Abstract
In this study we report on potential drug-drug interactions between drugs occurring in patient clinical data. Results are based on relationships in SemMedDB, a database of structured knowledge extracted from all MEDLINE citations (titles and abstracts) using SemRep. The core of our methodology is to construct two potential drug-drug interaction schemas, based on relationships extracted from SemMedDB. In the first schema, Drug1 and Drug2 interact through Drug1’s effect on some gene, which in turn affects Drug2. In the second, Drug1 affects Gene1, while Drug2 affects Gene2. Gene1 and Gene2, together, then have an effect on some biological function. After checking each drug pair from the medication lists of each of 22 patients, we found 19 known and 62 unknown drug-drug interactions using both schemas. For example, our results suggest that the interaction of Lisinopril, an ACE inhibitor commonly prescribed for hypertension, and the antidepressant sertraline can potentially increase the likelihood and possibly the severity of psoriasis. We also assessed the relationships extracted by SemRep from a linguistic perspective and found that the precision of SemRep was 0.58 for 300 randomly selected sentences from MEDLINE. Our study demonstrates that the use of structured knowledge in the form of relationships from the biomedical literature can support the discovery of potential drug-drug interactions occurring in patient clinical data. Moreover, SemMedDB provides a good knowledge resource for expanding the range of drugs, genes, and biological functions considered as elements in various drug-drug interaction pathways.
Keywords: Drug-drug interactions, MEDLINE, SemRep, SemMedDB, Natural language processing, Clinical data, Semantic predication
1 Introduction
Translational informatics is a relatively new field that emerged to bridge the gap between biomedical research and clinical practice. This gap is exacerbated by the rapid growth of knowledge contained in the biomedical literature and the relatively slow manual access to this information due to its unstructured nature. The growing gap between scientific knowledge and clinical practice makes the tasks of translational informatics all the more important and urgent.
Electronic health records (EHR) are being used by clinicians as primary tools for documentation and communication in clinical practice, and this trend can be expected to continue. Clinical data contain highly personalized patient information that has the potential to be explored for clinical research and especially the complex care of patients with multiple and chronic disorders. Many of these more complex patients have a long list of medications with new drugs added, existing drugs removed, or medication doses adjusted frequently due to the nature of their conditions and the need for disease management (e.g., medication titration or changes for a poorly controlled hypertensive patient). Even a single drug can have a diverse effect profile in individual patients, so the combination of multiple drugs increases the possibility of unexpected effects. One possible reason for unexpected medication effects are potential interactions between drugs within a patient’s medication list. Such interactions can make the therapeutic effect of one or more prescribed medications weaker (or stronger) than intended or make side effects more pronounced.
Drug-drug interactions (DDIs) are a serious concern in clinical practice, as physicians strive to provide the highest quality and safety in patient care. While DDI lists are commonly used in clinical practice to alert clinicians during prescribing, some DDIs result from combinations or various mechanistic pathways that are not widely known. The traditional model for DDIs consists of considering the effect of one drug on a protein or other targets that are involved in the metabolism or transport of a second. The effect of the second drug may have the same target as the first drug or a different target [1]. This can be considered a direct interaction between the two drugs and many examples of this type of interaction affect cytochrome P450 metabolism [2–5]. Significant interactions may result beyond this traditional schema and can be extended to different genes being affected within the same biological pathway [6]. These interactions can also extend from pathway to biological processes for a particular clinical application. In other words, when two drugs are not linked through a specific gene network but target the same biological function, they can produce an effect at the clinical level that is not evident at the level of gene expression or protein interaction, especially by compounding an effect that can be induced through distinct pathways. For example, dehydration can be caused by failure of the colon to reabsorb water leading to diarrhea. Dehydration can also be caused by increased water output in the urine, or diuresis. If one drug in a patient’s medication list induces diarrhea while another is a diuretic, these effects would be compounded, increasing the risk of dehydration and its complications. To our knowledge, no previous attempt at identifying DDIs includes these clinical-level physiological effects in searching for DDI interactions.
DDIs can be identified through several approaches, including in vitro pharmacology experiments [7, 8], in vivo clinical pharmacology studies [8, 9], and pharmacoepidemiology studies [10]. However, these methods are limited by the need to focus on a small set of target proteins and drugs and are, therefore, slow to elucidate an exhaustive set of DDIs while new drugs are continually added into the pharmacopeia. Because they depend on these methods of DDI discovery and anecdotal clinical reporting, current DDI databases do not include all of the potential DDIs. However, some of these interactions may be indirectly derived from the scientific literature [11] or drug-related documents [12] through informatics methods. Thus, a powerful literature-based discovery (LBD) tool that can extract DDI information from the biomedical literature has the potential to significantly enhance patient care.
In this paper, we propose a system rooted in natural language processing (NLP) that can find potential DDIs existing in the clinical data of an individual patient based on the knowledge transferred from the biomedical literature. Specifically, our system extracts patients’ medication lists from clinical data, extracts all relevant semantic predications related to these medications from SemMedDB [13] (i.e., a database of semantic predications generated by SemRep [14]), and, thereby, suggests potential DDIs based on our DDI pathway schemas (i.e., drug1→gene→drug2, and drug1→gene1→biological function←gene2←drug2) and physician selection. Our methodology identifies potential patient-specific DDIs that are supported by evidence in the literature but are not contained in standard databases.
2 Related work
We propose using schemas describing relationships between drugs, genes, and physiological conditions to extract relationships from the literature that reflect DDIs. Previous investigation has explored methods of identifying these types of relationships. Weeber et al. developed a tool to systematically analyze online literature, which uses concept cooccurence frequency coupled with expert review to identify promising “pathways” (or schemas) between a drug and a potential new target disease [15]. Wren et al. created three-concept drug-gene-disease schemas of cooccurring concept pairs in MEDLINE records with overlapping gene terms that serve as intermediates in an implicit relationship between drug and disease [16]. Frijters et al. have also reported CoPub, which couples relationships determined by co-occurrence of biomedical keywords in literature, to predict new relationships between genes, drugs, pathways and diseases. They validated several predicted relationships by using either independent literature sources or biological experiments [17].
Several investigators have extracted DDIs using NLP [11, 18–21]. Most have focused on a specific set of genes, especially cytochrome P450s [21, 22], or a focused set of drugs [21]. Percha et al. similarly predicted novel DDIs through the drug-gene-drug relationships by using text mining techniques on MEDLINE abstracts, though these were not applied to clinical data as in our method [11]. Their effort is shown to be effective on a limited set of 731 genes, heavily enriched in P450s known to be involved in drug metabolism. In the process of relationship extraction, their consideration of verbs and nominalized verbs as the sole relationship candidates for drug and gene entities misses relationships that can be reported using less explicit language. They make note of the limitation of using a constrained set of genes and not capturing gene-gene intermediate interactions or biological functions such as those that we have incorporated [11].
Similarly, Duke et al. combined cytochrome P450-based potential DDIs from the biomedical literature with EHR data to identify DDIs that might increase the risk of myopathy [18]. Their approach was even more focused since the literature mining was restricted to a group of P450s. The investigators did, however, combine their results with clinical data and were able to find 5 drug pairs with significant relative risks, thereby demonstrating some of the potential impact of their methodology. See Uzuner et al. [23] for a discussion of the detection of semantic relations between medical concepts within the context of the i2b2-2010 Challenges.
Our methodology builds on prior approaches by integrating literature-derived interactions not only between drugs and genes but also between genes and biological functions and by using actual medication combinations occurring in clinical data. This together specifies the potential interactions to individual patients while allowing for more complex interactions through multiple genes and pathways involved in biological functions.
3 Background
This study relied on several publicly available NLP tools that have been developed at the National Library of Medicine (NLM) including MetaMap and SemRep.
3.1 MetaMap
MetaMap [24] is an NLP system that maps biomedical text to concepts in the Unified Medical Language System (UMLS) Metathesaurus [25]. MetaMap processes input text using a series of lexical/syntactic analyses, followed by variant generation, candidate identification, mapping construction, and word sense disambiguation. It provides multiple processing options that allow users to choose vocabularies and the data model, control the algorithms, and select the output formats. MetaMap has been widely used for many applications including information retrieval [26], relation extraction [27], text mining [22], question answering [28], and knowledge discovery [29]. In this study, we use MetaMap to map medication lists extracted from clinical data to UMLS concepts for further information extraction.
3.2 SemRep and SemMedDB
SemRep is a semantic interpreter also developed at NLM. SemRep relies on the UMLS SPECIALIST Lexicon [30] and MedPost part-of-speech tagger [31] for lexical and underspecified syntactic analysis and MetaMap to access domain knowledge in the UMLS Metathesaurus and extracts semantic relationships from the biomedical literature in the form of semantic predications. To enhance SemRep, work has been done to link to other structured resources, such as Entrez Gene [32], to recognize and normalize protein/gene names.
Each semantic predication (i.e., a subject-PREDICATE-object triple) consists of UMLS Metathesaurus concepts as arguments (i.e., subject and object) and a semantic relationship from an extended version of the UMLS Semantic Network as a predicate. The predicates cover clinical medicine (e.g., TREATS, DIAGNOSES), substance interaction (e.g., INTERACTS_WITH, STIMULATES), genetic etiology of disease (e.g., ASSOCIATED_WITH, CAUSES), and pharmacogenomics (e.g., AFFECTS, AUGMENTS) [33]. For example, SemRep interprets the biomedical text in (1) as semantic predications in (2):
Treatment with fenofibrate for 24 h significantly increased the expression of leptin and TSHr genes. (PMID: 15291748)
-
Fenofibrate STIMULATES Leptin
Fenofibrate STIMULATES Thyrotropin Receptor
Note that TSHr is normalized to the UMLS Metathesaurus concept Thyrotropin Receptor, and the verb “increased” to the predicate STIMULATES.
The SemMedDB database [13] used for this study was generated by SemRep from all MEDLINE citations published as of June 30, 2012. The database contains over 21 million citations and 119 million sentences. About 57.6 million predication instances (12.9 million unique predications involving 58 predicates) were extracted from 37.2 million MEDLINE sentences. The database maintains links from each predication to its original sentence and PubMed entry through its PMID. It also contains the positional information regarding arguments and predicates in a given sentence.
