Abstract
The dopamine transporter (DAT), a member of the neurotransmitter:sodium symporter family, mediates the reuptake of dopamine at the synaptic cleft. DAT is the primary target for psychostimulants such as cocaine and amphetamine. We previously demonstrated that cocaine binding and dopamine transport alter the accessibility of Cys342 in the third intracellular loop (IL3). To study the conformational changes associated with the functional mechanism of the transporter, we made cysteine substitution mutants, one at a time, from Phe332 to Ser351 in IL3 of the background DAT construct, X7C, in which 7 endogenous cysteines were mutated. The accessibility of the 20 engineered cysteines to polar charged sulfhydryl reagents was studied in the absence and presence of cocaine or dopamine. Of the 11 positions that reacted with methanethiosulfonate ethyl ammonium, as evidenced by inhibition of ligand binding, 5 were protected against this inhibition by cocaine and dopamine (S333C, S334C, N336C, M342C and T349C), indicating that reagent accessibility is affected by conformational changes associated with inhibitor and substrate binding. In some of the cysteine mutants, transport activity is disrupted, but can be rescued by the presence of zinc, most likely because the distribution between inward- and outward-facing conformations is restored by zinc binding. The experimental data were interpreted in the context of molecular models of DAT in both the inward- and outward-facing conformations. Differences in the solvent accessible surface area for individual IL3 residues calculated for these states correlate well with the experimental accessibility data, and suggest that protection by ligand binding results from the stabilization of the outward-facing configuration. Changes in the residue interaction networks observed from the molecular dynamics simulations also revealed the critical roles of several positions during the conformational transitions. We conclude that the IL3 region of DAT undergoes significant conformational changes in transitions necessary for both cocaine binding and substrate transport.
Keywords: Dopamine transporter (DAT), Cocaine binding, Dopamine transport, Cysteine accessibility, Molecular dynamics simulations
1. Introduction
Eukaryotic members of the neurotransmitter: Na+ symporter (NSS) family, which include transporters for GABA, dopamine (DA), norepinephrine, and serotonin (GAT, DAT, NET, and SERT, respectively), terminate signaling by recapturing released neurotransmitter (Amara and Sonders, 1998; Rudnick, 2002; Sonders et al., 2005). These secondary active transporters enable the thermodynamically uphill transport of their respective substrates across the plasma membrane of the presynaptic neuron in a co-transport (symport) mechanism driven by the Na+ electrochemical gradient (Gu et al., 1994; Krause and Schwarz, 2005; Torres et al., 2003). Drugs that interfere with reuptake profoundly influence behavior and mood. DAT is the primary target for the psychostimulants cocaine and amphetamine (Koob, 1992) and is a target for treatment of attention deficit hyperactivity disorder and depression (Volkow et al., 2002). Genes encoding more than 200 putative NSS homologs have been identified computationally in prokaryotic genomes, thereby expanding the functional spectrum of this transporter family (Beuming et al., 2006).
A structural context for the information gathered from extensive experimental studies of DAT activity under various conditions has emerged even before the recent elucidation of a Drosophila DAT construct (Penmatsa et al., 2013), from the crystal structure of LeuT, a prokaryotic NSS homolog (Yamashita et al., 2005). In this structure, one Leu and two Na+ are bound in a pocket formed by the unwound portions of transmembrane segment (TM) 1 and TM6, and residues in TM3 and TM8 that are highly conserved among NSS. The LeuT structure has been invaluable for structure-based interrogation of functional mechanisms and ligand binding (Beuming et al., 2008; Celik et al., 2008; Forrest et al., 2006; Forrest et al., 2008; Huang et al., 2009; Kanner and Zomot, 2008; Kaufmann et al., 2009; Kniazeff et al., 2008; Rudnick, 2006; Shan et al., 2011; Shi et al., 2008; Zhao et al., 2010; Zhao et al., 2011).
In the transport cycle, substrate binding is thought to initiate a transition from an outward to an inward facing conformation, but the mechanism for this transition and how it is driven by the sodium gradient had been unclear. The first LeuT crystal structure, closed to both the outside and the inside (occluded), appeared to represent an intermediate conformation, and subsequently solved crystal structures of LeuT mutants have captured conformations thought to represent outward-open as well as inward-open configurations (Krishnamurthy and Gouaux, 2012; Singh et al., 2008; Yamashita et al., 2005).
Using steered molecular dynamics (SMD) simulations starting from the initial occluded structure of LeuT, the substrate translocation pathway in LeuT was explored computationally and a second substrate binding site (S2) was identified in the extracellular vestibule (Shi et al., 2008). In LeuT this site is also the binding site for tricyclic antidepressants (TCAs) (Singh et al., 2007; Zhou et al., 2007; Zhou et al., 2009). Substrate binding in the S2 site is thought to constitute an allosteric trigger for the mechanism of intracellular release of Na+ and substrate from the primary S1 site, whereas TCAs in the S2 site inhibit transport by binding in a way that prevents substrate release (Cheng and Bahar, 2013; Shi et al., 2008; Singh et al., 2007; Zhao et al., 2011). A similar role of the bound S2 substrate was also observed in the SMD simulations of a LeuT-based DAT homology model, where the stably bound dopamine molecule in the S2 site triggers the opening of the intracellular gate in the absence of sodium in the Na2 site, thereby enabling penetration of waters from the intracellular milieu and the gradual rearrangement of intracellular segments towards an inward-open conformation (Shan et al., 2011).
Binding of cocaine to DAT was shown to occur in the S1 site (Beuming et al., 2008). Interestingly, cocaine has been proposed to stabilize DAT in an outward-facing configuration (Chen et al., 2000; Chen and Justice, 1998). Extensive work in the homologous serotonin transporter (SERT) using the substituted-cysteine-accessibility method (SCAM) (Karlin and Akabas, 1998) has shown that cocaine modulates the conformation of this transporter, and has led to the proposal of a rocking-bundle model (Chen et al., 1997; Forrest et al., 2008; Henry et al., 2003; Zhang and Rudnick, 2006). Our combined computational and experimental studies of LeuT and DAT have brought to light detailed elements of the transport mechanism propagated through flexible rearrangements. The global rearrangements in DAT were found in computational simulations of substrate translocation in DAT (Shan et al., 2011) to be quite similar to those observed from similar simulations in LeuT (Shi et al., 2008), and to correspond substantially to inferences from the comparison of LeuT crystal structures. Notably, however, in LeuT some intracellular portions of TM segments exhibited either smaller-scale movements (TMs 4 and 8) or did not appear to move at all (TMs 3, 9 and 10). The structural rearrangements propagated in DAT by substrate translocation were shown (Shan et al., 2011) to take advantage of specific hinge regions and the rearrangement of local interaction networks that result in the conformational transitions observed both computationally and from smFRET measurements of LeuT (Shan et al., 2011; Shi et al., 2008; Zhao et al., 2010; Zhao et al., 2011). In DAT, one such region is the third intracellular loop (IL3), for which we found significant rearrangements associated with the TM1 and TM6 movements in the transition towards inward-facing conformations in mechanisms explored computationally and shown to reach beyond the inferences of rigid motions inherent in comparisons of crystal structures or whole bundle repositioning (Shan et al., 2011).
As described here, we used the substituted-cysteine accessibility method (SCAM) and computational modeling to identify the structural signature of ligand-dependent conformational rearrangements of IL3 in DAT, by evaluating the impact of cocaine binding and DA transport on changes in accessibility of cysteine-targeting reagents. The results are presented in the structural context of an established homology model for DAT based on the LeuT structure in which we simulated the outward and inward facing conformations (Shan et al., 2011), which was shown to be in excellent agreement with the recently determined structure of a construct from the drosophila DAT (Penmatsa et al., 2013). Our results suggest that the TM6-IL3-TM7 region plays an important role in functionally relevant conformational changes and thus shed light on conformational transitions during the transport cycle.
2. Material and methods
2.1. Mutagenesis and subcloning of cysteine mutants
We have shown previously that the accessibility of Cys342 in IL3 of DAT is transport-dependent (Chen et al., 2000) and sensitive to cocaine binding (Ferrer and Javitch, 1998), which is most consistent with this residue being buried in the TM bundle in the outward facing state and becoming intermittently accessible during the transport cycle. To investigate the properties and putative roles of residues in the TM6-IL3-TM7 region, we made a background DAT construct in which seven intra- and extracellular cysteines were mutated to other residues (X7C). Twenty cysteine substitution mutants were made in IL3 and the contiguous intracellular ends of TM6 and TM7 in the X7C background by replacing, one at a time, each residue from Phe332 to Ser351.
We used EM4 cells, which are human embryonic kidney 293 cells stably transfected with macrophage scavenger receptor to increase their adherence to tissue culture plastic (Robbins and Horlick, 1998). These cells were additionally stably transfected with a synthetic human DAT gene that was tagged at the amino terminus with a tandem epitope. The Flag epitope was fused to the hemagglutinin epitope (Flag-HA; 18 residues). This epitope replaced the first 22 amino acids of the protein and did not significantly affect expression or transport (Saunders et al., 2000). Mutations were made by PCR and confirmed by DNA sequencing. EM4 cells were grown in DMEM supplemented with 10% fetal bovine serum at 37 °C and 5% CO2. Thirty-five millimeter dishes of EM4 cells at 70-80% confluence were transfected with 2 μg of mutant DAT cDNA in the bicistronic expression vector pCIHyg (Saunders et al., 2000) with 9 μl of Lipofectamine (Invitrogen) and 500 μl of OPTIMEM (Invitrogen). Five hours after transfection the solution was removed and fresh media added. Twenty-four hours after transfection, the cells were split to a 100 mm-dish and 250 μg/ml hygromycin was added to select for a stably transfected pool of cells.
2.2. Indexing of residues
A generic numbering scheme for amino acid residues has been developed to facilitate direct comparison of sequences of the individual members in the Na+/Cl− family of transporters (Beuming et al., 2006). In this scheme the most conserved residue in each TM has been given the number 50, and each residue is numbered according to its position relative to this conserved residue. Each index number starts with the number of the TM, so a residue five positions carboxyl-terminal to the most conserved residue in TM6 of DAT (Gln3176.50) will be referred to as Leu3226.55. The most conserved residue in TM7 is Phe-356, and so the generic numbers for the residues mutated to cysteine in this study are Tyr3326.65, Ser3336.66, Ser3346.67, Tyr3356.68, Asn336IL3, Lys337IL3, Phe338IL3, Thr339IL3, Asn340IL3, Asn341IL3, Cys342IL3, Tyr3437.28, Arg3447.29, Asp3457.30, Ala3467.31, Ile3477.32, Val3487.33, Thr3497.34, Thr3507.35 and Ser3517.36.
