Abstract
The bone morphogenetic protein (BMP) signaling pathway is essential for normal development and tissue homeostasis. BMP signal transduction occurs when ligands interact with a complex of type 1 and type 2 receptors to activate downstream transcription factors. It is well established that a single BMP receptor may bind multiple BMP ligands with varying affinity, and this has been largely attributed to conformation at the amino acid level. However, all three type 2 BMP receptors (BMPR2, ACVR2A/B) contain consensus N-glycosylation sites in their extracellular domains (ECDs), which could play a role in modulating interaction with ligand. Here, we show a differential pattern of N-glycosylation between BMPR2 and ACVR2A/B. Site-directed mutagenesis reveals that BMPR2 is uniquely glycosylated near its ligand binding domain and at a position that is mutated in patients with heritable pulmonary arterial hypertension. We further demonstrate using a cell-free pulldown assay that N-glycosylation of the BMPR2-ECD enhances its ability to bind BMP2 ligand but has no impact on binding by the closely-related ACVR2B. Our results illuminate a novel aspect of BMP signaling pathway mechanics and demonstrate a functional difference resulting from post-translational modification of type 2 BMP receptors. Additionally, since BMPR2 is required for several aspects of normal development and defects in its function are strongly implicated in human disease, our findings are likely to be relevant in several biological contexts in normal and abnormal human physiology.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-013-1541-8) contains supplementary material, which is available to authorized users.
Keywords: Bone morphogenetic protein, BMPR2, ACVR2A, ACVR2B, Activin, Glycosylation, Pulmonary hypertension, Heritable pulmonary arterial hypertension
Introduction
The bone morphogenetic protein (BMP) signaling pathway has ancient origins in the evolution of metazoans [1, 2], where it plays an essential role in early developmental processes such as gastrulation and axis determination [3, 4]. BMP ligands exert their effects by interacting with a hetero-oligomeric complex of type 1 and type 2 receptors to activate downstream transcription factors [5, 6]. While the basic mechanics of BMP signal transduction are strikingly well conserved across all animals [1], gene duplication events are thought to have led to the sophisticated superfamily of ligands, receptors, and downstream effectors present in higher organisms [7]. For example, the mammalian genome contains approximately 20 BMP ligands, 7 type 1 receptors (ALK1/2/3/6), and 3 type 2 receptors (BMPR2, ACVR2A/B) that mediate their BMP activities through three canonical transcription factors (SMAD1/5/8) and a growing set of non-canonical effectors [5, 8, 9]. The importance of this fundamental pathway to human health is underscored by the fact that defects in every BMP receptor except ACVR2A have been implicated in a human disease [10–18].
Numerous studies have demonstrated that each BMP receptor has the capacity to interact with many BMP ligands with varying affinity (e.g., see Heinecke et al. [19]). This plasticity has largely been attributed to differences at the amino acid level affecting three-dimensional structures [20–27] or post-translational modifications (PTM) that occur on specific ligands [28]. However, considerably less attention has been paid to the role that PTMs in the receptor extra-cellular domain (ECD) might play in altering receptor: ligand interactions in the BMP pathway. The most common PTM in eukaryotes is the covalent addition of carbohydrate groups to asparagine residues, called N-glycosylation [29]. N-glycosylation substantially modifies the structure, localization, and function of glycoproteins [30], and each BMP type 2 receptor contains N-glycosylation sites in its ECD [31–38]. Moreover, three patients with the rare disease heritable pulmonary arterial hypertension (HPAH), which is linked to loss-of-function mutations in BMPR2 [10, 11], carry a point mutation in the same putative N-glycosylation site of BMPR2 (N126) [39, 40]. Yet, it is entirely unknown whether N-glycosylation impacts the function of type 2 BMP receptors.
In this study, we compare the putative glycosylation patterns for the ECD of each type 2 BMP receptor (BMPR2, ACVR2A, and ACVR2B) in light of their respective crystal structures, and find that differential N-glycosylation exists between ACVR2A/B and BMPR2: N-glycosylation of ACVR2A/B occurs distant to the ligand binding face while N-glycosylation of BMPR2 approximates the ligand binding domain. Using site-directed mutagenesis, we demonstrate that BMPR2 is glycosylated at three asparagine residues and that this N-glycosylation enhances the ability of the BMPR2-ECD to bind the ligand BMP2. In contrast, N-linked glycosylation of the ACVR2B-ECD is dispensable for BMP2 binding. Our results illuminate a novel aspect of the basic BMP pathway mechanics and demonstrate differences between the type 2 BMP receptors. Additionally, since BMPR2 is required for several aspects of normal development [41–43] and defects in its function are strongly implicated in disease [44], our findings are likely to be relevant to several contexts in normal and abnormal human physiology.
Materials and methods
Cell culture
W-20-17 (W20) cells [52] and HEK293T cells [53], obtained from ATCC, were cultured in DMEM supplemented with 10 % fetal bovine serum (Gibco) and grown at 37 °C in 5 % CO2. Transient transfection of plasmid DNA was achieved using X-tremeGENE 9 (Roche). Knockdown of Bmpr2 was performed using shRNA clone TRCN0000022530 (Sigma).
Plasmid construction
A plasmid containing the full-length human BMPR2 cDNA was obtained as a gift from Dr. James West (Vanderbilt). This was sub-cloned into pCRII-TOPO (Invitrogen) via TA cloning using the primers detailed in Table S1 to generate the shuttle vector pJL69-8. hBMPR2 cDNA was then sub-cloned into pcDNA3.1/V5-His-TOPO (Invitrogen) via TA cloning using primers detailed in Table S1 and GoTaq polymerase (Promega) to generate the pJL74-5 expression vector, which appends C-terminal V5 and His tags to hBMPR2. Mutant versions of hBMPR2 cDNA were then constructed via inverse PCR using AccuPrime Pfx polymerase (Invitrogen) followed by digestion with DpnI (New England Biolabs) using primers as outlined in Table S1. All plasmids were sequenced in forward and reverse directions at the Dana-Farber/Harvard Cancer Center DNA Resource Core to confirm fidelity.