4 Methods
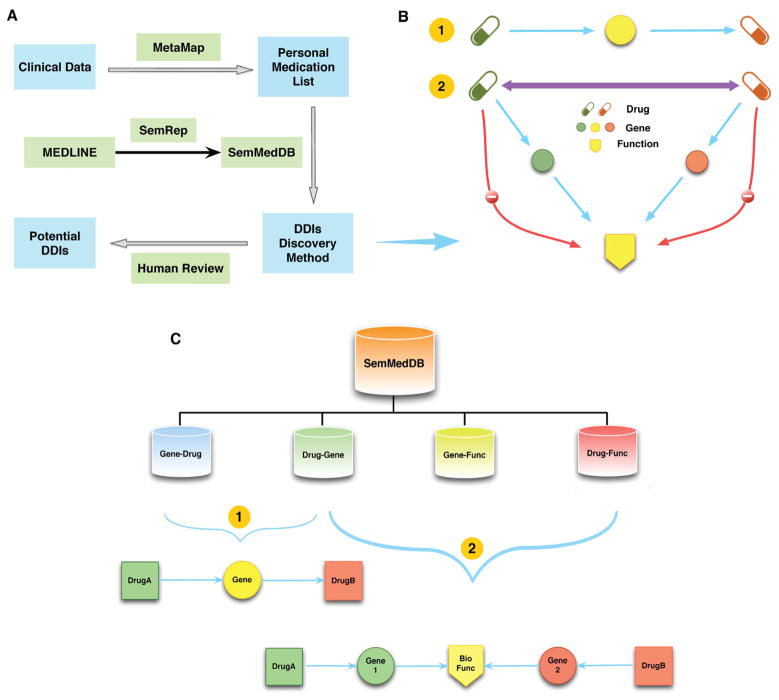
Our approach (Figure 1A) included six basic components: 1) extracting the personal medication list from clinical data and mapping to UMLS using MetaMap, 2) extracting all semantic predications relevant to these medications and the genes and biological functions that they affect from SemMedDB, 3) normalizing gene names to approved gene symbols, 4) discovering all possible DDIs based on combinations of semantic predications according to pathway schemas, 5) providing potential unknown DDIs after human review and exclusion of known DDIs, 6) evaluating semantic predications. These components are achieved through a series of steps detailed below.
Figure 1.
A) Overview of DDI discovery system. B) DDI discovery methodology for two drugs (details depicted in Step 5). Blue lines indicate allowed interactions. Red lines are prohibited. Purple double-arrowed line indicates the potential DDI. C) Semantic predication extraction process from SemMedDB.
Step 1: Medication list extraction
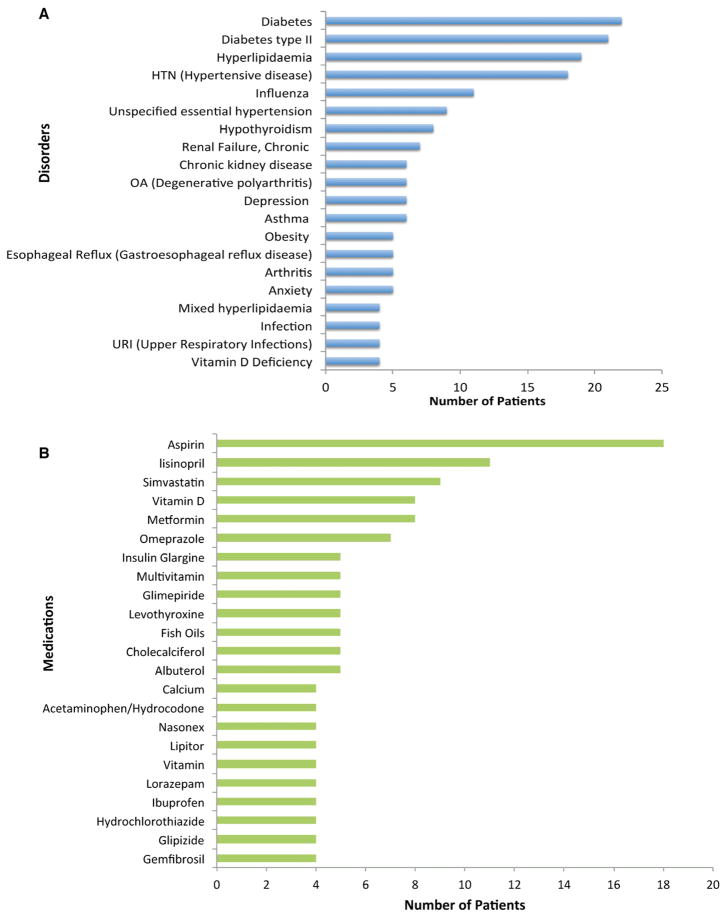
We randomly picked patient records from the Epic™ EHR system from 22 patients (13 females and 9 males) with outpatient clinical visits at Fairview Health Services (the integrated health system affiliated with the University of Minnesota Medical Center). The average age of patients was 63 with men slightly older than women, as shown in Figure 2. Most patients had chronic disorders (e.g., diabetes). Aspirin was the most commonly used medication in this cohort. Additional disorders and medication are depicted and ranked in Figure 3. The list of drugs was extracted from the EHR clinical data of each individual patient. Each drug was mapped to the appropriate concept in the UMLS Metathesaurus using MetaMap. The list of generic drug names for each office visit of an individual patient was used as the input to the DDI discovery system. A total of 224 unique drugs for all 22 patients were included in this study. Only interactions between two drugs with co-occurrence in the same patient record were examined.
Figure 2.
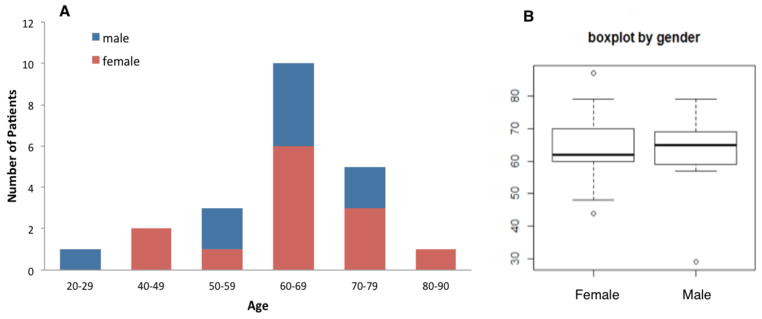
A) Histogram and B) boxplot of age ranges for female and male patients.
Figure 3.
Most common (A) disorders and (B) medications in patient notes.
Step 2: Predication extraction from SemMedDB (Figure 1C)
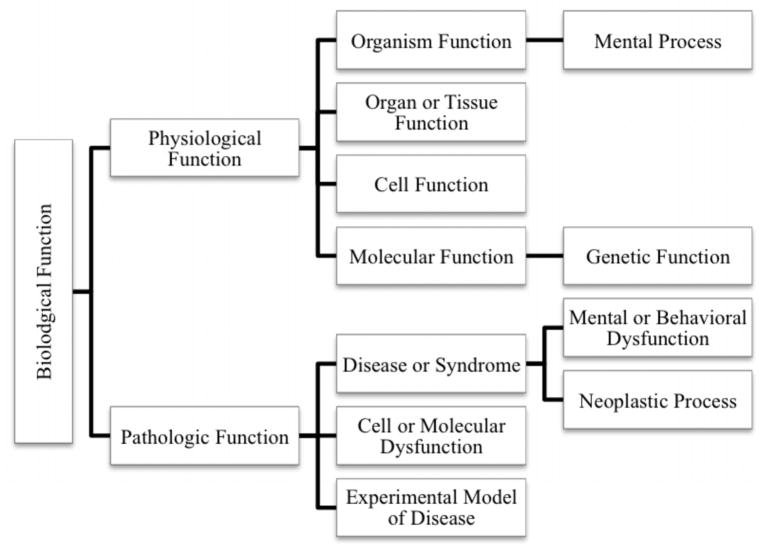
We extracted four types of predications from SemMedDB: drug-gene (i.e., predications with a drug as the subject and a gene as the object), gene-drug, gene-function, and drug-function. We extracted all predications describing an influence between a drug from the extracted medication list (Step 1) and a gene. Specifically, predications having a drug as the subject, a gene (including gene product or protein) as the object and three restricted predicate types (INHIBITS, INTERACTS_WITH, and STIMULATES) were extracted as drug-gene predications. The inverse of these, representing genes having an influence on drugs, with gene as the subject, drug as the object, and the same three predicate types were extracted as gene-drug predications. To identify genes influencing biological functions, we extracted gene-function predications with the genes as subjects, functions as objects, and predicate types including AFFECTS, AUGMENT, CAUSES, DISRUPTS, INHIBITS, and PREDISPOSES. Finally, to capture medications with an effect on biological functions, we generated drug-function predications having a drug as the subject and a function as the object. By function we are specifically referring to all descendants of the Biological Function semantic type in the UMLS semantic type hierarchy, as shown in Figure 4.
Figure 4.
Biological Function semantic types used for SemRep predications extraction.
Step 3: Gene name normalization
We used the downloadable Gene Nomenclature Committee dataset from HUGO [34] (the organization providing the international standard for gene names and symbols) as a dictionary to translate all gene and protein names to approved gene symbols (e.g., Sex Hormone-Binding Globulin to SHBG). 139 exact match genes were found in the gene-function predications from Step 2. Non-matches were usually non-specific terms such as classes of genes or proteins.
Step 4: DDI discovery pathways (Figure 1B)
Drug1→Gene→Drug2 (DGD) pathway (Figure 1B-1). We identified the potential DDIs between each pair of drugs in the medication list from clinical data for an individual patient using the drug-gene and gene-drug predications previously extracted in Step 2. Potential DDI candidates were generated by matching the object gene in a drug-gene predication with the subject gene in a gene-drug predication, requiring a different drug in each of the predications.
-
Drug1→Gene1→Biological_Function←Gene2←Drug2 (DGFGD) pathway (Figure 1B-2). We also identified the potential DDIs between each pair of drugs in the drug list using the drug-gene, gene-function, and drug-function predications. Potential DDI candidates were generated when the following constraints were satisfied:
-
1)
There exists a set of predications satisfying Drug1→Gene1→Biological_Function←Gene2←Drug2;
-
2)
There exists no direct Drug1→Biological_Function or Drug2→Biological_Function predications for this function.
-
1)
This ensures that both drugs have an independent effect on the function and the function is not an established effect of either drug.