2.3 [3H]tyramine uptake
The DAT substrate tyramine was used instead of DA for uptake assays since it is a substrate for DAT but not a substrate for endogenous catechol-O-methyl transferase, which has shown to be present in HEK 293 cells (Eshleman et al., 1997). EM4 cells stably expressing X7C or mutants thereof were seeded in poly-D-lysine coated 96-well plates. After 48 h, the cells were washed twice with serum free DMEM, and incubated in serum free DMEM for 4 h at 37 °C and 5% CO2. The cells were washed once with 200 μl uptake buffer (130 mM NaCl, 1.3 mM KCl, 1.2 mM MgSO4, 1.2 mM KH2PO4, 2.2 mM CaCl2, 10 mM glucose, 10 mM Hepes, pH 7.4) and incubated in uptake buffer for 30 min at 37 °C and 5% CO2 . The uptake assay was initiated by the addition of 60-80 nM [3H]tyramine (American Radiolabeled Chemicals, Inc.) in a final volume of 50 μl in uptake buffer. Nonspecific uptake was determined in the presence of 2 mM tyramine. Uptake kinetics were determined using a range of unlabeled tyramine (10 nM-10 μM) in competition with [3H]tyramine. After 5 min of incubation at 37 °C, uptake was terminated by aspiration and washing twice with 200 μl ice cold buffer. The cells were lysed in 50 μl of 1% Triton X-100 for 30 min, and 200 μl Optiphase Supermix scintillation fluid was added before counting the plate in a Wallac 1450 microbeta counter. Km and Vmax values were calculated from non-linear regression analysis (Prism 4, GraphPad software).
To measure the effect of Zn2+ on uptake, 10 μM ZnCl2 was added to the uptake buffer 15 min prior to [3H]tyramine; uptake measurements were otherwise performed as described above.
2.4. Membrane preparation and protein measurement
Stably transfected EM4 cells, expressing the IL3 cysteine mutants in the X7C background, were washed with PBS (1.9 mM NaH 2PO4;2H2O, 8.1 mM NaHPO4, 154 mM NaCl). The cells were scraped into PBS containing 1 mM EDTA and pelleted at 1000 × g for 5 min at 4 °C. Pellets were resuspended in PBS and disrupted on ice with a Polytron homogenizer for 10-15 s. The membranes were collected by centrifugation at 39,000 × g for 20 min at 4 °C and stored at −80 °C until use. Resuspended membranes were solubilized in 1% Triton and measured against a BSA standard curve using the bicinchoninic acid (BCA) protein assay (Pierce).
2.5. Binding of MFZ 2-12 and cocaine
Membrane pellets were thawed quickly, resuspended in binding buffer (130 mM NaCl, 1.3 mM KCl, 1.2 mM MgSO4, 1.2 mM KH2PO4, 2.2 mM CaCl2, 10 mM Hepes, pH 7.4) and 125 μl transferred to a prewetted 96 well Multiscreen-FB1 filter plate (Millipore) with 1.0 μm glass fiber filters. The binding assay was initiated by adding 2.5-3 nM [3H]MFZ 2-12 (Newman et al., 2001) to a total volume of 200 μl. Nonspecific binding was determined in the presence of 2 μM unlabeled MFZ 2-12. For determination of KD and Bmax values for MFZ 2-12 and Ki for cocaine binding, increasing concentrations of unlabeled ligand was added. When binding was performed in the presence of Zn2+, 10 μM ZnCl2 was added to the resuspended membranes 15 min prior to ligand. After 2 h incubation at RT, the binding was terminated by filtering the mixture through the plate on a vacuum manifold and washed three times with ice cold binding buffer. After removing the durapore backing, a 96 well plastic liner was fitted under the plate. Optiphase Supermix scintillation fluid (30 μl) was added to the radioactive samples 30 min prior to counting in a Wallac 1450 microbeta counter.
2.6. Treatment with MTS reagents
Methanethiosulfonate (MTS) reagents (Toronto Research Chemicals) were freshly prepared in water as 30× stocks and placed on ice prior to use. Resuspended membranes of X7C and the IL3 cysteine mutants were treated with either MTS ethylammonium (MTSEA) (0.25 mM), MTS ethyltrimethylammonium (MTSET) (1 mM) or MTS ethylsulfonate (MTSES) (10 mM), at room temperature, or 4 °C when stated, in a Multiscreen-FB1 filter plate. After 2 min incubation the reaction was terminated by filtering the mixture through the plate with vacuum suction and washing three times with binding buffer. The MTS treated membranes were resuspended in binding buffer and [3H]MFZ 2-12 binding activities were measured as described above. For estimation of rate of reaction, 6 concentrations of the MTS reagent were added.
2.7. Protection experiments with cocaine and DA
Resuspended membranes were treated with cocaine (0.1 mM, unless otherwise stated) or DA (1 mM) for 20 min at room temperature, or 4 °C when stated, prior to reaction with MTSEA. The MTSEA treatment was performed as described above, but with ligand present. To insure that all ligand was removed prior to the [3H]MFZ 2-12 binding step, the membranes were washed an additional 2 × 5 min with binding buffer on a plate shaker.
2.8. Electrophoresis and immunoblotting
Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) was performed as described in Laemmli (Laemmli, 1970) with separating gels consisting of 7.5% acrylamide. Immunoblotting was performed as described previously (Hastrup et al., 2001). Briefly, resolved proteins were transferred by electroblotting onto polyvinylidene fluoride (PVDF) membranes (Millipore) and blocked in a solution of 5% (w/v) dry milk, 1% BSA and 0.1% Tween-20 in TBS (50 mM Tris·HCl, 150 mM NaCl, pH 7.4). FLAG epitope-tagged DAT protein was detected by anti-FLAG M2 primary antibody (Sigma) and anti-mouse horseradish peroxidase (HRP) secondary antibody (Santa Cruz Biotechnology, Inc.) with ECL-Plus (Amersham). Chemiluminescence was detected and quantified on a FluorChem 8000 (Alpha Innotech Corp.).
2.9. Data calculations
Uptake data and binding data were analyzed by nonlinear regression analysis using Prism 4.0 from GraphPad Software. The Kmand Vmax for uptake and the KD and BMAX for binding were determined in Prism by fitting the data to the equation Y= (Bmax*L)/(L + C + Kd) + NS, where Y is binding or uptake, L is the concentration of hot ligand ([3 H]Tyramine or [3H]MFZ 2-12) and C is the concentration of cold ligand (tyramine or MFZ 2-12). The Ki for cocaine binding was calculated in Prism by fitting the data using the equations logEC50 = log(10^logKi*(1+HotNM/HotKdNM)) and Y= Bottom + (Top-Bottom)/(1+10^(XLogEC50)), where Y is binding, HotNM is the concentration of [3H]MFZ 2-12, HotKdNM is the KD for MFZ 2-12 and X is the logmolar concentration of cocaine. The rates of reaction (K) for MTSEA were calculated in Prism by fitting the data to the equation Y=Span*exp(−K*X) + Plateau, where Y is binding and X is the concentration of MTSEA.
Statistical significance was evaluated using one-way ANOVA with Dunnet's multiple comparison post hoc test using Graphpad Prism 4.0.
2.10. Solvent-Accessible Surface Area (SASA) calculations
The SASA values were calculated for the equilibrated structures obtained from the SMD simulations described recently (Shan et al., 2011). To this end, the 20 consecutive residues from F332 to S351 were mutated one at a time to cysteine with the structure building tool psfgen within the Visual Molecular Dynamics (VMD) visualization program (Humphrey et al., 1996). All the other residues in DAT, as well as the ligands including DA molecules, lipids, waters and ions were maintained at their original positions. The systems were minimized for 2000 steps, followed by short molecular dynamics (MD) simulations to take potential conformational changes into account.
For particular positions, in which the mutations to cysteines might affect the steric arrangement of the immediate environment, we sampled more fully the impact of the mutations with free energy perturbation (FEP) simulations. This control procedure was applied to two selected residues (F332 and T349) which were mutated to cysteines in both the occluded state homology model and the inward-facing conformation. For each FEP run, a total of 1.98 ns FEP calculation with 66 windows was carried out with explicit solvents using the molecular dynamics simulator NAMD (Henin et al., 2006). Each resulting model was further equilibrated for 5 ns MD after FEP. SASA values were evaluated along the entire MD trajectories and found to be consistent with the data obtained by the direct mutation method described above (Supplemental Fig. 1).
Other than for these controls, SASA values were calculated based on the last half of each MD simulation. In the SASA analysis, the calculated accessible area excludes the surface area exposed to lipids, which was treated as buried. The SASA percentage was obtained by dividing the SASA of the residue calculated in the environment of the protein and lipids, by the SASA reference value, calculated as usual for a residue X in the extended conformation of a Gly-X-Gly tripeptide (Miller et al., 1987; Newman et al., 2001).
3. Results
3.1. Transport activity of cysteine mutants
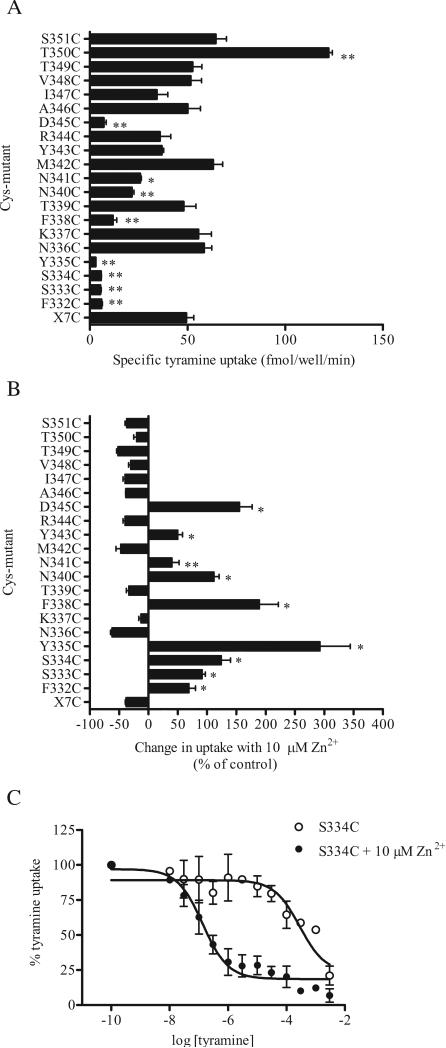
In order to generate a suitable human DAT background construct for studying the residues in IL3, all the reduced cysteines in predicted extracellular and intracellular loops were mutated. In the resulting construct, X7C DAT (ΔC6/C90A/C135A/C306A/C319M/ C342M/C581L), we mutated to cysteine, one at a time, each of the wild type residues in the predicted IL3 and the juxtamembrane regions of TM6 and TM7 from Phe332 to Ser351. After stable expression in EM4 cells, tyramine transport was determined in all the mutants (Fig. 1). Cysteine substitution of most of the highly conserved residues (Phe332, Ser333, Ser334, Tyr335, Phe338, Asn340, Asn341 and Asp345, Supplemental Table 1) led to significantly diminished transport activity, ranging from 6% to 52% of that for the X7C construct (Fig.1). Three conserved residues, Asn336, Tyr343 and Thr349, tolerated the mutation well, displaying transport activities similar to X7C. In T350C the uptake activity was significantly increased (250%), whereas in the remaining mutants it ranged from 70% to 130% that of X7C.