Western blotting
Western blots were performed on protein isolates from W20 or HEK293T cells after lysis in RIPA buffer (50 mM Tris Base, 150 mM NaCl, 1 % NP-40, 0.5 % sodium deoxycholate, 0.1 % SDS, pH 8.0) supplemented with Halt Protease & Phosphatase Inhibitor Cocktail (Thermo). Lysates were resolved on Novex Tris–Glycine gels (Invitrogen) and transferred to Amersham Hybond ECL nitrocellulose membranes (GE Healthcare). All samples were denatured by heating at 100 °C for 10 min after mixing with 6X reducing sample buffer (60 % glycerol, 300 mM Tris pH 6.8, 12 mM EDTA, 12 % SDS, 864 mM 2-mercaptoethanol, 0.05 % bromophenol blue). After blocking in 10 % milk in PBST (PBS + 0.1 % Tween-20), the following primary antibodies were applied in 5 % milk in PBST: anti-BMPR2 C-terminal domain (BD Biosciences, 612292), anti-His tag (Abcam, ab9108), anti-V5 tag (Abcam, ab27671), anti-HSP90 (Santa Cruz, sc-7947), and anti-BMP2 (R&D Systems, MAB3551). Appropriate HRP-conjugated, species-specific goat polyclonal secondary antibodies (anti-mouse: Kirkegaard & Perry Laboratories, 04-18-06; and anti-rabbit: Cell Signaling, 7074) were utilized and western blots were developed by chemiluminscence using SuperSignal West Femto Substrate (Thermo). Stripping of membranes for re-probing was accomplished using Restore Western Blot Stripping Buffer (Thermo). Western blots were visualized using a Syngene Pxi imager and quantified by ImageJ.
Receptor ECD pulldown assay
An amount of 500 ng native or de-glycosylated human BMPR2-ECD/Fc fusion (R&D Systems, #811-BR-100) or ACVR2B-ECD/Fc fusion (R&D Systems, #339-RB-100) was mixed with 10 μl Protein G-coupled Dynabeads (Invitrogen) at room temperature for 30 min in 200 μl total volume with rotation. The loaded beads were then washed twice with PBS and resuspended in 200 μl PBS ± 1 μg rhBMP2 produced in CHO cells (Genetics Institute/Wyeth) and incubated overnight at 4 °C with gentle shaking. After five washes with 200 μl PBS, the beads were resuspended in 100 μl PBS, transferred to a clean tube, aspirated, resuspended in 1× reducing sample buffer, and heated at 100 °C for 10 min.
Although the efficiency of ECD loading to the beads did not differ significantly between native and de-glycosylated isoforms (BMPR2: p = 0.8878; ACVR2B: p = 7434, by unpaired t test), some variability in loading due to unknown origin was observed across replicates. For this reason, our pulldown assays were quantified by determining the ratio of the BMP2 band density to the receptor ECD band density in order to account for any variability in loading for each independent sample. This ratio was then expressed as mean ± SEM percent relative to native receptor ECD pulldown ratio.
PNGase F digestion assay
Digestion with peptide-N-glycosidase-F (PNGase F; New England Biolabs) was performed on W20 and HEK293T cell lysates for 1 h as directed by the manufacturer. For digestion of the soluble hBMPR2-ECD and hACVR2B-ECD for use in pulldown assays, the initial denaturing step and denaturing buffer was left out to maintain native conformation.
In silico analyses
All in silico analyses were performed using the RefSeq amino acid sequences as follows: mouse BMPR2–NP_031587.1; human BMPR2–NP_001195.2; human ACVR2A–NP_001067.1; and human ACVR2B–NP_001097.2. Prediction of N-linked glycosylation sites was performed using NetNGlyc (http://www.cbs.dtu.dk/services/NetNGlyc). Prediction of transmembrane regions (J.W.L., data not shown) was performed using TMHMM (http://www.cbs.dtu.dk/services/TMHMM/). Amino acid alignments were performed using MUSCLE [54] in MEGA5 [55] on amino acid sequences as detailed in each respective figure. Images in Fig. 1c, d were generated using PyMOL (Schrodinger) on BMPR2 (PDB ID: 2HLQ), ACVR2A (PDB ID: 2H62) and ACVR2B (PDB ID: 1BTE).
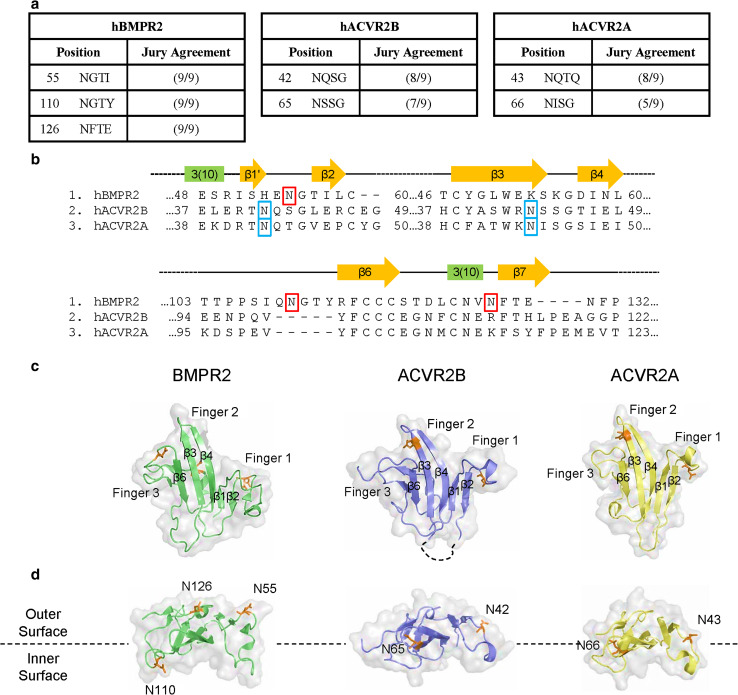
Fig. 1.