Step 5: Physician selection of salient predications and exclusion of known DDIs
We first retrieved the MEDLINE sentences that produced DDI candidate pairs based on DGD and DGFGD pathways from SemMedDB and contained drug pairs found in the patient medication lists. One of the authors (MJC, a physician) then selected the most promising candidates from the chains of predications which matched each of the schemas, considering the validity of the component predications relative to their source sentence, method of administration, location of administration for drugs with low systemic distribution, and similar concerns. The initial set consisted of 1,029,271 chains from which 2,839 were randomly selected and shown to the physician for initial review. From these, 111 chains were selected as interesting. Characteristics of interesting chains included common usage, systemic penetration, and specificity (e.g., Ascorbic Acid over Vitamins). Gene products with wide distribution systemically or in multiple pathways were preferred (e.g., hormones over immune receptors). For biological functions, specificity was preferred over general groups of disorders (e.g., Mood Disorders, Cardiomyopathies). The predications in the 111 most interesting chains were provided to the physician with the source sentence for each and evaluated again in the context of the sentence. At this stage the review was focused on the accuracy of the predications. Additionally, whether the context was exceptional and unlikely to be relevant to real patients (versus cell culture or model organism) was also considered. This resulted in a selection of 54 predications and we then identified all of the chains matching the schemas that were composed solely of these selected predications. Each candidate DDI was also checked against Drugs.com [36] to identify known DDIs and those found in their database were discarded.
Step 6: Evaluation of predications
Although the physician’s selection in Step 5 included validation of the predications against the source sentences as part of the filtering process, we also performed a separate linguistic evaluation of the predication set. For this evaluation, we selected 300 sentences that assert substance interactions --including drug-gene and gene-drug interactions-- (200 sentences) or drug-biological function relations (100 sentences) and manually annotated them with relevant semantic predications. The annotation process consisted of two steps.
In the first step, three authors of this paper (GR, HK, MF) annotated 100 sentences (60 substance interactions, 40 drug-function relations) randomly selected from the 300-sentence set for predications that involve relevant predicates. The relevant predicates are STIMULATES, INHIBITS, INTERACTS_WITH for substance interactions and AUGMENTS, DISRUPTS, AFFECTS, CAUSES, and PREDISPOSES for drug-function relations. The annotation process followed the guidelines developed in a previous annotation study [35]. The main difference was that the annotators were asked to limit themselves to 2006AA UMLS concepts, since this UMLS release underlies the SemMedDB database. The gene/gene product equivalence criterion from that study was also followed. This first step allowed us to compute interannotator agreement to determine the reliability of the annotations. We adopted the F-measure among pairs of annotators as the interannotator agreement measure.a In a previous study [35], we had found that agreement among annotators was relatively low for the aforementioned predicates and we also assessed whether annotation experience gained in that study helped with interannotator agreement.
In the second step, based on the same guidelines and the discussions among annotators, one of the authors (GR) annotated the remaining 200 sentences and adjudicated 100 sentences from the previous step. This set of annotations (all 300 sentences) was taken as the gold standard for the linguistic evaluation.
In assessing SemRep performance, we used standard information extraction evaluation metrics of precision, recall, and F-measure for this purpose. We also calculated precision/recall curves by using argument distance of predications as the inclusion criterion. We limited the precision/recall curve calculation to verbal predicates only, since argument distance is most meaningful with such predicates due to SemRep’s underspecified argument identification algorithm.
5 Results
5.1 DDIs discovered through Drug1→Gene→Drug2 (DGD) pathway schema
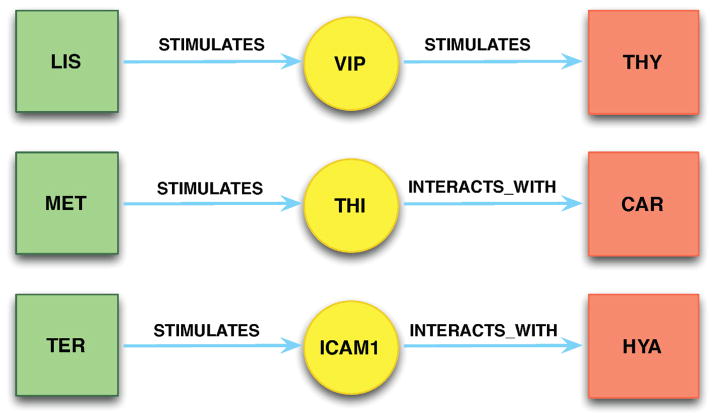
Using the DGD pathway schema (Step 4i), we found 14 unknown pairs of potentially interacting drugs (Table 1) in medication list of clinical data after physician selection of interesting predications (Step 5). Three DDI examples with corresponding citations are provided in Figure 5. The sentences that were used to generate three chains are listed in Table 2. The underlined words in sentences are extracted as subjects and objects in the predications. Highlighted words in the sentences indicate the relationships (predicates) between two biomedical concepts. In the first example, Lisinopril STIMULATES Vasoactive Intestinal Peptide (VIP), which in turn STIMULATES Thyroxine (both an endogenous hormone and given as a supplement for hypothyroidism). This chain indicates a potential DDI between lisinopril and thyroxine supplementation, suggesting that co-administration could lead to iatrogenic hyperthyroidism.
Table 1.
Potential DDIs extracted on DGD pathways.
| No. | Drug1 | → | Gene | → | Drug2 |
|---|---|---|---|---|---|
| 1 | Aspirin | INHIBITS | Epidermal Growth Factor | INTERACTS_WITH | Ascorbic Acid |
| 2 | Aspirin | INHIBITS | Epidermal Growth Factor | STIMULATES | Calcitriol |
| 3 | Aspirin | INHIBITS | Epidermal Growth Factor | STIMULATES | Hyaluronic Acid |
| 4 | Aspirin | INHIBITS | Epidermal Growth Factor | INTERACTS_WITH | Thyroxine |
| 5 | Insulin | INHIBITS | Glucagon | INTERACTS_WITH | Aspirin |
| 6 | Lisinopril | STIMULATES | Vasoactive Intestinal Peptide | STIMULATES | Thyroxine |
| 7 | Lisinopril | STIMULATES | Vasoactive Intestinal Peptide | INTERACTS_WITH | Ondansetron |
| 8 | Lovastatin | INHIBITS | Epidermal Growth Factor | INTERACTS_WITH | Thyroxine |
| 9 | Metformin | STIMULATES | Glucagon | INTERACTS_WITH | Aspirin |
| 10 | Metformin | STIMULATES | Sex Hormone-Binding Globulin | INTERACTS_WITH | Pioglitazone |
| 11 | Misoprostol | STIMULATES | Epidermal Growth Factor | INTERACTS_WITH | Thyroxine |
| 12 | Metoprolol | STIMULATES | Glucagon | INTERACTS_WITH | Aspirin |
| 13 | Simvastatin | STIMULATES | PLTP | INHIBITS | Vitamin E |
| 14 | Thyroxine | STIMULATES | Prolactin | INTERACTS_WITH | Clonidine |
Figure 5.
Three selected DDIs through Drug1→Gene→Drug2 pathway. (1) LIS = Lisinopril; VIP = Vasoactive Intestinal Peptide; THY = Thyroxine; (2) MET = Metformin; THI = Thioredoxin; CAR = Carvedilol; (3) TER = Terazosine; ICAM1 = Intercellular Adhesion Molecule 1; HYA = Hyaluronic Acid.
Table 2.
Predications and corresponding sentences that generated the chain in Figure 5. Arguments in sentences are underlined and predicate-indicating terms are bolded and italicized.
| Predication | Sentence (PMID) |
|---|---|
| Drug-Gene Relationships (alphabetic order of drug name) | |
| Lisinopril STIMULATES VIP | Increase in vasoactive intestinal polypeptides (VIP) by the angiotensin converting enzyme (ACE) inhibitor lisinopril in congestive heart failure. (PMID: 2822521) |
| Metformin STIMULATES Thioredoxin | Metformin increased Trx expression through the AMP-activated protein kinase (AMPK) pathway. (PMID: 20398632) |
| Terazosine STIMULATES Intercellular adhesion molecule 1 | Doxazosin, prazosin, and terazosin induced the expression of ICAM-1 and CD40 but had no effect on the expression of B7.1, B7.2, and CD40L. (PMID: 15614043) |
| Gene-Drug Relationships (alphabetic order of gene name) | |
| Intercellular adhesion molecule 1 INTERACTS_WITH Hyaluronic Acid | Effects of different hyaluronic acid products on synovial fluid levels of intercellular adhesion molecule-1 and vascular cell adhesion molecule-1 in knee osteoarthritis. (PMID: 15487709) |
| Thioredoxin INTERACTS_WITH Carvedilol | Among these proteins, actin in aortic smooth muscle (ACTA2), calmodulin, S100-A6, S100-A10, S100-A11, thioredoxin, lactadherin and heat-shock protein 105 kDa were found to be closely relevant with the clinical effects of Carvedilol. (PMID: 20403466) |
| VIP STIMULATES Thyroxine | Cch (10 microM) inhibits cellular cAMP accumulation and thyroxine (T4) release induced by vasoactive intestinal peptide (VIP), with or without a phosphodiesterase inhibitor. (PMID: 2829144) |
5.2 DDIs discovered through Drug1→Gene1→Biological Function←Gene2←Drug2 (DGFGD) pathway schema
Applying the DGFGD pathway schema (Step 4ii) to our predication set and the subsequent physician selection of 300 semantic predications (Step 5) yielded 48 unknown DDIs (Table 3). A selection of three pathways is detailed in Figure 6. When several drugs are the subjects of an otherwise consistent interaction, they are grouped together for clarity. In the left wing of the first pathway, Metformin STIMULATES gene Sex Hormone-Binding Globulin (SHBG), which CAUSES the function Benign Prostatic Hypertrophy (BPH). In the right wing of the chain, Cholecalciferol, Clonidine, and Sertraline each STIMULATES gene PRL which CAUSES the same biological function BPH. No interactions between Metformin and Cholecalciferol, Clonidine, Thyroxine, or Sertraline were reported in the DDI database, indicating four potential pairs of drug interactions as well as a compound effect by combining more than two of these medications. We also found examples of potential DDIs occurring through different pathways. For example, (Table 3, No. 33–38), Thyroxine can either INTERACTS_WITH Epidermal Growth Factor (EGF) which AUGMENTS Adipogenesis, or STIMULATES Prolactin (PRL) which CAUSES Adipogenesis. Each pair of drugs appeared in the same patient medication list. Some examples of the sentences and their extracted predications that generated these pathways are shown in Table 4. As in Table 2, the underlined words in sentences are extracted as subjects and objects in the predications and highlighted words in the sentences indicate the relationships (predicates) between two biomedical concepts.
