellular homeostasis and generation of pools of energy under stress conditions for the proper functioning of essential cellular process until the normal growth conditions resume (14). Autophagy is a cytoplasmic quality control mechanism in which cytosolic components, such as defective or surplus organelles and aggregated proteins are sequestered into double membrane structures, referred as autophagosomes (3, 4). The autophagosomes serve as vehicles for the selective transport of sequestered cytoplasmic cargo to lysosomes for degradation and clearance. Perturbations to this crucial mechanism results in variety of disease conditions, ranging from infectious diseases to cancer (15). Autophagy begins with the formation of isolated membranes that eventually elongate in size and enclose the cytoplasmic cargo, destined for lysosomal degradation. This multistep process involves an array of genes and is associated with complex membrane dynamics (16). While maintaining the cellular homeostasis, this mechanism protects host cells from invading pathogens, including bacteria and viruses. Enforced expression of Beclin 1, an autophagy gene involved in early stages of autophagy induction, reduces viral titers and protects mice against Sindbis virus encephalitis (17). Recently, P62, an autophagy adaptor protein has been reported to interact with Sindbis virus nucleocapsid protein and target the viral nucleocapsids to autophagosomes for degradation (22). Plants and insects rely on innate and intrinsic immune mechanisms for protection against invading pathogens. Recent studies have suggested that autophagy likely increases the degradation of tobacca mosaic virus (TMV) in infected plant cells (14, 18). Similar to plants, autophagy has been reported to protect Drosophila from vesicular stomatitis virus (VSV) infection (27).
Although autophagy is an innate cellular mechanism protecting cells from the attack of invading pathogens such as viruses which, in turn, have evolved strategies to fight back this cellular response for survival and persistence. The three herpesvirus subfamilies (α, β and γ) and lentiviruses either encode proteins with autophagy inhibitory potential or induce cellular signals to inhibit autophagy. For example, α-herpesvirus HSV-1-encoded ICP34.5 protein antagonizes autophagy by direct binding to Beclin 1 (21). All γ-herpesviruses encode at least one or two homologues of Bcl2 to counter the clearance of virus-infected cells by the host surveillance machineries (13). At least two of these virus-encoded homologues, KSHV vBcl2 and murine γ-HV68 M1, have been reported to directly bind to Beclin 1 for the inhibition of autophagy (13, 14).
Interestingly a growing list of viruses seem to induce autophagy or autophagosome like structures in virus infected cells during the course of infection (14). It is likely that induction of autophagy by these viruses facilitates virus replication and pathogenesis in infected hosts. Epsetin–Barr virus encoded latent membrane protein 1 (LMP1) has been reported to induce autophgy to regulate its own expression levels in cells (13). Elevated levels of this protein inhibit mRNA translation and perturb cellular homeostasis (13). A number of RNA viruses, such as influenza A, Dengue and hepatitis C virus have been reported to induce autophagy like events in infected cells and require autophagosome like structures for efficient replication. Pharmacological or genetic interventions to inhibit autophagy have resulted in decreased yields of these viruses in infected cells (1, 13, 32). However, further investigations are required to delineate precisely the roles of autophagy proteins, double membrane vesicles and classical autophagic pathways in the virus infectious cycle.
Hantaviruses are negative strand RNA viruses and members of the Bunyaviridae family. Pathogenic hantaviruses including Sin Nombre virus (SNV) use avβ3 integrins on endothelial cells for entry (6). These category A viruses cause serious illness when transmitted to humans through aerosolized excreta of infected rodent hosts (25). Two well characterized diseases caused by hantavirus infection are the hemorrhagic fever with renal syndrome (HFRS) and hantavirus cardiopulmonary syndrome (HCPS), having mortalities of 15% and 50% respectively (24, 25). The organs that suffer the pathogenic consequences during HFRS and HCPS are kidneys and lungs, respectively. Although HFRS and HCPS differ in the final organ depicting the diseased pathology, both the diseases primarily cause vascular leakage that leads to shock and hypotension (19). Recently, factor XII-dependent increased activation of the Kallikrein-Kinin System has been proposed to induce endothelial cell permeability during hantavirus infection (29). Annually, 150,000 to 200,000 hantavirus infections are reported worldwide (12). The three hantaviral genomic RNA segments S, M and L encode nucleocapsid protein (N), glycoprotein precursor (GPC) and viral RNA dependent RNA polymerase (RdRp), respectively. The GPC is post-translationally cleaved at a conserved WASA-motif, generating an N-terminal fragment (Gn) and a C-terminal fragment (Gc). Both Gn and Gc are packaged into viral envelop during virus assembly. The 142 amino acid long C-terminal tail domain of Gn carries out multiple functions for the successful establishment of viral infection in infected cells. The interaction between Gn cytoplasmic tail domain and the N protein has been proposed to play a role in virus assembly (10, 30).