Figure 1.
Tyramine transport by X7C and the TM6-IL3-TM7 cysteine substitution mutants. (A) [3H]Tyramine uptake was measured at 37 °C for 5 min, as described in Methods. Non-specific uptake was determined in the presence of 2 mM unlabeled tyramine. The data shown are the mean ± SEM from three separate experiments performed in triplicate. * and ** indicate significantly different (p<0.01 and p<0.001, respectively) from X7C by one way ANOVA and Dunnett's post hoc test. (B) Cells were incubated with 10 μM Zn2+ for 15 min at 37 °C prior to and during the tyramine uptake assay and the change in uptake from control (no Zn2+) is shown from 3 separate experiments performed in triplicate. Several mutants are potentiated (F332C through Y335C, F338C, N340C, N341C, Y343C and D345C) rather than inhibited (like wt (not shown here) and X7C) by the presence of Zn2+. * and ** indicate significantly different (p < 0.01 and p < 0.05, respectively) from X7C by one way ANOVA and Dunnett's post hoc test. (C) Impaired uptake kinetics for S334C were partially restored by the presence of 10 μM Zn2+ during uptake. Data shown are mean of 3 different uptake experiments +/- Zn2+.
Since these initial experiments were conducted at a single tracer concentration of [3H]tyramine, we also determined the tyramine concentration dependence of uptake for all 20 mutants (Table 1). We know from previous experiments that substituting Cys135 and Cys342 reduced the Km and Vmax for tyramine uptake (Chen et al., 2000)), and as expected, reinserting the endogenous Cys342 back into X7C increased both the Km and Vmax ~5-fold (Table 1). Interestingly, in N336C, T349C and T350C, the cysteine mutations had a similar effect, significantly increasing the Km and Vmax for tyramine uptake. The Km for tyramine transport by S333C, F338C, N340C, N341C and Y343C was 7-10 fold lower than that of X7C, and the Vmax of these mutants was markedly decreased (Table 1). Extremely low specific uptake precluded reliable determination of Km and Vmax for F332C, S334C and Y335C.
Table 1.
Kinetic characteristics of TM6-IL3-TM7 cysteine mutants in a X7C background
| X7C | 2.6 ± 0.6 | 5 | 29 ±8 | 5 |
| F332C | ND | ND | ||
| S333C | 0.35 ± 0.06 | 4 | 0.23 ± 0.16 | 4 |
| S334C | ND | ND | ||
| Y335C | ND | ND | ||
| N336C | 6.0 ± 0.9a | 3 | 109 ± 16b | 3 |
| K337C | 0.7 ± 0.2 | 5 | 18 ±5 | 5 |
| F338C | 0.3 ± 0.1 | 4 | 1.8 ± 0.8 | 4 |
| T339C | 1.5 ± 0.4 | 3 | 17 ±9 | 3 |
| N340C | 0.2 ± 0.1 | 4 | 1.2 ± 0.2 | 4 |
| N341C | 0.2 ± 0.1 | 5 | 2.2 ± 0.6 | 5 |
| M342C | 11 ± 4c | 3 | 188 ± 50c | 3 |
| Y343C | 0.4 ± 0.3 | 3 | 2 ± 1 | 3 |
| R344C | 0.8 ± 0.1 | 3 | 8 ±4 | 3 |
| D345C | 0.40 ± 0.26 | 4 | 0.8 ± 0.4 | 4 |
| A346C | 2.3 ± 0.2 | 3 | 24 ±6 | 3 |
| I347C | 1.3 ± 0.4 | 5 | 20 ±8 | 5 |
| V348C | 0.79 ± 0.04 | 3 | 8 ± 1 | 3 |
| T349C | 15.5 ± 0.3c | 3 | 137 ± 45c | 3 |
| T350C | 7 ± 1a | 3 | 135 ± 41c | 3 |
| S351C | 3.0 ± 0.3 | 3 | 39 ±5 | 3 |
|
+ 10 μM Zn2+ |
||||
| F332C | 26 ± 1 | 3 | 27 ±4 | 3 |
| S334C | 0.11 ± 0.04 | 4 | 0.6 ± 0.1 | 4 |
| Y335C | 0.59 ± 0.06 | 3 | 1.6 ± 0.2 | 3 |
The data shown are mean ± SEM of 3-5 experiments performed in duplicate. ND = not determined.
p < 0.05
p < 0.01
p < 0.001, vs. X7C by one-way ANOVA and Dunnett's multiple comparison post hoc test.
3.2. Effect of Zn2+ on tyramine uptake
It has previously been shown that the low uptake capacity caused by the mutation of Tyr335 to alanine is substantially restored by Zn2+ (Loland et al., 2002). This is in marked contrast to the effect seen in WT DAT, where binding of Zn2+ to its endogenous site inhibits dopamine uptake (Loland et al., 1999; Norregaard et al., 1998). We determined the effect of Zn2+ on the transport activity of all the IL3 cysteine mutants. Uptake by all the mutants with significantly lowered uptake capacity (indicated by * in Fig. 1A) was potentiated by the presence of Zn2+, as was uptake by Y343C (Fig. 1B). For all the remaining mutants as well as the background construct X7C, Zn2+ inhibited uptake, just as in WT.
Since uptake kinetics could not be reliably determined for F332C, S334C and Y335C, and transport activity was potentiated by Zn2+, we determined the Km and Vmax for tyramine uptake in the presence of 10 μM Zn2+ for these mutants (Table 1, Fig. 1C). The apparent substrate affinity of F332C in the presence of Zn2+ was 10-fold lower than for X7C. Both S334C and Y335C displayed a higher affinity for tyramine compared to X7C, which was accompanied by a marked reduction in the maximal rate of transport (23 and 48 fold lower, respectively, than X7C) (Table 1).
3.3. Binding properties of cysteine mutants
Since the substituted cysteines were in an intracellular loop, accessibility studies had to be performed in a membrane preparation to allow direct access of the MTS reagents, some of which are membrane impermeant, and also to overcome the reducing environment inside the cell. To characterize inhibitor binding to the mutants we determined the binding affinity (Kd) and binding capacity (B max) of the cocaine analog [3H]MFZ 2-12 (Newman et al., 2001) to each of the mutants (Table 2). The binding affinities of F332C, S334C, Y335C and D345C were dramatically impaired compared to X7C. In other mutants (S333C, F338C, N340C, N341C and Y343C), the cysteine mutation caused an intermediate 4-15 fold decrease in affinity for MFZ 2-12. The remaining mutants displayed affinities similar to that of X7C. The Bmax for many of the mutants was unchanged (Table 2), whereas the Bmax values for S334C, R344C and I347C were reduced 3-6 fold.
Table 2.
Apparent affinities for cocaine and MFZ 2-12 at DAT TM6-IL3-TM7 cysteine mutants
| MFZ 2-12 KD ± SEM | MFZ 2-12 Bmax ± SEM | Cocaine Ki ± SEM | ||||
|---|---|---|---|---|---|---|
| Mutant | (nM) | n | (pmol/mg) | n | (μM) | n |
| X7C | 2.2 ± 0.5 | 4 | 5 ± 2 | 4 | 4 ± 1 | 4 |
| F332C | ND | ND | ND | |||
| S333C | 21 ± 5b | 4 | 7 ± 1 | 4 | 10 ± 6 | 3 |
| S334C | 99 ± 7b | 3 | 1.7 ± 0.3 | 3 | 17 ± 10 | 2 |
| Y335C | ND | ND | ND | |||
| N336C | 4 ± 2 | 3 | 6 ± 2 | 3 | 4.6 ± 0.8 | 3 |
| K337C | 3 ± 1 | 3 | 3 ± 1 | 3 | 5.0 ± 0.8 | 3 |
| F338C | 31 ± 7b | 3 | 8 ± 3 | 3 | 13 ± 6 | 3 |
| T339C | 2.6 ± 0.6 | 4 | 3.8 ± 0.4 | 4 | 2.5 ± 0.4 | 3 |
| N340C | 26 ± 9b | 3 | 3.4 ± 0.3 | 3 | 14 ± 3c | 4 |
| N341C | 12 ± 2 | 3 | 3.1 ± 0.6 | 3 | 11.7 ± 0.7 | 3 |
| M342C | 2.0 ± 0.4 | 4 | 8 ± 4 | 4 | 1.3 ± 0.2 | 4 |
| Y343C | 9 ± 2 | 4 | 10 ± 3 | 4 | 10 ± 1 | 3 |
| R344C | 2.8 ± 0.3 | 3 | 0.8 ± 0.2 | 3 | 5 ± 1 | 3 |
| D345C | ND | ND | ND | |||
| A346C | 3 ± 1 | 3 | 9 ± 3 | 3 | 4.0 ± 0.9 | 3 |
| I347C | 5 ± 1 | 3 | 1.3 ± 0.2 | 3 | 5 ± 1 | 3 |
| V348C | 4.1 ± 0.9 | 3 | 4 ± 1 | 3 | 3.8 ± 0.8 | 3 |
| T349C | 2.2 ± 0.9 | 4 | 16 ± 6 | 4 | 3.0 ± 0.8 | 3 |
| T350C | 2.7 ± 0.9 | 3 | 8 ± 3 | 3 | 3.1 ± 0.3 | 3 |
| S351C | 4.3 ± 0.1 | 3 | 11 ± 4 | 3 | 3.8 ± 0.4 | 3 |
ND = not determined.
p < 0.001
p < 0.01, vs. X7C by one way ANOVA and Dunnett's multiple comparison post hoc test.