Table 3.
Potential DDIs extracted from DGFGD pathway.
| No. | Drug1 | → | Gene | → | Biological Function | ← | Gene | ← | Drug2 |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Atorvastatin | STI | Insulin | AUG | Adipogenesis | AUG | EGF | INH | Aspirin |
| 2 | Cholecalciferol | STI | Prolactin | AUG | Adipogenesis | AUG | EGF | INH | Aspirin |
| 3 | Clonidine | STI | Prolactin | AUG | Adipogenesis | AUG | EGF | INH | Aspirin |
| 4 | Clonidine | STI | Prolactin | AUG | Adipogenesis | AUG | Insulin | STI | Atorvastatin |
| 5 | Fenofibrate | STI | Leptin | AFF | Neoplasm Metastasis | AUG | EGF | INH | Aspirin |
| 6 | Glimepiride | INT | Glucagon | CAU | Pancreatitis | CAU | EGF | INH | Aspirin |
| 7 | Irbesartan | STI | Leptin | AFF | Neoplasm Metastasis | AUG | EGF | INH | Aspirin |
| 8 | Irbesartan | STI | Leptin | PRE | Insulin Resistance | PRE | Glucagon | INT | Glimepiride |
| 9 | Insulin | INH | Glucagon | PRE | Insulin Resistance | PRE | Leptin | STI | Glyburide |
| 10 | Lisinopril | STI | VIP | CAU | Catecholamine Secretion | AUG | EGF | INH | Aspirin |
| 11 | Lisinopril | STI | VIP | CAU | Psoriasis | CAU | Prolactin | STI | Cholecalciferol |
| 12 | Lisinopril | STI | VIP | CAU | Psoriasis | CAU | Prolactin | STI | Clonidine |
| 13 | Lisinopril | STI | VIP | CAU | Hyperglycemia | PRE | Glucagon | INT | Glimepiride |
| 14 | Lovastatin | INH | EGF | AUG | Adipogenesis | AUG | Prolactin | STI | Cholecalciferol |
| 15 | Metformin | STI | Glucagon | CAU | Pancreatitis | CAU | EGF | INH | Aspirin |
| 16 | Metformin | STI | SHBG | CAU | BPH | CAU | Prolactin | STI | Cholecalciferol |
| 17 | Metformin | STI | SHBG | CAU | BPH | CAU | Prolactin | STI | Clonidine |
| 18 | Metformin | STI | Glucagon | PRE | Insulin Resistance | PRE | Leptin | STI | Irbesartan |
| 19 | Metoprolol | STI | Glucagon | PRE | Hyperglycemia | CAU | VIP | STI | Lisinopril |
| 20 | Misoprostol | STI | EGF | AUG | Adipogenesis | AUG | Prolactin | STI | Cholecalciferol |
| 21 | Misoprostol | STI | EGF | AUG | Catecholamine Secretion | CAU | VIP | STI | Lisinopril |
| 22 | Misoprostol | STI | EGF | CAU | Pancreatitis | CAU | Glucagon | STI | Metformin |
| 23 | Sertraline | STI | Prolactin | CAU | BPH | CAU | SHBG | STI | Metformin |
| 24 | Simvastatin | STI | Leptin | AFF | Neoplasm Metastasis | AUG | EGF | INH | Aspirin |
| 25 | Simvastatin | STI | Leptin | AFF | Neoplasm Metastasis | AUG | EGF | STI | Misoprostol |
| 26 | Simvastatin | STI | Leptin | CAU | Hypogonadism | CAU | Prolactin | STI | Cholecalciferol |
| 27 | Simvastatin | STI | Leptin | CAU | Hypogonadism | CAU | Prolactin | STI | Clonidine |
| 28 | Simvastatin | STI | Leptin | PRE | Insulin Resistance | PRE | Glucagon | INH | Insulin |
| 29 | Simvastatin | STI | Leptin | PRE | Insulin Resistance | PRE | Glucagon | STI | Metformin |
| 30 | Simvastatin | STI | Leptin | PRE | Insulin Resistance | PRE | Glucagon | STI | Metoprolol |
| 31 | Simvastatin | STI | Leptin | PRE | Insulin Resistance | PRE | Glucagon | INH | Exenatide |
| 32 | Simvastatin | STI | Leptin | PRE | Insulin Resistance | PRE | Glucagon | INT | Glimepiride |
| 33 | Thyroxine | INT | EGF | AUG | Adipogenesis | AUG | EGF | INH | Aspirin |
| STI | Prolactin | CAU | |||||||
| 34 | Thyroxine | INT | EGF | AUG | Adipogenesis | AUG | Prolactin | STI | Cholecalciferol |
| STI | Prolactin | CAU | |||||||
| 35 | Thyroxine | INT | EGF | AUG | Adipogenesis | AUG | Prolactin | STI | Clonidine |
| STI | Prolactin | CAU | |||||||
| 36 | Thyroxine | INT | EGF | AUG | Catecholamine Secretion | CAU | VIP | STI | Lisinopril |
| STI | Prolactin | CAU | Psoriasis | ||||||
| 37 | Thyroxine | INT | EGF | AUG | Adipogenesis | AUG | EGF | STI | Misoprostol |
| STI | Prolactin | CAU | |||||||
| 38 | Thyroxine | INT | EGF | AUG | Adipogenesis | AUG | Insulin | STI | Atorvastatin |
| STI | Prolactin | CAU | |||||||
| 39 | Thyroxine | INT | EGF | CAU | Pancreatitis | CAU | Glucagon | INH | Exenatide |
| 40 | Thyroxine | INT | Glucagon | PRE | Insulin Resistance | PRE | Leptin | STI | Glimepiride |
| 41 | Valsartan | STI | Leptin | AFF | Neoplasm Metastasis | AUG | EGF | INH | Aspirin |
| 42 | Vitamin B 12 | INT | EGF | AUG | Neoplasm Metastasis | AFF | Leptin | STI | Glyburide |
| 43 | Vitamin B 12 | INT | EGF | CAU | Pancreatitis | CAU | Glucagon | INH | Glyburide |
| 44 | Vitamin B 12 | INT | EGF | CAU | Pancreatitis | CAU | Glucagon | INH | Insulin |
| 45 | Vitamin D | STI | Prolactin | AUG | Adipogenesis | AUG | EGF | INH | Aspirin |
| 46 | Vitamin D | STI | Prolactin | CAU | Psoriasis | CAU | VIP | STI | Lisinopril |
| 47 | Vitamin D | STI | Prolactin | CAU | BPH | CAU | SHBG | STI | Metformin |
| 48 | Vitamin D | STI | Prolactin | CAU | Hypogonadism | CAU | Leptin | STI | Simvastatin |
STI, STIMULATES; AUG, AUGMENTS; INH, INHIBITS; AFF, AFFECTS; CAU, CAUSES; INT, INTERACTS_WITH; PRE, PREDISPOSES; EGF, Epidermal Growth Factor; VIP, Vasoactive Intestinal Peptide; SHBG, Sex Hormone-Binding Globulin; BPH, Benign prostatic hypertrophy.
Figure 6.
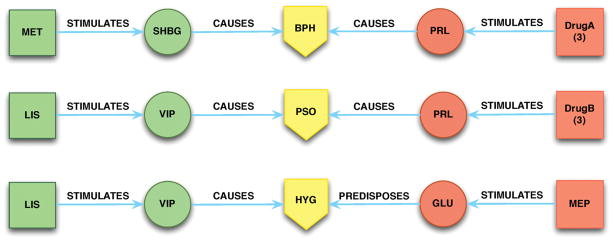
Three selected Drug1→Gene1→Function←Gene2←Drug2 pathways of potential DDIs. Only drugs co-existing in an individual’s medication list were selected. (1) MET = Metformin; SHBG = gene Sex Hormone-Binding Globulin; BPH = Benign Prostatic Hypertrophy (neoplastic process); PRL = gene Prolactin; DrugA = Cholecalciferol, Clonidine, or Sertraline; (2) LIS = Lisinoprol; VIP = Vasoactive Intestinal Peptide; PSO = Psoriasis; DrugB = Cholecalciferol, Clonidine, or Thyroxine; (3) HYG = Hyperglycemia; GLU = Glucagon; MEP = Metoprolol.
Table 4.
Predications and corresponding sentences that generated the chain in Figure 6. Arguments in sentences are underlined and predicate-indicating terms are bold and italic.