Type 2 BMP receptors are predicted to be glycoproteins. a Prediction of N-linked glycosylation sites in hBMPR2, hACVR2A, and hACVR2B using NetNGlyc. b Alignment comparison of type 2 BMP receptors. Human isoforms of BMPR2 (NP_031587.1), ACVR2B (NP_001097.2), and ACVR2A (NP_001067.1) were aligned using MUSCLE [54] in MEGA5 [55]. Secondary structures are annotated after Mace et al. [46] and Yin et al. [26]. Asparagine residues predicted to be glycosylated are indicated by red boxes in hBMPR2 and blue boxes in ACVR2A/B. Residue numbers indicate the position in each respective RefSeq sequence without gaps. c, d Structural comparison of BMPR2 to ACVR2A/B, highlighting the unique glycosylation sites in BMPR2. Views in (c) are the interior, ligand-binding faces of the extra-cellular domains. Select β-strands are annotated after Mace et al. [46] and Yin et al. [26]. Views in (d) are rotated 90° in the forward orientation (toward the reader) from (c). Interior Surface and Exterior Surface refer to the faces of the molecules with respect to the ternary complex
Statistical analysis
Statistical significance was determined by unpaired t test using GraphPad Prism. A p value of <0.05 was considered significant. Numbers of replicates are indicated in each respective figure legend.
Results
Differential N-glycosylation pattern between BMPR2 and ACVR2A/B
NetNGlyc analysis of the mouse and human RefSeq protein sequences for the type 2 BMP receptors predicts that three asparagine residues (N55, N110, and N126) in the ECD of BMPR2 are glycosylated while only two are glycosylated in ACVR2A/B (Fig. 1a). Amino acid sequence alignment (Fig. 1b) and crystal structure comparison (Fig. 1c) suggest that N55 in BMPR2 is analogous to N42 and N43 in ACVR2B and ACVR2A, respectively. In contrast, the putatively glycosylated residues N110 and N126 in BMPR2 and N66/N65 in ACVR2A/B raise the possibility that BMPR2 is glycosylated in a pattern distinct from ACVR2A/B (Fig. 1c, d). Moreover, the proximity of N110 in BMPR2 to the ligand binding hydrophobic patch [27] suggests that N-glycosylation of BMPR2 could be functionally significant.
Determination of glycosylation sites in BMPR2 ECD
In the clonal mouse bone marrow stromal cell line W-20-17 (W20), endogenous BMPR2 appears as a doublet of approximately 150 kDa (major band) and 140 kDa (minor band) (Fig. 2a, lane 1). Both bands are absent when Bmpr2 is knocked-down (Fig. 2a, lanes 3, 4), identifying each as an isoform of BMPR2. Digestion with the endoglycosidase PNGase F causes a shift in electrophoretic mobility of both bands, confirming that BMPR2 is glycosylated endogenously (Fig. 2a, lane 2).
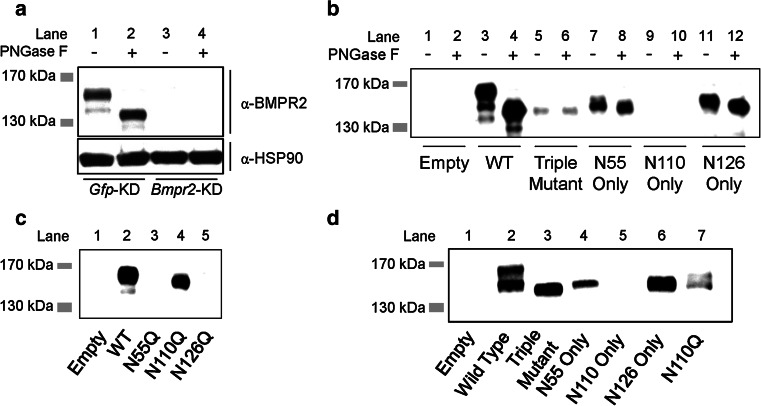
Fig. 2.
Determination of N-linked glycosylation sites in BMPR2. a Western blot for glycosylation status of endogenous BMPR2 in W20 cells compared to HSP90 loading control. Lysates were treated with vehicle or PNGase F. Specificity of anti-BMPR2 antibody is confirmed by lack of signal after Bmpr2 knockdown (Bmpr2-KD); shRNA against Gfp serves as negative control (Gfp-KD). Image is representative of two independent experiments. b Western blot for V5-tagged wild-type and triple or double mutant versions of hBMPR2 in W20 cell lysates treated with vehicle or PNGase F. c Expression analysis of V5-tagged wild-type and single glycosylation mutant versions of hBMPR2 in W20 cells. d Expression analysis of V5-tagged wild-type and mutant versions of hBMPR2 in HEK293T cells. Blots in b–d were performed using anti-V5 antibody; specificity of anti-V5 antibody is confirmed by lack of signal in lysates from cells transfected with empty plasmid control. Triple mutant is N55Q; N11Q; N126Q. N55 Only mutant is N110Q; N126Q. N110 Only mutant is N55Q; N126Q. N126 Only mutant is N55Q; N110Q
To determine which asparagine residues are glycosylated in hBMPR2, we expressed V5-tagged versions of wild-type and glycosylation-defective BMPR2 in W20 cells. Electrophoretic mobility of a triple mutant hBMPR2 (N55Q; N110Q; N126Q) is unaffected by PNGase F, indicating these are the only asparagines in full-length hBMPR2 that are glycosylated (Fig. 2b, lanes 5, 6). We then expressed double mutant versions of hBMPR2 where only one putative glycosylated asparagine residue is preserved in each and performed PNGase F digestion. This experiment revealed that N55 (Fig. 2b, lanes 7, 8) and N126 (Fig. 2b, lanes 11, 12) are both glycosylated; we were unable to achieve expression of the “N110 Only” hBMPR2 mutant (N55Q; N126Q) using this approach. Hence, we expressed an N110Q single mutant version of BMPR2, which demonstrated increased electrophoretic mobility compared to wild-type (Fig. 2c, lane 4). These findings confirm that hBMPR2 is glycosylated on residues N55, N110, and N126 in W20 cells.