We also measured the ability of cocaine to inhibit [3H]MFZ 2-12 binding to all of the mutants (Table 2). The impaired [3H]MFZ 2-12 binding in F332C, Y335C and D345C precluded reliable fits for cocaine inhibition. Binding in the presence of Zn did not improve reproducibility for F332C or Y335C. For D345C the presence of Zn2+ increased [3H]MFZ 2-12 binding and revealed a reduced affinity for cocaine (Ki(cocaine + Zn2+) = 96 ± 10 μM, n=3). The affinities for the remaining mutants were within 4 fold of that of X7C.
3.4. Reactivity towards sulfhydryl reagents
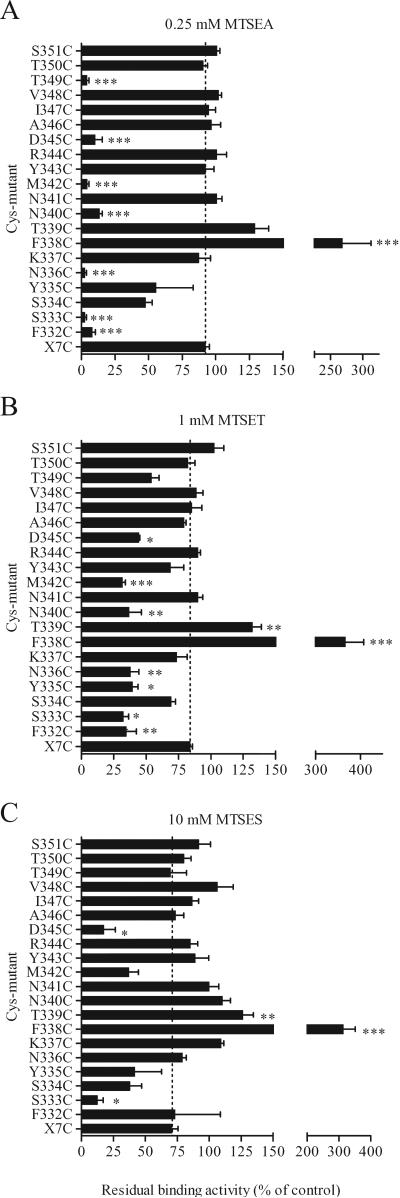
To study the accessibility of the loop residues, we assessed the effect of several charged hydrophilic MTS reagents on [3H]MFZ 2-12 binding to the cysteine mutants. A number of the mutants were reactive with MTSEA, MTSET or MTSES, as determined by their impact on subsequent [3H]MFZ 2-12 binding (Fig. 2). After treatment with the positively charged MTSEA (0.25 mM), the binding activity of F332C, S333C, N336C, N340C, M342C, D345C and T349C was inhibited by 87-98%, as compared to only 8% in X7C (Fig. 2A). The same mutants were also sensitive to the larger and positively charged MTSET, though to a lesser extent (Fig. 2B). S333C and D345C were significantly inhibited by the negatively charged MTSES (Fig. 2C). Binding of [3H]MFZ 2-12 to F338C and T339C was potentiated by all three MTS reagents.
Figure 2.
Effect of MTS-reagents on [3H]MFZ 2-12 binding. Membrane preparations of the cysteine mutants were incubated with 0.25 mM MTSEA, 1 mM MTSET or 10 mM MTSES for 2 min at RT, and washed, prior to [3H]MFZ2-12 binding assay, as described in Methods. The remaining binding activity is shown as % of control (no MTS-reagent). The binding activity of a large number of mutants in TM6-IL3-TM7 was greatly inhibited by the MTS-reagents. *, ** and *** indicate significantly different (p < 0.05, p < 0.01 and p< 0.001, respectively) from X7C by One-way ANOVA with Dunnett's post hoc test.
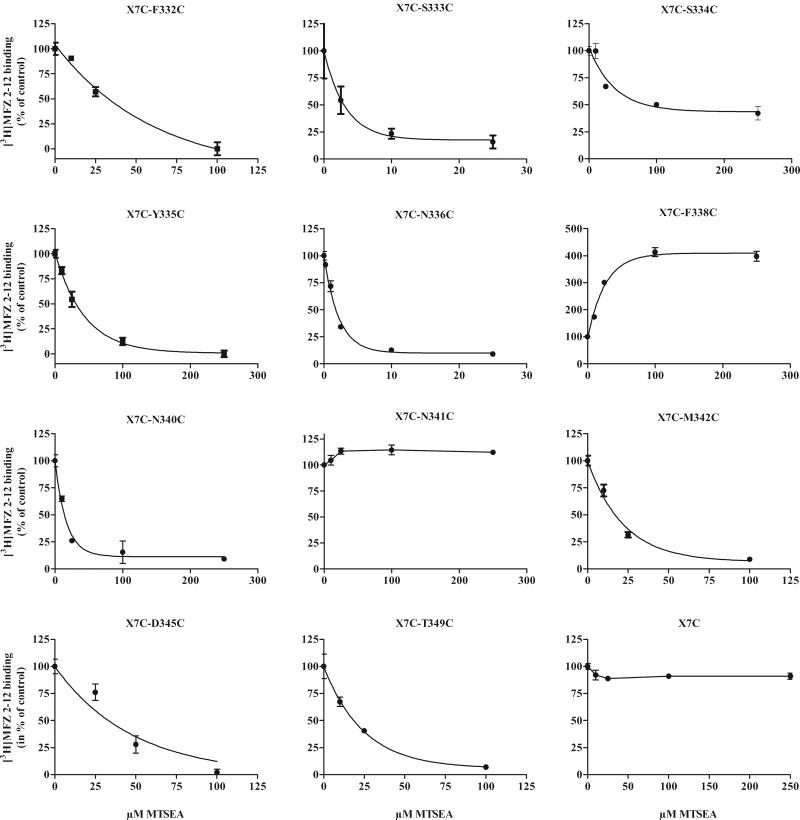
[3H]MFZ 2-12 binding to N336C, N340C and T349C was greatly inhibited by MTSEA, whereas MTSET and MTSES had much less of an effect on binding (Fig. 2). To determine whether these cysteines were less reactive with MTSET and MTSES or whether they reacted “silently” without a substantial impact on binding, membrane preparations of these mutants were treated with MTSES or MTSET followed by MTSEA. In T349C, MTSEA still inhibited after treatment with MTSET and MTSES (Supplemental Fig. 2), consistent with a decreased accessibility and lack of reaction of the larger reagents. Similarly, MTSET was less reactive with N336C (data not shown) and N340C (Supplemental Fig. 2B), as it did not prevent the subsequent effects of MTSEA. In contrast, the negatively charged MTSES slightly potentiated N340C (Supplemental Fig. 2A) and reacted silently with N336C (data not shown), in both cases preventing subsequent reaction with MTSEA. Thus, a measured effect on binding indicates that the substituted cysteine reacted, but the lack of an effect on binding is not conclusive evidence for lack of accessibility (as demonstrated by the lack of effect of MTSES despite its reaction with N336C).
The rate of reaction of MTSEA varied greatly among the sensitive cysteine mutants (Table 3 and Fig. 3). The most reactive mutant, N336C, had a rate constant of about 2000 M−1s−1. The reaction of MTSEA with F334C and Y335C was also quite rapid but with maximal inhibition of only about 50% (Figs. 1, 3, Table 3).
Table 3.
Rates of reaction of MTSEA
| KMTSEA ± SEM |
||
|---|---|---|
| Mutant | (M−1 s−1) | n |
| F332C | 455 ± 48 | 3 |
| S333C | 933 ± 110 | 5 |
| S334C | 353 ± 76 | 4 |
| Y335C | 243 ± 60 | 3 |
| N336C | 2113 ± 391 | 6 |
| F338C | 958 ± 373 | 3 |
| N340C | 649 ± 94 | 4 |
| M342C | 860 ± 134 | 7 |
| D345C | 90 ± 26 | 4 |
| T349C | 277 ± 33 | 6 |
n = number of experiments
Figure 3.
Rates of reaction of MTSEA. Membrane preparations of the mutants were incubated with increasing concentrations of MTSEA for 2 min at RT and washed, prior to the [3H]MFZ 2-12 binding assay, as described in Methods. The reactivity toward MTSEA varied greatly among the different cysteine mutants; note the 100-fold difference in the MTSEA concentrations for N336C and K337C. The data shown are from representative experiments performed in triplicate, and the summarized data are shown in Table 3.
3.5. The effect of cocaine and DA on MTSEA reaction
Ligand-dependent conformational changes that decrease access to a substituted cysteine or alter the microenvironment to reduce the reaction rate with MTSEA, would register as protection against the effect of MTSEA addition to the mutant constructs. Protection could also result from direct steric block by bound ligand, but since IL3 does not contribute directly to the DA and cocaine binding sites in DAT (Beuming et al., 2008), this seems unlikely for the mutants studied here. In contrast, increased reactivity is consistent only with an allosterically altered conformation.
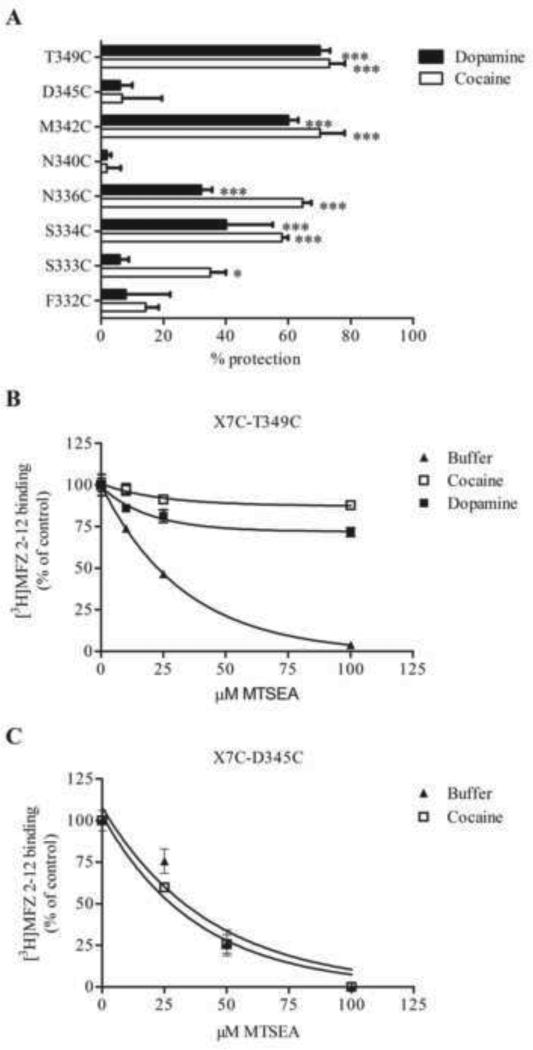
Membrane preparations of the MTSEA-sensitive mutants were treated with either cocaine or DA prior to and during treatment with MTSEA. A number of the mutants were significantly protected from MTSEA inhibition by cocaine. At S334C, N336C, M342C and T349C the presence of cocaine reduced the binding inhibition produced by MTSEA by 60-70% (Fig. 4A,B). The substrate DA also significantly protected against MTSEA reaction at S334C, N336C, M342C, and T349C (Fig. 4A,B) but failed to protect S333C, which was also less efficiently protected by cocaine (Fig. 4A). Neither DA nor cocaine protected F332C, N340C or D345C (Fig. 4A,C).