| Predication | Sentence (PMID) |
|---|---|
| Drug-Gene Relationships (alphabetic order of drug name) | |
| Atorvastatin SIMULATES Insulin | Atorvastatin increased fasting insulin, HOM-IR, NEFA and glycerol levels as well as reduced GIR. (PMID: 21889144) |
| Cholecalciferol SIMULATES Prolactin | Calcitonin inhibits and 1,25(OH)2-Vitamin D3 (1,25(OH)2D3) stimulates prolactin and thyrotropin secretion. (PMID: 2855317) |
| Clonidine SIMULATES Prolactin | Systemic (IV) administration of the alpha 2 receptor agonist clonidine is known to stimulate secretion of PRL and growth hormone (GH) suggesting a stimulatory role of the central alpha 2 receptors in the regulation of the two hormones. (PMID: 2575439) |
| Lisinopril SIMULATES VIP | Increase in vasoactive intestinal polypeptides (VIP) by the angiotensin converting enzyme (ACE) inhibitor lisinopril in congestive heart failure. (PMID: 2822521) |
| Metformin STIMULATES SHBG | Metformin also helps to increase SHBG, decrease androgen levels and induce ovulation. (PMID: 17426408) |
| Misoprostol SIMULATES EGF | Prostaglandin E1 and misoprostol increase epidermal growth factor production in 3D-cultured human annulus cells. (PMID: 19535298) |
| Sertraline SIMULATES Prolactin | RESULTS: Treatment with sertraline resulted in a comparable increase in prolactin secretion in male and female sheep. (PMID: 14634712) |
| Gene-Function Relationships (alphabetic order of gene name) | |
| Insulin AUGMENTS Adipogenesis | The biochemical mechanism by which insulin induces adipogenesis, converting fibroblast cells to adipocytes, is not clear. (PMID: 79227582) |
| Prolactin CAUSES BPH | The possible role of prolactin in the genesis of benign prostatic hypertrophy is discussed. (PMID: 6166478) |
| Prolactin CAUSES Psoriasis | Prolactin (PRL) may participate in the pathogenesis of psoriasis. (PMID: 19350575) |
| Prolactin AUGMENTS Adipogenesis | The lactogenic hormones (prolactin PRL and placental lactogen) also stimulate adipogenesis in preadipocyte cell lines but have variable lipolytic and lipogenic effects in mature adipose tissue. (PMID: 16735796) |
| SHBG CAUSES BPH | We present data that are consistent with a role for estradiol, and for a decrease in androgens and an increase in SHBG, in the pathogenesis of BPH. (PMID: 7515502) |
| VIP CAUSES Psoriasis | The imbalance of cutaneous VIP and SP and their disparate effects on the proliferation of normal human keratinocytes in culture would suggest that these peptides are involved in the pathogenesis of psoriasis and may exert different modulatory activities in the mechanisms underlying the psoriatic lesion. (PMID: 1372339) |
5.3 DDIs discovered through different pathway schema
By browsing the generated DDI results, we found five drug interactions that were included in both pathway schemas. For example, the interaction between Lisinopril and Thyroxine was found through DGD pathway (Table 1, No. 6) and DGFGD pathway (Table 3, No. 36). Others include Aspirin and Metformin (Table 1, No. 9; Table 3, No. 15), Aspirin and Thyroxine (Table 1, No. 4; Table 3, No. 33), Clonidine and Thyroxine (Table 1, No. 14; Table 3, No. 35), and Misoprostol and Thyroxine (Table 1, No. 11; Table 3, No. 37). The DGFGD pathway DDIs either provided further detail to the DGD pathway version (i.e., they contain the same gene in both wings) or they provided additional mechanisms of interaction (i.e., different genes in each wing).
5.4 Discovery of known DDIs
We checked all resulting drug interactions against the Drugs.com DDI database [36]. Three of the drug interactions discovered by DGD pathway schema were found in this database and 16 of the DDIs discovered by DGFGD pathway schema matched interactions in the database. All of the drug pairs identified as ‘known’ and their corresponding degree of severity are listed in Table 5.
Table 5.
Known drug-drug interactions found by our method.
| No. | Drug1 | Drug2 | Degree of interactions (provided by Drugs.com) |
|---|---|---|---|
| I. Drug1→Gene→Drug2 (DGD) pathway | |||
| 1 | Metformin | Carvedilol | moderate |
| 2 | Exenatide | Aspirin | moderate |
| 3 | Glimepiride | Aspirin | moderate |
| II. Drug1→Gene1→Biological function←Gene2←Drug2 (DGFGD) pathway | |||
| 1 | Metoprolol | Aspirin | minor |
| 2 | Thyroxine | Lovastatin | minor |
| 3 | Thyroxine | Metoprolol | minor |
| 4 | Thyroxine | Simvastatin | minor |
| 5 | Vitamin D | Misoprostol | minor |
| 6 | Insulin | Aspirin | moderate |
| 7 | Insulin | Citalopram | moderate |
| 8 | Lisinopril | Glycerol | moderate |
| 9 | Lisinopril | Insulin | moderate |
| 10 | Lisinopril | Exenatide | moderate |
| 11 | Metformin | Lisinopril | moderate |
| 12 | Sertraline | Aspirin | moderate |
| 13 | Thyroxine | Metformin | moderate |
| 14 | Vitamin D | Thyroxine | moderate |
| 15 | Exenatide | Aspirin | moderate |
| 16 | Glimepiride | Aspirin | moderate |
5.5 Annotation and evaluation of semantic predications
The number of predications annotated in the first phase of the annotation study and the agreement among annotators are provided in Table 6. The results show that an average of 2.22 predications were annotated per sentence, with a higher average (2.39) for substance interactions than that for drug-function relations (1.98). Agreement among annotators ranges from 0.650 to 0.753, with higher agreement for substance interactions than for drug-function relations (see Table 7). The distribution of the number of predications in the gold standard reference (by theme and by predicate type) is given in Tables 8 and 9, respectively.
Table 6.
Overall predication statistics for 100 sentences annotated by all three annotators
| Annotator | # of predications (per sentence) | Substance int. (per sentence) | Drug-function (per sentence) |
|---|---|---|---|
| GR | 236 (2.36) | 156 (2.60) | 80 (2.00) |
| HK | 208 (2.08) | 133 (2.22) | 75 (1.88) |
| MF | 223 (2.23) | 141 (2.35) | 82 (2.05) |
| Average | (2.22) | (2.39) | (1.98) |
Table 7.
Interannotator agreement
| Pair | Overall | Substance int. | Drug-function |
|---|---|---|---|
| GR-HK | 0.753 | 0.776 | 0.711 |
| GR-MF | 0.688 | 0.734 | 0.58 |
| HK-MF | 0.65 | 0.671 | 0.611 |
Table 8.
Gold standard reference
| Overall (per sentence) | Substance int. (per sentence) | Drug-function (per sentence) | |
|---|---|---|---|
| # predications | 689 (2.30) | 489 (2.45) | 200 (2.00) |
Table 9.
Distribution of predicate types in the gold standard reference
| Substance Interaction | Drug-Function | ||
|---|---|---|---|
| Predicate | # of predications | Predicate | # of predications |
| INTERACTS_WITH | 270 | AUGMENTS | 78 |
| STIMULATES | 111 | PREDISPOSES | 44 |
| INHIBITS | 108 | CAUSES | 36 |
| AFFECTS | 31 | ||
| DISRUPTS | 11 | ||
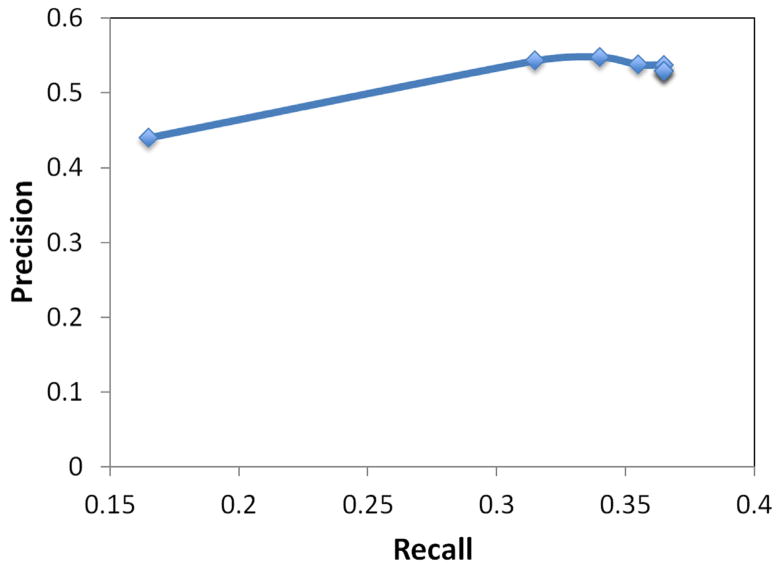
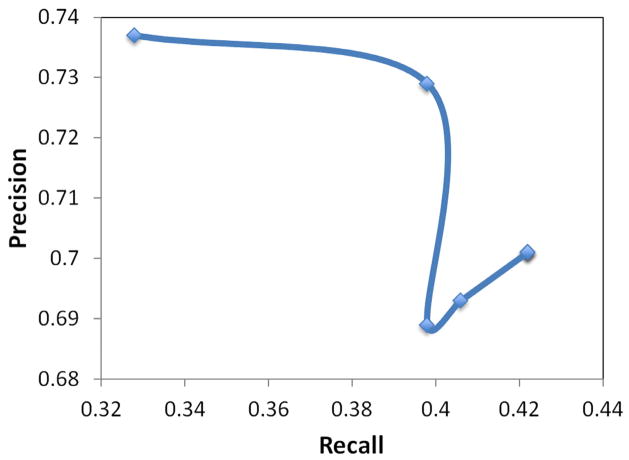
Evaluation was conducted on 300 sentences, from which SemRep extracted 524 relevant predications. Overall, 304 of these predications were deemed true positives, 220 false positives, and 384 false negatives, yielding precision of 0.580, recall of 0.442, and F-measure of 0.502. The evaluation metrics varied between the predication types: substance interactions (precision 0.585, recall 0.451, F-measure 0.509); drug-function interactions (precision 0.646, recall 0.420, F-measure 0.509). Additionally, variance in performance was noted between nominal and verbal predicates. For nominal predicates, SemRep precision was 0.574, recall 0.483, making F-measure 0.524. The F-measure for verbal predicates was lower at 0.473 with a slightly higher precision of 0.592 but much lower recall of 0.393.
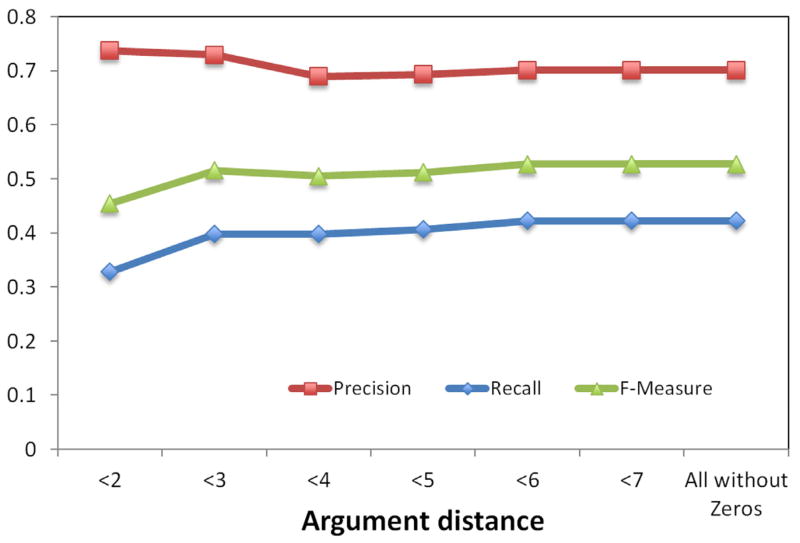
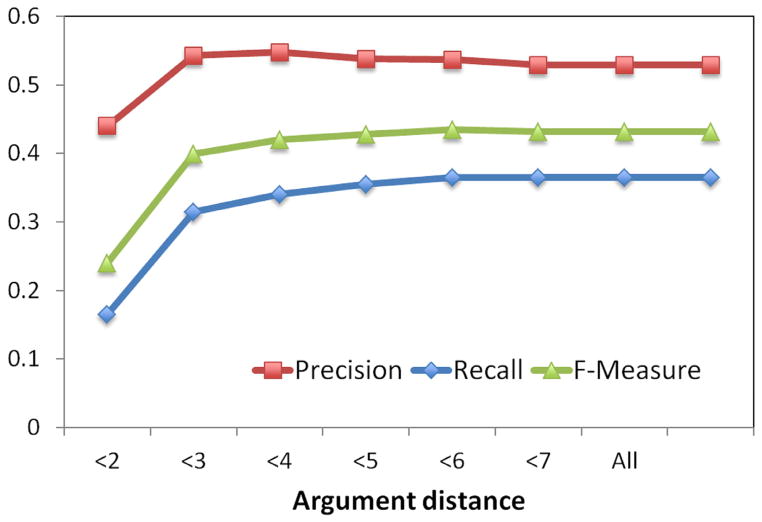
A variance in performance of verbal predicates by argument distance was also observed. A plot of precision, recall, and F-measure metrics for substance interactions and drug-function relationships are shown in Figures 7 and 9, respectively. Precision-recall curves for substance interactions and drug-function relationships are provided in Figures 8 and 10, respectively.