We have recently found that Gn expression in cells by either plasmid transfection or virus infection induces autophagy in cells. Interestingly, the activated autophagy response targets Gn for degradation by autophagy-lysosome pathway. The autophagic clearance of Gn was found to be required for efficient hantavirus replication in cells. Here, we demonstrate that co-expression of viral genomic RNA and hantavirus nucleocapsid protein in cells protect Gn from autophagic degradation. Although the virus infected cells are continuously undergoing degradative autophagy, it is still a mystery that how the elevated levels of nucleocapsid protein and viral genomic RNA facilitate the escape of Gn from autophagic degradation.
2. Results and discussion
To better understand the role of viral genomic RNA and nucleocapsid protein on the autophagic degradation of SNV Gn protein, we first provide an overview of our previously published results, followed by new data demonstrating that accumulation of viral genomic RNA and nucleocapsid protein in cells protect the degradation of Gn by the host autophagy machinery. To monitor the expression of SNV glycoprotein precursor (GPC) in cells, we previously fused the green fluorescence protein (GFP) at either N or C-terminus of the GPC and expressed the fusion protein in HeLa cells (11). The GFP expression was monitored by FACS analysis at increasing time intervals. Since both Gn and Gc are expressed from a single mRNA, it was expected that both proteins would be equally expressed in cells. Interestingly, we observed a negligible GFP signal in cells expressing GPC precursor containing an N-terminal GFP tag (11). In contrast, a significantly stronger GFP signal was observed in cells expressing GPC precursor containing a C-terminal GFP tag. To rule out the possibility that GFP fusion at the N-terminus of GPC precursor perturbed the GFP signal, we cloned Gn and Gc separately and expressed them in HeLa cells as GFP fusion proteins. Again, we observed a strong GFP signal in HeLa cells expressing Gc-GFP fusion protein in comparison to cells expressing Gn-GFP fusion protein (11). These observations suggested that Gn was likely degraded in cells, which was finally verified by western blot analysis using either anti-Gn or anti-GFP antibodies (11).
It has been previously reported that the cytoplasmic tail domain of Gn from New York hantavirus, expressed from a plasmid, is ubiquitinated and degraded by the proteasomal machinery in cells (7). Based on this information, we asked whether the cytoplasmic tail domain of SNV Gn is mediating the degradation of the entire Gn molecule in cells. Interestingly, we found that Gn mutant lacking the C-terminal tail domain was still degraded in cells similar to wild type Gn (11). To determine whether the wild type Gn and Gn mutant lacking the tail domain were degraded by the proteasome machinery, we added a proteasomal inhibitor (MG132) to cells four hours post-transfection and monitored the GFP signal over time. These studies revealed that degradation of both wild type and mutant Gn was not carried out by the proteasome, suggesting that signal required for the degradation of Gn is not present in its tail domain (11). In comparison, the MG132 treatment revealed that the cytoplasmic tail domain expressed separately from a plasmid was degraded by the proteasome machinery, consistent with similar observations from other investigators (7). However, a treatment with the autophagy inhibitors 3-methyladenine (3MA), LY-294002 (LY) or Wortmanin (Wort) showed that unlike the tail domain both wild type and mutant Gn were degraded by the host autophagy machinery (11). To rule out the possibility that GFP fusion at the N-terminus of Gn did not lead to the degradation of Gn in cells, we monitored the expression of both wild type Gn and GFP-Gn fusion proteins by western blot analysis using either anti GFP or anti-Gn