Figure 4.
Protection of MTSEA-induced binding inhibition by cocaine and dopamine. (A) Protection against MTSEA reaction by 0.1 mM cocaine or 1 mM dopamine was performed as described above. Percent protection is the change in MTSEA-inhibition seen with cocaine (white bars) or dopamine (black bars) present during MTSEA treatment relative to control (no ligand present). A MTSEA concentration that would give 70-80 % inhibition without ligand present was chosen for the individual mutants. (B, C) MTSEA effect on [3H]MFZ 2-12 binding in the absence (filled triangles) and presence of 0.1 mM (D345C) or 60 μM (T349C) cocaine (open squares), or 1 mM dopamine (filled squares). Membrane preparations of TM6-IL3-TM7 cysteine mutants were treated with ligand for 20 min, incubated with MTSEA, washed and assayed for [3H]MFZ 2-12 binding, as described in Methods. The results are from a representative experiment performed in triplicate. While the MTSEA-induced binding inhibition was greatly reduced in the presence of cocaine for several mutants (panels A and B), the MTSEA-sensitivity of some mutants was unaffected by the presence of ligand, as seen with D345C (panel C). The results are the mean ± SEM of 3-7 experiments. *, ** and *** indicate significantly different (p< 0.05, p<0.01 and p<0.001, respectively) from control (without ligand) by one way ANOVA and Dunnett's test. Y335C could not be tested due to poor binding of the radioligand.
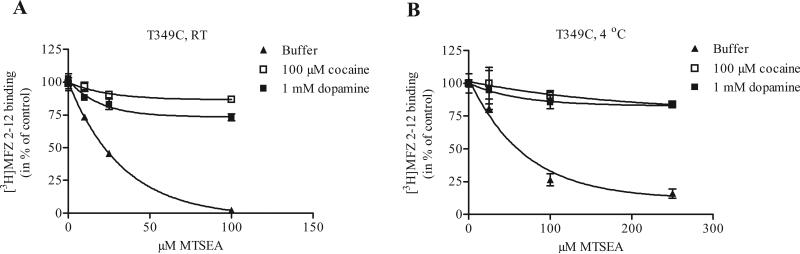
To test whether the presumed conformational changes induced by cocaine binding are restricted to higher temperatures, we carried out parallel protection experiments at 4 °C. Membrane preparations of N336C, M342C and T349C were incubated with cocaine and treated with MTSEA at 4 °C, with subsequent [3H]MFZ 2-12 binding at RT after removal of the MTS reagents at 4 °C. To compensate for the slower reaction rate at 4 °C, we increased the MTSEA concentration to achieve similar levels of inhibition. All three mutants were robustly protected by cocaine at 4 °C (shown for T349C in Fig. 5), consistent with an ability of DAT to undergo the conformational changes necessary to reduce reactivity with MTSEA at 4 °C, even though transport is extremely slow or absent at this temperature. This is in contrast to published inferences that at 4 °C protection of a substituted cysteine supports its location within the binding site where it must be sterically shielded by bound ligand.
Figure 5.
Protection by ligand is not temperature-dependent. The 4 °C- assay was performed just as for RT, except the membranes were kept at 4 °C during the cocaine and MTSEA incubations. The step with [3H]MFZ binding was performed at RT. To compensate for the slower reaction, 2.5-fold higher MTSEA concentrations were used in the 4 °C-assay. Similar protection by cocaine and dopamine at 4 °C was seen for N336C and M342C as well (not shown). Data displayed here are from a single representative experiment performed in triplicate.
3.6. Solvent accessible residues
As described in the Experimental Procedures, we created a homology model of DAT in the occluded state based on the LeuT structure and then established a model for the “inward-facing” conformation based on SMD simulation of DA movement towards the intracellular end of DAT from the S1 site, with the S2 site occupied by DA (Shan et al., 2011). The configuration of the TM domain on the intracellular side in the occluded model should reflect the properties of this region in an “outward facing” state since, like in the occluded state, the intracellular “gate” is expected to be closed. Indeed, the intracellular part of LeuT is almost identical in its occluded (PDB code: 2A65) structure and the Trp-bound outward facing (PDB code: 3F3A) structure (for this reason, the state of the investigated region of the intracellular side is referred to interchangeably as either “occluded conformation” or “outward-facing” conformation).
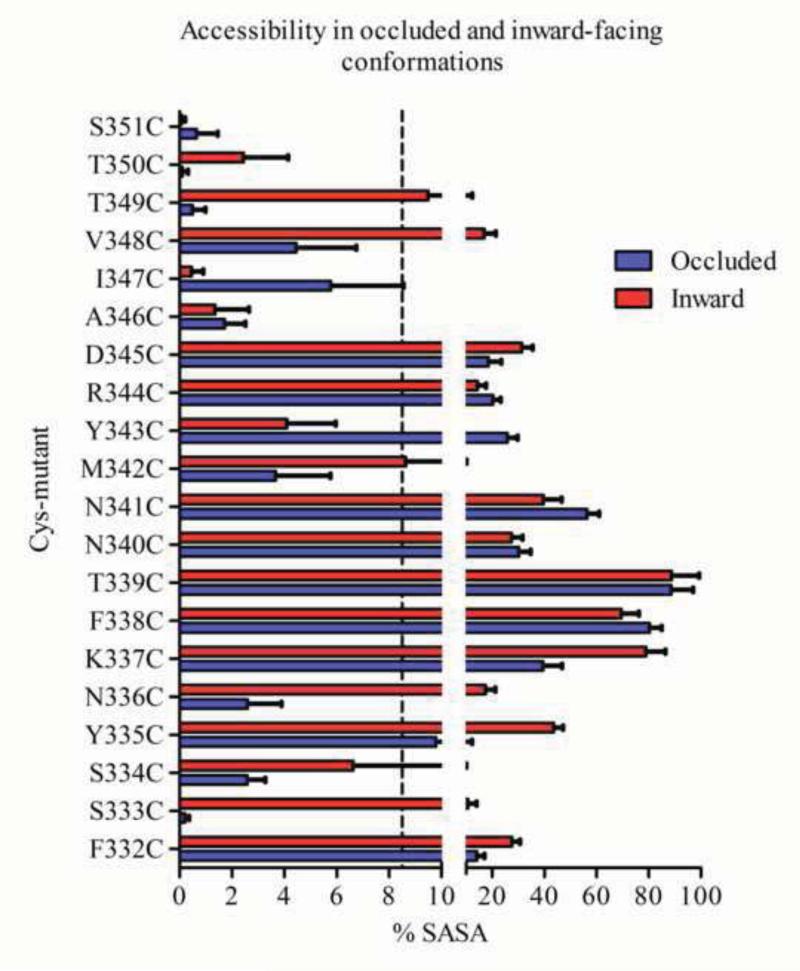
The solvent accessibility of the mutants was determined from SASA calculations for each residue in both the inward- and “outward-facing” conformations. We found that the predicted solvent accessibility correlates well with the effects of MTSEA on ligand binding. Thus the accessible residue positions identified by the SCAM study have larger SASA values than the buried positions in the corresponding conformation, with 8.5% as a threshold separating these two groups of positions (Supplemental Fig. 3). The 5 mutants S334C, A346C, I347C, T350C and S351C are buried, and their calculated SASA is <8.5% in both conformations (Fig. 6). As expected, ligand binding to these 5 mutants was not affected significantly by MTSEA (Fig. 2). All the other 15 mutants are predicted to be accessible with their SASA in either the outward or inward-facing conformation is greater than 8.5%. Among these 15 mutants, ligand binding to 10 of them (F332C, S333C, Y335C, N336C, F338C, T339C, N340C, M342C, D345C and T349C) was affected by the MTS reagents, consistent with their calculated accessibilities. Ligand binding to the remaining 5 mutants (K337C, N341C, Y343C, R344C and V348C) was not significantly affected by treatment with the MTS reagents. K337C and N341C are in the loop region and thus the MTS reagents might react “silently” without affecting ligand binding, as discussed above. Y343C, R344C and V348C face the lipid/solvent interface, so silent reaction might also explain the lack of functional effect at these positions. These findings from SASA calculations on a homology model of the human DAT agree substantially with the experimental accessibility measurements.
Figure 6.
SASA for TM6-IL3-TM7 in the occluded and inward-facing conformations of DAT. SASA values were calculated for each position after residues were mutated to cysteines, one at a time, and then minimized and equilibrated in a water/lipid environment. In the SASA analysis, only surface area accessible to solvents was counted; surface area exposed to lipids was treated as buried. The SASA percentage was obtained as described in Methods. The 8.5 % threshold chosen to define accessibility is indicated with a stippled line.