Figure 7.
Plot of precision, recall, and F-measure by argument distance for substance interaction predications with verbal predicates.
Figure 9.
Plot of precision, recall, and F-measure by argument distance for drug-function predications with verbal predicates.
Figure 8.
Precision-recall curve for substance interaction predications with verbal predicates.
Figure 10.
Precision-recall curve drug-function predications with verbal predicates.
6 Discussion
Our method of identifying drug-drug interactions from the biomedical literature is novel in two ways: (a) it makes use of the knowledge from the entire MEDLINE database (via semantic predications extracted by SemRep); and (b) it uses biological functions, as defined by the UMLS, in the definition of drug effects to allow for the detection of interactions that would not be noticed when considering only those linked through a single gene or protein. The use of SemMedDB predications expands the range of genes and drugs considered to all drugs contained in the UMLS and all genes and proteins within either the UMLS or Entrez Gene. The predications in SemMedDB are extracted from the complete set of biomedical citations contained in MEDLINE, and they are based on actual assertions in the text, not co-occurrence or similarity functions. The wide-open approach to drug list, gene list and drug interaction target offers more flexibility in finding potential interactions, while in its current incarnation our system can produce significant results even using a small clinical data set as done in this particular study.
6.1 Significance of results
An interesting example from the DGD pathways in Figure 5 is “Lisinopril STIMULATES VIP/VIP STIMULATES Thyroxine”. The source citation for the first predication (PMID: 2822521) asserts that lisinopril is found to induce increased plasma levels of vasoactive intestinal peptide (VIP). Although the stimulation of thyroxine in the second predication is referring to increased endogenous production (PMID: 2829144), clearly an increase in endogenous production could precipitate unwanted increases in plasma levels of thyroid hormone and repercussions of hyperthyroidism without adjustment of dosage administration.
The results shown in Figure 6 for the DGFGD pathway include “Lisinopril SIMULATES VIP/VIP CAUSES Psoriasis” coupled with “[Clonidine SIMULATES Prolactin, Cholecalciferol SIMULATES Prolactin, Sertraline SIMULATES Prolactin, Thyroxine SIMULATES Prolactin]/Prolactin CAUSES Psoriasis”. This demonstrates multiple drug-gene-function combinations having an effect on psoriasis. Their respective source citations (PMID: 19350575, 1372339) suggest that both VIP and prolactin are complicit in the pathogenesis of psoriasis. As noted above, lisinopril is noted to increase VIP (PMID: 2822521) while clonidine (PMID: 2575439), cholecalciferol (PMID: 2855317), and sertraline (PMID: 14634712) are all stated as increasing prolactin levels. Although the exacerbation of psoriasis is suggested for any of these medications individually, it is reasonable to conjecture that a combination of two or more could increase the likelihood and possibly the severity of psoriatic symptoms. It is not unlikely for patient to be taking a pair or multiple pairs of these medications. Indeed, the fact that this set is included in our results necessitates that at least one of the patients in our small set was taking all 5.
6.2 DDI network visualization
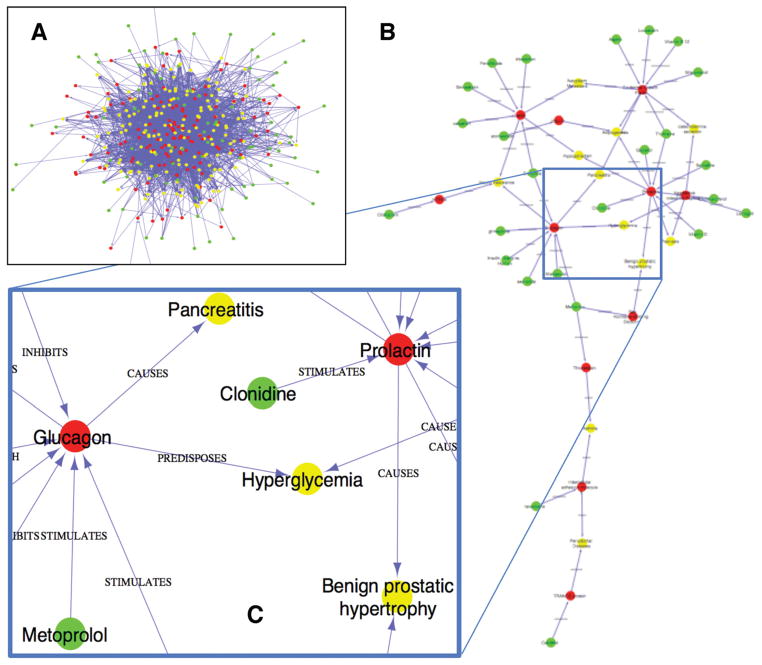
The dramatic reduction in network density that resulted from physician selection of significant relationships/predications is demonstrated in Figure 11. We used Cytoscape [37] to visualize a network of all DDIs obtained through the DGFGD pathway (box A). We also produced a focused DDI network after human review and known DDIs exclusion (box B).
Figure 11.
The DDI network (A) before and (B and C) after human review. Green, red and yellow dots represent drugs, genes and biological functions, respectively.
Before human review, the network of potential DDIs through the DGFGD pathway contains 620 nodes (including 192 drugs, 137 genes, and 291 biological functions) and 5,293 edges. The refined network, with 47 nodes (including 25 drugs, 11 genes and 11 biological functions) and 54 edges, is composed from the set of salient predications resulting from the physician selection process (Step 6). Details of this network provided in Figure 11 (box C) highlight some relationships provided as examples in Fig. 6.
6.3 Use of curated databases to validate drug interactions
It is common practice to use curated databases such as Drugs.com [36], DrugBank [38, 39], or PharmGKB [40] as a gold standard of interactions, but this deserves a note of caution. Although these may be extensive as a curated database of drug and gene interactions [41, 42], they do not contain all relationships contained in the literature as is true for all curated data sources, and are therefore, by definition, a subset of ‘known’ interactions. Therefore relationships that are not contained in the database may lead to true interactions being identified as false positives when using the database as a gold standard. Tari et al. demonstrate the pitfalls of using DrugBank as a gold standard with only 1.5–11.8% of interactions being found in the database though they manually validated 77.7–100% from supporting text [21].
6.4 Advantages of SemMedDB predications to find unknown DDIs
While interaction databases contain detailed information on gene and drugs based on published data, because these data are curated, the number of reviewed documents is subject to human limitations. SemMedDB contains not only interactions from the original documents reviewed in the generation of the databases but also millions of other documents not included in their formal review. This provides access to a wealth of knowledge that goes beyond the curated databases. The only limitation on which genes are included is whether they are contained in either the UMLS or Entrez Gene, which is used by SemRep as a supplementary resource for gene and protein terms. SemMedDB also contains predications regarding biological functions specifically linked to corresponding genes, thereby providing an additional framework for interactions beyond the traditional paradigm. Finally, SemRep considers a wider range of linguistic structures in identifying relationships; we take into account not only verbs and nominalizations, but also prepositions (e.g., in, for), allowing us to extract information that may not be retrievable through other NLP approaches.
6.5 Clinical usage: providing clinical information rather than giving alerts
Due to the theoretical nature of the extracted DDIs, this system is not intended to replace current approaches to DDI alert systems. There is a significant potential of inducing alert fatigue if the approach was to suggest any possible effect that may occur from all of the drug combinations in a patient’s medication list. Instead, we envision this system to suggest potential causes for unexplained patient symptoms or unusual responses to treatment. That is, by matching reported symptoms in the note or a search query to potential drug interactions in the same clinical record, we would be able to provide patient-specific, physician-sought information as opposed to an intrusive alert system.
Another potential application for this methodology is to facilitate curated database development. This system provides theoretical DDIs that can be investigated clinically to ascertain incidence and severity. Also, this mechanism could be incorporated into the development of new drugs, providing additional predictive capability for potential interactions.
Last, but not least, this approach may be used as a hypothesis generating step in the broader context of identifying potentially harmful drug-drug interactions. Hypotheses generated with this approach can then be tested either in in vivo (prospective patient observation) or in silico (prospective or retrospective examination of medical records) clinical studies.
6.6 SemRep analysis
6.6.1 Interannotator agreement analysis
Interannotator agreement rates in the range of 0.65–0.75 are considered acceptable, leading us to conclude that our annotations can be reliably used as reference for SemRep evaluation. The annotator responsible for adjudication and further annotation (of 200 sentences) had higher agreement with the other two annotators than those annotators had between themselves; therefore, it is also reasonable to say that the final annotations reflect the middle ground between all annotators.
At the predicate level, interannotator agreement rates are significantly higher than those obtained in our previous annotation study [35]. In that study, the agreement for all predicate types -- including but not limited to those discussed in this paper-- was found to be 0.536 between annotators GR and HK. The rate of agreement between the same two annotators is 0.753 in the current study. Even more interestingly, in the previous study, predicate types with lowest agreement were DISRUPTS (0.214), STIMULATES (0.238), and AFFECTS (0.308), all of which were considered in the current paper. We found that the agreement for these predicate types increased to 0.67, 0.80 and 0.60, respectively, in the current study. These results indicate that annotation experience leads to more consistent annotations over time. The fact that the current study was limited to a subset of the predicate types could have contributed to higher interannotator agreement as well; however, our study does not provide conclusive evidence for this.