antibody. This analysis revealed that both wild type Gn and GFP-Gn fusion proteins were degraded in cells to an undetectable level (11). However, addition of 3MA equally rescued both wild type Gn and GFP-Gn fusion proteins, suggesting that GFP tag at the N-terminus of Gn did not play any role in the degradation of Gn. The western blot analysis using anti-GFP antibody revealed that GFP tag was not cleaved from the GFP-Gn fusion protein by the N-terminal signal peptide of Gn (11). Our published results demonstrate that that addition of 3MA did not play any inhibitory role in the cleavage of N-terminal GFP tag. This is supported by the fact that no enhancement of GFP signal was observed in cells lacking 3MA. The cleaved GFP from the N-terminus of Gn in absence of 3MA would have easily been detected by either western blot analysis or fluorescence microscopy or FACS analysis, which was not the case. These observations suggest that the N-terminal signal peptide of wild type Gn is likely not cleaved after its translocation through the ER. Interestingly, it has been previously reported that N-terminal signal peptide of Hantaan virus Gn protein is cleaved in the mature protein (26), suggesting that Gn proteins from different hantavirus species may behave differently after expression. This is consistent with the significant difference in the N-terminal sequence of Gn from SNV and Hantaan virus.
We next found that Gn expression induced autophagosome formation in HeLa cells, consistent with the degradation of Gn by the autophagy (11). Using confocal imaging, we observed a strong colocalization of Gn and the autophagy marker LC3, which further supported the observation that Gn was degraded by the host autophagy machinery (11). An examination of P62 protein expression revealed that autophagic degradation was occurring in cells expressing Gn (11). Using siRNA we interrupted autophagy at both the nucleation and elongation phases by the simultaneous down-regulation of two critical genes, Bcln1 and ATG7, and monitored their effect upon the stability of Gn. Again, the inhibition of autophagosome formation significantly improved the stability of Gn (11). In addition, the use of lysosomal protease inhibitors E64d and pepstatin A (23) further confirmed that Gn was degraded by the autophagy-lysosome route.
We next wanted to determine whether autophagic degradation of Gn plays a role in hantavirus replication. Interestingly, we found that hantavirus infection rapidly induced autophagy in cells (11). We monitored SNV replication in Huh7 cells that were previously transfected with either control siRNA or siRNA against autophagy genes Bcln1 and ATG7 or treated with lysosome protease inhibitors E64d and pepstatin A. This experiment revealed that SNV replication was significantly inhibited by both of these treatments. It is highly likely that SNV replication was inhibited by the accumulation of Gn in virus infected cells.
Taken together these previously reported studies suggest that accumulation of Gn is likely inhibitory to virus replication and its clearance by the host autophagy machinery is required for the efficient virus replication in cells. However, it is unclear that why and how the accumulation of Gn, an important integral component of the virion inhibits virus replication. In addition, Gn is required at the later stages of virus replication cycle for the assembly of new virions. Thus, Gn must survive during the later stages of infection for proper assembly and production of new infectious progeny virions from the host cells. We hypothesize that a molecular switch exists between the autophagic degradative phase of Gn and the initiation of virus assembly phase at which Gn escapes autophagic degradation. It will be interesting to identify and characterize the molecular switch and define its regulation for maintaining the Gn homeostasis in cells during the course of virus infection.