Our observation that the “N110 Only” version of hBMPR2 is not expressed (Fig. 2b, lanes 9, 10; d, lane 5) suggested that glycosylation plays a role in BMPR2 stability. Consistent with this idea, we were also unable to achieve expression of the N55Q and N126Q mutants of BMPR2 (Fig. 2c, lanes 3 and 5, respectively). Interestingly, these results raise the possibility that glycosylation at N110 must be balanced by glycosylation at both N55 and N126 for proper expression and/or stability of BMPR2.
To examine whether glycosylation of BMPR2 differs between cell types, we extended our study to HEK293T cells and found a glycosylation pattern consistent with that seen in W20 cells (Fig. 2d).
In addition to the 150/140 kDa doublet observed for endogenous BMPR2 in W20 cells, we occasionally observed a minor band at approximately 135 kDa when wild-type BMPR2-V5 was over-expressed in W20 cells (Fig. 2b, lane 3). We are presently unable to comment on the exact nature of this isoform. However, BMPR2 is subject to additional PTMs which could contribute to alterations in electrophoretic mobility, including phosphorylation [45] and O-linked glycosylation (J.W.L., data not shown).
N-linked glycosylation of receptor ECD enhances ligand binding to BMPR2 but not ACVR2B
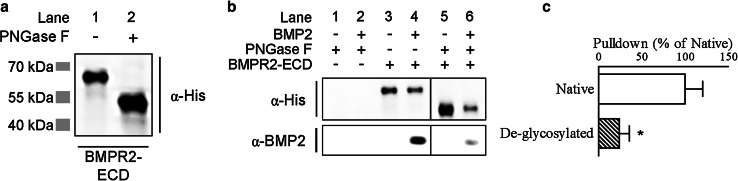
Having observed that BMPR2 is glycosylated on asparagines in its ECD, we sought to determine the functional significance of glycosylation to ligand binding. To circumvent any possible defects in BMPR2 due to aberrant localization, folding, and/or synthesis, we established a cell-free assay utilizing a soluble version of the hBMPR2-ECD fused to human IgG-Fc. We first confirmed that the soluble hBMPR2-ECD is glycosylated (Fig. 3a), then loaded Protein G beads with either native hBMPR2-ECD or de-glycosylated hBMPR2-ECD and mixed them with the prototypical BMP ligand BMP2. This revealed that BMP2 is efficiently pulled down by the native, glycosylated hBMPR2-ECD in this assay (Fig. 3b, lane 4). In contrast, ligand binding to de-glycosylated hBMPR2-ECD is greatly diminished (Figs. 3b, lane 6, 3c), indicating that asparagine glycosylation enhances the ligand binding ability of hBMPR2-ECD.
Fig. 3.
N-linked glycosylation enhances ligand binding to BMPR2-ECD. a Western blot for glycosylation status of the human His-tagged BMPR2 extracellular domain (hBMPR2-ECD)/Fc fusion protein using anti-His antibody. b, c Pulldown of BMP2 using native or de-glycosylated hBMPR2-ECD. Western blot images in (b) for His-tagged hBMPR2-ECD and BMP2 are from a single blot with intervening lane removed (vertical bars) and are representative of five independent experiments (quantified in c). Data in (c) are BMP2/BMPR2-ECD ratio (mean ± SEM) expressed as percent relative to native. *p < 0.05 by unpaired t test against pulldown using native hBMPR2-ECD
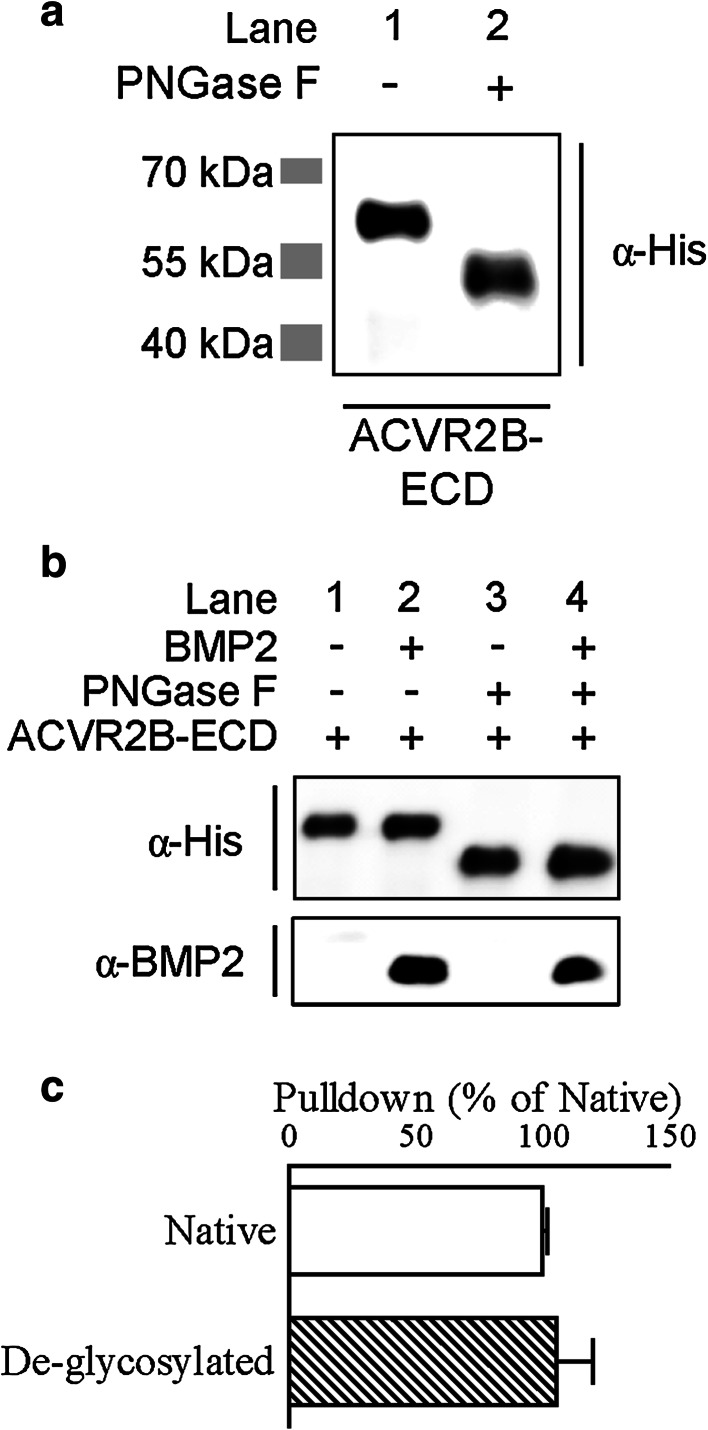
To determine if N-linked glycosylation plays a similar role in ACVR2B, we first demonstrated that the soluble hACVR2B-ECD/Fc fusion protein is N-glycosylated (Fig. 4a). Unlike hBMPR2-ECD, the pulldown efficiency of hACVR2B-ECD for BMP2 is unaffected by de-glycosylation (Fig. 4b, c), indicating that N-glycosylation is dispensable for the ligand binding ability of hACVR2B-ECD.