6.6.2 SemRep errors
Analysis of SemRep output for this evaluation set indicated that errors generally fell into two broad categories: (a) shortcomings in the knowledge sources that SemRep depends on and (b) shortcomings in SemRep’s processing of linguistic phenomena.
A significant number of false positive errors were due to incorrect mappings of gene/protein mentions to UMLS concepts or Entrez Gene terms. The two most frequent mapping errors involved Ca2+, the calcium ion, mapping to the gene CA2 (n=12) and LDC-C, low-density lipoprotein cholesterol, mapping to the gene COG2 (n=7).
Incorrect argument identification constituted another source of false positive errors or erroneous predications. Failure to identify the correct arguments was due either to missing concepts in the UMLS or to SemRep processing errors, as in the following sentence (3) and semantic predication (4), in which ’Hypoglycaemia,’ is the subject of AUGMENTS predicate, rather than Insulin:
-
(3)
Hypoglycaemia induced by insulin increased catecholamine secretion, with the adrenaline to noradrenaline ratio significantly higher than in the adrenal gland itself. (PMID: 5152027)
-
(4)
Insulin AUGMENTS catecholamine secretion
Two major classes of linguistic phenomena addressed inadequately by SemRep are negation and serial coordination. In the following example, SemRep fails to recognize negation in sentence (5) and produces predication (6) that asserts its polar opposite:
-
(5)
The Val66Met polymorphism of the brain-derived neurotrophic factor gene is not associated with risk for schizophrenia and tardive dyskinesia in Han Chinese population. (PMID: 20395113)
-
(6)
Brain-derived neurotrophic factor PREDISPOSES Schizophrenia
On the other hand, from sentence (7) SemRep extracts an erroneous predication (8) because it is unable to recognize that both subject and object arguments (Ro 25-1553 and vasoactive intestinal peptide (VIP) respectively) are coordinated elements within the same coordination structure.
-
(7)
Studies were conducted to compare the effect of native vasoactive intestinal peptide (VIP), Ro 25-1553 (a cyclic peptide analog of VIP) and salbutamol (a beta2-adrenoceptor agonist) on antigen-induced pathophysiological effects in the guinea pig. (PMID: 7932181)
-
(8)
Ro 25-1553 INTERACTS_WITH Vasoactive Intestinal Peptide
We used a rather strict criterion for validation of the extracted predications, requiring that the relationship be clearly asserted in the source sentence from the text to be considered a true positive. Inferences were considered to be false positives. This was especially relevant in titles. For example, from the title below (9) SemRep produces a predication (10) that may be true but it is not fully supported in the title.
-
(9)
Melanoma risk in association with serum leptin levels and lifestyle parameters: a case-control study (PMID: 17925285)
-
(10)
Leptin PREDISPOSES Melanoma
We identified this as a false positive because there is no explicit assertion in (9) that there is indeed increased melanoma risk with increased leptin levels. There may be no difference in risk, the risk may be increased or decreased, or the leptin level that is associated with greater risk may be an increase or decrease from normal.
We note that this rejection of inference may not be standard in similar work. For instance, the relationship (12) extracted from the sentence (11) by the system reported in Percha et al. [11] would not be considered a true positive by our system:
-
(11)
How atorvastatin could limit the pro-inflammatory response to thrombin was studied in cultured rat aortic smooth muscle cells. (PMID: 12921859)
-
(12)
Atorvastatin DECREASES F2 (i.e., the gene for Thrombin)
We would reject this assertion because there is no direct relationship asserted between atorvastatin and thrombin but, instead, to an inflammatory response to thrombin that does not necessitate that expression of the F2 gene or the activity of thrombin be decreased. Although such relations may be valid based on additional context from other sentences in the abstract or given prior knowledge, the source sentence itself is not sufficient to support this relationship.
6.6.3 Impact of argument distance
Although SemRep has been shown to perform well with pharmacogenomic relations [33], previous investigation has shown some indication that SemRep precision in predications related to genes can be improved by using only predications with low argument distance [44]. Argument distance is defined as the number of noun phrases between the predicate and the noun phrase selected as the argument of the predicate (subject or object).
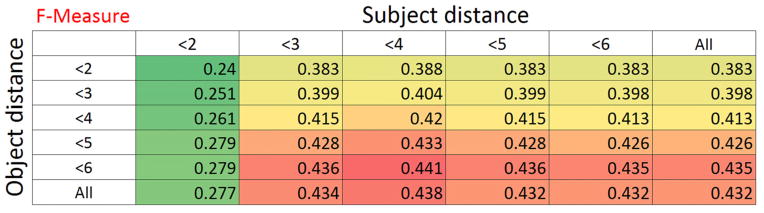
When we applied the same type of investigation using our gold standard reference, we found surprisingly different results. For substance interactions, although precision did increase when the maximum argument distance decreased from any size to 3, at smaller distances the trend was reversed and the lowest precision was noted at a distance of 1. Recall was much as expected, steadily decreasing as the distance threshold decreased. For drug-function predications performance was more in line with expectations as precision increased at distance thresholds below 4 and recall decreased generally as maximum distance was reduced. A surprising result of our analysis was that object distance had much more significance than subject distance. This is demonstrated in Figure 12. When subject distance is greater than 1, F-measure is generally consistent across all subject distance thresholds but steps up for each increase in object distance.
Figure 12.
Heatmap of F-measure for subject versus object argument distance for substance interaction predications with verbal predicates.
The increase in F-measure (and recall without a decline in precision) in high distances may seem counterintuitive. SemRep seems able to identify some of the long range dependencies between the indicators and the arguments due to its underspecified approach, leading to a precision increase at higher argument distances. Long range dependencies are a well-known feature of biomedical literature [43] and have, in fact, led to recent popularity of dependency parsing based approaches to relation extraction from biomedical literature.
6.7 Limitations and future work (application to larger clinical data set)
Our method currently depends on expert review of automatically extracted assertions. This is due in part to a low level of precision in this knowledge domain and expansion of SemRep capabilities continues to be a target for improvement, but at the same time it conveys a benefit in that it allows the expert to determine which types of interactions are most relevant to the patients in their care. After a semantic predication has been selected as interesting, it can be stored and it need not be judged again for the same user/care group combination, thereby allowing its incorporation into any additional relevant pathways.
Because this approach combines two separate assertions linked through a gene or biological function and does not search for stated drug-drug interactions directly, it produces theoretical DDIs that may not have been recognized before nor verified through clinical trials. Application of this methodology to a larger clinical dataset would not only allow for access to a greater range of theoretical DDIs, but resulting biological functions from the proposed interactions could be compared against presence of signs and symptoms in the clinical record to validate and determine incidence of the DDIs. Web-based healthcare data has been explored to discover interactions and could also be combined with our methodology. For instance, White et al. used web search log data to identify hyperglycemia-related terms combined with paroxetine and/or pravastatin in searches on Google, Bing, and Yahoo! [45]. Our method has the potential to automate validation of possible interactions suggested by web searches and other surveillance methods.
This method could also gain breadth by considering pharmacogenomics to personalize the effects of drugs for the genetic variants of specific patients. Rance et al. [46] describe a methodology for extracting drug-gene interactions specific to genetic variations that could be incorporated to extend the current results to variant-specific interactions. Though current clinical records may be rather sparse in regard to genetic variant assertions, current trends suggest this information is only likely to increase in prevalence.
Step 5 of our methodology requires the manual selection of candidate chains and their component predications. The use of such human intervention is limited not only by capacity (in our case less than 0.28% of potential schemas were actually reviewed), but also by inconsistencies between individual participants. In expanding this approach to a larger data set, this component may need to be replaced by a more automated approach using machine learning techniques. The chains and predications that have already been identified as interesting could serve as training data which might be augmented or supplanted by known DDI data.
7 Conclusion
We present a new methodology for detecting DDIs in clinical data that exploits semantic predications. We expand the search for interactions beyond the traditional paradigm by considering drug effects at the level of biological functions. Because SemRep extracts these from the over 20 million citations in MEDLINE, the search is not limited to a predetermined drug and gene set and has the potential to include any drug or gene contained in the UMLS or Entrez Gene. Several enhancements to SemRep, which can increase the potential of the DDI discovery system, are planned. These include extracting the degree of confidence expressed for a statement in text and the context in which the drug interactions appear.
Highlights.
Discovery of drug-drug interactions in patient data using literature knowledge
Structured knowledge extracted from MEDLINE and stored in SemMedDB
Direct effects on a gene or indirect through a common physiological function
Potential drug-drug interactions identified in medication lists from clinical data
Acknowledgments
This research was supported in part by an appointment to the NLM Research Participation Program, which is administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the U.S. Department of Energy and the National Library of Medicine. This project was supported in part by the Intramural Research Program of the NIH, National Library of Medicine.
Footnotes
See Kilicoglu et al. (2011) [35] for a discussion of kappa (κ) statistic vs. F-measure in interannotator agreement calculation.
Ethics statement
The patient data used for this project were de-identified and its use had IRB approval from the University of Minnesota.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Rui Zhang, Email: zhan1386@umn.edu.
Michael J. Cairelli, Email: cairellimj@mail.nih.gov.
Marcelo Fiszman, Email: fiszmanm@mail.nih.gov.
Graciela Rosemblat, Email: grosemblat@mail.nih.gov.
Halil Kilicoglu, Email: kilicogluh@mail.nih.gov.
Thomas C. Rindflesch, Email: trindflesch@mail.nih.gov.
Serguei V. Pakhomov, Email: pakh0002@umn.edu.
Genevieve B. Melton, Email: gmelton@umn.edu.