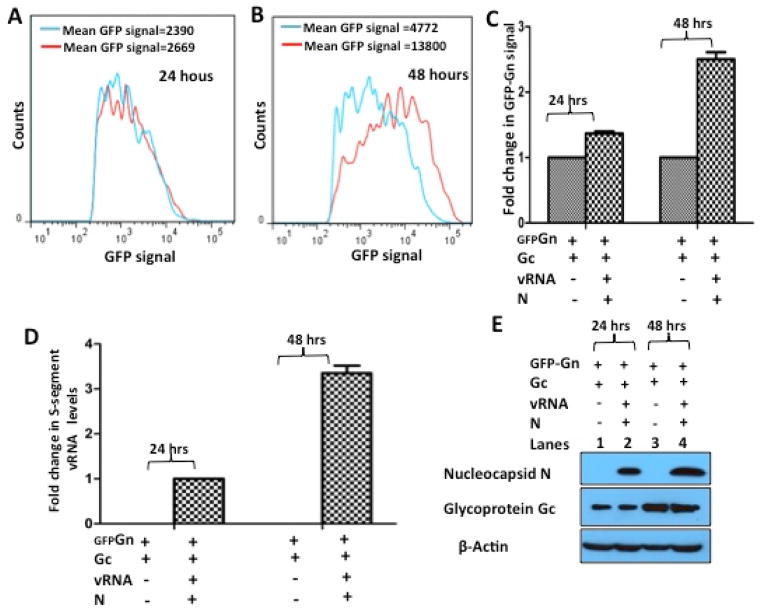
To gain insights into the hypothesis that initiation of virus assembly triggers the escape of Gn degradation from host autophagy machinery, HeLa cells in six well plates were co-transfected with plasmids expressing both Gn and Gc or Gn and Gc along with virus nucleocapsid protein and viral S-segment RNA. Each experiment was carried out in triplicates and repeated three times. In these co-transfection studies Gn was expressed as N-terminally fused GFP-Gn fusion protein. In contrast, both Gc and nucleocapsid protein were expressed as C-terminally His-tagged and HA-tagged fusion proteins, respectively. The plasmid expressing viral S-segment RNA was a gift from Antonoto Panganiban (University of New Mexico), it expresses S-segment vRNA from a Pol I promoter and nucleocapsid protein from a Pol II promoter. The two promoters are located opposite to each other on the plasmid holding the S-segment sequence in the middle. Cells were harvested 24 and 48 hours post transfection and examined for GFP-Gn expression by flow cytometry, as previously reported (11). As shown in Fig 1 (Panels A, B and C), there is a noticeable increase in GFP-Gn expression in HeLa cells at 48 hours post transfection due to the co-expression of N protein and viral S-segment RNA. To determine whether the increased expression of GFP-Gn fusion protein was due to the accumulation of viral S-segment RNA and nucleocapsid protein in cells at 48 hours post-transfection, the remaining cells from each experiment were lysed and total RNA was purified from one third of the lysate, using RNeasy kit (Promega). The purified RNA was reverse transcribed using a primer specific to the viral S-segment RNA and the resulting cDNA was used for quantitative estimation of S-segment RNA by real time PCR, as described in Materials and Methods. The remaining lysate was examined for the expression of Gc and N proteins by western blot analysis. It is evident from Fig 1D, that there is three fold increase in viral S-segment RNA levels in cells at 48 hour time point in comparison to 24 hour time point post-transfection. Similar increase in the intrinsic steady state levels of Gc and N proteins were also observed (Fig 1E).
Fig 1. The expression of Sin nombre hantavirus glycoprotein Gn increases due the accumulation of viral genomic RNA (vRNA), glycoprotein Gc and N protein in transfected cells.
HeLa cells grown in 6-well plates were co-transfected with 1μg of pTGFP-Gn and 1μg of pTGc along with either 2μg of empty vector or 1μg each of pADS and pSNV N TriEx 1.1. Each co-transfection was carried out in triplicates. Cells were harvested at 24 and 48 hours post-transfection and the expression of GFP-Gn fusion protein was determined by Flow cytometry. Overlays (A) and (B) represent GFP-Gn expression at 24 and 48 hours, respectively, in the presence (red) and absence (blue) of S-segment RNA and N protein. The experiment shown in panels A and B was carried out in triplicates, repeated three times and the results are shown in panel C. The total GFP-Gn expression in each replicate of an experiment was calculated as mentioned in Materials and Methods. The expression levels from three replicates of an experiment were averaged and the resulting average values from three independent experiments were further averaged and used for the calculation of standard deviation, shown as error bars. The expression levels of GFP-Gn fusion protein shown in the right bar at both 24 and 48 time points was normalized to the expression levels shown in the corresponding bar at the left (Panel C). The remaining cells from each flow cytometry experiment were collected and lysed. One third of the lysate was used for the purification of total RNA, as mentioned in Materials and Methods. The remaining lysate was used in western blot analysis. A total of 200 ng of purified RNA from each sample was used to generate cDNA for quantitative estimation of viral S-segment RNA by real time PCR (Panel D), as mentioned in Materials and Methods. Error bars represent the standard deviation from three independent experiments. Western blot analysis (Panel E) was carried out using either anti-HA tag antibody for N protein detection or anti His-tag antibody for the detection of glycoprotein Gc or anti-β actin antibody for the detection of β-actin, as internal control.