Fig. 4.
Ligand binding to ACVR2B is unaffected by N-linked glycosylation status. a Western blot for glycosylation status of the human His-tagged ACVR2B extracellular domain (hACVR2B-ECD)/Fc fusion protein using anti-His antibody. b, c Pulldown of BMP2 using native or de-glycosylated ACVR2B-ECD. Western blot images in (b) for ACVR2B-ECD and BMP2 are representative of four independent experiments (quantified in c). Data in (c) are BMP2/ACVR2B-ECD ratio (mean ± SEM) expressed as percent relative to native
Discussion
In the present study, we demonstrate that the N-glycosylation pattern differs between BMPR2 and the other type 2 BMP receptors, ACVR2A/B. BMPR2 is glycosylated on three asparagine residues (N55, N110, and N126) in its ECD, while the crystal structure of the ACVR2A-ECD shows glycosylation at N43 and N66 [33]; due to significant homology, it is reasonable to conclude that ACVR2B is also glycosylated at the related residues. Comparison of the available crystal structures indicates that N55 in BMPR2 is likely analogous to N42 and N43 in ACVR2B and ACVR2A, respectively. These residues lie on the outer face of Finger 1, away from the ligand binding region, making it unlikely that N-glycosylation at these sites would directly alter ligand binding. This supposition is supported by our experiments using the soluble ACVR2B-ECD, which revealed that BMP2 binding to native and de-glycosylated ACVR2B-ECD is identical. Comparable results were reported regarding the impact of N-glycosylation on the interaction between ACVR2A and Activin A [34, 36] and Inhibin [36].
In contrast, we found that N-glycosylation of the BMPR2-ECD enhances binding to BMP2. Insight into how N-glycosylation could alter this interaction might be gained from a previous report that non-glycosylated ovine BMPR2-ECD expressed in E. coli exhibits considerable structural mobility, especially in two of the domains that we show are N-glycosylated [46]. This raises the possibility that glycosylation of these flexible domains (Finger 1 and Finger 3) could direct proper folding and/or stabilize the global structure of the BMPR2-ECD, which is consistent with the well-known role of glycosylation in stabilizing protein conformation [47]. At the same time, since our ligand-binding experiments were performed in a cell-free system following PNGaseF treatment of fully synthesized BMPR2-ECD, our findings demonstrate a direct role for N-glycosylation in the function of BMPR2 irrespective of any additional role in trafficking, folding, and/or degradation of the protein (see below). Support for this idea comes from the fact that BMPR2 is uniquely glycosylated at N110 in Finger 3, which approximates the hydrophobic ligand binding domain [26, 48]. In fact, N110 is conserved as a consensus N-glycosylation site in every available full-length BMPR2 ortholog in the GenBank database (49/49 species, Fig. S1), suggestive that glycosylation at this site is a conserved mechanism that plays a role in interacting with ligand.
The other aspargine residue that is glycosylated in BMPR2, N126, lies exposed on the outer surface in a linker region between a short 3(10) helix and the seventh β-strand. The location of N126 and the glycosylation of Finger 1 shared between BMPR2 and ACVR2A/B suggests that glycosylation at these sites might play a role in protein–protein interactions, such as those with BMP co-receptors [49, 50], and/or may be involved in proper intracellular trafficking [30]. Future studies will evaluate these possibilities. Interestingly, point mutations at N126 are found in three different patients with the rare disease HPAH, which is linked to loss-of-function mutations in BMPR2 [10, 11], indicating that N126 is essential to the normal function of BMPR2. While we are unable to predict the exact role of the N126 mutation in human disease, it is likely telling that BMPR2-N126Q mutants were not expressed in either W20 or HEK293T cells, suggesting that glycosylation at N126 plays a role in stability and/or degradation of BMPR2. In fact, we were unable to express any BMPR2 mutant where N110 was available for glycosylation but either N55 or N126 were not. Strikingly, the un-glycosylated triple mutant BMPR2 was expressed, albeit at a lower level. All plasmids were sequenced for fidelity and these results were consistent across numerous plasmid purifications and transfections, suggesting they are not technical in nature.