References
- 1.Dale MM, Haylett DG. Pharmacology Condensed. Churchill Livingstone; 2009. [Google Scholar]
- 2.Kumar S, Sharma R, Roychowdhury A. Modulation of cytochrome-P450 inhibition (CYP) in drug discovery: a medicinal chemistry perspective. Curr Med Chem. 2012;19:3605–21. doi: 10.2174/092986712801323180. [DOI] [PubMed] [Google Scholar]
- 3.Li AP. Primary hepatocyte cultures as an in vitro experimental model for the evaluation of pharmacokinetic drug-drug interactions. Adv Pharmacol. 1997;43:103–30. doi: 10.1016/s1054-3589(08)60203-3. [DOI] [PubMed] [Google Scholar]
- 4.Nettleton DO, Einolf HJ. Assessment of cytochrome p450 enzyme inhibition and inactivation in drug discovery and development. Curr Top Med Chem. 2011;11:382–403. doi: 10.2174/156802611794480882. [DOI] [PubMed] [Google Scholar]
- 5.Yan Z, Caldwell GW. The current status of time dependent CYP inhibition assay and in silico drug-drug interaction predictions. Curr Top Med Chem. 2012;12:1291–7. doi: 10.2174/156802612800672871. [DOI] [PubMed] [Google Scholar]
- 6.Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, Sevier CS, et al. The genetic landscape of a cell. Science. 2010;327:425–31. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Quinney SK, Zhang X, Lucksiri A, Gorski JC, Li L, Hall SD. Physiologically based pharmacokinetic model of mechanism-based inhibition of CYP3A by clarithromycin. Drug Metab Dispos. 2010;38:241–8. doi: 10.1124/dmd.109.028746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang SM, Strong JM, Zhang L, Reynolds KS, Nallani S, Temple R, et al. New era in drug interaction evaluation: US Food and Drug Administration update on CYP enzymes, transporters, and the guidance process. J Clin Pharmacol. 2008;48:662–70. doi: 10.1177/0091270007312153. [DOI] [PubMed] [Google Scholar]
- 9.Maeda K, Ikeda Y, Fujita T, Yoshida K, Azuma Y, Haruyama Y, et al. Identification of the rate-determining process in the hepatic clearance of atorvastatin in a clinical cassette microdosing study. Clin Pharmacol Ther. 2011;90:575–81. doi: 10.1038/clpt.2011.142. [DOI] [PubMed] [Google Scholar]
- 10.Schelleman H, Bilker WB, Brensinger CM, Han X, Kimmel SE, Hennessy S. Warfarin with fluoroquinolones, sulfonamides, or azole antifungals: interactions and the risk of hospitalization for gastrointestinal bleeding. Clin Pharmacol Ther. 2008;84:581–8. doi: 10.1038/clpt.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Percha B, Garten Y, Altman RB. Discovery and explanation of drug-drug interactions via text mining. Pac Symp Biocomput. 2012:410–21. [PMC free article] [PubMed] [Google Scholar]
- 12.Tatonetti NP, Denny JC, Murphy SN, Fernald GH, Krishnan G, Castro V, et al. Detecting drug interactions from adverse-event reports: interaction between paroxetine and pravastatin increases blood glucose levels. Clin Pharmacol Ther. 2011;90:133–42. doi: 10.1038/clpt.2011.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kilicoglu H, Shin D, Fiszman M, Rosemblat G, Rindflesch TC. SemMedDB: a PubMed-scale repository of biomedical semantic predications. Bioinformatics. 2012;28:3158–60. doi: 10.1093/bioinformatics/bts591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rindflesch TC, Fiszman M. The interaction of domain knowledge and linguistic structure in natural language processing: interpreting hypernymic propositions in biomedical text. J Biomed Inform. 2003;36:462–77. doi: 10.1016/j.jbi.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 15.Weeber M, Vos R, Klein H, De Jong-Van Den Berg LT, Aronson AR, Molema G. Generating hypotheses by discovering implicit associations in the literature: a case report of a search for new potential therapeutic uses for thalidomide. J Am Med Inform Assoc. 2003;10:252–9. doi: 10.1197/jamia.M1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wren JD, Bekeredjian R, Stewart JA, Shohet RV, Garner HR. Knowledge discovery by automated identification and ranking of implicit relationships. Bioinformatics. 2004;20:389–98. doi: 10.1093/bioinformatics/btg421. [DOI] [PubMed] [Google Scholar]
- 17.Frijters R, van Vugt M, Smeets R, van Schaik R, de Vlieg J, Alkema W. Literature Mining for the Discovery of Hidden Connections between Drugs, Genes and Diseases. PLoS Comput Biol. 2010:6. doi: 10.1371/journal.pcbi.1000943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duke JD, Han X, Wang Z, Subhadarshini A, Karnik SD, Li X, et al. Literature based drug interaction prediction with clinical assessment using electronic medical records: novel myopathy associated drug interactions. PLoS Comput Biol. 2012;8:e1002614. doi: 10.1371/journal.pcbi.1002614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen H, Sharp BM. Content-rich biological network constructed by mining PubMed abstracts. BMC Bioinformatics. 2004;5:147. doi: 10.1186/1471-2105-5-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Segura-Bedmar I, Martinez P, de Pablo-Sanchez C. A linguistic rule-based approach to extract drug-drug interactions from pharmacological documents. BMC Bioinformatics. 2011;12 (Suppl 2):S1. doi: 10.1186/1471-2105-12-S2-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tari L, Anwar S, Liang S, Cai J, Baral C. Discovering drug-drug interactions: a text-mining and reasoning approach based on properties of drug metabolism. Bioinformatics. 2010;26:i547–53. doi: 10.1093/bioinformatics/btq382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fiszman M, Demner-Fushman D, Lang FM, Goetz P, Rindflesch TC. Interpreting comparative constructions in biomedical text. Proc BioNLP 2007. 2007:137–44. [Google Scholar]
- 23.Uzuner Ö, Solti I, Cadag E. Extracting medication information from clinical text. J Am Med Inform Assoc. 2010;17:514–8. doi: 10.1136/jamia.2010.003947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aronson AR, Lang FM. An overview of MetaMap: historical perspective and recent advances. J Am Med Inform Assoc. 2010;17:229–36. doi: 10.1136/jamia.2009.002733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bodenreider O. The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Res. 2004;32:D267–70. doi: 10.1093/nar/gkh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rindflesch TC, Aronson AR. Semantic processing in information retrieval. Proceedings/the Annual Symposium on Computer Application [sic] in Medical Care Symposium on Computer Applications in Medical Care; 1993. pp. 611–5. [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu X, Cherry C, Kiritchenko S, Martin J, de Bruijn B. Detecting concept relations in clinical text: insights from a state-of-the-art model. J Biomed Inform. 2013;46:275–85. doi: 10.1016/j.jbi.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 28.Demner-Fushman D, Humphrey SM, Ide NI, Loane RF, Mork JG, Ruch R, et al. Combining Resources to Find Answers to Biomedical Questions. Proc TREC. 2007:205–14. [Google Scholar]
- 29.Weeber M, Klein H, Aronson AR, Mork JG, de Jong-van den Berg LT, Vos R. Text-based discovery in biomedicine: the architecture of the DAD-system. AMIA Annu Symp Proc. 2000:903–7. [PMC free article] [PubMed] [Google Scholar]
- 30.McCray AT, Srinivasan S, Browne AC. Lexical methods for managing variation in biomedical terminologies. Proceedings/the Annual Symposium on Computer Application [sic] in Medical Care Symposium on Computer Applications in Medical Care; 1994. pp. 235–9. [PMC free article] [PubMed] [Google Scholar]
- 31.Smith L, Rindflesch T, Wilbur WJ. MedPost: a part-of-speech tagger for bioMedical text. Bioinformatics. 2004;20:2320–1. doi: 10.1093/bioinformatics/bth227. [DOI] [PubMed] [Google Scholar]
- 32.Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2007;35:D26–31. doi: 10.1093/nar/gkl993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ahlers CB, Fiszman M, Demner-Fushman D, Lang FM, Rindflesch TC. Extracting semantic predications from Medline citations for pharmacogenomics. Pac Symp Biocomput. 2007:209–20. [PubMed] [Google Scholar]
- 34.HUGO. Available from http://www.genenames.org/
- 35.Kilicoglu H, Rosemblat G, Fiszman M, Rindflesch TC. Constructing a semantic predication gold standard from the biomedical literature. BMC Bioinformatics. 2011;12:486. doi: 10.1186/1471-2105-12-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Drugs.com. Available from http://www.drugs.com/
- 37.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wishart DS, Knox C, Guo AC, Shrivastava S, Hassanali M, Stothard P, et al. DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006;34:D668–72. doi: 10.1093/nar/gkj067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Knox C, Law V, Jewison T, Liu P, Ly S, Frolkis A, et al. DrugBank 3.0: a comprehensive resource for ‘omics’ research on drugs. Nucleic Acids Res. 2011;39:D1035–41. doi: 10.1093/nar/gkq1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hewett M, Oliver DE, Rubin DL, Easton KL, Stuart JM, Altman RB, et al. PharmGKB: the Pharmacogenetics Knowledge Base. Nucleic Acids Res. 2002;30:163–5. doi: 10.1093/nar/30.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Whirl-Carrillo M, McDonagh EM, Hebert JM, Gong L, Sangkuhl K, Thorn CF, et al. Pharmacogenomics knowledge for personalized medicine. Clin Pharmacol Ther. 2012;92:414–7. doi: 10.1038/clpt.2012.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Klein TE, Chang JT, Cho MK, Easton KL, Fergerson R, Hewett M, et al. Integrating genotype and phenotype information: an overview of the PharmGKB project. Pharmacogenetics Research Network and Knowledge Base. Pharmacogenomics J. 2001;1:167–70. doi: 10.1038/sj.tpj.6500035. [DOI] [PubMed] [Google Scholar]
- 43.Fundel K, Kuffner R, Zimmer R. RelEx—Relation extraction using dependency parse trees. Bioinformatics. 2007;23(3):365–371. doi: 10.1093/bioinformatics/btl616. [DOI] [PubMed] [Google Scholar]
- 44.Masseroli M, Kilicoglu H, Lang FM, Rindflesch T. Argument-predicate distance as a filter for enhancing precision in extracting predications on the genetic etiology of disease. BMC Bioinformatics. 2006;7:291. doi: 10.1186/1471-2105-7-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.White RW, Tatonetti NP, Shah NH, Altman RB, Horvitz E. Web-scale pharmacovigilance: listening to signals from the crowd. J Am Med Inform Assoc. 2013 doi: 10.1136/amiajnl-2012-001482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rance B, Doughty E, Demner-Fushman D, Kann MG, Bodenreider O. A mutation-centric approach to identifying pharmacogenomic relations in text. J Biomed Inform. 2012;45:835–41. doi: 10.1016/j.jbi.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]