Since Gn is a membrane-associated protein, it is destined to enter endoplasmic reticulum (ER) after synthesis. Although the majority of Gn protein expressed in either transfected or virus infected cells has been reported to localize in Golgi, the SNV Gn protein has also been detected on the cell surface at later stages post-infection or post-transfection (28). These observations suggest that further study is required to verify whether SNV particles bud-off the Golgi or the plasma membrane. Although we observed that Gn is degraded by the host autophagy machinery in cells, it is still unclear how is Gn recognized in the ER membrane and selectively delivered to the host autophagy system for degradation. In addition, we previously reported that SNV infection immediately turns on the autophagy machinery of the infected host cell, which continues to stay on until later stages of virus replication cycle when mature virions are released (11). This observation invites interesting question that how Gn escapes degradation during packaging and assembly phase of the virus replication cycle in infected cells, which are continuously undergoing degradative autophagy. Since Gn enters ER after synthesis, it must be dislocated to cytosol for degradation by the autophagy machinery. The misfolded proteins in the ER are routinely dislocated into the cytosol and cleared in a series of events collectively referred as ER associated degradation (ERAD). The dislocated proteins in the cytosol are finally degraded by either the proteasome or the autophagy machinery depending upon the characteristics of the target protein such as, size, aggregation tendency and solubility etc. (5). It is possible that before the initiation of active packaging and assembly, the newly synthesized Gn accumulates in the ER, undergoes misfolding and is dislocated in the cell cytosol by the ERAD machinery. The dislocated Gn serves as a preferred autophagy target due to its tendency to form high molecular weight homo-tetramers (8), which are too big to be degraded by the proteasome.
The results shown in Fig 1 suggest that accumulation of viral genomic RNA, nucleocapsid protein and glycoprotein Gc facilitate the rescue of Gn from degradation. It is likely that accumulation of viral genomic RNA and nucleocapsid protein leads to the formation of nucleocapsids inside the cell. The resulting nucleocapsids might trigger the initiation of packaging and assembly, which in turn can facilitate the proper folding and rescue of Gn degradation. In addition, viral nucleocapsids have been proposed to directly interact with Gn (9, 31), suggesting that accumulated nucleocapsids might block the recognition of Gn by the autophagy adaptor molecule such as P62 protein. The failure of recognition due to the overwhelming abundance of nucleocapsids would also rescue Gn from degradation. This explanation is based on assumption that Gn contains a binding site for host autophagy adaptor.
An alternative explanation is that an intermediate molecule serves as a switch between the autophagic degradation phase during early stages of virus replication and survival of Gn during the assembly and packaging phase. The molecular switch regulates Gn expression levels in virus infected cells during the course of infection. The identification and characterization of such a molecular switch will provide more insights into the molecular mechanism of hantavirus pathogenesis.
3. Materials and Methods
3.1 Reagents
All Restriction enzymes were from New England Biolabs and all PCR primers were from Integrated DNA Technologies. Anti-His tag antibody was from Qiagen, anti-HA tag and anti-β actin antibodies were from Cell signaling Technologies. The real time PCR reagents were from Roche Diagnostics. All other reagents were from Sigma Aldrich.