Collectively, our findings illustrate divergence in the pattern of N-glycosylation between BMPR2 and the other type 2 BMP receptors, ACVR2A/B. Functional relevance comes from the fact that N-glycosylation enhances the ability of BMPR2-ECD, but not ACVR2B-ECD, to bind BMP2. Although significant functional redundancy occurs between type 2 BMP receptors in vivo (e.g., see Gamer et al. [51]), we speculate that differential N-glycosylation plays a role in segregating BMPR2, which is thought to act as a BMP-specific receptor in vivo, from ACVR2A/B, which signal both BMP and Activins (Activin/Activin-like ligands) with high affinity [7]. Because of the widespread actions of BMP signaling in development and tissue homeostasis, our findings may provide insight into the signaling mechanics underlying several human diseases and allow for new therapeutic approaches that exploit type 2 receptor–ligand interactions.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Figure S1 Conservation of N-linked glycosylation sites in BMPR2 orthologs. Alignment comparison of BMPR2 orthologs. All available amino acid sequences of full-length BMPR2 were obtained from the GenBank database and aligned using MUSCLE [54] in MEGA5 [55] (PDF 100 kb)
Supplementary Table S1 Outline of hBMPR2 expression plasmids utilized and their construction (PDF 17 kb)
Acknowledgments
We gratefully acknowledge James West (Vanderbilt) for the hBMPR2 cDNA; Mark de Caestecker (Vanderbilt) for early discussions; Giuseppe Intini (HSDM) for assistance with Bmpr2 knockdown. J.W.L. and J.M.A. are recipients of the Harvard School of Dental Medicine Dean’s Scholar Award and Research Science Institute fellowship, respectively. This work was supported by the NIH/NIAMS grant R01AR055904 awarded to V.R.
Conflict of interests
The authors declare that they have no competing interests.
References
- 1.Holstein TW, Watanabe H, Ozbek S. Signaling pathways and axis formation in the lower metazoa. Curr Top Dev Biol. 2011;97:137–177. doi: 10.1016/B978-0-12-385975-4.00012-7. [DOI] [PubMed] [Google Scholar]
- 2.Newfeld SJ, Wisotzkey RG, Kumar S. Molecular evolution of a developmental pathway: phylogenetic analyses of transforming growth factor-beta family ligands, receptors and Smad signal transducers. Genetics. 1999;152(2):783–795. doi: 10.1093/genetics/152.2.783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao GQ. Consequences of knocking out BMP signaling in the mouse. Genesis. 2003;35(1):43–56. doi: 10.1002/gene.10167. [DOI] [PubMed] [Google Scholar]
- 4.Kishigami S, Mishina Y. BMP signaling and early embryonic patterning. Cytokine Growth Factor Rev. 2005;16(3):265–278. doi: 10.1016/j.cytogfr.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 5.Miyazono K, Kamiya Y, Morikawa M. Bone morphogenetic protein receptors and signal transduction. J Biochem. 2010;147(1):35–51. doi: 10.1093/jb/mvp148. [DOI] [PubMed] [Google Scholar]
- 6.Lowery JW, Pazin D, Intini G, Kokabu S, Chappuis V, Capelo LP, Rosen V. The role of BMP2 signaling in the skeleton. Crit Rev Eukaryot Gene Expr. 2011;21(2):177–185. doi: 10.1615/CritRevEukarGeneExpr.v21.i2.60. [DOI] [PubMed] [Google Scholar]
- 7.Hinck AP. Structural studies of the TGF-betas and their receptors—insights into evolution of the TGF-beta superfamily. FEBS Lett. 2012;586(14):1860–1870. doi: 10.1016/j.febslet.2012.05.028. [DOI] [PubMed] [Google Scholar]
- 8.Zhang YE. Non-Smad pathways in TGF-beta signaling. Cell Res. 2009;19(1):128–139. doi: 10.1038/cr.2008.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lowery JW, de Caestecker MP. BMP signaling in vascular development and disease. Cytokine Growth Factor Rev. 2010;21(4):287–298. doi: 10.1016/j.cytogfr.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deng Z, Morse JH, Slager SL, Cuervo N, Moore KJ, Venetos G, Kalachikov S, Cayanis E, Fischer SG, Barst RJ, Hodge SE, Knowles JA. Familial primary pulmonary hypertension (gene PPH1) is caused by mutations in the bone morphogenetic protein receptor-II gene. Am J Hum Genet. 2000;67(3):737–744. doi: 10.1086/303059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lane KB, Machado RD, Pauciulo MW, Thomson JR, Phillips JA, 3rd, Loyd JE, Nichols WC, Trembath RC, The International PPH Consortium Heterozygous germline mutations in BMPR2, encoding a TGF-beta receptor, cause familial primary pulmonary hypertension. Nat Genet. 2000;26(1):81–84. doi: 10.1038/79226. [DOI] [PubMed] [Google Scholar]
- 12.Kosaki R, Gebbia M, Kosaki K, Lewin M, Bowers P, Towbin JA, Casey B. Left-right axis malformations associated with mutations in ACVR2B, the gene for human activin receptor type IIB. Am J Med Genet. 1999;82(1):70–76. doi: 10.1002/(SICI)1096-8628(19990101)82:1<70::AID-AJMG14>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 13.Shore EM, Xu M, Feldman GJ, Fenstermacher DA, Cho TJ, Choi IH, Connor JM, Delai P, Glaser DL, LeMerrer M, Morhart R, Rogers JG, Smith R, Triffitt JT, Urtizberea JA, Zasloff M, Brown MA, Kaplan FS. A recurrent mutation in the BMP type I receptor ACVR1 causes inherited and sporadic fibrodysplasia ossificans progressiva. Nat Genet. 2006;38(5):525–527. doi: 10.1038/ng1783. [DOI] [PubMed] [Google Scholar]
- 14.Howe JR, Bair JL, Sayed MG, Anderson ME, Mitros FA, Petersen GM, Velculescu VE, Traverso G, Vogelstein B. Germline mutations of the gene encoding bone morphogenetic protein receptor 1A in juvenile polyposis. Nat Genet. 2001;28(2):184–187. doi: 10.1038/88919. [DOI] [PubMed] [Google Scholar]