3.7. SASA differences of the protected positions
Because cocaine has been inferred to bind to the outward-facing conformation of the transporter (Beuming et al., 2008), the alternating-access model suggests that a residue position in the intracellular end of the transporter would be protected by cocaine if it was accessible in the inward-facing configuration but not in the outward-facing conformation. We found that the state-dependent accessibilities of the TM6-IL3-TM7 residues correlated well with the experimental protection data. The mutants that were significantly protected by cocaine, S333C, N336C, C342 and T349C (Fig. 4), are buried in the outward facing conformation with calculated SASA less than 4%. In contrast, in the inward facing conformation, residues in these positions are exposed with SASA greater than 8.5% (Fig. 6). The apparent increase in accessibility in moving from the outward to inward facing conformations is therefore >100% for these positions. Notably, the exposure of these residues in the inward-facing conformations is consistent with their reactivity with MTSEA when DAT is not stabilized by bound inhibitor so that it can interchange between the different conformations. Although the SASA of the Cys at position 334 was calculated to be <8.5% threshold, especially in the outward-facing conformation, binding to S334C was inhibited partially by MTSEA (Fig. 3) and was significantly protected by cocaine and dopamine (Fig. 4); the relative accessibility criterion is obeyed, however, as the calculated SASA in the inward-facing conformation was more than twice that in the outward-facing conformation (Fig. 6), consistent with the direction of the experimental observations. In contrast, the SASA values for F332C, N340C, and D345C were greater than 8.5% in both the outward and inward facing conformations (Fig. 6) and the changes in their SASA values are much smaller than for the protected residues. Since these residues are exposed to similar extents in both conformations, the corresponding substituted cysteine mutants react with MTSEA in the presence and absence of cocaine. This computational finding agrees with the experimental data, as cocaine failed to protect these mutants.
4. Discussion
The TM6-IL3-TM7 region is highly conserved across NSS, and, based on the LeuT structure and computational modeling and experimental studies of DAT (Beuming et al., 2008), it is not directly involved in ligand binding. Therefore, the accessibilities of residues in this region to the MTS reagents, and the ability of cocaine to alter these accessibilities, is likely indicative of the allosteric impact of ligand binding on the dynamic transitions between outward- and inward-facing states. Indeed, the present study provides evidence that residues in the TM6-IL3-TM7 region of DAT undergo conformational change upon ligand binding and presumably substrate translocation. Protection by DA was very similar to that by the inhibitor cocaine. The experiments were performed in membranes in the presence of high Na+ and the absence of a Na+ gradient. We have shown that under these conditions substrate stabilizes the inward-closed configuration, since inward opening requires loss of Na2 (Shi et al., 2008). Indeed, the LeuT crystal structure with Leu and Na+ bound (PDB 2A65) is in an inward-occluded configuration (Yamashita et al., 2005).
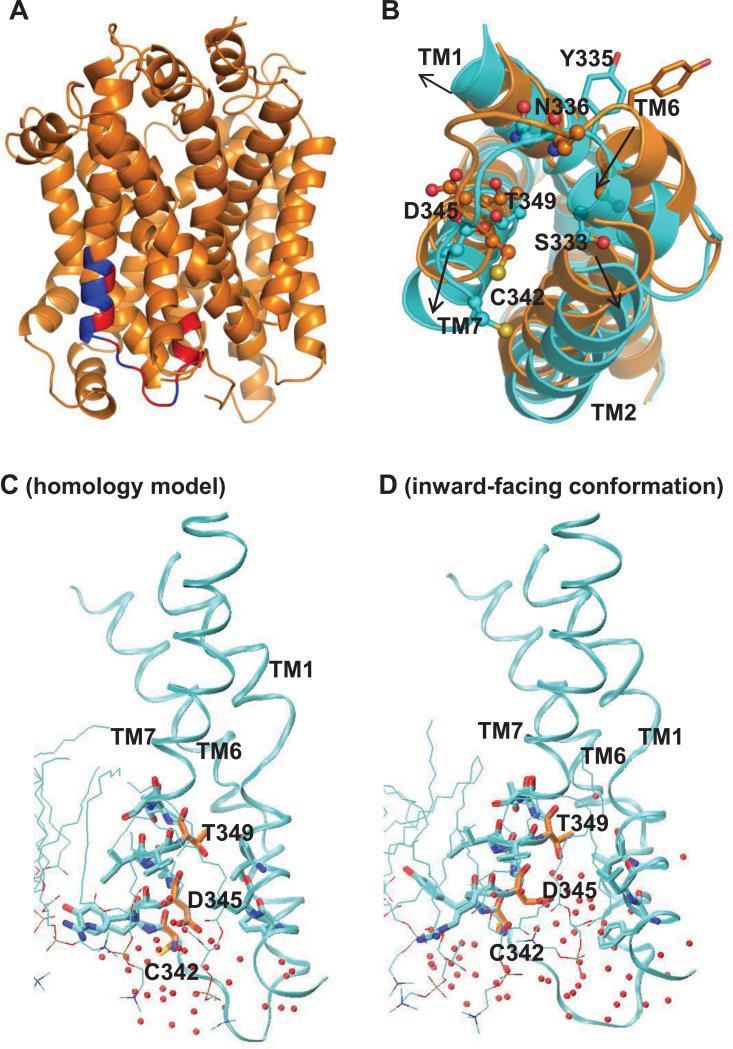
In our SMD simulations we observed significant rearrangements around TM6-IL3-TM7 when DA is translocated from the primary binding site to the intracellular side (Shan et al., 2011). These rearrangements, indicated by the arrows in Fig. 7b, increase the solvent exposure of Ser333, Asn336, Cys342, and Thr349 (Fig. 7b). Specifically, Thr349 forms close contacts with TM1, TM2 and TM6 in the outward-facing conformation; in the inward-facing conformation a cavity emerges between TM1, TM2, TM6 and TM7 due to their translations and rotations, and water molecules penetrate to the cavity from the intracellular side (Fig. 7c, 7d). The exposure of S333C, N336C, M342C, and T349C in the inward-facing conformation is consistent with our experimental data, which shows reactivity with MTS reagents (Fig. 2). The increased accessibilities for these residues in the inward-facing conformation also explain why the corresponding mutants were protected from reacting with MTSEA by cocaine pretreatment (Fig. 4), since inhibitor binding shifts the conformational equilibrium of DAT to the outward-facing conformation and makes these positions less accessible to MTS reagents.
Figure 7.
Global conformational changes around TM6-IL3-TM7 and accessibility changes of three TM7 residues comparing the occluded to inward-facing conformations. (A) Cartoon representation of the homology model of DAT. The experimental tested portion TM6-IL3-TM7 is colored in red and blue, representing, respectively, positions where [3H]MFZ 2-12 binding were affected by MTS reagents treatment (red) and those that were not (blue). (B) The global change in position of TMs 1, 2, 6 and 7, as well as loop IL3, from the occluded (orange) to inward facing conformation (cyan), which render Cys342, Asp345 and Thr349 (sticks) more exposed. Note that the SASA of Tyr335 and Asp345 is >8.5% in both the occluded and inward-facing conformations, and they are both more exposed in the inward-facing conformation. In addition, these residues are located on the same side of the helix as Cys 342 and Thr349. In the occluded state (C), residues Ser333, Ser334, Tyr335, and Asn336 are buried by interactions with TM1, but become more exposed to waters in the inward-facing conformation (D) where a water-filled cavity is created on the intracellular side between TMs 1, 2, 6 and 7 due to translational and rotational movements in that region.
Interestingly, the experimental data also correlate well with changes in the interaction networks of residues in this region of DAT, during conformational transitions between outward- and inward-facing conformations. For most mutants with intact uptake, the interaction networks do not change when DAT transitions from the outward- to inward-facing conformations. This is because they either interact with lipids or are buried in the protein without forming specific interactions or being exposed to the water phase. Specifically, residues Arg344, Ile347, Val348 and Ser351 face lipid; Ala346 is buried in the protein and forms non-specific hydrophobic interactions with a neighboring hydrophobic amino acid that can also be achieved by cysteine; Asn336, Lys337 and Thr339 are exposed to water and their interaction networks do not change during the transportation cycle.
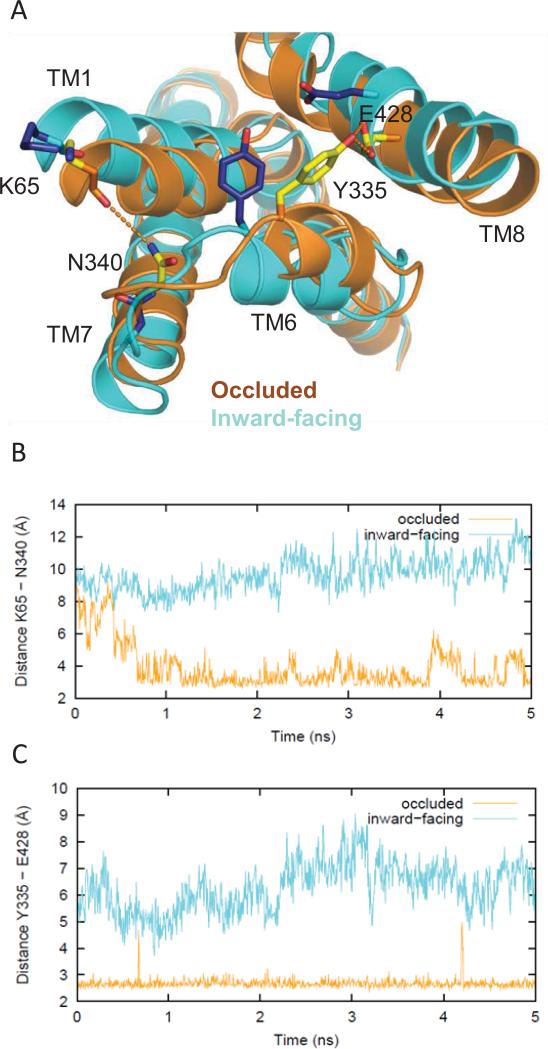
In contrast, most of the mutations that disrupt transport involve residues in interaction networks that are likely important for the conformational transition. Changes in their association with interacting residues were observed in our SMD/MD simulations when the transporter was steered toward an inward-facing conformation. Specifically, the mutations F332C, Y335C, F338C, N340C, N341C, and D345C destabilize interactions that are present in the outward-facing “starting” conformation, and thus are expected to disrupt transport. Thus, H-bonding of the pairs Tyr335-Glu4288.66 and Asn340-Lys651.31 seen in the occluded/outward-facing conformation, is no longer present in the inward-facing one (Fig. 8). In addition, residues Phe332 and Tyr335 are part of an aromatic cluster that also involves the neighboring Trp63NT, Phe691.35, and Phe761.42. When DA is translocated to the intracellular side in the simulation, the coordinated rotamer changes of these aromatic residues facilitate opening of the intracellular pathway (for details, see (Shan et al., 2011)). In particular, the rotation of Phe332 in TM6 pushes Phe69 in TM1 away, and thus TM1 swings out from the center of DAT; this creates a cavity between TM1 and TM6 which is detected by the calculated changes in SASA of TM6 residues from F332C to Y335C (Fig. 6).