3.2 Constructs
The plasmids pTGc and pTGFP-Gn were used for the expression of glycoproteins Gc and GFP-Gn fusion protein, respectively. The plasmids were constructed as previously reported (11). The plasmid pSNV N TriEx 1.1 was used for the expression of N protein in cells, as previously reported (20). The gene encoding the sin nombre hantavirus nucleocapsid protein was reverse transcribed from cells infected with hantavirus strain 77734, obtained from Brian Hjelle, University of New Mexico. The gene was cloned in pTriEX1.1 backbone (Novagen), as reported previously (20). The plasmid pADS was provided by Antonito Panganiban (University of New Mexico) and was used for the expression of SNV S-segment vRNA from pol I promoter. This plasmids contains S-segment gene in between Pol. I and Pol. II promoters and thus can express both S-segment genomic RNA and nucleocapsid protein in transfected cells.
3.3 Cells and Transfection
HeLa cells were maintained in Dulbecco’s modified eagle’s medium (DMEM) with 10% fetal bovine serum and 1% penicillin-streptomycin under 5% CO2 conditions. All transfections were done using Turbofect transfection reagent (Thermo Scientific) in a 6-well plate according to manufacturer’s instructions.
3.4 Western Blotting
The expression of GFP-Gn fusion protein, Gc and SNV-N in HeLa cells was determined by western blot analysis. Briefly, Hela cells were co-tranfected with either pTGFP-Gn and pT-Gc or pTGFP-Gn, pT-Gc, pSNV N TriEx 1.1 and pADS. Twenty-four and forty eight hours post tranfection, cells were washed with cold PBS and then lysed using radioimmunoprecipitation assay (RIPA) buffer containing protease inhibitor cocktail. Protein samples were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene difluiride (PVDF) membrane. The membrane was then blocked with blocking buffer (5% non-fat dry milk in 1X PBS containing 0.1% Tween 20) for 1 hour. Finally, the membrane was incubated overnight with primary antibody at 4 °C. Next day, membrane was washed three times with PBS containing 0.1%Tween 20, followed by further incubation with peroxidase conjugated secondary antibody for 1 hour at room temperature. The proteins of interest were detected with chemiluminescence detection reagents (Thermo Scientific) and the luminescence was detected by exposing the membrane to an X-ray film.
3.5 Real Time PCR
We used real time PCR analysis to quantitatively estimate the levels of viral S-segment RNA in cells at 24 and 48 hours post-transfection, as previously reported (11). Briefly, HeLa cells were co-transfected with either pTGFP-Gn and pTGC or pTGFP-Gn, pTGc, pSNV N TriEx 1.1 and pADS. Cells were harvested at either 24 or 48 hours post-transfection and total RNA was extracted using RNeasy kit (Qiagen), following manufacturer’s instructions. A total of 200ng of purified RNA from each sample was reversed transcribed using Moloney murine leukemia virus (MMLV) reverse transcriptase (Invitrogen) and a specific primer: 5’AGCACATTACAGAGCAGACGGGC3’ against viral S-segment RNA. Quantitative estimation of viral S-segment RNA level was carried out by Real Time PCR analysis using SYBR green PCR master Mix (Applied Biosystems). The primers used in Real Time PCR were forward primer: 5’GCAGACGGGCAGCTGTG3’ and reverse Primer: 5’AGATCAGCCAGTTCCCGCT3’. These primers have been previously validated and used for real time PCR analysis (2).
3.6 Flow Cytometry
Flow cytometry was used to examine the expression of GFP-Gn at different time points. Briefly, Hela cells were co-tranfected with either pTGFP-Gn and pTGC plasmids or pTGFP-Gn, pTGc, pSNV N TriEx 1.1 and pADS plasmids and the expression of GFP-Gn fusion protein was monitored by flow cytometry at 24 and 48 hours post transfection. The total GFP-Gn expression was calculated by multiplying the number of GFP positive cells with the mean fluorescence signal, as previously reported (11).
Highlights.