- 15.Howe JR, Sayed MG, Ahmed AF, Ringold J, Larsen-Haidle J, Merg A, Mitros FA, Vaccaro CA, Petersen GM, Giardiello FM, Tinley ST, Aaltonen LA, Lynch HT. The prevalence of MADH4 and BMPR1A mutations in juvenile polyposis and absence of BMPR2, BMPR1B, and ACVR1 mutations. J Med Genet. 2004;41(7):484–491. doi: 10.1136/jmg.2004.018598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lehmann K, Seemann P, Stricker S, Sammar M, Meyer B, Suring K, Majewski F, Tinschert S, Grzeschik KH, Muller D, Knaus P, Nurnberg P, Mundlos S. Mutations in bone morphogenetic protein receptor 1B cause brachydactyly type A2. Proc Natl Acad Sci USA. 2003;100(21):12277–12282. doi: 10.1073/pnas.2133476100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lehmann K, Seemann P, Boergermann J, Morin G, Reif S, Knaus P, Mundlos S. A novel R486Q mutation in BMPR1B resulting in either a brachydactyly type C/symphalangism-like phenotype or brachydactyly type A2. Eur J Hum Genet. 2006;14(12):1248–1254. doi: 10.1038/sj.ejhg.5201708. [DOI] [PubMed] [Google Scholar]
- 18.Demirhan O, Turkmen S, Schwabe GC, Soyupak S, Akgul E, Tastemir D, Karahan D, Mundlos S, Lehmann K. A homozygous BMPR1B mutation causes a new subtype of acromesomelic chondrodysplasia with genital anomalies. J Med Genet. 2005;42(4):314–317. doi: 10.1136/jmg.2004.023564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heinecke K, Seher A, Schmitz W, Mueller TD, Sebald W, Nickel J. Receptor oligomerization and beyond: a case study in bone morphogenetic proteins. BMC Biol. 2009;7:59. doi: 10.1186/1741-7007-7-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weber D, Kotzsch A, Nickel J, Harth S, Seher A, Mueller U, Sebald W, Mueller TD. A silent H-bond can be mutationally activated for high-affinity interaction of BMP-2 and activin type IIB receptor. BMC Struct Biol. 2007;7:6. doi: 10.1186/1472-6807-7-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keller S, Nickel J, Zhang JL, Sebald W, Mueller TD. Molecular recognition of BMP-2 and BMP receptor IA. Nat Struct Mol Biol. 2004;11(5):481–488. doi: 10.1038/nsmb756. [DOI] [PubMed] [Google Scholar]
- 22.Kotzsch A, Nickel J, Seher A, Heinecke K, van Geersdaele L, Herrmann T, Sebald W, Mueller TD. Structure analysis of bone morphogenetic protein-2 type I receptor complexes reveals a mechanism of receptor inactivation in juvenile polyposis syndrome. J Biol Chem. 2008;283(9):5876–5887. doi: 10.1074/jbc.M706029200. [DOI] [PubMed] [Google Scholar]
- 23.Allendorph GP, Vale WW, Choe S. Structure of the ternary signaling complex of a TGF-beta superfamily member. Proc Natl Acad Sci USA. 2006;103(20):7643–7648. doi: 10.1073/pnas.0602558103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hatta T, Konishi H, Katoh E, Natsume T, Ueno N, Kobayashi Y, Yamazaki T. Identification of the ligand-binding site of the BMP type IA receptor for BMP-4. Biopolymers. 2000;55(5):399–406. doi: 10.1002/1097-0282(2000)55:5<399::AID-BIP1014>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 25.Kirsch T, Nickel J, Sebald W. BMP-2 antagonists emerge from alterations in the low-affinity binding epitope for receptor BMPR-II. EMBO J. 2000;19(13):3314–3324. doi: 10.1093/emboj/19.13.3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yin H, Yeh LC, Hinck AP, Lee JC. Characterization of ligand-binding properties of the human BMP type II receptor extracellular domain. J Mol Biol. 2008;378(1):191–203. doi: 10.1016/j.jmb.2008.02.031. [DOI] [PubMed] [Google Scholar]
- 27.Yeh LC, Falcon WE, Garces A, Lee JC. A host-guest relationship in bone morphogenetic protein receptor-II defines specificity in ligand-receptor recognition. Biochemistry. 2012;51(35):6968–6980. doi: 10.1021/bi3003023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saremba S, Nickel J, Seher A, Kotzsch A, Sebald W, Mueller TD. Type I receptor binding of bone morphogenetic protein 6 is dependent on N-glycosylation of the ligand. FEBS J. 2008;275(1):172–183. doi: 10.1111/j.1742-4658.2007.06187.x. [DOI] [PubMed] [Google Scholar]
- 29.Mitra N, Sinha S, Ramya TN, Surolia A. N-linked oligosaccharides as outfitters for glycoprotein folding, form and function. Trends Biochem Sci. 2006;31(3):156–163. doi: 10.1016/j.tibs.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 30.Ohtsubo K, Marth JD. Glycosylation in cellular mechanisms of health and disease. Cell. 2006;126(5):855–867. doi: 10.1016/j.cell.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 31.Kawabata M, Chytil A, Moses HL. Cloning of a novel type II serine/threonine kinase receptor through interaction with the type I transforming growth factor-beta receptor. J Biol Chem. 1995;270(10):5625–5630. doi: 10.1074/jbc.270.10.5625. [DOI] [PubMed] [Google Scholar]
- 32.Mathews LS, Vale WW. Characterization of type II activin receptors. Binding, processing, and phosphorylation. J Biol Chem. 1993;268(25):19013–19018. [PubMed] [Google Scholar]
- 33.Greenwald J, Groppe J, Gray P, Wiater E, Kwiatkowski W, Vale W, Choe S. The BMP7/ActRII extracellular domain complex provides new insights into the cooperative nature of receptor assembly. Mol Cell. 2003;11(3):605–617. doi: 10.1016/S1097-2765(03)00094-7. [DOI] [PubMed] [Google Scholar]
- 34.Greenwald J, Le V, Corrigan A, Fischer W, Komives E, Vale W, Choe S. Characterization of the extracellular ligand-binding domain of the type II activin receptor. Biochemistry. 1998;37(47):16711–16718. doi: 10.1021/bi981939o. [DOI] [PubMed] [Google Scholar]