Figure 8.
Differences in the network of interactions in the occluded vs inward-facing states. (A) Tyr335 and Glu428 form an H-bond that helps stabilize the occluded conformation (yellow side chains, orange ribbons), but is not seen in the inward-facing conformation (blue side chains, cyan ribbons). Therefore, the Y335C mutant favors an inward-facing conformation over the occluded one, thus disrupting dopamine uptake. Similarly, Lys65 in TM1 and Asn340 in TM7 form an H-bond in the occluded conformation but do not interact in the inward-facing conformation (same color scheme as above). (B) Evolution of the changes in the H-bonding monitored by the minimum distances between the hydroxyl oxygen of Tyr335 and the carboxyl oxygens of Glu428 in the occluded (orange) and inward-facing conformations (cyan) in the last 5 ns of the equilibrated phase of the simulation. (C) The distances between the carbonyl oxygen of Lys65 and the side chain amide nitrogen of Asn340 in the occluded (orange) and inward-facing conformations (cyan) in the last 5 ns of the equilibrated phase of the simulation.
Previous studies have identified two residues (Tyr335 and Asp345) in TM6-IL3-TM7 where point-mutation leads to a great loss of inhibitor affinities, accompanied by an increased affinity for substrate and a substantial decrease in uptake velocity (Chen et al., 2004; Loland et al., 2004; Loland et al., 2002). While binding of Zn2+ to the endogenous Zn2+ binding site in WT DAT inhibits DA uptake—possibly by restricting a conformational change necessary for transport - DA uptake by Y335A and D345A is restored by micromolar concentrations of Zn2+ (Loland et al., 1999; Norregaard et al., 1998). Our occluded and inward-facing conformations of DAT provide a structural context for understanding these effects. Tyramine uptake in nine of the twenty cysteine-mutants was potentiated rather than inhibited by the presence of Zn2+. In our models for these mutants, either the interaction networks stabilizing the occluded conformation were disrupted, or networks stabilizing the inward-facing conformation were established, thereby explaining why uptake can be potentiated by Zn2+ in these constructs: the binding of Zn2+ helps restore the transition to the occluded or outward-facing state if the conformational equilibrium had shifted toward an inward-facing conformation, like in Y335A. Taken together, our findings indicate that the TM6-IL3-TM7 region plays a critical role in the transition toward the inward-facing conformation and its stabilization.
Supplementary Material
Supplemental Figure 1. SASA for F332C and T349C in the occluded and inward facing conformations of DAT. SASA values were calculated for each position after residues were mutated to cysteines either by using VMD or by using FEP simulations, one at a time, and then minimized and equilibrated in a water/lipid environment. In SASA analysis, only surface area accessible to solvents was counted; surface area exposed to lipids was treated as buried. The SASA percentage was obtained by dividing the SASA calculated in the environment of the protein and lipids, by a SASA reference value. The reference value was calculated for any residue side chain, X, in a Gly-X-Gly tripeptide in extended conformations (Miller et al., 1987). For positions F332C and T349C, SASA were also calculated after mutating these positions to cysteines using FEP followed by 5 ns equilibration molecular dynamics.
Supplemental Figure 2. Silent vs less reactive mutants. Some mutants were less inhibited by MTSES and/or MTSET than by MTSEA, as seen here for N340C and T349C. To distinguish between less affected by and less reactive to the MTS treatment, membrane preparations of these mutants were first treated with MTSES (A) or MTSET (B) for 2 min, washed, and then treated with MTSEA for 2 min, prior to [3H]MFZ binding. When MTSEA was able to inhibit binding following MTSET/MTSES binding, the mutants were considered less reactive to either MTSET (N340C and T349C) or MTSES (T349C). When MTSEA was not able to inhibit binding, the mutant was considered to react silently with MTSES (N340C).
Supplemental Figure 3. Calculated values of percent exposure (%SASA) are shown for the occluded- and inward-facing states. The red separator reveals the results obtained using 8.5% SASA exposed in its ability to collect the accessible residues discussed in the manuscript.
Highlights.
Using SCAM we probed the water accessibility of a stretch in TM6-IL3-TM7 of hDAT.
A subset of the water-accessible residues was protected by cocaine binding.
We infer that cocaine stabilizes an outward-facing, inward-occluded state.
Steered-molecular dynamics revealed a consistent rearrangement during transport.
A number of cysteine mutants with impaired transport were rescued by zinc binding.
Acknowledgements
This work was supported by the National Institutes of Health grants DA022413 and DA17293 to JAJ, DA012408 and U54GM087519 to H.W., and DA023694 to L.S. Computations were performed on Ranger at the Texas Advanced Computing Center (TG-MCB090022), and the Cofrin Center for Biomedical Information of the Institute for Computational Biomedicine at Weill Cornell Medical College.
Abbreviations
- ANOVA
analysis of variance
- BCA
bicinchoninic acid
- DA
dopamine
- DAT
dopamine transporter
- FEP
free energy perturbation
- GABA
gamma-amino butyric acid
- GAT
GABA transporter
- HRP
horseradish peroxidase
- IL3
intracellular loop 3
- MD
molecular dynamics
- MTS
methanethiosulfonate
- MTSEA
MTS ethyl ammonium
- MTSES
MTS ethylsulfonate
- MTSET
MTS ethyltrimethylammonium
- NET
norepinephrine transporter
- NSS
neurotransmitter: Na+ symporter
- PVDF
polyvinylidene fluoride
- S1
primary substrate binding site
- S2
second substrate binding site
- SASA
solvent accessible surface area
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- SMD
steered molecular dynamics
- SCAM
substituted cysteine accessibility method
- SERT
serotonin transporter
- TCA
tricyclic antidepressants
- TM
transmembrane segment
- VMD
Visual Molecular Dynamics
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary data
Supplementary data (supplemental figure 1-2 and table 1) associated with this article can be found...
References
- Amara SG, Sonders MS. Neurotransmitter transporters as molecular targets for addictive drugs. Drug Alcohol Depend. 1998;51:87–96. doi: 10.1016/s0376-8716(98)00068-4. [DOI] [PubMed] [Google Scholar]
- Beuming T, Kniazeff J, Bergmann ML, Shi L, Gracia L, Raniszewska K, Newman AH, Javitch JA, Weinstein H, Gether U, Loland CJ. The binding sites for cocaine and dopamine in the dopamine transporter overlap. Nat Neurosci. 2008;11:780–789. doi: 10.1038/nn.2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beuming T, Shi L, Javitch JA, Weinstein H. A comprehensive structure-based alignment of prokaryotic and eukaryotic neurotransmitter/Na+ symporters (NSS) aids in the use of the LeuT structure to probe NSS structure and function. Mol Pharmacol. 2006;70:1630–1642. doi: 10.1124/mol.106.026120. [DOI] [PubMed] [Google Scholar]
- Celik L, Sinning S, Severinsen K, Hansen CG, Moller MS, Bols M, Wiborg O, Schiott B. Binding of serotonin to the human serotonin transporter. Molecular modeling and experimental validation. J Am Chem Soc. 2008;130:3853–3865. doi: 10.1021/ja076403h. [DOI] [PubMed] [Google Scholar]
- Chen J-G, Sachpatzidis A, Rudnick G. The Third Transmembrane Domain of the Serotonin Transporter Contains Residues Associated with Substrate and Cocaine Binding. J Biol Chem. 1997;272:28321–28327. doi: 10.1074/jbc.272.45.28321. [DOI] [PubMed] [Google Scholar]
- Chen N, Ferrer JV, Javitch JA, Justice JB. Transport-dependent Accessibility of a Cytoplasmic Loop Cysteine in the Human Dopamine Transporter. J Biol Chem. 2000;275:1608–1614. doi: 10.1074/jbc.275.3.1608. [DOI] [PubMed] [Google Scholar]
- Chen N, Justice JB., Jr. Cocaine acts as an apparent competitive inhibitor at the outward-facing conformation of the human norepinephrine transporter: kinetic analysis of inward and outward transport. J Neurosci. 1998;18:10257–10268. doi: 10.1523/JNEUROSCI.18-24-10257.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N, Rickey J, Berfield JL, Reith MEA. Aspartate 345 of the Dopamine Transporter Is Critical for Conformational Changes in Substrate Translocation and Cocaine Binding. J Biol Chem. 2004;279:5508–5519. doi: 10.1074/jbc.M306294200. [DOI] [PubMed] [Google Scholar]
- Cheng, Mary H, Bahar I. Coupled Global and Local Changes Direct Substrate Translocation by Neurotransmitter-Sodium Symporter Ortholog LeuT. Biophys J. 2013;105:630–639. doi: 10.1016/j.bpj.2013.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshleman AJ, Stewart E, Evenson AK, Mason JN, Blakely RD, Janowsky A, Neve KA. Metabolism of catecholamines by catechol-O-methyltransferase in cells expressing recombinant catecholamine transporters. J Neurochem. 1997;69:1459–1466. doi: 10.1046/j.1471-4159.1997.69041459.x. [DOI] [PubMed] [Google Scholar]
- Ferrer JV, Javitch JA. Cocaine alters the accessibility of endogenous cysteines in putative extracellular and intracellular loops of the human dopamine transporter. Proc Natl Acad Sci U S A. 1998;95:9238–9243. doi: 10.1073/pnas.95.16.9238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrest LR, Tang CL, Honig B. On the Accuracy of Homology Modeling and Sequence Alignment Methods Applied to Membrane Proteins. Biophys J. 2006;91:508–517. doi: 10.1529/biophysj.106.082313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrest LR, Zhang YW, Jacobs MT, Gesmonde J, Xie L, Honig BH, Rudnick G. Mechanism for alternating access in neurotransmitter transporters. Proc Natl Acad Sci U S A. 2008;105:10338–10343. doi: 10.1073/pnas.0804659105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H, Wall SC, Rudnick G. Stable expression of biogenic amine transporters reveals differences in inhibitor sensitivity, kinetics, and ion dependence. J Biol Chem. 1994;269:7124–7130. [PubMed] [Google Scholar]