As previously reported that hantavirus nucleocapsid protein is degraded by the host autophagy machinery in cells
Autophagic degradation is required for efficient hantavirus replication
Accumulation of viral genomic RNA, glycoprotein GC and virus nucleocapsid protein facilitate the escape of glycoprotein from autophagic degradation
Acknowledgments
This work was supported by NIH grants RO1 AI095236-01 and 1R21 AI097355-01.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ait-Goughoulte M, Kanda T, Meyer K, Ryerse JS, Ray RB, Ray R. Hepatitis C virus genotype 1a growth and induction of autophagy. J Virol. 2008;82:2241–2249. doi: 10.1128/JVI.02093-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Botten J, Mirowsky K, Kusewitt D, Ye C, Gottlieb K, Prescott J, Hjelle B. Persistent Sin Nombre virus infection in the deer mouse (Peromyscus maniculatus) model: sites of replication and strand–specific expression. J Virol. 2003;77:1540–1550. doi: 10.1128/JVI.77.2.1540-1550.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deretic V. Autophagy in infection. Current opinion in cell biology. 2010;22:252–262. doi: 10.1016/j.ceb.2009.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deretic V. Autophagy of intracellular microbes and mitochondria: two sides of the same coin? F1000 biology reports. 2010;2 doi: 10.3410/B2-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ding WX, Yin XM. Sorting, recognition and activation of the misfolded protein degradation pathways through macroautophagy and the proteasome. Autophagy. 2008;4:141–150. doi: 10.4161/auto.5190. [DOI] [PubMed] [Google Scholar]
- 6.Gavrilovskaya IN, Shepley M, Shaw R, Ginsberg MH, Mackow ER. beta3 Integrins mediate the cellular entry of hantaviruses that cause respiratory failure. Proc Natl Acad Sci U S A. 1998;95:7074–7079. doi: 10.1073/pnas.95.12.7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geimonen E, Fernandez I, Gavrilovskaya IN, Mackow ER. Tyrosine residues direct the ubiquitination and degradation of the NY–1 hantavirus G1 cytoplasmic tail. J Virol. 2003;77:10760–10868. doi: 10.1128/JVI.77.20.10760-10768.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hepojoki J, Strandin T, Vaheri A, Lankinen H. Interactions and oligomerization of hantavirus glycoproteins. J Virol. 84:227–242. doi: 10.1128/JVI.00481-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hepojoki J, Strandin T, Wang H, Vapalahti O, Vaheri A, Lankinen H. Cytoplasmic tails of hantavirus glycoproteins interact with the nucleocapsid protein. The Journal of general virology. 2010;91:2341–2350. doi: 10.1099/vir.0.021006-0. [DOI] [PubMed] [Google Scholar]
- 10.Hepojoki J, Strandin T, Wang H, Vapalahti O, Vaheri A, Lankinen H. Cytoplasmic tails of hantavirus glycoproteins interact with the nucleocapsid protein. J Gen Virol. 91:2341–2350. doi: 10.1099/vir.0.021006-0. [DOI] [PubMed] [Google Scholar]
- 11.Hussein IT, Cheng E, Ganaie SS, Werle MJ, Sheema S, Haque A, Mir MA. Autophagic clearance of Sin Nombre hantavirus glycoprotein Gn promotes virus replication in cells. J Virol. 2012;86:7520–7529. doi: 10.1128/JVI.07204-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jonsson CB, Figueiredo LT, Vapalahti O. A global perspective on hantavirus ecology, epidemiology, and disease. Clin Microbiol Rev. 23:412–441. doi: 10.1128/CMR.00062-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ku B, Woo JS, Liang C, Lee KH, Hong HS, XE, Kim KS, Jung JU, Oh BH. Structural and biochemical bases for the inhibition of autophagy and apoptosis by viral BCL–2 of murine gamma–herpesvirus 68. PLoS pathogens. 2008;4:e25. doi: 10.1371/journal.ppat.0040025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kudchodkar SB, Levine B. Viruses and autophagy. Reviews in medical virology. 2009;19:359–378. doi: 10.1002/rmv.630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469:323–335. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liang XH, Kleeman LK, Jiang HH, Gordon G, Goldman JE, Berry G, Herman B, Levine B. Protection against fatal Sindbis virus encephalitis by beclin, a novel Bcl–2–interacting protein. J Virol. 1998;72:8586–8596. doi: 10.1128/jvi.72.11.8586-8596.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Y, Schiff M, Czymmek K, Talloczy Z, Levine B, Dinesh-Kumar SP. Autophagy regulates programmed cell death during the plant innate immune response. Cell. 2005;121:567–577. doi: 10.1016/j.cell.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 19.Macneil A, Nichol ST, Spiropoulou CF. Hantavirus pulmonary syndrome. Virus research. 2011;162:138–147. doi: 10.1016/j.virusres.2011.09.017. [DOI] [PubMed] [Google Scholar]
- 20.Mir MA, Panganiban AT. A protein that replaces the entire cellular eIF4F complex. EMBO J. 2008;27:3129–3139. doi: 10.1038/emboj.2008.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Orvedahl A, Alexander D, Talloczy Z, Sun Q, Wei Y, Zhang W, Burns D, Leib DA, Levine B. HSV–1 ICP34.5 confers neurovirulence by targeting the Beclin 1 autophagy protein. Cell host & microbe. 2007;1:23–35. doi: 10.1016/j.chom.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 22.Orvedahl A, MacPherson S, Sumpter R, Jr, Talloczy Z, Zou Z, Levine B. Autophagy protects against Sindbis virus infection of the central nervous system. Cell host & microbe. 2010;7:115–127. doi: 10.1016/j.chom.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sato K, Tsuchihara K, Fujii S, Sugiyama M, Goya T, Atomi Y, Ueno T, Ochiai A, Esumi H. Autophagy is activated in colorectal cancer cells and contributes to the tolerance to nutrient deprivation. Cancer Res. 2007;67:9677–9684. doi: 10.1158/0008-5472.CAN-07-1462. [DOI] [PubMed] [Google Scholar]
- 24.Schmaljohn CM. Molecular Biology of Hantaviruses. Plenum Press; New York: 1996. [Google Scholar]
- 25.Schmaljohn CS, Hooper JW. Bunyaviridae: The viruses and their replication. In: Howley Ka., editor. Fields Virology. Vol. 2. Lippencott, Williams, and Wilkins; Philadelphia: 2001. pp. 1581–1602. [Google Scholar]
- 26.Schmaljohn CS, Schmaljohn AL, Dalrymple JM. Hantaan virus M RNA: coding strategy, nucleotide sequence, and gene order. Virology. 1987;157:31–39. doi: 10.1016/0042-6822(87)90310-2. [DOI] [PubMed] [Google Scholar]
- 27.Shelly S, Lukinova N, Bambina S, Berman A, Cherry S. Autophagy is an essential component of Drosophila immunity against vesicular stomatitis virus. Immunity. 2009;30:588–598. doi: 10.1016/j.immuni.2009.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spiropoulou CF, Goldsmith CS, Shoemaker TR, Peters CJ, Compans RW. Sin Nombre virus glycoprotein trafficking. Virology. 2003;308:48–63. doi: 10.1016/s0042-6822(02)00092-2. [DOI] [PubMed] [Google Scholar]
- 29.Taylor SL, Wahl-Jensen V, Copeland AM, Jahrling PB, Schmaljohn CS. Endothelial Cell Permeability during Hantavirus Infection Involves Factor XII–Dependent Increased Activation of the Kallikrein–Kinin System. PLoS pathogens. 2013;9:e1003470. doi: 10.1371/journal.ppat.1003470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang H, Alminaite A, Vaheri A, Plyusnin A. Interaction between hantaviral nucleocapsid protein and the cytoplasmic tail of surface glycoprotein Gn. Virus Res. 151:205–212. doi: 10.1016/j.virusres.2010.05.008. [DOI] [PubMed] [Google Scholar]
- 31.Wang H, Alminaite A, Vaheri A, Plyusnin A. Interaction between hantaviral nucleocapsid protein and the cytoplasmic tail of surface glycoprotein Gn. Virus research. 2010;151:205–212. doi: 10.1016/j.virusres.2010.05.008. [DOI] [PubMed] [Google Scholar]
- 32.Zhou Z, Jiang X, Liu D, Fan Z, Hu X, Yan J, Wang M, Gao GF. Autophagy is involved in influenza A virus replication. Autophagy. 2009;5:321–328. doi: 10.4161/auto.5.3.7406. [DOI] [PubMed] [Google Scholar]