- 35.Donaldson CJ, Vaughan JM, Corrigan AZ, Fischer WH, Vale WW. Activin and inhibin binding to the soluble extracellular domain of activin receptor II. Endocrinology. 1999;140(4):1760–1766. doi: 10.1210/endo.140.4.6665. [DOI] [PubMed] [Google Scholar]
- 36.Daly R, Hearn MT. Expression of the human activin type I and II receptor extracellular domains in Pichia pastoris. Protein Expr Purif. 2006;46(2):456–467. doi: 10.1016/j.pep.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 37.Sako D, Grinberg AV, Liu J, Davies MV, Castonguay R, Maniatis S, Andreucci AJ, Pobre EG, Tomkinson KN, Monnell TE, Ucran JA, Martinez-Hackert E, Pearsall RS, Underwood KW, Seehra J, Kumar R. Characterization of the ligand binding functionality of the extracellular domain of activin receptor type IIb. J Biol Chem. 2010;285(27):21037–21048. doi: 10.1074/jbc.M110.114959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Frump AL, Lowery JW, Hamid R, Austin ED, de Caestecker M. Abnormal trafficking of endogenously expressed BMPR2 mutant allelic products in patients with heritable pulmonary arterial hypertension. PLoS ONE. 2013;8(11):e80319. doi: 10.1371/journal.pone.0080319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pfarr N, Szamalek-Hoegel J, Fischer C, Hinderhofer K, Nagel C, Ehlken N, Tiede H, Olschewski H, Reichenberger F, Ghofrani AH, Seeger W, Grunig E. Hemodynamic and clinical onset in patients with hereditary pulmonary arterial hypertension and BMPR2 mutations. Respir Res. 2011;12:99. doi: 10.1186/1465-9921-12-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Machado RD, Eickelberg O, Elliott CG, Geraci MW, Hanaoka M, Loyd JE, Newman JH, Phillips JA, 3rd, Soubrier F, Trembath RC, Chung WK. Genetics and genomics of pulmonary arterial hypertension. J Am Coll Cardiol. 2009;54(Suppl 1):S32–S42. doi: 10.1016/j.jacc.2009.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Beppu H, Kawabata M, Hamamoto T, Chytil A, Minowa O, Noda T, Miyazono K. BMP type II receptor is required for gastrulation and early development of mouse embryos. Dev Biol. 2000;221(1):249–258. doi: 10.1006/dbio.2000.9670. [DOI] [PubMed] [Google Scholar]
- 42.Beppu H, Malhotra R, Beppu Y, Lepore JJ, Parmacek MS, Bloch KD. BMP type II receptor regulates positioning of outflow tract and remodeling of atrioventricular cushion during cardiogenesis. Dev Biol. 2009;331(2):167–175. doi: 10.1016/j.ydbio.2009.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Delot EC, Bahamonde ME, Zhao M, Lyons KM. BMP signaling is required for septation of the outflow tract of the mammalian heart. Development. 2003;130(1):209–220. doi: 10.1242/dev.00181. [DOI] [PubMed] [Google Scholar]
- 44.Rabinovitch M. Molecular pathogenesis of pulmonary arterial hypertension. J Clin Investig. 2012;122(12):4306–4313. doi: 10.1172/JCI60658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schwappacher R, Weiske J, Heining E, Ezerski V, Marom B, Henis YI, Huber O, Knaus P. Novel crosstalk to BMP signalling: cGMP-dependent kinase I modulates BMP receptor and Smad activity. EMBO J. 2009;28(11):1537–1550. doi: 10.1038/emboj.2009.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mace PD, Cutfield JF, Cutfield SM. High resolution structures of the bone morphogenetic protein type II receptor in two crystal forms: implications for ligand binding. Biochem Biophys Res Commun. 2006;351(4):831–838. doi: 10.1016/j.bbrc.2006.10.109. [DOI] [PubMed] [Google Scholar]
- 47.Imperiali B, O’Connor SE. Effect of N-linked glycosylation on glycopeptide and glycoprotein structure. Curr Opin Chem Biol. 1999;3(6):643–649. doi: 10.1016/S1367-5931(99)00021-6. [DOI] [PubMed] [Google Scholar]
- 48.Mace PD, Cutfield JF, Cutfield SM. Bacterial expression and purification of the ovine type II bone morphogenetic protein receptor ectodomain. Protein Expr Purif. 2007;52(1):40–49. doi: 10.1016/j.pep.2006.07.025. [DOI] [PubMed] [Google Scholar]
- 49.Barbara NP, Wrana JL, Letarte M. Endoglin is an accessory protein that interacts with the signaling receptor complex of multiple members of the transforming growth factor-beta superfamily. J Biol Chem. 1999;274(2):584–594. doi: 10.1074/jbc.274.2.584. [DOI] [PubMed] [Google Scholar]
- 50.Lewis KA, Gray PC, Blount AL, MacConell LA, Wiater E, Bilezikjian LM, Vale W. Betaglycan binds inhibin and can mediate functional antagonism of activin signalling. Nature. 2000;404(6776):411–414. doi: 10.1038/35006129. [DOI] [PubMed] [Google Scholar]
- 51.Gamer LW, Tsuji K, Cox K, Capelo LP, Lowery J, Beppu H, Rosen V. BMPR-II is dispensable for formation of the limb skeleton. Genesis. 2011;49:719–724. doi: 10.1002/dvg.20761. [DOI] [PubMed] [Google Scholar]
- 52.Thies RS, Bauduy M, Ashton BA, Kurtzberg L, Wozney JM, Rosen V. Recombinant human bone morphogenetic protein-2 induces osteoblastic differentiation in W-20-17 stromal cells. Endocrinology. 1992;130(3):1318–1324. doi: 10.1210/endo.130.3.1311236. [DOI] [PubMed] [Google Scholar]
- 53.Graham FL, Smiley J, Russell WC, Nairn R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J Gen Virol. 1977;36(1):59–74. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- 54.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol. 2011;Evol28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1 Conservation of N-linked glycosylation sites in BMPR2 orthologs. Alignment comparison of BMPR2 orthologs. All available amino acid sequences of full-length BMPR2 were obtained from the GenBank database and aligned using MUSCLE [54] in MEGA5 [55] (PDF 100 kb)
Supplementary Table S1 Outline of hBMPR2 expression plasmids utilized and their construction (PDF 17 kb)