- Hastrup H, Karlin A, Javitch JA. Symmetrical dimer of the human dopamine transporter revealed by cross-linking Cys-306 at the extracellular end of the sixth transmembrane segment. Proc Natl Acad Sci U S A. 2001;98:10055–10060. doi: 10.1073/pnas.181344298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henin J, Maigret B, Tarek M, Escrieut C, Fourmy D, Chipot C. Probing a model of a GPCR/ligand complex in an explicit membrane environment: the human cholecystokinin-1 receptor. Biophys J. 2006;90:1232–1240. doi: 10.1529/biophysj.105.070599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry LK, Adkins EM, Han Q, Blakely RD. Serotonin and Cocaine-sensitive Inactivation of Human Serotonin Transporters by Methanethiosulfonates Targeted to Transmembrane Domain I. J Biol Chem. 2003;278:37052–37063. doi: 10.1074/jbc.M305514200. [DOI] [PubMed] [Google Scholar]
- Huang X, Gu HH, Zhan C-G. Mechanism for Cocaine Blocking the Transport of Dopamine: Insights from Molecular Modeling and Dynamics Simulations. J Phys Chem B. 2009;113:15057–15066. doi: 10.1021/jp900963n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33–38. 27–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Kanner BI, Zomot E. Sodium-Coupled Neurotransmitter Transporters. Chem Rev. 2008;108:1654–1668. doi: 10.1021/cr078246a. [DOI] [PubMed] [Google Scholar]
- Karlin A, Akabas MH. Substituted-cysteine accessibility method. Methods Enzymol. 1998;293:123–145. doi: 10.1016/s0076-6879(98)93011-7. [DOI] [PubMed] [Google Scholar]
- Kaufmann KW, Dawson ES, Henry LK, Field JR, Blakely RD, Meiler J. Structural determinants of species-selective substrate recognition in human and Drosophila serotonin transporters revealed through computational docking studies. Proteins. 2009;74:630–642. doi: 10.1002/prot.22178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kniazeff J, Shi L, Loland CJ, Javitch JA, Weinstein H, Gether U. An Intracellular Interaction Network Regulates Conformational Transitions in the Dopamine Transporter. J Biol Chem. 2008;283:17691–17701. doi: 10.1074/jbc.M800475200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koob GF. Drugs of abuse: anatomy, pharmacology and function of reward pathways. Trends Pharmacol Sci. 1992;13:177–184. doi: 10.1016/0165-6147(92)90060-j. [DOI] [PubMed] [Google Scholar]
- Krause S, Schwarz W. Identification and selective inhibition of the channel mode of the neuronal GABA transporter 1. Mol Pharmacol. 2005;68:1728–1735. doi: 10.1124/mol.105.013870. [DOI] [PubMed] [Google Scholar]
- Krishnamurthy H, Gouaux E. X-ray structures of LeuT in substrate-free outward-open and apo inward-open states. Nature. 2012;481:469–474. doi: 10.1038/nature10737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Loland CJ, Granas C, Javitch JA, Gether U. Identification of intracellular residues in the dopamine transporter critical for regulation of transporter conformation and cocaine binding. J Biol Chem. 2004;279:3228–3238. doi: 10.1074/jbc.M304755200. [DOI] [PubMed] [Google Scholar]
- Loland CJ, Norregaard L, Gether U. Defining Proximity Relationships in the Tertiary Structure of the Dopamine Transporter. J Biol Chem. 1999;274:36928–36934. doi: 10.1074/jbc.274.52.36928. [DOI] [PubMed] [Google Scholar]
- Loland CJ, Norregaard L, Litman T, Gether U. Generation of an activating Zn(2+) switch in the dopamine transporter: mutation of an intracellular tyrosine constitutively alters the conformational equilibrium of the transport cycle. Proc Natl Acad Sci U S A. 2002;99:1683–1688. doi: 10.1073/pnas.032386299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller S, Lesk AM, Janin J, Chothia C. The accessible surface area and stability of oligomeric proteins. Nature. 1987;328:834–836. doi: 10.1038/328834a0. [DOI] [PubMed] [Google Scholar]
- Newman AH, Zou MF, Ferrer JV, Javitch JA. [3H]MFZ 2-12: a novel radioligand for the dopamine transporter. Bioorg Med Chem Lett. 2001;11:1659–1661. doi: 10.1016/s0960-894x(01)00271-2. [DOI] [PubMed] [Google Scholar]
- Norregaard L, Frederiksen D, Nielsen EO, Gether U. Delineation of an endogenous zinc-binding site in the human dopamine transporter. EMBO J. 1998;17:4266–4273. doi: 10.1093/emboj/17.15.4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penmatsa A, Wang KH, Gouaux E. X-ray structure of dopamine transporter elucidates antidepressant mechanism. Nature. 2013;503:85–90. doi: 10.1038/nature12533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins AK, Horlick RA. Macrophage scavenger receptor confers an adherent phenotype to cells in culture. Biotechniques. 1998;25:240–244. doi: 10.2144/98252st04. [DOI] [PubMed] [Google Scholar]
- Rudnick G. Mechanisms of biogenic amine neurotransmitter transporters. In: Reith ME, editor. Neurotransmitter Transporters: Structure, Function, and Regulation. 2 ed Humana Press Inc.; Totowa, New Jersey: 2002. pp. 25–52. [Google Scholar]
- Rudnick G. Serotonin transporters--structure and function. J Membr Biol. 2006;213:101–110. doi: 10.1007/s00232-006-0878-4. [DOI] [PubMed] [Google Scholar]
- Saunders C, Ferrer JV, Shi L, Chen J, Merrill G, Lamb ME, Leeb-Lundberg LMF, Carvelli L, Javitch JA, Galli A. Amphetamine-induced loss of human dopamine transporter activity: An internalization-dependent and cocaine-sensitive mechanism. Proc Natl Acad Sci U S A. 2000;97:6850–6855. doi: 10.1073/pnas.110035297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan J, Javitch JA, Shi L, Weinstein H. The Substrate-Driven Transition to an Inward-Facing Conformation in the Functional Mechanism of the Dopamine Transporter. PLoS ONE. 2011;6:e16350. doi: 10.1371/journal.pone.0016350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Quick M, Zhao Y, Weinstein H, Javitch JA. The mechanism of a neurotransmitter:sodium symporter--inward release of Na+ and substrate is triggered by substrate in a second binding site. Mol Cell. 2008;30:667–677. doi: 10.1016/j.molcel.2008.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SK, Piscitelli CL, Yamashita A, Gouaux E. A Competitive Inhibitor Traps LeuT in an Open-to-Out Conformation. Science. 2008;322:1655–1661. doi: 10.1126/science.1166777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SK, Yamashita A, Gouaux E. Antidepressant binding site in a bacterial homologue of neurotransmitter transporters. Nature. 2007;448:952–956. doi: 10.1038/nature06038. [DOI] [PubMed] [Google Scholar]
- Sonders MS, Quick M, Javitch JA. How did the neurotransmitter cross the bilayer? A closer view. Curr Opin Neurobiol. 2005;15:296–304. doi: 10.1016/j.conb.2005.05.009. [DOI] [PubMed] [Google Scholar]
- Torres GE, Gainetdinov RR, Caron MG. Plasma membrane monoamine transporters: structure, regulation and function. Nat Rev Neurosci. 2003;4:13–25. doi: 10.1038/nrn1008. [DOI] [PubMed] [Google Scholar]
- Volkow ND, Fowler JS, Wang G, Ding Y, Gatley SJ. Mechanism of action of methylphenidate: insights from PET imaging studies. J Atten Disord. 2002;6(Suppl 1):S31–43. doi: 10.1177/070674370200601s05. [DOI] [PubMed] [Google Scholar]
- Yamashita A, Singh SK, Kawate T, Jin Y, Gouaux E. Crystal structure of a bacterial homologue of Na+/Cl--dependent neurotransmitter transporters. Nature. 2005;437:215–223. doi: 10.1038/nature03978. [DOI] [PubMed] [Google Scholar]
- Zhang Y-W, Rudnick G. The Cytoplasmic Substrate Permeation Pathway of Serotonin Transporter. J Biol Chem. 2006;281:36213–36220. doi: 10.1074/jbc.M605468200. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Terry D, Shi L, Weinstein H, Blanchard SC, Javitch JA. Single-molecule dynamics of gating in a neurotransmitter transporter homologue. Nature. 2010;465:188–193. doi: 10.1038/nature09057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Terry DS, Shi L, Quick M, Weinstein H, Blanchard SC, Javitch JA. Substrate-modulated gating dynamics in a Na+-coupled neurotransmitter transporter homologue. Nature. 2011;474:109–113. doi: 10.1038/nature09971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Zhen J, Karpowich NK, Goetz RM, Law CJ, Reith MEA, Wang D-N. LeuT-Desipramine Structure Reveals How Antidepressants Block Neurotransmitter Reuptake. Science. 2007;317:1390–1393. doi: 10.1126/science.1147614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Zhen J, Karpowich NK, Law CJ, Reith MEA, Wang D-N. Antidepressant specificity of serotonin transporter suggested by three LeuT-SSRI structures. Nat Struct Mol Biol. 2009;16:652–657. doi: 10.1038/nsmb.1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. SASA for F332C and T349C in the occluded and inward facing conformations of DAT. SASA values were calculated for each position after residues were mutated to cysteines either by using VMD or by using FEP simulations, one at a time, and then minimized and equilibrated in a water/lipid environment. In SASA analysis, only surface area accessible to solvents was counted; surface area exposed to lipids was treated as buried. The SASA percentage was obtained by dividing the SASA calculated in the environment of the protein and lipids, by a SASA reference value. The reference value was calculated for any residue side chain, X, in a Gly-X-Gly tripeptide in extended conformations (Miller et al., 1987). For positions F332C and T349C, SASA were also calculated after mutating these positions to cysteines using FEP followed by 5 ns equilibration molecular dynamics.
Supplemental Figure 2. Silent vs less reactive mutants. Some mutants were less inhibited by MTSES and/or MTSET than by MTSEA, as seen here for N340C and T349C. To distinguish between less affected by and less reactive to the MTS treatment, membrane preparations of these mutants were first treated with MTSES (A) or MTSET (B) for 2 min, washed, and then treated with MTSEA for 2 min, prior to [3H]MFZ binding. When MTSEA was able to inhibit binding following MTSET/MTSES binding, the mutants were considered less reactive to either MTSET (N340C and T349C) or MTSES (T349C). When MTSEA was not able to inhibit binding, the mutant was considered to react silently with MTSES (N340C).
Supplemental Figure 3. Calculated values of percent exposure (%SASA) are shown for the occluded- and inward-facing states. The red separator reveals the results obtained using 8.5% SASA exposed in its ability to collect the accessible residues discussed in the manuscript.